Abstract
L1 cell adhesion molecule (L1CAM) is well-known for its importance in nervous system development and cancer progression. In addition to its role as a plasma membrane protein in cytoskeletal organization, recent in vitro studies have revealed that both transmembrane and cytosolic fragments of proteolytically cleaved vertebrate L1CAM translocate to the nucleus. In vitro studies indicate that nuclear L1CAM affects genes with functions in DNA post-replication repair, cell cycle control, and cell migration and differentiation, but its in vivo role and how its nuclear levels are regulated is less well-understood. Here, we report that mutations in the conserved ankyrin-binding domain affect nuclear levels of the sole Drosophila homolog neuroglian (Nrg) and that it also has a noncanonical role in regulating transcript levels of the oncogene Myc in the adult nervous system. We further show that altered nuclear levels of Nrg correlate with altered transcript levels of Myc in neurons, similar to what has been reported for human glioblastoma stem cells. However, whereas previous in vitro studies suggest that increased nuclear levels of L1CAM promote tumor cell survival, we found here that elevated levels of nuclear Nrg in neurons are associated with increased sensitivity to oxidative stress and reduced life span of adult animals. We therefore conclude that these findings are of potential relevance to the management of neurodegenerative diseases associated with oxidative stress and cancer.
Keywords: transcription regulation, ankyrin, cancer, Drosophila, Myc (c-Myc), oxidative stress, L1-CAM, neuroglian, nuclear function, ankyrin-binding motif
Introduction
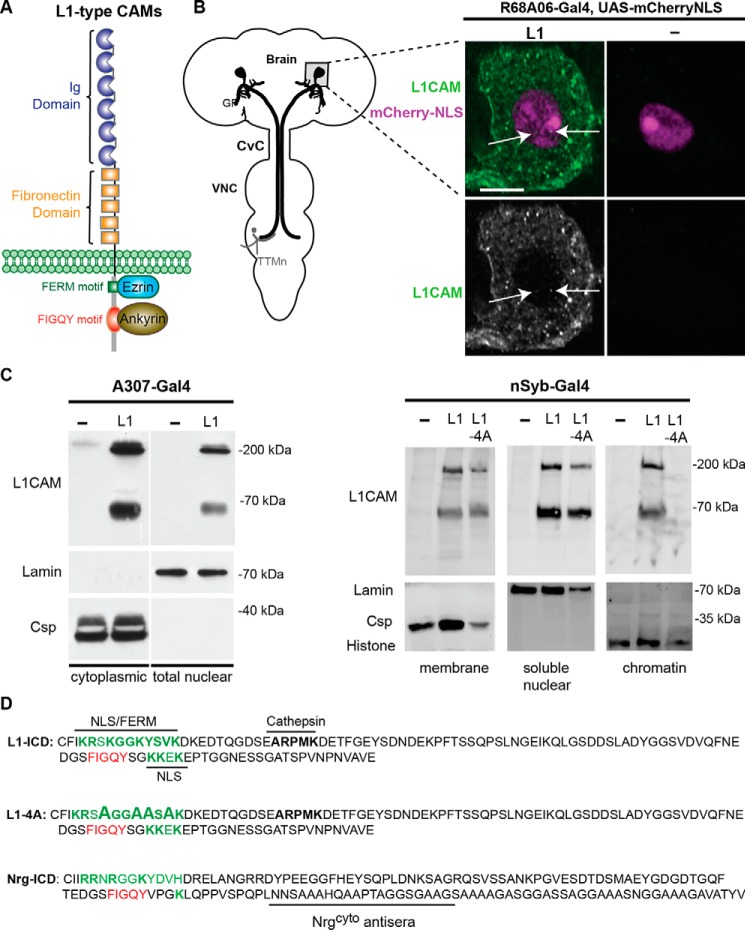
L1-type cell adhesion molecules (CAMs)4 are single-pass transmembrane glycoproteins belonging to the immunoglobulin family of receptors that are highly conserved from invertebrates to vertebrates (1). The structure of L1CAM consists of extracellular immunoglobulin and fibronectin type III domains, a transmembrane domain, and a cytoplasmic tail harboring an ezrin-binding FERM domain as well as an ankyrin-binding FIGQY domain (Fig. 1A). In its unphosphorylated state, the highly conserved FIGQY domain reversibly binds to ankyrin (Fig. 1A), which couples L1-type CAMs to actin. This interaction is known to mediate neuritogenesis, synapse growth, and stability (2–4). In contrast, phosphorylation of the tyrosine in the FIGQY domain inhibits ankyrin binding (5, 6).
Figure 1.
Subcellular localization of human L1CAM in the Drosophila nervous system. A, cartoon of conserved domains in L1-type CAMs. The extracellular domain contains Ig and fibronectin type III repeats. The cytoplasmic tail contains a FERM-binding domain (green) and a FIGQY motif (red) for ankyrin binding. B, subcellular localization of human L1CAM in the GF soma. Left, schematic representation of the giant fiber circuit in the Drosophila nervous system. The GF soma (gray square) is in the brain, and the GF terminals synapse with the tergo-trochanteral motoneuron (TTMn) in the ventral nerve cord (VNC). For visualization of the GF morphology, fluorescent dyes are injected into the GFs at the cervical connective (CvC). Right panels, UAS-mCherry-NLS was co-expressed with and without UAS-L1 in the GFs using the R68A06-Gal4 line. A single confocal slice intersecting the GF nucleus and probed with L1CAM antibodies against the cytoplasmic domain is shown. Anti-L1CAM labeling is shown with (green, top panels) and without (gray, bottom panels) nuclear labeling by mCherry-NLS (magenta). In addition to labeling of L1CAM at the plasma membrane and vesicular compartments in the cytosol, L1CAM puncta can also be seen in the nucleus (arrows). No specific L1CAM labeling is detected in GF somas that do not express L1CAM. C, subcellular localization of human L1CAM (L1) in the Drosophila nervous system using two distinct fractionation kits. Left, the A307-Gal4 driver was used to express UAS-L1 broadly in the nervous system inclusive of the GF circuit. The Thermo Fisher Scientific NE-PER® kit was used to obtain cytoplasmic and nuclear fractions. The cytoplasmic fraction of ∼7 brains and the nuclear fraction of ∼21 brains were loaded per lane. The full-length (200-kDa) as well as the proteolytically cleaved L1CAM (65–70-kDa) isoforms were detected in the cytoplasmic as well as nuclear fractions. Anti-lamin and anti-Csp labeling served as loading controls for the nuclear and cytoplasmic fractionation. Right, human WT L1CAM (L1) and L1CAM carrying four alanine missense mutations (L1–4A) in the FERM motif were panneuronally (nSyb-Gal4) expressed in Drosophila. The Thermo Fisher Scientific Subcellular Protein Fractionation Kit was used to obtain membrane, soluble nuclear as well as chromatin fractions. The membrane fraction of 10 heads, nuclear fraction of 100 heads and chromatin fraction of 100 heads were loaded per lane. Anti-Csp, anti-lamin, and anti-histone served as loading controls for the membrane, nuclear, and chromatin fractions, respectively. D, amino acid sequences of the ICDs of human L1, L1–4A, and Nrg180. The putative NLS and ankryin-binding motifs are labeled in green and red, respectively. Peptide sequence used to generate the Nrgcyto antiserum is indicated (3).
In addition to its function as a cytoskeleton-organizing protein at the plasma membrane, vertebrate L1CAM can be proteolytically cleaved with fragments translocating to the nucleus (7–11). The 200-kDa full-length L1CAM is cleaved proximal to the plasma membrane by metalloproteases to a 32-kDa fragment (12, 13). The 32-kDa fragment is further cleaved by γ-secretase/presenilin, releasing a 28-kDa cytosolic fragment containing the intracellular domain (ICD), which translocates to the nucleus (10, 11). Similar to full-length L1CAM, recombinant expression of L1-ICD in nonneuronal cell lines affects gene expression of CRABPII and β3-integrin (11). In addition, it was shown that nuclear L1-ICD also led to up-regulation of NBS1 (Nijmegen breakage syndrome gene) via c-Myc, which plays a role in DNA damage checkpoint responses and renders human glioblastoma stem cells chemo- and radioresistant (7, 14). However, little is known about the normal role of nuclear L1-type CAMs in the nervous system.
In the nervous system, it was shown that an endocytosed sumoylated 70-kDa L1CAM transmembrane fragment is released from endosomes into the cytosol and translocates into the nuclei of cerebellar neurons with potential functions in development, regeneration, neurodegeneration, and synaptic plasticity (8). The 70-kDa L1CAM fragment can also be cleaved into a sumoylated 30-kDa cytosolic fragment (Fig. 1D) that translocates to the nucleus in neurons and glia, which was shown to promote migration and myelination in vitro (9). A mutation of Lys-1147 at the beginning of the conserved FERM binding domain (Fig. 1D) was shown to prevent the presence of the 70- and 30-kDa L1CAM fragments in chromatin fractions (8, 9), although a second putative nuclear localization motif (NLS) adjacent to the FIGQY motif was unaltered (Fig. 1D). However, it remains to be determined whether other motifs have a role in regulating nuclear levels of L1CAM, whether altered nuclear levels of L1-type CAMs also affect c-Myc transcript levels in neurons, and what the in vivo effects are of altered nuclear levels of L1-type CAMs.
Here, we use Drosophila as a model to address the above questions in the adult nervous system. The domain structure of the sole Drosophila homolog neuroglian (Nrg) is conserved, and the gene produces a neuron-specific isoform, Nrg180, and a ubiquitous isoform, Nrg167, via alternative splicing allowing for genetic alteration of the FIGQY motif for each individual isoform (3).
This study involves transgenic overexpression of Nrg180 and L1CAM constructs as well as animals carrying mutations specific for the neuronal isoform of Nrg (Nrg180) to determine the domain requirements and in vivo effects. We previously used nrg pacman mutants to determine the functional role of various domains in Nrg180 in nervous system development and maintenance (3, 15). In these mutants, the loss-of-function allele nrg14 was rescued with gene constructs that carry the entire WT nrg gene locus (nrg14;Pac[WT]) or the nrg gene locus in which the neuron-specific exon was mutated (3). WT and mutant Nrg180 proteins are expressed at endogenous levels in these pacman mutants (3); thus, they are a valuable tool to determine domain-specific functions in regulation of nuclear levels of Nrg in vivo.
Results
Nuclear localization of human L1CAM in Drosophila neurons
Cytosolic as well as transmembrane L1CAM fragments translocate to the nucleus in vertebrates (7–9, 11). To test whether the mechanism for nuclear localization of human L1CAM is also conserved in Drosophila, we transgenically expressed L1CAM in the giant fiber (GF) neurons. The somas of the GFs are located in the brain (Fig. 1B) and, due to their large nuclear size (≥5 μm), allow for the determination of subcellular localization in vivo. In addition, we previously demonstrated that human L1CAM can rescue developmental loss of function phenotypes of Nrg in the GFs and that the 200-kDa full-length isoform is proteolytically cleaved into 65–70-kDa fragments in the fly nervous system (16, 17).
We used the R68A06-Gal4 line to selectively drive expression of human L1CAM in the GFs (18). In addition to L1CAM labeling of the plasma membrane and vesicular compartments in the cytoplasm, we also observed L1CAM labeling in the nucleus (Fig. 1B, right panels, green). Whereas a single confocal slice of the GF soma intersecting the nucleus demonstrated the presence of L1CAM in the nucleus, the absence of immunohistochemical labeling in a soma that does not transgenically express L1CAM demonstrated the specificity of the antibody directed against the intracellular domain of L1CAM (Fig. 1B, right panels). We quantified L1CAM labeling in six somas for the entire nuclear volumes and observed on average 3.2 ± 0.47 L1CAM puncta. In addition, we found that fluorescent sum intensity of the entire nuclear areas of L1CAM expression was statistically significant (p = 0.0002, two-tailed Student's t test) from background labeling in nuclear areas that did not express L1CAM (n = 5) and showed no specific L1CAM puncta in the nucleus. This suggests that L1CAM is imported to the nucleus in Drosophila neurons.
To further confirm that L1CAM translocates to nuclei in Drosophila and to determine the relevant isoforms, we employed two distinctive fractionation kits (Fig. 1C). The Thermo Fisher Scientific NE-PER® kit (Fig. 1C, left) allows the separation of the cytoplasm and of the whole nucleus excluding the nuclear membrane with high yields. To further distinguish between soluble nuclear and chromatin-bound content, we used the Thermo Fisher Scientific Subcellular Protein Fractionation Kit (Fig. 1C, right). Although this kit also generates cytosolic, membrane, and cytoskeletal fractions, we only used the membrane-specific fraction as a nonnuclear control. In addition to WT L1, we also tested the effects of four alanine missense mutations (L1–4A) in the FERM domain on nuclear localization and chromatin binding (19). For fractionations of nervous systems, we used the nSyb-Gal4 and A307-Gal4 lines to pan-neuronally or broadly express L1CAM throughout the central nervous system, including the GF, respectively.
With both fractionation kits, we detected not only the 65–70-kDa L1CAM fragment but also the full-length 200-kDa L1CAM in the nuclear/chromatin fractions (Fig. 1C). Interestingly, both isoforms were detected in fractions (n = 3) that contained the whole nucleoplasm but were absent in chromatin fractions in animals expressing L1–4A. The synaptic vesicle–associated cysteine string protein (Csp), nuclear cytoskeleton protein lamin, and histone were only detected in the respective membrane, soluble nuclear, or chromatin fractions, demonstrating subcellular specificity of the fractions (Fig. 1C). In summary, our findings suggest that nuclear import of L1CAM is conserved in Drosophila and that full-length L1CAM as well as L1CAM fragments translocate to the nucleus. In addition, our results suggest that the mutated amino acids in the FERM motif are required for chromatin binding in vivo.
Mutations in the FIGQY motif affect nuclear levels of Drosophila Nrg180
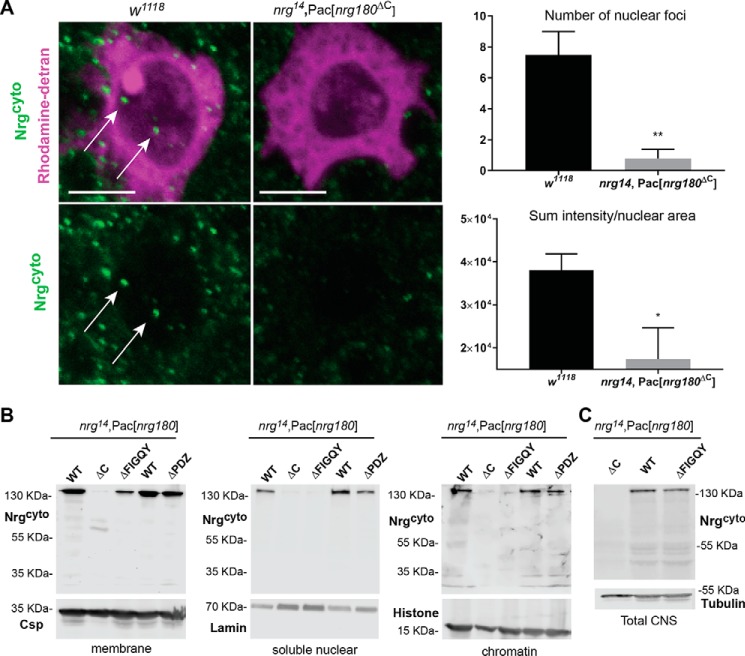
We previously demonstrated a developmental role for Nrg180 in the GFs (3, 16, 17). To further investigate whether Nrg180, similar to vertebrate L1CAM, translocates to the nucleus in adult neurons, we analyzed the subcellular Nrg180 localization in GF somas using the Nrg180cyto antiserum. Nrg180cyto is directed against a C-terminal sequence of the neuronal Nrg180 isoform (Fig. 1D) (3). As a control for the specific labeling of antibodies, we used nrg14; Pac[nrg180ΔC] animals in which the neuron-specific exon of Nrg180 is deleted (3). On average, 7.5 (n = 6) Nrg puncta (Fig. 2A, green) were labeled with Nrg180cyto in the GF nucleus (Fig. 2A, magenta). Analysis of the sum fluorescent intensity labeled by Nrg180cyto in the nuclei areas showed a significant 2-fold difference when compared with nrg14; Pac[nrg180ΔC] (Fig. 2A), suggesting that Nrg is imported to the nucleus.
Figure 2.
Subcellular localization of Drosophila Nrg in WT and Nrg mutants. A, subcellular localization of Nrg180 in the WT (w1118) and nrg14;Pac[nrg180ΔC] negative control flies with Nrg180cyto labeling. WT and negative control animals were immunohistochemically processed in parallel and scanned with the same settings. Shown is the image of single confocal slice through the nucleus of the giant fiber soma dye filled with rhodamine-dextran (magenta), which is preferentially excluded from the nucleus. Nrg180 (green) labeled puncta were seen in the cytoplasm and the nucleus (arrows) of w1118 flies. Scale bar, 5 μm. Right, quantification of Nrg180 labeling with Nrg180cyto antibodies in the nucleus. The number of puncta (p < 0.003, two-tailed Student's t test) and the sum of fluorescent intensity levels (p < 0.026, two-tailed Student's t test) normalized to the total nuclear areas is significantly different in WT (n = 6) and negative control (n = 5) animals. B, Western blots of the membrane (10 heads), soluble nuclear (100 heads), and chromatin (100 heads) fractions (Subcellular Protein Fractionation Kit) of WT (nrg14;Pac[WT]), negative controls (nrg14;Pac[nrg180ΔC]), and Nrg pacman mutants (nrg14;Pac[nrg180ΔPDZ], nrg14;Pac[nrg180ΔFIGQY]) lacking the FIGQY or C-terminal PDZ motifs probed with Nrg180cyto antibodies. Anti-Csp, anti-lamin, and anti-histone were used as controls for the membrane, soluble nuclear, and chromatin fractions, respectively. C, Western blotting of total protein extracts of 20 heads from nrg14;Pac[nrg180ΔC], nrg14;Pac[WT], and nrg14;Pac[nrg180ΔFIGQY] animals probed with Nrg180cyto antibodies. Anti-tubulin served as a loading control. Error bars represent standard error of the mean. Two-tailed Student′s t test was used to determine statistically significant differences (*, p ≤ 0.05, **, p < 0.01).
Canonical nuclear localization motifs typically contain a series of positively charge amino acids, such as arginine and lysine, and the FERM domain has previously been shown to play a role in nuclear localization of L1CAM (8, 9). However, in three independent soluble nuclear fractions, we detected L1–4A (Fig. 1C), suggesting that other domains play a role in regulating nuclear levels of L1-type CAMs. Therefore, to confirm that Nrg180 localizes to nuclei in the nervous system and to test for the involvement of other domains required for regulation of nuclear levels of Nrg180, we performed cellular fractions of available nrg pacman mutants in which the FIGQY motif and the c-terminal PDZ motifs of Nrg180 are deleted (3).
In WT (nrg14;Pac[WT]) and nrg14;Pac[nrg180ΔPDZ] mutants but not in nrg14; Pac[nrg180ΔC] and nrg14;Pac[nrg180ΔFIGQY] animals, we observed full-length Nrg180 as well as smaller fragments of very low abundance in nuclear fractions, whereas WT and mutant Nrg180 proteins were detected in membrane fractions (Fig. 2B). We previously showed that expression levels of nrg14;Pac[nrg180ΔFIGQY] and nrg14;Pac[WT] are equal in larval nervous systems (3). Here, we confirmed that total protein levels of Nrg180 and Nrg180 lacking its FIGQY motif in the mutant animals are also similar in protein extracts of adult nervous systems (Fig. 2C). Thus, these results suggest that the FIGQY motif but not the PDZ motif has a role in regulating nuclear levels of Nrg180.
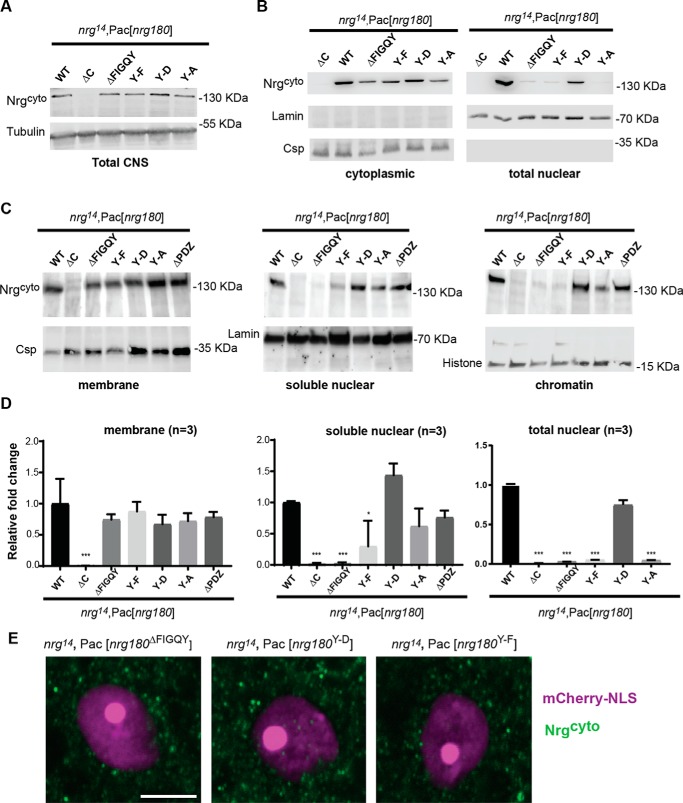
To further investigate the role of the FIGQY motif in regulating nuclear levels of Nrg180, we took advantage of pacman mutants that carry missense mutations of the tyrosine residue within the FIGQY motif. These mutants mimic either constitutively phosphorylated (Y-D) or nonphosphorylated (Y-F) isoforms of Nrg180 (3). We previously showed that all rescue constructs express WT and mutant Nrg180 at endogenous protein levels in larval brains (3). Similarly, we find that total expression levels of WT and mutant Nrg180 proteins are comparable with control animals in the adult nervous system as well (Fig. 3A). With the NE-PER® kit, we found that the nuclear levels of full-length Nrg180 in nrg14;Pac[nrg180Y-F], nrg14;Pac[nrg180Y-A], and nrg14;Pac[nrg180ΔC] but not in nrg14;Pac[nrg180Y-D] mutants were significantly reduced when compared with control animals, whereas the Lamin loading control was similar for all genotypes (Fig. 3, B and D). Further analyses with the Thermo Fisher Scientific Subcellular Protein Fractionation Kit showed similar levels of Nrg180ΔFIGQY, Nrg180Y-F, Nrg180Y-A, and Nrg180Y-D proteins in membrane fractions when compared with WT Nrg180 (Fig. 3, C and D). In contrast, significantly less or no Nrg180 was detected in the soluble nuclear and chromatin fractions of nrg14;Pac[nrg180Y-F] and nrg14;Pac[nrg180ΔFIGQY] mutants, whereas no significant difference from control animals was observed for nrg14; Pac[nrg180Y-D] animals (Fig. 3, C and D). Consistent with this, we found visibly more abundant labeling of Nrg180Y-D than Nrg180Y-F and Nrg180ΔFIGQY proteins in the GF nucleus (Fig. 3E). These results further support a critical role for the tyrosine in FIGQY motif in regulating nuclear levels of Nrg180.
Figure 3.
Subcellular localization of Nrg FIGQY mutants. A, Western blotting of 10 heads of WT controls (nrg14;Pac[WT]) and Nrg loss-of-function mutants (nrg14;Pac[nrg180ΔC], nrg14;Pac[nrg180ΔFIGQY], nrg14;Pac[nrg180Y-A], nrg14;Pac[nrg180Y-D], and nrg14;Pac[nrg180Y-F]). Tubulin served as a loading control. B, Western blots of cytoplasmic and nuclear fractions of WT controls and Nrg180 mutants using Thermo Fisher Scientific NE-PER® kit. The cytoplasmic fraction of ∼20 brains and the nuclear fraction of ∼130 brains were loaded per lane. Anti-Csp and anti-lamin labeling was used as loading control. C, Western blotting of the membrane (10 heads), soluble nuclear (100 heads), and chromatin (100 heads) fractions from WT controls (nrg14;Pac[WT]) and Nrg loss-of-function mutants (nrg14; Pac[nrg180ΔC], nrg14;Pac[nrg180ΔFIGQY], nrg14;Pac[nrg180Y-A], nrg14;Pac[nrg180Y-D], nrg14;Pac[nrg180Y-F], and nrg14;Pac[nrg180ΔPDZ]) probed with Nrg180cyto antibodies. Anti-Csp, anti-lamin, and anti-histone served as controls for the membrane, soluble nuclear, and the chromatin fractions, respectively. D, quantitative densitometry analysis of three independent Western blots of membrane (Subcellular Protein Fractionation Kit), soluble nuclear (Subcellular Protein Fractionation Kit), and nuclear (Thermo Fisher Scientific NE-PER®) fractions from WT controls (nrg14;Pac[WT]) and Nrg180 mutants probed with Nrg180cyto antibodies. Error bars, S.E. Statistical significance was assessed using Student's t test (*, p ≤ 0.05; **, p < 0.01; ***, p < 0.001). E, GF somas of nrg14;Pac[nrg180ΔFIGQY], nrg14;Pac[nrg180Y-D], and nrg14;Pac[nrg180Y-F] animals were labeled by expression of mCherry-NLS (magenta) with the R68A06-Gal4 line. Images of single confocal slices through the nucleus of the giant fiber somas labeled with Nrg180cyto (green) are shown. Scale bar, 5 μm.
However, Nrg180Y-A protein was present in soluble nuclear and chromatin fractions, whereas its levels were only mildly reduced in both fractions (Fig. 3, C and D). Whereas a Y-A mutation is often used to mimic the “unphosphorylated” form, we previously found that this is not the case for binding of the FIGQY motif to ankyrin; in contrast to the Y-F mutation, the Y-A mutation severely disrupted ankyrin binding (3). Because an alanine is structurally very distinctive from a phenylalanine and because it does not carry a negative charge, Nrg180Y-A protein may not act as the unphosphorylated or phosphorylated isoform in vivo. This may be a cause for its inconsistent subcellular localization.
C-terminal GFP tag but not HA tag affects nuclear levels of full-length Nrg180
Overexpression of full-length L1CAM or expression of the intracellular domain of L1CAM in nonneuronal cell lines results in altered target gene expression, such as the oncogene c-Myc (7, 11). To further study the functional role of Nrg in the nucleus and to determine whether it also affects Myc transcript levels in the Drosophila nervous system in vivo, we generated a UAS construct that allows the expression of the Nrg180 intracellular domain with an N-terminal HA tag (hereafter referred to as (NrgICD-HA). In addition to NrgICD-HA, we also analyzed the subcellular localization of C-terminally HA- and GFP-tagged full-length Nrg180 (hereafter referred to as NrgHA and NrgGFP). We were able to detect panneuronally (nSyb-Gal4) expressed NrgHA and NrgICD-HA in nuclear fractions of 80 Drosophila brains (Fig. 4A), and the HA-tagged Nrg180 proteins were also detected in the nucleus when selectively expressed in the GF neurons with the R68A06 Gal4 driver (Fig. 4C). In contrast to NrgHA, in nuclear fractions of 80 brains, NrgGFP was not detected (Fig. 4B), and immunohistochemical labeling of NrgGFP in the GF somas showed that it was present in the cytoplasm and the perinuclear region but not inside the nucleus (Fig. 4C). This suggests that the presence of a GFP but not an HA tag at the C terminus prevents or significantly reduces localization of Nrg in the nucleus.
Figure 4.
Subcellular localization of tagged Nrg180 constructs and in vivo effects of Nrg180 alterations on Myc expression, sensitivity to oxidative stress, and life span. A (left panel), Western blotting heads of controls (nSyb-Gal4) and animals expressing NrgHA (nSyb-Gal4,UAS-NrgHA). Csp served as a loading control. Right panels, Western blots of the membrane (5 heads) and soluble nuclear (80 heads) fractions (Subcellular Protein Fractionation Kit) of flies panneuronally (nSyb-Gal4) expressing NrgHA or NrgICD-HA probed with anti-HA antibodies. Csp and lamin labeling served as fraction-specific loading controls. B, Western blots of the cytoplasmic (5 heads) and nuclear (80 heads) fractions (NE-PER® kit) of flies panneuronally (nSyb-Gal4) expressing NrgGFP probed with anti-GFP antibodies. Csp and lamin labeling served as fraction-specific loading controls. C, images of single confocal slices through the nucleus of the giant fiber soma of flies expressing tagged NrgHA, NrgICD-HA, or NrgGFP (green) with the R68A06-Gal4 line. GF nucleus (magenta) was labeled by expression of UAS-mCherry-NLS. Scale bars, 5 μm. D, quantitative real-time PCR data showing the mean -fold expression of Drosophila Myc normalized to GAPDH in flies panneuronally overexpressing Nrg180 constructs (left) and in Nrg180 pacman mutants (right). Each sample was run in triplicate simultaneously with a nontemplate control. Error bars, S.E. of multiple independent experiments. *, p ≤ 0.05; **, p < 0.01; ***, p < 0.001 compared with nrg14;Pac[WT] using multiple-comparison one-way ANOVA. E, percent survival of animals panneuronally overexpressing NrgHA, NrgICD-HA, NrgGFP, or untagged Nrg normalized to control flies (nSyb-Gal4/+). Male flies (3–5 days old) were kept in vials containing 20 mm paraquat, and survival was assessed after 72 h. The mean of 10 vials (20 flies/vial) was calculated for each genotype. The difference in the mean of each genotype was compared with nSyb-Gal4/+ control flies using multiple-comparison ANOVA (*, p ≤ 0.05). F, Kaplan–Meier survival analysis of animals panneuronally (nSyb-Gal4) overexpressing NrgHA, NrgICD-HA, NrgGFP, or untagged Nrg180. Animals overexpressing Nrg180 (p = 0.0002), NrgHA (p = 0.002), and NrgICD-HA (p = 0.01) were statistically significant (Holm–Sidak method) from WT controls (w1118) and NrgGFP gain-of-function animals.
Nrg180 FIGQY motif affects Myc transcript levels in the Drosophila nervous system
Expression of full-length L1CAM or of the intracellular domain of L1CAM resulted in up-regulation of Myc transcript levels in vitro (7, 11). Here, we used quantitative real-time PCR to determine whether Nrg180 gain of function and mutations in the FIGQY motif affect Myc transcript levels also in nervous systems. We observed reduced Myc transcripts in nrg14;Pac[nrg180ΔC], nrg14; Pac[nrg180ΔFIGQY], and nrg14; P[nrg180Y-F] mutants but not in nrg14; Pac[nrg180Y-D] when compared with control animals (nrg14;Pac[WT]; Fig. 4D). These results correlate well with the nuclear but not the membrane levels of Nrg180 in the different mutants (Fig. 3). Similar to the in vitro studies with L1CAM, we observed the opposite in Nrg gain of function animals (7, 11). The Myc transcript levels were increased 3–5-fold when NrgICD-HA, NrgHA, or untagged Nrg180 was overexpressed panneuronally (Fig. 4D). However, overexpression of NrgGFP did not affect Myc transcript levels, which is consistent with our observation that NrgGFP was not detected in the nucleus (Fig. 4D). Although we cannot exclude the possibility that the failure of NrgGFP to not affect Myc transcript levels is not derived from a dysfunction at that plasma membrane that is distinct from its failure to localize to the nucleus, our results nevertheless suggest a novel noncanonical function for the FIGQY motif in regulating Myc transcript levels and that this function is likely to be associated with Nrg levels in the nucleus.
Nrg180 overexpression increases sensitivity to oxidative stress and reduces life span
Expression of the intracellular domain of L1CAM in glioblastoma stem cells up-regulates c-Myc expression, which in turn was shown to result in up-regulation of the DNA damage checkpoint response gene NBS1, suggesting that nuclear L1CAM renders these tumor cells radioresistant (7). Although these findings imply a protective function in cell survival in vitro in cancer cells, the impact of excess nuclear L1-type CAMs has not been studied in vivo or in noncancerous cells. Numerous studies suggest that overexpression of c-Myc increases the reactive oxygen species levels sufficiently to induce DNA damage and apoptosis (20–24). Therefore, we analyzed both the sensitivity to oxidative stress and the life span of flies overexpressing NrgICD-HA, NrgHA, NrgGFP, or untagged Nrg180 panneuronally. We observed that the survival rate was inversely proportional to the Myc transcript levels, when flies were exposed to 20 mm paraquat. Significantly fewer NrgHA or Nrg180 gain-of-function animals were alive after 72 h when compared with WT controls (nSyb-Gal/+) or NrgGFP-expressing flies (Fig. 5B). Although not significant, expression of NrgICD-HA also increased the sensitivity to oxidative stress. Similarly, we found that the life span of animals overexpressing Nrg180, NrgICD-HA, or NrgHA was significantly shorter than that of control animals (nSyb/+ and NrgGFP; Fig. 5C). Alterations of Myc transcript levels, sensitivity to oxidative stress, and longevity correlated with the overexpression of Nrg constructs that localized to the nucleus but not with an Nrg180 construct (NrgGFP) that did localize to the nucleus. This suggests that the impact of Nrg180 gain of function on an animal's viability is likely due to elevated levels of nuclear Nrg180 and the consequence of increased reactive oxygen species associated with elevated Myc transcript levels.
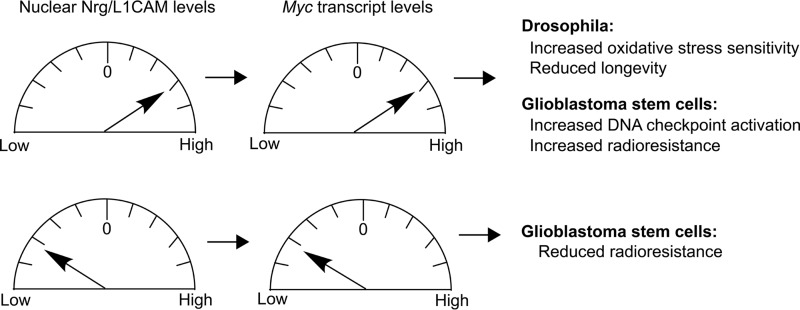
Figure 5.
Summary of gain- and loss-of-function effects of nuclear levels of L1-type CAMs.
Discussion
Previous studies showed that cytoplasmic as well as transmembrane fragments of L1CAM translocate to the nucleus in vertebrate neurons and glia (8, 9). Here, we found that, in addition to fragments, full-length Nrg180 and L1CAM also localize to the nucleus in Drosophila nervous systems. Whereas nuclear translocation of the proteolytically generated intracellular domains of receptors is very common, there is accumulating evidence showing that numerous holoreceptors, including tyrosine kinase receptors, epidermal growth factor receptors, and fibroblast growth factor receptors, translocate to the nucleus and potentially serve as transcription activators (25–31). A mechanism for the release of the sumoylated transmembrane 70-kDa L1CAM fragment from an endosomal compartment to the cytosol and its subsequent nuclear import has been described (8). It is thus conceivable that full-length L1-type CAMs use a similar mechanism.
The FERM-binding motif contains a putative nuclear localization motif (Fig. 1D), and in vitro studies showed that the lysine (Lys-1147) proximal to the transmembrane domain is required for the presence of the sumoylated 70- and 30-kDa vertebrate L1CAM fragments in the chromatin fraction (8, 9). We identified four other amino acids in the FERM-binding motif (19) that were also required for chromatin binding of full-length transmembrane fragments of L1CAM but did not affect the presence of L1CAM in soluble nuclear fractions (Fig. 1C). This suggests that other motifs play a role in regulating nuclear levels of L1-type CAMS via canonical or noncanonical mechanisms (32–35), and here, we reveal a novel role for the ankyrin-binding FIGQY motif in the regulation of nuclear levels of full-length Nrg180.
One mechanism by which the FIGQY motif can affect nuclear levels of Nrg is by regulating nuclear import directly, as a noncanonical NLS motif, or indirectly. Phosphorylation has been well-documented to regulate nuclear translocation and transcription activation (34). A prior study suggests that the ICD of L1CAM exists in an open and folded conformation and that phosphorylation of Tyr-1151 or Tyr-1229 within the FERM domain and FIGQY motif promotes opening of the cytoplasmic domain to mediate inside-out signaling of L1CAM (36). Therefore, phosphorylation of the FIGQY motif may have an indirect role in regulating nuclear levels of Nrg180 by inducing a conformation change of the intracellular domain as a mechanism to unmask motifs that regulate nuclear import or export. Alternatively, to regulating nuclear import, it is also conceivable that the FIGQY motif has a role in nuclear export. Here, the lack of the FIGQY motif or the unphosphorylated motif would promote Nrg180 nuclear export via an unknown mechanism.
Intriguingly, we find that the lack of the C-terminal PDZ motif or the addition of a small HA tag had no effect on nuclear levels of Nrg, but the addition of a GFP tag did (Figs. 2 and 4). It is likely that the large size of the GFP tag (∼27 kDa) or its tendency to dimerize (37) prevents posttranslational modification, such as phosphorylation or sumoylation required for nuclear trafficking sterically, or that it affects the normal conformation of the intracellular domain, which promotes such modifications.
The finding that decreased Myc transcript levels correlated with decreased Nrg180 levels in the nucleus but not in the membrane, which is unaltered in Nrg mutants (Figs. 3–5) suggests that Nrg180 exerts its effect on Myc transcript levels in the nucleus and has a promotive effect on Myc transcript abundance directly or indirectly. A role for Nrg in up-regulation of Myc transcript levels is further supported by the finding that, like for L1CAM and L1-ICD in vitro (7), the overexpression of full-length or ICD Nrg constructs that do localize to the nucleus had the reverse effect and increased Myc transcript levels (Figs. 4 and 5).
The fact that NrgGFP overexpression did not affect Myc transcript levels, while also not localizing to the nucleus, provides further evidence that up- and down-regulation of Nrg180 levels in the nucleus alters Myc transcript levels. Therefore, NrgGFP is also a valuable additional control to assess whether in vivo effects of Nrg180 gain-of-function constructs correlate with Myc transcript levels and nuclear levels of Nrg180.
In contrast to previous studies suggesting that elevated nuclear levels of L1CAM in glioblastoma stem cells promote cell survival by the activation of a DNA damage checkpoint gene, which renders tumor cell radio-resistant (7, 38), we found that only expression of Nrg180 constructs that localized to the nucleus and were associated with elevated Myc transcript levels correlated with an increase of an animal's sensitivity to oxidative stress and with reduced life span. Our in vivo findings are consistent with numerous previous studies in Drosophila and in vertebrates that show that increased expression of Myc is associated with an increase in reactive oxygen species, induction of DNA damage and apoptosis, and reduced life span (20, 23, 39–43). One possible cause for the differences between our in vivo studies and the previous in vitro studies may stem from the Warburg effect; metabolism of cancer cells is distinct from WT cells (44, 45).
In conclusion, aberrant expression of L1CAM correlates with poor prognosis of various cancers, including ovarian, pancreatic, glioma, and nonsmall cell lung cancer (46–48), and high nuclear L1CAM levels play an important mechanistic role in rendering tumor cells radioresistant (7, 38). Our findings that altered nuclear levels of Nrg180 associated with altered Myc transcript levels result in increased sensitivity to oxidative stress and reduced longevity (Fig. 5) suggest that abnormally high nuclear L1-type CAM levels may contribute to the pathology of neurodegenerative diseases, whereas low levels in noncancerous cells are likely to reduce oxidative stress and thereby promote cell survival. Thus, post-developmental knockdown of L1CAM selectively in the nucleus may be an effective approach to increase chemo- and radiosensitivity of tumor cells while providing protection against oxidative stress in noncancerous cells. Thus, the identification of a novel role for the FIGQY motif in regulating nuclear L1-type CAM levels and Myc transcript levels allows the exploration of new avenues to identify putative therapeutic reagents for cancer and degenerative diseases associated with oxidative stress.
Experimental procedures
Fly stocks
The w1118 (stock no. 38323), UAS-HuL1/Tm6B (stock no. 24171), UAS-mCherry-NLS (stock no. 38424), nSyb-Gal4 (stock no. 51635), OK307-Gal4 (stock no. 6488, referred to as A307), Da-Gal4 (stock no. 55850), R68A06-Gal4 (stock no. 39449), and R91H05-Gal4 (stock no. 40594) fly stocks were obtained from the Bloomington Stock Center (Bloomington, IN). The nrg14;Pac[WT], nrg14;Pac[nrg180ΔFIGQY], nrg14;Pac[nrg180ΔC], nrg14;Pac[nrg180Y-F], nrg14;Pac[nrg180Y-A], nrg14;Pac[nrg180Y-D], nrg14;Pac[nrg180ΔPDZ] pacman stocks and the 10× pUAST Nrg180, Nrg180-GFP, and Nrg180-HA (inserted at attP40) lines have been described previously (3). UAS-L1 and UAS-L1–4A lines have been described previously (16, 17, 19). Da-Gal4 and nSyb-Gal4 were used to drive expression ubiquitously or pan-neuronally, respectively. A307 and R68A06 Gal4 lines were used to drive expression broadly, including the Giant Fiber circuitry or more exclusively in the GF, respectively (18, 49). All genetic crosses were performed on standard fly media at 25 °C, and 2–5-day-old flies were used for all of the experiments unless indicated otherwise.
Generation of HA-tagged NrgICD construct
The intracellular domain of Nrg180 (nucleotides 3761–4207) was amplified with an HA tag included in the forward primer (forward primer, CAC CAT GTA CCC ATA CGA TGT TCC AGA TTA CGC TCG ACG CAA TCG TGG CGG AAA; reverse primer, TAG ACG TAG GTG GCC ACG GC (Integrated DNA Technologies, Coralville, IA)) and inserted into the pENTR vector (Invitrogen) via TOPO cloning. The construct was subsequently recombined into pUASTattB-10xUAS destination vector (3) using the standard gateway cloning protocol. The UAS-NrgICD-HA construct was verified by sequencing (Genewiz LLC). For comparability with other Nrg180 UAS constructs, transgenic lines were established at the attP40 site (BestGene Inc.).
Real-time PCR
Total RNA was isolated from 15 heads for each genotype using the RNAeasy Mini Kit (Qiagen, Valencia, CA) and quantified using NanoDropTM. RNA was then reverse-transcribed to cDNA according to the manufacturer's protocol for the Bio-Rad iScript cDNA synthesis kit. Quantitative expression was performed with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) using the Bio-Rad CFX96 Real-Time PCR Machine 7300 Gene Expression System. Primers against Drosophila Myc (forward primer, GAG CAA CAA CAG GCC ATC GAT ATA G; reserve primer, CCT TCA GAC TGG ATC GTT TGC G) and GAPDH (forward primer, AGC CAT CAC AGT CGA TTC; reverse primer, CCG ATG CGA CCA AAT CCA T) were used. GAPDH expression was used to normalize each sample. Each experiment was done in triplicates with a nontemplate control run simultaneously. The statistical significance (p ≤ 0.05) across various genotypes was assessed by multiple-comparison ANOVA (GraphPad Prism version 7.02; GraphPad Software Inc., La Jolla, CA).
Behavioral studies
The paraquat assay was performed similarly to previously described protocols (50–53). Twenty 3–5-day-old flies were transferred to vials containing standard cornmeal media with a final concentration of 20 mm paraquat (methyl viologen hydrate (98%), ACROS Organics). The survival of 100 flies (20 flies/vial) per genotype was determined after 72 h, and the percentages of survivors were plotted. The statistical significance (p ≤ 0.05) of the survivability across various genotypes was assessed by multiple-comparison ANOVA (GraphPad Prism version 7.02, GraphPad Software).
For life span studies, 20 males (10 vials/genotype) were kept in vials containing standard cornmeal medium at 25 °C. The surviving flies were transferred to the new vials every 2 days, and the number of dead flies was recorded. The percentages of surviving flies were plotted on the x axis versus the age in days on the y axis. For statistical analyses, Kaplan–Meier survival analyses with pairwise comparison using the Holm–Sidak method was used (GraphPad Prism version 7.02; GraphPad Software).
Western blotting and fractionation
For Western blots, dissected nervous systems or whole heads were homogenized in Laemmli buffer with HaltTM protease and phosphatase inhibitor mixture (catalog no. 87786, Thermo Scientific; 1:100 dilution). The samples were separated on SDS-polyacrylamide gels and subsequently blotted onto nitrocellulose using standard protocols. The Western blots were incubated with the following primary and secondary antibodies for 60 min at room temperature: Nrg180cyto (1:500 dilution (3)), L1-C20 (Santa Cruz Biotechnology, Inc., sc-c20; preabsorbed 1:200 dilution), α-tubulin (12G10, DSHB; 1:8000 dilution), anti-HA (3F10 (Roche Applied Science; 1:100 dilution) or C29F4 (Cell Signaling; 1:1000 dilution)), goat anti-rabbit (1:3000 dilution; Santa Cruz Biotechnology sc-2004), goat anti-mouse (Santa Cruz Biotechnology sc-516102 (1:3000 dilution) or LI-COR IRDye680RD (1:1000 dilution)), goat anti-rat (Jackson ImmunoResearch (1:3000 dilution) or LI-COR IRDye 800CW (1:1000 dilution)), or donkey anti-goat (Santa Cruz Biotechnology sc-2020 (1:3000 dilution) or LI-COR IRDye 800CW (1:1000 dilution)). Clarity ECL (Bio-Rad), X-ray films, and the LI-COR Odyssey Fx system were used for visualization of the protein bands. Dissected brains or whole heads were homogenized, and fractions were obtained with the subcellular fractionation (Thermo Fisher Scientific NE-PER® or the Thermo Fisher Scientific Subcellular Protein Fractionation Kit) as per the manufacturer's instructions. Nuclear and cytoplasmic fractions (NE-PER® kit) or the membrane, soluble nuclear, and chromatin-bound fractions (Subcellular Protein Fractionation Kit) were analyzed by Western blotting using L1-C20 or Nrg180cyto antibodies as described above. In addition, the following primary antibodies were used for validation of the distinct fractions: cysteine string protein (Csp 6D6 or ab49, DSHB, 1:200 dilution), lamin Dmo (ADL67.10, DSHB, 1:500 dilution), and Ac-histone H3 (Santa Cruz Biotechnology, sc-56616, 1:10 dilution) for the membrane, nuclear, and chromatin fractions, respectively. For analyses of differences in nuclear import of Nrg in the diverse pacman mutants, the images of three independent fractions were subjected to densitometry analysis using ImageJ (Wayne Rasband, National Institutes of Health) and normalized to fraction-specific loading controls and then to nrg14;Pac[WT]. Relative band densities were compared with nrg14;Pac[WT] using Student's t test to determine statistically significant differences (p ≤ 0.05).
GF preparation and immunohistochemistry
The procedure for adult Drosophila nervous system dissection and subsequent dye injection has been described previously in detail (54, 55). To visualize the localization of protein in the giant fiber nucleus, either a 10 mm Alexa Fluor 555 hydrazide (Molecular Probes) in 200 mm KCl or TRITC-dextran (Invitrogen) in 2 m potassium acetate was injected into the GF axons by passing hyperpolarizing or depolarizing current, respectively. Alternatively, UAS-mCherry-NLS was expressed with the R68A06 Gal4 driver. The samples were fixed with 4% paraformaldehyde and incubated with the following primary and secondary antibodies: L1CAM (Santa Cruz Biotechnology, c-20, 1:50 dilution), Nrg180cyto (1:500 dilution), GFP (ab13970, Abcam, 1:2000 dilution), donkey anti-goat Cy2 (Jackson ImmunoResearch, 1:500 dilution), goat anti-rabbit Cy2 (Jackson ImmunoResearch, 1:200 dilution), donkey anti-rabbit Alexa Fluor 488 (Invitrogen, 1:1000), or anti-HA (3F10 Roche Applied Science, 1:100 dilution). Samples were scanned at a resolution of 1024 × 1024 pixels, ×2.5 zoom, and 0.2-μm step size with a Nikon C1 or a Nikon A1R confocal microscope using a Plan Apo λ ×60 oil immersion objective. Images were processed using Nikon Elements Advanced Research version 4.4 and Adobe Photosuite CS5 software. For analysis of nuclear labeling, the Binary Editor of Nikon Elements Advanced Research 4.4 was used to trace and three-dimensionally reconstruct the GF nuclei, and the ND Images Arithmetic function was used to extract Nrg180cyto labeling. The sum fluorescent intensity was normalized to the trace nuclei area, and the average for each genotype with the S.E. was plotted. Two-tailed Student's t test was used to determine statistically significant differences.
Author contributions
P. P. K., T. P., B. H., and T. A. G. formal analysis; P. P. K. and T. A. G. validation; P. P. K., T. P., B. P. D., B. H., J. B., A. R., and T. A. G. investigation; P. P. K., T. P., B. P. D., J. B., J. P., and T. A. G. writing-review and editing; T. P. and T. A. G. visualization; B. H. and T. A. G. funding acquisition; J. P. and T. A. G. resources; T. A. G. conceptualization; T. A. G. data curation; T. A. G. software; T. A. G. supervision; T. A. G. writing-original draft.
Acknowledgments
We thank Julie Freund for excellent technical assistance as well as the Bloomington Stock Center (Bloomington, IN; supported by National Institutes of Health Grant P40OD018537), the Vienna Drosophila Resource Center (Vienna, Austria), and the Developmental Studies Hybridoma Bank (Iowa City, IA) for fly stocks and antibodies. We especially thank the A. Keene laboratory for the use of the Nikon A1 confocal microscope.
This work was supported by NINDS, National Institutes of Health, Grant R15 NS090043 (to T. A. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CAM
- cell adhesion molecule
- L1CAM
- L1-type cell adhesion molecule
- TRITC
- tetramethylrhodamine isothiocyanate
- FERM
- 4.1 protein-ezrin-radixin-moesin
- ICD
- intracellular domain
- NLS
- nuclear localization motif
- Nrg
- neuroglian
- Csp
- cysteine string protein
- HA
- hemagglutinin
- UAS
- upstream activating sequence
- ANOVA
- analysis of variance.
References
- 1. Hortsch M. (2000) Structural and functional evolution of the L1 family: are four adhesion molecules better than one? Mol. Cell. Neurosci. 15, 1–10 10.1006/mcne.1999.0809 [DOI] [PubMed] [Google Scholar]
- 2. Davis J. Q., McLaughlin T., and Bennett V. (1993) Ankyrin-binding proteins related to nervous system cell adhesion molecules: candidates to provide transmembrane and intercellular connections in adult brain. J. Cell Biol. 121, 121–133 10.1083/jcb.121.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Enneking E. M., Kudumala S. R., Moreno E., Stephan R., Boerner J., Godenschwege T. A., and Pielage J. (2013) Transsynaptic coordination of synaptic growth, function, and stability by the L1-type CAM neuroglian. PLoS Biol. 11, e1001537 10.1371/journal.pbio.1001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nishimura K., Yoshihara F., Tojima T., Ooashi N., Yoon W., Mikoshiba K., Bennett V., and Kamiguchi H. (2003) L1-dependent neuritogenesis involves ankyrinB that mediates L1-CAM coupling with retrograde actin flow. J. Cell Biol. 163, 1077–1088 10.1083/jcb.200303060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tuvia S., Garver T. D., and Bennett V. (1997) The phosphorylation state of the FIGQY tyrosine of neurofascin determines ankyrin-binding activity and patterns of cell segregation. Proc. Natl. Acad. Sci. U.S.A. 94, 12957–12962 10.1073/pnas.94.24.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whittard J. D., Sakurai T., Cassella M. R., Gazdoiu M., and Felsenfeld D. P. (2006) MAP kinase pathway-dependent phosphorylation of the L1-CAM ankyrin binding site regulates neuronal growth. Mol. Biol. Cell 17, 2696–2706 10.1091/mbc.e06-01-0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng L., Wu Q., Huang Z., Guryanova O. A., Huang Q., Shou W., Rich J. N., and Bao S. (2011) L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. EMBO J. 30, 800–813 10.1038/emboj.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lutz D., Wolters-Eisfeld G., Joshi G., Djogo N., Jakovcevski I., Schachner M., and Kleene R. (2012) Generation and nuclear translocation of sumoylated transmembrane fragment of cell adhesion molecule L1. J. Biol. Chem. 287, 17161–17175 10.1074/jbc.M112.346759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lutz D., Wolters-Eisfeld G., Schachner M., and Kleene R. (2014) Cathepsin E generates a sumoylated intracellular fragment of the cell adhesion molecule L1 to promote neuronal and Schwann cell migration as well as myelination. J. Neurochem. 128, 713–724 10.1111/jnc.12473 [DOI] [PubMed] [Google Scholar]
- 10. Maretzky T., Schulte M., Ludwig A., Rose-John S., Blobel C., Hartmann D., Altevogt P., Saftig P., and Reiss K. (2005) L1 is sequentially processed by two differently activated metalloproteases and presenilin/γ-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol. Cell Biol. 25, 9040–9053 10.1128/MCB.25.20.9040-9053.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riedle S., Kiefel H., Gast D., Bondong S., Wolterink S., Gutwein P., and Altevogt P. (2009) Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/γ-secretase activity. Biochem. J. 420, 391–402 10.1042/BJ20081625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalus I., Schnegelsberg B., Seidah N. G., Kleene R., and Schachner M. (2003) The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J. Biol. Chem. 278, 10381–10388 10.1074/jbc.M208351200 [DOI] [PubMed] [Google Scholar]
- 13. Mechtersheimer S., Gutwein P., Agmon-Levin N., Stoeck A., Oleszewski M., Riedle S., Postina R., Fahrenholz F., Fogel M., Lemmon V., and Altevogt P. (2001) Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 155, 661–673 10.1083/jcb.200101099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Q. E. (2015) DNA damage responses in cancer stem cells: implications for cancer therapeutic strategies. World J. Biol. Chem. 6, 57–64 10.4331/wjbc.v6.i3.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siegenthaler D., Enneking E. M., Moreno E., and Pielage J. (2015) L1CAM/Neuroglian controls the axon-axon interactions establishing layered and lobular mushroom body architecture. J. Cell Biol. 208, 1003–1018 10.1083/jcb.201407131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Godenschwege T. A., Kristiansen L. V., Uthaman S. B., Hortsch M., and Murphey R. K. (2006) A conserved role for Drosophila neuroglian and human L1-CAM in central-synapse formation. Curr. Biol. 16, 12–23 10.1016/j.cub.2005.11.062 [DOI] [PubMed] [Google Scholar]
- 17. Kudumala S., Freund J., Hortsch M., and Godenschwege T. A. (2013) Differential effects of human L1CAM mutations on complementing guidance and synaptic defects in Drosophila melanogaster. PLoS One 8, e76974 10.1371/journal.pone.0076974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T. T., Misra S., Murphy C., Scully A., Carlson J. W., Wan K. H., Laverty T. R., Mungall C., Svirskas R., Kadonaga J. T., Doe C. Q., Eisen M. B., Celniker S. E., and Rubin G. M. (2008) Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 9715–9720 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng L., Itoh K., and Lemmon V. (2005) L1-mediated branching is regulated by two ezrin-radixin-moesin (ERM)-binding sites, the RSLE region and a novel juxtamembrane ERM-binding region. J. Neurosci. 25, 395–403 10.1523/JNEUROSCI.4097-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffman B., and Liebermann D. A. (2008) Apoptotic signaling by c-MYC. Oncogene 27, 6462–6472 10.1038/onc.2008.312 [DOI] [PubMed] [Google Scholar]
- 21. Tanaka H., Matsumura I., Ezoe S., Satoh Y., Sakamaki T., Albanese C., Machii T., Pestell R. G., and Kanakura Y. (2002) E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappa B activity that facilitates MnSOD-mediated ROS elimination. Mol. Cell 9, 1017–1029 10.1016/S1097-2765(02)00522-1 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka K., Nagaoka S., Takemura T., Arai T., Sawabe M., Takubo K., Sugihara K., Kitagawa M., and Hirokawa K. (2002) Incidence of apoptosis increases with age in colorectal cancer. Exp. Gerontol. 37, 1469–1479 10.1016/S0531-5565(02)00167-5 [DOI] [PubMed] [Google Scholar]
- 23. Vafa O., Wade M., Kern S., Beeche M., Pandita T. K., Hampton G. M., and Wahl G. M. (2002) c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol. Cell 9, 1031–1044 10.1016/S1097-2765(02)00520-8 [DOI] [PubMed] [Google Scholar]
- 24. Yu L., Hitchler M. J., Sun W., Sarsour E. H., Goswami P. C., Klingelhutz A. J., and Domann F. E. (2009) AP-2α inhibits c-MYC induced oxidative stress and apoptosis in HaCaT human keratinocytes. J. Oncol. 2009, 780874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brand T. M., Iida M., Luthar N., Starr M. M., Huppert E. J., and Wheeler D. L. (2013) Nuclear EGFR as a molecular target in cancer. Radiother. Oncol. 108, 370–377 10.1016/j.radonc.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 26. Carpenter G. (2003) ErbB-4: mechanism of action and biology. Exp. Cell Res. 284, 66–77 10.1016/S0014-4827(02)00100-3 [DOI] [PubMed] [Google Scholar]
- 27. Carpenter G., and Liao H. J. (2009) Trafficking of receptor tyrosine kinases to the nucleus. Exp. Cell Res. 315, 1556–1566 10.1016/j.yexcr.2008.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carpenter G., and Liao H. J. (2013) Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 5, a008979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., and Hung M. C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 10.1038/ncb0901-802 [DOI] [PubMed] [Google Scholar]
- 30. Schlessinger J., and Lemmon M. A. (2006) Nuclear signaling by receptor tyrosine kinases: the first robin of spring. Cell 127, 45–48 10.1016/j.cell.2006.09.013 [DOI] [PubMed] [Google Scholar]
- 31. Wang S. C., and Hung M. C. (2009) Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15, 6484–6489 10.1158/1078-0432.CCR-08-2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cai M., Wang S., Xing J., and Zheng C. (2011) Characterization of the nuclear import and export signals, and subcellular transport mechanism of varicella-zoster virus ORF9. J. Gen. Virol. 92, 621–626 10.1099/vir.0.027029-0 [DOI] [PubMed] [Google Scholar]
- 33. Freitas N., and Cunha C. (2009) Mechanisms and signals for the nuclear import of proteins. Curr. Genomics 10, 550–557 10.2174/138920209789503941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nardozzi J. D., Lott K., and Cingolani G. (2010) Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8, 32 10.1186/1478-811X-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wagstaff K. M., and Jans D. A. (2009) Importins and beyond: non-conventional nuclear transport mechanisms. Traffic 10, 1188–1198 10.1111/j.1600-0854.2009.00937.x [DOI] [PubMed] [Google Scholar]
- 36. Chen M. M., Leland H. A., Lee C. Y., and Silletti S. (2009) Tyrosine and serine phosphorylation regulate the conformation and subsequent threonine phosphorylation of the L1 cytoplasmic domain. Biochem. Biophys. Res. Commun. 389, 257–264 10.1016/j.bbrc.2009.08.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Costantini L. M., Fossati M., Francolini M., and Snapp E. L. (2012) Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic 13, 643–649 10.1111/j.1600-0854.2012.01336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rached J., Nasr Z., Abdallah J., and Abou-Antoun T. (2016) L1-CAM knock-down radiosensitizes neuroblastoma IMR-32 cells by simultaneously decreasing MycN, but increasing PTEN protein expression. Int. J. Oncol. 49, 1722–1730 10.3892/ijo.2016.3625 [DOI] [PubMed] [Google Scholar]
- 39. de la Cova C., Abril M., Bellosta P., Gallant P., and Johnston L. A. (2004) Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116 10.1016/S0092-8674(04)00214-4 [DOI] [PubMed] [Google Scholar]
- 40. Greer C., Lee M., Westerhof M., Milholland B., Spokony R., Vijg J., and Secombe J. (2013) Myc-dependent genome instability and lifespan in Drosophila. PLoS One 8, e74641 10.1371/journal.pone.0074641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hofmann J. W., Zhao X., De Cecco M., Peterson A. L., Pagliaroli L., Manivannan J., Hubbard G. B., Ikeno Y., Zhang Y., Feng B., Li X., Serre T., Qi W., Van Remmen H., Miller R. A., et al. (2015) Reduced expression of MYC increases longevity and enhances healthspan. Cell 160, 477–488 10.1016/j.cell.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Montero L., Müller N., and Gallant P. (2008) Induction of apoptosis by Drosophila Myc. Genesis 46, 104–111 10.1002/dvg.20373 [DOI] [PubMed] [Google Scholar]
- 43. Sedivy J. M. (2015) Abstract IA20: reduced expression of MYC increases longevity and enhances healthspan. Mol. Cancer Res. 13, IA20 10.1158/1557-3125.MYC15-IA20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liberti M. V., and Locasale J. W. (2016) The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218 10.1016/j.tibs.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwartz L., Supuran C. T., and Alfarouk K. O. (2017) The Warburg effect and the hallmarks of cancer. Anticancer Agents Med. Chem. 17, 164–170 10.2174/1871520616666161031143301 [DOI] [PubMed] [Google Scholar]
- 46. Bondong S., Kiefel H., Hielscher T., Zeimet A. G., Zeillinger R., Pils D., Schuster E., Castillo-Tong D. C., Cadron I., Vergote I., Braicu I., Sehouli J., Mahner S., Fogel M., and Altevogt P. (2012) Prognostic significance of L1CAM in ovarian cancer and its role in constitutive NF-κB activation. Ann. Oncol. 23, 1795–1802 10.1093/annonc/mdr568 [DOI] [PubMed] [Google Scholar]
- 47. Fogel M., Gutwein P., Mechtersheimer S., Riedle S., Stoeck A., Smirnov A., Edler L., Ben-Arie A., Huszar M., and Altevogt P. (2003) L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 362, 869–875 10.1016/S0140-6736(03)14342-5 [DOI] [PubMed] [Google Scholar]
- 48. Li S., Jo Y. S., Lee J. H., Min J. K., Lee E. S., Park T., Kim J. M., and Hong H. J. (2009) L1 cell adhesion molecule is a novel independent poor prognostic factor of extrahepatic cholangiocarcinoma. Clin. Cancer Res. 15, 7345–7351 10.1158/1078-0432.CCR-09-0959 [DOI] [PubMed] [Google Scholar]
- 49. Kudumala S. R., Penserga T., Börner J., Slipchuk O., Kakad P., Lee L. H., Qureshi A., Pielage J., and Godenschwege T. A. (2017) Lissencephaly-1 dependent axonal retrograde transport of L1-type CAM neuroglian in the adult Drosophila central nervous system. PLoS One 12, e0183605 10.1371/journal.pone.0183605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cassar M., Issa A. R., Riemensperger T., Petitgas C., Rival T., Coulom H., Iché-Torres M., Han K. A., and Birman S. (2015) A dopamine receptor contributes to paraquat-induced neurotoxicity in Drosophila. Hum. Mol. Genet. 24, 197–212 10.1093/hmg/ddu430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hosamani R., Ramesh S. R., and Muralidhara (2010) Attenuation of rotenone-induced mitochondrial oxidative damage and neurotoxicty in Drosophila melanogaster supplemented with creatine. Neurochem. Res. 35, 1402–1412 10.1007/s11064-010-0198-z [DOI] [PubMed] [Google Scholar]
- 52. Jumbo-Lucioni P. P., Hopson M. L., Hang D., Liang Y., Jones D. P., and Fridovich-Keil J. L. (2013) Oxidative stress contributes to outcome severity in a Drosophila melanogaster model of classic galactosemia. Dis. Model. Mech. 6, 84–94 10.1242/dmm.010207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Minois N., Carmona-Gutierrez D., Bauer M. A., Rockenfeller P., Eisenberg T., Brandhorst S., Sigrist S. J., Kroemer G., and Madeo F. (2012) Spermidine promotes stress resistance in Drosophila melanogaster through autophagy-dependent and -independent pathways. Cell Death Dis. 3, e401 10.1038/cddis.2012.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boerner J., and Godenschwege T. A. (2010) Application for the Drosophila ventral nerve cord standard in neuronal circuit reconstruction and in-depth analysis of mutant morphology. J. Neurogenet. 24, 158–167 10.3109/01677063.2010.489624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boerner J., and Godenschwege T. A. (2011) Whole mount preparation of the adult Drosophila ventral nerve cord for giant fiber dye injection. J. Vis. Exp. 2011, 3080 10.3791/3080 [DOI] [PMC free article] [PubMed] [Google Scholar]