Abstract
In vertebrate cells, mitochondrial Ca2+ uptake by the mitochondrial calcium uniporter (MCU) leads to Ca2+-mediated stimulation of an intramitochondrial pyruvate dehydrogenase phosphatase (PDP). This enzyme dephosphorylates serine residues in the E1α subunit of pyruvate dehydrogenase (PDH), thereby activating PDH and resulting in increased ATP production. Although a phosphorylation/dephosphorylation cycle for the E1α subunit of PDH from nonvertebrate organisms has been described, the Ca2+-mediated PDP activation has not been studied. In this work, we investigated the Ca2+ sensitivity of two recombinant PDPs from the protozoan human parasites Trypanosoma cruzi (TcPDP) and T. brucei (TbPDP) and generated a TcPDP-KO cell line to establish TcPDP's role in cell bioenergetics and survival. Moreover, the mitochondrial localization of the TcPDP was studied by CRISPR/Cas9-mediated endogenous tagging. Our results indicate that TcPDP and TbPDP both are Ca2+-sensitive phosphatases. Of note, TcPDP-KO epimastigotes exhibited increased levels of phosphorylated TcPDH, slower growth and lower oxygen consumption rates than control cells, an increased AMP/ATP ratio and autophagy under starvation conditions, and reduced differentiation into infective metacyclic forms. Furthermore, TcPDP-KO trypomastigotes were impaired in infecting cultured host cells. We conclude that TcPDP is a Ca2+-stimulated mitochondrial phosphatase that dephosphorylates TcPDH and is required for normal growth, differentiation, infectivity, and energy metabolism in T. cruzi. Our results support the view that one of the main roles of the MCU is linked to the regulation of intramitochondrial dehydrogenases.
Keywords: CRISPR/Cas9, energy metabolism, Trypanosoma cruzi, bioenergetics, calcium, calcium signaling, mitochondrial calcium uniporter, mitochondrial dehydrogenase, pyruvate dehydrogenase phosphatase
Introduction
Mitochondrial Ca2+ uptake in vertebrate cells is important for regulating the activity of three mitochondrial dehydrogenases (1). Intramitochondrial Ca2+ stimulates pyruvate, 2-oxoglutarate, and isocitrate dehydrogenases, leading to stimulation of oxidative phosphorylation and ATP production (2).
The pyruvate dehydrogenase complex (PDC)5 catalyzes the conversion of pyruvate to acetyl-CoA and links glycolysis with several pathways such as the Krebs cycle and the synthesis of fatty acids and cholesterol. PDC is a multienzyme complex consisting of multiple copies of E1 (subunits α and β, pyruvate dehydrogenase), E2 (dihydrolipoyl transacetylase), and E3 (dihydrolipoyl dehydrogenase) subunits, along with an E3-binding protein (3). In vertebrate cells the PDC is regulated through the interconversion of phosphorylated and dephosphorylated forms (4). Inactivation of PDC results by phosphorylation of three serine residues on the α chain of the E1 subunit named sites 1 (Ser-264), 2 (Ser-271), and 3 (Ser-203) (numbers correspond to the mature human enzyme) (5, 6). This phosphorylation is catalyzed by a pyruvate dehydrogenase kinase (PDK) whereas their dephosphorylation and reactivation, which is stimulated by Ca2+, is catalyzed by a pyruvate dehydrogenase phosphatase (PDP). Of the two PDP catalytic subunits present in mammalian mitochondria, PDP1c is the one activated by Ca2+ (7).
There is evidence that the PDC is regulated by phosphorylation in yeast, plants, and invertebrates (8–11) but their sensitivity to Ca2+ is unknown (2). Early work suggested that the acquisition of Ca2+ sensitivity by mitochondrial dehydrogenases was linked to the acquisition of a mitochondrial Ca2+ uniporter (MCU) (1, 12).
The finding of MCU activity in trypanosomatids, such as Trypanosoma cruzi (13, 14), the agent of Chagas disease, and T. brucei (15), which belongs to the group of parasites that cause sleeping sickness, was important for the discovery of a gene encoding a modulator of the uniporter (mitochondrial calcium uptake 1, or MICU1) (16) and the pore subunit of the channel or MCU (17–19). After the molecular identification of MCU, other components of the MCU complex were described (20–23), but only orthologs of MCU, MCUb, MICU1, and MICU2 are present in trypanosomes, whereas MCU regulator (MCUR1) and essential MCU regulator (EMRE) are absent (24, 25). Mitochondrial Ca2+ transport in trypanosomes is critical for shaping the dynamics of cytosolic Ca2+ increases, for the bioenergetics of the cells, and for viability and infectivity (26, 27). Interestingly, the PDH E1α subunit of trypanosomes exhibits the putative phosphorylation sites present in the mammalian enzyme (28). This enzyme is active in all stages of T. cruzi (29) and T. brucei (26, 30) and our previous results (26) suggested that Ca2+ could stimulate it, although a direct stimulation was not demonstrated.
In this study, we report the identification of the mitochondrial PDP of T. cruzi (TcPDP) and T. brucei (TbPDP), demonstrate their activation by Ca2+, and evaluate the function of the T. cruzi enzyme in growth, differentiation, infectivity, and bioenergetics by CRISPR/Cas9-driven ablation of its gene. The results provide evidence of the presence of a Ca2+-activated PDP in organisms other than vertebrates and support the view (1) that one of the main roles of the mitochondrial Ca2+ uniporter is linked to the regulation of intramitochondrial dehydrogenases.
Results
Sequence analysis
Putative T. cruzi pyruvate dehydrogenase phosphatase (TcPDP) is a 415-aa protein with an expected size of 46 kDa encoded by a single copy gene (TcCLB.506739.200). The gene is located on chromosome 3 of T. cruzi CL Brener Esmeraldo-like genome sequence (curated reference strain, TriTrypDB) (31). This is the gene ortholog of T. brucei putative protein phosphatase 2C (PP2C, gene ID: Tb427.05.1660) that has been proposed to be the specific phosphatase of TbPDH E1α subunit (TbPDP) in a proteomic study using stable isotope labeling of amino acids in cell culture (SILAC) in combination with MS (32). TbPDP is a homolog of the PDH-activating enzyme in other systems and is up-regulated at the protein level upon transition to the insect form (32). TbPDP and TcPDP belong to the PP2C/PPM family, which is a group of Mg2+/Mn2+-dependent serine/threonine phosphatases essential for regulation of cell cycle and stress signaling pathways in different cell types (InterPro protein sequence analysis and classification) (33). Both enzymes exhibit 31% identity with human PDP catalytic subunit 1 at the amino acid level. Using ScanProsite tool (34) for detection of signature matches on TcPDP amino acid sequence we found a PPM-type phosphatase domain (aa 143–414). This type of domain is also present in TbPDP (aa 155–424) and in the catalytic subunit of the mammalian Ca2+-sensitive PDP1c (35).
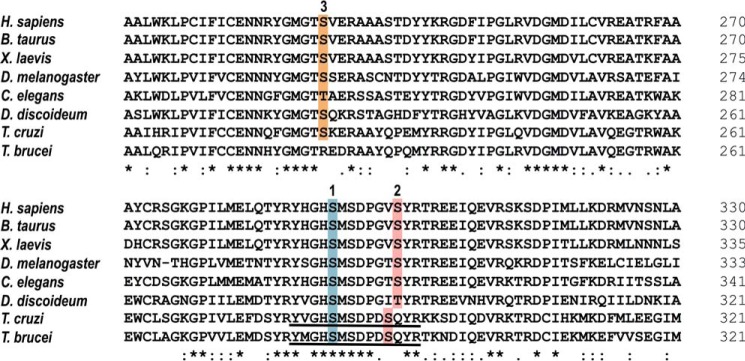
Studies of purified PDH from bovine kidney and heart mitochondria (36) and pig heart mitochondria (37) have identified the Ser-5 and Ser-12 of the PDH E1α peptide Tyr-His-Gly-His-Ser-Met-Ser-Asn-Pro-Gly-Val-Ser-Tyr-Arg, as sites responsible for inactivation by phosphorylation by their intrinsic kinase (indicated in bold). These phosphorylation sites correspond to sites 1 and 2 of human PDH E1α. Similar peptides, Tyr-Val-Gly-His-Ser-Met-Ser-Asp-Pro-Asp-Ser-Gln-Tyr-Arg and Tyr-Met-Gly-His-Ser-Met-Ser-Asp-Pro-Asp-Ser-Gln-Tyr-Arg, are present in the T. cruzi (TcCLB.507831.70) and T. brucei (Tb927.10.12700) PDH E1α orthologs, respectively (Fig. 1). A third phosphorylation site is present in the mammalian orthologs and conserved in T. cruzi but absent in T. brucei (Fig. 1). The presence of these specific serine residues could account for a phosphorylation/dephosphorylation model resembling mammalian E1α.
Figure 1.
Phosphorylation sites of PDH E1α orthologs. Shown is an amino acid sequence comparison of PDH E1α orthologs across several species, where putative phosphorylation sites 1 (blue), 2 (pink), and 3 (orange) are highlighted. Sequence of the phosphopeptides used as substrates in PDP activity assays are underlined in T. cruzi and T. brucei orthologs. Accession numbers: Homo sapiens (NP_000275.1), Bos taurus (NP_001094516.1), Xenopus laevis (AAI06671.1), Drosophila melanogaster (NP_726946.1), Caenorhabditis elegans (NP_871953.1), Dictyostelium discoideum (XP_629349.1), T. cruzi (XP_814071.1), and T. brucei (XP_823475.1). Protein alignment was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (81) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.). Asterisk indicates positions which have a single, fully conserved residue; colon indicates conservation between groups of strongly similar properties (Clustal scoring >0.5); period indicates conservation between groups of weakly similar properties (Clustal scoring ≤0.5).
TcPDP localization
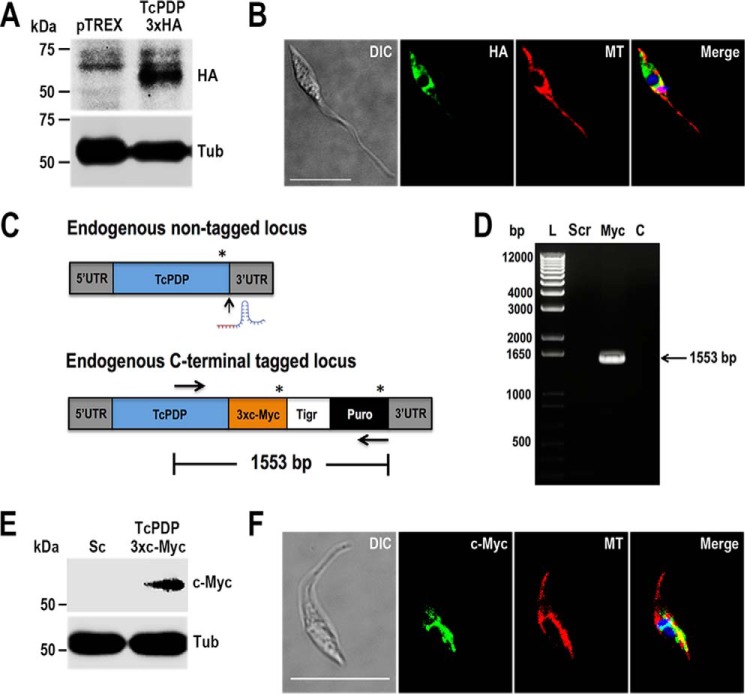
In eukaryotic cells, pyruvate dehydrogenase is a conserved enzyme that catalyzes the decarboxylation of pyruvate into acetyl-CoA, a reaction that takes place in the mitochondrial matrix. To determine the cellular localization of putative TcPDP we generated a mutant cell line that overexpresses a tagged version of the protein (TcPDP-3xHA) by cloning the PCR-amplified ORF of the gene into pTREX-n/3xHA vector (38). After selection, G418-resistant cells were analyzed by Western blotting and immunofluorescence analyses (IFA) using monoclonal anti-HA antibodies (Fig. 2, A and B). Fluorescence microscopy images showed TcPDP localized to mitochondria, as observed by partial co-localization with the mitochondrial marker MitoTracker (Fig. 2B).
Figure 2.
TcPDP localization. A, TcPDP-3xHA overexpression was confirmed by Western blot analysis using anti-HA monoclonal antibodies, with pTREX empty vector as control cell line. Tubulin (Tub) was used as loading control. B, IFA showed partial co-localization between TcPDP-3xHA (green) and MitoTracker (MT, red) in the merged image (yellow). C, schematic representation of CRISPR/Cas9-mediated TcPDP endogenous tagging. Asterisks indicate stop codons. Vertical arrow indicates Cas9 cut site at the 3′ end of the gene. Horizontal arrows represent annealing sites for primers used to verify TcPDP endogenous tagging by PCR analysis shown in D. D, lanes on 1% agarose gel: L, 1 kb plus ladder; Scr, control cells transfected with scramble sgRNA; Myc, TcPDP-3xc-Myc; C, PCR negative control. E and F, TcPDP endogenous tagging was verified by Western blotting and IFA using antibodies anti-c-Myc. Merged image on F shows co-localization (yellow) of TcPDP-3xc-Myc (green) and MT (red). DAPI staining (blue) and differential interference contrast (DIC) images are also shown. Scale bars = 10 μm.
We have observed previously that protein overexpression can lead to mislocalization of proteins (39). Therefore, to obtain an accurate localization pattern we used CRISPR/Cas9 system to generate an endogenously tagged mutant cell line (TcPDP-3xc-Myc), as described previously (39, 40). TcPDP tagging was confirmed by PCR (Fig. 2, C and D) and also by Western blotting and IFA using monoclonal anti–c-Myc antibodies (Fig. 2, E and F). Our results indicate that TcPDP is a mitochondrial enzyme, as shown by co-localization with MitoTracker (Fig. 2F, merged image).
Calcium sensitivity of PDP in trypanosomes
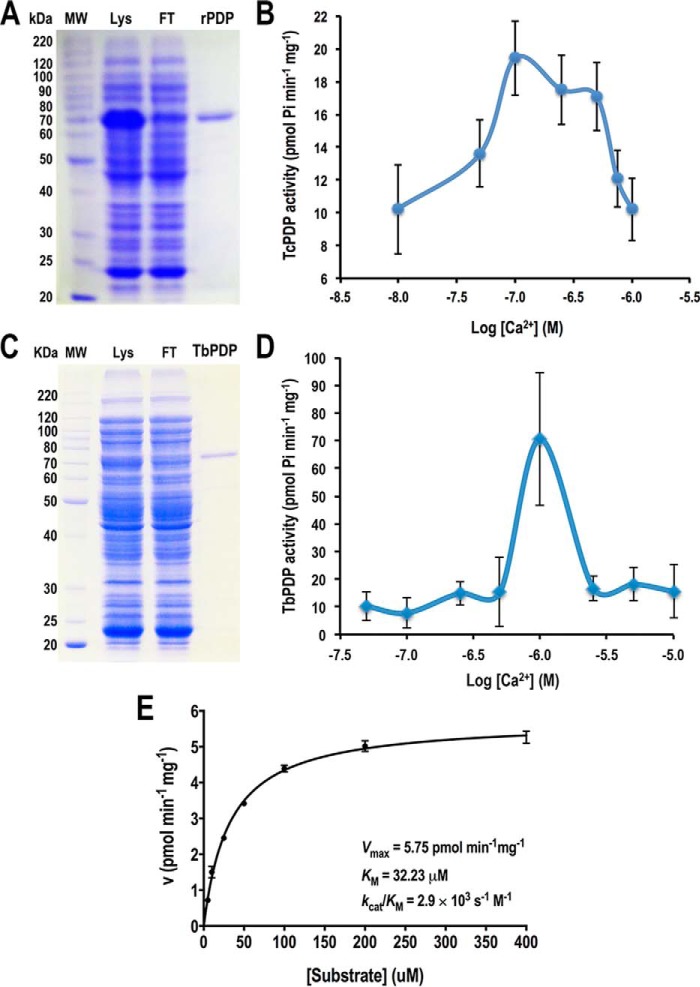
None of the dehydrogenases stimulated by Ca2+ in vertebrates has been studied in detail in trypanosomes. However, the PDH E1α subunit of T. cruzi and T. brucei possesses putative phosphorylation sites that are similar to those of the mammalian homologs (28), suggesting that, as the mammalian enzyme, they could be activated by Ca2+-stimulated dephosphorylation. To further investigate the role of mitochondrial putative TcPDP and TbPDP we induced their heterologous expression in Escherichia coli and assayed their activity under different free Ca2+ concentrations. Gene cloning, protein expression, and purification under native conditions were performed as described under “Experimental Procedures.” Both recombinant proteins were expressed as fusion proteins with an N-terminal polyhistidine tag. In Fig. 3, A and C, we analyzed different fractions from TcPDP and TbPDP protein purification by SDS-PAGE on 10% gels. In each image, the band corresponding to the recombinant protein is visualized in the total lysate (Lys), but is absent in the flow through fraction (FT), and then the purified protein is observed in the eluted fraction as a single band (TcPDP and TbPDP lanes). Both proteins exhibit molecular weights close to their expected sizes (63 kDa and 64 kDa, respectively), according to the protein molecular weight standard (MW). Purified recombinant PDPs were quantified and used for enzymatic assays. PDP activity was assayed by measuring phosphate release from a synthetic phosphopeptide from either TcPDH or TbPDH E1α subunits containing the phosphorylated sites 1 and 2 that regulate PDH activity (41), as described in “Experimental Procedures.” A colorimetric assay with malachite green/ammonium molybdate solution allowed the quantification of total Pi released during substrate dephosphorylation under different Ca2+ concentrations. Our results indicate that TcPDP and TbPDP exhibit maximal activity at 100 nm Ca2+ and 1 μm Ca2+, respectively (Fig. 3, B and D), which suggests a physiological Ca2+ response in vivo for the putative phosphatases that activate the mitochondrial pyruvate dehydrogenases of T. cruzi and T. brucei.
Figure 3.
Calcium sensitivity of recombinant TcPDP and TbPDP. A–D, purification of TcPDP (A) and TbPDP (C) recombinant proteins by immobilized metal affinity chromatography. Total lysate (Lys), flow through (FT) and eluted/desalted fractions (rPDP and TbPDP) were analyzed by SDS-PAGE on 10% gel stained with Coomassie Brilliant Blue. Molecular weight (MW) markers are shown on the left side of panels A and C. Recombinant TcPDP (B) and TbPDP (D) enzymatic activities were assayed at different Ca2+ concentrations. E, enzyme kinetics of TcPDP. Recombinant TcPDP phosphatase activity was assayed in buffer containing 50 mm Hepes, pH 7.0, 50 μm MgCl2, 2 mm DTT, 100 nm free Ca2+, 120 μg of BSA and different concentrations of TcPDH E1α synthetic phosphopeptide as substrate. Error bars are smaller than the symbols used for some data points. Kinetic parameters of recombinant TcPDP obtained from linear regression using Michaelis-Menten equation are shown in the plot. Values are mean ± S.D. of three independent experiments (B, D, and E).
Enzyme kinetics of recombinant TcPDP
Free Ca2+ concentration found to render optimal TcPDP activity (100 nm) was used to assay the enzyme kinetics at different substrate concentrations as described under “Experimental Procedures.” TcPDP kinetic parameters were determined by linear regression using the Michaelis-Menten equation (Fig. 3E). Recombinant TcPDP exhibits a Vmax of 5.75 ± 0.1 pmol min−1 mg−1, a Km of 32.23 ± 2.04 μm, and a catalytic efficiency (kcat/Km) of 2.9 × 103 s−1 m−1.
TcPDP knockout phenotype
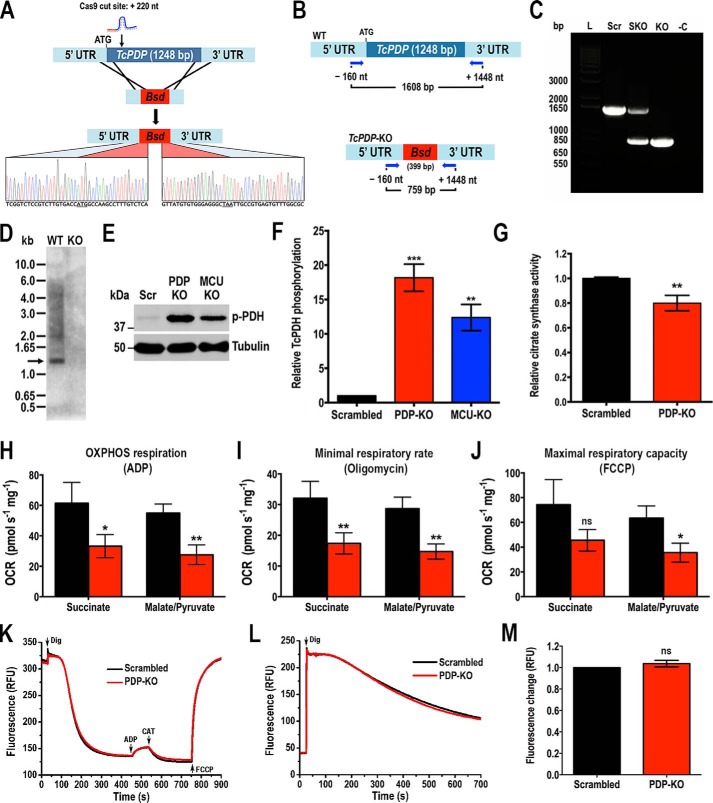
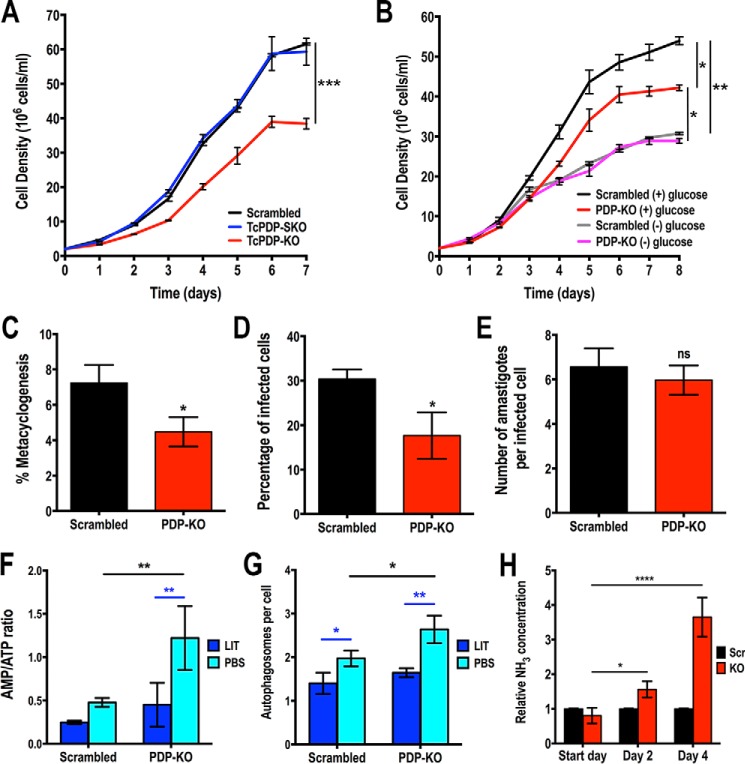
To further investigate the role of putative TcPDP we generated a TcPDP knockout (TcPDP-KO) cell line using the CRISPR/Cas9 system we have adapted and successfully used in T. cruzi (38, 42). Using this methodology a DNA donor template is provided to promote double-strand break repair by homologous recombination and therefore gene replacement with a resistance marker (the gene encoding blasticidin-S deaminase, Bsd) at the genomic locus (Fig. 4A) (43). We generated a molecular construct for the constitutive expression of Cas9 and a specific sgRNA targeting TcPDP gene of T. cruzi, as described under “Experimental Procedures.” Alternatively, we generated a version of this construct containing an enhanced Cas9 (eSpCas9) with nuclear localization signal and fused to GFP (44), cloned in pTREX-n vector. Each plasmid was co-transfected with the DNA donor cassette into T. cruzi Y strain epimastigotes. After 5 weeks of selection, genomic DNA from double-resistant parasites was analyzed by PCR using a primer set designed to discriminate between the intact TcPDP locus and the one replaced by Bsd (Fig. 4B). PCR products were resolved in a 1% agarose gel and a band of 1608 bp was detected in the control cell line transfected with a scrambled sgRNA (Scr) corresponding to the intact TcPDP locus (Fig. 4C). A single band corresponding to the size of the replaced gene (759 bp) was amplified in the transfectant cell line obtained with regular Cas9, indicating the generation of a homogeneous knockout population where both TcPDP alleles were ablated. Interestingly, using the eSpCas9 enhanced endonuclease, a single knockout (SKO) cell line was generated, as indicated by the amplification of both bands with the same intensity from its gDNA (Fig. 4C). Subsequently, we obtained clonal populations by serial dilutions from this cell line and confirmed the replacement of only one TcPDP allele in these clones (data not shown). The absence of TcPDP in the TcPDP-KO cell line was confirmed by sequencing and Southern blot analysis (Fig. 4, A and D), and then we proceeded to evaluate the phenotype of this mutant. We first analyzed the level of PDH phosphorylation in these parasites by Western blotting using commercial antibodies anti-PDH E1α subunit (phosphopeptide Ser-293) of human cells, which recognizes the phosphorylated site 1 (Ser-264 in the mature enzyme). As the phosphopeptide of the T. cruzi PDH E1α subunit homolog is highly conserved, these antibodies also detect the T. cruzi phosphorylated protein (expected size: 42.8 kDa). We also used commercial antibodies against the entire mammalian PDH E1α subunit, but they did not recognize the TcPDH E1α subunit (data not shown), probably because of the low identity (45%) in the amino acid sequence of both homolog proteins. Our results indicate that TcPDH phosphorylation in TcPDP-KO is significantly higher than in parasites transfected with a scrambled sgRNA (Fig. 4, E and F). Interestingly, knockout parasites for the mitochondrial calcium uniporter (TcMCU-KO) we studied previously (38) also exhibited an increased level of PDH phosphorylation (Fig. 4, E and F), which suggests a link between mitochondrial Ca2+ uptake, PDP activation and PDH stimulation, as parasites with low mitochondrial Ca2+ levels (TcMCU-KO) exhibit a similar pattern of PDH phosphorylation to TcPDP-KO parasites (Fig. 4, E and F). We also evaluated mitochondrial integrity of TcPDP-KO by measuring citrate synthase activity in these parasites and found it significantly lower than in control cells (Fig. 4G). Then we evaluated oxygen consumption rates (OCR) under ADP-stimulated (state 3), oligomycin-inhibited (state 4), and FCCP-stimulated (state 3u) conditions in control (scrambled) and TcPDP-KO digitonin-permeabilized cells, in the presence of succinate or malate and pyruvate as respiratory substrates. Control and PDP-KO mitochondria showed well-coupled respiration, although OCR in the presence of ADP, oligomycin, and FCCP were significantly lower in TcPDP-KO mitochondria (Fig. 4, H–J). This difference was more significant in the presence of malate and pyruvate as mitochondrial substrates. The lower OCR of TcPDP-KO mitochondria correlated with lower citrate synthase activity, suggesting a mitochondrial defect in these cells. We also evaluated the mitochondrial membrane potential (ΔΨm) of knockout and control parasites by measuring Safranine O fluorescence in digitonin-permeabilized epimastigotes in the presence of succinate as the mitochondrial substrate, as described previously (38). When using Safranine O, a decrease in fluorescence after addition of digitonin indicates stacking of the dye to the energized inner mitochondrial membrane (Fig. 4K). Addition of ADP produced the expected small dissipation of membrane potential (observed as a small increase in fluorescence), indicating ADP phosphorylation. ΔΨm returned to its initial level after addition of the adenine nucleotide translocator inhibitor carboxyatractyloside (CAT). Addition of FCCP collapsed the membrane potential. Ablation of TcPDP did not affect the ΔΨm at the steady state or ADP phosphorylation. Under the same conditions, we evaluated the capacity of TcPDP-KO to take up mitochondrial calcium by monitoring the fluorescence of Calcium Green-5N probe in digitonin-permeabilized epimastigotes. In this assay a decrease in fluorescence correlates with a decrease of extramitochondrial Ca2+, which is concomitant with mitochondrial Ca2+ uptake. The results indicate that the mitochondrial capacity to take up calcium was unaffected in parasites lacking TcPDP (Fig. 4, L and M).
Figure 4.
Phenotype analysis of TcPDP-KO epimastigotes. A, schematic representation of the strategy used to generate a TcPDP-KO cell line by CRISPR/Cas9-mediated genome editing. A double-stranded gDNA break was produced by Cas9 at nt +220 of the TcPDP ORF. DNA was repaired with a blasticidin-S deaminase (Bsd) cassette containing 100-bp homologous regions from TcPDP 5′ and 3′ UTRs. Gene replacement was confirmed by Sanger sequencing. B, primers (arrows) that were used to verify gene replacement by PCR. The intact locus generates a PCR product of 1608 bp, whereas the replaced locus generates a fragment of 759 bp. C, PCR analysis showing that TcPDP was ablated at its genomic locus and replaced in genomic DNA of the KO cell line. Lanes: L, 1 kb plus ladder; Scr, control parasites; SKO, TcPDP single knockout parasites generated using eCas9; KO, TcPDP knockout epimastigotes obtained with Cas9; -C, PCR negative control with ultrapure water. D, Southern blot analysis of WT and TcPDP-KO (KO) epimastigotes. The blot was hybridized with a 340-bp chemiluminescent probe amplified from TcPDP 5′ end. E, representative Western blotting of TcPDH E1α subunit phosphorylation in control (Scr), TcPDP-KO and TcMCU-KO cell lines. Tubulin was used as loading control. F, densitometry analysis of three Western blots (as in E). Values are mean ± S.D.; n = 3; **, p < 0.01, ***, p < 0.001 (one-way ANOVA with Dunnett's multiple comparisons test). G, relative citrate synthase activity of control (scrambled) and TcPDP-KO epimastigotes. Values are mean ± S.D.; n = 3; **, p < 0.01 (Student's t test). H–J, respiration of TcPDP knockout epimastigotes. OCR of digitonin-permeabilized control (scrambled, black bars) and TcPDP-KO (red bars) epimastigotes in the presence of succinate or malate/pyruvate as substrates. Bar charts show OCR after addition of (H) 100 μm ADP (respiration stimulated by oxidative phosphorylation or OXPHOS), (I) 1 μg/ml oligomycin (oligo) (minimal respiratory rate), and (J) 1.0 μm FCCP (maximal respiratory capacity). Values are mean ± S.D.; n = 3; *, p < 0.05, **, p < 0.01, ns, no significant differences (two-way ANOVA with Sidak's multiple comparisons test). K, changes in mitochondrial membrane potential (ΔΨm) of digitonin-permeabilized epimastigotes as detected by changes in Safranine O fluorescence in parasites transfected with scrambled sgRNA/Cas9/pTREX-n (Scrambled) or TcPDP-KO (PDP-KO). Cells (5 × 107) were added to the reaction buffer (2 ml) containing 0.2% BSA, 5 mm succinate, 50 μm EGTA, and 5 μm Safranine O. The reaction was started with 50 μm digitonin, and 250 μm ADP, 20 μm carboxyatractyloside (CAT), and 4 μm FCCP were added where indicated. L, Ca2+ uptake by digitonin-permeabilized TcPDP-KO and control (scrambled) epimastigotes in relative fluorescence units (RFU). The reaction was started after adding 50 μm digitonin in the presence of 20 μm free Ca2+, 5 mm succinate, and 0.5 μm Calcium Green-5N probe. M, quantification of data in panel L. Relative Ca2+ uptake at 600 s compared with control (scrambled) epimastigotes. Values are mean ± S.D. (n = 3; ns, no significant differences; Student's t test).
To investigate the biological relevance of TcPDP we evaluated the growth in vitro of TcPDP-KO and TcPDP-SKO epimastigotes compared with the control (scrambled) cells. We observed that TcPDP-KO parasites exhibited a significantly slower growth rate in rich (LIT) medium than the TcPDP-SKO and the control cells (Fig. 5A), indicating that this protein is required for normal proliferation in vitro. However, when this cell line was grown in low-glucose medium, there was no difference in growth rate compared with control parasites under the same conditions (Fig. 5B). We also investigated the ability of TcPDP-KO epimastigotes to differentiate in vitro into metacyclic trypomastigotes. Metacyclogenesis was significantly impaired in these parasites (Fig. 5C), as well as the ability of trypomastigotes to infect tissue-cultured cells (Fig. 5D). However, the replication of intracellular amastigotes was not affected in TcPDP-KO parasites (Fig. 5E).
Figure 5.
Phenotypic changes of TcPDP-KO cells in different life cycle stages. A, growth of control (scrambled), TcPDP-SKO, and TcPDP-KO epimastigotes in LIT medium. B, growth of scrambled and TcPDP-KO epimastigotes in normal (+) glucose and low-glucose (−) glucose LIT medium. In A and B, one-way ANOVA with multiple comparisons was applied to growth rates calculated from each growth curve (*, p < 0.05, **, p < 0.01,***, p < 0.001). C, percentage of metacyclic trypomastigotes observed after incubation in TAU 3AAG medium. Differentiation of epimastigotes to metacyclic trypomastigotes was quantified by DAPI staining to distinguish the position of the kinetoplast related to the nucleus by fluorescence microscopy. D and E, TcPDP-KO trypomastigote infection of Vero cells. There was a significant difference in the percentage of infected Vero cells (D) but not in the number of intracellular amastigotes per infected host cell observed 48 h post infection (E). Values are mean ± S.D.; n = 3; *, p < 0.05; ns, not significant (Student's t test). F, AMP/ATP ratios of control (scrambled) and TcPDP-KO epimastigotes incubated in LIT medium or PBS for 16 h. G, number of autophagosomes per cell observed by fluorescence microscopy images of scrambled and TcPDP-KO epimastigotes labeled with anti-TcATG8.1 antibody after incubation in LIT medium or PBS for 16 h. H, relative ammonia concentration determined in culture medium of control (Scr) and TcPDP-KO epimastigotes at start day, day 2, and day 4 of growth in LIT medium. Cell cultures were started at 2 × 106 cells/ml. Values are mean ± S.D.; n = 3; *, p < 0.05, **, p < 0.01, ***, p < 0.001; ****, p < 0.0001. Statistical analyses were performed using two-way ANOVA with Sidak's multiple comparisons test (F and G) or Tukey's multiple comparisons test (H).
Finally, we analyzed the effect of TcPDP down-regulation on cell bioenergetics by measuring the adenine nucleotide levels of knockout and control parasites under normal culture (incubation in LIT medium) and starvation conditions (incubation in PBS), as described previously (38). The results indicate that AMP/ATP ratio is significantly increased under starvation conditions in TcPDP-KO parasites, as compared with control epimastigotes (Fig. 5F). This result is consistent with the increased number of autophagosomes per cell observed in knockout parasites under starvation (PBS), as detected by immunofluorescence microscopy using antibodies against ATG8.1 (Fig. 5G). This is a marker of autophagosomes that has been used previously to evaluate autophagy in T. cruzi (38, 45).
Although TcPDP-KO epimastigotes were found to be bioenergetically affected under starvation conditions, they exhibited similar AMP/ATP ratio than control cells in normal culture medium, suggesting the existence of a compensatory mechanism that overcomes TcPDH inactivation and, as a consequence, the decrease in acetyl-CoA levels, a substrate of the tricarboxylic acid (TCA) cycle. Thus, we investigated whether TcPDP-KO epimastigotes, which showed increased levels of phosphorylated (inactive) PDH, could compensate the decreased synthesis of acetyl-CoA by up-regulating the oxidative metabolism of amino acids, with the production of ammonia that is excreted to the extracellular medium (46). Hence, we monitored ammonia concentration in the culture media of knockout and control parasites at days 2 and 4 of growth in LIT medium. The results indicate that ammonia concentration in the culture medium of TcPDP-KO epimastigotes was significantly higher than that of control cells at day 2 of growth, and this relative increase was much higher after 4 days of cell culture (Fig. 5H), suggesting that a compensatory mechanism involving oxidative metabolism of amino acids is taking place in this mutant cell line.
Taken together our results indicate that TcPDP is a mitochondrial phosphatase sensitive to physiological Ca2+ concentrations in vitro, which could be also Ca2+ stimulated in vivo to specifically activate mitochondrial pyruvate dehydrogenase. Moreover, TcPDP dephosphorylates TcPDH E1α subunit in vivo and is required for normal growth, differentiation, infectivity, and energy metabolism in T. cruzi.
Discussion
Our studies have shown that the pyruvate dehydrogenase phosphatase of T. cruzi (TcPDP) and T. brucei (TbPDP) are stimulated by low Ca2+ concentrations, as occurs with vertebrate PDPs, supporting the link between the evolutionary acquisition of a mitochondrial Ca2+ uniporter (MCU) and the presence of Ca2+-activated mitochondrial dehydrogenases. TcPDP localizes to the mitochondria and is important for normal growth of epimastigotes in rich medium, for differentiation to metacyclic trypomastigotes, and for trypomastigote invasion of host cells in vitro. TcPDP-KO epimastigotes showed increased levels of phosphorylated PDH E1α subunit indicating its requirement for in vivo dephosphorylation of this subunit. As compared with control epimastigotes, TcPDP-KO cells have a lower oxygen consumption rate and citrate synthase activity, but higher AMP/ATP ratios and autophagosome formation under starvation conditions. These cells have a compensatory increase in the oxidative metabolism of amino acids to overcome the limited production of acetyl-CoA.
Although the presence of a Ca2+-regulated PDH complex has been reported in animals, which, together with fungi, belong to the eukaryotic supergroup Ophistokonta, our work demonstrates its presence in trypanosomes, which belong to the supergroup Excavata, one of the earliest branches of eukaryotic evolution. These results reveal an early origin of this Ca2+ signaling pathway and the possibility of its occurrence in the last eukaryotic common ancestor (LECA).
Early binding studies using purified mammalian PDP1c have shown that it possesses an intrinsic Ca2+-binding site, and a second Ca2+-binding site that is produced in the presence of PDH E2 subunit (47). Later studies failed to detect binding of Ca2+ to PDP1c alone (48). The T. cruzi lipoylated PDH E2 subunit has been recently identified by Western blot analysis with a polyclonal antibody that detects lipoyl moieties (49), but the gene encoding this protein has not been identified yet, and we could not investigate whether association of TcPDP with TcPDH E2 increased the sensitivity of the enzyme to Ca2+. However, in agreement with the presence of an intrinsic Ca2+-binding site (47), our results indicate that Ca2+ can activate TcPDP alone although with only a 2-fold increase in activity. This level of activation is lower than that observed for the PDP isolated from different mammalian tissues and assayed in the presence of EGTA-Ca2+ buffers, where at least 5-fold phosphatase activation by Ca2+ has been reported with a K0.5 value of about 1 μm (50–52). Interestingly, we did not observe a saturable Ca2+ activation with either TcPDP or TbPDP recombinant proteins, as enzyme activation was not observed at concentrations higher than 500 nm and 1 μm for TcPDP and TbPDP, respectively. Free Ca2+ concentrations found to stimulate recombinant TcPDP and TbPDP (100–500 nm and 1 μm, respectively) are higher than the cytosolic free Ca2+ concentrations ([Ca2+]cyt) reported for T. cruzi (10–20 nm in amastigotes and trypomastigotes, 40–45 nm in epimastigotes (53, 54)) and T. brucei (90–100 nm in procyclic forms, 20–30 nm in bloodstream forms (54, 55)), respectively. Mitochondrial basal Ca2+ concentrations ([Ca2+]mit) are usually in the same range as [Ca2+]cyt. Therefore, in T. cruzi a mitochondrial Ca2+ concentration of 100 nm corresponds to a 2- to 10-fold increase of basal Ca2+ concentration, whereas in T. brucei a free Ca2+ concentration of 1 μm corresponds to a 10- to 50-fold increase in Ca2+ concentration, which appears to be very high. It thus looks as if Ca2+ regulation is more likely to occur in T. cruzi than in T. brucei. Experiments using the photoprotein aequorin targeted to the mitochondria and cytosol of T. brucei procyclic forms and treated with melittin (a plasma membrane–permeabilizing agent) or the acidocalcisome Ca2+-releasing agent monensin found increases in [Ca2+]mit up to 7- to 12-fold higher than [Ca2+]cyt reaching micromolar values (56). These increases are compatible with the free Ca2+ concentration observed to stimulate recombinant TbPDP and a role of the enzyme in these stages.
Using the CRISPR/Cas9 system we developed for T. cruzi (42), we generated a knockout cell line for TcPDP. We also used an enhanced Cas9 (eSpCas9) (44) to reduce the possibility of off-target effects. However, it was not possible to generate a double knockout using the eSpCas9 nuclease. After three attempts, only single knockout cell lines were detected by PCR analysis of clonal populations. Sequence analysis of both alleles did not show nucleotide differences at the chosen protospacer for sgRNA targeting. It seems that eSpCas9 is less efficient than regular Cas9 in T. cruzi, or at least it was for this specific locus. However, a double knockout homogeneous population for TcPDP was obtained using the conventional Cas9, with a specific sgRNA that is predicted not to generate off-target effects in the T. cruzi genome. Phenotypic evaluation of this mutant cell line indicates that TcPDP is required for TcPDH dephosphorylation. Interestingly, a TcMCU-KO cell line we generated in a previous study (38) also exhibited a significant higher level of PDH E1α subunit phosphorylation, although lower than that of TcPDP-KO parasites. An explanation for this difference is that basal Ca2+ could slightly activate TcPDP in TcMCU-KO cell line, thus resulting in some PDH dephosphorylation that does not occur in TcPDP-KO parasites. The reduced citrate synthase activity observed in these mutant epimastigotes evidences a down-regulation of the TCA cycle, which could be the consequence of higher levels of inactive (phosphorylated) TcPDH. A direct consequence of a defective TCA cycle is the decrease in the oxygen consumption rate observed in TcPDP-KO cells. Our results show that TcPDP-KO–permeabilized epimastigotes exhibit a lower oxygen consumption rate than control cells when pyruvate (the PDH substrate) and malate are used as mitochondrial substrates in the respiration medium. However, respiration was less impaired in TcPDP-KO cells when succinate was used as mitochondrial substrate, probably because in this way the need of acetyl-CoA from pyruvate is being bypassed. A similar result was observed for Saccharomyces cerevisiae Δptc6 mutant, which lacks the gene encoding PDP (57). This strain exhibited lower OCR than WT cells when grown on pyruvate-based media.
The impaired respiration observed in TcPDP knockout epimastigotes correlates with the growth deficiency observed in these cells. This phenomenon was observed previously in epimastigotes lacking the MCUb subunit of the T. cruzi MCU complex (38), indicating that this protein is important for normal proliferation in vitro. However, no difference in growth rate compared with control cells was observed when TcPDP-KO was grown in low glucose medium, probably because under these conditions a slower glycolysis rate provides less pyruvate as PDH substrate. So, growth differences resulting from PDH inactivation in TcPDP-KO cells (LIT medium) are not observed in low-glucose medium and parasites grow as control cells. Moreover, epimastigotes have a large pool of free amino acids that is used to maintain cell osmolarity (58). Increased amino acid oxidation through the TCA cycle would counteract the lack of PDH-catalyzed formation of acetyl-CoA from pyruvate. In this regard, the increased amount of ammonia detected in the culture medium of TcPDP-KO parasites after 2–4 days, compared with control epimastigotes, evidences an increase in amino acid catabolism to compensate the acetyl-CoA deficiency in this mutant cell line.
Ca2+ signaling is required for T. cruzi epimastigote differentiation (59) and for host cell invasion by trypomastigotes (60). In agreement with this requirement, a defect in the differentiation of epimastigotes into infective metacyclic trypomastigotes, and also in the infection of Vero cells by trypomastigotes, was observed in TcPDP-KO parasites, probably because Ca2+ stimulates TcPDP and consequently activates PDH and energy metabolism. However, replication of intracellular amastigotes was unaffected. Amastigotes face a low-glucose condition in the host cell cytosol (29), and pyruvate is equally scarce in either TcPDP-KO or control parasites; therefore, no differences in intracellular replication were observed. The predominance of fatty acid oxidation over glycolysis in the intracellular stages, as suggested by the increased levels of fatty acid oxidation enzymes (61), would also explain that TcPDP is not required for the replication of amastigotes. Moreover, amino acid catabolism has been shown to be a main energy source in intracellular T. cruzi stages (61, 62). Amino acid consumption leads to the production of ammonia (NH3), which needs to be excreted to avoid toxicity. In this regard, two proteins involved in NH3 detoxification have been recently shown to be required for normal proliferation of T. cruzi amastigotes, the ammonium transporter (TcAMT) (63) and the glutamine synthetase (64). Thus, their combined activity could contribute to the normal replication phenotype observed in TcPDP-KO amastigotes. Nevertheless, changes in the calcium regulation of TcPDP cannot be ruled out in different T. cruzi stages. In general, the biological defects observed in TcPDP knockout parasites highlight the importance of phosphorylation-mediated TcPDH regulation for parasite survival at different stages of the T. cruzi life cycle.
An increase in AMP/ATP ratio was observed in TcPDP-KO epimastigotes, indicating a bioenergetic imbalance that was stronger under starvation. Concomitantly, autophagy was also increased in knockout parasites and this effect was more evident under nutrient deprivation. In mammalian cells, down-regulation of MCU promotes autophagy (65). This has also been reported in T. brucei (26) and T. cruzi (38). A defective TCA cycle produced by PDP down-regulation could have a similar effect. The increase in the AMP/ATP ratio produced by down-regulation of the TCA cycle could promote autophagy by stimulation of AMP-dependent kinase (AMPK) (65). However, this correlation was not previously observed in T. cruzi when characterizing MCU and MCUb subunits of the MCU complex (38), suggesting an AMPK-independent autophagy pathway similar to that of T. brucei (66). Further studies should be performed to elucidate the mechanism of autophagy induction in T. cruzi.
In summary, we have identified a mitochondrial pyruvate dehydrogenase phosphatase in T. cruzi that is sensitive to physiological Ca2+ concentrations, suggesting that it could also be Ca2+ stimulated in vivo to specifically activate mitochondrial PDH. Moreover, TcPDP dephosphorylates TcPDH, stimulates energy metabolism through TCA cycle activation, and is required for parasite normal development at different stages of its life cycle. These findings provide new insights into the role of Ca2+ signaling in T. cruzi cell bioenergetics.
Experimental procedures
Chemicals and reagents
Blasticidin S HCl, BenchMark prestained protein ladder, BenchMark protein ladder, Alexa Fluor–conjugated secondary antibodies, and HRP-conjugated secondary antibodies were purchased from Life Technologies. Benzonase® nuclease was from Novagen (EMD Millipore, Billerica, MA). Wizard® Plus SV Miniprep Purification System, Wizard® SV Gel and PCR Clean-Up System, GoTaq DNA Polymerase, and T4 DNA Ligase were from Promega (Madison, WI). Antarctic Phosphatase, restriction enzymes and Q5® High-Fidelity DNA Polymerase were from New England Biolabs (Ipswich, MA). Fluoromount-G® was from SouthernBiotech (Birmingham, AL). Anti-HA High Affinity rat mAb (clone 3F10) was purchased from Roche Applied Science. The pMOTag23M vector (67) was from Dr. Thomas Seebeck (University of Bern, Bern, Switzerland). DNA oligonucleotides were purchased from Exxtend Biotecnologia Ltda. (Campinas, Brazil). Custom synthesized peptides were from New England Peptide (Gardner, MA). Precision Plus ProteinTM Dual Color Standards and nitrocellulose membranes were from Bio-Rad. ATP Determination Kit, BCA Protein Assay Kit, North2SouthTM Biotin Random Prime Labeling Kit and North2SouthTM Chemiluminescent Hybridization and Detection Kit were from Thermo Fisher Scientific. PD-10 columns were purchased from GE Healthcare Life Sciences. Anti-c-Myc mAb (clone 9E10) was from Santa Cruz Biotechnology (Dallas, TX). Antibodies anti–serine 293 phosphorylated PDH-E1α were from Abcam (Cambridge, MA). Anti-tubulin mAb, puromycin, G418, mammalian cell protease inhibitor mixture (Sigma P8340), other protease inhibitors, and all other reagents of analytical grade were from Sigma.
Cell culture
T. cruzi Y strain epimastigotes were cultured in liver infusion tryptose (LIT) medium containing 10% heat-inactivated fetal bovine serum (FBS) at 28 °C (68). CRISPR/Cas9 mutant cell lines were maintained in medium containing 250 μg/ml G418 and 10 μg/ml blasticidin or 5 μg/ml puromycin. Epimastigotes overexpressing TcPDP-3xHA were cultured in medium containing 250 μg/ml G418. The growth rate of epimastigotes was determined by counting cells in a Muse® Cell Analyzer. Tissue culture cell–derived trypomastigotes were obtained from Vero cells infected with metacyclic trypomastigotes obtained as described below. T. cruzi trypomastigotes were collected from the culture medium of infected host cells, using a modification of the method of Schmatz and Murray (69) as described previously (60). Vero cells were grown in RPMI supplemented with 10% fetal bovine serum and maintained at 37 °C with 5% CO2.
TcPDP overexpression
TcPDP ORF (1248 nt) was PCR amplified (primers 1 and 2, Table 1) and cloned into the pTREX-n/3xHA vector (38) by restriction sites XbaI/XhoI and subsequently used to transfect T. cruzi epimastigotes. Gene cloning was confirmed by PCR and sequencing. TcPDP overexpression was confirmed by Western blot analysis using anti-HA antibodies.
Table 1.
Oligonucleotides used in this study
Bold, specific protospacer; underlined, restriction site.
| No. | Primer | Sequence (5′ → 3′) |
|---|---|---|
| 1 | Fw_TcPDP-3xHA_OE | ACTGTCTAGATGCCGAGCAGCAGCAAAAAG |
| 2 | Rv_TcPDP-3xHA_OE | TGACCTCGAGCCCCTTAAGATCGACGATCATGATG |
| 3 | Fw_sgRNA_TcPDP-Ctag | GATCGGATCCAGGCTGAGTAAAAATTAACCGTTTTAGAGCTAGAAATAGC |
| 4 | Rv_sgRNA | CAGTGGATCCAAAAAAGCACCGACTCGGTG |
| 5 | Fw_TcPDP-Ctag_ultramer | TATTTGCCGTGAGTGTTTGGCGCCACCGGCGGAGGGCGGTGGTCGCAGCTCACGCGCCGAGGGCACAGACAACATGACCATCATGATCGTCGATCTTAAGGGTACCGGGCCCCCCCTCGAG |
| 6 | Rv_TcPDP-Ctag_ultramer | CACACAAAGACAGACAGACAGACAGACACACACACACACACACACACACACAGAGAGAGAGAGAGAGAGAGATACAAACACAAAAATAGAATCACCTGGTTGGCGGCCGCTCTAGAACTAGTGGAT |
| 7 | Fw_TcPDP_Ctag_check | GAACCGCGTAAATGGCCAAC |
| 8 | Rv_Puro_Ctag_check | TCAGGCACCGGGCTTGCGGG |
| 9 | Fw_TcPDP-pET32 Ek/LIC | GACGACGACAAGATGCCGAGCAGCAGCAAAAA |
| 10 | Rv_TcPDP-pET32 Ek/LIC | GAGGAGAAGCCCGGTTACTTAAGATCGACGATCATG |
| 11 | Fw_TbPDP-pET32_KpnI | GACTGGTACCATGCCACCCAAAAATAGGAA |
| 12 | Rv_TbPDP-pET32_XhoI | GATCCTCGAGCTTAAGATCCACAATCATTATTG |
| 13 | Fw_sgRNA_TcPDP-KO | GATCGGATCCGCGTGATGCGAAGATGCGGGGTTTTAGAGCTAGAAATAGC |
| 14 | Fw_sgRNA_Scrambled | GATCGGATCCGCACTACCAGAGCTAACTCAGTTTTAGAGCTAGAAATAGC |
| 15 | Fw_TcPDP-KO_ultramer | CGCAGCAAGGAAGTGACATTGGCAGCAGCGCATTTTGAATAAATTTGGATATATTTATTTATTTTTAGATTGTGTGTGTATCGGTCTCCGTCTTGTGACCATGGCCAAGCCTTTGTCTCA |
| 16 | Rv_TcPDP-KO_ultramer | TTACTTAAGATCGACGATCATGATGGTCATGTTGTCTGTGCCCTCGGCGCGTGAGCTGCGACCACCGCCCTCCGCCGGTGGCGCCAAACACTCACGGCAATTAGCCCTCCCACACATAAC |
| 17 | Rv_HX1-pTREX | TAATTTCGCTTTCGTGCGTG |
| 18 | Fw_TcPDP-KO_check | CTGGCTGCGGTTTGAGTGA |
| 19 | Rv_TcPDP-KO_check | CATGGGTGAGTGAATGTTTCTG |
| 20 | Fw_TcPDP_probe | ATGCCGAGCAGCAGCAAAAAG |
| 21 | Rv_TcPDP_probe | GAAGAAGATCCTGGAACTGC |
TcPDP endogenous C-terminal tagging
To achieve the C-terminal tagging of endogenous TcPDP we used the method we developed for T. cruzi (39, 40) to clone a specific sgRNA sequence targeting the 3′ end of TcPDP gene (TriTrypDB ID: TcCLB.506739.200) into Cas9/pTREX-n vector (42). The construct (3′end-sgRNA/Cas9/pTREX-n), together with a DNA donor cassette to induce homology-directed repair, was used to co-transfect T. cruzi epimastigotes and to insert a specific 3xc-Myc tag sequence at the 3′ end of the gene. The sgRNA targeting TcPDP 3′ end was obtained by PCR from plasmid pUC_sgRNA as described previously (42) using specific oligonucleotides (primers 3 and 4, Table 1).
To avoid Cas9 off-targeting, protospacer was analyzed with EuPaGDT Design Tool. A DNA donor cassette containing the 3xc-Myc tag sequence and the puromycin resistance gene was amplified using the pMOTag23M vector (67). Template for homologous-directed repair was amplified by PCR with 120 bp ultramers, of which 100 bp correspond to regions located right upstream of the stop codon and downstream of the Cas9 target site of the TcPDP gene, and 20 bp for annealing on the pMOTag23M vector (primers 5 and 6, Table 1). PCR reaction was carried out using the following cycling conditions: initial denaturation for 2 min at 95 °C; followed by 40 cycles of 20 s at 95 °C, 20 s at 63 °C, and 1 min 40 s at 72 °C; then a final extension for 10 min at 72 °C. Epimastigotes co-transfected with 3′end-sgRNA/Cas9/pTREX-n and DNA donor were cultured for 5 weeks with G418 and puromycin for selection of double-resistant parasites. Endogenous gene tagging was verified by PCR from gDNA using a specific primer set (primers 7 and 8, Table 1) and by Western blot analysis.
Gene cloning and protein heterologous expression
TcPDP and TbPDP open reading frames (gene IDs: TcCLB.506739.200 and Tb427.05.1660) were PCR-amplified from T. cruzi Y strain gDNA and T. brucei Lister 427 gDNA, respectively, using specific primer sets (primers 9–12, Table 1), and separately cloned into vector pET32 Ek/LIC (Novagen) for heterologous expression in bacteria. The sequence of recombinant clones was verified, and then they were independently transformed by heat shock into E. coli BL21 Codon Plus (DE3)-RIPL chemically competent cells. Expression of recombinant TcPDP and TbPDP proteins was induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside (IPTG) in LB broth for 3 h at 37 °C. Total lysates from induced and uninduced recombinant bacteria were analyzed by SDS-PAGE to confirm the expression of both enzymes.
Purification of recombinant PDPs under native conditions
Cell pellet from 500-ml cultures of recombinant E. coli BL21 Codon Plus (DE3)-RIPL cells expressing TcPDP or TbPDP were resuspended and incubated for 30 min on ice in 40 ml of cold lysis buffer: 50 mm Tris-HCl, pH 7.6, 150 mm sodium chloride, 10 mm imidazole, 0.1% Triton X-100, 0.1 mg/ml lysozyme, 25 units/ml Benzonase nuclease, and protease inhibitor mixture for purification of histidine-tagged proteins (Sigma P8849; 50 μl/g of cell paste). Then three sonication pulses (40% amplitude, 30 s, on ice) were applied to ensure the complete disruption of cells. After centrifugation at 20,000 × g for 30 min at 4 °C, supernatant was clarified by passing it through a 0.2 μm pore nitrocellulose filter. Protein extract was kept on ice and used for immediate purification of recombinant TcPDP or TbPDP. Protein purification was performed at 4 °C using HisPur Ni-NTA Chromatography Cartridges, following the manufacturer's protocol for histidine-tagged protein purification under native conditions. Tris-buffered saline (TBS: 50 mm Tris-HCl, pH 7.6, 150 mm sodium chloride) replaced PBS in all the steps during the purification process to avoid phosphate contamination in the PDP enzymatic reaction. One-ml fractions were eluted (elution buffer: 250 mm imidazole in Tris-buffered saline), and buffer exchange was performed immediately using PD-10 desalting columns to finally obtain the protein in 50 mm HEPES, pH 7.0. All purification steps were verified by SDS-PAGE. Desalted recombinant proteins were quantified using the Pierce BCA Protein Assay Kit.
PDP activity assay
TcPDP activity was assayed by quantifying phosphate release from a synthetic phosphopeptide corresponding to a 14 amino acid long segment of TcPDH E1α subunit surrounding phosphorylation sites (Ser) 1 and 2: Tyr-Val-Gly-His-Ser-Met-Ser-Asp-Pro-Asp-Ser-Gln-Tyr-Arg (Fig. 1), as described previously for the mammalian enzyme (41, 70, 71). Enzymatic reactions were performed for 1 h at room temperature in a 100-μl reaction containing 200 μm substrate peptide, 50 mm Hepes, pH 7.0, 50 μm MgCl2, 2 mm DTT, and 120 μg of BSA. Different Ca2+ concentrations (from 10−9 to 10−4 m) were achieved using a CaCl2-EGTA buffer and calculated with the freely distributed software MaXChelator based on the Bers stability constants, as described previously (72). The reaction was stopped by adding freshly mixed malachite green reagent (3 parts 0.045% malachite green and 1 part 4.2% ammonium molybdate, 4 m HCl), and absorbance at 660 nm was immediately read on a microplate spectrophotomer (Molecular Devices). A potassium phosphate standard curve was included in the assay for quantification purposes. TbPDP activity was assayed following the same methodology, but using a homolog synthetic phosphopeptide corresponding to a 14 amino acid long segment of TbPDH E1α subunit surrounding phosphorylation sites (Ser) 1 and 2: Tyr-Met-Gly-His-Ser-Met-Ser-Asp-Pro-Asp-Ser-Gln-Tyr-Arg, as TbPDP substrate (Fig. 1).
TcPDP-KO
A single guide RNA sequence to target the gene coding for the hypothetical TcPDP protein was PCR-amplified from plasmid pUC_sgRNA, as previously described (42). Selection of protospacer was performed using EuPaGDT (Eukaryotic Pathogen CRISPR guide RNA Design Tool, http://grna.ctegd.uga.edu/)6 (80). The protospacer sequence was included into the forward primer while using a common reverse primer for sgRNA amplification. These primers also contained a BamHI restriction site for cloning into Cas9/pTREX-n (42) to generate TcPDP-sgRNA/Cas9/pTREX-n construct. The sgRNA orientation was verified by PCR using the specific TcPDP-sgRNA forward primer and the HX1 reverse primer (42). Positive clones that generate a 190-bp PCR fragment were also sequenced. A scrambled sgRNA (Scr-sgRNA/Cas9/pTREX-n) was used as control. A DNA donor cassette designed to promote homologous directed repair and replacement of TcPDP ORF was obtained by PCR using a set of long primers (ultramers) containing 120 nucleotides, from which 100 nucleotides correspond to the first 100 nt (forward ultramer) and the last 100 nt (reverse ultramer) of TcPDP ORF, and 20 nt annealing on Bsd gene. Circular construct TcPDP-sgRNA/Cas9/pTREX-n and linear Bsd cassette were used to co-transfect T. cruzi epimastigotes. After 5 weeks of selection with 250 μg/ml G418 and 10 μg/ml blasticidin, TcPDP gene replacement was verified by PCR. Primers used to generate TcPDP-KO are listed in Table 1.
Cell transfections
Transfections were performed as described previously (38). Briefly, T. cruzi Y strain epimastigotes in early exponential phase (4 × 107 cells) were washed with PBS, pH 7.4, at room temperature (RT) and transfected in ice-cold CytoMix (120 mm KCl, 0.15 mm CaCl2, 10 mm K2HPO4, 25 mm HEPES, 2 mm EDTA, 5 mm MgCl2, pH 7.6) containing 25 μg of each plasmid construct and 25 μg of donor DNA in 4-mm electroporation cuvettes with three pulses (1500 volts, 25 microfarads) delivered by a Gene Pulser XcellTM Electroporation System (Bio-Rad). Stable cell lines were established and maintained under drug selection (250 μg/ml G418 and 10 μg/ml blasticidin or 5 μg/ml puromycin, or 250 μg/ml G418 alone). Transfectant epimastigotes were cultured in LIT medium supplemented with 20% heat-inactivated FBS until stable cell lines were obtained.
Southern blot analysis
To confirm TcPDP gene knockout 25 μg of gDNA from control (transfected with scramble sgRNA) and TcPDP-KO epimastigotes were digested with PvuII enzyme and resolved on a 0.8% agarose gel. Restriction fragments were transferred to nylon membrane and hybridized with a biotin-labeled probe corresponding to the first 340 nt of TcPDP ORF that was amplified by PCR using primers 20 and 21 (Table 1) and labeled using the North2SouthTM Biotin Random Prime Labeling Kit. Hybridization, posthybridization washes, and detection were performed with North2SouthTM Chemiluminescent Hybridization and Detection Kit, following manufacturer's recommendations. Signal detection was performed using UVItec Alliance Gel Documentation System (UVItec, Cambridge, UK).
Western blot analysis
Western blotting analyses were performed using standard procedures used in our laboratory (38, 73, 74). Briefly, parental and mutant cell lines were harvested separately. Parasites were washed twice in PBS and resuspended in radio-immunoprecipitation assay buffer (RIPA: 150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1% SDS, 0.1% Triton X-100) plus a mammalian cell protease inhibitor mixture (diluted 1:250), 1 mm phenylmethylsulfonyl fluoride, 2.5 mm tosyl phenylalanyl chloromethyl ketone (TPCK), 100 μm N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E64), and Benzonase Nuclease (25 units/ml of culture). The cells were then incubated for 1 h on ice and protein concentration was determined by BCA protein assay. Thirty micrograms of protein from each cell lysate were mixed with 6× Laemmli sample buffer (125 mm Tris-HCl, pH 7, 10% (w/v) β-mercaptoethanol, 20% (v/v) glycerol, 4.0% (w/v) SDS, 4.0% (w/v) bromphenol blue) before application to 10% SDS-polyacrylamide gels. Separated proteins were transferred onto nitrocellulose membranes with a Bio-Rad trans-blot apparatus. Membranes were blocked with 5% nonfat dried skim milk in PBS-T (PBS containing 0.1% v/v Tween 20) overnight at 4 °C. Next, blots were incubated for 1 h at RT with a primary antibody, i.e. monoclonal anti-HA (1:5000), monoclonal anti-c-Myc-tag (1:100), rabbit polyclonal anti PDH-E1α (phospho–Ser-293) (1:1000), and monoclonal anti-tubulin (1:40,000). After three washes with PBS-T, blots were incubated with the secondary antibody (goat anti-mouse IgG or goat anti-rabbit IgG, HRP-conjugated antibody, diluted 1:10,000). Membranes were washed three times with PBS-T, and Western blot images were obtained and processed with a C-DiGit Blot Scanner (LI-COR Biosciences).
Immunofluorescence analysis
Epimastigotes were incubated with 100 nm MitoTracker® Deep Red FM for 30 min at 28 °C in culture medium. Then cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 1 h at RT. Cells were allowed to adhere to poly-l-lysine–coated coverslips and then permeabilized for 5 min with 0.1% Triton X-100. Permeabilized cells were blocked with PBS containing 3% BSA, 1% fish gelatin, 50 mm NH4Cl, and 5% goat serum overnight at 4 °C. Then, cells were incubated with a primary antibody (monoclonal anti-HA-Tag (1:500) or monoclonal anti-c-Myc-tag (1:10)), diluted in 1% BSA in PBS (pH 8.0) for 1 h at RT. Cells were washed three times with 1% BSA in PBS (pH 8.0), and then incubated for 1 h at RT in the dark with Alexa Fluor 488–conjugated goat anti-mouse secondary antibody (1:1000). Then, cells were washed and mounted on slides using Fluoromount-G mounting medium containing 5 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) to stain DNA. Controls were performed as described above but in the absence of a primary antibody. Differential interference contrast and fluorescence optical images were captured with a confocal microscope Leica TCS SP5 II, with a 100× objective (1.44 aperture) under nonsaturating conditions, that uses photomultiplier tubes for detection of emission, and LAS AF software (Leica, Wetzlar, Germany) for acquisition and processing of digital images.
Metacyclogenesis
We followed the protocol described by Bourguignon et al. (75) with minor modifications. Epimastigotes were obtained after 4 days in LIT medium and incubated for 2 h in triatome artificial urine (TAU) medium (190 mm NaCl, 17 mm KCl, 2 mm MgCl2, 2 mm CaCl2, 0.035% sodium bicarbonate, 8 mm phosphate, pH 6.9) at room temperature). Then, parasites were incubated for 96 h in TAU 3AAG medium (TAU medium supplemented with 10 mm l-proline, 50 mm sodium l-glutamate, 2 mm sodium l-aspartate, and 10 mm glucose). To increase the number of metacyclic forms to infect Vero cells, the contents of the flask were collected and resuspended in medium containing fresh fetal bovine serum and incubated at 37 °C for 20 h. The complement in fresh FBS kills epimastigotes whereas metacyclic trypomastigotes survive. Samples were harvested from the TAU 3AAG plus FBS-containing medium at days 5 and 10 of cultivation.
In vitro infection assay
Gamma-irradiated (2000 radiation-absorbed doses) Vero cells (4.5 × 105 cells) were plated onto sterile coverslips in a 12-well plate and incubated overnight at 35 °C, 7% CO2, in RPMI medium plus 10% fresh fetal bovine serum. Tissue culture-derived trypomastigote collections were incubated at 4 °C overnight to allow amastigotes to settle from swimming trypomastigotes. Trypomastigotes from the supernatants of these collections were counted and used to infect the coverslips at a 10:1 ratio of parasites to host cells. At 4 h post infection, coverslips were washed extensively with Hanks' balanced salt solution, followed by PBS, pH 7.4, to remove any extracellular parasites. Coverslips were fixed immediately in 4% paraformaldehyde in PBS, pH 7.4, at 4 °C for 30 min. Coverslips were washed once with PBS and mounted onto glass slides in Fluoromount-G containing 15 μg/ml DAPI, which stains host and parasite DNA. Coverslips were viewed on an Olympus BX60 microscope to quantify the number of host cells that contained intracellular parasites and the number of intracellular parasites per cell in randomly selected fields. Three hundred host cells were counted per sample in three independent experiments. To quantify amastigote replication, the following modifications were used: host cells were infected at a ratio of 10 parasites to 1 host cell, and coverslips were allowed to incubate for 48 h post infection at 35 °C, 7% CO2, prior to fixation and DAPI staining.
Cellular respiration
The OCR of digitonin-permeabilized epimastigotes was measured using a high-resolution respirometer (Oroboros Oxygraph-2k, Innsbruck, Austria) with DatLab 4 software for data acquisition and analysis, and calibrated as reported by their manufacturers. Cells (1 × 108) were incubated at 28 °C in a 2 ml chamber containing 125 mm sucrose, 65 mm KCl, 10 mm Hepes-KOH, pH 7.2, 2.5 mm K2PO4, 1 mm MgCl2, 50 μm EGTA, 5 mm succinate, and 25 μm digitonin. Alternatively, 5 mm malate and 5 mm pyruvate were used as mitochondrial substrates, instead of succinate. OCR was calculated as the negative time derivative of oxygen concentration measured in the closed respirometer chambers and expressed per milligram of protein. Data were recorded at 2 s intervals, and 10 data points were used to calculate the slope of the OCR plot through a polynomial fit with DatLab 4 software, as described (76).
Citrate synthase activity
Citrate synthase activity was measured using a previously described protocol (77) adapted to trypanosomes (38). Briefly, the conversion of oxaloacetate and acetyl-CoA to citrate and SH-CoA was monitored by quantification of the colorimetric product thionitrobenzoic acid (78). T. cruzi epimastigotes in early exponential phase (∼1 × 108 cells) were washed twice with PBS and incubated in lysis buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 0.1% Triton X-100, and 25 units of Benzonase nuclease) for 10 min on ice. Then, proteins were quantified by BCA protein assay and 260 μl reactions were set up in buffer containing 5 μg protein, 250 μm oxaloacetate, 50 μm acetyl-CoA, 100 μm 5,5′-dithio-bis(2-nitrobenzoic acid), and 10 mm Tris-HCl, pH 8.0. The increase in absorbance at 412 nm was monitored for 20 min at 28 °C using a microplate reader (PowerWave XS 2, BioTek Instruments, Winooski, VT). Values of Vmax were normalized taking scrambled cell line as reference value.
Mitochondrial membrane potential
Assessment of mitochondrial membrane potential in situ was done spectrofluorometrically using the indicator dye Safranine O, as described previously (38, 79). Briefly, T. cruzi epimastigotes (5 × 107 cells) were incubated at 28 °C in reaction buffer (125 mm sucrose, 65 mm KCl, 10 mm Hepes-KOH buffer, pH 7.2, 1 mm MgCl2, 2.5 mm potassium phosphate; 1.95 ml) containing 5 mm succinate, 0.2% BSA, 50 μm EGTA, and 5 μm Safranine O, and the reaction was started with digitonin (50 μm). ADP (250 μm), carboxyatractyloside (20 μm), and FCCP (4 μm) were added to the media at different time points. Fluorescence changes were monitored on a Hitachi 4500 spectrofluorometer (excitation = 495 nm; emission = 586 nm).
Mitochondrial calcium uptake
The uptake of Ca2+ by permeabilized T. cruzi epimastigotes was assayed by fluorescence of Calcium Green-5N probe at 28 °C, as described previously (38). Briefly, cells were collected by centrifugation at 1000 × g for 7 min and washed twice with Buffer A with glucose (116 mm NaCl, 5.4 mm KCl, 0.8 mm MgSO4, 5.5 mm d-glucose, and 50 mm HEPES, pH 7.0). Epimastigotes were resuspended to a final density of 1 × 109 cells/ml in Buffer A with glucose and kept on ice. Before each experiment, a 50-μl aliquot of T. cruzi epimastigotes (5 × 107 cells) was added to the reaction buffer (125 mm sucrose, 65 mm KCl, 10 mm Hepes-KOH buffer, pH 7.2, 1 mm MgCl2, 2.5 mm potassium phosphate; 1.95 ml) containing 5 mm succinate, 20 μm free Ca2+, and 0.5 μm Calcium Green-5N. Mitochondrial Ca2+ uptake was initiated by the addition of 50 μm digitonin. Fluorescence changes were monitored in a F-4500 Fluorescence Spectrophotometer (Hitachi) with excitation at 506 nm and emission at 532 nm.
Autophagy assay
Expression of the TcAtg8.1 autophagy marker and autophagosome formation in T. cruzi epimastigotes grown in LIT medium and under starvation conditions was estimated by immunofluorescence analyses using anti-TcATG8.1 antibody as described (45). For starvation induction, mid-log parasites were washed twice with PBS, resuspended in the same buffer at a concentration of 5 × 107 cells/ml, and incubated for 16 h at 28 °C as described previously (45).
Adenine nucleotide levels
Control (transfected with scrambled sgRNA) and TcPDP-KO epimastigotes were harvested and washed once with Buffer A (116 mm NaCl, 5.4 mm KCl, 0.8 mm MgSO4 and 50 mm HEPES at pH 7.0). After washing, 1 × 108 cells per treatment were centrifuged and resuspended in 100 μl of Buffer A, and then lysed on ice for 30 min by addition of 150 μl of 0.5 m HClO4. The lysates were neutralized (pH 6.5) by addition of 60 μl of 0.72 m KOH/0.6 m KHCO3. Samples were centrifuged at 1000 × g for 5 min and the supernatant was separated for adenine nucleotide determination. ATP, ADP, and AMP in extracted samples were quantified by a luciferin-luciferase bioluminescence assay in a plate reader as described (38). Briefly, we used an ATP Determination Kit according to manufacturer's instructions, with adenylate kinase and/or nucleoside-diphosphate kinase (NDK) (Sigma). To determine the amount of adenine nucleotides, four measurements were taken of three different reactions for each sample by end point determination of the ATP concentration: One reaction without addition of any ATP-generating enzyme (for ATP), another reaction adding NDK (for ATP + ADP), and the third reaction adding both adenylate kinase and NDK (for ATP + ADP + AMP). The amount of ADP was obtained by subtracting the ATP value from the (ATP + ADP) value and the amount of AMP was calculated from the difference between the (ATP + ADP + AMP) content and the (ATP + ADP) content.
Ammonia determination in culture medium
Cell cultures of T. cruzi epimastigotes were started at 2 × 106 cells/ml (day 0). Ammonia concentration in culture medium was determined using an Ammonia Assay Kit (Sigma, catalogue no. AA0100) at days 0, 2, and 4 following manufacturer's instructions. The assay was adapted to be performed in 96-well plates. Briefly, 1 ml of cell culture was centrifuged at 1000 × g for 5 min and supernatant was transferred to a new tube. Then, supernatants were diluted 1:10 in ultrapure water and filtered through 0.2 μm pore nitrocellulose membrane. Then 20 μl from each filtered sample were distributed in triplicate into 96-well plates, including water as reagent blank and ammonia standard solution as assay control. Two hundred microliters of Ammonia Assay Reagent were added to each well and samples were incubated 5 min at RT. Absorbance at 340 nm was immediately measured in a plate reader (Molecular Devices). Subsequently, 2 μl of l-Glutamate Dehydrogenase solution (Sigma, catalogue no. G2294) were added to each well, and after 5 min incubation at RT absorbance at 340 nm was measured again. Ammonia concentration of samples was calculated according to the kit's protocol.
Statistical analysis
Statistical analyses were performed with GraphPad Prism software (La Jolla, CA), version 6.0. Reported values are mean ± S.D. of n biological experiments, as indicated in the figure legends. The level of significance was evaluated by Student's t test for comparisons between two cell lines, one-way analysis of variance (ANOVA) for comparisons between more than two cell lines, and two-way ANOVA with multiple comparison tests for analyses of grouped data.
Author contributions
N. L., M. A. C., M. S. B., M. S., A. E. V., and R. D. conceptualization; N. L., M. A. C., M. S. B., M. S., and A. E. V. data curation; N. L. and M. A. C. software; N. L., M. A. C., M. S. B., M. S., A. E. V., and R. D. formal analysis; N. L., M. A. C., M. S. B., M. S., and R. D. validation; N. L., M. A. C., M. S. B., M. S., A. E. V., and R. D. investigation; N. L., M. A. C., M. S. B., M. S., and R. D. visualization; N. L., M. A. C., M. S. B., M. S., A. E. V., and R. D. methodology; N. L. writing-original draft; A. E. V. and R. D. resources; A. E. V. and R. D. supervision; A. E. V. and R. D. project administration; R. D. funding acquisition; R. D. writing-review and editing.
Acknowledgments
We thank Thomas Seebeck for pMOTag23M vector, Vanina Alvarez for TcATG8.1 antibodies, and the staff of the Life Sciences Core Facility (LaCTAD) from State University of Campinas (UNICAMP), for the acquisition of the confocal microscopy images.
This work was funded by the São Paulo Research Foundation (FAPESP) Grant 2013/50624-0 (to R. D.) and National Institutes of Health Grant AI107663 (to R. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PDC
- pyruvate dehydrogenase complex
- MCU
- mitochondrial calcium uniporter
- PDP
- pyruvate dehydrogenase phosphatase
- PDH
- pyruvate dehydrogenase
- TcPDP
- Trypanosoma cruzi PDP
- TbPDP
- Trypanosoma brucei PDP
- aa
- amino acid
- IFA
- immunofluorescence analysis
- sgRNA
- single guide RNA
- Scr
- scrambled sgRNA
- SKO
- single knockout
- gDNA
- genomic DNA
- OCR
- oxygen consumption rates
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- TCA
- tricarboxylic acid
- PBS-T
- PBS containing 0.1% v/v Tween 20
- NDK
- nucleoside-diphosphate kinase
- ANOVA
- analysis of variance
- RT
- room temperature.
References
- 1. Denton R. M., and McCormack J. G. (1990) Ca2+ as a second messenger within mitochondria of the heart and other tissues. Annu. Rev. Physiol. 52, 451–466 10.1146/annurev.ph.52.030190.002315 [DOI] [PubMed] [Google Scholar]
- 2. Denton R. M. (2009) Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 1787, 1309–1316 10.1016/j.bbabio.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 3. Reed L. J. (2001) A trail of research from lipoic acid to α-keto acid dehydrogenase complexes. J. Biol. Chem. 276, 38329–38336 10.1074/jbc.R100026200 [DOI] [PubMed] [Google Scholar]
- 4. Roche T. E., Baker J. C., Yan X., Hiromasa Y., Gong X., Peng T., Dong J., Turkan A., and Kasten S. A. (2001) Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog. Nucleic Acids Res. Mol. Biol. 70, 33–75 [DOI] [PubMed] [Google Scholar]
- 5. Linn T. C., Pettit F. H., and Reed L. J. (1969) α-keto acid dehydrogenase complexes, X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc. Natl. Acad. Sci. U.S.A. 62, 234–241 10.1073/pnas.62.1.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korotchkina L. G., and Patel M. S. (2001) Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J. Biol. Chem. 276, 37223–37229 10.1074/jbc.M103069200 [DOI] [PubMed] [Google Scholar]
- 7. Huang B., Gudi R., Wu P., Harris R. A., Hamilton J., and Popov K. M. (1998) Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J. Biol. Chem. 273, 17680–17688 10.1074/jbc.273.28.17680 [DOI] [PubMed] [Google Scholar]
- 8. Uhlinger D. J., Yang C. Y., and Reed L. J. (1986) Phosphorylation-dephosphorylation of pyruvate dehydrogenase from bakers' yeast. Biochemistry 25, 5673–5677 10.1021/bi00367a049 [DOI] [PubMed] [Google Scholar]
- 9. Tovar-Méndez A., Miernyk J. A., and Randall D. D. (2003) Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur. J. Biochem. 270, 1043–1049 10.1046/j.1432-1033.2003.03469.x [DOI] [PubMed] [Google Scholar]
- 10. Patel M. S., and Korotchkina L. G. (2006) Regulation of the pyruvate dehydrogenase complex. Biochem. Soc. Trans. 34, 217–222 10.1042/BST20060217 [DOI] [PubMed] [Google Scholar]
- 11. McCormack J. G., and Denton R. M. (1981) A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex and NAD+-isocitrate dehydrogenase from a variety of sources. Biochem. J. 196, 619–624 10.1042/bj1960619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCormack J. G., and Denton R. M. (1986) Ca2+ as a second messenger within mitochondria. Trends Biochem. Sci. 11, P258–P262 10.1016/0968-0004(86)90190-8 [DOI] [Google Scholar]
- 13. Docampo R., and Vercesi A. E. (1989) Characteristics of Ca2+ transport by Trypanosoma cruzi mitochondria in situ. Arch. Biochem. Biophys. 272, 122–129 10.1016/0003-9861(89)90202-6 [DOI] [PubMed] [Google Scholar]
- 14. Docampo R., and Vercesi A. E. (1989) Ca2+ transport by coupled Trypanosoma cruzi mitochondria in situ. J. Biol. Chem. 264, 108–111 [PubMed] [Google Scholar]
- 15. Vercesi A. E., Docampo R., and Moreno S. N. (1992) Energization-dependent Ca2+ accumulation in Trypanosoma brucei bloodstream and procyclic trypomastigotes mitochondria. Mol. Biochem. Parasitol. 56, 251–257 10.1016/0166-6851(92)90174-I [DOI] [PubMed] [Google Scholar]
- 16. Perocchi F., Gohil V. M., Girgis H. S., Bao X. R., McCombs J. E., Palmer A. E., and Mootha V. K. (2010) MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 467, 291–296 10.1038/nature09358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baughman J. M., Perocchi F., Girgis H. S., Plovanich M., Belcher-Timme C. A., Sancak Y., Bao X. R., Strittmatter L., Goldberger O., Bogorad R. L., Koteliansky V., and Mootha V. K. (2011) Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Stefani D., Raffaello A., Teardo E., Szabò I., and Rizzuto R. (2011) A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Docampo R., and Lukeš J. (2012) Trypanosomes and the solution to a 50-year mitochondrial calcium mystery. Trends Parasitol. 28, 31–37 10.1016/j.pt.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mallilankaraman K., Cárdenas C., Doonan P. J., Chandramoorthy H. C., Irrinki K. M., Golenár T., Csordás G., Madireddi P., Yang J., Müller M., Miller R., Kolesar J. E., Molgó J., Kaufman B., Hajnóczky G., Foskett J. K., and Madesh M. (2012) MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat. Cell Biol. 14, 1336–1343 10.1038/ncb2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plovanich M., Bogorad R. L., Sancak Y., Kamer K. J., Strittmatter L., Li A. A., Girgis H. S., Kuchimanchi S., De Groot J., Speciner L., Taneja N., Oshea J., Koteliansky V., and Mootha V. K. (2013) MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8, e55785 10.1371/journal.pone.0055785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sancak Y., Markhard A. L., Kitami T., Kovács-Bogdán E., Kamer K. J., Udeshi N. D., Carr S. A., Chaudhuri D., Clapham D. E., Li A. A., Calvo S. E., Goldberger O., and Mootha V. K. (2013) EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382 10.1126/science.1242993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raffaello A., De Stefani D., Sabbadin D., Teardo E., Merli G., Picard A., Checchetto V., Moro S., Szabò I., and Rizzuto R. (2013) The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 32, 2362–2376 10.1038/emboj.2013.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lander N., Chiurillo M. A., Bertolini M. S., Docampo R., and Vercesi A. E. (2018) The mitochondrial calcium uniporter complex in trypanosomes. Cell Biol. Int. 42, 656–663 10.1002/cbin.10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Docampo R., Vercesi A. E., and Huang G. (2014) Mitochondrial calcium transport in trypanosomes. Mol. Biochem. Parasitol. 196, 108–116 10.1016/j.molbiopara.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang G., Vercesi A. E., and Docampo R. (2013) Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat. Commun. 4, 2865 10.1038/ncomms3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang G., and Docampo R. (2018) The mitochondrial Ca2+ uniporter complex (MCUC) of Trypanosoma brucei is a hetero-oligomer that contains novel subunits essential for Ca2+ uptake. MBio 9, e01700-18 10.1128/mBio.01700-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buscaglia C. A., Pollevick G. D., Veloso C., Lorca M., Frasch A. C., and Sánchez D. O. (1996) A putative pyruvate dehydrogenase α subunit gene from Trypanosoma cruzi. Biochim. Biophys. Acta 1309, 53–57 10.1016/S0167-4781(96)00140-6 [DOI] [PubMed] [Google Scholar]
- 29. Maugeri D. A., Cannata J. J., and Cazzulo J. J. (2011) Glucose metabolism in Trypanosoma cruzi. Essays Biochem. 51, 15–30 10.1042/bse0510015 [DOI] [PubMed] [Google Scholar]
- 30. Zhuo Y., Cordeiro C. D., Hekmatyar S. K., Docampo R., and Prestegard J. H. (2017) Dynamic nuclear polarization facilitates monitoring of pyruvate metabolism in Trypanosoma brucei. J. Biol. Chem. 292, 18161–18168 10.1074/jbc.M117.807495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B. P., Carrington M., Depledge D. P., Fischer S., Gajria B., Gao X., Gardner M. J., Gingle A., Grant G., Harb O. S., Heiges M., et al. (2010) TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38, D457–D462 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gunasekera K., Wüthrich D., Braga-Lagache S., Heller M., and Ochsenreiter T. (2012) Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics 13, 556 10.1186/1471-2164-13-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finn R. D., Attwood T. K., Babbitt P. C., Bateman A., Bork P., Bridge A. J., Chang H. Y., Dosztányi Z., El-Gebali S., Fraser M., Gough J., Haft D., Holliday G. L., Huang H., Huang X., et al. (2017) InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., Gasteiger E., Bairoch A., and Hulo N. (2006) ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vassylyev D. G., and Symersky J. (2007) Crystal structure of pyruvate dehydrogenase phosphatase 1 and its functional implications. J. Mol. Biol. 370, 417–426 10.1016/j.jmb.2007.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yeaman S. J., Hutcheson E. T., Roche T. E., Pettit F. H., Brown J. R., Reed L. J., Watson D. C., and Dixon G. H. (1978) Sites of phosphorylation on pyruvate dehydrogenase from bovine kidney and heart. Biochemistry 17, 2364–2370 10.1021/bi00605a017 [DOI] [PubMed] [Google Scholar]
- 37. Sugden P. H., Kerbey A. L., Randle P. J., Waller C. A., and Reid K. B. (1979) Amino acid sequences around the sites of phosphorylation in the pig heart pyruvate dehydrogenase complex. Biochem. J. 181, 419–426 10.1042/bj1810419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chiurillo M. A., Lander N., Bertolini M. S., Storey M., Vercesi A. E., and Docampo R. (2017) Different roles of mitochondrial calcium uniporter complex subunits in growth and infectivity of Trypanosoma cruzi. MBio 8, e00574-17 10.1128/mBio.00574-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lander N., Chiurillo M. A., Storey M., Vercesi A. E., and Docampo R. (2016) CRISPR/Cas9-mediated endogenous C-terminal tagging of Trypanosoma cruzi genes reveals the acidocalcisome localization of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 291, 25505–25515 10.1074/jbc.M116.749655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lander N., Chiurillo M. A., Vercesi A. E., and Docampo R. (2017) Endogenous C-terminal tagging by CRISPR/Cas9 in Trypanosoma cruzi. Bio Protoc. 7, e2299 10.21769/BioProtoc.2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caruso M., Maitan M. A., Bifulco G., Miele C., Vigliotta G., Oriente F., Formisano P., and Beguinot F. (2001) Activation and mitochondrial translocation of protein kinase Cδ are necessary for insulin stimulation of pyruvate dehydrogenase complex activity in muscle and liver cells. J. Biol. Chem. 276, 45088–45097 10.1074/jbc.M105451200 [DOI] [PubMed] [Google Scholar]
- 42. Lander N., Li Z. H., Niyogi S., and Docampo R. (2015) CRISPR/Cas9-induced disruption of paraflagellar rod protein 1 and 2 genes in Trypanosoma cruzi reveals their role in flagellar attachment. MBio 6, e01012 10.1128/mBio.01012-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lander N., Chiurillo M. A., and Docampo R. (2016) Genome editing by CRISPR/Cas9: a game change in the genetic manipulation of protists. J. Eukaryot. Microbiol. 63, 679–690 10.1111/jeu.12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Slaymaker I. M., Gao L., Zetsche B., Scott D. A., Yan W. X., and Zhang F. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarez V. E., Kosec G., Sant'Anna C., Turk V., Cazzulo J. J., and Turk B. (2008) Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 283, 3454–3464 10.1074/jbc.M708474200 [DOI] [PubMed] [Google Scholar]
- 46. Alonso G. D., Pereira C. A., Remedi M. S., Paveto M. C., Cochella L., Ivaldi M. S., Gerez de Burgos N. M., Torres H. N., and Flawiá M. M. (2001) Arginine kinase of the flagellate protozoa Trypanosoma cruzi. Regulation of its expression and catalytic activity. FEBS Lett. 498, 22–25 10.1016/S0014-5793(01)02473-5 [DOI] [PubMed] [Google Scholar]
- 47. Teague W. M., Pettit F. H., Wu T. L., Silberman S. R., and Reed L. J. (1982) Purification and properties of pyruvate dehydrogenase phosphatase from bovine heart and kidney. Biochemistry 21, 5585–5592 10.1021/bi00265a031 [DOI] [PubMed] [Google Scholar]
- 48. Turkan A., Hiromasa Y., and Roche T. E. (2004) Formation of a complex of the catalytic subunit of pyruvate dehydrogenase phosphatase isoform 1 (PDP1c) and the L2 domain forms a Ca2+ binding site and captures PDP1c as a monomer. Biochemistry 43, 15073–15085 10.1021/bi048901y [DOI] [PubMed] [Google Scholar]
- 49. Vacchina P., Lambruschi D. A., and Uttaro A. D. (2018) Lipoic acid metabolism in Trypanosoma cruzi as putative target for chemotherapy. Exp. Parasitol. 186, 17–23 10.1016/j.exppara.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 50. McCormack J. G., Halestrap A. P., and Denton R. M. (1990) Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol. Rev. 70, 391–425 10.1152/physrev.1990.70.2.391 [DOI] [PubMed] [Google Scholar]
- 51. Denton R. M., Randle P. J., and Martin B. R. (1972) Stimulation by calcium ions of pyruvate dehydrogenase phosphate phosphatase. Biochem. J. 128, 161–163 10.1042/bj1280161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Denton R. M., Randle P. J., Bridges B. J., Cooper R. H., Kerbey A. L., Pask H. T., Severson D. L., Stansbie D., and Whitehouse S. (1975) Regulation of mammalian pyruvate dehydrogenase. Mol. Cell Biochem. 9, 27–53 10.1007/BF01731731 [DOI] [PubMed] [Google Scholar]
- 53. Moreno S. N., Vercesi A. E., Pignataro O. P., and Docampo R. (1992) Calcium homeostasis in Trypanosoma cruzi amastigotes: Presence of inositol phosphates and lack of an inositol 1,4,5-trisphosphate-sensitive calcium pool. Mol. Biochem. Parasitol. 52, 251–261 10.1016/0166-6851(92)90057-Q [DOI] [PubMed] [Google Scholar]
- 54. Vercesi A. E., Moreno S. N., Bernardes C. F., Meinicke A. R., Fernandes E. C., and Docampo R. (1993) Thapsigargin causes Ca2+ release and collapse of the membrane potential of Trypanosoma brucei mitochondria in situ and of isolated rat liver mitochondria. J. Biol. Chem. 268, 8564–8568 [PubMed] [Google Scholar]
- 55. Moreno S. N., Docampo R., and Vercesi A. E. (1992) Calcium homeostasis in procyclic and bloodstream forms of Trypanosoma brucei. Lack of inositol 1,4,5-trisphosphate-sensitive Ca2+ release. J. Biol. Chem. 267, 6020–6026 [PubMed] [Google Scholar]
- 56. Xiong Z. H., Ridgley E. L., Enis D., Olness F., and Ruben L. (1997) Selective transfer of calcium from an acidic compartment to the mitochondrion of Trypanosoma brucei. Measurements with targeted aequorins. J. Biol. Chem. 272, 31022–31028 10.1074/jbc.272.49.31022 [DOI] [PubMed] [Google Scholar]
- 57. Guo X., Niemi N. M., Coon J. J., and Pagliarini D. J. (2017) Integrative proteomics and biochemical analyses define Ptc6p as the Saccharomyces cerevisiae pyruvate dehydrogenase phosphatase. J. Biol. Chem. 292, 11751–11759 10.1074/jbc.M117.787341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rohloff P., Rodrigues C. O., and Docampo R. (2003) Regulatory volume decrease in Trypanosoma cruzi involves amino acid efflux and changes in intracellular calcium. Mol. Biochem. Parasitol. 126, 219–230 10.1016/S0166-6851(02)00277-3 [DOI] [PubMed] [Google Scholar]
- 59. Lammel E. M., Barbieri M. A., Wilkowsky S. E., Bertini F., and Isola E. L. (1996) Trypanosoma cruzi: Involvement of intracellular calcium in multiplication and differentiation. Exp. Parasitol. 83, 240–249 10.1006/expr.1996.0070 [DOI] [PubMed] [Google Scholar]
- 60. Moreno S. N., Silva J., Vercesi A. E., and Docampo R. (1994) Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J. Exp. Med. 180, 1535–1540 10.1084/jem.180.4.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Atwood J. A. 3rd, Weatherly D. B., Minning T. A., Bundy B., Cavola C., Opperdoes F. R., Orlando R., and Tarleton R. L. (2005) The Trypanosoma cruzi proteome. Science 309, 473–476 10.1126/science.1110289 [DOI] [PubMed] [Google Scholar]
- 62. Silber A. M., Tonelli R. R., Lopes C. G., Cunha-e Silva N., Torrecilhas A. C., Schumacher R. I., Colli W., and Alves M. J. (2009) Glucose uptake in the mammalian stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 168, 102–108 10.1016/j.molbiopara.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 63. Cruz-Bustos T., Potapenko E., Storey M., and Docampo R. (2018) An intracellular ammonium transporter is necessary for replication, differentiation, and resistance to starvation and osmotic stress in Trypanosoma cruzi. mSphere 3, e00377–18 10.1128/mSphere.00377-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Crispim M., Damasceno F. S., Hernández A., Barisón M. J., Pretto Sauter I., Souza Pavani R., Santos Moura A., Pral E. M. F., Cortez M., Elias M. C., and Silber A. M. (2018) The glutamine synthetase of Trypanosoma cruzi is required for its resistance to ammonium accumulation and evasion of the parasitophorous vacuole during host-cell infection. PLoS Negl. Trop. Dis. 12, e0006170 10.1371/journal.pntd.0006170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cárdenas C., Miller R. A., Smith I., Bui T., Molgó J., Müller M., Vais H., Cheung K. H., Yang J., Parker I., Thompson C. B., Birnbaum M. J., Hallows K. R., and Foskett J. K. (2010) Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142, 270–283 10.1016/j.cell.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li F. J., Xu Z. S., Soo A. D., Lun Z. R., and He C. Y. (2017) ATP-driven and AMPK-independent autophagy in an early branching eukaryotic parasite. Autophagy 13, 715–729 10.1080/15548627.2017.1280218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oberholzer M., Morand S., Kunz S., and Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 10.1016/j.molbiopara.2005.09.002 [DOI] [PubMed] [Google Scholar]
- 68. Bone G. J., and Steinert M. (1956) Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature 178, 308–309 [DOI] [PubMed] [Google Scholar]
- 69. Schmatz D. M., and Murray P. K. (1982) Cultivation of Trypanosoma cruzi in irradiated muscle cells: Improved synchronization and enhanced trypomastigote production. Parasitology 85, 115–125 [DOI] [PubMed] [Google Scholar]
- 70. Huang B., Wu P., Popov K. M., and Harris R. A. (2003) Starvation and diabetes reduce the amount of pyruvate dehydrogenase phosphatase in rat heart and kidney. Diabetes 52, 1371–1376 10.2337/diabetes.52.6.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Love L. K., LeBlanc P. J., Inglis J. G., Bradley N. S., Choptiany J., Heigenhauser G. J., and Peters S. J. (2011) The relationship between human skeletal muscle pyruvate dehydrogenase phosphatase activity and muscle aerobic capacity. J. Appl. Physiol. 111, 427–434 10.1152/japplphysiol.00672.2010 [DOI] [PubMed] [Google Scholar]
- 72. Furuya T., Kashuba C., Docampo R., and Moreno S. N. (2000) A novel phosphatidylinositol-phospholipase C of Trypanosoma cruzi that is lipid modified and activated during trypomastigote to amastigote differentiation. J. Biol. Chem. 275, 6428–6438 10.1074/jbc.275.9.6428 [DOI] [PubMed] [Google Scholar]
- 73. Lander N., Bernal C., Diez N., Añez N., Docampo R., and Ramírez J. L. (2010) Localization and developmental regulation of a dispersed gene family 1 protein in Trypanosoma cruzi. Infect. Immun. 78, 231–240 10.1128/IAI.00780-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lander N., Ulrich P. N., and Docampo R. (2013) Trypanosoma brucei vacuolar transporter chaperone 4 (TbVtc4) is an acidocalcisome polyphosphate kinase required for in vivo infection. J. Biol. Chem. 288, 34205–34216 10.1074/jbc.M113.518993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bourguignon S. C., de Souza W., and Souto-Padrón T. (1998) Localization of lectin-binding sites on the surface of Trypanosoma cruzi grown in chemically defined conditions. Histochem. Cell Biol. 110, 527–534 10.1007/s004180050314 [DOI] [PubMed] [Google Scholar]
- 76. Pesta D., and Gnaiger E. (2012) High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol. Biol. 810, 25–58 10.1007/978-1-61779-382-0_3 [DOI] [PubMed] [Google Scholar]
- 77. Figueira T. R., Castilho R. F., Saito A., Oliveira H. C., and Vercesi A. E. (2011) The higher susceptibility of congenital analbuminemic rats to Ca2+-induced mitochondrial permeability transition is associated with the increased expression of cyclophilin D and nitrosothiol depletion. Mol. Genet. Metab. 104, 521–528 10.1016/j.ymgme.2011.08.031 [DOI] [PubMed] [Google Scholar]
- 78. Shepherd D., and Garland P. B. (1969) The kinetic properties of citrate synthase from rat liver mitochondria. Biochem. J. 114, 597–610 10.1042/bj1140597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Figueira T. R., Melo D. R., Vercesi A. E., and Castilho R. F. (2012) Safranine as a fluorescent probe for the evaluation of mitochondrial membrane potential in isolated organelles and permeabilized cells. Methods Mol. Biol. 810, 103–117 10.1007/978-1-61779-382-0_7 [DOI] [PubMed] [Google Scholar]
- 80. Peng D., and Tarleton R. (2015) EuPaGDT: A web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. Genom. 1, e000033 10.1099/mgen.0.000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chojnacki S., Cowley A., Lee J., Foix A., and Lopez R. (2017) Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553 10.1093/nar/gkx273 [DOI] [PMC free article] [PubMed] [Google Scholar]