Abstract
The epithelial Na+ channel (ENaC) possesses a large extracellular domain formed by a β-strand core enclosed by three peripheral α-helical subdomains, which have been dubbed thumb, finger, and knuckle. Here we asked whether the ENaC thumb domains play specific roles in channel function. To this end, we examined the characteristics of channels lacking a thumb domain in an individual ENaC subunit (α, β, or γ). Removing the γ subunit thumb domain had no effect on Na+ currents when expressed in Xenopus oocytes, but moderately reduced channel surface expression. In contrast, ENaCs lacking the α or β subunit thumb domain exhibited significantly reduced Na+ currents along with a large reduction in channel surface expression. Moreover, channels lacking an α or γ thumb domain exhibited a diminished Na+ self-inhibition response, whereas this response was retained in channels lacking a β thumb domain. In turn, deletion of the α thumb domain had no effect on the degradation rate of the immature α subunit as assessed by cycloheximide chase analysis. However, accelerated degradation of the immature β subunit and mature γ subunit was observed when the β or γ thumb domain was deleted, respectively. Our results suggest that the thumb domains in each ENaC subunit are required for optimal surface expression in oocytes and that the α and γ thumb domains both have important roles in the channel's inhibitory response to external Na+. Our findings support the notion that the extracellular helical domains serve as functional modules that regulate ENaC biogenesis and activity.
Keywords: epithelial sodium channel (ENaC), acid sensing ion channel (ASIC), protein domain, protein denaturation, ion channel, amiloride, channel gating, self-inhibition
Introduction
Epithelial Na+ channel (ENaC)3-mediated Na+ transport regulates extracellular fluid volume and K+ homeostasis, as well as blood pressure (1). Three homologous subunits typically constitute a functional trimeric ENaC complex (1–4). Distinct from most ion channels composed of subunits with two transmembrane domains, ENaC/Degenerin family members possess a large extracellular domain (ECD) composed of over 400 residues per subunit. The resolved structure of an acid-sensing ion channel (ASIC1), a member of the ENaC/Degenerin family, revealed a highly organized ECD with five distinct subdomains. These include a central core formed by β strand-structured palm and β-ball domains, and peripheral α-helical thumb, finger, and knuckle domains. Sequence similarities suggest that the ECDs of other members of the ENaC/Degenerin family have a similar structure (2–5). Previous studies also suggest that the well-organized and tightly packed ECDs sense a variety of external cues that modulate channel open probability (6–18).
ENaCs in both native tissues and heterologous expression systems are constitutively open, albeit with considerably variable open probability. The latter can be attributed to factors that regulate ENaC gating, including ions, pH, proteases, temperature, mechanic forces, acidic phospholipids, and palmitoylation (4). ENaC has been suggested to function as a ligand-regulated ion channel (9, 19). ENaC open probability is reduced by extracellular Na+, a process referred to as Na+ self-inhibition (7, 8, 18, 20). Many of the identified residues where substitutions either enhance or suppress Na+ self-inhibition are located within the ECDs, including a region in the α subunit containing a putative Na+-binding site that mediates the inhibitory effect of external Na+ (18). We hypothesized that the independently folded helical domains (thumb, finger, and knuckle) serve as functional modules to regulate gating and mediate the interactions of ENaC with various factors, including extracellular Na+. In support of this hypothesis, we reported that deletion of the α subunit knuckle domain leads to hyperactive channels with a loss of the Na+ self-inhibition response, whereas deletion of either the β or γ subunit knuckle domain diminished channel expression and activity (21, 22). In this study, we sought to determine specific functional roles of the helical thumb domains within ENaC subunits. Our data indicate that the thumb domain supports various ENaC-specific activities. Surprisingly the thumb domain in each subunit exhibits distinct functional roles. These results emphasize the complex inter- and intra-protein interactions to which this regulated channel is exposed.
Results
Deletion of the thumb domain alters ENaC activity in a subunit-specific manner
The thumb domain in ASIC1 is composed of two α helices (α4 and α5) connected by a loop and by a series of five disulfide bridges. Two additional loops connect the α helices to adjacent β strands of the palm domain (β9 and β10) (2). The disulfide-bridged Cys residues are strictly conserved among ENaC/Degenerin family members. In this study, we deleted all 86 to 88 residues between (and including) the most proximal and distal Cys residues in the thumb domain in each mouse ENaC subunit, leaving an 8-residue loop that provides a transition between the β9 and β10 strands of the palm domain (Fig. 1). To examine the effects of thumb domain deletions, we expressed either wildtype (WT) ENaC subunits (α, β, and γ) or two WT subunits together with a mutant subunit lacking a thumb domain (αΔT, βΔT, or γΔT) in oocytes. Amiloride-sensitive whole cell Na+ currents measured in oocytes expressing channels with either αΔT or βΔT were dramatically less than in cells expressing WT ENaC (normalized currents, 0.02 ± 0.02 for αΔT, 0.07 ± 0.03 for βΔT, mean ± S.D., p < 0.001 versus WT, n = 29–30, Fig. 2). Currents in oocytes expressing channels with γΔT were similar to WT channels (0.89 ± 0.39, p > 0.05, n = 30, Fig. 2).
Figure 1.
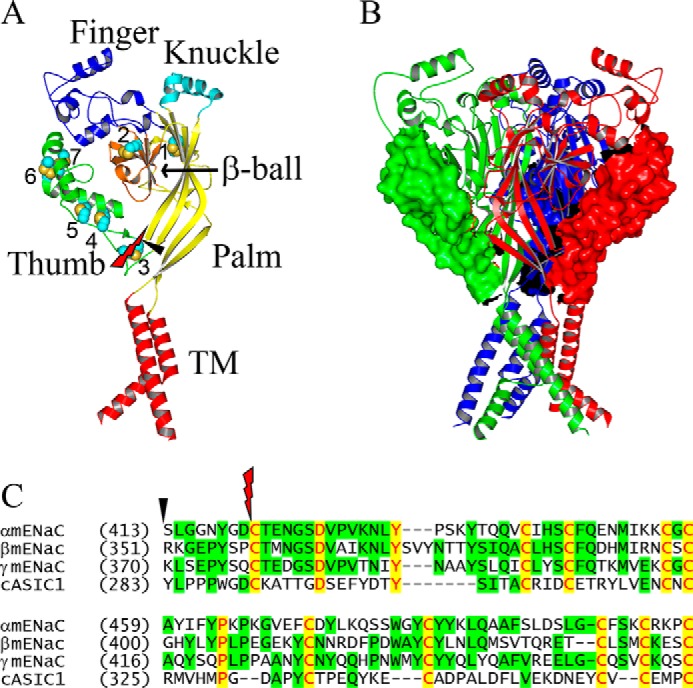
Thumb domains in monomeric and trimeric ASIC1 models. A, a monomer ASIC1 structural model is shown as ribbons with individual domains colored and labeled (TM, transmembrane domain). Seven disulfide bridges are numbered and displayed as space-filled spheres. The beginnings and ends of the two sets of thumb domain deletions are identified by a red knife and a black arrowhead. B, a trimeric ASIC1 model is shown as three colored ribbons with the deletion targets as surface rendering. Both monomeric and trimeric ASIC1 models were built from the coordinates of the closed state structure of chicken ASIC1 (PDB entry code 5WKV (55)), using PyMol (59). C, sequence alignments showing the targeted residues in the two sets of thumb domain deletions. Alignments were done with Vector NTI 11.0 (Invitrogen) with manual adjustment from protein sequences with GenBankTM accession numbers AAD21244 (αmENaC), AAD21245 (βmENaC), AAD21246 (γmENaC), and AY956393 (cASIC1). The first residue number in each line of sequence is shown in parentheses. Identical and conserved residues are shown in red letters on yellow background and black letters on green background, respectively. The percentage of identical residues between a mouse ENaC subunit and cASIC1 is 22–24 for the full-length subunits and 17–24 for the thumb domains alone. The black arrowhead and red knife show the beginnings of two sets of thumb domain deletion.
Figure 2.
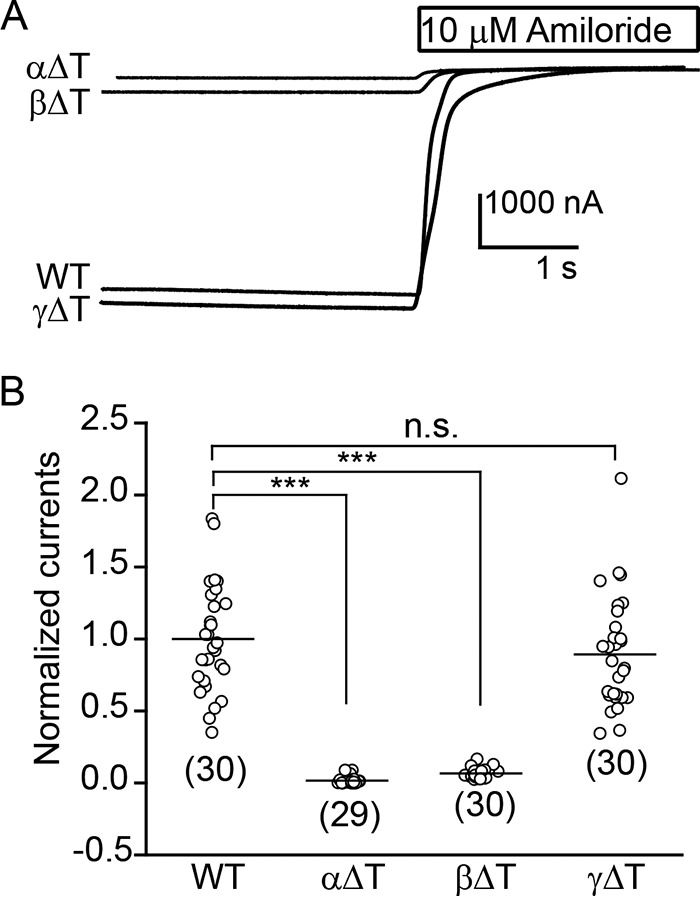
α or β subunit thumb domain deletions significantly reduce ENaC activity. Whole cell Na+ currents were measured in oocytes injected with WT or mutant mouse αβγ cRNAs (2 ng/subunit) at a clamping voltage of −100 mV (intracellular potential). Mutant channels contained one subunit with a thumb domain deletion (ΔT) and two WT subunits. A, representative current recordings in the absence or presence of 10 μm amiloride from oocytes expressing αβγ (WT), αΔTβγ, αβΔTγ, or αβγΔT ENaCs. The traces are superimposed with the same scale bars. B, scatter plots of normalized currents in groups. Normalized currents represent amiloride-sensitive currents from individual oocytes, divided by the mean of the amiloride-sensitive currents in oocytes expressing WT channels from the same batch of oocytes. Data were collected in two batches of oocytes. The amiloride-sensitive currents from the two batches of oocytes expressing WT ENaC were −3.8 ± 1.6 μA (mean ± S.D., n = 15) and −5.6 ± 1.6 μA (n = 15). Numbers of oocytes for each group are shown below data points. Horizontal bars are mean values. Statistical significances between mutant and WT channels were obtained from one-way ANOVA followed by a Tukey post hoc test (***, p < 0.001, n.s., no significant difference).
As the thumb deletion constructs retained an 8-residue loop to link the β9 and β10 strands, it was possible that these linking residues contribute to the functional changes described above. We generated a set of thumb deletions in which all thumb domain residues, including the 8-residue linking tracts, were deleted. Channels with an individual subunit bearing a thumb deletion lacking an 8-residue linker exhibited levels of activity in oocytes similar to channels with a thumb deletion bearing an 8-residue linker. Normalized currents were 0.03 ± 0.04 (p < 0.001 versus WT, n = 39) for α mutant, 0.07 ± 0.06 (p < 0.001 versus WT, n = 38) for β mutant, and 1.12 ± 0.93 (p > 0.05 versus WT, n = 75) for γ mutant. These observations suggest that both the α and β subunit thumb domains are required for maximal functional ENaC expression in oocytes.
Thumb domain deletions reduce ENaC surface expression
To determine whether a thumb domain deletion in the α or β subunit reduced whole-cell currents by reducing channel expression at the plasma membrane, we examined surface protein levels of WT and mutant channels with a chemiluminescence-based assay, as previously described (22, 23). As shown in Fig. 3, channels with αΔT or βΔT had levels of surface expression that were markedly lower compared with WT channels (relative expression, 0.06 ± 0.04 for αΔT, 0.09 ± 0.11 for βΔT, p < 0.001 versus WT, n = 46–53). In contrast, channels with γΔT had levels of surface expression that were moderately, but significantly lower than WT channels (0.70 ± 0.46, p < 0.05 versus WT, n = 46).
Figure 3.
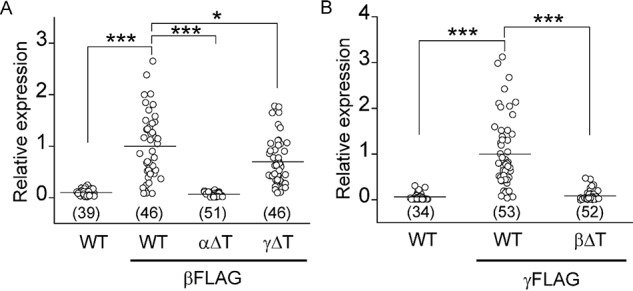
Thumb domain deletions reduce ENaC surface expression. A, relative surface expression levels in cells expressing αβγ (WT, negative control), αβFLAGγ (WT, βFLAG), αΔTβFLAGγ (αΔT, βFLAG), or αβFLAGγΔT (γΔT, βFLAG) mouse ENaCs. Relative expression levels were obtained by normalizing measured relative light units from individual oocytes to the mean of the relative light units from the same batch of oocytes expressing the FLAG-tagged WT control (WT, βFLAG). Data were from two batches of oocytes. The relative light units in βFLAG-tagged WT control cells were 1.18 ± 0.86 million (mean ± S.D., n = 46). Oocytes were injected with the same amount of cRNAs in each batch (4 ng/subunit). B, relative surface expression levels in cells expressing αβγ (WT, negative control), αβγFLAG (WT, γFLAG), or αβΔTγFLAG (βΔT, γFLAG) mouse ENaCs. Data were from three batches of oocytes. The relative light units in γFLAG-tagged WT control cells were 2.09 ± 1.74 million (n = 53). Oocytes were injected with the same amount of cRNAs (4 ng/subunit). The numbers of oocytes (n) assayed are indicated in parentheses. Statistical significances were analyzed with one-way ANOVA followed by a Tukey post hoc test (*, p < 0.05; ***, p < 0.001 versus WT/βFLAG in A or WT/γFLAG in B).
Deletion of either the α or γ subunit thumb domain reduces the Na+ self-inhibition response
ENaC lacking a γ subunit thumb domain had current levels similar to the WT channel, but there were also lower levels of surface expression. This discrepancy suggests that the mutant channel has a higher channel open probability, compared with WT. A key factor regulating ENaC open probability is Na+ self-inhibition. We have previously shown that, for selected mutants, channel open probability correlates with the magnitude of the Na+ self-inhibition response (24). Therefore, we hypothesized that channels with a thumb domain deletion in the γ subunit have a reduced Na+ self-inhibition response. Indeed, γΔT channels showed a significantly reduced Na+ self-inhibition response, as shown by an increase in the ratio of the steady state to peak current (Iss/Ipeak, 0.93 ± 0.04 (γΔT) versus 0.65 ± 0.08 (WT), p < 0.001 versus WT, n = 24, Fig. 4, C and F).
Figure 4.
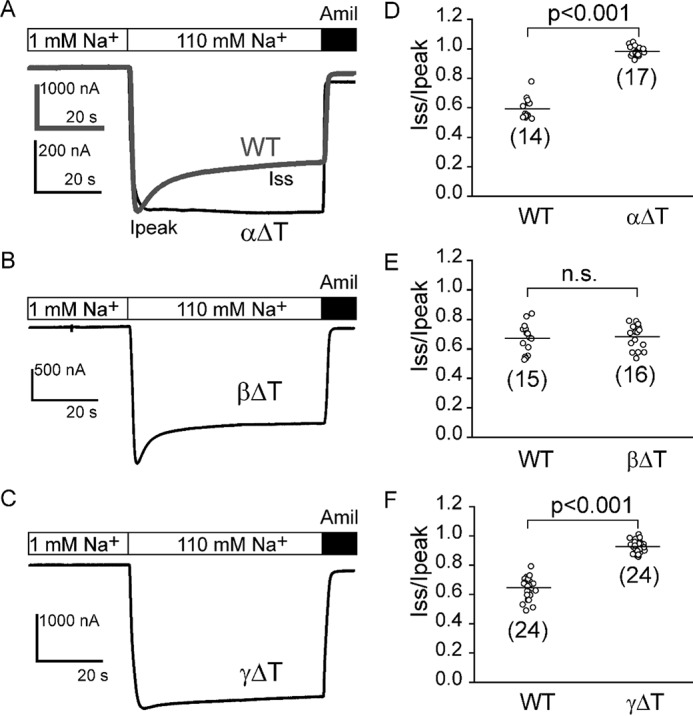
Deletion of either the α or γ thumb domain diminishes the Na+ self-inhibition response. Na+ self-inhibition responses were examined in oocytes expressing WT or mutant mouse αβγ ENaCs. Representative recordings are shown in A for WT αβγ (WT, gray trace) and αΔTβγ (αΔT, black trace), in B for αβΔTγ (βΔT), and in C for αβγΔT (γΔT) ENaCs. Bath Na+ concentrations and amiloride (10 μm) addition are shown above the traces. Ipeak and Iss in A show where peak current and steady state currents were measured. D–F, scatter plots of individual datum points of Iss/Ipeak (representing the magnitude of Na+ self-inhibition) for WT and mutant channels. The ratio was determined from amiloride-sensitive Iss and Ipeak. Number of oocytes in each group is shown in parentheses. Data were collected from 2 to 5 batches of oocytes. For WT and αβγΔT, 1 ng/subunit of cRNAs was injected, whereas for αΔTβγ and αβΔTγ, 4 ng/subunit of cRNAs were injected to increase expressed currents to levels that permit analyses of the Na+ self-inhibition response. Bar, mean for each group. Statistical comparisons were analyzed with two group Student's t test (p < 0.001, or n.s., no significant difference, as indicated).
It was formally possible that oocytes injected with αβγΔT cRNAs express both channels with three subunits (i.e. αβγΔT) as well as channels lacking the γΔT subunit (i.e. αβ). Rat αβ channels have been reported to have a very high open probability in the presence of >100 mm extracellular Na+ or Li+ (25, 26), indicating that αβ channels lack a Na+ self-inhibition response. When we expressed only α and β subunits in oocytes, amiloride-sensitive currents were generally too low to permit an examination of the Na+ self-inhibition response. Na+ currents in cells injected with α and β cRNAs were 0.01 ± 0.01, relative to WT (i.e. αβγ) channels (n = 65, p < 0.001 versus WT, n = 65–74). Nevertheless, a small number of αβ cRNA-injected oocytes expressed currents at levels sufficient to examine the Na+ self-inhibition response. Not surprisingly, Na+ self-inhibition was absent (Iss/Ipeak, 0.98 ± 0.01, p < 0.001 versus WT, n = 7–18, Fig. 5). In contrast, cells injected with αγ cRNAs showed a robust Na+ self-inhibition response that was similar to that in cells injected with cRNAs corresponding to α, β, and γ (Iss/Ipeak, 0.74 ± 0.06, p > 0.05 versus WT, n = 18–22, Fig. 5). These results suggest that loss of the Na+ self-inhibition response seen with αβγΔT channels directly reflects the response of this channel, and not contributions from αβ channels.
Figure 5.
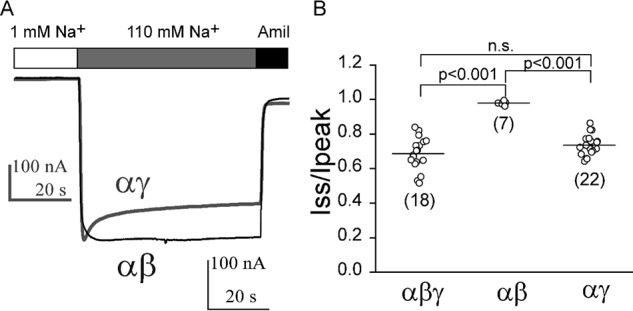
Na+ self-inhibition is absent in αβ channels. A, representative current recordings in oocytes expressing αβ (black trace) or αγ (gray trace), superimposed with different scale bars as shown. Bath Na+ concentrations and amiloride (Amil; 10 μm) are shown above the traces. B, scatter plots of individual data points of Iss/Ipeak from oocytes injected with αβγ (1 ng/subunit), αβ or αγ (4 ng/subunit) mouse ENaC cRNAs. Data were collected from three batches of oocytes. Number of oocytes in each group is shown in parentheses. Statistical significances were analyzed with one-way ANOVA followed by a Tukey post hoc test (p < 0.001, or n.s., not significant as indicated).
To examine whether the α and β thumb domains also play a role in the Na+ self-inhibition response, we examined the response in channels expressing either αΔTβγ or αβΔTγ. As oocytes expressing these mutants exhibited very low Na+ currents, we injected 4 ng of cRNA/subunit for the mutant channels, rather than 1 ng/subunit for the WT channel. A Na+ self-inhibition response was absent in the recordings corresponding to the mutant channels lacking the α thumb domain (Iss/Ipeak, 0.98 ± 0.03, p < 0.001 versus WT, n = 14–17, Fig. 4, A and D). These data suggest that the α thumb domain also plays a role in the Na+ self-inhibition response. As βγ channels lack function (27, 28), the loss of Na+ self-inhibition likely reflects the assembled αΔTβγ channels.
Mutant channels lacking the β thumb domain (αβΔTγ) had a Na+ self-inhibition response that was similar to that of αβγ WT channels, suggesting that the β thumb domain is dispensable for Na+ self-inhibition. However, as αγ channels exhibit modest currents when expressed in oocytes (0.03 ± 0.03, relative to WT, p < 0.001 versus WT, n = 69) (27), it is possible that oocytes injected with αβΔTγ cRNAs express both αγ as well as αβΔTγ channels. As noted above, αγ channels exhibited a Na+ self-inhibition response similar to WT channels (Fig. 5). To confirm that βΔT subunits reside in the functional channel complex, we introduced a S518C mutation into the βΔT construct. The sulfhydryl reactive reagent MTSET specifically activates αβS518Cγ channels, increasing channel open probability to near 1 (29, 30). As shown in Fig. 6A, αβS518Cγ channels responded to 1 mm MTSET with an increase in channel current and a loss of Na+ self-inhibition, consistent with a transition to a fully open state. In a majority of cells expressing αβΔT-S518Cγ, MTSET increased the current and reduced Na+ self-inhibition, suggesting that currents are mediated predominately by the αβΔT-S518Cγ channels. The effect of MTSET on αβΔT-S518Cγ was similar to that on αβS518Cγ (IMTSET/I, 1.73 ± 0.38 ((αβΔT-S518Cγ) versus 1.85 ± 0.32 (αβS518Cγ), p > 0.05, n = 9–10). Therefore, most channels harbored a β subunit. In contrast, MTSET had no effect on αβγ channels (31, 32). There was also no statistically significant difference in the Na+ self-inhibition responses of αβΔT-S518Cγ and αβS518Cγ prior to MTSET treatment, and following MTSET treatment (Fig. 6, A–C). Taken together, our observations suggest that a β thumb domain deletion does not significantly affect Na+ self-inhibition.
Figure 6.
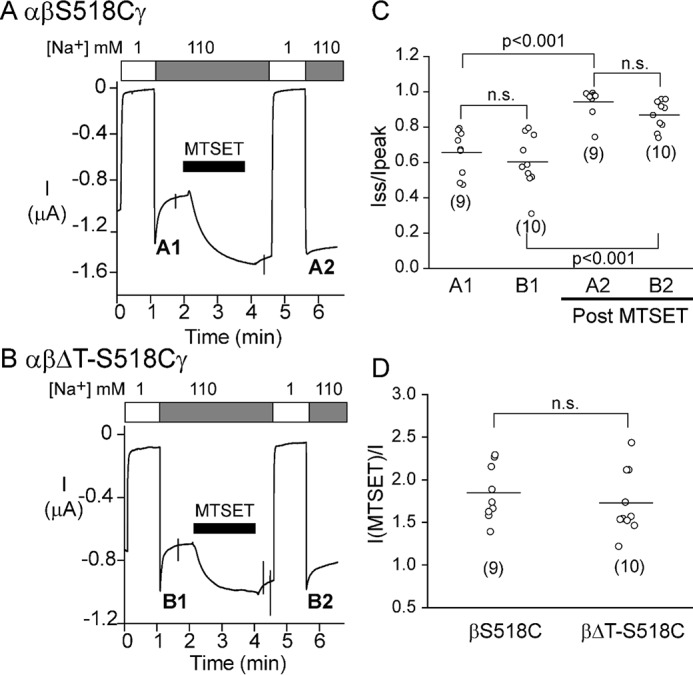
Oocytes injected with αβΔTγ cRNAs predominantly express channels containing three subunits. A, representative recording showing the effect of 1 mm MTSET and the Na+ self-inhibition response prior to (A1) and after (A2) MTSET treatment. Cells were injected with αβS518Cγ cRNAs (2 ng/subunit). B, representative recording showing the effect of 1 mm MTSET and Na+ self-inhibition responses prior to (B1) and after (B2) MTSET treatment. Cells were injected with αβΔT-S518Cγ ENaC cRNAs (2 ng/subunit). C, scatter plot of individual Iss/Ipeak values. A1 and A2 are from trace A and B1 and B2 are from trace B. Data were collected from two batches of oocytes. Number of oocytes in each group is shown in parentheses. Statistical significances were analyzed with two-way ANOVA followed by a Tukey post hoc test (p < 0.001, or n.s., not significant as indicated). D, IMTSET/I, ratio of amiloride-sensitive currents measured at the end of the 2-min MTSET treatment relative to the current measured immediately prior to MTSET treatment. Data were collected from two batches of oocytes. Number of oocytes in each group is shown in parentheses. Statistical significance was analyzed with a Student's t test.
Thumb domain deletions differentially affect channel protein maturation and degradation in a subunit-specific manner
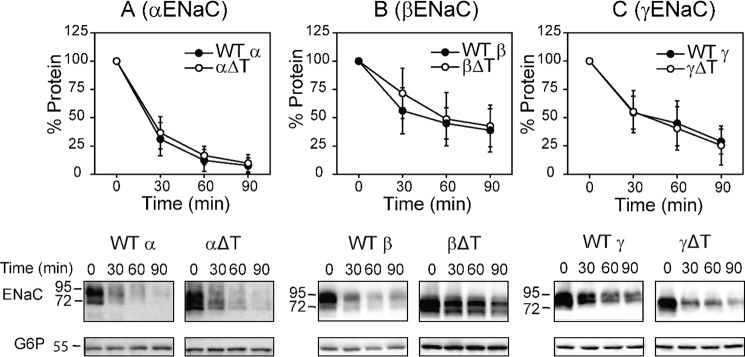
ENaC matures inefficiently, which leads to robust targeting via the endoplasmic reticulum-associated degradation pathway (33, 34). Given the low level of surface expression of channels lacking the α or β subunit thumb domain, we examined whether these individual subunits underwent accelerated endoplasmic reticulum-associated degradation. To test this possibility, we performed cycloheximide chase experiments in yeast, because this model has served as a readout for inefficient channel folding and chaperone-mediated selection of orphaned ENaC subunits (34–37). As shown in Fig. 7, there was no significant difference in rates of degradation between mutant and WT subunits when individual subunits were expressed in Saccharomyces cerevisiae.
Figure 7.
Deletion of a thumb domain does not alter rates of degradation of individual ENaC subunits expressed in yeast. Cycloheximide chase experiments were performed in WT yeast strains expressing a mouse ENaC subunit with a C-terminal HA epitope tag: A, WT α (closed circles) or αΔT (open circles); B, WT β (closed circles) or βΔT (open circles); and C, WT γ (closed circles) or γΔT (open circles). Representative blots are shown below each graph. Chase reactions were performed at 37 °C and lysates were immunoblotted with anti-HA antisera (ENaC) and with anti-glucose-6-phosphate dehydrogenase (G6P) as a loading control. Data are shown as percentages of proteins representing normalized protein levels to time 0, mean ± S.D. Data were collected in 6–13 experiments. No significant difference at each time point between the WT and mutant subunits was found (two-way ANOVA with Tukey post hoc test).
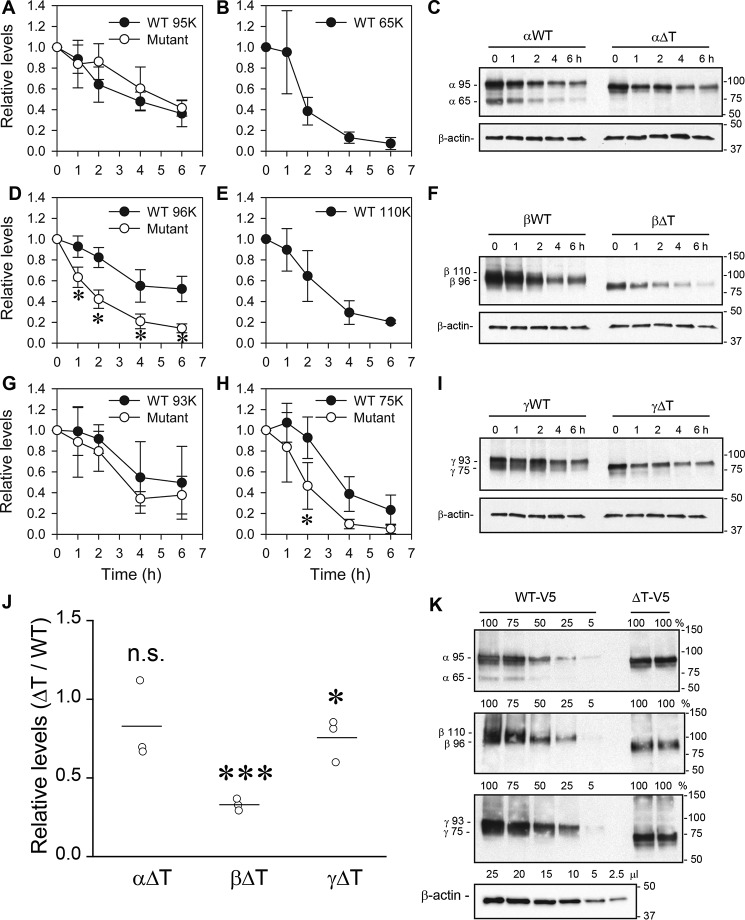
To determine the stabilities of the subunits in a system in which the trimeric ENaC channel can assemble and transit beyond the ER, we performed cycloheximide chase experiments in Fisher rat thyroid (FRT) cells expressing either WT αβγ or mutant ENaCs lacking a thumb domain in a single subunit. Only the mutant subunit or the corresponding WT control contained an HA epitope tag at the N terminus and V5 tag at the C terminus (Fig. 8). Our previous studies showed that ENaC subunits processed in the biosynthetic pathway acquire mature N-linked glycans, and that the α and γ subunits are cleaved at specific sites in their extracellular domains (38–41). As a result of these maturation steps, the α, β, and γ subunits appear on immunoblots as bands at the following molecular weights using antibodies against the C-terminal V5 tag: immature α (95 kDa) and mature α (65 kDa); immature β (96 kDa) and mature β (110 kDa); immature γ (93 kDa) and mature γ (75 kDa). We observed that deletion of the α subunit thumb domain had no effect on the degradation rate when compared with that of the immature, noncleaved α subunit. We also failed to detect a cleaved, mature α subunit when the thumb domain was deleted (Fig. 8, A–C). Deletion of the thumb domain in the β subunit also prevents its maturation, but in this case the β subunit thumb domain deletion resulted in significantly faster degradation of the immature β subunit (Fig. 8, D–F). In turn, a modest degree of proteolytic processing of the γ subunit was detectable in the γ subunit thumb deletion mutant. Although the degradation of the immature γ subunit did not differ between WT and mutant channels with a γΔT subunit, degradation of the mature cleaved γΔT subunit was significantly faster at the 2-h time point compared with the degradation of the cleaved WT γ subunit (75 kDa, Fig. 8, G–I). Whole cell protein expression levels of both βΔT and γΔT at the start of the chase (i.e. t = 0) were also significantly lower than those of the respective WT subunits (p < 0.05, Fig. 8J, n = 3), consistent with faster degradation rates of the ΔT subunits. In contrast, whole cell expression of αΔT was similar to the WT α subunit, consistent with its WT-like degradation rate.
Figure 8.
Thumb domain deletions differentially affect subunit maturation and degradation. FRT cells were transfected with αβγ where the α (A–C), β (D–F), or γ (G–I) subunit had both an N-terminal HA and C-terminal V5 tag. Epitope-tagged subunits were either wildtype (WT) or a thumb domain deletion mutant (ΔT) as indicated. Cycloheximide (100 μg/ml) was added to cells growing on plastic (12-well size) at t = 0. Cells lysates were prepared at varying times after cycloheximide addition (t = 0, 1, 2, 4, or 6 h) in the presence of protease inhibitors. ENaC was immunoprecipitated with anti-V5 antibodies conjugated to beads and immunoblotted with anti-V5 antibodies. Aliquots of the initial cell extracts were subjected to immunoblotting with anti-β-actin antibodies. The relative levels of proteins (compared with t = 0) are presented as the mean ± S.D. (A, B, D, E, G, and H) from three experiments, and a representative immunoblot is shown in each case (C, F, and I). Molecular weights of the immature α (95,000), β (96,000), and γ (93,000) subunits, and the mature α (65,000), β (110,000), and γ (75,000) subunits are indicated to the left of the immunoblots for the WT subunits. The mobility of the mutant subunits in each case were slightly faster than for the WT subunits as expected for the thumb deletions (∼10 kDa). The mature form of the γΔT subunit was observed (H and I), but mature forms of the αΔT and βΔT subunits were absent. Degradation rates of the immature α and γ subunits were similar between WT and ΔT (A and G, p > 0.05, two-way ANOVA). The immature βΔT subunit had significantly faster degradation than the WT β subunit (D, *, p < 0.001, two-way ANOVA with Tukey post hoc test). The mature γΔT level was significantly less than the mature WT γ subunit level at t = 2 h (H, *, p < 0.01, two-way ANOVA with Tukey post hoc test). J, relative levels of mutant versus WT subunit proteins at t = 0 (ΔT/WT, n = 3). Protein levels were normalized to β-actin expression. Although there was no significant difference in α subunit level in cells transfected with WT αβγ and αΔTβγ (0.83 ± 0.25, n.s., no significant difference, Student's t test) cDNAs, both αβΔTγ- and αβγΔT-transfected cells had significantly less β subunit (0.33 ± 0.04, ***, p < 0.001, Student's t test) and γ subunit (0.76 ± 0.14, *, p < 0.05, Student's t test) protein levels than WT-transfected cells, respectively. Both mature and immature forms were included in the quantification. K, representative examples (n = 2) of immunoblots are shown for anti-V5 immunoprecipitates from 10 to 100% cell extract expressing WT αβγ (for comparison to 100% cell extract expressing ΔT subunits), and for relevant amounts of extract for immunoblotting β-actin.
Finally, we examined whether the γ thumb domain deletion affects the protein levels of the α or β subunit. As shown in Fig. 9, γΔT reduced whole cell β subunit expression levels. No effect on whole cell α subunit expression was evident. However, there was a loss of α (65 kDa) and β subunit (110 kDa) maturation in the presence of γΔT. Overall, our results suggest that the presence of a thumb domain on each of the three subunits is required for channel maturation, and the absence of the β or γ subunit thumb domain enhances β (immature) or γ (mature) subunit degradation, respectively.
Figure 9.
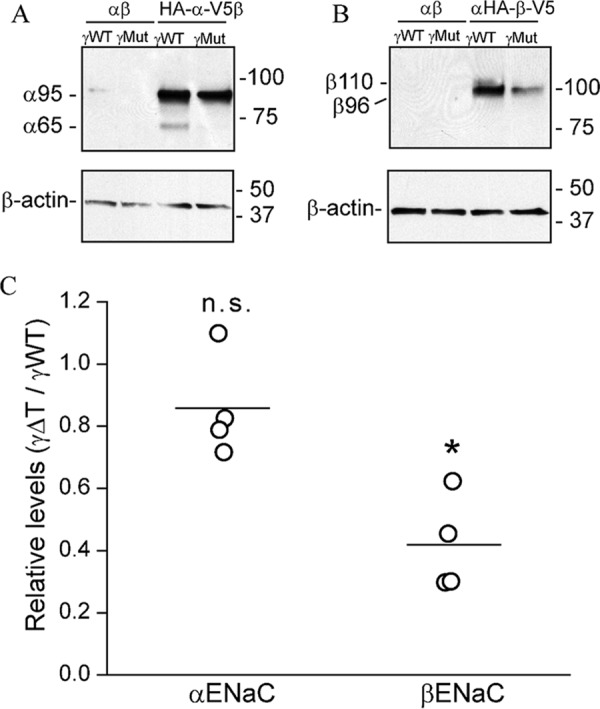
Deletion of γ thumb domain reduces β subunit expression. To examine the effect of the γ thumb domain deletion on stability of the α subunit, FRT cells were transfected with WT α or HA-α-V5 and WT β cDNAs with either WT γ or γΔT cDNA (A). To examine the effect of γ thumb domain deletion on β subunit stability, cells were transfected with WT β or HA-β-V5, and WT α cDNAs with either WT γ or γΔT cDNA (B). ENaC was immunoprecipitated with anti-V5 antibodies conjugated to beads and immunoblotted with anti-V5 antibodies. Representative blots are shown with mobility of immature (α95K and β96K) and mature (α65K and β110K) subunits shown to the left of the panels, and molecular mass markers shown to the right. Aliquots of the initial cell extracts were subjected to immunoblotting with anti-β-actin antibodies. Cells transfected with nontagged α and β cDNAs were used as negative control in both experiments. C, relative levels represent the protein levels of either α or β subunit in cells expressing γΔT, relative to their levels in the presence of the WT γ subunit (n = 4). Protein levels were normalized to β-actin expression. There was no significant difference in relative α subunit level when expressed with WT γ or γΔT (n.s., no significant difference, Student's t test). However, β subunit protein level was significantly lower when expressed with γΔT than with the WT γ subunit (p < 0.001, Student's t test).
Discussion
In this study we sought to dissect the functional roles of the extracellular thumb domains within each of the ENaC subunits. Our results suggest that thumb domains in each subunit are required for optimal ENaC surface expression in Xenopus oocytes. Channels with an α or β subunit thumb domain deletion had a very low level of surface expression, whereas the absence of the γ subunit thumb domain resulted in only a moderate reduction in channel surface expression. Furthermore, deletions of the α or γ subunit thumb domain were associated with a loss of the Na+ self-inhibition response, whereas channels with a β subunit thumb domain deletion had a Na+ self-inhibition response that was similar to the WT channel. These results indicate that the thumb domains within the three ENaC subunits have distinct and nonequivalent roles for ENaC activity and surface expression. Although deletion of the γ subunit thumb domain did not significantly change macroscopic Na+ currents in oocytes, we found that γΔT channels had reduced surface expression as well as a loss of the Na+ self-inhibition response that should increase channel open probability.
Previous studies analyzing channels with specific point mutations in the α or γ subunit thumb domain suggested that these domains are involved in the Na+ self-inhibition response. A reduction in the Na+ self-inhibition response was seen with channels bearing individual mutations in six of the 10 Cys residues in the γ subunit thumb domain (42). Mutation of a Met residue (γMet-438) within α5 helix was also associated with a loss of the Na+ self-inhibition response (24). Although oocytes expressing αΔTβγ channels had very low currents in oocytes, reflecting markedly reduced ENaC surface expression, we observed that these channels lost Na+ self-inhibition. Previous findings have also suggested that the α subunit thumb domain has a role in this response. Specifically, channels with an individual mutation in five of the 10 α subunit thumb domain Cys residues exhibited altered responses to extracellular Na+ (42). Interestingly, mutations at four sites led to a reduction of the Na+ self-inhibition response, whereas one mutation enhanced the response (42). Other work indicated that mutation of a Cys residue (αC479R) in the α subunit thumb domain of human ENaC was associated with Liddle syndrome in a sibling pair (43). As a mutation of the homologous residue in mouse αENaC (αC506A) eliminated Na+ self-inhibition (42), the phenotype seen with the human αC579R mutant activity likely reflects suppressed Na+ self-inhibition and an increase in channel open probability. Interestingly, most mutations in the region linking α helices in the thumb domain of the α subunit dramatically enhanced Na+ self-inhibition and reduced ENaC activity (42, 44). In contract to channels with an αΔT or γΔT subunit, channels with a βΔT subunit had a Na+ self-inhibition response that was similar to WT channels. This result is also in agreement with several previous studies where β subunit mutations did not affect Na+ self-inhibition (8, 15, 16, 22, 24, 42).
ENaCs are cleaved and activated by proteases within the biosynthetic pathway and at the cell surface (4, 45). ENaC proteolysis appears to have a role in activating the channel under certain physiologic conditions, including volume depletion and hyperkalemia (46, 47). Studies of ENaC activation by proteases have provided clues regarding a mechanism by which the α and γ thumb domains influence the Na+ self-inhibition response. Channel activation by proteases involves cleavage at sites flanking imbedded inhibitory tracts in the α and γ subunit finger domains (4, 45, 48–50). Release of these inhibitory tracts transitions channels to a higher open probability state with an associated loss of Na+ self-inhibition (48–50). Peptides corresponding to the released tracts are reversible channel inhibitors, and a putative inhibitory binding region for the α subunit-based peptide mapped to sites in α helices (α1 and α2) in the finger domain, and the loop connecting the thumb domain α helices in the α subunit (5, 51). It was proposed that the α subunit thumb-finger domain interface is dynamic, and that the α subunit-imbedded inhibitory tract stabilizes the thumb-finger domain interface, conferring a low activity channel state (5, 44, 51). In support of this model, cross-linking the finger domain α1 helix and the loop connecting the thumb domain in the α subunit stabilized ENaC in a low activity state (51). As this model was based on functional studies, a resolved ENaC structure including the finger and thumb domains is needed to confirm key components of the model. We predict that the γ subunit imbedded inhibitory tract stabilizes the channel in a lower activity channel state by a similar mechanism.
Our model suggests that the α and γ subunit thumb domains help restrain the activity of noncleaved channels. As ENaC activation by proteases occurs in association with reductions in (or a loss of) the Na+ self-inhibition response (7, 48), the model also predicts that channels lacking an α and γ subunit thumb domain will exhibit a reduced Na+ self-inhibition response. This is exactly what was observed (Fig. 4). We also found that the Na+ self-inhibition response of βΔT channels was similar to WT channels. This is notable as β subunits are not processed by proteases (38–40), and there is no evidence that β subunits have imbedded inhibitory tracts.
Thumb domain interactions with other extracellular components likely impact channel gating. For example, intersubunit cross-linking between thumb and palm domains alters ENaC activity in a length-dependent manner (52). Furthermore, there is evidence that ASIC1 thumb domain interactions with the β-ball and palm domains influence channel gating (53, 54). The recently published closed state structure of ASIC1 highlights a prominent role of the thumb domains in channel gating (55).
Although the reduced Na+ self-inhibition response in channels with an αΔT or γΔT subunit should result in an increase in channel activity, thumb domain deletions were also associated with significant reductions in ENaC surface expression. Expression of WT or mutant channels in FRT cells suggested that the α and β subunit thumb domains are required for subunit maturation. Cycloheximide-chase studies suggested that the β subunit thumb domain has a role in stabilizing the immature β subunit (Fig. 7). In addition, the γ thumb domain appears to have a role in processing leading to the mature forms of the α and β subunits, or stabilizing the mature form of the γ subunit (Figs. 8 and 9). These findings are in agreement with previous studies of ENaCs with deletions of specific extracellular domains, where reductions in subunit maturation and channel surface expression were seen. For example, deletion of either the β or γ subunit knuckle domain, but not the α subunit knuckle domain, dramatically reduced ENaC surface expression and prevented subunit maturation (22).
In summary, we found that ENaC extracellular thumb domains are important regulators of ENaC gating by extracellular Na+, steady state levels of channels at the cell surface, and channel subunit maturation. Our observations support the notion that specific extracellular domain structures serve as key functional modules (3, 21, 22).
Experimental procedures
Site-directed mutagenesis
Point mutations and deletions were introduced into mouse α, β, and γ ENaC cDNAs using the QuikChange II XL Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA). One set of thumb domain deletions was generated where all residues between (and including) the first and last Cys residues within the thumb domains were deleted leaving an 8-residue linking tract to allow proper transition between the β9 and β10 strands of the palm domain (referred to as ΔT). There 8 residue tracts were SLGGNYGD for αΔT, RKGEPYSP for βΔT, and KLSEPYSQ for γΔT. Deleted residues are identified in Fig. 1C. A second set of thumb domain deletions was generated where all thumb domain residues, including the 8-residue tracts, were deleted. Target mutations were verified by DNA sequencing. WT and mutant ENaC cRNAs were synthesized by in vitro transcription using T3 RNA polymerase (Ambion, Inc.), purified by an RNA purification kit (Qiagen), and quantified by spectrophotometry.
ENaC expression and two-electrode voltage clamp
ENaC expression in Xenopus oocytes and current measurements by two-electrode voltage clamp were performed as previously reported (13). Stage V and VI oocytes with the follicle cell layer removed were injected with between 0.5 and 4 ng of each subunit (α, β and γ) cRNA, as indicated, in a volume of 50 nl, and incubated at 18 °C in modified Barth's solution (MBS, 88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 15 mm HEPES, 0.3 mm Ca(NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 10 μg/ml of streptomycin sulfate, 100 μg/ml of gentamycin sulfate, pH 7.4). All experiments were performed at room temperature (20–24 °C) 20–52 h following injection. Oocytes were placed in a recording chamber from Warner Instruments (Hamden, CT) and perfused with a constant flow rate of 12–15 ml/min. The perfusion solution contained 110 mm NaCl, 2 mm KCl, 2 mm CaCl2, and 10 mm HEPES and had the pH adjusted at 7.4. Voltage clamp was performed using Axoclamp 900A Computer-controlled Microelectrode Amplifier and DigiData 1440A interface controlled by pClamp 9.2 (Molecular Devices Corp., Sunnyvale, CA). The protocol for harvesting oocytes from Xenopus laevis was approved by the University of Pittsburgh's Institutional Animal Care and Use Committee.
Na+ self-inhibition
Na+ self-inhibition was examined as previously reported (8). Briefly, a low [Na+] bath solution (NaCl-1; containing 1 mm NaCl, 109 mm N-methyl-d-glucamine, 2 mm KCl, 2 mm CaCl2, 10 mm HEPES, pH 7.4) was rapidly replaced with a high [Na+] bath solution (NaCl-110; containing 110 mm NaCl, 2 mm KCl, 2 mm CaCl2, 10 mm HEPES, pH 7.4). Oocytes expressing ENaCs were continuously clamped to a membrane potential of −60 or −100 mV. Bath solution exchange was done with a Teflon valve computer-controlled perfusion system (AutoMate Scientific Inc., Berkeley, CA). At the end of each experiment, 10 μm amiloride was added to the bath solution to determine the amiloride-insensitive portion of the whole-cell current. High perfusion rate (10 ml/min) and small bath volume (0.1 ml) in a linear flow chamber permitted rapid solution exchange (<1 s for a full exchange). The magnitude of Na+ self-inhibition was represented by the ratio of the steady state current (Iss) to peak current (Ipeak). Ipeak was obtained by visual inspection as the maximal inward current after bath [Na+] increase, typically within seconds. Iss was determined as the current at 40 s following the Ipeak. Both Iss and Ipeak were amiloride-sensitive currents, obtained by subtracting the current in the presence of 10 μm amiloride. To minimize complications due to variability in the Na+ self-inhibition response between batches of oocytes, the responses of WT channels were always tested in an alternating manner with mutants in the same batch of oocytes.
Surface expression in oocytes
Surface expression of mouse ENaC in oocytes was determined using a β or γ subunit with an extracellular FLAG epitope tag, as previously reported (23). Briefly, oocytes were injected with 4 ng/subunit of the three ENaC cRNAs with either the β or γ subunit containing an extracellular FLAG epitope tag (DYKDDDDK). The FLAG tag was placed immediately following residue Thr137 in the β subunit, as previously described (56). The γ subunit tag was generated by replacing residues Val131–Thr138 with a FLAG sequence. Robust amiloride-sensitive Na+ currents confirmed that ENaCs containing a FLAG-tagged β or γ subunit were functional. Two days after cRNA injection, a surface expression assay was performed on ice, except for the last step that was at room temperature (20–24 °C). Following a 30-min incubation in antibiotic-free MBS supplemented with 1% BSA (MBS/BSA), oocytes were incubated for 1 h with MBS/BSA supplemented with 1 μg/ml of a mouse monoclonal anti-FLAG antibody (M2, Sigma). Oocytes were then washed in MBS/BSA and incubated with MBS/BSA supplemented with 1 μg/ml of a horseradish peroxidase coupled to a secondary antibody (peroxidase-conjugated AffiniPure F(ab′)2 fragment goat anti-mouse IgG, Jackson ImmunoResearch, West Grove, PA) for 1 h. Cells were washed and transferred to MBS without BSA. Individual oocytes were placed in a 96-well plate, and 100 μl of SuperSignal ELISA Femto Maximum Sensitivity Substrates (Thermo Scientific, Rockford, IL) was added to each well. Cells were then incubated at room temperature for 1 min, and chemiluminescence in relative light units was quantified in a GloMax-Multi+ Detection System (Promega, Madison, WI).
ENaC degradation in yeast. Yeast S. cerevisiae strains were grown at 26 °C using standard methods, and media preparation and transformation were performed as described (57). The WT yeast strain used was BY4742 from Open Biosystems (Thermo Scientific). Construction of pRS426GPD-αENaC-HA, pRS426GPD-βENaC-HA, and pRS426GPD-γENaC-HA was previously described (34). The cloning strategy and restriction enzymes used to construct pRS426GPD-αENaCΔT-HA, pRS426GPD-βENaCΔT-HA, and pRS426GPD-γENaCΔT-HA were similar to the strategy used to generate knuckle deletion mutants (22). Cycloheximide chase analyses of the ENaC subunits were performed as published (34), and cell lysates from chase samples were generated using alkaline lysis, followed by TCA precipitation (58). Proteins were resolved by SDS-PAGE before Western blot analysis. The ENaC subunits were detected using horseradish peroxidase-conjugated anti-HA antibodies (clone 3F10; Roche Applied Science) at a dilution of 1:5000. Western blots were also probed with anti-glucose-6-phosphate dehydrogenase (Glc-6-P; Sigma) rabbit antisera, which served as a loading control. The Glc-6-P primary antibody was detected with a donkey horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (GE Healthcare). Immunoblot signals were imaged using enhanced chemiluminescence (Pierce) and visualized on a Bio-Rad ChemiDoc XRS+ system. Quantitation was carried out using ImageJ software (version 1.44, NIH).
ENaC degradation in FRT cells
FRT cells, provided by Dr. Michael Myerburg (University of Pittsburgh), were cultured in Dulbecco's modified Eagles's medium/F-12 medium supplemented with 7.5% decomplimented fetal bovine serum (FBS heated to 50 °C for 50 min). Cells were plated in plastic wells (12-well size from Costar, Corning, NY) to form a uniform monolayer. The next day (18–20 h), cells were transfected using Lipofectamine 2000 with 0.5 μg of plasmids encoding α, β, and γ mouse ENaC subunits, with or without a thumb domain, as described in the figure legends. Thumb domains were deleted as indicated in subunits containing N-terminal HA and C-terminal V5 epitope tags, and co-expressed with the other two WT subunits with no epitope tag. 20–24 h post-transfection cells were washed with Dulbecco's PBS with Mg2+ and Ca2+ (DPBS) (Corning Cellgro, Manassas, VA), and fresh media containing cycloheximide (diluted from ×1000 stock in DMSO) was added to cells at a final concentration of 100 μg/ml (Calbiochem EMD Millipore, Billerica, MA). At 0, 1, 2, 4, or 6 h following the addition of cycloheximide, the cells were washed with DPBS, and then solubilized at 4 °C in a detergent solution (0.4% sodium deoxycholate, 1% Nonidet P-40, 63 mm EDTA, and 50 mm Tris-HCl, pH 8) with 1% protease inhibitor mixture III (EMD Millipore, Billerica, MA; pH 7.4). ENaC was immunoprecipitated with anti-V5 antibodies conjugated to beads, as described previously (38). Supernatant (10%) from the immunoprecipitation was saved and used for immunoblotting for β-actin. Immunoprecipitated proteins were eluted from beads with Laemmli buffer followed by SDS-PAGE and immunoblotting with anti-V5 antibodies. To confirm that the bands quantified by scanning film were within a linear range with regard to band density, we examined band intensities of anti-V5 immunoprecipitates from a dilution series (between 5 and 100%) of extracts of cells expressing WT αβγ where only one subunit had N-terminal HA and C-terminal V5 epitope tags (Fig. 8K). The average band intensities of αΔT, βΔT, and γΔT (Fig. 8K) were within the band intensities of the dilution series for the WT subunits.
To study the effect of the γ subunit thumb domain deletion on maturation and processing of the α and β subunits, FRT cells were plated in 12-well plastic plates (Costar, Corning, NY) to form a uniform monolayer. The next day (18–20 h) cells were transfected using Lipofectamine 2000 with 0.5 μg of plasmids encoding mouse α and β subunits, and a γ subunit with or without the thumb domain, as described in the figure legends. α or β subunit processing and maturation were examined with N-terminal HA and C-terminal V5-tagged α or β subunits, respectively, where the other two subunits had no epitope tag. 18–20 h post-transfection, cells were solubilized at 4 °C in a detergent solution (0.4% sodium deoxycholate, 1% Nonidet P-40, 63 mm EDTA, and 50 mm Tris-HCl, pH 8) with 1% protease inhibitor mixture III (EMD Millipore, Billerica, MA; pH 7.4). ENaC was immunoprecipitated with anti-V5 antibodies conjugated to beads, as described previously (38). Supernatant (10%) from the immunoprecipitation was saved and used for immunoblotting for β-actin. Immunoprecipitated proteins were eluted from beads with Laemmli buffer, followed by SDS-PAGE and immunoblotting with anti-V5 antibodies.
Statistical analyses
Data are presented as mean ± S.D. Significance comparisons between WT and mutant groups were performed with a Student's t test for two group data, a one-way or two-way ANOVA with Tukey post hoc test for multiple group comparisons.
Author contributions
S. S. and T. R. K. conceptualization; S. S., J. C., and T. M. B. data curation; S. S., J. L. B., and T. R. K. formal analysis; S. S., J. L. B., and T. R. K. supervision; S. S. validation; S. S., J. C., A. M., M. E. Y., T. M. B., M. A. T., and R. P. H. investigation; S. S., J. C., T. M. B., R. P. H., and T. R. K. methodology; S. S., R. P. H., and T. R. K. writing-original draft; S. S. and T. R. K. project administration; S. S., J. C., A. M., M. E. Y., T. M. B., J. L. B., M. A. T., R. P. H., and T. R. K. writing-review and editing; J. L. B. and T. R. K. funding acquisition.
This work was supported by National Institutes of Health Grants P30 DK079307, K01 DK90195, R03 DK109024, R37 DK051391, and R01 GM075061. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ENaC
- epithelial Na+ channel
- WT
- wildtype
- ECD
- extracellular domain
- ASIC
- acid-sensing ion channel
- MTSET
- 2-(trimethylammonium)ethyl methanethiosulfonate, bromide
- FRT
- Fisher rat thyroid
- MBS
- modified Barth's solution
- HA
- hemagglutinin
- Glc-6-P
- glucose-6-phosphate dehydrogenase
- ANOVA
- analysis of variance
- cRNA
- complementary RNA.
References
- 1. Warnock D. G., Kusche-Vihrog K., Tarjus A., Sheng S., Oberleithner H., Kleyman T. R., and Jaisser F. (2014) Blood pressure and amiloride-sensitive sodium channels in vascular and renal cells. Nat. Rev. Nephrol. 10, 146–157 10.1038/nrneph.2013.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jasti J., Furukawa H., Gonzales E. B., and Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9-Å resolution and low pH. Nature 449, 316–323 10.1038/nature06163 [DOI] [PubMed] [Google Scholar]
- 3. Kashlan O. B., and Kleyman T. R. (2011) ENaC structure and function in the wake of a resolved structure of a family member. Am. J. Physiol. Renal Physiol. 301, F684–F696 10.1152/ajprenal.00259.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kleyman T. R., Kashlan O. B., and Hughey R. P. (2018) Epithelial Na+ channel regulation by extracellular and intracellular factors. Annu. Rev. Physiol. 80, 263–281 10.1146/annurev-physiol-021317-121143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kashlan O. B., Adelman J. L., Okumura S., Blobner B. M., Zuzek Z., Hughey R. P., Kleyman T. R., and Grabe M. (2011) Constraint-based, homology model of the extracellular domain of the epithelial Na+ channel α subunit reveals a mechanism of channel activation by proteases. J. Biol. Chem. 286, 649–660 10.1074/jbc.M110.167098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheng S., Perry C. J., and Kleyman T. R. (2002) External nickel inhibits epithelial sodium channel by binding to histidine residues within the extracellular domains of α and γ subunits and reducing channel open probability. J. Biol. Chem. 277, 50098–50111 10.1074/jbc.M209975200 [DOI] [PubMed] [Google Scholar]
- 7. Chraïbi A., and Horisberger J. D. (2002) Na self-inhibition of human epithelial Na channel: temperature dependence and effect of extracellular proteases. J. Gen. Physiol. 120, 133–145 10.1085/jgp.20028612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheng S., Bruns J. B., and Kleyman T. R. (2004) Extracellular histidine residues crucial for Na+ self-inhibition of epithelial Na+ channels. J. Biol. Chem. 279, 9743–9749 10.1074/jbc.M311952200 [DOI] [PubMed] [Google Scholar]
- 9. Sheng S., Perry C. J., and Kleyman T. R. (2004) Extracellular Zn2+ activates epithelial Na+ channels by eliminating Na+ self-inhibition. J. Biol. Chem. 279, 31687–31696 10.1074/jbc.M405224200 [DOI] [PubMed] [Google Scholar]
- 10. Collier D. M., and Snyder P. M. (2009) Extracellular protons regulate human ENaC by modulating Na+ self-inhibition. J. Biol. Chem. 284, 792–798 10.1074/jbc.M806954200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collier D. M., and Snyder P. M. (2009) Extracellular chloride regulates the epithelial sodium channel. J. Biol. Chem. 284, 29320–29325 10.1074/jbc.M109.046771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winarski K. L., Sheng N., Chen J., Kleyman T. R., and Sheng S. (2010) Extracellular allosteric regulatory subdomain within the γ subunit of the epithelial Na+ channel. J. Biol. Chem. 285, 26088–26096 10.1074/jbc.M110.149963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J., Myerburg M. M., Passero C. J., Winarski K. L., and Sheng S. (2011) External Cu2+ inhibits human epithelial Na+ channels by binding at a subunit interface of extracellular domains. J. Biol. Chem. 286, 27436–27446 10.1074/jbc.M111.232058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J., Winarski K. L., Myerburg M. M., Pitt B. R., and Sheng S. (2012) Probing the structural basis of Zn2+ regulation of the epithelial Na+ channel. J. Biol. Chem. 287, 35589–35598 10.1074/jbc.M112.394734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi S., Ghosh D. D., Okumura S., Carattino M. D., Kashlan O. B., Sheng S., and Kleyman T. R. (2011) Base of the thumb domain modulates epithelial sodium channel gating. J. Biol. Chem. 286, 14753–14761 10.1074/jbc.M110.191734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi S., Carattino M. D., and Kleyman T. R. (2012) Role of the wrist domain in the response of the epithelial sodium channel to external stimuli. J. Biol. Chem. 287, 44027–44035 10.1074/jbc.M112.421743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi S., Blobner B. M., Kashlan O. B., and Kleyman T. R. (2012) Extracellular finger domain modulates the response of the epithelial sodium channel to shear stress. J. Biol. Chem. 287, 15439–15444 10.1074/jbc.M112.346551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kashlan O. B., Blobner B. M., Zuzek Z., Tolino M., and Kleyman T. R. (2015) Na+ inhibits the epithelial Na+ channel by binding to a site in an extracellular acidic cleft. J. Biol. Chem. 290, 568–576 10.1074/jbc.M114.606152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horisberger J. D., and Chraïbi A. (2004) Epithelial sodium channel: a ligand-gated channel? Nephron. Physiol. 96, p37–41 10.1159/000076406 [DOI] [PubMed] [Google Scholar]
- 20. Fuchs W., Larsen E. H., and Lindemann B. (1977) Current-voltage curve of sodium channels and concentration dependence of sodium permeability in frog skin. J. Physiol. 267, 137–166 10.1113/jphysiol.1977.sp011805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J., Kleyman T. R., and Sheng S. (2014) Deletion of alpha-subunit exon 11 of the epithelial Na+ channel reveals a regulatory module. Am. J. Physiol. Renal Physiol. 306, F561–F567 10.1152/ajprenal.00587.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J., Ray E. C., Yates M. E., Buck T. M., Brodsky J. L., Kinlough C. L., Winarski K. L., Hughey R. P., Kleyman T. R., and Sheng S. (2015) Functional roles of clusters of hydrophobic and polar residues in the epithelial Na+ channel Knuckle domain. J. Biol. Chem. 290, 25140–25150 10.1074/jbc.M115.665398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carattino M. D., Hill W. G., and Kleyman T. R. (2003) Arachidonic acid regulates surface expression of epithelial sodium channels. J. Biol. Chem. 278, 36202–36213 10.1074/jbc.M300312200 [DOI] [PubMed] [Google Scholar]
- 24. Maarouf A. B., Sheng N., Chen J., Winarski K. L., Okumura S., Carattino M. D., Boyd C. R., Kleyman T. R., and Sheng S. (2009) Novel determinants of epithelial sodium channel gating within extracellular thumb domains. J. Biol. Chem. 284, 7756–7765 10.1074/jbc.M807060200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fyfe G. K., and Canessa C. M. (1998) Subunit composition determines the single channel kinetics of the epithelial sodium channel. J. Gen. Physiol. 112, 423–432 10.1085/jgp.112.4.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kellenberger S., Gautschi I., and Schild L. (2003) Mutations in the epithelial Na+ channel ENaC outer pore disrupt amiloride block by increasing its dissociation rate. Mol. Pharmacol. 64, 848–856 10.1124/mol.64.4.848 [DOI] [PubMed] [Google Scholar]
- 27. Canessa C. M., Schild L., Buell G., Thorens B., Gautschi I., Horisberger J. D., and Rossier B. C. (1994) Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature 367, 463–467 10.1038/367463a0 [DOI] [PubMed] [Google Scholar]
- 28. Harris M., Garcia-Caballero A., Stutts M. J., Firsov D., and Rossier B. C. (2008) Preferential assembly of epithelial sodium channel (ENaC) subunits in Xenopus oocytes: role of furin-mediated endogenous proteolysis. J. Biol. Chem. 283, 7455–7463 10.1074/jbc.M707399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Snyder P. M., Bucher D. B., and Olson D. R. (2000) Gating induces a conformational change in the outer vestibule of ENaC. J. Gen. Physiol. 116, 781–790 10.1085/jgp.116.6.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kellenberger S., Gautschi I., and Schild L. (2002) An external site controls closing of the epithelial Na+ channel ENaC. J. Physiol. 543, 413–424 10.1113/jphysiol.2002.022020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheng S., Li J., McNulty K. A., Kieber-Emmons T., and Kleyman T. R. (2001) Epithelial sodium channel pore region: structure and role in gating. J. Biol. Chem. 276, 1326–1334 10.1074/jbc.M008117200 [DOI] [PubMed] [Google Scholar]
- 32. Snyder P. M., Olson D. R., and Bucher D. B. (1999) A pore segment in DEG/ENaC Na+ channels. J. Biol. Chem. 274, 28484–28490 10.1074/jbc.274.40.28484 [DOI] [PubMed] [Google Scholar]
- 33. Valentijn J. A., Fyfe G. K., and Canessa C. M. (1998) Biosynthesis and processing of epithelial sodium channels in Xenopus oocytes. J. Biol. Chem. 273, 30344–30351 10.1074/jbc.273.46.30344 [DOI] [PubMed] [Google Scholar]
- 34. Buck T. M., Kolb A. R., Boyd C. R., Kleyman T. R., and Brodsky J. L. (2010) The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell 21, 1047–1058 10.1091/mbc.e09-11-0944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buck T. M., Plavchak L., Roy A., Donnelly B. F., Kashlan O. B., Kleyman T. R., Subramanya A. R., and Brodsky J. L. (2013) The Lhs1/GRP170 chaperones facilitate the endoplasmic reticulum-associated degradation of the epithelial sodium channel. J. Biol. Chem. 288, 18366–18380 10.1074/jbc.M113.469882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buck T. M., Jordan R., Lyons-Weiler J., Adelman J. L., Needham P. G., Kleyman T. R., and Brodsky J. L. (2015) The expression of three topologically distinct membrane proteins elicits unique stress response pathways in the yeast Saccharomyces cerevisiae. Physiol. Genomics 47, 198–214 10.1152/physiolgenomics.00101.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buck T. M., Jordahl A. S., Yates M. E., Preston G. M., Cook E., Kleyman T. R., and Brodsky J. L. (2017) Interactions between intersubunit transmembrane domains regulate the chaperone-dependent degradation of an oligomeric membrane protein. Biochem. J. 474, 357–376 10.1042/BCJ20160760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., and Kleyman T. R. (2003) Maturation of the epithelial Na+ channel involves proteolytic processing of the α- and γ-subunits. J. Biol. Chem. 278, 37073–37082 10.1074/jbc.M307003200 [DOI] [PubMed] [Google Scholar]
- 39. Hughey R. P., Bruns J. B., Kinlough C. L., and Kleyman T. R. (2004) Distinct pools of epithelial sodium channels are expressed at the plasma membrane. J. Biol. Chem. 279, 48491–48494 10.1074/jbc.C400460200 [DOI] [PubMed] [Google Scholar]
- 40. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., and Kleyman T. R. (2004) Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 279, 18111–18114 10.1074/jbc.C400080200 [DOI] [PubMed] [Google Scholar]
- 41. Kashlan O. B., Kinlough C. L., Myerburg M. M., Shi S., Chen J., Blobner B. M., Buck T. M., Brodsky J. L., Hughey R. P., and Kleyman T. R. (2018) N-Linked glycans are required on epithelial Na+ channel subunits for maturation and surface expression. Am. J. Physiol. Renal Physiol. 314, F483–F492 10.1152/ajprenal.00195.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheng S., Maarouf A. B., Bruns J. B., Hughey R. P., and Kleyman T. R. (2007) Functional role of extracellular loop cysteine residues of the epithelial Na+ channel in Na+ self-inhibition. J. Biol. Chem. 282, 20180–20190 10.1074/jbc.M611761200 [DOI] [PubMed] [Google Scholar]
- 43. Salih M., Gautschi I., van Bemmelen M. X., Di Benedetto M., Brooks A. S., Lugtenberg D., Schild L., and Hoorn E. J. (2017) A missense mutation in the extracellular domain of αENaC causes Liddle syndrome. J. Am. Soc. Nephrol. 28, 3291–3299 10.1681/ASN.2016111163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kashlan O. B., Boyd C. R., Argyropoulos C., Okumura S., Hughey R. P., Grabe M., and Kleyman T. R. (2010) Allosteric inhibition of the epithelial Na+ channel through peptide binding at peripheral finger and thumb domains. J. Biol. Chem. 285, 35216–35223 10.1074/jbc.M110.167064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kleyman T. R., Carattino M. D., and Hughey R. P. (2009) ENaC at the cutting edge: regulation of epithelial sodium channels by proteases. J. Biol. Chem. 284, 20447–20451 10.1074/jbc.R800083200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang L., Xu S., Guo X., Uchida S., Weinstein A. M., Wang T., and Palmer L. G. (2018) Regulation of renal Na transporters in response to dietary K. Am. J. Physiol. Renal Physiol. 10.1152/ajprenal.00117.2018 10.1152/ajprenal.00117.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frindt G., Yang L., Bamberg K., and Palmer L. G. (2018) Na restriction activates epithelial Na channels in rat kidney through two mechanisms and decreases distal Na+ delivery. J. Physiol. 596, 3585–3602 10.1113/JP275988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sheng S., Carattino M. D., Bruns J. B., Hughey R. P., and Kleyman T. R. (2006) Furin cleavage activates the epithelial Na+ channel by relieving Na+ self-inhibition. Am. J. Physiol. Renal Physiol. 290, F1488–F1496 10.1152/ajprenal.00439.2005 [DOI] [PubMed] [Google Scholar]
- 49. Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., and Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the γ-subunit. J. Biol. Chem. 282, 6153–6160 10.1074/jbc.M610636200 [DOI] [PubMed] [Google Scholar]
- 50. Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., and Kleyman T. R. (2006) The epithelial Na+ channel is inhibited by a peptide derived from proteolytic processing of its α subunit. J. Biol. Chem. 281, 18901–18907 10.1074/jbc.M604109200 [DOI] [PubMed] [Google Scholar]
- 51. Kashlan O. B., Blobner B. M., Zuzek Z., Carattino M. D., and Kleyman T. R. (2012) Inhibitory tract traps the epithelial Na+ channel in a low activity conformation. J. Biol. Chem. 287, 20720–20726 10.1074/jbc.M112.358218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collier D. M., Tomkovicz V. R., Peterson Z. J., Benson C. J., and Snyder P. M. (2014) Intersubunit conformational changes mediate epithelial sodium channel gating. J. Gen. Physiol. 144, 337–348 10.1085/jgp.201411208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krauson A. J., and Carattino M. D. (2016) The thumb domain mediates acid-sensing ion channel desensitization. J. Biol. Chem. 291, 11407–11419 10.1074/jbc.M115.702316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vullo S., Bonifacio G., Roy S., Johner N., Bernèche S., and Kellenberger S. (2017) Conformational dynamics and role of the acidic pocket in ASIC pH-dependent gating. Proc. Natl. Acad. Sci. U.S.A. 114, 3768–3773 10.1073/pnas.1620560114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoder N., Yoshioka C., and Gouaux E. (2018) Gating mechanisms of acid-sensing ion channels. Nature 555, 397–401 10.1038/nature25782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Firsov D., Schild L., Gautschi I., Mérillat A. M., Schneeberger E., and Rossier B. C. (1996) Cell surface expression of the epithelial Na channel and a mutant causing Liddle syndrome: a quantitative approach. Proc. Natl. Acad. Sci. U.S.A. 93, 15370–15375 10.1073/pnas.93.26.15370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Adams A., and Gottschling D. E. (1997) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 58. Zhang Y., Nijbroek G., Sullivan M. L., McCracken A. A., Watkins S. C., Michaelis S., and Brodsky J. L. (2001) Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12, 1303–1314 10.1091/mbc.12.5.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schrödinger L. (2010) The PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC., New York [Google Scholar]