Abstract
Protein arginine methyltransferase 5 (PRMT5) is a member of the arginine methyltransferase protein family that critically mediates the symmetric dimethylation of Arg-3 at histone H4 (H4R3me2s) and is involved in many key cellular processes, including hematopoiesis. However, the post-translational modifications (PTMs) of PRMT5 that may affect its biological functions remain less well-understood. In this study, using MS analyses, we found that PRMT5 itself is methylated in human erythroleukemia Lys-562 cells. Biochemical assays revealed that coactivator-associated arginine methyltransferase 1 (CARM1) interacts directly with and methylates PRMT5 at Arg-505 both in vivo and in vitro. Substitutions at Arg-505 significantly reduced PRMT5's methyltransferase activity, decreased H4R3me2s enrichment at the γ-globin gene promoter, and increased the expression of the γ-globin gene in Lys-562 cells. Moreover, CARM1 knockdown consistently reduced PRMT5 activity and activated γ-globin gene expression. Importantly, we show that CARM1-mediated methylation of PRMT5 is essential for the intracellular homodimerization of PRMT5 to its active form. These results thus reveal a critical PTM of PRMT5 that represses human γ-globin gene expression. We conclude that CARM1-mediated asymmetric methylation of PRMT5 is critical for its dimerization and methyltransferase activity leading to the repression of γ-globin expression. Given PRMT5's crucial role in diverse cellular processes, these findings may inform strategies for manipulating its methyltransferase activity for managing hemoglobinopathy or cancer.
Keywords: protein arginine N-methyltransferase 5 (PRMT5), protein methylation, hemoglobin, histone methylation, gene expression, CARM1
Introduction
Post-translational modifications (PTMs)3 of histone proteins play important roles in defining chromatin structure and controlling gene activities such as globin gene expression (1–6). Among the various modifications, arginine methylation is particularly critical for several cellular processes, including signal transduction, DNA repair, transcription, protein subcellular localization, and RNA processing (7, 8). In eukaryotes, arginine methylation is catalyzed by a family of enzymes called protein arginine methyltransferases (PRMTs). In humans, this family currently consists of nine members subdivided into three categories based on differences in primary sequences and substrate specificity: Type I PRMTs, which catalyze monomethyl arginine formation and the asymmetric dimethylation (me2a) of arginine residues (PRMT1, 2, 3, 4, 6, and 8); type II PRMTs, which catalyze monomethyl arginine formation and the symmetric dimethylation (me2s) of arginine residues (PRMT5 and 9); and type III PRMTs, which catalyze only the monomethylation of arginine residues (PRMT7) (9, 10). Although some of the PRMTs appear histone specific, many have now been found to modify both histone and nonhistone proteins (10).
PRMT4, more commonly known as CARM1 (coactivator-associated arginine methyltransferase 1), was originally identified through its binding to GRIP1, the p160 steroid receptor coactivator in a yeast two-hybrid screen (11). CARM1 is a type I arginine methyltransferase that asymmetrically dimethylates proteins at arginine residues involved in the regulation of transcription, pre-mRNA splicing, cell cycle progression, and the DNA damage response (9). CARM1 methylates histone H3 at Arg-17, Arg-26, and Arg-42, which is normally linked to gene activation (12, 13). CARM1 can also methylate nonhistone substrates acting as gene coregulators, such as the transcription factors p/CIP, pRB, SOX2, and the NF-κB subunit p65 (14–17); the transcriptional coactivators CBP/p300 and SRC3 (18, 19); the chromatin remodeling factor BAF155 (20), PAX7 (21); the mediator Med12 (22); the RNA-binding proteins PABP1 and HuD (23, 24); the splicing factors CA150, SAP49, U1C, and SmB; and RNA pol II (25, 26).
PRMT5 is a type II arginine methyltransferase that can symmetrically methylate histone H2A/H4 N-terminal arginine residues to modulate chromatin structure and to epigenetically regulate the transcription of target genes in cells (8). In particular, PRMT5 is essential for embryonic stem cell function, adult hematopoietic cell maintenance, and globin gene switching (27–29). Similar to CARM1, PRMT5 can also methylate nonhistone protein substrates to regulate ribosome biogenesis; the assembly of the Golgi apparatus; cellular differentiation; and cell growth, migration, and development (8, 9). Of note, PRMT5 dysfunction or alteration (including amplification, aberrant expression, deletion, and mutation) has been associated with various cancers and other diseases, such as gastric, colorectal, lung, and prostate cancer; lymphoma; leukemia; renal and cardiovascular disease; Huntington's disease; and Alzheimer's disease (9, 30–33). Thus, the screening of functional PRMT5 inhibitors as potential chemotherapeutics has been a highly pursued and promising strategy for preventing cancers and other diseases (34).
Although the crystal structure of PRMT5 was resolved 6 years ago (35), the PTMs affecting its functions, including is enzymatic activities, remain poorly understood. Thus far, there have only been reports that phosphorylation or ubiquitination of PRMT5 affects its activity (36, 37). Whether methylation of PRMT5 is involved in its functional modulation remains unknown. In the current study, we found that PRMT5 could be methylated by CARM1 at the highly conserved residue arginine 505 (Arg-505). CARM1 repressed human γ-globin gene expression indirectly through the methylation of PRMT5, which increased histone H4R3me2s enrichment at the γ-promoter in Lys-562 cells. We have demonstrated that methylation at Arg-505 of PRMT5 is essential for its dimerization and is required for its optimal methyltransferase activity in Lys-562 cells. Our study thus reveals a novel post-translational modification of PRMT5 by CARM1 that modulates human γ-globin gene expression.
Results
PRMT5 is dimethylated at Arg-505
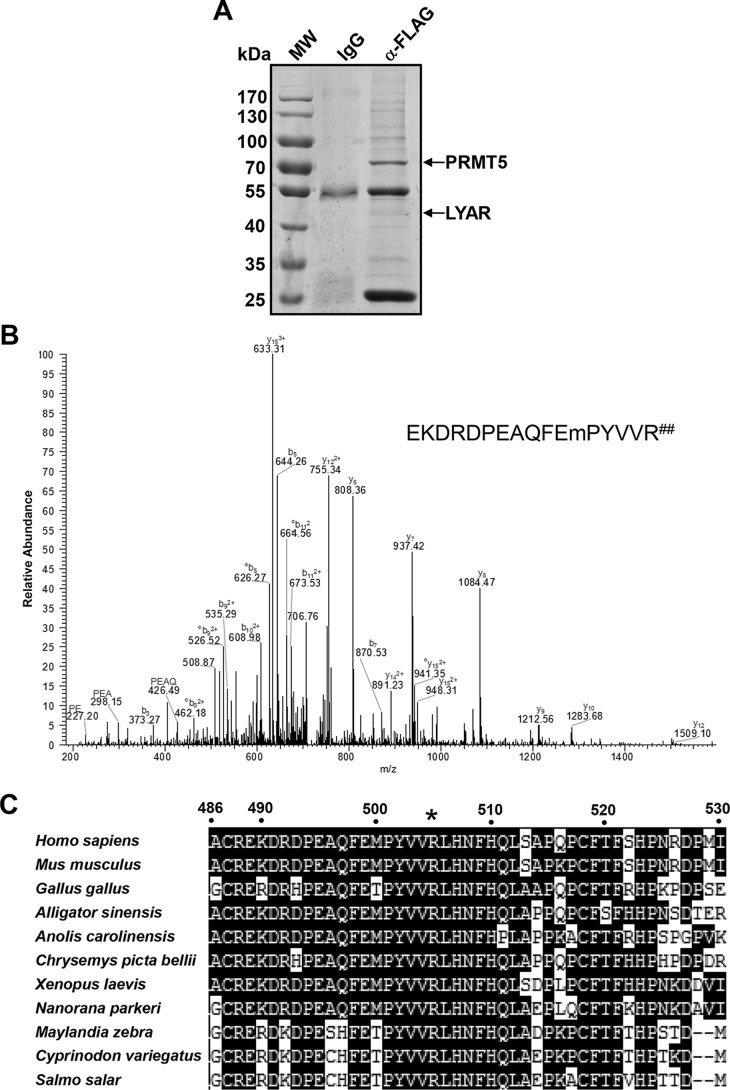
In previous studies, we have shown that the PRMT5-mediated symmetric dimethylation of H4R3 induces the direct binding of DNMT3A, resulting in the methylation of adjacent CpG dinucleotides and in γ-globin gene silencing in erythroid cells (6, 29). In the current study, we used an anti-FLAG antibody conjugated to Sepharose beads to precipitate PRMT5 and its associated proteins from cellular extracts of human Lys-562 cells stably overexpressing FLAG-tagged PRMT5; in immunoprecipitation experiments, we found that LYAR (human homologue of mouse Ly-1 antibody reactive clone) interacts with PRMT5, acting as a novel transcription factor that binds the γ-globin gene, and is essential for silencing γ-globin gene expression (Fig. 1A) (38–40). Other potential PRMT5-interacting proteins identified are shown in Table S1. Interestingly, after the protein band corresponding to PRMT5 was cut out and subjected to MS analysis, we found a polypeptide with the sequence EKDRDPEAQFEPYVVR (positions 489 to 505 of PRMT5) in which the last arginine was dimethylated (Fig. 1, A and B). This last arginine residue corresponds to Arg-505 in human PRMT5. Alignment of PRMT5 sequences from different mammal species showed that this Arg-505 site is evolutionarily well-conserved (Fig. 1C).
Figure 1.
PRMT5 is dimethylated at Arg-505. A, SimplyBlue SafeStain of an SDS-PAGE gel of FLAG antibody immunoprecipitates from Lys-562 cells stably overexpressing PRMT5-FLAG before analysis by MS. Protein bands corresponding to PRMT5 and LYAR (with peptide sequences: KKGQEADLEAGGEEVPEANGSAGKR; VHNESILDQVWNIFSEASNSEPVNKEQDQRPLHPVANPHAEISTK) are indicated. B, ESI MS/MS spectrum of the tryptic peptide EKDRDPEAQFEmPYVVR## (where m is oxidized methionine and R## represents dimethylated arginine). All y ions in the dimethylated peptide spectrum have a mass shift of +28 Da compared with the y ions from the nonmethylated peptide spectrum. C, amino acid sequence alignment of PRMT5 proteins among mammals. The Homo sapiens PRMT5 amino acid sequence from residues 486 to 530 is indicated on the top, and the conserved arginine (Arg-505 in Homo sapiens) is indicated by an asterisk.
Methylation of PRMT5 at Arg-505 is essential for its methyltransferase activity
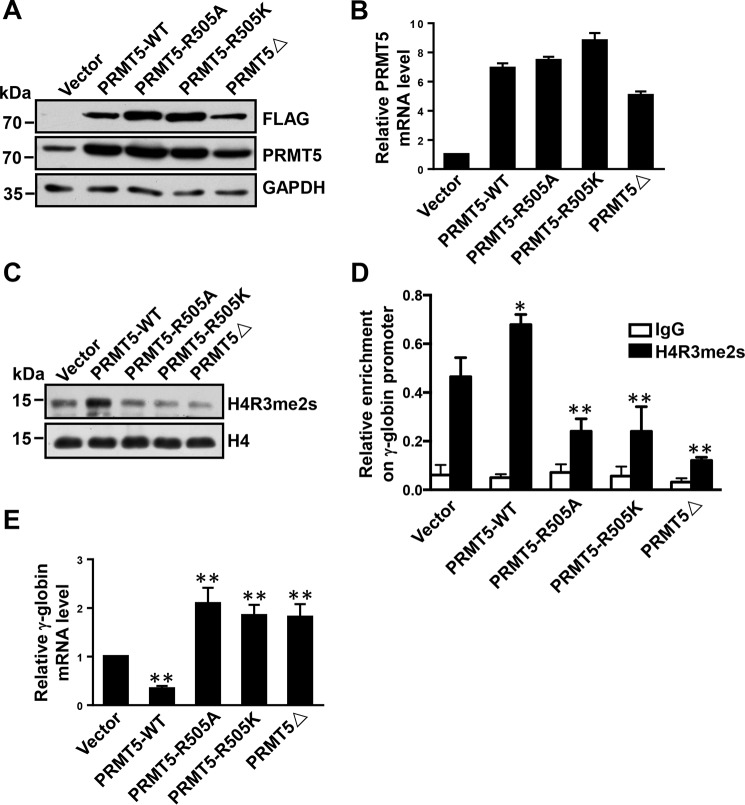
To examine the role of Arg-505 in PRMT5, we constructed two PRMT5 mutants, PRMT5-R505A and PRMT5-R505K, by mutagenesis and generated stable FLAG-tagged Lys-562 cell lines overexpressing these mutants as well as cell lines expressing WT PRMT5 and PRMT5Δ (the methyltransferase-dead mutant form of PRMT5 in which five amino acids in the S-adenosyl-l-methionine–binding motif have been deleted) using lentiviral vectors (6, 41). PRMT5 protein levels were assessed by Western blotting, and PRMT5 gene expression in the cells was quantified from total RNA by Q-RT-PCR. PRMT5 expression levels were significantly increased in these cell lines compared with the vector controls (Fig. 2, A and B). Subsequently, we performed Western blot analyses to measure the pancellular levels of the histone mark H4R3me2s, which is mediated by PRMT5. We found that the levels of H4R3me2s were significantly reduced in cells overexpressing either PRMT5-R505A or PRMT5-R505K compared with cells expressing WT PRMT5 and were similar to the levels of H4R3me3s in PRMT5Δ-expressing cells (Fig. 2C). Because MEP50 is a key component in the PRMT5-containing complex and is essential for the methyltransferase activity of PRMT5 (35), we tested whether the PRMT5 mutants bound MEP50 differently than the WT PRMT5. We found that PRMT5 mutants could immunoprecipitate MEP50 as well as the WT PRMT5 (Fig. S1), excluding the possibility that the mutants exhibited a decreased MEP50-binding capability in Lys-562 cells. Previously, we have found that the repressive H4R3me2s mark triggered by PRMT5 is enriched at the γ-globin gene promoter to repress γ-globin expression in Lys-562 cells (29). Thus, we analyzed the enrichment of the histone mark H4R3me2s at the γ-promoter in these cells by ChIP analysis. We found that the levels of H4R3me2s enrichment at the γ-promoter in cells overexpressing mutant PRMT5 were significantly lower than in the cells overexpressing WT PRMT5 (Fig. 2D). Consistently, γ-globin gene expression was significantly increased in cells overexpressing the two mutant forms of PRMT5 compared with the WT cells (Fig. 2E). Therefore, these results suggest that the methylation of PRMT5 at Arg-505 is essential for its methyltransferase activity in cells and is important for the repression of γ-globin gene expression in Lys-562 cells.
Figure 2.
Methylation of PRMT5 at Arg-505 is essential for its methyltransferase activity. A, Western blot analysis of extracts from Lys-562 cells containing vector or overexpressing PRMT5-WT, PRMT5-R505A, PRMT5-R505K, or PRMT5Δ using FLAG and PRMT5 antibodies. GAPDH was used as a loading control. Blots are representative of three independent experiments. B, quantitative real-time PCR analysis of PRMT5 mRNA normalized to β-actin in Lys-562 cells containing vector or overexpressing PRMT5-WT, PRMT5-R505A, PRMT5-R505K, or PRMT5Δ. The results are shown as the mean ± S.D. from three independent experiments. C, Western blot analysis of extracted histones from Lys-562 cells containing vector or overexpressing PRMT5-WT, PRMT5-R505A, PRMT5-R505K, or PRMT5Δ using H4R3me2s antibody. Histone H4 was used as a loading control. Blots are representative of three independent experiments. D, ChIP analysis of H4R3me2s enrichment at the γ-promoter in Lys-562 cells containing vector or overexpressing PRMT5-WT, PRMT5-R505A, PRMT5-R505K, or PRMT5Δ. The results are shown as the mean ± S.D. from three independent experiments. Two-tailed Student's t-tests were used to compare means. *, p < 0.05, **, p < 0.01 compared with the vector control. E, quantitative real-time PCR analysis of γ-globin mRNA normalized to β-actin in Lys-562 cells containing vector or overexpressing PRMT5-WT, PRMT5-R505A, PRMT5-R505K, or PRMT5Δ. The results are shown as the mean ± S.D. from three independent experiments. Two-tailed Student's t-tests were used to compare means. **, p < 0.01 compared with the vector control.
CARM1 methylates PRMT5 in vitro
Although we found that PRMT5 was dimethylated at Arg-505 in MS analysis, whether the dimethylation was symmetric or asymmetric could not be determined. To identify the methyltransferase responsible for the methylation of PRMT5, we purified three major arginine methyltransferases—the recombinant fusion proteins GST-PRMT1 and GST-CARM1 from Escherichia coli, and the FLAG-tagged PRMT5 (PRMT5-f) from Lys-562 cells (Fig. 3, B–D, left)—as well as their potential substrate, the recombinant PRMT5 protein (rPRMT5) from E. coli (Fig. 3A). Of note, the recombinant PRMT5 protein from E. coli has never been observed to exhibit methyltransferase activity in vitro (35). We used free histones as substrates to confirm the enzymatic activity of GST-PRMT1, GST-CARM1, and PRFT5-f in Western blot analysis using specific antibodies against histone H4R3me2a, H3R17me2a, and H4R3me2s, respectively (Fig. 3, B–D, left). Subsequently, the purified recombinant PRMT5 protein from E. coli was incubated with each methyltransferase independently in the presence of 3H-SAM, the methyl donor. After SDS-PAGE and autoradiography, we found that there was a 3H-labeled protein band corresponding to the size of PRMT5 only when the recombinant PRMT5 was incubated with GST-CARM1 (Fig. 3C, right). No signals were detectable after incubation with GST-PRMT1 or PRMT5-f (Fig. 3, B and D, right). These results demonstrate that PRMT5 may be methylated by CARM1 in vitro and that there is no automethylation of PRMT5.
Figure 3.
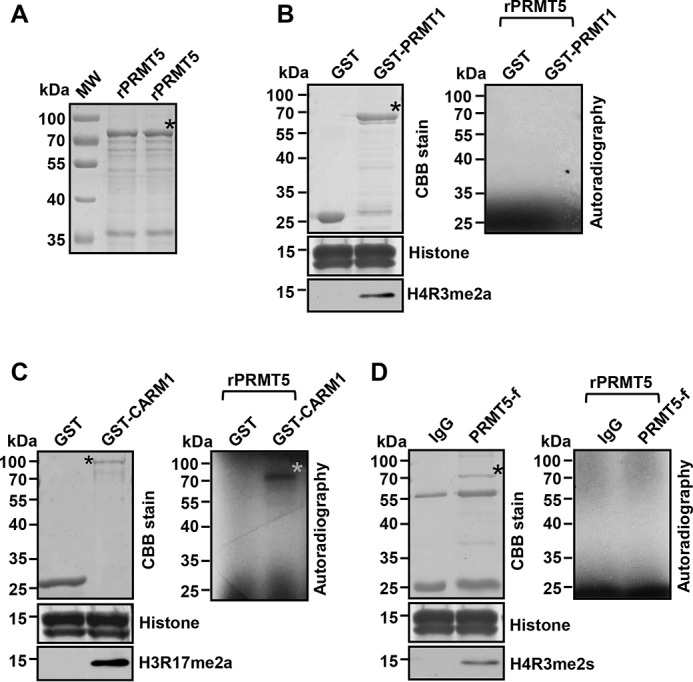
CARM1 methylates PRMT5. A, SDS-PAGE analysis of purified recombinant PRMT5 (rPRMT5) from E. coli as the substrate for following in vitro methylation assays. B, SDS-PAGE analysis of purified recombinant GST-PRMT1 and GST control from E. coli (top left). SDS-PAGE analysis of free histones stained by Coomassie Brilliant Blue (CBB) (middle left). Western blot analysis of free histones from an in vitro methylation assay with H4R3me2a antibody (bottom left). Autoradiographic image from an in vitro methylation assay with GST-PRMT1 or GST control (right). C, SDS-PAGE analysis of purified recombinant GST-CARM1 and GST control from E. coli (top left). SDS-PAGE analysis of free histones stained by Coomassie Brilliant Blue (CBB) (middle left). Western blot analysis of free histones from an in vitro methylation assay with H3R17me2a antibody (bottom left). Autoradiographic image from an in vitro methylation assay with GST-CARM1 or GST control (right). A gray asterisk denotes the positive 3H-labeled PRMT5 band. D, SDS-PAGE analysis of IgG control (top left) and of FLAG-PRMT5 immunoprecipitated from Lys-562 cells overexpressing FLAG-tagged PRMT5. SDS-PAGE analysis of free histones stained by Coomassie Brilliant Blue (CBB) (middle left). Western blot analysis of free histones from an in vitro methylation assay with H4R3me2s antibody (bottom left). Autoradiographic image from an in vitro methylation assay with FLAG-PRMT5 or IgG control (right). Black asterisks denote the fusion proteins in assays.
CARM1 directly interacts with PRMT5
To test whether CARM1 and PRMT5 interact in vitro, a GST pulldown experiment was performed. As shown in Fig. 4A, GST-CARM1 could readily pull down the purified recombinant PRMT5, whereas GST did not interact with PRMT5. Further PRMT5 fragmentation pulldown assays demonstrated that only amino acids 454–637 of PRMT5 interacted with CARM1 (Fig. 4B). Of note, Arg-505 is located in this region (Fig. 1C), which is consistent with its methylation by CARM1. Indeed, we found that PRMT5 coimmunoprecipitated with CARM1 protein from a Lys-562 cellular extract (Fig. 4C). Thus, these results indicate that PRMT5 and CARM1 interact in cells and provide support for CARM1-mediated PRMT5 methylation.
Figure 4.
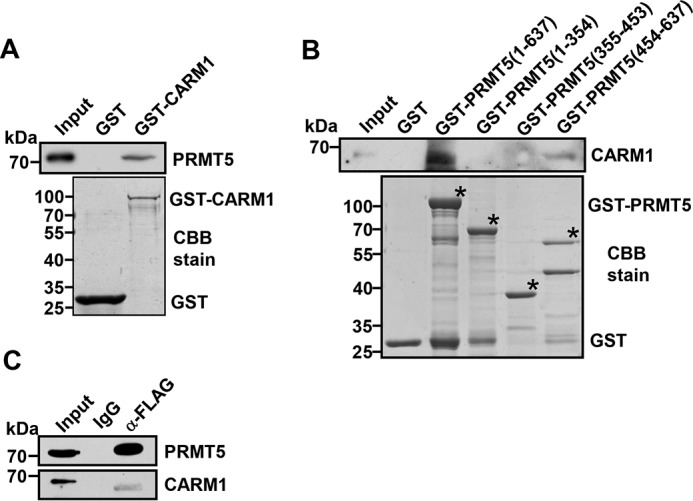
CARM1 directly interacts with PRMT5. A, Western blot analysis of PRMT5 binding to purified GST or GST-CARM1 using PRMT5 antibody (top). SDS-PAGE analysis of purified GST or GST-CARM1 from E. coli stained by Coomassie Brilliant Blue (CBB) (bottom). B, Western blot analysis of CARM1 binding to purified GST, GST-PRMT5(1–637), GST-PRMT5(1–354), GST-PRMT5(355–453) or GST-PRMT5(454–637) using CARM1 antibody (top). SDS-PAGE analysis of purified GST, GST-PRMT5(1–637), GST-PRMT5(1–354), GST-PRMT5(355–453), or GST-PRMT5(454–637) from E. coli stained by Coomassie Brilliant Blue (CBB) (bottom). Black asterisks denote the GST fusion proteins in assays. C, coimmunoprecipitation of PRMT5 and CARM1 from FLAG-PRMT5 overexpressing Lys-562 cell lysate. IgG was used as the negative control.
CARM1 represses globin gene expression through methylation of PRMT5
To examine whether Arg-505 in PRMT5 is asymmetrically dimethylated, we used the peptide MPYVVR (me2a) LHNFH, which corresponds to amino acids 500–510 of the human PRMT5 protein, where arginine is asymmetrically demethylated, as an antigen to generate a rabbit polyclonal antibody. Dot blot and peptide competition assays confirmed that this anti-PRMT5-R505me2a antibody specifically recognized the peptide in which Arg-505 was asymmetrically dimethylated, not the nonmethylated peptide (Fig. S2, A and B).
To determine the role of CARM1 in the methylation of PRMT5, we generated two stable CARM1 knockdown (CARM1-KD) Lys-562 cell lines using lentiviral vectors containing specific shRNAs. CARM1 protein levels were assessed by Western blotting (Fig. 5A), and gene expression in these cells was quantified from total RNA by Q-RT-PCR (Fig. 5B). CARM1 expression levels were reduced in these two cell lines to ∼30% of the expression in the scrambled control. Indeed, the methylation of PRMT5 was greatly reduced in CARM1-KD Lys-562 cell lines compared with scrambled cells as revealed by Western blot analysis using the PRMT5-R505me2a antibody (Fig. 5A). Interestingly, this reduction was accompanied by significantly decreased levels of the histone mark H4R3me2s, which is triggered by PRMT5, although the total levels of cellular PRMT5 were not changed (Fig. 5A). We consistently observed significantly elevated expression of γ-globin in CARM1-KD cells compared with Scr Lys-562 cells (Fig. 5C). ChIP experiments demonstrated that the enrichment of the histone mark H4R3me2s at the γ-promoter was significantly decreased in CARM1-KD cells compared with Scr Lys-562 cells, whereas the enrichment of H3R17me2a, which is normally triggered by CARM1, remained unchanged (Fig. 5D).
Figure 5.
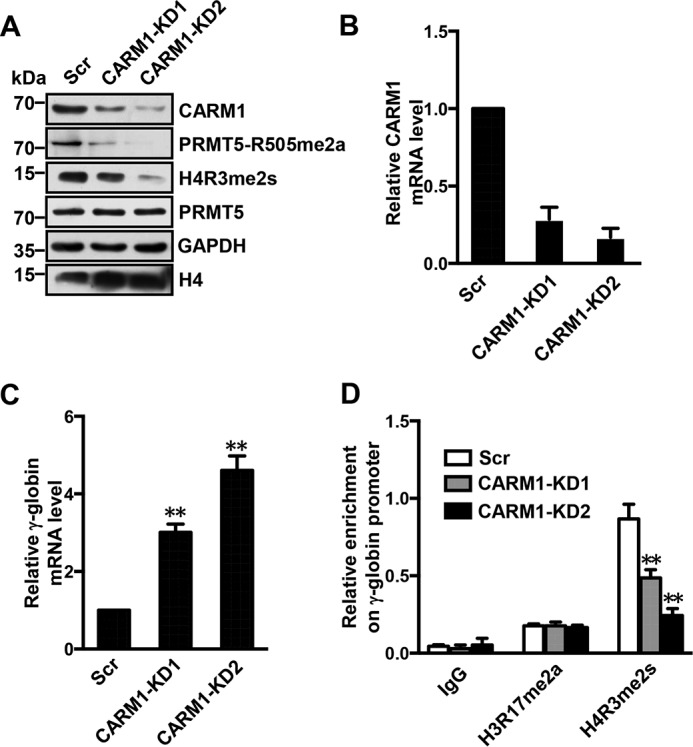
CARM1 represses γ-globin expression. A, Western blot analysis of indicated protein levels in cell lysates from the scramble control (Scr), CARM1-KD1, and CARM1-KD2 Lys-562 cells. GAPDH and histone H4 were used as loading controls. Blots are representative of three independent experiments. B and C, quantitative real-time PCR analysis of CARM1 (B) and γ-globin (C) mRNA level in Scr, CARM1-KD1, and CARM1-KD2 Lys-562 cells normalized to β-actin mRNA. The results are shown as the mean ± S.D. from three independent experiments. Two-tailed Student's t-tests were used to compare means. **, p < 0.01 compared with the Scr control. D, ChIP analysis of H3R17me2a and H4R3me2s enrichment at the γ-promoter in Scr, CARM1-KD1, and CARM1-KD2 Lys-562 cells. IgG was used as the negative control. The results are shown as the mean ± S.D. from three independent experiments. Two-tailed Student's t-tests were used to compare means. **, p < 0.01 compared with the Scr control.
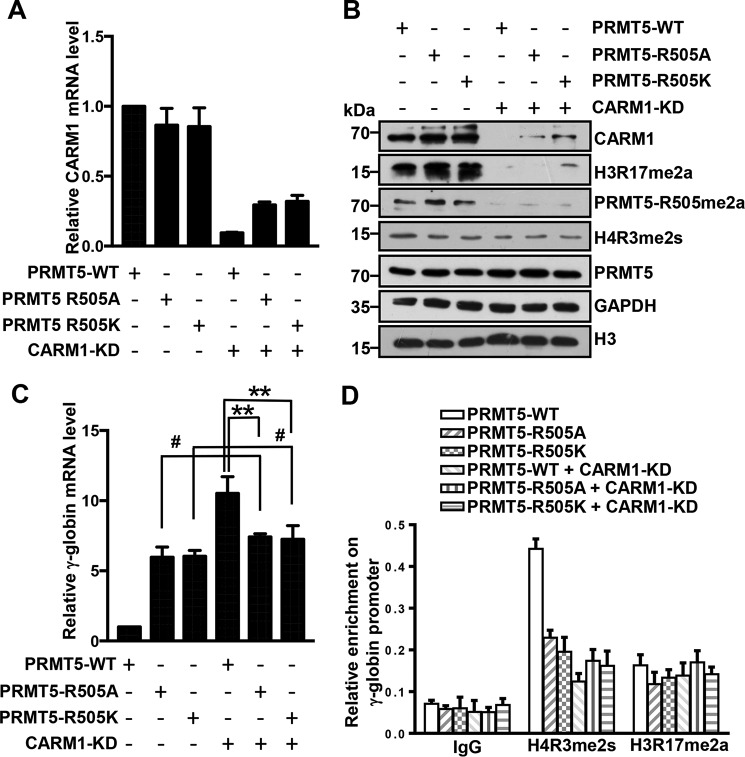
To further confirm that CARM1 influences γ-globin expression by affecting the methylation of histone H4R3 rather than through direct enzymatic activity on histone H3R17, we analyzed γ-globin expression in Lys-562 cells with WT or PRMT5 mutants (PRMT5-R505A or -R505K) with or without CARM1 knockdown. The expression levels of CARM1 in these cells were verified by Q-RT-PCR (from total RNA) and by Western blot analyses (Fig. 6, A and B). PRMT5 levels and the methylation changes mediated by PRMT5, histone H4R3me2s, and H3R17me2a were also confirmed by Western blotting with specific antibodies (Fig. 6B). As expected, γ-globin expression was significantly activated in cells overexpressing PRMT5 mutants (PRMT5-R505A or -R505K) compared with those overexpressing WT PRMT5, as determined by Q-RT-PCR (Fig. 6C), which suggests that PRMT5 mutants (R505A or R505K) may confer dominant negative effects on PRMT5 activity. Interestingly, no further increases in γ-globin expression were observed in CARM1-KD Lys-562 cells overexpressing PRMT5 mutants compared with native cells overexpressing the mutants (Fig. 6C). In contrast, there was a significant increase in γ-globin expression in CARM1-KD cells overexpressing WT PRMT5 compared with native cells or CARM1-KD cells overexpressing PRMT5 mutants (Fig. 6C). The ChIP assay also indicated that the enrichment of the histone mark H4R3me2s at the γ-promoter was negatively correlated with the expression of the γ-globin gene in these cells (Fig. 6D). However, the enrichment of H3R17me2a at the γ-promoter in those cells was not correlated with γ-globin gene expression (Fig. 6D). These results further demonstrate that CARM1 methylates PRMT5 to repress γ-globin gene expression in Lys-562 cells.
Figure 6.
CARM1 methylates PRMT5 Arg-505 to repress γ-globin expression. A and C, quantitative real-time PCR analysis of CARM1 (A) and γ-globin (C) mRNA levels normalized to β-actin in Lys-562 cells overexpressing PRMT5-WT, PRMT5-R505A, or PRMT5-R505K with or without CARM1 knockdown. The results are shown as the mean ± S.D. from three independent experiments. Two-tailed Student's t-tests were used to compare means. **, p < 0.01, #, p > 0.05 compared with indicated controls. B, Western blot analysis of indicated protein levels in cell lysates from Lys-562 cells overexpressing PRMT5-WT, PRMT5-R505A, or PRMT5-R505K with or without CARM1 knockdown. GAPDH and histone H3 were used as negative controls. D, ChIP analysis of H4R3me2s and H3R17me2a enrichment at the γ-promoter in Lys-562 cells overexpressing PRMT5-WT, PRMT5-R505A, or PRMT5-R505K with or without CARM1 knockdown. The results are shown as the mean ± S.D. from three independent experiments.
Arg-505 methylation is essential for PRMT5 homodimerization
Post-translational modifications of proteins often affect protein oligomerization. The crystal structure of the human PRMT5-MEP50 complex has revealed that the PRMT5 subunits form a core tetramer, and the MEP50 subunits are arranged peripherally in complex with PRMT5, which is critical for methyltransferase activity (35). To investigate whether Arg-505 methylation can affect the oligomerization of PRMT5, we treated Lys-562 cells expressing either WT PRMT5 or PRMT5 mutants (R505A or R505K) with the “zero length” cross-linking agent EDC/NHS (42, 43). As shown in Fig. 7B, a Western blot assay with a PRMT5 antibody revealed that WT PRMT5 was able to form a dimer or a tetramer in the presence of the cross-linker, whereas PRMT5 mutants (R505A or R505K) were not able to oligomerize under the same conditions. No dimers or tetramers were detected in the PBS treatment (negative control; Fig. 7A). These results suggest that PRMT5 mutants (R505A or R505K) confer dominate negative effects on PRMT5 homodimerization and that Arg-505 methylation may be essential for PRMT5 tetramer complex and its optimal methyltransferase activity in Lys-562 cells.
Figure 7.
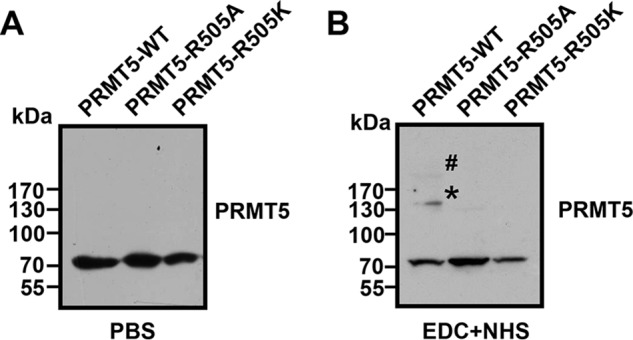
Arg-505 methylation is essential for PRMT5 oligomerization. A and B, Western blot analysis using PRMT5 antibody to detect PRMT5 oligomerization in Lys-562 cells overexpressing PRMT5-WT, PRMT5-R505A, or PRMT5-R505K treated with PBS control (A) or the cross-linker EDC+NHS (B). The asterisk and the number sign indicate the PRMT5 dimer and PRMT5 tetramer, respectively.
Discussion
Protein post-translational modifications, including arginine methylation, play important roles in determining protein functions (1). Although PRMT5 is an arginine methyltransferase whose function is, on most occasions, to methylate other proteins, it can also be methylated by other methyltransferases. In this study, we demonstrated that PRMT5 was methylated at arginine 505 and that this modification affected the methyltransferase activity of PRMT5. CARM1 interacted with PRMT5 directly and resulted in the asymmetric dimethylation of PRMT5 Arg-505. The methylation of PRMT5 by CARM1 facilitated the homodimerization of PRMT5 and repressed γ-globin expression in Lys-562 cells. Interestingly, we found that PRMT5 is also methylated by CARM1 in two other cell lines, SGC7901 and BGC823 (Fig. S3, A and B), which suggests that PRMT5 methylation might be a general phenomenon.
PRMT5 is the most important type II protein arginine methyltransferase in eukaryotes. Although regulation of PRMT5 expression may occur at various stages from transcription to post-translation, the catalytic activity of PRMT5 can be modulated by protein binding and post-translational modifications (35–37). MEP50 is one of the important interacting partners of PRMT5 and is essential for PRMT5-mediated histone methylation. Previously, two tyrosine residues (Tyr-304 and Tyr-307) of PRMT5 were found to be phosphorylated by the JAK2 kinase mutant V617F, which disrupted the interaction between PRMT5 and MEP50 and thus impaired the methyltransferase activity of PRMT5 (36). However, our study shows that the methylation of PRMT5 Arg-505 had no effect on the association of PRMT5 with MEP50, indicating that Arg-505 may not be located at the interface between PRMT5 and MEP50 or that Arg-505 is not a key residue affecting the PRMT5-MEP50 interaction. Instead, we found that the Arg-505 methylation of PRMT5 was essential for the homodimerization of PRMT5. According to the resolved crystal structure of the PRMT5-MEP50 complex, the complex is a hetero-octamer in which the PRMT5 subunits form a core tetramer and four MEP50 subunits are arranged peripherally (35). In the current study, the impairment of PRMT5 methyltransferase activity because of mutations of PRMT5 at Arg-505 may be a consequence of the disruption of PRMT5 dimerization. Interestingly, there was a naturally occurring PRMT5 mutant, R505W (at position 505, the residue Arg was mutated to Trp), in a melanoma patient with a resistant tumor (44). It would be interesting to investigate whether the R505W mutation of PRMT5 is associated with reduced methyltransferase activity or altered target sequence preferences and whether the mutation contributes to drug resistance.
Unfortunately, we show no evidence that PRMT5 is auto–arginine methylated similar to PRMT6, PRMT7, PRMT8, and CARM1 (45–48), although the biological roles of such modifications are currently unclear. We found that CARM1 asymmetrically dimethylated Arg-505 of PRMT5 and indirectly regulated γ-globin expression in Lys-562 cells. In fact, CARM1 has been identified as a key regulator of fetal hematopoiesis and thymocyte development (49). In addition, alternative splicing of CARM1 is involved in regulating gene expression during terminal erythropoiesis (50). Thus, CARM1 is a crucial epigenetic modulator that contributes to proper differentiation of hematopoietic lineages. CARM1 is known as a transcriptional coactivator because it heavily methylates histone H3 to deposit H3R17me2a (12). We found that knockdown of CARM1 reduced global H3R17me2a levels but had no effect on the histone mark enrichment at the γ-globin gene promoter in Lys-562 cells. These results suggest that CARM1-mediated histone modifications in gene regulation may be context-dependent and gene-specific. Our study unveils a novel role of CARM1 in globin gene regulation as a transcriptional corepressor.
In conclusion, our study demonstrates that CARM1-mediated asymmetric methylation of PRMT5 is critical for methyltransferase activity and for the repression of γ-globin expression. Given that PRMT5 plays crucial roles in diverse cellular processes, these findings may provide an alternative strategy for manipulating the methyltransferase activity of PRMT5 in the context of hemoglobinopathy or cancer.
Experimental procedures
Cell cultures, viral infection, and plasmids
Lys-562 cells and HEK293T cells were maintained as described previously (6). Retrovirus or lentivirus production in HEK293T cells and the infection of Lys-562 cells were also performed as described previously (29). Transduced cells were selected for GFP expression by fluorescence-activated cell sorting (FACS).
PRMT5 or PRMT5Δ was inserted into the retroviral vector MSCV-HA-IRES-GFP as described in a previous study (6, 41). PRMT5 mutants were constructed using a site-directed mutagenesis kit (SBS Genetech, Shanghai, China). The shRNA target sequences for RNAi of CARM1 were inserted into the XhoI/HpaI sites in the pLL3.7 lentiviral vector according to the manufacturer's instructions (American Type Culture Collection). The oligonucleotides used were as follows: Human CARM1 shRNA KD1, 5′-AGAACATGATGCAGGACTA-3′; human CARM1 shRNA KD2, 5′-ATTTCTGTTCCTTCTACAA-3′.
Mass spectrometry analysis
FLAG immunoprecipitates from Lys-562 cells expressing PRMT5-FLAG were resolved as described previously (6). Protein bands of interest were excised and analyzed by electrospray ion trap (ESI-IT) tandem mass spectrometry (MS/MS) (LCQ Deca, Thermo Finnigan).
Western blot analysis and protein interaction studies
Cellular proteins were extracted by radioimmune precipitation assay buffer (RIPA) lysis buffer at a high salt concentration (420 mm NaCl), and Western blot analysis was performed as described previously (6). The following antibodies were used for Western blotting: FLAG (Sigma-Aldrich), PRMT5 (Sigma-Aldrich), CARM1 (Cell Signaling Technology), GAPDH (MBL International), H4R3me2s (Abcam), H4R3me2a (Active Motif), H3R17me2a (Abcam), histone H4 (GenScript), and histone H3 (GenScript).
Human PRMT1, CARM1, and full-length or truncated PRMT5 cDNA sequences were cloned into a pGEX6p-1 vector. GST fusion proteins were induced in E. coli BL21 (DE3) by isopropyl 1-thio-β-d-galactopyranoside (IPTG) and purified with GST Sepharose beads (GenScript). Immunoprecipitation, immunoblotting, and GST pulldown assays were performed as described previously (6). We used the following antibodies in the immunoprecipitations: FLAG (Sigma-Aldrich), PRMT5 (Sigma-Aldrich), and CARM1 (Cell Signaling Technology).
Generation of methylated PRMT5-specific antibody (PRMT5-R505me2a)
The KLH (keyhole limpet hemocyanin)-conjugated PRMT5 peptide MPYVVR (me2a) LHNFH, with R asymmetrically dimethylated, was synthesized by Abmart. This peptide, corresponding to the human PRMT5 sequence from amino acids 500 to 510, was used to immunize rabbits. The IgG fraction from the resulting serum was purified by Abmart.
In vitro methylation assay
Purified GST-PRMT1, GST-CARM1, and FLAG-PRMT5 were used as the enzyme sources for in vitro methyltransferase assays as described previously (6). Briefly, we incubated these enzymes with 5 μg of purified recombinant PRMT5 and 2 mCi of the methyl donor, S-adenosyl-l-methyl-3H-methionine (3H-SAM, PerkinElmer) in 20 μl of HMTase buffer (25 mm NaCl, 25 mm Tris-HCl, pH 8.8) for 2 h at 30 °C. Proteins were resolved on a 10% (w/v) SDS-PAGE gel, dried, and then subjected to autoradiography.
Cross-linking reactions
Lys-562 cell lysates were cross-linked using N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) at final concentrations of 5 mm and 10 mm, respectively (42, 43). After cross-linking with gentle agitation at room temperature for 2 h, the reactions were quenched with the addition of 20 mm DTT. The samples were then analyzed with Western blotting assays.
RNA isolation, quantitative real-time PCR, and ChIP analysis
RNA was isolated from cells with TRIzol reagent (Life Technologies) according to the manufacturer's instructions. cDNA was synthesized with a HiScript 1st Strand cDNA Synthesis Kit (Vazyme Biotech, China). Quantitative RT-PCR was performed using FastStart Universal SYBR Green Master (Roche) in a Rotor-Gene 6000 (Corbett Research) in a final volume of 20 μl. Cycling conditions were 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Each reaction was performed in triplicate. The primers used are listed in Table S2.
ChIP assays were performed with Lys-562 cells as described previously. The antibodies used were H3R17me2a (Abcam), H4R3me2s (Abcam), and FLAG (Sigma-Aldrich). Normal rabbit IgG served as the control. The ChIP samples were analyzed by quantitative RT-PCR using FastStart Universal SYBR Green Master (Roche). A standard curve was prepared for each set of primers using serial titration of the input DNA. The percentage of ChIP DNA was calculated relative to the input DNA from the primer-specific standard curves using the Rotor-Gene 6000 Series Software 1.7. Each reaction was performed in triplicate. The primers used are listed in Table S3.
Statistical analysis
Data analysis was performed with the statistical program GraphPad Prism (v. 6.01, La Jolla, CA). The results are presented as the mean ± S.D. Differences between two groups were analyzed using two-tailed Student's t-tests.
Author contributions
M. N., Yadong Wang, and C. G. data curation; M. N., Yadong Wang, C. G., X. L., Ying Wang, Y. D., B. Y., T. G., C. M., M. L., P. W., R. W., and R. T. formal analysis; M. N. supervision; M. N., M. L., P. W., R. T., M. F., and Q. Z. funding acquisition; M. N., Yadong Wang, Y. H., D. C. S. H., J. J., and Q. Z. investigation; M. N., Yadong Wang, C. G., M. L., P. W., R. T., M. F., B. C., Y. H., D. C. S. H., and J. J. methodology; M. N. writing-original draft; M. N. and Q. Z. project administration; Q. Z. conceptualization.
Supplementary Material
Acknowledgments
We thank Dr. Wei Xu for providing the CARM1 cDNA and the Zhao laboratories for helpful discussions.
This work was supported by National Natural Science Foundation of China NSFC 31770809 and 31470750 (to Q. Z.), 81700108 (to J. J.), and 31701191 (to M. L.), China Postdoctoral Science Foundation 2017M621706 (to J. J.) and 2016M590442 (to M. L.), and State Key Laboratory (SKL) fund 01KF-201703 (to M. F.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1–S3.
- PTM
- post-translational modifications
- PRMT
- protein arginine methyltransferase
- Q-RT-PCR
- quantitative RT-PCR
- Scr
- scramble control.
References
- 1. Allis C. D., and Jenuwein T. (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- 2. Xu J., Bauer D. E., Kerenyi M. A., Vo T. D., Hou S., Hsu Y. J., Yao H., Trowbridge J. J., Mandel G., and Orkin S. H. (2013) Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc. Natl. Acad. Sci. U.S.A. 110, 6518–6523 10.1073/pnas.1303976110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cui S., Lim K. C., Shi L., Lee M., Jearawiriyapaisarn N., Myers G., Campbell A., Harro D., Iwase S., Trievel R. C., Rivers A., DeSimone J., Lavelle D., Saunthararajah Y., and Engel J. D. (2015) The LSD1 inhibitor RN-1 induces fetal hemoglobin synthesis and reduces disease pathology in sickle cell mice. Blood 126, 386–396 10.1182/blood-2015-02-626259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krivega I., Byrnes C., de Vasconcellos J. F., Lee Y. T., Kaushal M., Dean A., and Miller J. L. (2015) Inhibition of G9a methyltransferase stimulates fetal hemoglobin production by facilitating LCR/γ-globin looping. Blood 126, 665–672 10.1182/blood-2015-02-629972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Renneville A., Van Galen P., Canver M. C., McConkey M., Krill-Burger J. M., Dorfman D. M., Holson E. B., Bernstein B. E., Orkin S. H., Bauer D. E., and Ebert B. L. (2015) EHMT1 and EHMT2 inhibition induces fetal hemoglobin expression. Blood 126, 1930–1939 10.1182/blood-2015-06-649087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., and Jane S. M. (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311 10.1038/nsmb.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., and Sif S. (2011) Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 10.1016/j.tibs.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanc R. S., and Richard S. (2017) Arginine methylation: The coming of age. Mol. Cell 65, 8–24 10.1016/j.molcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- 9. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- 10. Yang Y., Hadjikyriacou A., Xia Z., Gayatri S., Kim D., Zurita-Lopez C., Kelly R., Guo A., Li W., Clarke S. G., and Bedford M. T. (2015) PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 6, 6428 10.1038/ncomms7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., and Stallcup M. R. (1999) Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 10.1126/science.284.5423.2174 [DOI] [PubMed] [Google Scholar]
- 12. Bauer U. M., Daujat S., Nielsen S. J., Nightingale K., and Kouzarides T. (2002) Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3, 39–44 10.1093/embo-reports/kvf013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casadio F., Lu X., Pollock S. B., LeRoy G., Garcia B. A., Muir T. W., Roeder R. G., and Allis C. D. (2013) H3R42me2a is a histone modification with positive transcriptional effects. Proc. Natl. Acad. Sci. U.S.A. 110, 14894–14899 10.1073/pnas.1312925110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naeem H., Cheng D., Zhao Q., Underhill C., Tini M., Bedford M. T., and Torchia J. (2007) The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell Biol. 27, 120–134 10.1128/MCB.00815-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim K. Y., Wang D. H., Campbell M., Huerta S. B., Shevchenko B., Izumiya C., and Izumiya Y. (2015) PRMT4-mediated arginine methylation negatively regulates retinoblastoma tumor suppressor protein and promotes E2F-1 dissociation. Mol. Cell Biol. 35, 238–248 10.1128/MCB.00945-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao H. Y., Zhang Y. J., Dai H., Zhang Y., and Shen Y. F. (2011) CARM1 mediates modulation of Sox2. PLoS One 6, e27026 10.1371/journal.pone.0027026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Covic M., Hassa P. O., Saccani S., Buerki C., Meier N. I., Lombardi C., Imhof R., Bedford M. T., Natoli G., and Hottiger M. O. (2005) Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO J. 24, 85–96 10.1038/sj.emboj.7600500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daujat S., Bauer U. M., Shah V., Turner B., Berger S., and Kouzarides T. (2002) Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12, 2090–2097 10.1016/S0960-9822(02)01387-8 [DOI] [PubMed] [Google Scholar]
- 19. Feng Q., Yi P., Wong J., and O'Malley B. W. (2006) Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell Biol. 26, 7846–7857 10.1128/MCB.00568-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang L., Zhao Z., Meyer M. B., Saha S., Yu M., Guo A., Wisinski K. B., Huang W., Cai W., Pike J. W., Yuan M., Ahlquist P., and Xu W. (2014) CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21–36 10.1016/j.ccr.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawabe Y., Wang Y. X., McKinnell I. W., Bedford M. T., and Rudnicki M. A. (2012) Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11, 333–345 10.1016/j.stem.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L., Zeng H., Wang Q., Zhao Z., Boyer T. G., Bian X., and Xu W. (2015) MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci. Adv. 1, e1500463 10.1126/sciadv.1500463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujiwara T., Mori Y., Chu D. L., Koyama Y., Miyata S., Tanaka H., Yachi K., Kubo T., Yoshikawa H., and Tohyama M. (2006) CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell Biol. 26, 2273–2285 10.1128/MCB.26.6.2273-2285.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee J., and Bedford M. T. (2002) PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 3, 268–273 10.1093/embo-reports/kvf052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng D., Côté J., Shaaban S., and Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 10.1016/j.molcel.2006.11.019 [DOI] [PubMed] [Google Scholar]
- 26. Sims R. J. 3rd, Rojas L. A., Beck D. B., Bonasio R., Schüller R., Drury W. J. 3rd, Eick D., and Reinberg D. (2011) The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332, 99–103 10.1126/science.1202663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tee W. W., Pardo M., Theunissen T. W., Yu L., Choudhary J. S., Hajkova P., and Surani M. A. (2010) Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 24, 2772–2777 10.1101/gad.606110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu F., Cheng G., Hamard P. J., Greenblatt S., Wang L., Man N., Perna F., Xu H., Tadi M., Luciani L., and Nimer S. D. (2015) Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J. Clin. Invest. 125, 3532–3544 10.1172/JCI81749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rank G., Cerruti L., Simpson R. J., Moritz R. L., Jane S. M., and Zhao Q. (2010) Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood 116, 1585–1592 10.1182/blood-2009-10-251116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu Z., Li Y., Lee P., Liu T., Wan C., and Wang Z. (2012) Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One 7, e44033 10.1371/journal.pone.0044033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bode-Böger S. M., Scalera F., Kielstein J. T., Martens-Lobenhoffer J., Breithardt G., Fobker M., and Reinecke H. (2006) Symmetrical dimethylarginine: A new combined parameter for renal function and extent of coronary artery disease. J. Am. Soc. Nephrol. 17, 1128–1134 10.1681/ASN.2005101119 [DOI] [PubMed] [Google Scholar]
- 32. Ratovitski T., Arbez N., Stewart J. C., Chighladze E., and Ross C. A. (2015) PRMT5-mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD). Cell Cycle 14, 1716–1729 10.1080/15384101.2015.1033595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quan X., Yue W., Luo Y., Cao J., Wang H., Wang Y., and Lu Z. (2015) The protein arginine methyltransferase PRMT5 regulates Aβ-induced toxicity in human cells and Caenorhabditis elegans models of Alzheimer's disease. J. Neurochem. 134, 969–977 10.1111/jnc.13191 [DOI] [PubMed] [Google Scholar]
- 34. Richters A. (2017) Targeting protein arginine methyltransferase 5 in disease. Future Med. Chem. 9, 2081–2098 10.4155/fmc-2017-0089 [DOI] [PubMed] [Google Scholar]
- 35. Antonysamy S., Bonday Z., Campbell R. M., Doyle B., Druzina Z., Gheyi T., Han B., Jungheim L. N., Qian Y., Rauch C., Russell M., Sauder J. M., Wasserman S. R., Weichert K., Willard F. S., Zhang A., and Emtage S. (2012) Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 17960–17965 10.1073/pnas.1209814109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu F., Zhao X., Perna F., Wang L., Koppikar P., Abdel-Wahab O., Harr M. W., Levine R. L., Xu H., Tefferi A., Deblasio A., Hatlen M., Menendez S., and Nimer S. D. (2011) JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 19, 283–294 10.1016/j.ccr.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H. T., Zeng L. F., He Q. Y., Tao W. A., Zha Z. G., and Hu C. D. (2016) The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim. Biophys. Acta 1863, 335–346 10.1016/j.bbamcr.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ju J., Wang Y., Liu R., Zhang Y., Xu Z., Wang Y., Wu Y., Liu M., Cerruti L., Zou F., Ma C., Fang M., Tan R., Jane S. M., and Zhao Q. (2014) Human fetal globin gene expression is regulated by LYAR. Nucleic Acids Res. 42, 9740–9752 10.1093/nar/gku718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen D., Zuo Y., Zhang X., Ye Y., Bao X., Huang H., Tepakhan W., Wang L., Ju J., Chen G., Zheng M., Liu D., Huang S., Zong L., Li C., Chen Y., Zheng C., Shi L., Zhao Q., Wu Q., Fucharoen S., Zhao C., and Xu X. (2017) A genetic variant ameliorates β-thalassemia severity by epigenetic-mediated elevation of human fetal hemoglobin expression. Am. J. Hum. Genet. 101, 130–138 10.1016/j.ajhg.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Breveglieri G., Bianchi N., Cosenza L. C., Gamberini M. R., Chiavilli F., Zuccato C., Montagner G., Borgatti M., Lampronti I., Finotti A., and Gambari R. (2017) An Aγ-globin G→A gene polymorphism associated with β039 thalassemia globin gene and high fetal hemoglobin production. BMC Med. Genet. 18, 93 10.1186/s12881-017-0450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., and Clarke S. (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276, 32971–32976 10.1074/jbc.M105412200 [DOI] [PubMed] [Google Scholar]
- 42. El-Shafey A., Tolic N., Young M. M., Sale K., Smith R. D., and Kery V. (2006) “Zero-length” cross-linking in solid state as an approach for analysis of protein-protein interactions. Protein Sci. 15, 429–440 10.1110/ps.051685706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pieper J. S., Oosterhof A., Dijkstra P. J., Veerkamp J. H., and van Kuppevelt T. H. (1999) Preparation and characterization of porous crosslinked collagenous matrices containing bioavailable chondroitin sulphate. Biomaterials 20, 847–858 10.1016/S0142-9612(98)00240-3 [DOI] [PubMed] [Google Scholar]
- 44. Van Allen E. M., Wagle N., Sucker A., Treacy D. J., Johannessen C. M., Goetz E. M., Place C. S., Taylor-Weiner A., Whittaker S., Kryukov G. V., Hodis E., Rosenberg M., McKenna A., Cibulskis K., Farlow D., et al. (2014) The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 10.1158/2159-8290.CD-13-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., and Bedford M. T. (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277, 3537–3543 10.1074/jbc.M108786200 [DOI] [PubMed] [Google Scholar]
- 46. Geng P., Zhang Y., Liu X., Zhang N., Liu Y., Liu X., Lin C., Yan X., Li Z., Wang G., Li Y., Tan J., Liu D. X., Huang B., and Lu J. (2017) Automethylation of protein arginine methyltransferase 7 and its impact on breast cancer progression. FASEB J. 31, 2287–2300 10.1096/fj.201601196R [DOI] [PubMed] [Google Scholar]
- 47. Sayegh J., Webb K., Cheng D., Bedford M. T., and Clarke S. G. (2007) Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 282, 36444–36453 10.1074/jbc.M704650200 [DOI] [PubMed] [Google Scholar]
- 48. Kuhn P., Chumanov R., Wang Y., Ge Y., Burgess R. R., and Xu W. (2011) Automethylation of CARM1 allows coupling of transcription and mRNA splicing. Nucleic Acids Res. 39, 2717–2726 10.1093/nar/gkq1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li J., Zhao Z., Carter C., Ehrlich L. I., Bedford M. T., and Richie E. R. (2013) Coactivator-associated arginine methyltransferase 1 regulates fetal hematopoiesis and thymocyte development. J. Immunol. 190, 597–604 10.4049/jimmunol.1102513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pimentel H., Parra M., Gee S., Ghanem D., An X., Li J., Mohandas N., Pachter L., and Conboy J. G. (2014) A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 42, 4031–4042 10.1093/nar/gkt1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.