Abstract
Background
Early identification of prognostic superior characteristics in glioma patients such as isocitrate dehydrogenase (IDH) mutation and O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation status is of great clinical importance. The study purpose was to investigate the non-invasive predictability of IDH mutation status, MGMT promoter methylation, and differentiation of low-grade versus high-grade glioma (LGG vs HGG) in newly diagnosed patients employing relaxation-compensated multipool chemical exchange saturation transfer (CEST) MRI at 7.0 Tesla.
Methods
Thirty-one patients with newly diagnosed glioma were included in this prospective study. CEST MRI was performed at a 7T whole-body scanner. Nuclear Overhauser effect (NOE) and isolated amide proton transfer (APT; downfield NOE-suppressed APT = dns-APT) CEST signals (mean value and 90th signal percentile) were quantitatively investigated in the whole tumor area with regard to predictability of IDH mutation, MGMT promoter methylation status, and differentiation of LGG versus HGG. Statistics were performed using receiver operating characteristic (ROC) and area under the curve (AUC) analysis. Results were compared with advanced MRI methods (apparent diffusion coefficient and relative cerebral blood volume ROC/AUC analysis) obtained at 3T.
Results
dns-APT CEST yielded highest AUCs in IDH mutation status prediction (dns-APTmean = 91.84%, P < 0.01; dns-APT90 = 97.96%, P < 0.001). Furthermore, dns-APT metrics enabled significant differentiation of LGG versus HGG (AUC: dns-APTmean = 0.78, P < 0.05; dns-APT90 = 0.83, P < 0.05). There was no significant difference regarding MGMT promoter methylation status at any contrast (P > 0.05).
Conclusions
Relaxation-compensated multipool CEST MRI, particularly dns-APT imaging, enabled prediction of IDH mutation status and differentiation of LGG versus HGG and should therefore be considered as a non-invasive MR biomarker in the diagnostic workup.
Keywords: amide proton transfer imaging, APT, biomarkers, chemical exchange saturation transfer, CEST, glioma, isocitrate dehydrogenase, O(6)-methylguanine-DNA methyltransferase
Importance of the study
Non-invasive early identification of prognostic superior characteristics in glioma patients such as IDH mutation and MGMT promoter methylation status is of great clinical importance for determination of the appropriate therapy. Our study demonstrates that relaxation-compensated multipool CEST MRI at 7.0T enables non-invasive prediction of IDH mutation status and differentiation of lower- versus higher-grade tumors in patients with newly diagnosed untreated glioma. Consequently, the study forms a fundament for further clinical application of multipool CEST MRI as a non-invasive imaging biomarker in the preoperative diagnostic workup of glioma patients.
Gliomas account for most of primary brain tumors in adults.1 Patients with high-grade glioma (HGG), especially World Health Organization (WHO) grade IV glioblastomas (GBMs), still have a dismal prognosis, even after resection and additive radiochemotherapy.2 Therefore, early identification of patients with prognostic superior characteristics is of utmost importance. Survival in general, radiosensitivity, and chemosensitivity are highly dependent on both WHO grade and histopathological characteristics, particularly isocitrate dehydrogenase (IDH) mutation and O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation status.3–6 Nevertheless, this information can currently only be provided by resection or biopsy, through histopathological analysis, although clinicians are in desperate need of this information from the very beginning of the disease. However, gadolinium contrast-enhanced (GdCE) multiparametric MRI, the key element of today’s diagnostic routine, is not capable of gaining reliable data about histopathological characteristics.7–10 Promising results have been reported on in vivo detection of 2-hydroxyglutarate (2-HG) using magnetic resonance spectroscopy (MRS), possibly enabling determination of IDH status in glioma patients.11,12 However, MRS-based detection of 2-HG is still challenging and therefore not widely used in clinical practice. Consequently, alternative MRI sequences that non-invasively provide reliable information on histopathological characteristics are urgently needed.
Chemical exchange saturation transfer (CEST) imaging might have the potential to cope with this problem. CEST contrasts are based on the spontaneous chemical exchange between solute-bound protons and protons of free water that to a large extent depends on the concentration of endogenous cellular proteins. Furthermore, CEST signals are sensitive to tissue microenvironment, such as pH, affecting proton exchange properties.13,14 Consequently, through its sensitivity to local protein concentration,13,15 intracellular pH,16–18 and protein states,19,20 CEST imaging provides complementary information to current standard MRI sequences. Recently, Jiang et al reported the predictability of IDH mutation status in grade II gliomas employing amide proton transfer-weighted (APTw) CEST MRI at 3T in a retrospectively analyzed study cohort.21 Furthermore, Jiang et al found a significant correlation between MGMT promoter methylation status and APTw CEST metrics.22 Similar to the aforementioned studies, most previous CEST approaches were based on magnetization transfer ratio asymmetry (MTRasym) analyses.13,23,24 However, the limitation of these approaches is that the MTRasym contrasts have multiple contributions, including upfield and downfield relayed nuclear Overhauser effect (rNOE) signals, T1 and T2 relaxation times, and conventional semisolid magnetization transfer (MT) asymmetry.25–28 Consequently, a proper separation of different CEST pools is essential to better understand the role of each effect in brain tumor imaging and to investigate its clinical diagnostic value. For CEST data with high spectral resolution obtained at 7T, it has been shown that this separation is feasible using Lorentzian fit analysis.26,27 Furthermore, using the relaxation-compensated metric yields the apparent exchange-dependent relaxation rate (AREX), which is corrected for contributions of semisolid MT, T2 relaxation (spillover), and T1 relaxation times.27 Ultimately, it has been shown that downfield rNOE signals can be eliminated, yielding the downfield rNOE suppressed amide proton CEST contrast (dns-APT).
In this prospective study, we investigated isolated APT- and NOE-mediated CEST effects and the predictability of histopathological features using the obtained multipool CEST metrics, in a large glioma patient study cohort at 7.0T. We hypothesized that the CEST contrasts may enable prediction of the molecular parameters IDH mutation, MGMT promoter methylation status, and differentiation of lower- versus higher-grade WHO tumors in patients with newly diagnosed gliomas.
Materials and Methods
Patients
This prospective study received approval by the local institutional review board committee. Thirty-eight patients with newly diagnosed untreated intracranial expanding lesions were included after the nature and possible consequences of the study were explained and written informed consent was obtained. Inclusion criteria were patients with MRI findings suspicious for glioma, previously untreated, age 18 years or older, and eligibility for 7T MRI.
In total, 31 patients had histopathological diagnosis of glioma (WHO grades II–IV) subsequent to the study examination; 5 patients had tumors of a nonglial entity (metastasis, lymphoma), and 2 patients with typical low-grade glioma (LGG) lesions did not undergo surgical resection or biopsy, since a watch-and-wait strategy was chosen. Consequently, those patients were excluded from statistical analyses (Table 1). Eleven patients of the study cohort had been previously included in methodical publications.27,28
Table 1.
Glioma patient cohort included in this study*
| Patient | Age | Sex | Diagnosis* | WHO Grade | MGMT | IDH |
|---|---|---|---|---|---|---|
| #1 | 46 | f | GBM | IV | unmet | wt |
| #2 | 51 | f | GBM | IV | n/a | n/a |
| #3 | 69 | m | GBM | IV | met | wt |
| #4 | 60 | m | GBM | IV | met | wt |
| #5 | 53 | m | GBM | IV | met | wt |
| #6 | 53 | f | GBM | IV | IND | wt |
| #7 | 52 | m | GBM | IV | unmet | wt |
| #8 | 75 | m | GBM | IV | IND | wt |
| #9 | 65 | m | GBM | IV | unmet | wt |
| #10 | 65 | m | GBM | IV | met | wt |
| #11 | 71 | m | GBM | IV | IND | wt |
| #12 | 81 | f | GBM | IV | met | wt |
| #13 | 54 | m | GBM | IV | unmet | wt |
| #14 | 53 | f | GBM | IV | met | wt |
| #15 | 63 | f | GBM | IV | unmet | wt |
| #16 | 59 | m | Gliosarcoma | IV | unmet | wt |
| #17 | 59 | m | GBM | IV | unmet | wt |
| #18 | 61 | m | GBM | IV | unmet | wt |
| #19 | 48 | f | GBM | IV | met | wt |
| #20 | 50 | m | GBM | IV | IND | wt |
| #21 | 50 | m | GBM | IV | unmet | wt |
| #22 | 86 | m | GBM | IV | met | wt |
| #23 | 41 | f | GBM | IV | n/a | mut |
| #24 | 42 | f | AA | III | met | mut |
| #25 | 27 | f | AA | III | met | mut |
| #26 | 48 | f | ODG | II | met | mut |
| #27 | 25 | m | Astrocytoma | II | n/a | mut |
| #28 | 49 | f | ODG | II | n/a | mut |
| #29 | 74 | m | Astrocytoma | II | n/a | wt |
| #30 | 29 | f | ODG | II | met | mut |
| #31 | 39 | f | Astrocytoma | II | met | mut |
*All diagnoses were histopathologically proven after surgical resection or tumor biopsy subsequent to CEST MRI at 7.0 T (AA = anaplastic astrocytoma, GBM = glioblastoma, ODG = oligodendroglioma). n/a = data not available, IND = result of histopathological analysis indeterminate. WHO grade II tumors were considered LGG and WHO grades III‒IV as HGG.
Histopathology
All diagnoses were histopathologically proven after surgical resection or tumor biopsy, according to the 2016 WHO classification of CNS tumors, by neuropathologists who were blinded to the MRI data. WHO tumor grade was obtained for all patients (6/31 = 19.4% LGG; 25/31 = 80.6% HGG), IDH1-R132H mutation status for 30/31 patients (8/30 = 26.7% IDH mutant [mut]; 22/30 = 73.3% IDH wildtype [wt]), and MGMT promoter methylation status in 26/31 cases (13/26 = 50.0% methylated [met]; 9/26 = 34.6% unmethylated [unmet]; 4/26 = 15.4% indeterminate). All histopathological findings of the study cohort are described in Table 1.
MRI Acquisition
Conventional MRI at 3T
The acquisition of conventional MRI exams was performed at 3T, employing the following protocol parameters: T1-weighted GdCE MRI (GdCE-T1) (echo time [TE] = 4.04 ms; repetition time [TR] = 1710 ms; field of view [FoV] in mm2: 256 × 256; matrix: 512 × 512; slice thickness: 1 mm), T2 fluid attenuated inversion recovery (TE = 135 ms; TR = 8500 ms; FoV: 230 × 172 mm2; matrix: 256 × 192; slice thickness: 5 mm), and T2-weighted MRI turbo spin echo (TSE) (TE = 86 ms; TR = 5550 ms; FoV: 229 × 172 mm2; matrix: 384 × 230; slice thickness: 5 mm). Apparent diffusion coefficient (ADC) maps were generated from diffusion-weighted imaging (DWI) employing a single-shot spin-echo echo-planar imaging (EPI) sequence, relative cerebral blood volume (rCBV) maps were generated from dynamic susceptibility contrast-enhanced (DSC) MRI performed after gadolinium prebolus injection using a T2*-weighted gradient-echo EPI sequence; both techniques have been described previously in detail.29 DSC-MRI and DWI data were available in 26 of the 31 (~84%) glioma patients included. In 5 patients these sequences were not acquired due to time constraints in daily clinical practice. Coregistration of all images was performed using an automatic multimodal rigid registration algorithm of the Medical Imaging Interaction Toolkit.30
CEST MRI at 7T and data post processing
CEST MRI was performed on a 7T whole body MRI scanner (Magnetom 7.0T; Siemens Healthineers) employing a custom-developed sequence based on a centric-reordered 2D single slice gradient echo (TE = 3.76 ms; TR = 7.6 ms; FoV: 200 × 175 mm2; matrix: 128 × 112; resolution: 1.56 × 1.56 × 5 mm3; flip angle: 10°) and a single channel transmit (Tx)/24-channel receive (Rx) 1H head coil as previously described.27 In vivo CEST images were obtained following saturation by a train of 150 Gaussian-shaped radiofrequency pulses (pulse width = 15 ms, duration time = 10 ms, duty cycle = 60%, saturation time = 3.75 s) for 2 different B1 amplitudes (1.0 μT and 0.6 μT) at 65 unevenly sampled saturation offsets dubbed Δω.27 To achieve separation of APT- and NOE-mediated CEST effects and correction for conventional MT asymmetry, Z-spectra were fitted pixelwise by a multi-Lorentzian fit approach as recently reported by Windschuh et al.31 The fit model provides a specifically labeled (Zlab) and a reference Z-spectrum (Zref) for each CEST effect, which are used to calculate the T1 and T2 relaxation-compensated and MT-corrected contrasts:
| (1) |
An explicit derivation of the formula has been previously reported.27 Ultimately, the downfield-rNOE-suppressed (dns)-APT imaging technique additionally removes downfield resonating rNOE contributions from the amide signal. The contrast was calculated at = 3.5 ppm by using the ratio rrNOE = 0.2 as proposed by Zaiss et al28:
| (2) |
B0 and B1 maps were acquired simultaneously by means of the water shift and B1 (WASABI) method.32 B1 correction was achieved employing the B1-correction method proposed by Windschuh et al, yielding CEST contrasts corresponding to 0.6 μT saturation amplitude.31 The CEST scans at 2 B1 amplitudes required approximately 4 min each, the WASABI scan about 2 min, and an additional T1 map 1:20 min, yielding total measurement duration of approximately 11 min for B0- and B1-corrected, relaxation-compensated, multipool CEST MRI.
Data Analysis
The tumor region, defined as the whole area of abnormal signal intensity on T1-GdCE and T2-weighted images, was manually segmented (by A.R., with 9 years of experience in neuroradiology) for quantitative analysis in each patient. Necrotic tumor regions were thoroughly excluded. The approach of whole tumor segmentation and subsequent determination of signal mean and 90th percentile of all included voxel intensities allowed reproducibility, in contrast to manual selection of multiple regions of interest. For NOE CEST contrasts, the 10th percentile of included voxels was determined, since NOE-mediated effects are known to be decreased in tumor lesions.26,33–35 Accordingly, mean value and 10th percentile of all included voxels were determined for the ADC, while mean signal intensity and 90th percentile were calculated for rCBV.
Statistics
Receiver operating characteristic (ROC) area under the curve (AUC) analyses were performed for the investigated CEST metrics (NOE mean, NOE 10th percentile [NOE10], APT mean, APT 90th percentile [APT90], dns-APT mean, and dns-APT 90th percentile [dns-APT90]) and the advanced imaging methods acquired at 3T (ADC mean, ADC 10th percentile, rCBV mean, rCBV 90th percentile) in order to assess the contrasts’ ability to predict (i) WHO tumor grade (LGG = grades I–II vs HGG = grades III–IV), (ii) IDH mutation (IDH1-R132H wt vs mut), and (iii) MGMT promoter methylation status (met vs unmet). Best sensitivity and specificity pairs were determined for each contrast and parameter with stronger weighting of specificity (condition: specificity >75%). Furthermore, mean signal intensity and standard deviation were calculated for each contrast and parameter category over all patients of the group (WHO grade: HGG/LGG, IDH: wt/mut, and MGMT promoter: unmet/met). Statistics were performed with SigmaPlot version 12.5 (SystatSoftware). For statistical analyses the level of significance was set to P < 0.05.
Results
Predictability of WHO Tumor Grade
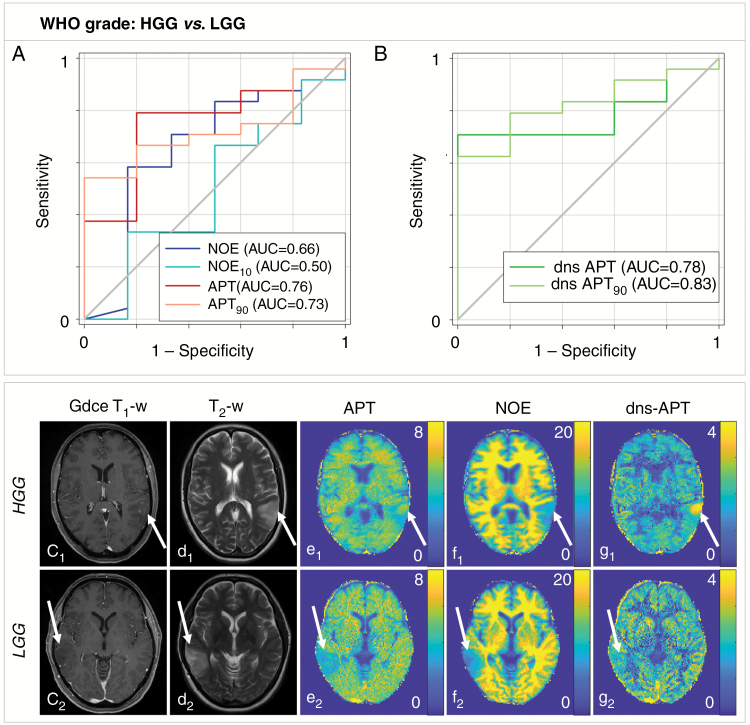
Reliable prediction of WHO tumor grade (LGG vs HGG) was only possible using the dns-APT metric yielding areas under the ROC curve of 0.78 (P < 0.05, mean dns-APT) and 0.83 (P < 0.05, dns-APT90) (Fig. 1). Best classification was achieved using the mean dns-APT metric with a test sensitivity and specificity of 71% and 100% (cutoff value = 1.88). All other CEST contrasts and rCBV analysis showed a tendency toward higher values in HGG tumors but did not allow for significant differentiation of lower- versus higher-grade glioma (P > 0.05). No trend was observed for the investigated ADC metrics.
Fig. 1.
Predictability of WHO tumor grade (HGG vs LGG) in newly diagnosed untreated glioma. (A, B) dns-APT allowed prediction of WHO tumor grade with highest AUC for the dns-APT90 metric (0.83) and a test sensitivity/specificity of 0.63 (0.41–0.81)/1.00 (0.48–1.00) (P < 0.05). Two exemplary patients with HGG (GBM, c1–g1) and LGG (oligodendroglioma II, c2–g2) shown: ci: GdCE T1-w, di: T2-w (TSE), relaxation-compensated multipool CEST MRI at 7T with separated APT (ei), NOE (fi), and dns-APT (gi) effects (unit: %). The GBM patient shows only a small spot-like contrast enhancement in the tumor region (c1), while a clear hyperintensity displays within the tumor at dns-APT imaging (g1, white arrow).
Predictability of IDH Mutation Status
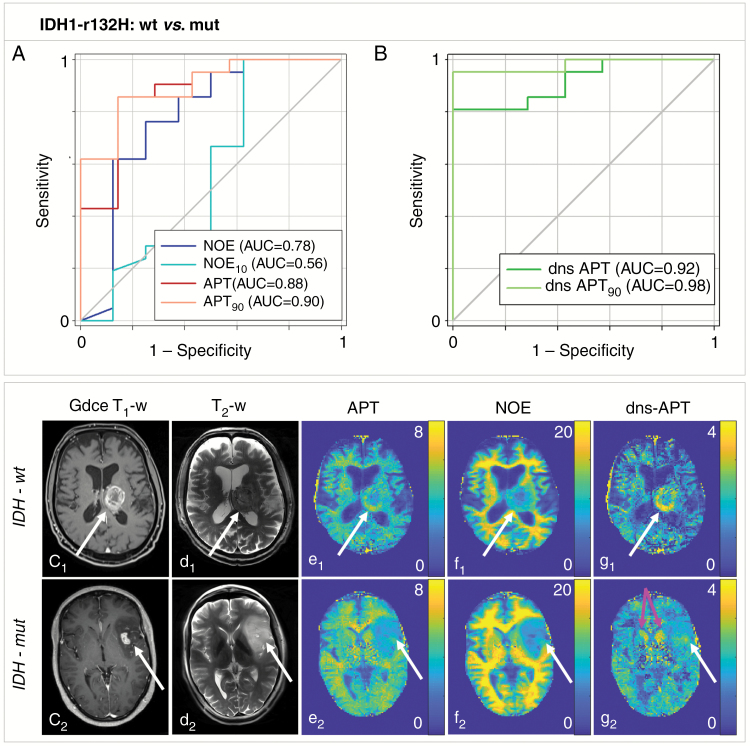
Higher signal intensity in tumors of patients with IDH-wt status allowed significant prediction of IDH-mut status at multipool CEST imaging with highest AUCs at dns-APT contrasts (mean APT = 0.88; P < 0.05, APT90 = 0.90; P < 0.05, mean dns-APT = 0.92; P < 0.01, and dns-APT90 = 0.98; P < 0.001). Mean NOE signal intensity yielded an AUC of 0.78 (P < 0.05) (Fig. 2). Furthermore, perfusion MRI showed significantly increased rCBV in IDH-wt tumors, yielding an AUC of 0.79 (P < 0.05). ADC values did not enable significant prediction of IDH-mut status. Best test performance was achieved using the dns-APT90 metric with sensitivity and specificity of 95% and 100%, respectively, at a cutoff value of 2.86.
Fig. 2.
Predictability of IDH mutation status (IDH1-R132H wt versus mutation) in newly diagnosed untreated glioma. (A, B) APT and dns-APT CEST metrics allowed prediction of IDH mutation status with highest AUC for the dns-APT90 metric (0.98) with a test sensitivity/specificity of 0.95 (0.77–1.00)/1.00 (0.59–1.00) (P < 0.0001). Two exemplary patients with newly diagnosed GBM IDH-wt (c1 –g1) and IDH-mut (c2 –g2) shown: ci: GdCE T1-w, di: T2-w (TSE), relaxation-compensated multipool CEST MRI at 7T with separated APT (ei), NOE (fi), and dns-APT (gi) effects (unit: %). A ring-like hyperintensity can be delineated in the periphery of the IDH-wt glioblastoma at dns-APT imaging (g1, white arrow), while the IDH-mut GBM displays barely hyperintense at dns-APT (g2, white arrow). The head of the caudate nucleus also displays hyperintense on dns-APT images (g2, pink arrows).
Predictability of MGMT Promoter Methylation Status
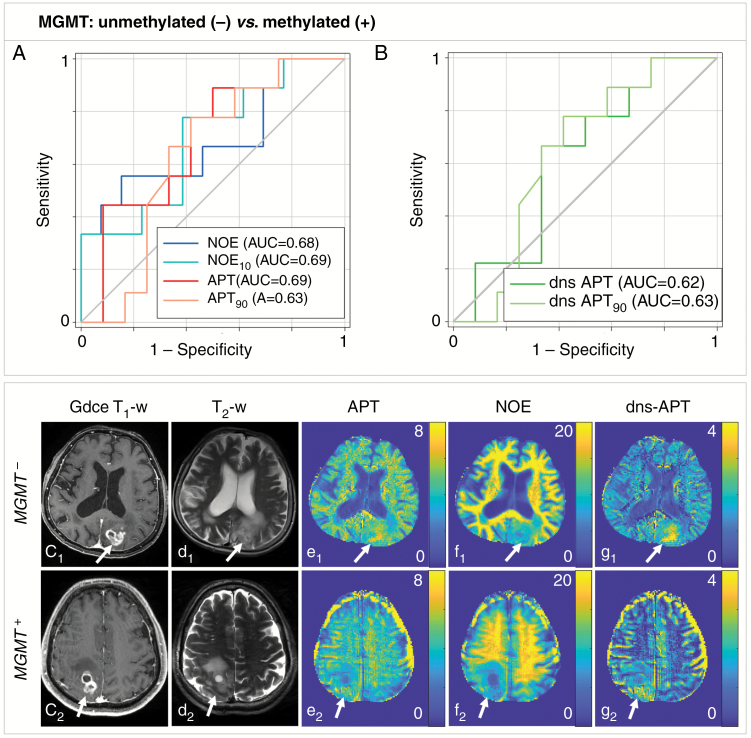
None of the investigated CEST metrics at 7T and none of the advanced MRI methods at 3T allowed for significant prediction of MGMT promoter methylation in glioma patients (P > 0.05) (Fig. 3). Trends toward higher signal intensities in patients with unmethylated MGMT promoter were observed at all investigated CEST contrasts, while no trend could be found for ADC and rCBV values. The results of all ROC curve analyses are provided in Table 2.
Fig. 3.
Predictability of MGMT promoter methylation status (unmethylated vs methylated) in newly diagnosed untreated glioma patients. (A, B) None of the investigated CEST contrasts allowed for significant prediction of the MGMT promoter methylation in glioma patients. Two GBM patients with unmethylated (GBM MGMT−: c1–g1) and methylated (GBM MGMT+: c2–g2) promoter shown: ci: GdCE T1-w, di: T2-w (TSE), relaxation-compensated multipool CEST MRI at 7T with separated APT (ei), NOE (fi), and dns-APT (gi) effects (unit: %). A tendency toward higher signal intensities for patients with unmethylated MGMT promoter was observed for all investigated CEST contrasts.
Table 2.
Results of quantitative analyses for multipool CEST MRI at 7.0T and advanced MRI methods at 3.0T
| WHO Tumor Grade (LGG vs HGG) | |||||||
|---|---|---|---|---|---|---|---|
| CEST MRI @ 7T | LGG (mean ± SD) |
HGG (mean ± SD) |
Cutoff | Sensitivity (95% CI) |
Specificity (95% CI) |
AUC | P-val. |
| NOE mean | 7.69 ± 3.96 | 9.06 ± 3.21 | 8.95 | 0.61 (0.39–0.80) | 0.83 (0.36–1.00) | 0.66 | 0.24 |
| NOE 10th pc | 4.24 ± 2.59 | 4.22 ± 2.01 | 5.18 | 0.33 (0.16–0.55) | 0.83 (0.36–1.00) | 0.50 | 1.00 |
| APT mean | 3.07 ± 1.50 | 3.96 ± 1.32 | 3.66 | 0.79 (0.58–0.93) | 0.80 (0.28–0.99) | 0.76 | 0.07 |
| APT 90th pc | 4.54 ± 2.15 | 6.03 ± 2.26 | 5.42 | 0.67 (0.45–0.84) | 0.80 (0.28–0.99) | 0.73 | 0.11 |
| dns-APT mean | 1.47 ± 0.68 | 2.14 ± 0.85 | 1.88 | 0.71 (0.49–0.87) | 1.00 (0.48–1.00) | 0.78 | 0.0497 |
| dns-APT 90th pc | 2.37 ± 1.20 | 4.01 ± 1.85 | 3.62 | 0.63 (0.41–0.81) | 1.00 (0.48–1.00) | 0.83 | 0.0234 |
| Advanced MRI methods @ 3T | |||||||
| rCBV | 462 ± 405 | 914 ± 581 | 606.8 | 0.68 (0.45–0.86) | 0.75 (0.19–0.99) | 0.73 | 0.17 |
| rCBV 90th pc | 1069 ± 863 | 1945 ± 1024 | 1317 | 0.73 (0.50–0.89) | 0.75 (0.19–0.99) | 0.74 | 0.16 |
| ADC mean | 1168 ± 231 | 1117 ± 146 | 1189 | 0.36 (0.17–0.59) | 0.80 (0.28–0.99) | 0.53 | 0.85 |
| ADC 10th pc | 772 ± 220 | 749 ± 145 | 876.9 | 0.29 (0.11–0.52) | 0.80 (0.28–0.99) | 0.53 | 0.87 |
| IDH status (IDH1-R132H wildtype vs mutation) | |||||||
| CEST MRI @ 7T | IDH mut (mean ± std) | IDH wt (mean ± std) | Cutoff |
Sensitivity
(95% CI) |
Specificity
(95% CI) |
AUC | P-val. |
| NOE mean | 6.03 ± 4.55 | 9.68 ± 2.15 | 8.95 | 0.62 (0.41–0.83) | 0.88 (0.47–1.00) | 0.78 | 0.02 |
| NOE 10th pc | 3.63 ± 2.94 | 4.29 ± 1.69 | 5.18 | 0.29 (0.11–0.50) | 0.75 (0.35–0.97) | 0.56 | 0.64 |
| APT mean | 2.30 ± 1.77 | 4.30 ± 0.80 | 3.66 | 0.86 (0.65–0.97) | 0.86 (0.42–1.00) | 0.88 | 0.0032 |
| APT 90th pc | 3.36 ± 2.43 | 6.67 ± 1.64 | 5.22 | 0.86 (0.65–0.97) | 0.86 (0.42–1.00) | 0.90 | 0.0019 |
| dns-APT mean | 1.10 ± 0.81 | 2.36 ± 0.61 | 1.88 | 0.81 (0.60–0.95) | 1.00 (0.59–1.00) | 0.92 | 0.0011 |
| dns-APT 90th pc | 1.69 ± 1.13 | 4.45 ± 1.53 | 2.86 | 0.95 (0.77–1.00) | 1.00 (0.59–1.00) | 0.98 | 0.0001 |
| Advanced MRI methods @ 3T | |||||||
| rCBV | 465 ± 390 | 1002 ± 567 | 851 | 0.67 (0.43–0.87) | 0.86 (0.42–1.00) | 0.79 | 0.0251 |
| rCBV 90th pc | 1133 ± 871 | 2104 ± 974 | 2072 | 0.61 (0.38–0.84) | 0.86 (0.42–1.00) | 0.79 | 0.0293 |
| ADC mean | 1190 ± 239 | 1107 ± 142 | 911 | 0.10 (0.01–0.30) | 0.83 (0.36–1.00) | 0.63 | 0.33 |
| ADC 10th pc | 833 ± 108 | 721 ± 167 | 737 | 0.53 (0.32–0.77) | 0.83 (0.36–1.00) | 0.72 | 0.11 |
| MGMT promoter methylation status (methylated [+] vs unmethylated [−]) | |||||||
| CEST MRI @ 7T | MGMT + (mean ± std) | MGMT - (mean ± std) | Cutoff |
Sensitivity
(95% CI) |
Specificity
(95% CI) |
AUC | P-val. |
| NOE mean | 7.34 ± 3.76 | 9.97 ± 3.09 | 10.12 | 0.56 (0.30–0.80) | 0.84 (0.54–0.98) | 0.68 | 0.15 |
| NOE 10th pc | 3.38 ± 1.88 | 4.86 ± 2.04 | 4.60 | 0.44 (0.20–0.70) | 0.77 (0.46–0.95) | 0.69 | 0.13 |
| APT mean | 3.35 ± 1.85 | 4.34 ± 0.95 | 4.73 | 0.44 (0.20–0.70) | 0.75 (0.43–0.95) | 0.68 | 0.17 |
| APT 90th pc | 5.01 ± 2.74 | 6.18 ± 1.10 | 6.59 | 0.44 (0.20–0.70) | 0.75 (0.43–0.95) | 0.62 | 0.34 |
| dns-APT mean | 1.86 ± 1.11 | 2.35 ± 0.69 | 2.71 | 0.22 (0.07–0.52) | 0.75 (0.43–0.95) | 0.61 | 0.39 |
| dns-APT 90th pc | 3.20 ± 1.93 | 4.03 ± 1.00 | 6.59 | 0.44 (0.20–0.70) | 0.75 (0.43–0.95) | 0.62 | 0.34 |
| Advanced MRI methods @ 3T | |||||||
| rCBV | 730 ± 582 | 986 ± 688 | 1167 | 0.50 (0.19–0.75) | 0.83 (0.52–0.98) | 0.58 | 0.60 |
| rCBV 90th pc | 1561 ± 1065 | 2087 ± 1165 | 2453 | 0.50 (0.19–0.75) | 0.92 (0.62–1.00) | 0.59 | 0.56 |
| ADC mean | 1161 ± 176 | 1087 ± 195 | 955 | 0.25 (0.07–0.52) | 0.91 (0.48–0.98) | 0.59 | 0.51 |
| ADC 10th pc | 779 ± 114 | 787 ± 115 | 884 | 0.13 (0.02–0.40) | 0.82 (0.48–0.98) | 0.52 | 0.90 |
Receiver operating characteristic (ROC) area under the curve (AUC) analyses performed for NOE mean, NOE 10th percentile (NOE10), APT mean, APT 90th percentile (APT90), dns-APT mean, and dns-APT 90th percentile (dns-APT90) in order to assess the contrasts’ ability to predict (1) WHO tumor grade (grades III‒IV versus grade II), (2) IDH mutation status (IDH1-R132H wildtype versus mutation), and (3) MGMT promoter methylation status (methylated versus unmethylated) in untreated glioma patients. Same analyses were performed for ADC (mean, 10th percentile) and rCBV (mean, 90th percentile) obtained from DWI and DSC MRI at 3T. Best sensitivity and specificity with 95% CI shown for each contrast and category with stronger weighting of specificity (condition: specificity >75%). Mean signal intensity and standard deviation for each contrast and category over all patients additionally provided (CEST MRI [%], ADC [µm2/s], rCBV [dimensionless])
Discussion
In this study we investigated novel relaxation-compensated multipool CEST MRI contrasts at 7.0T and demonstrated the non-invasive predictability of IDH mutation status and WHO tumor grade in untreated glioma patients, with the best test performance using the dns-APT metric. None of the investigated CEST contrasts and none of the advanced MRI methods at 3.0T allowed significant differentiation between glioma patients with methylated and unmethylated MGMT promoter.
Prediction of IDH Mutation Status
Relaxation-compensated multipool CEST MRI, particularly APT and dns-APT imaging, performed markedly better in predicting IDH mutation status than WHO tumor grade in the investigated study cohort. Therefore, we infer that genetic status, by means of IDH mutation status, is more reflective than the histologic class, regarding APT-mediated CEST effects.
The latest WHO classification of CNS tumors termed gliomas of WHO grades I‒III together as “lower grade gliomas” with a great majority of IDH-mut and a wide range of overall survival within this group.36,37 Glioblastomas were divided into GBM IDH-wt, most frequently de novo tumors (~90% of cases), and GBM IDH-mut (~10% of cases), closely corresponding to so-called secondary glioblastoma.36 Consequently, the 2016 WHO CNS definition allows for dynamic classification based on both phenotype and genotype, taking into account the prognostic importance of IDH mutation status.36 Generally, histologically low-grade diffuse IDH-wt tumors are known to clinically often resemble aggressive grade IV GBMs.38 Mutations in IDH gene-encoded enzymes are expected to cause widespread disturbances of cellular metabolism, including alteration of amino acid concentrations and global downregulation of protein expression.21 Hence, APT-mediated CEST signal is expected to be significantly lower in IDH-mut tumors, which is in agreement with our study findings. Alterations of tissue microenvironment (eg, pH, lactate acid production), particularly in IDH-wt tumors, may additionally affect the amide CEST signal. Therefore, CEST MRI is an alternative indirect method for determining IDH mutation status, significantly extending the existing repertoire of non-invasive imaging biomarkers used in the diagnostic workup of brain tumor patients.
Best test performance of CEST MRI based preoperative prediction of IDH mutation status was achieved using APT and dns-APT metrics with highest sensitivity/specificity (95%/100%, AUC = 0.98) for the dns-APT90 contrast. This result outperforms the findings of Jiang et al, who reported a sensitivity and specificity of 57% and 100%, respectively (AUC = 0.89), using APTw MR imaging at 3T.21 However, Jiang et al investigated APTw CEST metrics in LGG patients, thereby limiting the comparability of the study cohorts. NOE-mediated CEST imaging yielded inferior performance in IDH status prediction (sensitivity, 61%; specificity, 83%) compared with all investigated APT CEST metrics in our study.
Compared with rCBV and ADC ROC analyses, the amide CEST metrics, particularly dns-APT, yielded clearly better results in IDH mutation status prediction. However, results of rCBV ROC analysis were significant too, which is in agreement with previous studies reporting significantly increased rCBV in IDH-wt tumors of glioma patients.39,40 ADC values tended to be lower in IDH-wt tumors, but differences were not statistically significant.
Prediction of WHO Tumor Grade
Preoperative assessment of glioma employing APTw CEST MRI based on MTRasym at clinical field strength (3.0T) has previously been shown in several studies to positively correlate with the tumors’ histopathological grade.24,41–43 Moreover, the ability of APTw imaging to distinguish non-enhancing high-grade lesions from LGG was demonstrated.44 Our study confirms these findings and additional proofs that these previous results particularly hold true for isolated APT effects, employing relaxation-compensated dns-APT MRI. For conventional (non-dns) APT effects at 7T, a positive trend but no significant differences according to WHO tumor grade were observed, which is in agreement with previous results obtained in smaller patient cohorts (n = 10).34 Regarding NOE-mediated CEST effects at 7T, Heo et al reported significantly decreased NOE signals in HGGs compared with LGGs (grade IV vs grades I–III, grade III vs grades I–II),34 which could not be confirmed in our study cohort. In this work, NOE effects were not significantly different according to tumor grade. Perfusion imaging by means of rCBV did not enable significant prediction of LGG versus HGG but revealed a clear trend of increased rCBV in HGG tumors, while no trend was observed for the investigated ADC metrics.
MGMT Promoter Methylation Status Prediction
None of the investigated CEST contrasts and none of the advanced MRI methods at 3T allowed for prediction of MGMT promoter methylation status in glioma patients. This result is in contrast to the findings of Jiang et al, who recently reported discriminability of MGMT promoter methylation status in patients with GBM using APTw MRI metrics based on MTRasym.22 However, a trend toward higher signal intensity in tumors with unmethylated MGMT promoter was also observed in our data. Moreover, Jiang et al employed the APTw sequence at 3.0T with imaging parameters different from our multipool CEST approach at 7.0T.
Generally, prediction of histopathological parameters using MRI parameters has previously been assessed employing radiomics analysis. Radiomics approaches combine information obtained from multiple conventional MR sequences and have so far been tested in relatively small study cohorts with promising results for prediction of IDH mutation status and MGMT promoter methylation.45–48 CEST, however, is fundamentally different, since the new MRI contrasts provide additional information on a subcellular protein level. Therefore, both approaches are promising and may be combined in the future to most accurately predict histologic features non-invasively. While MRI techniques can currently not replace histopathological analysis, the presented approach may be of major value in surgery planning when performing biopsy and in the follow-up setting for therapy monitoring, as serial tissue biopsies are usually not feasible.
Limitations
Some limitations of this study need to be acknowledged.
(1) Although this study included the so far largest glioma patient cohort undergoing relaxation-compensated multipool CEST MRI at 7T, the results need to be validated in independent study cohorts. Moreover, the demonstrated relationship of CEST MRI and IDH mutation status in glioma needs to be confirmed in HGG patient cohorts of the same WHO grade. The significant results of this work can justify and motivate such studies.
(2) The employed multipool CEST MRI approach used a single-slice readout sequence. For routine clinical application we suggest the implementation of three-dimensional readout sequences, which can be combined with the multipool CEST preparation module.
(3) MR units at 7T are currently not widely available in most clinical settings, which therefore limits the immediate applicability of the presented approach. However, vendors are pushing toward the increase of 7T units on the market with the goal to include them in the clinical routine. Moreover, several studies reported separation of different CEST pools also at clinical field strength (3.0T),49,50 and the knowledge gained in this study can thus be used as prior knowledge for the preparation and evaluation of those CEST approaches.
Conclusions
In this prospective study we investigated NOE- and APT-mediated CEST effects at 7T and their non-invasive predictability of histopathological parameters in newly diagnosed untreated glioma. Relaxation-compensated multipool CEST MRI allowed for prediction of IDH mutation status and differentiation of LGG versus HGG tumors. Our results suggest that the genetic subtype, by means of IDH mutation status, has a greater effect on the amide CEST signal than the histologic grade of the tumor, particularly at dns-APT imaging. Therefore dns-APT CEST may be considered an imaging biomarker to non-invasively assess genetic features in the preoperative workup and to monitor treatment response of glioma patients.
Funding
This study did not receive funding from third parties.
Conflict of interest statement. None declared.
Acknowledgments
We acknowledge authorship as: study concepts/study design (all authors), data analysis and interpretation (all authors), manuscript writing (all authors), implementation (D.P., M.Z., A.R., J.W.), clinical/experimental studies (D.P, J.W, J.O., C.D., J.E.M., S.G., P.S., S.R., M.Z., A.R.), statistical analysis (D.P.), guarantors of integrity of entire study (D.P., A.R., H.P.S.).
References
- 1. Porter KR, McCarthy BJ, Freels S, Kim Y, Davis FG. Prevalence estimates for primary brain tumors in the United States by age, gender, behavior, and histology. Neuro Oncol. 2010;12(6):520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Roila F; ESMO Guidelines Working Group Malignant glioma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2009;20(Suppl 4):126–128. [DOI] [PubMed] [Google Scholar]
- 3. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 4. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 5. Platten M, Bunse L, Wick W, Bunse T. Concepts in glioma immunotherapy. Cancer Immunol Immunother. 2016;65(10):1269–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henson JW, Gaviani P, Gonzalez RG. MRI in treatment of adult gliomas. Lancet Oncol. 2005;6(3):167–175. [DOI] [PubMed] [Google Scholar]
- 8. Ellingson BM, Chung C, Pope WB, Boxerman JL, Kaufmann TJ. Pseudoprogression, radionecrosis, inflammation or true tumor progression? Challenges associated with glioblastoma response assessment in an evolving therapeutic landscape. J Neurooncol. 2017;134(3):495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi YS, Ahn SS, Kim DW, et al. Incremental prognostic value of ADC histogram analysis over MGMT promoter methylation status in patients with glioblastoma. Radiology. 2016;281(1):175–184. [DOI] [PubMed] [Google Scholar]
- 10. Pope WB, Lai A, Mehta R, et al. Apparent diffusion coefficient histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. AJNR Am J Neuroradiol. 2011;32(5):882–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi C, Ganji SK, DeBerardinis RJ, et al. 2-Hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pope WB, Prins RM, Albert Thomas M, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PC. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50(6):1120–1126. [DOI] [PubMed] [Google Scholar]
- 14. Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9(8):1085–1090. [DOI] [PubMed] [Google Scholar]
- 15. Jin T, Wang P, Zong X, Kim SG. MR imaging of the amide-proton transfer effect and the pH-insensitive nuclear overhauser effect at 9.4 T. Magn Reson Med. 2013;69(3):760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J, Payen JF, Wilson DA, Traystman RJ, van Zijl PC. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9(8):1085–1090. [DOI] [PubMed] [Google Scholar]
- 17. Sun PZ, Benner T, Copen WA, Sorensen AG. Early experience of translating pH-weighted MRI to image human subjects at 3 Tesla. Stroke. 2010;41(10 Suppl):S147–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zaiss M, Xu J, Goerke S, et al. Inverse Z-spectrum analysis for spillover-, MT-, and T1-corrected steady-state pulsed CEST-MRI–application to pH-weighted MRI of acute stroke. NMR Biomed. 2014;27(3):240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zaiss M, Kunz P, Goerke S, Radbruch A, Bachert P. MR imaging of protein folding in vitro employing nuclear-Overhauser-mediated saturation transfer. NMR Biomed. 2013;26(12):1815–1822. [DOI] [PubMed] [Google Scholar]
- 20. Goerke S, Zaiss M, Kunz P, et al. Signature of protein unfolding in chemical exchange saturation transfer imaging. NMR Biomed. 2015;28(7):906–913. [DOI] [PubMed] [Google Scholar]
- 21. Jiang S, Zou T, Eberhart CG, et al. Predicting IDH mutation status in grade II gliomas using amide proton transfer-weighted (APTw) MRI. Magn Reson Med. 2017;78(3):1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang S, Rui Q, Wang Y, et al. Discriminating MGMT promoter methylation status in patients with glioblastoma employing amide proton transfer-weighted MRI metrics. Eur Radiol. 2018;28(5):2115–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60(4):842–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Togao O, Yoshiura T, Keupp J, et al. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro Oncol. 2014;16(3):441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PC, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med. 2007;58(4):786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones CK, Huang A, Xu J, et al. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. Neuroimage. 2013;77:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zaiss M, Windschuh J, Paech D, et al. Relaxation-compensated CEST-MRI of the human brain at 7T: unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage. 2015;112:180–188. [DOI] [PubMed] [Google Scholar]
- 28. Zaiss M, Windschuh J, Goerke S, et al. Downfield-NOE-suppressed amide-CEST-MRI at 7 Tesla provides a unique contrast in human glioblastoma. Magn Reson Med. 2017;77(1):196–208. [DOI] [PubMed] [Google Scholar]
- 29. Deike K, Wiestler B, Graf M, et al. Prognostic value of combined visualization of MR diffusion and perfusion maps in glioblastoma. J Neurooncol. 2016;126(3):463–472. [DOI] [PubMed] [Google Scholar]
- 30. Nolden M, Zelzer S, Seitel A, et al. The Medical Imaging Interaction Toolkit: challenges and advances: 10 years of open-source development. Int J Comput Assist Radiol Surg. 2013;8(4):607–620. [DOI] [PubMed] [Google Scholar]
- 31. Windschuh J, Zaiss M, Meissner JE, et al. Correction of B1-inhomogeneities for relaxation-compensated CEST imaging at 7T. NMR Biomed. 2015;28(5):529–537. [DOI] [PubMed] [Google Scholar]
- 32. Schuenke P, Windschuh J, Roeloffs V, Ladd ME, Bachert P, Zaiss M. Simultaneous mapping of water shift and B1 (WASABI)-application to field-inhomogeneity correction of CEST MRI data. Magn Reson Med. 2017;77(2):571–580. [DOI] [PubMed] [Google Scholar]
- 33. Paech D, Zaiss M, Meissner JE, et al. Nuclear overhauser enhancement mediated chemical exchange saturation transfer imaging at 7 Tesla in glioblastoma patients. PLoS One. 2014;9(8):e104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heo HY, Jones CK, Hua J, et al. Whole-brain amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging in glioma patients using low-power steady-state pulsed chemical exchange saturation transfer (CEST) imaging at 7T. J Magn Reson Imaging. 2016;44(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paech D, Burth S, Windschuh J, et al. Nuclear overhauser enhancement imaging of glioblastoma at 7 Tesla: region specific correlation with apparent diffusion coefficient and histology. PLoS One. 2015;10(3):e0121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 37. The Cancer Genome Atlas Research N. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delfanti RL, Piccioni DE, Handwerker J, et al. Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol. 2017;135(3):601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan W, Xiong J, Huang W, Wu J, Zhan S, Geng D. Noninvasively detecting isocitrate dehydrogenase 1 gene status in astrocytoma by dynamic susceptibility contrast MRI. J Magn Reson Imaging. 2017;45(2):492–499. [DOI] [PubMed] [Google Scholar]
- 40. Leu K, Ott GA, Lai A, et al. Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II-III diffuse gliomas. J Neurooncol. 2017;134(1):177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bai Y, Lin Y, Zhang W, et al. Noninvasive amide proton transfer magnetic resonance imaging in evaluating the grading and cellularity of gliomas. Oncotarget. 2017;8(4):5834–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sakata A, Okada T, Yamamoto A, et al. Grading glial tumors with amide proton transfer MR imaging: different analytical approaches. J Neurooncol. 2015;122(2):339–348. [DOI] [PubMed] [Google Scholar]
- 43. Choi YS, Ahn SS, Lee SK, et al. Amide proton transfer imaging to discriminate between low- and high-grade gliomas: added value to apparent diffusion coefficient and relative cerebral blood volume. Eur Radiol. 2017;27(8):3181–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Togao O, Hiwatashi A, Yamashita K, et al. Grading diffuse gliomas without intense contrast enhancement by amide proton transfer MR imaging: comparisons with diffusion- and perfusion-weighted imaging. Eur Radiol. 2017;27(2):578–588. [DOI] [PubMed] [Google Scholar]
- 45. Kickingereder P, Bonekamp D, Nowosielski M, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. 2016;281(3):907–918. [DOI] [PubMed] [Google Scholar]
- 46. Hsieh KL, Chen CY, Lo CM. Radiomic model for predicting mutations in the isocitrate dehydrogenase gene in glioblastomas. Oncotarget. 2017;8(28):45888–45897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kickingereder P, Götz M, Muschelli J, et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22(23):5765–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xi YB, Guo F, Xu ZL, et al. Radiomics signature: a potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. 2018;47(5):1380–1387. [DOI] [PubMed] [Google Scholar]
- 49. Heo HY, Zhang Y, Jiang S, Lee DH, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semisolid magnetization transfer reference (EMR) signals: II. Comparison of three EMR models and application to human brain glioma at 3 Tesla. Magn Reson Med. 2016;75(4):1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu J, Yadav NN, Bar-Shir A, et al. Variable delay multi-pulse train for fast chemical exchange saturation transfer and relayed-nuclear overhauser enhancement MRI. Magn Reson Med. 2014;71(5):1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]