Abstract
Mesenchymal stem cells (MSC) have been derived from a variety of tissues, and cultured either in animal serum-containing (SC) or serum-free (SF) media. We have previously derived MSC from human embryonic stem cells via an intermediate trophoblast step (named EMSC), which also have immunosuppressive and therapeutic effects on animal models of autoimmune disease. To promote the clinical application of this new source of MSC, we report here EMSC derived and cultured in a SF medium MesenCult (SF-EMSC) in comparison with a SC medium (SC-EMSC). SF-EMSC derived in MesenCult also expressed typical MSC markers CD73, CD90, and CD105, and manifested multipotency to differentiate to osteocytes, chondrocytes, and adipocytes. Comparably, CD105+ cells reached 90% about one week slower in the SF than SC conditions, and the proliferation rate was slightly faster for SF-EMSC than SC-EMSC at later passages. Both SF- and SC-EMSC responded similarly to the inflammatory stimulus IFNγ. However, the inflammatory cytokines IL-6 and IL-8 were expressed much less in SF-EMSC than SC-EMSC. Furthermore, knockdown of P16INK4A in both SF- and SC-EMSC reduced replicative senescence. Together, our results suggest that EMSC can be generated in a complete SF condition, and SF-EMSC are largely similar to SC-EMSC. However, it takes longer time to derive EMSC in the SF than SC conditions, and the SF-EMSC proliferate faster at later passages and produce less of the inflammatory cytokines IL-6 and IL-8 than SC-EMSC. This study provides important information for production of clinically applicable EMSC.
Keywords: Serum, mesenchymal stem cells, human embryonic stem cells, derivation, and culture
Introduction
Mesenchymal stem cells (MSC) are a population of stromal cells with the capability of self-renewal and multipotent differentiation 1, 2. MSC are present in most, if not all, tissue types, and have been isolated from a variety of tissues such as amniotic membrane, umbilical cord, bone marrow and adipose 2. MSC can also be derived from pluripotent stem cells (PSC) including embryonic stem (ES) and induced pluripotent stem (iPS) cells 3-9. In addition to their potential for replacing damaged and diseased tissues by differentiating into tissue-specific cells or providing reparative and nutrient factors, human PSC-derived MSC have been shown to modulate functions of immune cells and exert disease-modifying efficacy in animal models of many autoimmune and inflammatory diseases 9-12. MSC-based clinical trials have been ongoing worldwide although the efficacy of MSC has not yet been conclusive.
Many culture media have been used for MSC isolation and expansion, and can be categorized into two types, i.e., serum-containing (SC) and serum-free (SF). Almost all sera used for mammalian cell culture are derived from animals. Xenogeneic and undefined components contained in animal sera cause not only complexity and uncertainty when used for studying MSC, but also safety concerns for their clinical application. To overcome this problem, many SF media have been developed. Among them, MesenCult medium is not only SF, but also xeno-free (free of animal products) and contains only defined components.
The methods for MSC derivation from human ES cells (hESC) or iPS cells include direct differentiation, embryoid body formation, co-culturing with mouse OP9 cells, sorting, scraping or handpicking of cells 3-8. Although various advantages are associated with some of the methods, they are all limited by low efficiency and tedious procedures, and all depend on use of SC media. We have recently found that MSC can be derived via an intermediate step - trophoblast formation (named EMSC) in monolayer 9 or a 3-dimentional method starting from hESC spheres 13. The first step of EMSC derivation (trophoblast differentiation from hESC) is induced by BMP4 and A83-01 (an inhibitor of TGFβ signaling) in the commercially available SF medium, mTeSR Minus Select Factors, in which lithium chloride, bFGF, and TGFβ were removed 14, 15, however the second step (MSC differentiation from the trophoblasts) has to be completed in a SC medium (called MSC medium).
Replicative senescence is an irreversible process that leads to the loss of proliferative potential and permanent growth arrest of any differentiated or partially differentiated cells including MSC 16. It has been well known that P16INK4A (P16) plays an important role in the regulation of proliferation and senescence of human MSC. More senescent cells and higher expression of P16INK4A have been found in human bone marrow MSC (BM-MSC) at high passages than low passages, and inhibition of P16 delays senescence of human BM-MSC 17.
This study was attempted to remove this hurdle by generating EMSC in a completely SF medium such as MesenCult. We found that MesenCult supported the derivation and proliferation of EMSC. EMSC derived in the SF medium (SF-EMSC) used longer time for derivation, proliferated faster after derivation, and expressed less the inflammatory cytokines IL-6 and IL-8 than EMSC derived in the serum-containing (SC) condition (SC-EMSC). P16 was involved in the proliferation and senescence of both SF- and SC-EMSC.
Results
Generation of EMSC in SF media
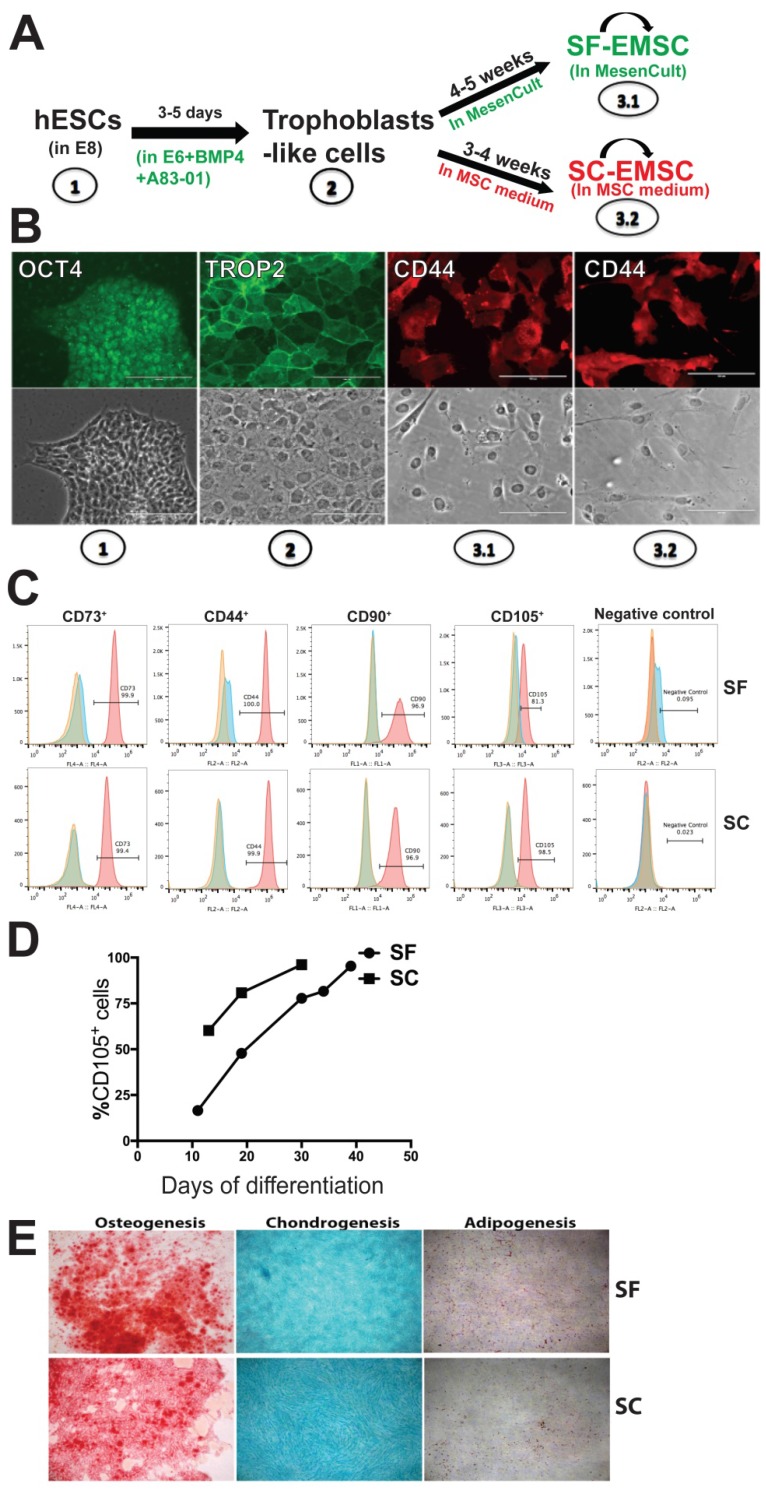
Human ESC lines H9 and CT3 were used in this study 18. SF- and SC-EMSC were derived from hESC as shown in a schematic (Fig. 1A). hESC were routinely propagated in E8 (Stemcell Technologies), which is a SF, xeno-free, albumin-free, and all-defined medium 19. We first split hESC with small colonies and cultured the cells in E8. On day 2-3, consumed medium was removed and cells were induced to differentiate into trophoblasts via treatment with 10-ng/ml BMP4 and 1-μM A83-01 in E6, which lacked FGF2 and TGFβ1 compared to E8, as we reported previously except that mTeSR Minus Select Factors was replaced with E6 9. Within 3-5 days, hESC uniformly became flattened and acquired trophoblast-like cell morphology and positive for the trophoblast marker TROP2, while untreated hESC remaining positive for the pluripotency marker OCT4 (Fig. 1B).
Figure 1.
MSC generated from H9 hESC in SF media. (A) Schematic of the generation of SF-EMSC and SC-EMSC. (B) Representative micrographs showing the typical immunostaining for OCT4, TROP2 and CD44 during the differentiation of hESC to EMSC. Bars, 100 μm. (C) The phenotypes of passage-1 SF- and SC-EMSC were analyzed via flow cytometry. The percentage of each indicated cell surface marker was shown on each plot. (D) The percentage of CD105+ cells during hESC differentiation to EMSC in either SC- and SF condition. (E) Tri-lineage differentiation potentials of SF- and SC-EMSC were detected by culturing EMSC in appropriate differentiation induction media. Osteocytes, chondrocytes and adipocytes were detected via Alizarin Red, Alcian Blue, and oil red O staining, respectively.
On day 5, further differentiation from trophoblast-like cells to MSC was initiated by replacing the spent medium with either MesenCult (SF) or the MSC medium (SC), i.e., αMEM supplemented with 20% fetal bovine serum and 1-mM L-glutamine. The medium was refreshed every 2-3 days and mesenchymal-like cells emerged in 1-2 and 2-3 weeks in the SC and SF medium, respectively. By day 19 (in SC) or 35 (in SF), over 90% of the cells were stained positive for the MSC marker CD44, CD73, CD90, and CD105 (with only CD44 shown in Fig. 1B), which was confirmed via flow cytometry (Fig. 1C). However, the cells were negative for a cocktail of hematopoietic markers (Fig. 1C). The mesenchymal-like cells were split weekly and finally became MSC with homogeneous phenotypes within 3-4 (for SC) or 4-5 (for SF) weeks, and are named SC- and SF-EMSC, respectively (Fig. 1A). CD105 appeared slower during generation of SF-EMSC than SC-EMSC, especially at the early times (Fig. 1D). MesenCult and MSC medium were continuously used to maintain SF- and SC-EMSC, respectively.
To simplify the differentiation system, we also tried to use MesenCult, instead of E6, to differentiate hESC into trophoblasts in the presence of BMP4 and A83-01. However, cells in this condition quickly detached and died. Thus, MesenCult could not replace E6 in the initial step of the MSC derivation. Furthermore, we confirmed the multipotency of SF- and SC-EMSC continuously cultured in the corresponding medium and found both could differentiate into osteocytes, chondrocytes or adipocytes within two weeks or so (Fig. 1E). Therefore, both SC- and SF-EMSC meet the minimal standards for MSC 20. EMSC were also derived from another hESC line CT3 in either SF- or SC condition, and expressed the typical MSC markers and demonstrated the capability of the tri-lineage differentiation (Fig. S1).
Proliferation of EMSC in the SF- and SC-media
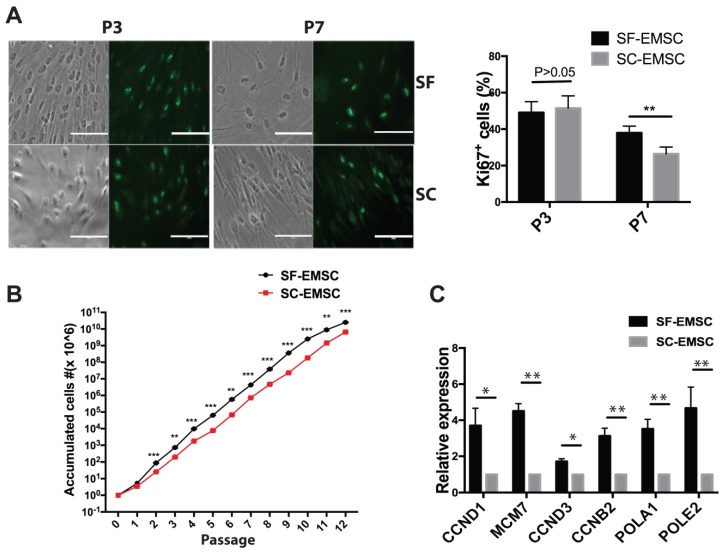
Next, we evaluated the proliferation rate of both SF- and SC-EMSC by detecting Ki67, a marker for cell division, by immunostaining. As shown in Fig. 2A, the ratio of Ki67+ cells were 49.1±2.2% (p3) and 38.0±1.3% (p7) for SF-EMSC and 51.5±2.6% (p3) and 26.4±1.4% (p7) for SC-EMSC, respectively. The proliferation capacity of SF- and SC-EMSC was similar to each other at earlier passage (p3). However, the proliferation of SC-EMSC started to slow down at p7, while SF-EMSC still kept high proliferation capacity. SF- and SC-EMSC could continuously expand in the corresponding medium for up to 12 passages exponentially with accumulated cell number increased about 1,012- and 1,010-fold in the SF and SC groups, respectively (Fig. 2B). QPCR results showed that the expression of cell cycle related genes (CCND1, MCM7, CCND3 and CCNB2) and DNA replication related genes (POLA1 and POLE2) were higher in SF-EMSC than SC-EMSC (Fig. 2C). Thus, SF-EMSC proliferated faster than SC-EMSC in later passage. After p15 or so, the proliferation of both SF- and SC-EMSC remarkably declined (data not shown).
Figure 2.
Analyses of proliferation of SF- and SC-EMSC at various passages. (A) Immunofluorescent staining analyzed Ki67 expression in both SF- and SC-EMSC at passage 3 and 7. Bars, 100 μm. **P < 0.01. (B) The total number of cells recovered at each subsequent passage was calculated and plotted. **P < 0.01, ***P < 0.001. (C) The expression of cell cycle related genes and DNA replication related genes were analyzed by qPCR. Gene expression was normalized to the value for GAPDH.
Replicative senescence of SF-EMSC
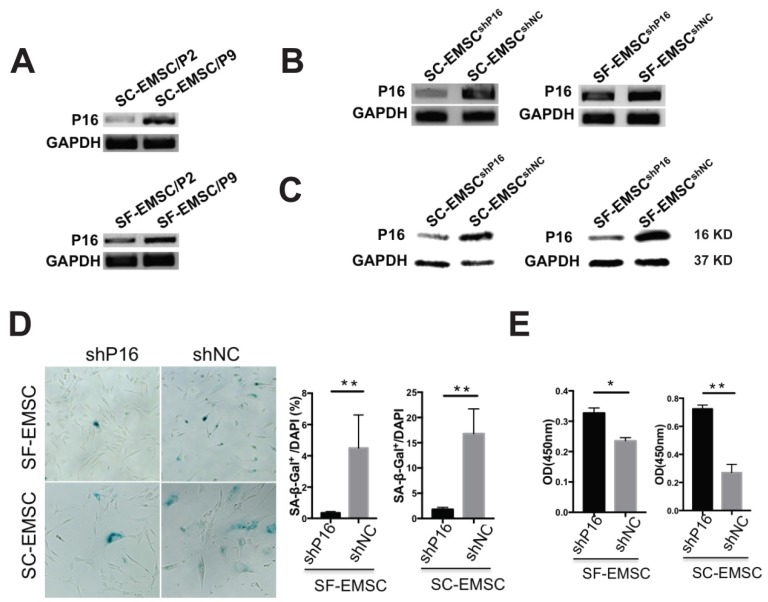
Since replicative senescence develops during passaging of MSC, we compared the development of replicative senescence in both SF- and SC-EMSC. We found that P16 expression was higher in both SF- and SC-EMSC at a high passage (p9) than a low passage (p2) (Fig. 3A). Therefore, we decided to test whether knockdown of P16 can rescue EMSC from senescence. We established stable SC- and SF-EMSC lines with P16 knockdown using a lentiviral vector to co-express green fluorescent protein (GFP) and P16 shRNA (shP16) or a scrambled shRNA as a negative control (shNC). GFP+ cells were sorted and used for the following analyses. P16 expression was remarkably reduced in both SC-EMSC and SF-EMSC with shP16 than shNC as detected via RT-PCR (Fig. 3B), and confirmed via Western blotting (Fig. 3C). On the other hand, senescence-associated β-galactosidase (SA-β-gal) staining showed that P16 knockdown reduced cellular senescence (Fig. 3D) and promoted proliferation (Fig.3E) of both SC-EMSC and SF-EMSC.
Figure 3.
Knockdown of P16 rescued SF- and SC-EMSC from senescence. (A) P16 mRNA expression in an indicated passage of SF- and SC-EMSC was detected by RT-PCR. GAPDH was used as a loading control. (B) SF- and SC-EMSC were transduced with lentivirus co-expressing GFP and shP16 or shNC, GFP+ cells were sorted and tested for P16 expression via RT-PCR. GAPDH was used as a loading control. (C) P16 protein in GFP+ SF- and SC-EMSC was detected via western blotting. GAPDH was used as a loading control. (D) The senescence of GFP+ SF- and SC-EMSC were detected by senescence-associated beta-galactosidase assays. Total cell number was counted by DAPI staining. (E) Cell proliferation was evaluated GFP+ SF- and SC-EMSC using the CCK-8 assay. *P < 0.05, **P < 0.01.
Immunomodulatory cytokines generated by SF- and SC-EMSC
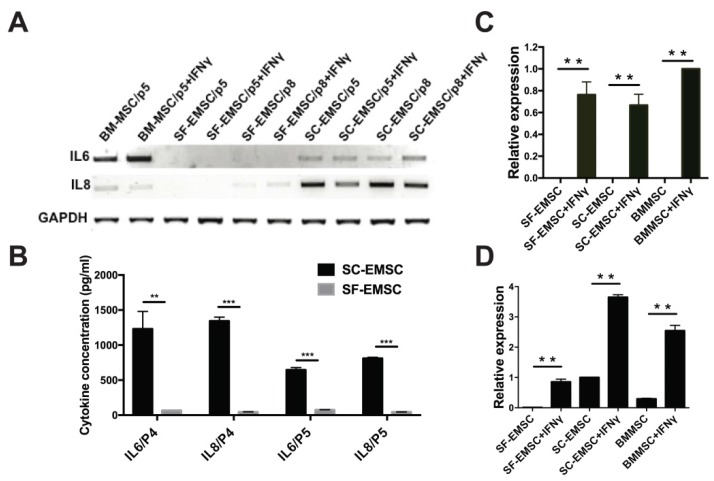
Cytokines including interferons, tumor necrosis factor, and interleukins play critical role in mediating MSC effects 21. We have previously observed that MSC derived from hESC via other methods had higher secretion of the anti-inflammatory cytokine IL-8 and lower secretion of the pro-inflammatory cytokine IL-6 than BM-MSC 9, 22. Using RT-PCR (Fig. 4A) and cytokine bead array (CBA) (Fig. 4B), we found here that the basal levels of both IL-6 and IL-8 secreted by SF-EMSC were much lower than SC-EMSC. It has been known that priming of MSC with pro-inflammatory cytokines such as IFNγ can change the profile of cytokine expression and enhance the immunosuppressive effects of the cells 23. Upon stimulation with IFNγ, the expression of IL-6 was increased and IL-8 was decreased in BM-MSC; IL-6 and IL-8 were not changed in SF-EMSC; IL-8 was reduced in SC-EMSC but IL-6 was not changed. It has been reported that immunosuppression by human- or monkey-derived MSC is mediated by indoleamine 2,3-dioxygenase (INDO or IDO) and PDL1 24. IDO (Fig. 4C) and PDL1 (Fig. 4D) expression dramatically increased in both SC- and SF-EMSC following IFNγ treatment. These data suggest that both SC- and SF-EMSC can generate immunomodulatory cytokines in response to the potent immunological stimulus IFNγ.
Figure 4.
Comparison of inflammatory cytokine expression between SF- and SC-EMSC. (A) Gene expression of IL-6 and IL8 in SF- and SC-EMSC were detected by RT-PCR. GAPDH was used as loading control. (B) IL-6 and IL-8 protein were analyzed by Cytometric Bead Array. (C and D) The expression of IDO1 (C) and PDL1 (D) in EMSC with or without IFNγ treatment was detected via qPCR. Gene expression was normalized to GAPDH. **P < 0.01, ***P < 0.001.
Discussion
MSC derived from hPSC share great similarities with MSC obtained from other sources. As established pluripotent cell lines, hPSC also have unique advantages. They are an unlimited source for generation of MSC and many other cell types, easy for quality control for pathogen-free and standardized production, and appropriate for genetic manipulation, for example, to reduce immunogenicity and tumorigenicity 25. As a result, hPSC-derived MSC are potentially a better candidate than other sources for clinical applications to treat many degenerative and autoimmune diseases. However, in order to use these cells in clinical settings, it is mandatory to establish a consistently reproducible protocol for large-scale production and define a standard operating procedure compliable with good manufacturing practice (GMP) including selection of an optimal cell therapy-compliant culture medium. We have recently reported that MSC can be derived from hESC via a trophoblast-like stage and expanded to large scale in a reasonable time without altering their basic characteristics 9. Although the differentiation into trophoblasts was conducted in a SF medium, the subsequent differentiation into MSC and expansion of the resultant MSC were in a SC medium, thus not ideal for GMP-compliable generation of clinical-grade of MSC.
Here we have attempted to develop a completely SF system to generate and expand EMSC. We first differentiated hESC into trophoblast-like cells in the SF medium E6 supplemented with BMP4 and A83-01, and then differentiated the trophoblast-like cells into MSC in another SF medium MesenCult, then maintained and expanded the SF-EMSC in MesenCult. It took a bit longer time (4-7 days) to establish SF-EMSC than SC-EMSC due to slower appearance of CD105. SF- and SC-EMSC proliferated at similar rates up to 15 passages and then SF-EMSC proliferated slightly faster than SC-EMSC. Knockdown of P16INK4A rescued senescence and promoted proliferation in both SF-EMSC and SC-EMSC. Similar results were observed with another SF medium StemPro (data not shown).
MesenCult was designated to help MSC researchers to standardize their culture systems and reduce variability of MSC cultures. Although it was originally developed for isolation and expansion of human BM-MSC 26, it has also been used for isolation and expansion of MSC from many other sources such as human umbilical cord blood 27, 28, Wharton's jelly 29, adipose 30, corneal limbus 31, and even teratomas formed by hESC 32. The cell morphology, proliferation rate, senescence development, differentiation capability, immunosuppressive effect, cytogenetic stability, tumorigenicity, etc., of MSC cultured in different media have been carefully analyzed. Given the general recognition of the capability of MesenCult (as well as some other SF media) to support isolation and expansion of MSC from various sources, controversial observations have been reported between MSC cultured in the SF media versus conventional SC media.
For example, MSC derived from bone marrow 26, umbilical cord blood (UCB) 27, Wharton's jelly 29, and adipose 30 all have higher proliferation rate and yield in MesenCult than SC media. However, Gottipamula, et al., 33 found that BM-MSC cultured in SF media including MesenCult and the BD Mosaic Mesenchymal Stem Cell Serum-Free Medium (BD-SFM) grow much slower than those in a control SC medium. Comparably, we found that EMSC had similar and steady proliferation up to p15 in both the SC and SF media (faster in the SF medium than the SC medium at later passage), with a short PD time at around 20 h (Fig. 2B). Post-thaw viability of BM-MSC derived from both the SF and SC media were found to be above 90% 33. We also observed over 90% post-thaw viability of EMSC that had been cultured in either the SF or SC medium and frozen at various passages (data not shown).
Furthermore, IFN-γ treatment increases the expression level of the immunosuppressive molecule IDO in BM-MSC cultured in either the SC or SF conditions 33, which is consistent with our observations with EMSC.
In summary, we have demonstrated an array of common and different features between EMSC derived and maintained in SC and SF conditions. Both conditions allowed derivation of EMSC. However, it took a bit longer time to derive SF-EMSC than SC-EMSC, and SF-EMSC proliferated faster at later passages and produced less of the inflammatory cytokines IL-6 and IL-8 than SC-EMSC. These differences might be relevant to differential expression of some genes essential for development of inflammation, senescence and immunosuppression between SF- and SC-EMSC. The exact mechanism regarding the culture-related differences awaits further studies.
Methods
Culture of hESC and Generation of EMSC in SF media
Under approval of the Research Ethics Panel of University of Macau, hESC lines H9 and CT3 were used in this study. hESC were split into a 6 well plate coated with Matrigel (BD Bioscience) at about 5 x 104 cells/well in E8 medium 19 (Stemcell Technologies) at 37 °C and 5% CO2, and cultured overnight. For trophoblast differentiation, the spent media was removed, each well was washed with PBS twice, and E6 (E8 minus FGF2 and TGFβ1) (Stemcell Technologies) was added at 2 ml/well together with 10 ng/ml BMP4 and 1 μM A83-01 (Stemgent). Cells were cultured at 37 °C and 5% CO2, and monitored under microscope every day.
At day 3-5, cells were washed with PBS. Without passaging, the cells were directly cultured in MesenCult (Stemcell Technologies) or MSC medium, i.e., αMEM medium supplemented with 20% fetal bovine serum (GIBCO), L-glutamine (GIBCO) and 1x non-essential amino acids (GIBCO) at 2 ml/well. The medium was refreshed every two days and the cells were checked for expression of the MSC markers CD73, CD44, CD90, and CD105 periodically. When over 90% of the cells became positive for all the four markers, we designated the cells as passage-1 EMSC. The cells were split when they reached 80-90% confluence in each passage, which took about every 5-7 days.
Flow cytometry for cell surface markers
Flow cytometry staining was performed using standard methods and antibodies against TROP2, CD31, CD34, CD73, CD90, CD105, and CD44 (BD Bioscience). Data were collected on Accuri C6 flow cytometer (BD Bioscience). Data analysis was performed with Accuri C6 software.
Immunocytochemistry
Immunocytochemistry was performed as described before 34 using anti-OCT4 (sc-5279, Santa Cruz Biotechnology) and anti-TROP2 (BD Bioscience) antibodies. Cell nuclei were counterstained with DAPI.
Tri-lineage Differentiation of EMSC
EMSC were differentiated into osteogenic, adipogenic and chondrogenic lineages using StemPro differentiation kits (Life Technologies), according to the manufacturer's protocols.
Cytometric Bead Array (CBA)
Spent medium from cultures of EMSC with or without IFNγ treatment was collected and subjected to CBA assay to analyze levels of multiple cytokines in the medium using CBA Flex Sets (BD Bioscience), according to the manufacturer's instructions. The concentration of each cytokine was calculated based on standard curves generated from serial dilutions of cytokine standards provided by the manufacturer.
β-Galactosidase staining for senescent cells
The Senescence β-Galactosidase Staining Kit (Cell Signaling Technology) was used according to the manufacturer's instructions. Briefly, following removal of culture medium, cells were rinsed one time with PBS, and fixed with 1x Fixative Solution (1 ml/well) for 10-15 min at room temperature. Following removal of the Fixative Solution, cells were rinsed twice with PBS, and treated with the β-Galactosidase Staining Solution (2 ml/well). The plate was sealed with parafilm to prevent evaporation, and incubated at 37°C overnight in a dry incubator (without CO2). While the β-Galactosidase Staining Solution was still on the plate, the cells were checked under a microscope for the development of blue color. Cell nuclei were counterstained with DAPI.
RT-PCR and qPCR
RNA was isolated from cells using TRIzol reagent (Invitrogen), and cDNA was synthesized from RNA using Superscript II (Invitrogen), according to the manufacturer's instructions. Gene expression was assessed through PCR and qPCR with primers for specific genes (Table S1). QPCR was performed using the CFX96 Real-Time System (BioRad). The relative amount of each mRNA to GAPDH was calculated using the 2-ΔΔCt method.
shRNA-mediated P16 Knockdown
To generate the pLVTHM-shP16 vector, we cloned oligonucleotides sequences targeting ACC AGA GGC AGT AAC CAT G into the MluI and ClaI sites of the lentiviral vector pLVTHM that expressed GFP. As a negative control, the pLVTHM-shNC vector was generated using a scramble sequence CGG AGG CTT ACA GTC TGG T cloned into the MluI and ClaI sites of pLVTHM. Either pLVTHM-shP16 or pLVTHM-shNC was co-transfected together with Delta R 8.74 and pMDG-VSVG to 293T cells with X-tremeGENE HP DNA Transfection Reagent (Roche) to package the lentiviruses. The lentiviruses were then transduced into SC-EMSC or SF-EMSC. GFP+ positive cells were sorted via flow cytometry BD FACSARIA III (BD Bioscience) to establish stably transduced EMSC lines.
Western blot
Cellular total proteins were extracted using RIPA buffer supplemented with proteinase inhibitor (Thermo Fisher Scientific). Proteins were separated on 12% SDS-PAGE gels and electroblotted onto PVDF membrane. The membranes were incubated with antibodies against P16 (BD Bioscience) and GAPDH (SANTA CRUZ). The membranes were then blotted with HRP-conjugated secondary antibodies.
CCK-8 assay
For CCK-8 assay, SF-EMSC or SC-EMSC was plated into 96-well plates. Three days later, the cells were incubated in a CCK-8 solution for 2 h. The amount of formazan dye was measured using absorbance at 450 nm with a microplate reader.
Statistical analyses
Data are presented as mean ± SD. Student's t test was used for statistical analysis. Percentage data was arcsine transformed prior to analysis. P < 0.05 was considered significant, and P < 0.01 highly significant.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by University of Macau Research Committee funds MYRG #2015-00169-FHS, #2016-00070-FHS, and #2017-00124-FHS, and Macau Science and Technology Development Fund (FDCT) #028/2015/A1 and #095/2017/A1 to R.X.
Author contributions
E.L., and R.-H.X. conceived and designed the research. E.L., Z.Z., B.J., and L.Y. performed the experiments. E.L., Z.Z., and R.-H.X. analyzed the data and wrote the manuscript. J.P. helped revise the manuscript. R.X gave the final approval of the manuscript.
References
- 1.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD. et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2:e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivier EN, Rybicki AC, Bouhassira EE. Differentiation of human embryonic stem cells into bipotent mesenchymal stem cells. Stem Cells. 2006;24:1914–22. doi: 10.1634/stemcells.2005-0648. [DOI] [PubMed] [Google Scholar]
- 5.Brown SE, Tong W, Krebsbach PH. The derivation of mesenchymal stem cells from human embryonic stem cells. Cells Tissues Organs. 2009;189:256–60. doi: 10.1159/000151746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang NS, Varghese S, Lee HJ, Zhang Z, Ye Z, Bae J. et al. In vivo commitment and functional tissue regeneration using human embryonic stem cell-derived mesenchymal cells. Proc Natl Acad Sci U S A. 2008;105:20641–6. doi: 10.1073/pnas.0809680106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruenloh W, Kambal A, Sondergaard C, McGee J, Nacey C, Kalomoiris S. et al. Characterization and in vivo testing of mesenchymal stem cells derived from human embryonic stem cells. Tissue Eng Part A. 2011;17:1517–25. doi: 10.1089/ten.tea.2010.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vodyanik MA, Yu J, Zhang X, Tian S, Stewart R, Thomson JA. et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell stem cell. 2010;7:718–29. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Lazorchak AS, Song L, Li E, Zhang Z, Jiang B. et al. Immune modulatory mesenchymal stem cells derived from human embryonic stem cells through a trophoblast-like stage. Stem Cells. 2016;34:380–91. doi: 10.1002/stem.2242. [DOI] [PubMed] [Google Scholar]
- 10.Kimbrel EA, Kouris NA, Yavanian GJ, Chu J, Qin Y, Chan A. et al. Mesenchymal stem cell population derived from human pluripotent stem cells displays potent immunomodulatory and therapeutic properties. Stem Cells Dev. 2014;23:1611–24. doi: 10.1089/scd.2013.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan L, Jiang B, Niu Y, Wang H, Li E, Yan Y. et al. Intrathecal delivery of human ESC-derived mesenchymal stem cell spheres promotes recovery of a primate multiple sclerosis model. Cell Death Discovery. doi: 10.1038/s41420-018-0091-0. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao H, Wang H, Rong X, Li E, Xu RH, Peng Y. Mesenchymal Stem Cells Attenuate Radiation-Induced Brain Injury by Inhibiting Microglia Pyroptosis. Biomed Res Int. 2017;2017:1948985. doi: 10.1155/2017/1948985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan L, Jiang B, Li E, Wang X, Ling Q, Zheng D. et al. Scalable Generation of Mesenchymal Stem Cells from Human Embryonic Stem Cells in 3D. Int J Biol Sci. 2018;14:5. doi: 10.7150/ijbs.25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C. et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature biotechnology. 2002;20:1261–4. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 15.Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N. et al. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. The Journal of biological chemistry. 2008;283:24991–5002. doi: 10.1074/jbc.M803893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata KR, Aoyama T, Shima Y, Fukiage K, Otsuka S, Furu M. et al. Expression of the p16INK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem Cells. 2007;25:2371–82. doi: 10.1634/stemcells.2007-0225. [DOI] [PubMed] [Google Scholar]
- 17.Shibata KR, Aoyama T, Shima Y, Fukiage K, Otsuka S, Furu M. et al. Expression of the p161NK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem cells. 2007;25:2371–82. doi: 10.1634/stemcells.2007-0225. [DOI] [PubMed] [Google Scholar]
- 18.Lin G, Martins-Taylor K, Xu RH. Human embryonic stem cell derivation, maintenance, and differentiation to trophoblast. Methods in molecular biology. 2010;636:1–24. doi: 10.1007/978-1-60761-691-7_1. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD. et al. Chemically defined conditions for human iPSC derivation and culture. Nature methods. 2011;8:424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 21.Yan L, Zheng D, Xu RH. Critical Role of Tumor Necrosis Factor Signaling in Mesenchymal Stem Cell-Based Therapy for Autoimmune and Inflammatory Diseases. Front Immunol. 2018;9:1658. doi: 10.3389/fimmu.2018.01658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Kimbrel EA, Ijichi K, Paul D, Lazorchak AS, Chu J. et al. Human ESC-derived MSCs outperform bone marrow MSCs in the treatment of an EAE model of multiple sclerosis. Stem Cell Reports. 2014;3:115–30. doi: 10.1016/j.stemcr.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Su J, Roberts AI, Shou P, Rabson AB, Ren G. How mesenchymal stem cells interact with tissue immune responses. Trends Immunol. 2012;33:136–43. doi: 10.1016/j.it.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren G, Su J, Zhang L, Zhao X, Ling W, L'Huillie A. et al. Species variation in the mechanisms of mesenchymal stem cell-mediated immunosuppression. Stem Cells. 2009;27:1954–62. doi: 10.1002/stem.118. [DOI] [PubMed] [Google Scholar]
- 25.Zheng D, Wang X, Xu RH. Concise Review: One Stone for Multiple Birds: Generating Universally Compatible Human Embryonic Stem Cells. Stem Cells. 2016;34:2269–75. doi: 10.1002/stem.2407. [DOI] [PubMed] [Google Scholar]
- 26.Miwa H, Hashimoto Y, Tensho K, Wakitani S, Takagi M. Xeno-free proliferation of human bone marrow mesenchymal stem cells. Cytotechnology. 2012;64:301–8. doi: 10.1007/s10616-011-9400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain I, Magd SA, Eremin O, El-Sheemy M. New approach to isolate mesenchymal stem cell (MSC) from human umbilical cord blood. Cell Biol Int. 2012;36:595–600. doi: 10.1042/CBI20110336. [DOI] [PubMed] [Google Scholar]
- 28.Vasaghi A, Dehghani A, Khademalhosseini Z, Khosravi Maharlooei M, Monabati A, Attar A. Parameters that influence the isolation of multipotent mesenchymal stromal cells from human umbilical cord blood. Hematol Oncol Stem Cell Ther. 2013;6:1–8. doi: 10.1016/j.hemonc.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Swamynathan P, Venugopal P, Kannan S, Thej C, Kolkundar U, Bhagwat S. et al. Are serum-free and xeno-free culture conditions ideal for large scale clinical grade expansion of Wharton's jelly derived mesenchymal stem cells? A comparative study. Stem cell research & therapy. 2014;5:88. doi: 10.1186/scrt477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Saqi SH, Saliem M, Asikainen S, Quezada HC, Ekblad A, Hovatta O. et al. Defined serum-free media for in vitro expansion of adipose-derived mesenchymal stem cells. Cytotherapy. 2014;16:915–26. doi: 10.1016/j.jcyt.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Bray LJ, Heazlewood CF, Munster DJ, Hutmacher DW, Atkinson K, Harkin DG. Immunosuppressive properties of mesenchymal stromal cell cultures derived from the limbus of human and rabbit corneas. Cytotherapy. 2014;16:64–73. doi: 10.1016/j.jcyt.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 32.An SM, Zeng Q, Teng XY, Long ZG, Li J, Pan Q. et al. [Growing of human embryonic stem cells on feeders derived from themselves] Yi Chuan. 2008;30:1567–73. doi: 10.3724/sp.j.1005.2008.01567. [DOI] [PubMed] [Google Scholar]
- 33.Gottipamula S, Ashwin KM, Muttigi MS, Kannan S, Kolkundkar U, Seetharam RN. Isolation, expansion and characterization of bone marrow-derived mesenchymal stromal cells in serum-free conditions. Cell Tissue Res. 2014;356:123–35. doi: 10.1007/s00441-013-1783-7. [DOI] [PubMed] [Google Scholar]
- 34.Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW. et al. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS ONE. 2010;5:e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.