Summary.
Background:
The human genome encodes two paralogs of vitamin-K-epoxide reductase, VKORC1 and VKORL1, that support blood coagulation and other vitamin-K-dependent processes. Warfarin inhibits both enzymes, but VKORL1 is relatively resistant to warfarin.
Objectives:
To understand the difference between VKORL1 and VKORC1, and the cause of warfarin-resistant (WR) mutations in VKORC1.
Methods:
We performed systematic mutagenesis and analyzed warfarin responses with a cell-based activity assay. Mass spectrometry analyses were used to detect cellular redox state.
Results:
VKORC1 and VKORL1 adopt a similar intracellular redox state with four-transmembrane-helix topology. Most WR mutations identified in VKORC1 also confer resistance in VKORL1, indicating that warfarin inhibits these paralogs at a common binding site. A group of WR mutations, distant from the warfarin-binding site, show significantly less resistance in VKORL1 than in VKORC1, implying that their different warfarin responses are determined by peripheral interactions. Remarkably, we identify a critical peripheral region in which single mutations, Glu37Lys or His46Tyr, drastically increase the warfarin sensitivity of VKORL1. In the background of these warfarin-sensitive VKORL1 mutants, WR mutations showing relative less resistance in wild-type VKORL1 become much more resistant, suggesting a structural conversion to resemble VKORC1. At this peripheral region, we also identified a human SNP that confers warfarin sensitivity of VKORL1.
Conclusions:
Peripheral regions of VKORC1 and VKORL1 primarily maintain the stability of their common warfarin-binding pocket, and differences of such interactions determine their relative sensitivity to warfarin inhibition. This new model also explains most WR mutations located at the peripheral regions of VKORC1.
Keywords: Blood Coagulation, Drug Resistance, Membrane proteins, Vitamin K, Vitamin K Epoxide Reductases, Warfarin
Introduction
Warfarin is an oral anticoagulant taken by ~1% of the US population to treat and prevent deep vein thrombosis, pulmonary embolism, stroke, and myocardial infarction. Warfarin targets vitamin K epoxide reductase in the vitamin K cycle [1–3], which supports the activity of vitamin-K-dependent proteins including several coagulation factors. The cycle begins with the γ-carboxylation of selected glutamic acids in these proteins, a post-translational modification that allows their membrane association and activation. The γ-carboxylase activity is driven by the epoxidation of the vitamin K hydroquinone, which is regenerated by vitamin K epoxide reductases to complete the vitamin K cycle.
The epoxide reductase activity has been identified for two paralogs found in the human genome, VKORC1 and VKORL1, which share ~74% similarity (48% identity) in their protein sequences [3–7] (Fig. 1). VKORC1 is highly expressed in liver where coagulation factors are produced, whereas VKORL1 is more abundant in extrahepatic tissues where other vitamin-K-dependent proteins, such as osteocalcin and matrix Gla protein, are made [8,9]. Compared to VKORC1, VKORL1 shows 30–50-fold more resistance to warfarin in an in vitro assay [8]. Thus, at a warfarin dose administered to inhibit VKORC1-supported blood coagulation, the VKORL1 activity is largely retained, possibly explaining the small effects that short-term warfarin administration usually has against the bone mineralization and the inhibition of vascular calcification [8,9]. The mechanism underlying the relative resistance of VKORL1 to warfarin inhibition, however, is unclear.
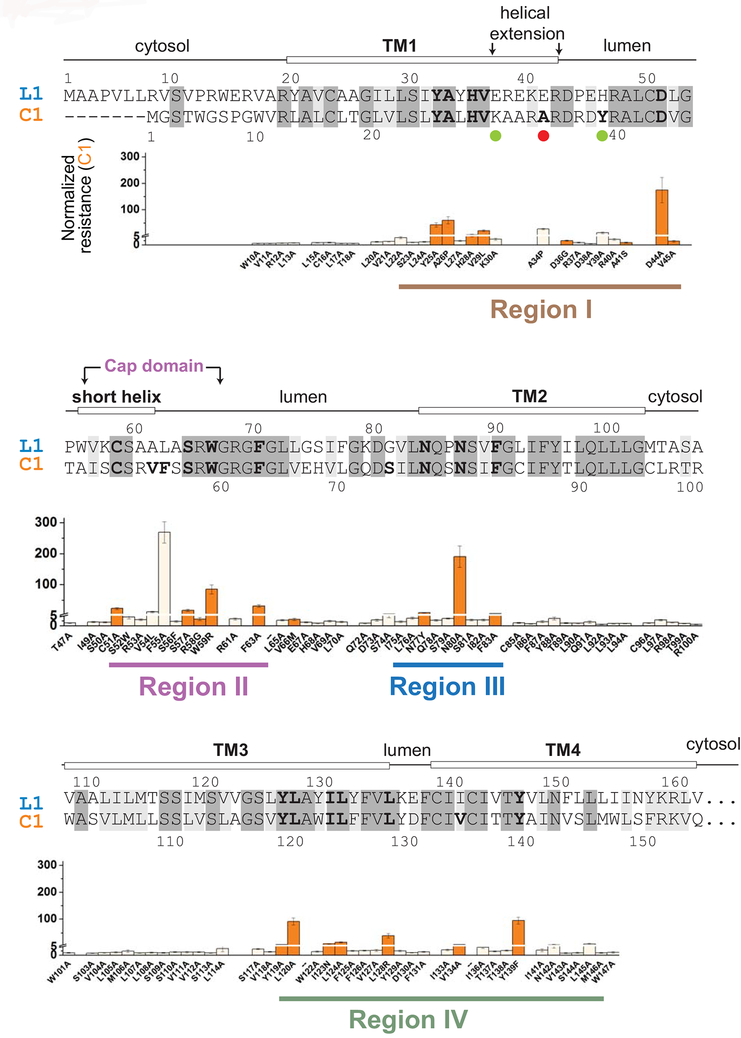
Figure 1. Mapping of WR mutations in VKORC1 with sequence alignment of VKORC1 (C1) and VKORL1 (L1).
Identical residues (48%) are shadowed in dark grey, and similar residues (26%) in grey. Prediction of secondary structures (top) are based on the crystal structure of a bacterial VKOR homolog [22]. Regions I–IV are identified (bottom) based on the distribution of WR mutations in VKORC1. The underlying panels show the resistant level of WRs in VKORC1 [15], with a Y-axis break at NRwar = 5. Residues with NRwar > 5 mutations are shown in bold letters in the sequence alignment. The orange-colored WR mutations in VKORC1 are selected for mutagenesis analysis of the corresponding residues in VKORL1 (Fig. 3A–C). Matching mutations (Fig. 4B) and the human SNP (Fig. 6) that increase the warfarin sensitivity of VKORL1 are indicated by green and red dots, respectively.
Warfarin resistance (WR) is a well-known phenomenon for VKORC1, caused by mutations in patients requiring much higher warfarin dosage [4,10,11]. The WR mutants of VKORC1 are also found in rodents, after decades of usage of warfarin-derived pesticides [12–14]. Moreover, alanine mutagenesis scan of human VKORC1 has identified many novel WR mutations [15]. Although numerous mutational sites are known to cause warfarin resistance in VKORC1 (Fig. 1), WR mutations have not been reported in VKORL1.
Understanding warfarin resistance in VKORC1 and VKORL1 requires understanding their folding topology. We recently provided an unequivocal demonstration that native human VKORC1 adopts a conformation with four transmembrane helices (TM) [15] and settled earlier debates about its membrane topology [6,16–20]. In this four-TM conformation, all of the WR mutations cluster at the luminal portion of VKORC1, whereas these mutations cannot be sensibly interpreted if VKORC1 is a three-TM protein [21,22]. VKORL1 is also a four-TM protein, with the same active site location as VKORC1 [3,6,8,23]. This common four-TM topology is consistent with the crystal structures of a bacterial VKORC1 homolog [22,24]. The homolog structure suggests that warfarin binds at the active sites of VKORL1 and VKORC1 and shows that many WR mutations are located near the VKORC1 active site [22,25]. This model, however, cannot explain a group of strongly resistant mutations (e.g., Asp44Ala) that are located in peripheral regions distant from the active site.
Here we identify a peripheral region that determines the different sensitivity of VKORC1 and VKORL1 to warfarin inhibition. Substitution of a single residue in VKORL1 to match the VKORC1 sequence in this region increases the warfarin sensitivity of VKORL1 to a similar level as that of VKORC1. This critical region maintains the structural stability of the common warfarin-binding pocket, explaining the effects of WR mutations found in the peripheral region of VKORC1.
Materials and Methods
Plasmids and stable cell lines
For the cell-based activity assay, wild-type and mutant VKORL1 and VKORC1 were cloned into an engineered pBudCE4.1 expression vector containing a luciferase gene. The VKORC1 or VKORL1 constructs contain a Flag tag (DYKDDDDK) at their C-terminus, followed by an ER retention signal (KAKRH). Site-directed mutagenesis was performed by Quikchange™ and the nucleotide sequences of all the constructs were verified by DNA sequencing.
For mass spectrometry analyses, stable cell lines were established in 293TRex cells to express the wild-type human VKORC1 and VKORL1 proteins. The VKORL1 and VKORC1 genes, with the same tags as above, were cloned into an expression vector pcDNA5/FRT/TOTOPO. The 293TRex was a gift from S. Wanrooij. The cell lines were generated as previously described [26].
Cell-based activity assay and analysis of warfarin responses
Assays of the epoxide reductase activity were performed as previously described [5] using a cell line established with a chimeric FIXgla-Protein C gene and with endogenous VKORC1 and VKORL1 genes knocked out. Wild-type and mutant VKORL1 clones, along with a luciferase gene, were transfected into this double-knockout cell line. The carboxylation level of secreted FIXgla-PC was measured by a sandwich ELISA by using the cell-culture medium, with luciferase activity serving as the control for transfection efficiency. The chimeric FIXgla-Protein C was used because antibody that specifically recognizes fully carboxylated FIX-gla is commercially available (FIXgla mAb from Green Mountain Antibodies) [27]. In addition, over 90% of the uncarboxylated protein C is degraded and not secreted into the culture medium, significantly reducing the possible background. The ELISA assay was conducted following a protocol reported by the Stafford group [27]. Briefly, ELISA plates were coated with FIXgla mAb overnight at 4°C, and subsequently blocked by bovine serum albumin. Samples of secreted FIXgla-PC and protein standards with 5 mM CaCl2 were added and incubated for 2 hours at room temperature. After washing with TBST buffer (20mM Tris-HCl, pH 7.6, 150mM NaCl, and 0.1% Tween 20) containing CaCl2, the HRP conjugated anti–human protein C IgG (Affinity Biologicals) was added to each well and incubated for 45 minutes at room temperature. The HRP activity was analyzed by incubating with 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) and the absorbance was measured at 405 nm using a ELISA plate reader (Molecular Devices).
To measure IC50s of warfarin (and 4-HC) in the VKORL1 constructs, the transfected cells were treated with 11 different concentrations of warfarin, with the concentration range optimized according to the warfarin response of each construct. The IC50 of each construct was analyzed using GraphPad Prism. The normalized warfarin resistance (nRwar) was determined by dividing IC50s of the mutants to the IC50 of wild type VKORL1, measured in the same set of experiments. Error propagations were calculated accordingly. In our experience with this assay, nRwar > 3 is generally a reliable result and nRwar > 5 indicates substantial resistance (Fig. 1). The relative normalized resistance of VKORL1 and VKORC1 constructs to warfarin (Table S1) was given by (nRwar of VKORL1 / nRwar of VKORC1), with error propagations calculated accordingly.
Analysis of VKORL1 redox state
The intracellular redox states of VKORL1 were analyzed by a mass spectrometry (MS)-based method with isotope-encoded N-ethylmaleimide (NEM) labeling as previously described [15]. Product-ion (MS/MS) spectra afforded two peptides each that contained Cys23, Cys50, and Cys58, and one peptide each for Cys142 and Cys139/Cys142. For peptides containing one cysteine, the apparent oxidized fraction was calculated by dividing the intensity (i) of NEM-d5 modified peptide by the summed peak areas of NEM and NEM-d5 modified peptides, i.e., apparent oxidation fraction = i(NEM-d5) / [i(NEM-d5 + i(NEM)]. For peptides containing two cysteines, the percentage of each redox state (both cysteines oxidized, both reduced, or one reduced/one oxidized) was obtained by dividing the peak area of a given state by the summed peak areas of the three states. Because a peptide containing a single Cys139 was not found, we deduced the apparent oxidized fraction of this cysteine (Ox139) by the following equation: Ox139 = 1 – Red139/142 + Ox139/142 - Ox142, where Red139/142 is the fraction for which both Cys139/Cys142 are reduced in the peptide containing Cys139 and Cys142; Ox139/142 is the fraction for which both residues are oxidized; and Ox142 is fraction of oxidized Cys142 in the peptide containing only Cys142. Peptides from two repeats of separate experiments (from sample preparation to MS analysis) were averaged to give the final values.
Generation of VKORL1 homology model
The initial homology model of VKORL1 was generated with I-TASSER [28] using the bacterial VKORC1 structure (PDB code 4NV5). This structure contains a disulfide corresponding to Cys58-Cys139 in VKORL1, a major cellular state adopted by VKORL1. The warfarin molecule was docked by Swissdock [29] into the hVKOR homology model with its 4-HC group facing the cap domain and its phenyl butanone side group facing the TMs, a binding topology established from our previous VKORC1 footprinting study [15].
Results
VKORL1 and VKORC1 adopt a similar cellular redox state
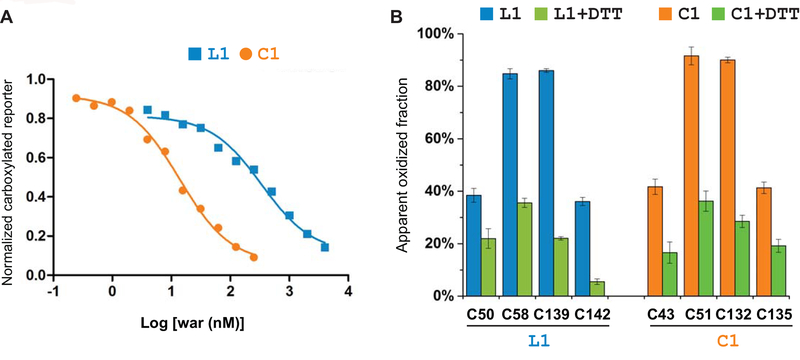
The relative resistance levels of VKORL1 and VKORC1 to warfarin are known from a conventional in vitro assay [8]. This assay, however, is unable to detect the resistance of most WR mutations in VKORC1 [12,13], unlike the cell-based assays [5,30]. To avoid the complications arising from the in vitro system, we used a well-established cell system [5] to determine the warfarin responses of all VKORL1 and VKORC1 [15] constructs. We find that, compared to VKORC1, VKORL1 is ~ 25-fold more resistant to warfarin inhibition (Fig. 2A), consistent with the in vitro assay that shows a 30–50 fold higher resistance of VKORL1 [8].
Figure 2. Different warfarin sensitivity of VKORL1 and VKORC1 is not owing to their redox-state change.
(A) Warfarin inhibition curves of VKORL1 and VKORC1. (B) Cellular redox status of conserved cysteines in VKORL1 (left) and VKORC1 (right), analyzed by MS-based footprinting before and after DTT reduction. Calculation of the error bars is described in Materials and Methods.
Warfarin is known to inhibit preferably VKORC1 in its oxidized form [15,31]. Therefore, we investigated whether the cellular redox state of VKORL1 is different from that of VKORC1, and this difference may cause the apparent resistance of VKORL1 to warfarin inhibition. VKORL1 contains four functional cysteines: Cys139 and Cys142 catalyze vitamin K epoxide reduction at the active site, and Cys50 and Cys58 mediate an electron-transfer process to maintain this reductase activity [6]. MS footprinting shows that, in a cellular environment, ~37% of Cys50 and Cys142 and 85% of Cys58 and Cys139 in VKORL1 are oxidized (Fig. 2B). DTT treatment of the cells, which reduces the disulfide bond formed between these cysteines, significantly lowers the fraction of these oxidized cysteines (Fig. 2B), but has no effect on Cys23, a cysteine not forming disulfide bond (Supplemental Data). As a comparison, in VKORC1 , ~40% of Cys43 and Cys135 and 90% of Cys51 and Cys132 are oxidized [15] (Fig. 2B); the corresponding residue number in VKORC1 is minus 7 from that in VKORL1 according to the sequence alignment of VKORC1 and VKORL1 (Fig. 1). Because the disulfide bonds are formed between these cysteines in a sequential manner, owing to structural restraints [15,22,24], distribution of oxidative states in VKORL1 and VKORC1 can be estimated from the oxidized fraction of individual cysteines. We find that the cellular VKORL1 adopts two major oxidative states: ~37% of VKORL1 is fully oxidized with two disulfides, Cys50-Cys58 and Cys139-Cys142, and 48% VKORL1 is partially oxidized with one disulfide, Cys58-Cys139. For VKORC1, ~40% is in the fully oxidized form with Cys43-Cys51 and Cys132-Cys135, and 50% is in the partially oxidized state with Cys51-Cys132 [15,31]. Because the redox states of VKORC1 are very similar to those of VKORL1 [15], their different warfarin responses are likely caused by other factors.
Warfarin inhibits VKORL1 and VKORC1 at the same site
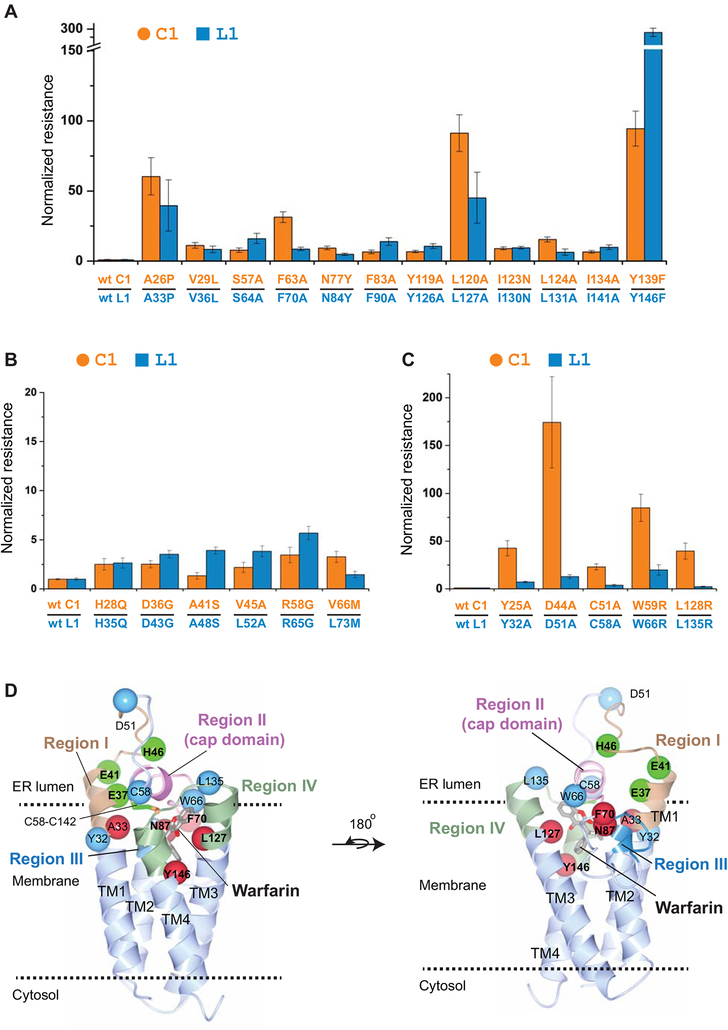
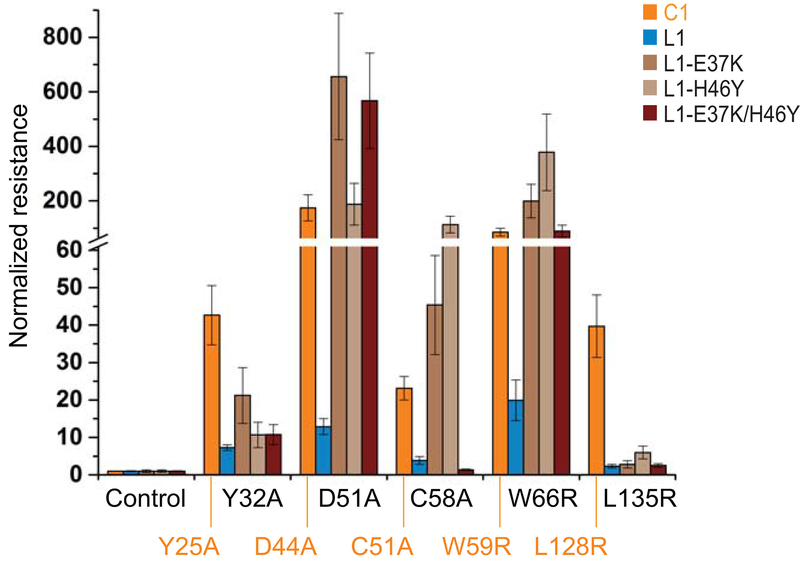
To understand the difference between VKORL1 and VKORC1, we asked whether warfarin inhibits them at the same site, which can be inferred from the location of WR mutations. WR mutations have been identified for numerous residues on VKORC1, most of which are conserved in VKORL1 (Fig. 1). Therefore, we mutated in VKORL1 all the conserved residues showing strong resistance (nRwar > 5) in VKORC1 [15] (Fig. 1). In addition, we included a few naturally occurring mutations that cause weak resistance (nRwar < 5) in VKORC1. Most of these mutations retained resistance in VKORL1 (Fig. 3A–C). Several strongly resistant mutations in VKORC1 also conferred strong resistance in VKORL1 (e.g., Ala33Pro, Leu127Ala, and Tyr146Phe (Fig. 3A)). On the other hand, the weakly resistant mutations in VKORC1 are also weak in VKORL1 (Fig. 3B). Interestingly, Asn87Ala completely diminishes catalysis in VKORL1, suggesting that this mutation also affects catalysis; the corresponding Asn80Ala mutation in VKORC1 shows strong WR and retains 39% activity (Table S1). Clustering of the strongly resistant mutations in both VKORC1 and VKORL1 can nearly define their common warfarin-binding pockets (Fig. 3D, Video S1), which overlap with their active sites [15,22] according to the crystal structures of the bacterial VKOR homolog [22,24] (Fig. S1).
Figure 3. The relative resistance of WR mutations in VKORL1 and VKORC1 correlates with the structural location of these mutations.
(A) WR mutations showing strong resistance in both VKORL1 and VKORC1. The IC50s of VKORL1 and VKORC1 mutants are normalized to those of wild-type VKORL1 and VKORC1, respectively. Error bars are s.e.m. with error propagation calculated for the normalization. The data of normalized resistance of VKORC1 and VKORL1 mutations, and their relative ratios are reported in Table S1. (B) WR mutations showing weak resistance in both VKORL1 and VKORC1. (C) WR mutations showing much lower resistance in VKORL1. (D) Homology model of VKORL1 based on the structure of the bacterial VKOR homolog [28]. The warfarin-binding pocket is surrounded by residues (red spheres) that, when mutated, cause strong resistance in both VKORL1 and VKORC1 (as in A). The cap domain (pink) is maintained by peripheral residues (cyan) showing different resistance in VKORL1 and VKORC1 (as in C), and by peripheral residues (green) increasing the warfarin sensitivity of VKORL1. Regions I to IV are indicated by different colors.
We built a homology model of VKORL1 to illustrate this warfarin-binding site (Fig. 3D, Video S1). The WR mutations in VKORL1 and VKORC1 cluster into four regions in this model (Fig. 1), which closely resembles the bacterial structure (Fig. S1). Region I represents a helical extension from transmembrane helix 1 (TM1) and a loop following this helix; most of this region is not part of the active site (Fig. S1). Region II constitutes a short amphipathic helix and a few residues after this helix, which form a cap domain above the active site. The rest of strong WR mutations are located at the luminal half of TM2 (Region III), TM3, and TM4 (Region IV), with most residues surrounding the active site. Importantly, the cap domain is saddled between region I and region IV. Thus, conformation of the cap domain is primarily maintained by these two peripheral regions.
A group of WR mutations confer lower resistance for VKORL1 than for VKORC1
Despite the general trend of mutations that cause similar resistance, several mutations in VKORL1 show 5 to 20-fold lower resistance than those in VKORC1 (Fig. 3C). These include Tyr32Ala and Asp51Ala in region I, Cys58Ala and Trp66Arg in region II, and Leu135Arg in a loop between TM3 and TM4 at region IV (Fig. 1). The homology model of VKORL1 shows that these residues, except Trp66, are located in regions that do not directly contact bound warfarin. Instead, these peripheral residues interact with the cap domain [24] that forms part of the warfarin-binding pocket (Fig. 3D). Mutations of these peripheral residues cause strong resistance of VKORC1, but not of VKORL1, suggesting that the cap domain is maintained by different interactions in these proteins.
Single mutations render VKORL1 sensitive to warfarin
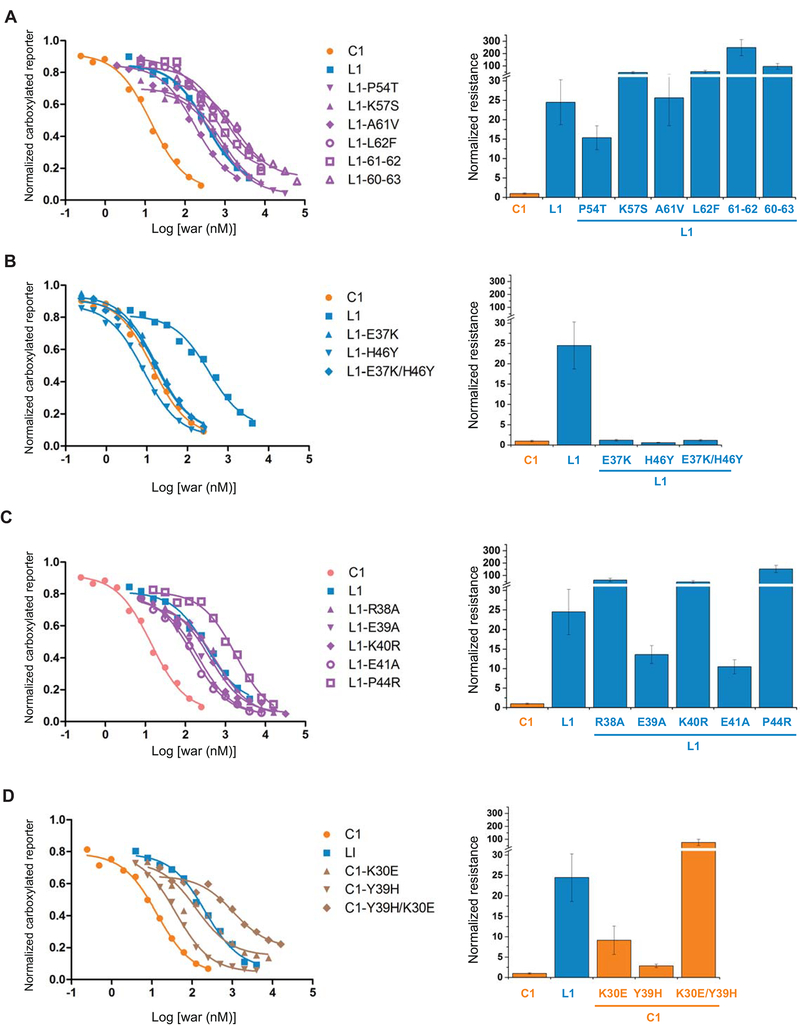
The 25-fold higher resistance in VKORL1 may be due to sequence differences from that of VKORC1, given that single mutations in VKORC1 can induce strong WR (up to a few hundred fold [15] (Fig. 1)). To identify the key residues causing the resistance of VKORL1 compared to VKORC1, we focused on the four regions where mutations that cause strong resistance are clustered (Fig. 1). The entire protein sequences of VKORC1 and VKORL1, including these regions, can be well aligned without gap. The corresponding residues are highly conserved in region III and IV, whereas they show large variations in regions I and II. At region II, Phe55Ala is a particularly strong WR mutation in VKORC1 [15,32], but this residue corresponds to a leucine (Leu62) in VKORL1, making this non-conserved residue an obvious candidate to explain the relative resistance of VKORL1. A Leu62Phe substitution, however, did not render VKORL1 sensitive to warfarin inhibition (Fig. 4A). We then substituted all other non-conserved residues in the entire region II of VKORL1 to match the VKORC1 sequence. The single mutations, Pro54Thr, Lys57Ser, Ala61Vla, and the combined mutations, Ala61Vla/Leu62Phe, Ala60Arg/Ala61Vla/Leu62Phe/Ala63Ser, do not increase the warfarin sensitivity of VKORL1 (Fig. 4A). Thus, the sequence difference in region II (i.e., the cap domain), which forms part of the warfarin-binding pocket, is likely not to be the cause of VKORL1’s resistance to warfarin inhibition.
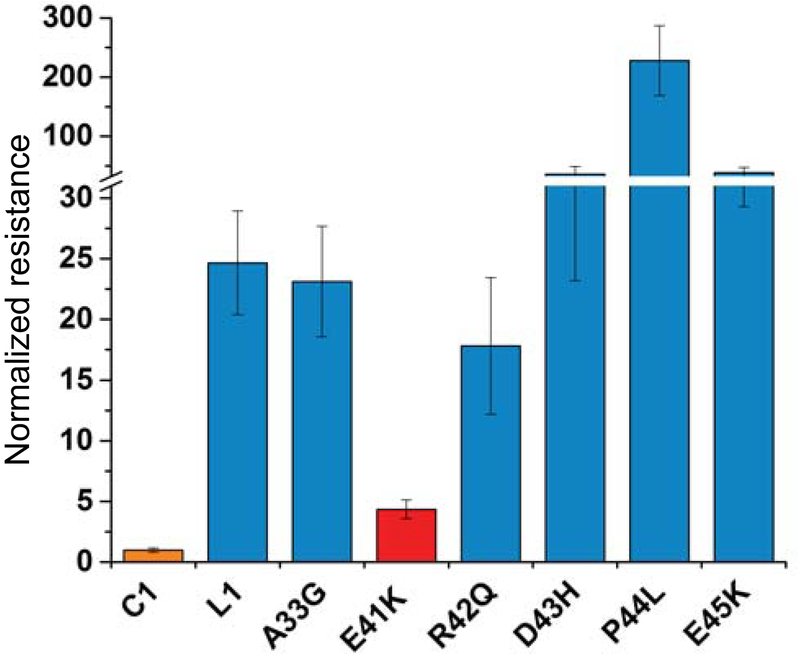
Figure 4. Region I determines the different warfarin sensitivity of VKORC1 and VKORL1.
Left: Inhibition curves of wild-type and mutant VKORC1 and VKORL1 (A–D). Right: relative resistance levels (A–D). The IC50s are normalized to that of the wild-type VKORC1. The error bars are s.e.m. with error propagation calculated for the normalization. (A) Matching mutations at region II. L1–61–62 stands for the double mutation of Ala61Val/Leu62Phe in VKORL1, and L1–60–63 stands for combined mutations of Ala60Arg/Ala61Val/Leu62Phe/Ala63Ser. (B) Matching mutations at region I that increase the warfarin sensitivity of VKORL1. (C) Other matching mutations at region I. (D) Corresponding mutations at the region I of VKORC1, which match with the VKORL1 sequence, confer warfarin resistance.
In contrast, two single matching mutations in region I, Glu37Lys or His46Tyr, and their combination, changed the IC50 of VKORL1 to a similar level as that of VKORC1 (Fig. 4B). Other matching mutations in region I, Arg38Ala, Glu39Ala, Lys40Arg, Glu41Ala, and Pro44Arg, however, did not increase the warfarin sensitivity of VKORL1 (Fig. 4C). Thus, only Glu37Lys and His46Tyr make critically differences between the region I of VKORL1 and that of VKORC1. To confirm this, we made substitutions on the corresponding residues in VKORC1 to match its sequence to VKORL1. These mutations, Lys30Glu and/or Tyr39His, generated warfarin resistance in VKORC1 (Fig. 4D). Thus, region I interacts differently in VKORC1 than in VKORL1 to stabilize the nearby cap domain (Fig. 3D). Changing a single residue in region I converts the warfarin responses of VKORC1 and VKORL1 back-and-forth (Fig. 4B, D).
A group of WR mutations in the converted VKORL1 show similar resistance as in VKORC1
Another difference between VKORL1 and VKORC1 is the group of WR mutations (e.g., Asp51Ala) that shows much lower resistance in VKORL1 (Fig. 3C). Given that the Glu37Lys or His46Tyr mutation can convert VKORL1 to show similar warfarin sensitivity as VKORC1 does, we investigated whether these sensitive mutations can also convert this group of WR mutants in VKORL1 to have similar resistance as the corresponding WR mutants in VKORC1. Under the background of Glu37Lys and/or His46Tyr, most WR mutations, which cause less resistance in the background of wild-type VKORL1, become highly resistant, reaching a level similar to the corresponding WR mutations in VKORC1 (Fig. 5). Such changes were observed for Asp51Ala, Cys58Ala, and Trp66Arg, but to a lesser extent for Tyr32Ala and Leu135Arg. Thus, the sensitive mutations in region I may have converted its interactions with cap domain in VKORL1 to resemble those in VKORC1, allowing the WRs in the converted VKORL1 to generate similar resistance as in VKORC1.
Figure 5. Mutants showing relatively lower resistance in VKORL1 than in VKORC1 can be converted to show higher resistance under the background of warfarin-sensitive VKORL1.
The IC50s of resistant mutations are normalized to the different backgrounds, i.e., IC50s of wild-type VKORC1, wild-type VKORL1, and VKORL1 with the sensitive mutations, respectively. The error bars are s.e.m. with error propagation calculated for the normalization.
A human SNP in region I renders VKORL1 sensitive to warfarin
Many missense mutations were reported in the NCBI database of SNPs. The above results allow us to focus on investigating the mutations in region I, which controls the warfarin response of VKORL1. Interestingly, among the six SNPs in region I, we find that Glu41Lys significantly increases the warfarin sensitivity of VKORL1 (Fig. 6) to a similar level as that of VKORC1. Thus, there exists a warfarin-sensitive variant of VKORL1 in the human population.
Figure 6. A human SNP in Region I of VKORL1 drastically increases its warfarin sensitivity.
Glu41Lys (red) is identified among the six SNPs reported in region I. The IC50s are normalized to that of the wild-type VKORC1. The error bars are s.e.m. with error propagation calculated for the normalization.
Discussion
Structures of VKORC1 and VKORL1 are the basis to understand their different sensitivity to warfarin inhibition. These paralogous proteins share 74% sequence similarity and almost certainly adopt a similar structural fold, given that there is a relatively small number (~1,000) of different protein folds for hundreds of millions of protein sequences [33]. On the other hand, it is conceptually difficult to rationalize a previous proposal that the folding topology of VKORC1 is three-TM and that of VKORL1 is four-TM [6]. Such an unlikely proposal would require the first 100 residues of VKORC1, which constitutes almost two thirds of the protein (Fig. 1), to be completely rearranged and yet retain the same epoxidase activity as VKORL1 and also retain the inhibition by warfarin. Experimentally, we disproved the three-TM model of VKORC1 by using MS-based footprinting of the native VKORC1 protein in live cells, a strategy that avoids perturbation of the structure or function of this unstable protein [34]. A major cellular fraction of native VKORC1 was found to contain Cys51-Cys132, a disulfide bond existing only in the four-TM topology. In the current study, we show that the corresponding Cys58-Cys139 disulfide is also a major cellular form of VKORL1. Therefore, there is no essential difference between the overall structures of VKORC1 and VKORL1.
Effective warfarin inhibition of VKORC1 is highly dependent on its redox status [15,31], which may also change the warfarin response of VKORL1. To test this possibility, we compared the redox status of VKORL1 with that of VKORC1 and found that the cellular redox states of these proteins are similar (Fig. 2B). Therefore, their different sensitivity to warfarin inhibition cannot be explained by the change in redox state.
Existing in similar structural and redox states, VKORL1 and VKORC1 bind warfarin at a common pocket that overlaps with their active sites. For the WR mutations identified in VKORC1, the same mutations also show resistance in VKORL1, exhibiting a general trend that strong WR mutations remain strong, and weak WR mutations remain weak (Fig. 3A, B). All the mutations showing strong WR in both VKORL1 and VKORC1 are located around the active site (Fig. 3D). Among these, Leu127/Leu120 and Tyr146/Tyr139 (numbering in VKORL1/VKORC1) are near the middle of TM3 and TM4, respectively (Fig. 1), suggesting that the warfarin-binding pocket is partly buried in the membrane to accommodate the hydrophobic warfarin molecule. Warfarin is long thought to mimic a transition state of VKORC1 catalysis [35–37]. In fact, most previous studies suggest that warfarin is bound at the active site of VKORC1 [25,31,35–39]. Our current study shows that the same mutations giving strong WR in both VKORC1 and VKORL1 define their common warfarin-binding pocket, which is between their cap domain and TM domain, thereby overlapping with the active sites of these paralogous proteins (Fig. 3D). Thus, the different warfarin sensitivities of VKORC1 and VKORL1 are not caused by their binding of warfarin at different sites.
To identify the cause of different warfarin sensitivity, we made mutations to match the protein sequence between VKORC1 and VKORL1 at their common warfarin-binding pockets and at peripheral regions interacting with this pocket. Sequence alignment (Fig. 1) shows that the major sequence difference surrounding the warfarin-binding pocket (Fig. 3D) is in region II (i.e., cap domain). All the tested substitutions at this region, however, did not increase the warfarin sensitivity of VKORL1 (Fig. 4A). Thus, sequence differences at the cap domain are not the cause of the different warfarin response of VKORL1 and VKORC1. Their warfarin-binding pockets are capped by different residues (e.g., Ala61/Val54 and Leu62/Phe55), but both pockets can accommodate warfarin, probably because its binding involves an induced-fit mechanism that can tolerate minor changes in the size of these hydrophobic residues.
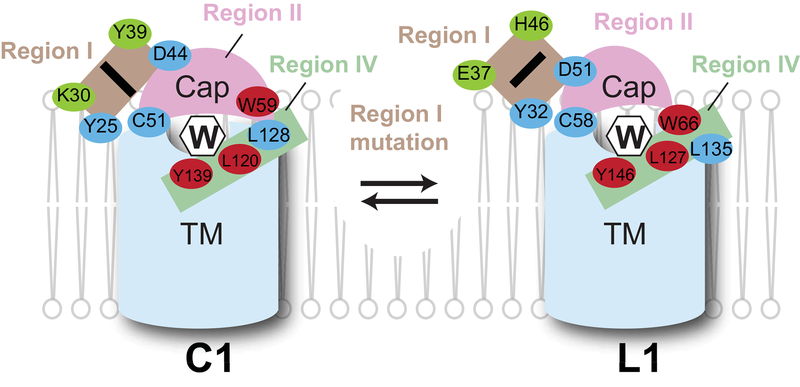
Remarkably, we find that the different sensitivity of VKORL1 and VKORC1 to warfarin is due to two corresponding residues in region I, Glu37 and His46 for VKORL1 and Lys30 and Tyr39 for VKORC1 (Fig. 4B, D). The homology model shows that both residues are far from the warfarin-binding pocket and located at a peripheral region I that interacts with the cap domain (Fig. 3D). The stability of this cap domain is different in VKORL1 than in VKORC1, as implied from their relative different responses to warfarin and 4HC (Fig. S2). Thus, our data support that this region I maintains the stability of the cap domain by using different interactions in VKORL1 and VKORC1 (Fig. 7), with those residues that we substituted (i.e., Glu37/Lys30 and His46/Tyr39) being the key contributors to these different peripheral interactions.
Figure 7. Different peripheral interactions in VKORL1 and VKORC1 maintain their common warfarin-binding pockets.
Cartoon shows that the cap domain (pink) and the TM domain (blue) together form the warfarin (W) binding pocket. Residues making direct contact with warfarin are shown as red spheres. Stabilization of the cap domain is highly dependent on region I (brown). Two key residues (green) differ in region I may induce alternative conformations between VKORC1 and VKORL1. Other peripheral residues (blue) may also contribute to these different conformations.
The different peripheral interactions used by VKORL1 and VKORC1 are supported by the peripheral location of several WR mutations that cause much lower resistance in VKORL1 than in VKORC1, including Tyr32Ala/Tyr25Ala and Asp51Ala/Asp44Ala in region I, Cys58Ala/Cys51Ala and Trp66Arg/Trp59Arg in region II (cap domain), and Leu135Arg/Leu128Arg in region IV. Indeed, these are the exact structural regions that form the peripheral interactions, because the homology model shows that the cap domain is saddled between regions I and IV (Fig. 3D). Remarkably, most of these WR mutations can be converted to show high relative resistance under the background of warfarin-sensitive mutations, Glu37Lys or His46Tyr. Presumably, these sensitive mutations can induce an alternative conformation in VKORL1 to resemble that of VKORC1. With VKORC1 and warfarin-sensitive VKORL1 in similar conformations, their peripheral interactions supporting this conformation also become similar. Consequently, when these peripheral interactions are disrupted by WR mutations, their relative resistance to warfarin becomes similar in warfarin-sensitive VKORL1 and VKORC1.
Taken together, our data support a model whereby warfarin inhibits VKORL1 and VKORC1 by binding at the same active site. Furthermore, the peripheral regions in these proteins, especially region I, maintain the stability of this binding pocket and determine their relative sensitivity and resistance to warfarin inhibition (Fig. 7). This new warfarin-binding model, compared to previous models [15,25], takes into account the peripheral interactions maintaining the warfarin-binding pocket and offers a clear explanation of the WR mutations found in the peripheral regions of VKORC1. In particular, many mutations in region I, including Val29Leu, Ala34Pro, Tyr39Ala, Asp44Ala, may disrupt interactions required to maintain the conformation of region I or its interaction with the cap domain. Consequently, the stability of cap domain is changed, thereby resulting in warfarin resistance. This model explains not only most WR mutations in VKORC1, but also the different warfarin sensitivity of VKORC1 and VKORL1. Making a prediction from this model, we identified a human SNP in the region I of VKORL1 that confers warfarin sensitivity. In the population carrying this SNP, VKORL1 may be severely inhibited under warfarin treatment, thus motivating further investigation of its physiological consequences [40–43].
Supplementary Material
Same as in Fig. 3D.
Essentials:
VKORL1 and VKORC1 have a similar overall structure and warfarin-binding pocket.
A peripheral region stabilizing this pocket controls warfarin sensitivity of the VKOR paralogs.
A human SNP in this peripheral region renders VKORL1 sensitive to warfarin.
A group of warfarin-resistant mutations in VKORC1 acts by disrupting peripheral interactions.
Acknowledgements
G. S. is supported by National Natural Science Foundation of China (81770140) and Henan Department of Science & Technology (182102410079). M.L.G. and the MS measurements are supported by NIH NIGMS (P41 GM103422). W.L. is supported by NEI (R21 EY028705) and NHLBI (R01 HL121718).
Footnotes
Disclosure of Conflict of Interests
The authors declare no competing financial interests.
References
- 1.Stafford DW. The vitamin K cycle. J Thromb Haemost 2005; 3: 1873–8. [DOI] [PubMed] [Google Scholar]
- 2.Oldenburg J, Marinova M, Müller-Reible C, Watzka M. The vitamin K cycle. Vitam Horm 2008; 78: 35–62. [DOI] [PubMed] [Google Scholar]
- 3.Van Horn WD. Structural and functional insights into human vitamin K epoxide reductase and vitamin K epoxide reductase-like1. Crit Rev Biochem Mol Biol 2013; 48: 357–72. [DOI] [PubMed] [Google Scholar]
- 4.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004; 427: 537–41. [DOI] [PubMed] [Google Scholar]
- 5.Tie JK, Jin DY, Tie K, Stafford DW. Evaluation of warfarin resistance using transcription activator-like effector nucleases-mediated vitamin K epoxide reductase knockout HEK293 cells. J Thromb Haemost 2013; 11: 1556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tie JK, Jin DY, Stafford DW. Conserved Loop Cysteines of Vitamin K Epoxide Reductase Complex Subunit 1-Like 1 (VKORC1L1) Are Involved in Its Active Site Regeneration. J Biol Chem 2014; 289: 9396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westhofen P, Watzka M, Marinova M, Hass M, Kirfel G, Müller J, Bevans CG, Müller CR, Oldenburg J. Human vitamin K 2,3-epoxide reductase complex subunit 1-like 1 (VKORC1L1) mediates vitamin K-dependent intracellular antioxidant function. J Biol Chem 2011; 286: 15085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammed A, Matagrin B, Spohn G, Prouillac C, Benoit E, Lattard V. VKORC1L1, an enzyme rescuing the vitamin K 2,3-epoxide reductase activity in some extrahepatic tissues during anticoagulation therapy. J Biol Chem 2013; 288: 28733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeer C, Hamulyak K. Vitamin K: lessons from the past. J Thromb Haemost 2004; 2: 2115–7. [DOI] [PubMed] [Google Scholar]
- 10.Watzka M, Geisen C, Bevans CG, Sittinger K, Spohn G, Rost S, Seifried E, Müller CR, Oldenburg J. Thirteen novel VKORC1 mutations associated with oral anticoagulant resistance: insights into improved patient diagnosis and treatment. J Thromb Haemost 2011; 9: 109–18. [DOI] [PubMed] [Google Scholar]
- 11.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 2005; 352: 2285–93. [DOI] [PubMed] [Google Scholar]
- 12.Rost S, Pelz H, Menzel S, MacNicoll AD, León V, Song K, Jäkel T, Oldenburg J, Müller CR. Novel mutations in the VKORC1 gene of wild rats and mice--a response to 50 years of selection pressure by warfarin? BMC Genet 2009; 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodroge A, Longin-Sauvageon C, Fourel I, Benoit E, Lattard V. Biochemical characterization of spontaneous mutants of rat VKORC1 involved in the resistance to antivitamin K anticoagulants. Arch Biochem Biophys Elsevier Inc; 2011; 515: 14–20. [DOI] [PubMed] [Google Scholar]
- 14.Pelz H, Rost S, Hünerberg M, Fregin A, Heiberg A, Baert K, MacNicoll AD, Prescott CV, Walker A, Oldenburg J, Müller CR. The genetic basis of resistance to anticoagulants in rodents. Genetics 2005; 170: 1839–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen G, Cui W, Zhang H, Zhou F, Huang W, Liu Q, Yang Y, Li S, Bowman GR, Sadler JE, Gross ML, Li W. Warfarin traps human vitamin K epoxide reductase in an intermediate state during electron transfer. Nat Struct Mol Biol 2017; 24: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tie JK, Nicchitta C, Heijne G Von, Stafford DW, Von Heijne G. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J Biol Chem 2005; 280: 16410–6. [DOI] [PubMed] [Google Scholar]
- 17.Tie J-K, Jin D-Y, Stafford DW. Human vitamin k epoxide reductase and its bacterial homologue have different membrane topologies and reaction mechanisms. J Biol Chem 2012; 287: 33945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Cousins E, Sandford G, Nicholas J. Human Herpesvirus 8 Viral Interleukin-6 Interacts with Splice Variant 2 of Vitamin K Epoxide Reductase Complex Subunit 1. J Virol 2012; 86: 1577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulman S, Wang B, Li W, Rapoport T a. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci U S A 2010; 107: 15027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Z, van Lith M, Mitchell LJ, Pringle MA, Inaba K, Bulleid NJ. The membrane topology of vitamin K epoxide reductase is conserved between human isoforms and the bacterial enzyme. Biochem J 2016; 473: 851–8. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Shen G, Li W. Intramembrane Thiol Oxidoreductases: Evolutionary Convergence and Structural Controversy. Biochemistry 2018; 57: 258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 2010; 463: 507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matagrin B, Hammed A. Identification of Key Functional Residues in the Active Site of Vitamin K Epoxide Reductase-like Protein (VKORC1L1). Biochem Mol Biol J 2016; 2. [Google Scholar]
- 24.Liu S, Cheng W, Fowle Grider R, Shen G, Li W. Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer. Nat Commun 2014; 5: 3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czogalla KJ, Biswas A, Wendeln A-C, Westhofen P, Müller CR, Watzka M, Oldenburg J. Human VKORC1 mutations cause variable degrees of 4-hydroxycoumarin resistance and affect putative warfarin binding interfaces. Blood 2013; 122: 2743–50. [DOI] [PubMed] [Google Scholar]
- 26.Wanrooij S, Goffart S, Pohjoismäki JLO, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res 2007; 35: 3238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tie J-K, Jin D-Y, Straight DL, Stafford DW. Functional study of the vitamin K cycle in mammalian cells. Blood 2011; 117: 2967–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010; 5: 725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 2011; 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fregin A, Czogalla KJ, Gansler J, Rost S, Taverna M, Watzka M, Bevans CG, Müller CR, Oldenburg J. A new cell culture-based assay quantifies vitamin K 2,3-epoxide reductase complex subunit 1 function and reveals warfarin resistance phenotypes not shown by the dithiothreitol-driven VKOR assay. J Thromb Haemost 2013; 11: 872–80. [DOI] [PubMed] [Google Scholar]
- 31.Fasco MJ, Principe LM, Walsh W a, Friedman P a.. Warfarin inhibition of vitamin K 2,3-epoxide reductase in rat liver microsomes. Biochemistry 1983; 22: 5655–60. [DOI] [PubMed] [Google Scholar]
- 32.Czogalla KJ, Biswas A, Höning K, Hornung V, Liphardt K, Watzka M, Oldenburg J. Warfarin and vitamin K compete for binding to Phe55 in human VKOR. Nat Struct Mol Biol 2017; 24: 77–85. [DOI] [PubMed] [Google Scholar]
- 33.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 1995; 247: 536–40. [DOI] [PubMed] [Google Scholar]
- 34.Shen G, Cui W, Zhang H, Zhou F, Huang W, Liu Q, Yang Y, Li S, Bowman GR, Sadler JE, Gross ML, Li W, Guomin S, Weidong C, Hao Z, Fengbo Z, Wei H, Qian L, Yihu Y, Bowman GR, et al. Warfarin prevents blood coagulation by trapping human vitamin K epoxide reductase in an intermediate state during electron transfer. Nat Struct Mol Biol 2016; 24: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman RB. Model Studies for a Molecular Mechanism of Action of Oral Anticoagulants. J Am Chem SOC 1981; 103: 3910–5. [Google Scholar]
- 36.Gebauer M Synthesis and structure-activity relationships of novel warfarin derivatives. Bioorg Med Chem 2007; 15: 2414–20. [DOI] [PubMed] [Google Scholar]
- 37.Davis CH, Deerfield D, Wymore T, Stafford DW, Pedersen LG, Transition II. A quantum chemical study of the mechanism of action of Vitamin K epoxide reductase ( VKOR ). J Mol Graph Model 2007; 26: 401–8. [DOI] [PubMed] [Google Scholar]
- 38.Silverman RB. Chemical Model Studies for the Mechanism of Vitamin K Epoxide Reductase. J Am Chem SOC 1981; 103: 5939–41. [Google Scholar]
- 39.Fasco MJ, Preusch PC, Hildebrandt E, Suttie JW. Formation of hydroxyvitamin K by vitamin K epoxide reductase of warfarin-resistant rats. J Biol Chem 1983; 258: 4372–80. [PubMed] [Google Scholar]
- 40.Rennenberg RJMW, Van Varik BJ, Schurgers LJ, Hamulyak K, Ten Cate H, Leiner T, Vermeer C, De Leeuw PW, Kroon AA. Chronic coumarin treatment is associated with increased extracoronary arterial calcification in humans. Blood 2010; 115: 5121–3. [DOI] [PubMed] [Google Scholar]
- 41.Poterucha TJ, Goldhaber SZ. Warfarin and Vascular Calcification. American Journal of Medicine. 2016. p. 635e1–635e4. [DOI] [PubMed] [Google Scholar]
- 42.Tantisattamo E, Han KH, Charles O’Neill W. Increased vascular calcification in patients receiving warfarin. Arterioscler Thromb Vasc Biol 2015; 35: 237–42. [DOI] [PubMed] [Google Scholar]
- 43.Namba S, Yamaoka-Tojo M, Hashikata T, Ikeda Y, Kitasato L, Hashimoto T, Shimohama T, Tojo T, Takahira N, Masuda T, Ako J. Long-term warfarin therapy and biomarkers for osteoporosis and atherosclerosis. BBA Clin 2015; 4: 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Same as in Fig. 3D.