Abstract
The purpose of this study was to investigate individually and in combination the association between the ACE (I/D), NOS3 (Glu298Asp), BDKRB2 (-9/+9), UCP2 (Ala55Val) and AMPD1 (Gln45Ter) variants with endurance performance in a large, performance-homogenous cohort of elite Polish half marathoners. The study group consisted of 180 elite half marathoners: 76 with time < 100 minutes and 104 with time > 100 minutes. DNA of the subjects was extracted from buccal cells donated by the runners and genotyping was carried out using an allelic discrimination assay with a C1000 Touch Thermal Cycler (Bio-Rad, Germany) instrument with TaqMan® probes (NOS3, UCP2, and AMPD1) and a T100™ Thermal Cycler (Bio-Rad, Germany) instrument (ACE and BDKRB2). We found that the UCP2 Ala55Val polymorphism was associated with running performance, with the subjects carrying the Val allele being overrepresented in the group of most successful runners (<100 min) compared to the >100 min group (84.2 vs. 55.8%; OR = 4.23, p < 0.0001). Next, to assess the combined impact of 4 gene polymorphisms, all athletes were classified according to the number of 'endurance' alleles (ACE I, NOS3 Glu, BDKRB2 -9, UCP2 Val) they possessed. The proportion of subjects with a high (4-7) number of 'endurance' alleles was greater in the better half marathoners group compared with the >100 min group (73.7 vs. 51.9%; OR = 2.6, p = 0.0034). These data suggest that the likelihood of becoming an elite half marathoner partly depends on the carriage of a high number of endurance-related alleles.
Key words: half marathoners, endurance performance, gene polymorphism, gene-gene interaction
Introduction
The probability of becoming an elite athlete is influenced by genetic factors (Eynon et al., 2013). A PubMed base has publicized that at least 120 genetic markers are linked to elite athlete status (Ahmetov and Fedotovskaya, 2015). However, only few of genes have been associated with endurance performance (Ahmetov and Fedotovskaya, 2015). Among them genetic markers such as ACE (Montgomery et al., 1998), AMPD1 (Ginevičienė et al., 2014; Rubio et al., 2005), NOS3 (Drozdovska et al., 2009; Saunders et al., 2006), UCP2 (Buemann et al., 2001), BDKRB2 (Williams et al., 2004) have shown positive associations with endurance.
NOS3 (Glu298Asp) (rs1799983)
Nitric oxide (NO) is a gaseous free radical that is the most potent endothelium-derived relaxation factor synthesized by nitric oxide synthase (NOS). Among three different forms of NOS i.e. neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), exactly endothelial NOS (eNOS or NOS3) are predominantly expressed in vascular endothelial cells. The 135 kDa functional homodimer, which function is to preclude neuronal damage by generating small volumes of NO to expand blood vessels, maintain cerebral blood, inhibit platelet aggregation and relaxation, and prevent oxidative destruction Popp et al., 1998), is encoded by the human NOS3 gene (localization: 7q35-36). The NOS3 has been extensively investigated for single nucleotide polymorphisms (SNIPs) and several SNPs have been recognized i.e. G894T (Glu298Asp, rs1799983) and -786T/C (rs2070744). Glu298Asp SNP seems to be associated with several health/fitness, or exercise response phenotypes. Saunders et al. (2006) found a tendency of the NOS3 Glu298 allele combined with a BDKRB2 -9/-9 genotype to be overrepresented in the elite triathletes compared with control individuals.
BDKRB2 (-9/+9) (rs5810761)
Bradykinin is a potent endothelium-dependent vasodilator, significantly reducing blood pressure via the bradykinin B2 receptors encoded by the BDKRB2 gene (location: 14q32.1–q32.2). Polymorphism of 9 bp repeat sequences in exon 1 is associated with altered BDKRB2 mRNA expression. -9/-9 genotype of the BDKRB2 gene is overrepresented in endurance athletes (Triathletes and Ironman triathletes) compared to male controls (Saunders et al., 2006). However, these observations were not confirmed in two studies involving 334 endurance Russian and Polish athletes (Sawczuk et al., 2013). Likewise Eynon et al. (2011) did not find significant differences in the frequencies of the -9 allele and -9/-9 genotype between 74 Israeli endurance athletes and 240 controls.
UCP2 (Ala55Val) (rs660339)
The uncoupling protein 2 (UCP2) is a member of the mitochondrial anioncarrier proteins family (MCAPs). The UCP2 (location: 11q13) is widely expressed in the muscles, heart, kidneys, lungs, spleen, central nervous system and white adipose tissue (Fleury and Sanchis, 1999). In adipose tissue, energy coupling with ADP phosphorylation is only partial, because uncoupling proteins induce a proton leak, releasing the energy stored in ATP as heat (Holdys et al., 2013). However, the physiological role of UCP2 is not clear. Aerobic training leads to increased intensity in expression of the UCP2 in skeletal muscles and the heart. Holdys et al. (2013) observed the association of maximum oxygen uptake (VO2max) with I/D polymorphism in exon 8 of the UCP2 gene. Additionally, the Val allele of another important and common Ala55Val polymorphism (rs660339 C/T) in the UCP2 gene has been also associated with higher VO2max (Ahmetov et al., 2008), higher exercise efficiency (Buemann et al., 2001), enhanced metabolic efficiency, physical activity (Astrup et al., 1999) and endurance athlete status (Ahmetov et al., 2009). On the contrary, Sessa et al. (2011) found an increased frequency of the Ala55 allele in power-oriented athletes.
AMPD1 (Gln45Ter) (rs17602729)
Adenosine monophosphate deaminase 1 (AMPD1) is one of the significant enzymes used to utilize the energy from ATP. AMPD1 catalyzes the deamination of adenosine monophosphate (AMP) to inosine monophosphate (IMP). Deficiency of the AMPD1 is one of the most common cause of exercise-induced myopathy (Fishbein et al., 1978) and it is associated with the presence of a 34C/T transition in exon 2 (rs17602729C/T) of the AMPD1 gene (location: 1p13), which creates a nonsense codon (Gln45X) that prematurely terminates translation.
Thomaes et al. (2011) found that carriers of the X allele characterized significantly lower maximal oxygen uptake (VO2max), in details, a relative lower increase in peak VO2 after 12 weeks of endurance training. This observation was confirmed by Cięszczyk et al. (2011) and Rubio et al. (2005) in a group of 127 Polish rowers and 104 top-level Spanish male endurance athletes (cyclists and runners), respectively. However, Ginevičienė et al. (2014) reported opposite results when 84 Lithuanian athletes were compared with 260 controls, which make these observations ambiguous.
ACE (I/D) (rs4340)
Via cleavage an angiotensin I at a particular location, the angiotensin-converting enzyme (ACE) converts this protein to angiotensin II which entails blood vessels to narrow the diameter, and results in increased blood pressure. Lower serum and tissue ACE activity is a consequence of presence of a 287 bp Alu sequence insertion fragment (I allele) rather than the absence (deletion, D allele) in a polymorphism mutation site in intron 16 of the human ACE gene (location: 17q23.3). Lower blood pressure in I/I genotype individuals causes that the ACE remains the most frequently analysed gene in contexts of elite athletes’ performance, especially endurance performance (Ahmetov et al., 2008; Ahmetov and Fedotovskaya, 2015; Montgomery et al., 1998).
I allele is overrepresented among triathletes (Shenoy et al., 2010), successful marathon runners (scoring better than 150th place) (Hruskovicova et al., 2006), long-distance (25 km) swimmers (Tsianos et al., 2004), and rowers (Cięszczyk et al., 2009).
One of the limitations of most studies mentioned above, investigating the association between a certain genotype and endurance performance, is grouping together highly specialized athletes and amateurs. This approach, while understandable given the very low number of world-class marathoners, reduces the accuracy of the phenotype. In our approach, we sought to address these limitations and provide deeper insights into the influence of the association between the NOS3, UCP2, BDKRB2, AMPD1 and ACE variants on endurance performance by studying the influence of the genotype on actual endurance performance (half marathon personal times less than 100 min or >100 min). Therefore, the purpose of this study was to investigate individually and in combination the association between the ACE (I/D), NOS3 (Glu298Asp), BDKRB2 (-9/+9), UCP2 (Ala55Val) and AMPD1 (Gln45Ter) variants with endurance performance in a large, performance-homogenous cohort of highly trained Polish half marathoners. We also examined multifactor dimensionality reduction for detecting gene-gene interaction (ACE, AMPD1, NOS3, UCP2 and BDKRB2 variants) in half marathoners.
Methods
Subjects
One hundred eighty healthy and experienced half marathon runners (all Polish Caucasians) volunteered to participate in this investigation. Potential participants were contacted from a group of runners that had taken part in previous investigations or they were enrolled at the registration desk of the competition in the Half Marathon in Szczecin. Inclusion criteria were as follows: sex men, age between 18 and 65 years, participating with success in the half marathon and having running experience of at least 3 years. The study was approved by the Local Ethics Committee and was performed according to the Declaration of Helsinki (20110714).
Genetic Analyses
All genetic analyses were performed at the Centre for Human Structural and Functional Research, University of Szczecin. The buccal cells donated by the subjects were collected in Resuspension Solution (GenElute Mammalian Genomic DNA Miniprep Kit, Sigma, Germany) with the use of sterile foam-tipped applicators (Puritan, USA). DNA was extracted from the buccal cells using a GenElute Mammalian Genomic DNA Miniprep Kit (Sigma, Germany) according to the manufacturer’s protocol. All DNA samples were then stored in the same conditions at −25°C until subsequent processes were performed. The samples were genotyped in duplicate.
NOS3, UCP2, and AMPD1 genotyping
The samples were genotyped using an allelic discrimination assay with a C1000 Touch Thermal Cycler (Bio-Rad, Germany) instrument with TaqMan® probes. To discriminate NOS3 Glu298Asp (NP_000594.2:p.Asp298Glu), UCP2 Ala55Val (NP_003346.2:p.Ala55Val), and AMPD1 Gln45Ter (NP_000027.2:p.Gln45Ter) alleles, TaqMan® Pre-Designed SNP Genotyping Assays were used (Applied Biosystems, USA) (assay ID: C_590093_1, C_3219460_20, C_903746_1, and C_33603912_10 respectively), including primers and fluorescently labelled (FAM and VIC) MGBTM probes to detect alleles. Genotypes were assigned using all of the data from the study simultaneously.
ACE genotyping
PCR amplification of the polymorphic region of the ACE gene containing either the insertion (I) or deletion (D) fragment was performed using a T100™ Thermal Cycler (Bio-Rad, Germany) instrument. One pair of primers (forward: CTG GAG ACC ACT CCC ATC CTT TCT and reverse: GAT GTG GCC ATC ACA TTC GTC AGA) was used to determine the ACE genotype, yielding amplification products of approximately 490 bp (for the I allele) and 190 bp (for the D allele), as it has been described earlier (Rigat et al., 1992). The PCR mixture and thermal-time profile were coequal as described by Cięszczyk et al. (2009). The amplified DNA fragments were visualized using 1.5% agarose gels stained with ethidium bromide.
BDKRB2 genotyping
The BDKRB2 -9/+9 polymorphism was genotyped by PCR using a T100™ Thermal Cycler (Bio-Rad, Germany) instrument. The 100 (refers to the +9 allele) and 91 (refers to the 29 allele) bp fragments of the gene were amplified by PCR using the forward primer 50-TCTGGCTTCTGGGCTCCGAG-30 and the reverse primer 50-AGCGGCATGGGCACTTCAGT-30, as recommended by Williams et al. (2004). The PCR mixture and thermal-time profile were coequal, as described by Sawczuk et al. (2013). The amplified DNA fragments were visualized using 7.5% polyacrylamide gel electrophoresis.
Statistical Analysis
Hardy-Weinberg equilibrium was checked with the Chi-square test. The allelic frequencies were calculated using genotype counts. Genotype and allele frequencies were compared between groups using the Chi-square test. For analysis of gene-gene interaction the multifactor dimensionality reduction (MDR) algorithm was used followed by entropy-based quantification of epistasis. An in-depth presentation of the MDR method can be found in Gui et al. (2013). Entropy estimates were used to construct an interaction map that showed the percentages of entropy removed (eg. Information gain) by SNPs and pairwise interactions (Moore et al., 2006). For evaluation of interaction models we used 10-fold cross-validation, where the dataset was divided into a training set (9/10 of the dataset) and a testing set (1/10 of the dataset). The cross-validation training score, cross-validation testing score, and cross validation consistency (CVC, the number of times the same model was chosen in the training set) were calculated; p values were determined by the permutation test and obtained from the empirical distribution of cross-validation testing scores. The MDR software package (version 3.0.2) available on http://sourceforge.net was used.
Results
Except for AMPD1 and UCP2, all polymorphisms conformed to Hardy-Weinberg equilibrium, both in the subgroups (<100 and >100 min) and the whole group. For the AMPD1 and UCP2 variants deviations from HWE were observed in the runners with finish time <100 min. A distribution of the genotypes and alleles in half marathon runners with respect to finish times is presented in Table 1. There were significant differences in the AMPD1 and UCP2 genotype and allele distribution between runners. We did not detect any subjects with the mutant AMPD1 TT genotype in both groups. We found that the UCP2 Ala55Val polymorphism was associated with running performance, with the subjects carrying the Val allele being overrepresented in the group of more successful runners (<100 min) compared to the >100 min group (84.2 vs. 55.8%; odds ratio (OR) = 4.23, p < 0.0001). Next, to assess the combined impact of 4 gene polymorphisms (AMPD1 was excluded from this analysis due to the absence of the unfavourable TT genotype and conflict with the literature data), all athletes were classified according to the number of 'endurance' alleles (ACE I, NOS3 Glu, BDKRB2 -9, UCP2 Val) they possessed. The proportion of subjects with a high (4-7) number of 'endurance' alleles was greater in the better half marathoners group compared with the >100 min group (73.7 vs. 51.9%; OR = 2.6, p = 0.0034).
Table 1.
Genotypes and alleles in the half-marathon runners with respect to finish time.
| ACE (HWE, χ2 = 1.8, p = 0.180) | |||||
|---|---|---|---|---|---|
| Group (HWE) | II ( n = 50) | ID (n = 81) | DD (n = 49) | I | D |
| <100 (χ2 = 0.38, p = 0.538) | |||||
| 24 (31.6%) | 35 (46.1%) | 17 (22.4%) | 83 (54.6%) | 69 (45.4%) | |
| (n = 76) | |||||
| >100 (χ2 = 1.31, p = 0.252) | 26 (25.0%) | 46 (44.2%) | 32 (30.8%) | 98 (47.1%) | 110 (52.9%) |
| (n = 104) | p = 0.396 | p =0.160 | |||
| BDKRB2 (HWE, χ2 = 0.13, p = 0.718) | |||||
| -/- (n = 32) | +/- (n = 85) | +/+ (n = 63) | - | + | |
| <100 (χ2 = 0.32, p = 0.572) | 14 (18.4%) | 40 (52.6%) | 22 (29.0%) | 68 (44.7%) | 84 (55.3%) |
| >100 (χ2 = 0.84, p = 0.359) | 18 (17.3%) | 45 (43.3%) | 41 (39.4%) | 81 (38.9%) | (61.1127%) |
| p = 0.329 | p = 0.270 | ||||
| NOS3 (HWE, χ2 = 0.01, p = 0.920) | |||||
| Glu/Glu (n = 88) | Glu/Asp (n = 76) | Asp/Asp (n = 16) | Glu | Asp | |
| <100 (χ2 = 0.32, p = 0.572) | 38 (50.0%) | 30 (39.5%) | 8 (10.5%) | (69.7106 %) | 46 (30.3%) |
| >100 (χ2 = 0.34, p = 0.560) | 50 (48.1%) | 46 (44.2%) | 8 (7.7%) | (70.2146 %) | 62 (29.8%) |
| p = 0.717 | p = 0.926 | ||||
| AMPD1 (HWE, χ2 = 3.49, p = 0.062) | |||||
| CC (n = 136) | CT (n = 44) | TT (n = 0) | C | T | |
| <100 (χ2 = 4.99, p = 0.025) | 45 (59.2%) | 31 (40.8%) | 0 (0) | (79.6121 %) | 31 (20.4%) |
| >100 (χ2 = 2.58, p = 0.108) | 91 (87.5%) | 13 (12.5%) | 0 (0) | (93.8195 %) | 13 (6.3%) |
| p = 0.00001 | p = 0.00005 | ||||
| UCP2 (HWE, χ2 = 1.85, p = 0.174) | |||||
| CC (n = 58) | CT (n = 96) | TT (n = 26) | C | T | |
| <100 (χ2 = 13.54, p=0.0002) = | 12 (15.8%) | 54 (71.1%) | 10 (13.2%) | 78 (51.3%) | 74 (48.7%) |
| >100 (χ2 = 1.47, p = 0.225) | 46 (44.2%) | 42 (40.4%) | 16 (15.4%) | (64.4134 %) | 74 (35.6%) |
| p = 0.00008 | p = 0.0125 | ||||
The interaction map for the five-locus model (Figure 1) revealed two main effects independent of other loci (AMPD1 and UCP2 with information gain of 7.65% and 7.93%, respectively) as well as an interaction effect between NOS3 and BDKRB2 (information gain of 1.43%) in the absence of main effects (information gain of 0.27% and 0.90% for the NOS3 and BDKRB2, respectively). Thus, in the next step, a new (single, multilocus) attribute was constructed and evaluated using the MDR method for mentioned above polymorphisms (AMPD1, UCP2, NOS3 and BDKRB2) (Table 2). The best model (Figure 2) consisted of three variants: BDKRB2, AMPD1, UCP2 with cross-validation consistency of 10, testing prediction error of 21% and p value of 0.001. The two-locus and four-locus models (the one with interacting loci, NOS3 and BDKRB2) were also significant and consistent (perfect cross-validation consistency), but had greater prediction error (30.5% and 23.1% for the two-locus and three-locus model, respectively). The interaction map for the best model is shown in Figure 3. The odds ratio for completing a half-marathon <100 min using the single multilocus attribute based on the best model (BDKRB2, AMPD1, UCP2) was 18.8 (8.4 – 42.0) (p < 0.00001).
Figure 1.
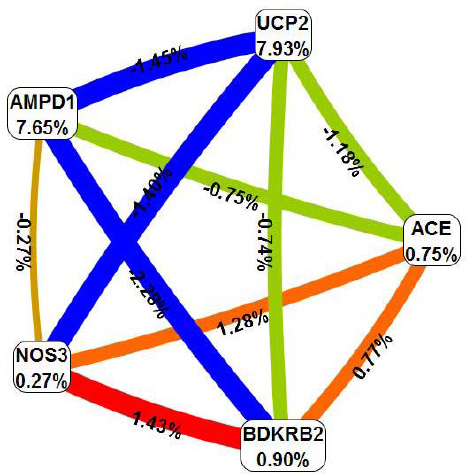
Interaction map using entropy-based measure of information gain. Percentages indicate the amount of entropy removed by each polymorphism and each pairwise combination of polymorphisms. Positive values (orange, red lines) reflect synergistic interaction, negative values (blue, green lines) indicate redundancy. Near zero values (brown line) indicate independence.
Table 2.
Multilocus interaction models constructed using the MDR method
| Model | CV Consistency | Testing accuracy | p* |
|---|---|---|---|
| AMPD1, UCP2 | 10 | 69.5% | 0.001 |
| BDKRB2, AMPD1, UCP2 | 10 | 79% | 0.001 |
| BDKRB2, NOS3, AMPD1, UCP2 | 10 | 76.9% | 0.001 |
CV – cross-validation,
empirical p value based on 1000 permutations
Figure 2.
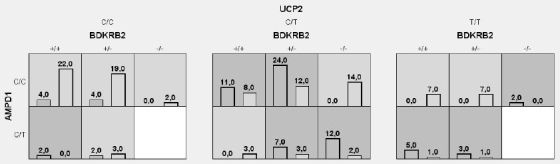
Graphical model of multilocus association between BDKRB2, AMPD1, UCP2 polymorphisms and half-marathon finish time (<100 vs >100 min). Dark-shaded areas represent <100 level of the new attribute, while light-shaded areas represent runners with finish time >100. Left bars - the number of runners <100, right bars - the number of runners >100.
Figure 3.
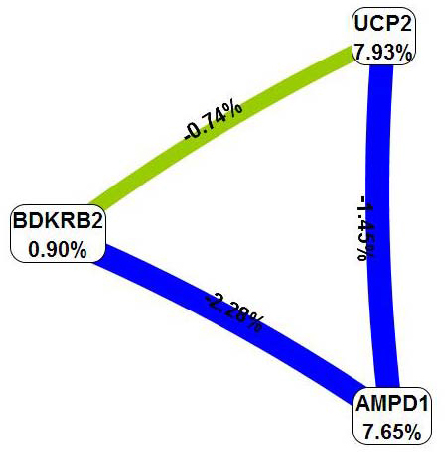
Interaction map using entropy-based measure of information gain of the best multilocus model selected using the MDR method
Discussion
This study estimated the association between human endurance performance and variants of 5 genes previously linked to elite athlete status. We conducted an analysis of genetic markers such as ACE (I/D) (rs4340), NOS3 (Glu298Asp) (rs1799983), BDKRB2 (-9/+9) (rs5810761), UCP2 (Ala55Val) (rs660339), AMPD1 (Gln45Ter) (rs17602729) on a large, performance-homogenous cohort of elite half marathoners. These variants were chosen based on previous association studies on different populations (Ahmetov et al., 2008; Montgomery et al., 1998) and because of their impact on human variability in one or more endurance phenotypic traits (Cięszczyk et al., 2009; Holdys et al., 2013; Sawczuk et al., 2013).
Most studies analyze just one polymorphism (Gronek and Holdys, 2013) and populations of endurance athletes that are generally from aerobic-anaerobic sporting disciplines (Holdys et al., 2011, 2013) and different ethnic/geographical origins (Zarebska et al., 2017), or combined with amateurs (Wilkinson et al., 2013). We decided to compare data of Polish half marathoners with personal time less and more than 100 min.
For the AMPD1 and UCP2 variants, deviations from HWE were observed in the runners with a finish time < 100 min. This may be caused by diverse reasons (Salanti et al., 2005). In our project, the analyzed group was a selected population with selection based on a phenotype (participation in a very exhausting endurance event) that reflects aerobic endurance performance (Tsianos et al., 2010). In our study group, there were no participants with the genotype TT (XX) for AMPD1 (Gln45X) polymorphism. The study showed significant differences in AMPD1 genotypes distribution between subgroups of half marathoners with times below and above 100 min. In the < 100 min group, the genotype CT was significantly more frequent compared with the > 100 min group (p = 0.00001). In the > 100 min group, the genotype CC was overrepresented and sevenfold more frequent than the genotype CT, compared with the < 100 min group where the genotype CC was about one third more common than the genotype CT. Also alleles distribution between these groups differed. In the < 100 min group, frequency of allele T was significantly higher compared to the > 100 min group (p = 0.00005). The (Gln45Ter) polymorphism of the adenosine monophosphate deaminase 1 gene is involved in the salvage of adenine nucleotides and regulation of muscle glycolysis during intense exercise (Rubio et al., 2005). Transition C > T in exon 2 of AMPD1 gene induces exchange codon Gln on nonsense codon X (Ter), which terminates translation of the AMPD1 enzyme earlier than the correct isoform (Morisaki et al., 1992). Transition Gln45Ter predominantly causes exercise-induced myopathy (Fishbein et al., 1978). Additionally the X codon carriers also characterize significantly lower maximal oxygen uptake (VO2max) (Thomaes et al., 2011). These findings may explain the low frequency of T allele in the runners’ population and lack of runners with the genotype TT. However, it must be surprising that we found more individuals with the CT genotype in the group of faster half marathoners. Conversely, in the general population the frequency of TT genotype remains very low, which is confirmed by research studies conducted on a group of Spaniards (Rubio et al., 2005). Individuals with a TT genotype in the Spanish population accounted for 1%, whereas among Spanish elite endurance athletes the frequency of the TT genotype was 0%. Thus, we may conclude that the carriage of the AMPD1 CT genotype is not an unfavourable factor for half marathon running performance.
Our study also demonstrated differences in genotypes and alleles distribution of polymorphism Ala55Val of the UCP2 gene between runners. In faster half marathoners the genotype CT (Ala/Val) was most common compared to slower half marathoners wherein the genotype CT was smaller by 30%. The genotype CC in the < 100 min group was distinctly rarer compared to the > 100 min group, where the genotype CC was threefold more frequent and likewise most common (p = 0.00008). We observed an overrepresentation of subjects carrying the Val allele (i.e. Ala/Val and Val/Val) in the group of more successful runners (< 100 min) compared to the >100 min group (p < 0.0001). It is worth mentioning that in faster runners deviation from HWE for variants of the UCP2 gene was observed. In analysis of alleles distribution of this polymorphism between runners we also observed significant differences (p = 0.0125). In the < 100 min group allele C was significantly rarer than in the > 100 min group, and allele T was distinctly more common in the faster half marathoners compared to their slower peers. It may be concluded that the genotype CT and allele T are associated with the achievement of better results in half marathons and this assumption is consistent with previous literature data (Ahmetov et al., 2008, 2009).
Given the polygenic nature of endurance performance, we assessed the combined effect of genes polymorphisms on running performance. We found that the proportion of subjects with a high (4-7) number of 'endurance' alleles (ACE I, NOS3 Glu, BDKRB2 -9, UCP2 Val) was greater in the better half marathoners compared with the >100 min group (p = 0.0034). These data suggest that the likelihood of becoming an elite half marathoner partly depends on the carriage of a high number of endurance-related alleles.
In this study, we also performed the analysis of the interaction of all five genes. The five-gene model showed two main effects independent of other loci for genes AMPD1 and UCP2, which is consistent with the results presented in Table 1. In addition, we found an interaction between NOS3 and BDKRB2 (entropy removed 1.43%) in the absence of main effects. Smaller interaction effects were observed between genes NOS3 and ACE, as well as ACE and BDKR2, also in the absence of main effects between these genes. A three-gene model (BDKRB2, AMPD1, UCP2) was selected as the best one based on the highest value of testing accuracy (79%) using the MDR method (Gui et al., 2013). In this model, there was no gene NOS3, which has a significant effect on loss of interaction observed in the five-gene model. Further analysis carried out on the three-gene model did not show any interaction between any pair of 3 tested genes, while maintaining the main effect for AMPD1 and UCP2. The four-gene model comprising NOS3 was not chosen as the best one because of the 2% decreased testing accuracy as compared with the three-gene model.
Although the full model revealed some degree of interaction between NOS3 and BDKRB2 genes, the best model selected by cross validation consistency and prediction error did not include the NOS3 locus, thereby eliminating the gene-gene interaction. It should be noted that the model-free data mining method that we used in the current study can only detect (as many other tools) the statistical epistasis which does not guarantee any kind of interaction between biomolecules (biological epistasis) underlying the statistical phenomenon (Moore et al., 2006). Interestingly, Saunders et al. (2006) found a tendency for the −9/−9 BDKRB2 genotype combined with an NOS3 G allele to be overrepresented in the fastest finishing triathletes. However, the authors did not determine any biological meaning of this finding. As suggested by Oliveira-Paula et al. (2017), molecular mechanisms explaining the interactions among NOS3 and BDKRB2 could involve NOS3 activity and NO bioavailability.
In conclusion, our data suggest that the likelihood of becoming an elite half marathoner partly depends on the carriage of a high number of endurance-related alleles. Our results may help explain individual variations in human endurance performance.
Acknowledgements
The study was supported by the Czech Ministry of Sport (grant no. PROGRES Q41)Charles University (UNCE/HUM/032) and the Academy of Physical Education (grant no. 112/BS/IS/2016).
References
- Ahmetov II, Fedotovskaya ON. Current Progress in Sports Genomics. Adv Clin Chem. 2015;70:247. doi: 10.1016/bs.acc.2015.03.003. –. [DOI] [PubMed] [Google Scholar]
- Ahmetov II, Williams AG, Popov DV, Lyubaeva EV, Hakimullina AM, Fedotovskaya ON, Mozhayskaya IA, Vinogradova OL, Astratenkova IV, Montgomery HE, Rogozkin VA. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum Genet. 2009;126:751. doi: 10.1007/s00439-009-0728-4. –. [DOI] [PubMed] [Google Scholar]
- Ahmetov II, Popov DV, Astratenkova IV, Druzhevskaia AM, Missina SS, Vinogradova OL, Rogozkin VA. The use of molecular genetic methods for prognosis of aerobic and anaerobic performance in athletes. Hum Physiol. 2009;34:338. –. [PubMed] [Google Scholar]
- Alvarez R, Terrados N, Ortolano R, Iglesias-Cubero G, Reguero JR, Batalla A, Cortina A, Fernandez-Garcıa B, Rodrıguez C, Braga S, Alvarez V, Coto E. Genetic variation in the rennin angiotensin system and athletic performance. Eur J Appl Physiol. 2000;82:117. doi: 10.1007/s004210050660. –. [DOI] [PubMed] [Google Scholar]
- Astrup A, Toubro S, Dalgaard LT, Urhammer SA, Sorensen TI, Pedersen O. Impact of the v/v 55 polymorphism of the uncoupling protein 2 gene on 24-h energy expenditure and substrate oxidation. Int J Obesity Related Metab Disorder. 1999;23:1030. doi: 10.1038/sj.ijo.0801040. –. [DOI] [PubMed] [Google Scholar]
- Buemann B, Schierning B, Toubro S, Bibby BM, Sørensen T, Dalgaard L, Pedersen O, Astrup A. The association between the val/ala-55 polymorphism of the uncoupling protein 2 gene and exercise efficiency. Int J Obesity Related Metab Disorder. 2001;25:467. doi: 10.1038/sj.ijo.0801564. –. [DOI] [PubMed] [Google Scholar]
- Casas JP, Cavalleri GL, Bautista LE, Smeeth L, Humphries SE, Hingorani AD. Endothelial nitric oxide synthase gene polymorphisms and cardiovascular disease: a HuGE review. Am J Epidemiol. 2006;164:921. doi: 10.1093/aje/kwj302. –. [DOI] [PubMed] [Google Scholar]
- Cieszczyk P, Eider J, Ostanek M, Leońska-Duniec A, Ficek K, Kotarska K, Girdauskas G. Is the C34T polymorphism of the AMPD1 gene associated with athlete performance in rowing? Int J Sports Med. 2009;32:987. doi: 10.1055/s-0031-1283186. –. [DOI] [PubMed] [Google Scholar]
- Cieszczyk P, Krupecki K, Maciejewska A, Sawczuk M. The angiotensin converting enzyme gene I/D polymorphism in polish rowers. Int J Sports Med. 2009;30:624. doi: 10.1055/s-0029-1202825. –. [DOI] [PubMed] [Google Scholar]
- Eynon N, Hanson ED, Lucia A, Houweling PJ, Garton F, North KN, Bishop DJ. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013;43:803. doi: 10.1007/s40279-013-0059-4. –. [DOI] [PubMed] [Google Scholar]
- Eynon N, Meckel Y, Alves AJ, Nemet D, Eliakim A. Is there an interaction between BDKRB2-9/+9 and GNB3 C825T polymorphisms and elite athletic performance? Scand J Med Sci Sports. 2011;21:242. doi: 10.1111/j.1600-0838.2010.01261.x. –. [DOI] [PubMed] [Google Scholar]
- Fishbein WN, Armbrustmacher VW, Griffin JL. Myoadenylate deaminase deficiency: a new T. disease of muscle. Science. 1987;200:545. doi: 10.1126/science.644316. –. [DOI] [PubMed] [Google Scholar]
- Fleury C, Sanchis D. The mitochondrial uncoupling protein-2: current status. Int J Biochem Cell Biol. 1999;31:1261. doi: 10.1016/s1357-2725(99)00049-7. –. [DOI] [PubMed] [Google Scholar]
- Gayagay G, Yu B, Hambly B, Boston T, Hahn A, Celermajer DS, Trent RJ. Elite endurance athletes and the ACE I allele—the role of genes in athletic performance. Hum Genet. 1998;103:48. doi: 10.1007/s004390050781. –. [DOI] [PubMed] [Google Scholar]
- Ginevičienė V, Jakaitiene A, Pranculis A, Milašius K, Tubelis L, Utkus A. AMPD1 rs17602729 is associated with physical performance of sprint and power in elite Lithuanian athletes. BMC Genet. 2014;15:58. doi: 10.1186/1471-2156-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronek P, Holdys J. Genes and physical fitness. Trends Sport Sci. 2013;20:16. –. [Google Scholar]
- Gronek P, Holdys J, Konarski J, Kryściak J, Wolc A. ACE I/D genotype in Professional field hockey players. TRENDS Sport Sci. 2013;20:36. –. [Google Scholar]
- Gui J, Moore JH, Williams SM, Andrews P, Hillege HL, van der Harst P, Navis G, Van Gilst WH, Asselbergs FW, Gilbert-Diamond D. A Simple and Computationally Efficient Approach to Multifactor Dimensionality Reduction Analysis of Gene-Gene Interactions for Quantitative Traits. PLoS One. 2013;8:e66545. doi: 10.1371/journal.pone.0066545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdys J, Gronek P, Krysciak J, Stanisławski D. Genetics variants of uncoupling proteins-2 and-3 in relations to maximal oxygen uptake in different sports. Acta Biochim Pol. 2013;60:71. –. [PubMed] [Google Scholar]
- Holdys J, Kryściak J, Stanisławski D, Gronek P. Polymorphism of the ACTN3 Gene in Individuals Practising Different Sports Disciplines. Biol Sport. 2011;28:101. –. [Google Scholar]
- Hruskovicova H, Dzurenkova D, Selingerova M, Bohus B, Timkanicova B, Kovacs L. The angiotensin converting enzyme I/D polymorphism in long distance runners. J Sports Med Phys Fit. 2006;46:509. –. [PubMed] [Google Scholar]
- Jelakovic B, Kuzmanic D, Milicic D. Influence of angiotensin converting enzyme (ACE) gene polymorphism and circadian blood pressure (BP) changes on left ventricle (LV) mass in competitive oarsmen. Am J Hypertens. 2000;13:182A. [Google Scholar]
- Lucıa A, Gomez-Gallego F, Chicharro JL, Hoyos J, Celaya K, Cordova A, Villa C, Alonso JM, Barriopedro M. Perez M, Earnest CP. Is there an association between ACE and CKMM polymorphisms and cycling performance status during 3-week races? Int J Sports Med. 2005;26:442. doi: 10.1055/s-2004-821108. –. [DOI] [PubMed] [Google Scholar]
- Massidda M, Bachis V, Corrias L, Piras F, Scorcu M, Culigioni C, Masala D, Calò CM. ACTN3 R577X polymorphism is not associated with team sport athletic status in Italians. Sports Med Open. 2015;1:6. doi: 10.1186/s40798-015-0008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery HE, Marshall R, Hemingway H, Myerson S, Clarkson P, Dollery C, Hayward M, Holliman DE, Jubb M, World M, Thomas EL, Brynes AE, Saeed N, Barnard M, Bell JD, Prasad K, Rayson M, Talmud PJ, Humphries SE. Human gene for physical performance. Nature. 1998;393:221. doi: 10.1038/30374. –. [DOI] [PubMed] [Google Scholar]
- Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252. doi: 10.1016/j.jtbi.2005.11.036. –. [DOI] [PubMed] [Google Scholar]
- Morisaki T, Gross M, Morisaki H, Pongratz D, Zöllner N, Holmes EW. Molecular basis of AMP deaminase deficiency in skeletal muscle. Proc Natl Acad Sci U S A. 1992;89:6457. doi: 10.1073/pnas.89.14.6457. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Paula GH, Luizon MR, Lacchini R, Fontana V, Silva PS, Biagi C, Tanus-Santos JE. Gene-Gene Interactions Among PRKCA, NOS3 and BDKRB2 Polymorphisms Affect the Antihypertensive Effects of Enalapril. Basic Clin Pharmacol. 2017;120:284. doi: 10.1111/bcpt.12682. –. [DOI] [PubMed] [Google Scholar]
- Pe´russe L, Bouchard C. Endothelial nitric oxide synthase gene polymorphism and elite endurance athlete status: the Genathlete study. Scand J Med Sci Sports. 2008;18:485. doi: 10.1111/j.1600-0838.2007.00717.x. –. [DOI] [PubMed] [Google Scholar]
- Popp R, Fleming I, Busse R. Pulsatile stretch in coronary arteries elicits release of endothelium- derived hyperpolarizing factor: a modulator of arterial compliance. Circ Res. 1998;82:696. doi: 10.1161/01.res.82.6.696. –. [DOI] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Corvol P, Soubrier F. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidyl carboxypeptidase 1) Nucleic Acids Res. 1992;20:1433. doi: 10.1093/nar/20.6.1433-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio JC, Martin MA, Rabadan MF, Gómez-Gallego F, San Juan AF, Alonso JM, Chicharro JL, Pérez M, Arenas J, Lucia A. Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: does this mutation impair performance? J Applied Physiol. 2005;98:2108. doi: 10.1152/japplphysiol.01371.2004. –. [DOI] [PubMed] [Google Scholar]
- Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13:840. doi: 10.1038/sj.ejhg.5201410. –. [DOI] [PubMed] [Google Scholar]
- Saunders CJ, Xenophontos SL, Cariolou MA, Anastassiades LC, Noakes TD, Collins M. The bradykinin beta 2 receptor (BDKRB2) and endothelial nitric oxide synthase 3 (NOS3) genes and endurance performance during Ironman Triathlons. Hum Mol Genet. 2006;15:979. doi: 10.1093/hmg/ddl014. –. [DOI] [PubMed] [Google Scholar]
- Sawczuk M, Timshina YI, Astratenkova IV, Maciejewska-Karlowska A, Leońska-Duniec A, Ficek K, Mustafina LJ, Cięszczyk P, Klocek T, Ahmetov II. The -9/+9 polymorphism of the bradykinin receptor beta 2 gene and athlete status: A study involving two European cohorts. Hum Biol. 2013;85:741. doi: 10.3378/027.085.0511. –. [DOI] [PubMed] [Google Scholar]
- Scanavini D, Bernardi F, Castoldi E, Conconi F, Mazzoni G. Increased frequency of the homozygous II ACE genotype in Italian Olympic endurance athletes. Eur J Hum Genet. 2002;10:576. doi: 10.1038/sj.ejhg.5200852. –. [DOI] [PubMed] [Google Scholar]
- Sessa F, Chetta M, Petito A, Franzetti M, Bafunno V, Pisanelli D, Sarno M, Iuso S, Margaglione M. Gene polymorphisms and sport attitude in Italian athletes. Genet Test Mol Bioma. 2011;15:285. doi: 10.1089/gtmb.2010.0179. –. [DOI] [PubMed] [Google Scholar]
- Shenoy S, Tandon S, Sandhu J, Bhanwer AS. Association of converting enzyme gene polymorphism and Indian army triathletes performance. Asian Journal of Sports Medicine. 2010;1(3):143. doi: 10.5812/asjsm.34855. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomaes T, Thomis M, Onkelinx S, Fagard R, Matthijs G, Buys R, Schepers D, Cornelissen V, Vanhees L. A genetic predisposition score for muscular endophenotypes predicts the increase in aerobic power after training: the CAREGENE study. BMC Genet. 2011;12:84. doi: 10.1186/1471-2156-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsianos GI, Evangelou E, Boot A, Zillikens MC, van Meurs JBJ, Uitterlinden AG, Ioannidis JPA. Associations of polymorphisms of eight muscle- or metabolism-related genes with performance in Mount Olympus marathon runners. J Applied Physiol. 2010;108:567. doi: 10.1152/japplphysiol.00780.2009. –. [DOI] [PubMed] [Google Scholar]
- Tsianos G, Sanders J, Dhamrait S, Humphries S, Grant S, Montgomery H. The ACE gene insertion/deletion polymorphism and elite endurance swimming. Eur J Applied Physiol. 2004;92:360. doi: 10.1007/s00421-004-1120-7. –. [DOI] [PubMed] [Google Scholar]
- Wilkinson AV, Gabriel KP, Wang J, Bondy ML, Dong Q, Wu X, Shete S, Spitz MR. Sensation seeking genes and physical activity in youth. Genes Brain Behav. 2013;12:181. doi: 10.1111/gbb.12006. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AG, Dhamrait SS, Wootton PTE, Day SH, Hawe E, Payne JR, Myerson SG, World M, Budgett R, Humphries SE, Montgomery HE. Bradykinin receptor gene variant and human physical performance. J Applied Physiol. 2004;96:938. doi: 10.1152/japplphysiol.00865.2003. –. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhu Z, Fang X, Yin L, Liu Y, Xu S, Li A. The Association between NOS3 Gene Polymorphisms and Hypoxic-Ischemic Encephalopathy Susceptibility and Symptoms in Chinese Han Population. BioMed Research Int. 2016. Article ID 1957374. [DOI] [PMC free article] [PubMed]
- Zarebska A, Jastrzebski Z, Ahmetov II, Zmijewski P, Cięszczyk P, Leonska-Duniec A, Sawczuk M, Leznicka K, Trybek G, Semenova EA, Maciejewska-Skrendo A. GSTP1 c.313A>G polymorphism in Russian and Polish athletes. Physiol Genomics. 2017;49:127. doi: 10.1152/physiolgenomics.00014.2016. –. [DOI] [PubMed] [Google Scholar]


