Abstract
The comparative proteomic data presented in this article provide supporting information to the related research article "Proteomic identification of elevated saliva kallikrein levels in the mdx-4cv mouse model of Duchenne muscular dystrophy " (Murphy et al., 2018). Here we provide additional datasets on the comparative proteomic analysis of saliva and serum proteins and the mass spectrometric identification of kallikrein isoform Klk-1 in wild type versus mdx-4cv saliva specimens. The data article presents the systematic identification of the assessable saliva proteome and the differential presence of proteins in saliva versus serum samples. Representative mass spectrometric scans of unique peptides that were employed to identify the kallikrein isoform Klk-1 in wild type versus mdx-4cv saliva specimens are provided. The dataset contains typical saliva-associated marker proteins, including alpha-amylase and albumin, as well as distinct isoforms of cystatin, serpin, kallikrein, cathepsin, glutathione transferase, carbonic anhydrase, mucin, pyruvate kinase, and aldolase.
Specifications table
| Subject area | Biology |
| More specific subject area | Biomedicine |
| Type of data | Tables, MS/MS scans, Venn diagram |
| How data was acquired | LC-MS/MS analysis, using an Ultimate 3000 NanoLC system (Dionex Corporation, Sunnyvale, CA, USA) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific) |
| Data format | Analyzed |
| Experimental factors | Protein was extracted from whole saliva and pre-fractionated serum specimens from wild type versus dystrophic mdx-4cv mice. |
| Experimental features | Comparative mass spectrometry-based proteomic profiling of the saliva and serum fraction. |
| Data source location | Maynooth, Ireland |
| Data accessibility | The data are available with this article |
| Related research article | Murphy S, Zweyer M, Mundegar RR, Swandulla D, Ohlendieck K. Proteomic identification of elevated saliva kallikrein levels in the mdx-4cv mouse model of Duchenne muscular dystrophy. Biochem Biophys Rep. (2018) In press [1] |
Value of the data
-
•
Proteomic data presented here provide an overview of biofluid changes in the mdx-4cv mouse model of X-linked muscular dystrophy.
-
•
This data provide comparative listings of proteins in saliva versus serum specimens, as well as their mass spectrometric identification.
-
•
The mass spectrometric data are valuable to serve as a pathobiochemical biofluid signature of the dystrophin-deficient mdx-4cv mouse.
1. Data
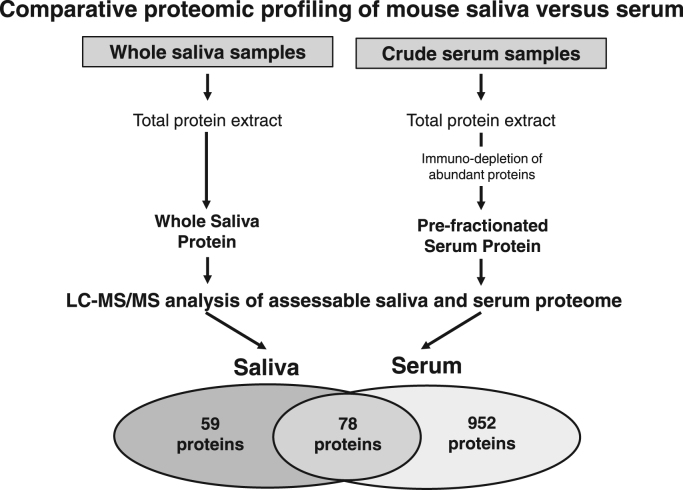
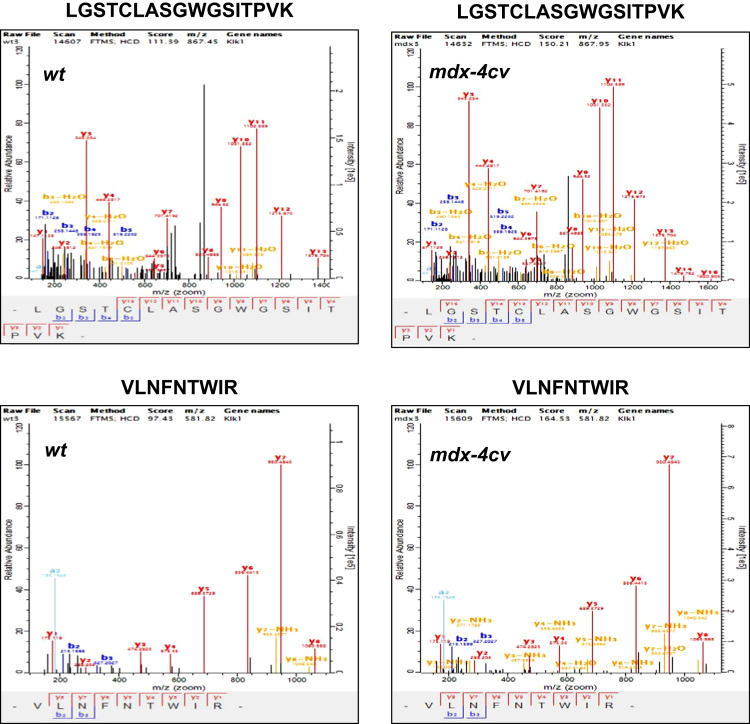
The data presented relate to the systematic survey of whole saliva using mass spectrometry-based proteomics of the mdx-4cv mouse model of Duchenne muscular dystrophy [1]. This accompanying article lists the proteomic identification of the total saliva protein population and the differential presence of protein species in saliva versus serum samples, as well as representative MS/MS scans of unique peptides that were used to identify the kallikrein isoform Klk-1 in wild type versus mdx-4cv saliva specimens. Table 1 lists the mass spectrometric profiling of the mouse saliva proteome. Listed are the protein name, gene name, the number of unique peptides, the number of total peptides, the relative molecular mass, and the estimated isoelectric point of the identified protein species. A set of typical marker proteins of whole saliva were identified, including alpha-amylase and albumin, as well as distinct isoforms of cystatin, serpin, kallikrein, cathepsin, glutathione transferase, carbonic anhydrase, mucin, pyruvate kinase, and aldolase [2], [3], [4], [5]. The identified protein species in saliva were compared with the previously established serum proteome [6]. Fig. 1 shows a Venn diagram of the distribution of proteins that are shared between saliva and serum, and protein species that are uniquely associated with saliva versus serum samples. Table 2, Table 3 list the mass spectrometric identification of proteins identified in saliva only or are shared between serum and saliva. In Table 2 are listed 59 proteins found in wild-type saliva, but not serum, including carbonic anhydrase 6, BPI fold-containing family A members 1 and 2, cystatin 10, cardiomyopathy-associated protein 5, mucin-19, and desmoplakin. Table 3 lists 78 proteins found in both serum and saliva, including alpha-amylase, cathepsin D, serum albumin, and fructose-bisphosphate aldolase A, as well as kallikrein-1 and Klk1-related peptidases b1, b3, b4, b5, b8, b9, b11, b16, b21, b22, b24, b26, and b27. In addition to the MS/MS scans of the unique peptide NNFLEDEPSAQHR shown in the accompanying research manuscript [1], Fig. 2 displays additional MS/MS scans of the unique peptides LGSTCLASGWGSITPVK and VLNFNTWIR that were used to identify the Klk-1 isoform in both wild type and mdx-4cv samples.
Table 1.
Mass spectrometry-based proteomic identification of proteins in whole saliva from wild type mouse.
| Protein name | Gene | Number of unique peptides | Number of peptides | Molecular mass kDa | Isoelectric point pI |
|---|---|---|---|---|---|
| Mucin-19 | Muc19 | 4 | 4 | 693.1 | 5.54 |
| Cardiomyopathy-associated protein 5 | Cmya5 | 1 | 1 | 412.8 | 4.75 |
| Desmoplakin | Dsp | 1 | 1 | 332.7 | 6.80 |
| Hornerin | Hrnr | 1 | 1 | 247.4 | 10.33 |
| Ovostatin | Ovos | 6 | 6 | 162.2 | 5.26 |
| WD repeat-containing protein 7 | Wdr7 | 1 | 1 | 160.2 | 7.01 |
| Calcium-dependent secretion activator 2 | Cadps2 | 2 | 2 | 143.8 | 6.14 |
| Pro-epidermal growth factor | Egf | 10 | 10 | 133.0 | 6.46 |
| Repetin | Rptn | 1 | 1 | 128.5 | 7.61 |
| Collagen alpha-1(I) chain | Col1a1 | 1 | 1 | 117.7 | 5.72 |
| Lysosomal alpha-mannosidase | Man2b1 | 14 | 14 | 114.6 | 8.13 |
| Aminopeptidase N | Anpep | 4 | 4 | 109.6 | 5.90 |
| Zinc finger CCHC domain-containing protein 14 | Zcchc14 | 1 | 1 | 98.6 | 8.25 |
| Dipeptidyl peptidase 4 | Dpp4 | 1 | 1 | 87.4 | 6.42 |
| Neprilysin | Mme | 2 | 2 | 85.6 | 5.81 |
| Heat shock protein 75 kDa, mitochondrial | Trap1 | 1 | 1 | 80.2 | 6.68 |
| Cytosolic carboxypeptidase-like protein 5 | Agbl5 | 1 | 1 | 80.1 | 8.24 |
| Solute carrier family 15 member 1 | Slc15a1 | 1 | 1 | 78.5 | 7.93 |
| Amyloid beta A4 protein | App | 1 | 1 | 78.4 | 4.83 |
| Lactotransferrin | Ltf | 1 | 1 | 77.8 | 8.53 |
| Protein-glutamine gamma-glutamyltransferase E | Tgm3 | 2 | 2 | 77.3 | 6.81 |
| Galactocerebrosidase | Galc | 2 | 2 | 77.2 | 6.74 |
| Stress-70 protein, mitochondrial | Hspa9 | 1 | 1 | 73.4 | 6.07 |
| Keratin, type II cytoskeletal 2, epidermal | Krt2 | 4 | 5 | 70.9 | 8.06 |
| Heat shock cognate 71 kDa protein | Hspa8 | 1 | 1 | 70.8 | 5.52 |
| Serum albumin | Alb | 3 | 3 | 68.6 | 6.07 |
| Keratin, type II cytoskeletal 1 | Krt1 | 5 | 7 | 65.6 | 8.15 |
| Sulfhydryl oxidase 1 | Qsox1 | 2 | 2 | 63.3 | 7.93 |
| Keratin, type II cytoskeletal 2, oral | Krt76 | 8 | 11 | 62.8 | 8.43 |
| Vomeromodulin | Bpifb9a | 15 | 15 | 62.4 | 5.68 |
| Keratin, type II cytoskeletal 5 | Krt5 | 3 | 9 | 61.7 | 7.75 |
| Prosaposin | Psap | 1 | 1 | 61.4 | 5.19 |
| Beta-hexosaminidase subunit beta | Hexb | 9 | 9 | 61.1 | 8.12 |
| Keratin, type II cytoskeletal 6B | Krt6b | 1 | 16 | 60.3 | 8.32 |
| Keratin, type II cytoskeletal 6A | Krt6a | 2 | 17 | 59.3 | 7.94 |
| Keratin, type II cytoskeletal 73 | Krt73 | 1 | 3 | 58.9 | 8.09 |
| Biotinidase | Btd | 1 | 1 | 58.1 | 5.80 |
| Pyruvate kinase | Pkm | 3 | 3 | 57.8 | 7.47 |
| N-acetylgalactosamine-6-sulfatase | Galns | 1 | 1 | 57.6 | 6.52 |
| Alpha-amylase 1 | Amy1 | 10 | 10 | 57.6 | 6.96 |
| Keratin, type II cytoskeletal 79 | Krt79 | 1 | 3 | 57.5 | 7.69 |
| Keratin, type I cytoskeletal 10 | Krt10 | 9 | 11 | 57.0 | 5.07 |
| Keratin, type II cytoskeletal 4 | Krt4 | 19 | 21 | 56.2 | 8.15 |
| Podocalyxin | Podxl | 1 | 1 | 53.4 | 4.97 |
| Aldehyde dehydrogenase family 3 member B2 | Aldh3b2 | 2 | 2 | 52.9 | 6.09 |
| Keratin, type I cytoskeletal 14 | Krt14 | 3 | 7 | 52.8 | 5.17 |
| Acidic mammalian chitinase | Chia | 3 | 3 | 52.0 | 5.06 |
| Angiotensinogen | Agt | 1 | 1 | 52.0 | 5.44 |
| Keratin, type I cytoskeletal 16 | Krt16 | 4 | 7 | 51.6 | 5.20 |
| Carboxypeptidase Q | Cpq | 4 | 4 | 50.5 | 6.40 |
| Aldehyde dehydrogenase, dimeric NADP-preferring | Aldh3a1 | 2 | 2 | 50.4 | 6.95 |
| Keratin, type I cytoskeletal 42 | Krt42 | 1 | 5 | 50.1 | 5.16 |
| Elongation factor 1-alpha 1 | Eef1a1 | 3 | 3 | 50.1 | 9.01 |
| Serpin B12 | Serpinb12 | 1 | 1 | 47.8 | 5.17 |
| Keratin, type I cytoskeletal 13 | Krt13 | 14 | 18 | 47.7 | 4.86 |
| Transcobalamin-2 | Tcn2 | 1 | 1 | 47.6 | 6.33 |
| Alpha-N-acetyl-galactosaminidase | Naga | 1 | 1 | 47.2 | 6.44 |
| Alpha-enolase | Eno1 | 2 | 2 | 47.1 | 6.80 |
| Rab GDP dissociation inhibitor beta | Gdi2 | 1 | 1 | 46.6 | 6.90 |
| Chitinase-like protein 4 | Chil4 | 4 | 4 | 44.9 | 6.19 |
| Cathepsin D | Ctsd | 4 | 4 | 44.9 | 7.15 |
| Phosphoglycerate kinase 2 | Pgk2 | 1 | 1 | 44.8 | 6.80 |
| MANSC domain-containing protein 1 | Mansc1 | 1 | 1 | 44.8 | 9.11 |
| Renin-1 | Ren1 | 2 | 2 | 44.3 | 7.17 |
| Prostatic acid phosphatase | Acpp | 1 | 1 | 43.7 | 6.24 |
| Serpin B11 | Serpinb11 | 1 | 1 | 43.5 | 8.94 |
| Synaptic vesicle membrane protein VAT-1 | Vat1 | 1 | 1 | 43.1 | 6.37 |
| Serpin B6 | Serpinb6 | 4 | 4 | 42.6 | 5.74 |
| Actin, cytoplasmic 1 | Actb | 4 | 4 | 41.7 | 5.48 |
| Adenosine deaminase | Ada | 3 | 3 | 40.0 | 5.72 |
| Fructose-bisphosphate aldolase A | Aldoa | 2 | 2 | 39.3 | 8.09 |
| Annexin A1 | Anxa1 | 1 | 1 | 38.7 | 7.37 |
| Protein LEG1 | Leg1 | 2 | 2 | 38.3 | 4.36 |
| Guanine nucleotide-binding protein subunit beta-4 | Gnb4 | 1 | 1 | 37.4 | 6.16 |
| Malate dehydrogenase, cytoplasmic | Mdh1 | 3 | 3 | 36.5 | 6.58 |
| L-lactate dehydrogenase A chain | Ldha | 2 | 2 | 36.5 | 7.74 |
| Carbonic anhydrase 6 | Ca6 | 7 | 7 | 36.5 | 6.60 |
| Gamma-glutamyl hydrolase | Ggh | 3 | 3 | 35.4 | 8.29 |
| Polyubiquitin-B | Ubb | 1 | 1 | 34.3 | 7.53 |
| Triosephosphate isomerase | Tpi1 | 2 | 2 | 32.2 | 5.74 |
| Deoxyribonuclease-1 | Dnase1 | 3 | 3 | 32.0 | 4.92 |
| Phospholipid phosphatase 1 | Plpp1 | 1 | 1 | 31.9 | 7.02 |
| Syntaxin-3 | Stx3 | 1 | 1 | 30.9 | 5.63 |
| Syntaxin-7 | Stx7 | 3 | 3 | 29.8 | 5.78 |
| Kallikrein 1-related peptidase b1 | Klk1b1 | 4 | 8 | 29.0 | 8.10 |
| Kallikrein 1-related peptidase b3 | Klk1b3 | 4 | 7 | 29.0 | 6.84 |
| Kallikrein 1-related peptidase b24 | Klk1b24 | 3 | 9 | 28.9 | 8.16 |
| Kallikrein 1-related peptidase b9 | Klk1b9 | 5 | 9 | 28.9 | 7.64 |
| Kallikrein-1 | Klk1 | 2 | 6 | 28.8 | 5.12 |
| Kallikrein 1-related peptidase b5 | Klk1b5 | 4 | 7 | 28.7 | 5.59 |
| Kallikrein 1-related peptidase b27 | Klk1b27 | 3 | 9 | 28.7 | 8.56 |
| Kallikrein 1-related peptidase b11 | Klk1b11 | 5 | 9 | 28.7 | 7.14 |
| Kallikrein 1-related peptidase b16 | Klk1b16 | 7 | 9 | 28.7 | 5.64 |
| Kallikrein 1-related peptidase b21 | Klk1b21 | 2 | 8 | 28.7 | 7.37 |
| BPI fold-containing family A member 1 | Bpifa1 | 1 | 1 | 28.6 | 6.51 |
| Kallikrein 1-related peptidase-like b4 | Klk1b4 | 6 | 7 | 28.5 | 4.86 |
| Kallikrein 1-related peptidase b8 | Klk1b8 | 7 | 11 | 28.5 | 8.00 |
| Kallikrein 1-related peptidase b26 | Klk1b26 | 3 | 9 | 28.4 | 6.86 |
| Kallikrein 1-related peptidase b22 | Klk1b22 | 5 | 6 | 28.4 | 6.65 |
| 14-3-3 protein zeta/delta | Ywhaz | 1 | 1 | 27.8 | 4.79 |
| Cysteine-rich secretory protein 1 | Crisp1 | 3 | 4 | 27.7 | 6.87 |
| Glutathione S-transferase omega-1 | Gsto1 | 2 | 2 | 27.5 | 7.36 |
| Cysteine-rich secretory protein 3 | Crisp3 | 1 | 2 | 27.3 | 8.37 |
| Beta-nerve growth factor | Ngf | 3 | 3 | 27.1 | 9.47 |
| Ras-related protein Rab-27A | Rab27a | 1 | 1 | 25.0 | 5.36 |
| BPI fold-containing family A member 2 | Bpifa2 | 4 | 4 | 24.7 | 5.01 |
| Ras-related protein Rab-2A | Rab2a | 1 | 1 | 23.5 | 6.54 |
| Rho GDP-dissociation inhibitor 1 | Arhgdia | 1 | 1 | 23.4 | 5.20 |
| Synaptosomal-associated protein 23 | Snap23 | 1 | 1 | 23.2 | 4.98 |
| Ras-related protein Rab-10 | Rab10 | 1 | 3 | 22.5 | 8.38 |
| Ras-related protein Rab-1B | Rab1b | 1 | 3 | 22.2 | 5.73 |
| Peroxiredoxin-1 | Prdx1 | 1 | 1 | 22.2 | 8.12 |
| Major urinary protein 3 | Mup3 | 1 | 1 | 21.5 | 4.81 |
| Vomeronasal secretory protein 2 | Lcn4 | 1 | 1 | 21.4 | 5.73 |
| Ras-related protein Rap-1A | Rap1a | 1 | 1 | 21.0 | 6.67 |
| Major urinary protein 5 | Mup5 | 3 | 3 | 20.9 | 4.86 |
| Placenta-expressed transcript 1 protein | Plet1 | 1 | 1 | 20.8 | 6.14 |
| Vomeronasal secretory protein 1 | Lcn3 | 1 | 1 | 20.6 | 4.60 |
| Major urinary protein 4 | Mup4 | 4 | 4 | 20.5 | 5.80 |
| Tumor protein D52 | Tpd52 | 1 | 1 | 20.0 | 4.88 |
| Odorant-binding protein 2a | Obp2a | 2 | 2 | 20.0 | 6.42 |
| Odorant-binding protein 1b | Obp1b | 3 | 3 | 19.4 | 6.29 |
| Protein MAL2 | Mal2 | 1 | 1 | 19.1 | 6.49 |
| Destrin | Dstn | 1 | 1 | 18.5 | 7.97 |
| Odorant-binding protein 1a | Obp1a | 4 | 4 | 18.5 | 5.67 |
| Peptidyl-prolyl cis-trans isomerase A | Ppia | 1 | 1 | 18.0 | 7.90 |
| Nucleoside diphosphate kinase B | Nme2 | 3 | 3 | 17.4 | 7.50 |
| Prolactin-inducible protein | Pip | 4 | 4 | 16.8 | 4.78 |
| Calmodulin-4 | Calm4 | 1 | 1 | 16.8 | 4.89 |
| Cystatin 10 | Cst10 | 2 | 2 | 16.4 | 7.72 |
| Superoxide dismutase [Cu-Zn] | Sod1 | 3 | 3 | 15.9 | 6.51 |
| Submaxillary gland androgen-regulated protein 3A | Smr3a | 1 | 1 | 15.5 | 9.09 |
| Profilin-1 | Pfn1 | 1 | 1 | 14.9 | 8.28 |
| Protein S100-A9 | S100a9 | 1 | 1 | 13.0 | 7.17 |
| Secretoglobin family 2B member 2 | Scgb2b2 | 1 | 1 | 12.8 | 5.95 |
| Vesicle-associated membrane protein 8 | Vamp8 | 2 | 2 | 11.4 | 8.19 |
| Protein S100-A1 | S100a1 | 1 | 1 | 10.5 | 4.50 |
Fig. 1.
Overview of the comparative proteomic profiling of mouse saliva and serum. Shown is the flow chart of the preparation of saliva and serum protein populations for the mass spectrometry-based proteomic identification of biofluid markers. The Venn diagram illustrates the distribution of protein species between saliva and serum.
Table 2.
Mass spectrometry-based proteomic identification of proteins present in whole saliva from wild type mouse, but not serum.
| Accession Number | Protein name | Gene name |
|---|---|---|
| P07744 | Keratin, type II cytoskeletal 4 | Krt4 |
| Q80XI7 | Vomeromodulin | Bpifb9a |
| P08730 | Keratin, type I cytoskeletal 13 | Krt13 |
| P18761 | Carbonic anhydrase 6 | Ca6 |
| Q9Z331 | Keratin, type II cytoskeletal 6B | Krt6b |
| P07743 | BPI fold-containing family A member 2 | Bpifa2 |
| Q9D3H2 | Odorant-binding protein 1a | Obp1a |
| P11590 | Major urinary protein 4 | Mup4 |
| P11591 | Major urinary protein 5 | Mup5 |
| P02535-2 | Isoform 2 of Keratin, type I cytoskeletal 10 | Krt10 |
| A2AEP0 | Odorant-binding protein 1b | Obp1b |
| P61027 | Ras-related protein Rab-10 | Rab10 |
| Q9JM84 | Cystatin 10 | Cst10 |
| Q6UGQ3 | Secretoglobin family 2B member 2 | Scgb2b2 |
| Q91Z98 | Chitinase-like protein 4 | Chil4 |
| Q8C6C9 | Protein LEG1 homolog | Leg1 |
| Q91XA9 | Acidic mammalian chitinase | Chia |
| P06281 | Renin-1 | Ren1 |
| Q9JM83 | Calmodulin-4 | Calm4 |
| P49183 | Deoxyribonuclease-1 | Dnase1 |
| Q61900 | Submaxillary gland androgen-regulated protein 3A | Smr3a |
| O09044 | Synaptosomal-associated protein 23 | Snap23 |
| Q62472 | Vomeronasal secretory protein 2 | Lcn4 |
| P38647 | Stress-70 protein, mitochondrial | Hspa9 |
| Q62471 | Vomeronasal secretory protein 1 | Lcn3 |
| Q62465 | Synaptic vesicle membrane protein VAT-1 homolog | Vat1 |
| Q8BI08 | Protein MAL2 | Mal2 |
| P53994 | Ras-related protein Rab-2A | Rab2a |
| Q9ERI2 | Ras-related protein Rab-27A | Rab27a |
| P97361 | BPI fold-containing family A member 1 | Bpifa1 |
| Q61469 | Phospholipid phosphatase 1 | Plpp1 |
| Q09M02–6 | Isoform 6 of Cytosolic carboxypeptidase-like protein 5 | Agbl5 |
| Q62393-2 | Isoform 2 of Tumor protein D52 | Tpd52 |
| Q3UU35 | Ovostatin homolog | Ovos |
| Q8BND5-3 | Isoform 3 of Sulfhydryl oxidase 1 | Qsox1 |
| Q8VIG0–2 | Isoform 2 of Zinc finger CCHC domain-containing protein 14 | Zcchc14 |
| P47739 | Aldehyde dehydrogenase, dimeric NADP-preferring | Aldh3a1 |
| P10107 | Annexin A1 | Anxa1 |
| E9Q3E1 | Aldehyde dehydrogenase family 3 member B2 | Aldh3b2 |
| Q9R0M4 | Podocalyxin | Podxl |
| P09041 | Phosphoglycerate kinase 2 | Pgk2 |
| Q08189 | Protein-glutamine gamma-glutamyltransferase E | Tgm3 |
| P54818 | Galactocerebrosidase | Galc |
| Q9CR33 | MANSC domain-containing protein 1 | Mansc1 |
| Q9D7P9 | Serpin B12 | Serpinb12 |
| Q64704-3 | Isoform 3C of Syntaxin-3 | Stx3 |
| Q8CE08 | Prostatic acid phosphatase | Acpp |
| Q920I9-2 | Isoform 2 of WD repeat-containing protein 7 | Wdr7 |
| P29387 | Guanine nucleotide-binding protein subunit beta-4 | Gnb4 |
| Q9QWR8 | Alpha-N-acetylgalactosaminidase | Naga |
| Q8VHD8 | Hornerin | Hrnr |
| Q9CQN1 | Heat shock protein 75 kDa, mitochondrial | Trap1 |
| Q8BYR5-5 | Isoform 5 of Calcium-dependent secretion activator 2 | Cadps2 |
| Q9JIP7 | Solute carrier family 15 member 1 | Slc15a1 |
| P12023-2 | Isoform APP695 of Amyloid beta A4 protein | App |
| P97347 | Repetin | Rptn |
| Q70KF4 | Cardiomyopathy-associated protein 5 | Cmya5 |
| Q6PZE0 | Mucin-19 | Muc19 |
| E9Q557 | Desmoplakin | Dsp |
Table 3.
Mass spectrometry-based proteomic identification of proteins that are present in both saliva and serum from wild type mouse.
| Accession number | Protein name | Gene name |
|---|---|---|
| P05064 | Fructose-bisphosphate aldolase A | Aldoa |
| P00756 | Kallikrein 1-related peptidase b3 | Klk1b3 |
| P07724 | Serum albumin | Alb |
| Q01768 | Nucleoside diphosphate kinase B | Nme2 |
| P15946 | Kallikrein 1-related peptidase b11 | Klk1b11 |
| P00755 | Kallikrein 1-related peptidase b1 | Klk1b1 |
| P35700 | Peroxiredoxin-1 | Prdx1 |
| P06151 | L-lactate dehydrogenase A chain | Ldha |
| P15948 | Kallikrein 1-related peptidase b22 | Klk1b22 |
| P07628 | Kallikrein 1-related peptidase b8 | Klk1b8 |
| P15949 | Kallikrein 1-related peptidase b9 | Klk1b9 |
| P17751 | Triosephosphate isomerase | Tpi1 |
| P60710 | Actin, cytoplasmic 1 | Actb |
| P52480 | Pyruvate kinase PKM | Pkm |
| P04071 | Kallikrein 1-related peptidase b16 | Klk1b16 |
| P62962 | Profilin-1 | Pfn1 |
| Q9JM71 | Kallikrein 1-related peptidase b27 | Klk1b27 |
| P17182 | Alpha-enolase | Eno1 |
| P36369 | Kallikrein 1-related peptidase b26 | Klk1b26 |
| P08228 | Superoxide dismutase [Cu-Zn] | Sod1 |
| P14152 | Malate dehydrogenase, cytoplasmic | Mdh1 |
| P0CG49 | Polyubiquitin-B | Ubb |
| Q61759 | Kallikrein 1-related peptidase b21 | Klk1b21 |
| P17742 | Peptidyl-prolyl cis-trans isomerase A | Ppia |
| P63017 | Heat shock cognate 71 kDa protein | Hspa8 |
| P15945 | Kallikrein 1-related peptidase b5 | Klk1b5 |
| P63101 | 14-3-3 protein zeta/delta | Ywhaz |
| O88968 | Transcobalamin-2 | Tcn2 |
| P00757 | Kallikrein 1-related peptidase-like b4 | Klk1b4 |
| Q61754 | Kallikrein 1-related peptidase b24 | Klk1b24 |
| P00687 | Alpha-amylase 1 | Amy1 |
| P15947 | Kallikrein-1 | Klk1 |
| Q61598-2 | Isoform 2 of Rab GDP dissociation inhibitor beta | Gdi2 |
| Q8CIF4 | Biotinidase | Btd |
| Q99PT1 | Rho GDP-dissociation inhibitor 1 | Arhgdia |
| Q03401 | Cysteine-rich secretory protein 1 | Crisp1 |
| O09131 | Glutathione S-transferase omega-1 | Gsto1 |
| P04939 | Major urinary protein 3 | Mup3 |
| O09159 | Lysosomal alpha-mannosidase | Man2b1 |
| P20060 | Beta-hexosaminidase subunit beta | Hexb |
| P11859 | Angiotensinogen | Agt |
| Q9WVJ3-2 | Isoform 2 of Carboxypeptidase Q | Cpq |
| P10126 | Elongation factor 1-alpha 1 | Eef1a1 |
| P56565 | Protein S100-A1 | S100a1 |
| Q9R0P5 | Destrin | Dstn |
| Q922U2 | Keratin, type II cytoskeletal 5 | Krt5 |
| Q6IFX2 | Keratin, type I cytoskeletal 42 | Krt42 |
| Q61781 | Keratin, type I cytoskeletal 14 | Krt14 |
| P50446 | Keratin, type II cytoskeletal 6A | Krt6a |
| P08071 | Lactotransferrin | Ltf |
| P01132 | Pro-epidermal growth factor | Egf |
| Q9Z2K1 | Keratin, type I cytoskeletal 16 | Krt16 |
| Q03402 | Cysteine-rich secretory protein 3 | Crisp3 |
| P01139 | Beta-nerve growth factor | Ngf |
| Q9D1G1 | Ras-related protein Rab-1B | Rab1b |
| P97449 | Aminopeptidase N | Anpep |
| Q3TTY5 | Keratin, type II cytoskeletal 2 epidermal | Krt2 |
| P31725 | Protein S100-A9 | S100a9 |
| Q60854 | Serpin B6 | Serpinb6 |
| Q8VEN2-2 | Isoform 2 of Placenta-expressed transcript 1 protein | Plet1 |
| O70404 | Vesicle-associated membrane protein 8 | Vamp8 |
| P18242 | Cathepsin D | Ctsd |
| Q61207 | Prosaposin | Psap |
| Q9CQV3 | Serpin B11 | Serpinb11 |
| P11087-2 | Isoform 2 of Collagen alpha-1(I) chain | Col1a1 |
| P28843 | Dipeptidyl peptidase 4 | Dpp4 |
| Q6NXH9 | Keratin, type II cytoskeletal 73 | Krt73 |
| Q3UV17 | Keratin, type II cytoskeletal 2 oral | Krt76 |
| Q8VED5 | Keratin, type II cytoskeletal 79 | Krt79 |
| P62835 | Ras-related protein Rap-1A | Rap1a |
| P04104 | Keratin, type II cytoskeletal 1 | Krt1 |
| P02816 | Prolactin-inducible protein homolog | Pip |
| Q8K1H9 | Odorant-binding protein 2a | Obp2a |
| Q571E4 | N-acetylgalactosamine-6-sulfatase | Galns |
| Q9Z0L8-2 | Isoform II of Gamma-glutamyl hydrolase | Ggh |
| P03958 | Adenosine deaminase | Ada |
| Q61391 | Neprilysin | Mme |
| O70439 | Syntaxin-7 | Stx7 |
Fig. 2.
Proteomic identification of kallikrein isoform Klk1 in saliva from the wild type versus the mdx-4cv mouse model of Duchenne muscular dystrophy. Shown are representative MS/MS scans of the unique Klk-1 peptides LGSTCLASGWGSITPVK and VLNFNTWIR, which were identified and compared in wild type versus mdx-4cv saliva, respectively.
2. Experimental design, materials, and methods
Details of the methodological approach used in this study are available in [1], [6].
2.1. Sample collection and processing
For the proteomic profiling of easily assessable biofluids, saliva and serum specimens were obtained from 6-month-old dystrophic mdx-4cv and age-matched wild type C57BL/6 mice through the Bioresource Unit of the University of Bonn [6], where mice were kept under standard conditions according to German legislation on the use of animals in experimental research. Sample collection and preparation of protein extracts were carried out as previously described in detail [1], [6]. The collected saliva and serum specimens were transported to Maynooth University on dry ice in accordance with the Department of Agriculture (animal by-product register number 2016/16 to the Department of Biology, National University of Ireland, Maynooth).
2.2. Mass spectrometric analysis of saliva and serum proteins
Serum samples were processed as previously described [6]. For the proteomic analysis of saliva samples, 30 µg of protein was processed by the filter-aided sample preparation (FASP) method, as described in detail by Wiśniewski et al. [7], using a trypsin to protein ratio of 1:25 (protease:protein). Following overnight digestion and elution of peptides from the spin filter, 2% trifluoroacetic acid (TFA) in 20% acetonitrile (ACN) was added to the filtrates (3:1 (v/v) dilution). Peptides were analyzed by label-free liquid chromatography mass spectrometry (LC-MS/MS) by a standardized method using an Ultimate 3000 NanoLC system (Dionex Corporation, Sunnyvale, CA, USA) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific) as previously described in detail [1], [6], [8], [9].
2.3. Protein identification and quantification
Proteins present in the wild type and the mdx-4cv salivary and serum proteomes were initially identified using Proteome Discoverer 1.4 against Sequest HT (SEQUEST HT algorithm, licence Thermo Scientific, registered trademark University of Washington, USA) using the UniProtKB/Swiss-Prot database, with 25,041 sequences for Mus musculus [1], [6]. Identified saliva peptides were then filtered using a minimum XCorr score of 1.5 for 1, 2.0 for 2, 2.25 for 3, and 2.5 for 4 charge states, with peptide probability set to high confidence. For quantitative analysis, samples were evaluated with MaxQuant software (version 1.6.1.0) and the Andromeda search engine used to explore the detected features against the UniProtKB/SwissProt database for Mus musculus. The following search parameters were used: (i) first search peptide tolerance of 20 ppm, (ii) main search peptide tolerance of 4.5 ppm, (iii) cysteine carbamidomethylation set as a fixed modification, (iv) methionine oxidation set as a variable modification, (v) a maximum of two missed cleavage sites, and (vi) a minimum peptide length of seven amino acids. The false discovery rate (FDR) was set to 1% for both peptides and proteins using a target-decoy approach. Relative quantification was performed using the MaxLFQ algorithm [10]. The “proteinGroups.txt” file produced by MaxQuant was further analysed in Perseus (version 1.5.1.6). Proteins that matched to the reverse database or a contaminants database or that were only identified by site were removed. The LFQ intensities were log2 transformed, and only proteins found in all eight replicates in at least one group were used for further analysis. Data imputation was performed to replace missing values with values that simulate signals from peptides with low abundance chosen from a normal distribution specified by a downshift of 1.8 times the mean standard deviation of all measured values and a width of 0.3 times this standard deviation [11]. A two-sample t-test was performed using p<0.05 on the post imputated data to identify statistically significant differentially abundant proteins.
Acknowledgements
Research was supported by a Hume Scholarship from Maynooth University, and project grants from Muscular Dystrophy Ireland and the Irish Health Research Board (HRB/MRCG-2016-20). The Q-Exactive quantitative mass spectrometer was funded under the Research Infrastructure Call 2012 by Science Foundation Ireland (SFI-12/RI/2346/3).
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.10.082.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Murphy S., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic identification of elevated saliva kallikrein levels in the mdx-4cv mouse model of Duchenne muscular dystrophy. Biochem. Biophys. Rep. 2018 doi: 10.1016/j.bbrep.2018.05.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karn R.C., Chung A.G., Laukaitis C.M. Shared and unique proteins in human, mouse and rat saliva. Proteomes: Footprints Funct. Adapt. Proteomes. 2013;1:275–289. doi: 10.3390/proteomes1030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivadasan P., Kumar Gupta M., Sathe G.J., Balakrishnan L., Palit P., Gowda H., Suresh A., Abraham Kuriakose M., Sirdeshmukh R. Data from human salivary proteome: A resource of potential biomarkers for oral cancer. Data Brief. 2015;4:374–378. doi: 10.1016/j.dib.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loo J.A., Yan W., Ramachandran P., Wong D.T. Comparative human salivary and plasma proteomes. J. Dent. Res. 2010;89:1016–1023. doi: 10.1177/0022034510380414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grassl N., Kulak N.A., Pichler G., Geyer P.E., Jung J., Schubert S., Sinitcyn P., Cox J., Mann M. Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 2016;8:44. doi: 10.1186/s13073-016-0293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy S., Dowling P., Zweyer M., Henry M., Meleady P., Mundegar R.R., Swandulla D., Ohlendieck K. Proteomic profiling of mdx-4cv serum reveals highly elevated levels of the inflammation-induced plasma marker haptoglobin in muscular dystrophy. Int. J. Mol. Med. 2017;39:1357–1370. doi: 10.3892/ijmm.2017.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 8.Murphy S., Henry M., Meleady P., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Simultaneous pathoproteomic evaluation of the dystrophin-glycoprotein complex and secondary changes in the mdx-4cv mouse model of duchenne muscular dystrophy. Biology (Basel) 2015;4:397–423. doi: 10.3390/biology4020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy S., Ohlendieck K. Mass spectrometric identification of dystrophin, the protein product of the Duchenne muscular dystrophy gene, in distinct muscle surface membranes. Int. J. Mol. Med. 2017;40:1078–1088. doi: 10.3892/ijmm.2017.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deslyper G., Colgan T.J., Cooper A.J., Holland C.V., Carolan J.C. A proteomic investigation of hepatic resistance to ascaris in a murine model. PLoS Negl. Trop. Dis. 2016;10:e0004837. doi: 10.1371/journal.pntd.0004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material