Abstract
Background and Aims
Vedolizumab (VDZ) is effective for Crohn’s disease (CD) but costly and is slow to produce remission. Early knowledge of whether vedolizumab is likely to succeed is valuable for physicians, patients, and insurers.
Methods
Phase 3 clinical trial data on VZD for CD were used to predict outcomes. Random forest modeling on the training cohort was used to predict the outcome of corticosteroid-free biologic remission at week 52 on the testing cohort. Models were constructed using baseline data, or data through week 6 of VDZ therapy.
Results
The clinical trial included 594 subjects who received VDZ with baseline active inflammation [elevated C-reactive protein (>5 mg/L)]. Subjects with missing predictor variables (N = 120) or missing outcome data (N = 2) were excluded to produce a modeling dataset of 472 subjects. The Area Under the Receiver Operating Characteristic curve (AuROC) for corticosteroid-free biologic remission at week 52 using baseline data was only 0.65 (95% CI: 0.53 – 0.77), but was 0.75 (95% CI: 0.64 – 0.86) with data through week 6 of VDZ . Patients predicted to be in corticosteroid-free biologic remission at week 52 by the model achieved this endpoint 35.8% of the time, whereas patients predicted to fail only succeeded 6.7% of the time.
Conclusions
An algorithm using laboratory data through week 6 of VDZ therapy was able to identify which CD patients with baseline inflammation would achieve corticosteroid-free biologic remission on VDZ at week 52. A majority of patients can be identified by week 6 as very unlikely to achieve remission.
Keywords: inflammatory bowel disease, Crohn’s disease, vedolizumab, machine learning algorithm
BACKGROUND
Crohn’s disease (CD) affects over 600,000 people in the United States and over 1 million people in Europe.1 The treatment of CD is often quite expensive, and many therapies used to treat CD are slow to produce remission. New clinical responses continue to accumulate even after many weeks of therapy. Physicians, patients, and insurers want to know whether a given patient with CD will eventually succeed when starting a new therapy, or whether early discontinuation to switch to a different mechanism of action could save time and money in a patient likely to fail therapy.
Vedolizumab (VDZ), a gut-selective alpha-4-beta-7 integrin therapy, has been shown to be effective at inducing remission, but concerns about its ability to induce rapid remission have been raised.2, 3 A study by Dulai et al suggests that VDZ is very well tolerated and few individuals discontinue this drug due to adverse events. However, CD patients often respond slowly to VDZ, and a majority of patients will require a switch to another therapy to achieve remission.3 Given the cost and the concerns about the speed of induction of remission with VDZ, insurers, patients, and providers are often reluctant to pay for or continue this therapy, which produces biologic remission in fewer than 1 in 6 of the CD patients treated.
Clinical trial data could not only provide us with the ability to identify which CD patients are most likely to benefit from each therapy but also affect clinical decisions about starting or continuing therapies. Through the Clinical Study Data Request (CSDR) website,4 we obtained access to the phase 3 clinical trial data for the induction and maintenance treatment of CD using VDZ (NCT00783692).2 We hypothesized that the early clinical trial data could allow prediction of biologic remission at week 52. These data were used to predict whether baseline data, or data through week 6, could predict week 52 biologic remission in CD patients treated with VDZ.
METHODS
Overview
Our Institutional Review Board (IRB) confirmed that IRB approval was not necessary to perform post hoc analyses of previously collected and deidentified clinical trial data from the CSDR website (https://www.clinicalstudydatarequest.com) for the phase 3 clinical trial data for the induction and maintenance of CD using VDZ (HUM00118527). Predictors and outcomes from the clinical trial dataset were used to construct and validate predictive models of treatment response.
Modeling Cohort and Demographics
The trial had a complex design where individuals were stratified in the maintenance phase based on a week 6 response. In our analysis, we evaluated all the arms where drug was provided and used in both the induction and maintenance arms of the study, and included those on every 4-week and every 8-week regimes, illustrated in Supplementary Figure 1. The original study included 813 subjects on VDZ throughout the induction and maintenance periods. Subjects without evidence of active inflammation, determined by elevated C-reactive protein (CRP) (>5 mg/L) at baseline (N = 219), with missing predictor variables [N = 120; 113 of these subjects were missing week 6 CRP, week 6 drug level, or fecal calprotectin (FCP], or with missing outcome data (N = 2) were excluded for a final dataset of 472 subjects, which was later split into training (70%) and testing (30%). Table 1 shows the comparison of demographics from the original cohort (VDZ treatment arms) and the final modeling cohort (subjects with missing data or missing outcomes removed). A CONSORT diagram of the final dataset has been provided as Supplementary Figure 1.
Table 1:
Subject Demographics
| Variable | Original Cohort (No. = 813) | Final Modeling Cohort (No. = 472) |
|---|---|---|
| Mean Age in Years | 35.5 ± 11.9 | 34.2 ± 11.4 |
| Percentage Male Sex | 46.5% | 48.9% |
| Percentage White Race | 89.8% | 87.7% |
| Mean Body Weight (kg) | 70.1 ± 19.7 | 69.6 ± 19.9 |
| Percentage Current Smoker | 26.4% | 26.5% |
| Mean Duration of Disease in Years | 9.1 ± 7.5 | 9.0 ± 7.1 |
| Mean CRP at Baseline (mg/L) | 22.1 ± 28.3 | 26.8 ± 27.2 |
| Median FCP at Baseline | 689 (226–1221) | 799 (343–1450) |
| Site of Disease (%) | ||
| Colon | 22.4% | 22.5% |
| Colon, other | 5.8% | 5.1% |
| Ileum | 15.1% | 12.3% |
| Ileum, colon | 42.3% | 44.3% |
| Ileum, colon, other | 12.2% | 12.9% |
| Ileum, other | 2.2% | 3.0% |
| Median prednisone equivalent dose in people who were on prednisone (mg) | 20.0 (13.0–30.0) (No. = 357) | 20.0 (10.0–30.0) (No. = 209) |
| Median budesonide equivalent dose in people who were on budesonide (mg) | 9.0 (6.0–9.0) (No. = 73) | 9.0 (6.0–9.0) (No. = 35) |
| Percentage with prior anti- TNF therapy | 65.7% | 65.9% |
| Baseline concomitant medications (%) | ||
| Glucocorticoids only | 34.4% | 34.5% |
| Immunosuppressants only | 16.4% | 14.8% |
| Glucocorticoids and Immunosuppressants | 16.9% | 17.2% |
| No Glucocorticoids or Immunosuppressants | 32.3% | 33.5% |
For normally distributed continuous variables, we report mean ± SD and for skewed continuous variables, we report medians with interquartile range (IQR).
Predictor Variables
Baseline predictor variables included all available quantitative laboratory test values at baseline, patient demographics (age, sex, and race), patient height and weight, and medication status (VDZ interval dosing arm, immunomodulator and steroid use at the start of the trial, and previous exposure to anti-tumor necrosis therapy).
The week 6 model included the above baseline variables with the following exception: quantitative laboratory tests except FCP were from week 6 (or nearest earlier date if week 6 data were unavailable), instead of from baseline. Note that FCP was included by using the baseline value, since it was collected at week 0, but not at week 6. The VDZ drug level at week 6 and calculated longitudinal variables were also included as predictors.
Longitudinal variables that were calculated included the slope of CRP (CRP at week 6 minus CRP before initiation of medication)/6, and the slope of the VDZ drug level (VDZ drug level at week 6 minus VDZ drug level at week 2)/4. The slope, acceleration, mean, and maximum of each of the other laboratory predictors were tested but provided no improvement to the AuROC (Area Under the Receiver Operating Characteristic Curve) of the week 6 model and were removed from the final model. Additional clinical predictors, including disease extent and disease duration, were tested, but added no improvement to the AuROC and were removed from the final model.
A list of all predictors used for each model, including those that were tested and removed due to no improvement of AuROC, can be found in Table 2.
Table 2:
Predictors for the Baseline Model
| Baseline variables | Labs: FCP, CRP, Albumin (ALB), Alkaline Phosphatase (ALP), Alanine Aminotransferase (ALT), Amylase, Aspartate Aminotransferase (AST), Direct Bilirubin, Bilirubin, Blood Urea Nitrogen (BUN), Calcium, Chloride, CO2, Creatinine, Glucose, Potassium, Lipase, Magnesium (Mg), Sodium, Phosphorous, Protein, Uric Acid, Basophils, Eosinophils, Hematocrit (HCT), Hemoglobin (HGB), Lymphocytes, Monocytes, Neutrophils, Platelet, Red blood cell (RBC), White blood cell (WBC) Others: Age, Gender, Race, Height, Weight, VDZ interval (VDZ Interval: dosing every 4 or 8 weeks), Immunomodulator at start of trial (ImmAtStart), SteroidAtStart, Previous exposure to TNF (PriorTNF) |
| Predictors for the model through week 6: | |
| Baseline variables |
Labs: Fecal Calprotectin (FCP) Others: Age, Gender, Race, Height, Weight, Vedolizumab interval (VDZ Interval, dosing every 4 or 8 weeks), Immunomodulator at start of trial (ImmAtStart), Steroid use at the start of the trial (SteroidAtStart), Previous exposure to TNF (PriorTNF) |
| Week 6 variables | Labs: CRP, ALB, ALP, ALT, Amylase, AST, Direct Bilirubin, Bilirubin, BUN, Calcium, Chloride, CO2, Creatinine, Glucose, Potassium, Lipase, Mg, Sodium, Phosphorous, Protein, Uric Acid, Basophils, Eosinophils, HCT, HGB, Lymphocytes, Monocytes, Neutrophils, Platelet, RBC, WBCVDZ level: VDZ drug level (VDZ drug level) |
| Longitudinal variables | Slope of CRP (CRP at week 6 minus CRP before initiation of medication)/6), Slope of VDZ drug level (VDZ drug level at week 6 minus VDZ drug level at week 2)/4), |
| Predictors that were tested and removed due to no improvement of AuROC | |
| Variables deleted |
Baseline variables: disease extent, disease duration Longitudinal variables: mean, max, mean of slope, max of slope, and mean of acceleration of labs except FCP and CRP up to week 6 |
Definition of Outcomes
The primary outcome was corticosteroid-free biologic remission with VDZ at week 52, defined by no use of corticosteroid medications (including prednisone and budesonide) at week 52, and reduction of CRP from >5 mg/L at baseline to ≤ 5 mg/L at week 52. Subjects without a visit at week 52, having CRP greater than 5 at week 52, or using steroids (Prednisone EQ Dose or Budesonide EQ Dose > 0 at week 52) were defined as failures. Patients who had a visit but did not have CRP at week 52 were defined as having a missing outcome, and they were removed from the cohort (N = 2).
Statistical Analysis and Model Development
We developed 2 random forest models using the above predictor variables, 1 using only baseline data and 1 using data through week 6. The random forest method of prediction uses many decision trees to classify patients by outcome.5, 6 To classify a new observation, the observation is run through each of the decision trees and each tree gives a classification (vote). The ‘forest’ combines the votes from all the trees to compute a predicted score of the outcome. One can select a desired sensitivity or specificity by choosing a particular value of the predicted score as a cutoff. To allow internal validation of the predictive models, each dataset was split into a 70% training dataset and a 30% testing dataset.
A random forest model with baseline variables was trained on the training set, and a forest of 1000 trees was grown to produce the predictions. An additional random forest model using data at week 6 and calculated longitudinal variables (from week 0 – week 6) was trained on the training set and tested on the test set. The results of these models were compared to each other.
Training and testing cohorts
Training and testing datasets were determined by splitting the data randomly into 70% and 30% subsets. This process was repeated 50 times. Each model was fit on the training dataset and tested on the testing dataset each time. The AuROC values from the 50 trials were averaged to produce a mean AuROC. Out of all 50 splits, the one that produced an AuROC closest to the average for the week 6 model was used as a representative split (training and testing cohorts) for each of the 2 final prediction models (baseline and week 6) to allow (1) production of the ROC (receiver operating characteristic) plots, (2) calculation of the representative AuROCs, (3) cutoff point selections, and (4) misclassification tables. To develop the most accurate model for future use, we constructed a random forest model using the entire dataset, which enables us to calculate variable importance and partial dependence plots.
Model performance
The AuROC was used to assess the performance of each model. All statistical methods were performed using the statistical language R (version 3.3), with the packages randomForest and pROC.2
Cutoff point selection
An optimal cutoff point was determined for each model to minimize the following criterion: (1 - sensitivity)2 + (1- specificity).2 This cutoff is defined as the point on each ROC plot closest to the perfect point where sensitivity and specificity are both equal to 1.
Variable importance
We assessed the importance of each predictor based on a random forest model built on the entire dataset. To determine the relative importance of each predictor variable, nodes in which the individual predictor variable appeared were identified in the ensemble of trees. The relative information content was summed over all of the nodes containing that variable. The predictor variables with the greatest combined relative information content have a greater importance.
Partial dependence plots
Partial dependence plots were constructed to show how individual predictor variables can affect the probability of success, while controlling for all of the other predictor variables. We used the random forest model on the entire dataset to predict the probability of success for each patient. Each predictor variable was set to a given value for each patient and then averaged over all patients to produce a mean probability of success. This process was repeated for each value of that variable in the data, or for its 50 quantiles if more than 50 values were available.
Simplified models
Based on the information in the variable importance plot for the week 6 model and individual predictors that have been shown to be predictors of response, such as high VDZ,7 decreased clearance of VDZ based on weight, and albumin level,8 we hypothesized post hoc that the following single predictor at week 6 might be useful: the quotient (HGB * ALB * VDZ level) / (CRP * weight in kg). We evaluated the simplified model on the entire dataset to see if it could predict corticosteroid-free biologic remission at week 52.
Reproducible Research Code Repository
The code used to produce this analysis in R is available in a public Github repository at: https://github.com/higgi13425/vedoCD. Access to the trial data can be obtained only through the CSDR website at https://clinicalstudydatarequest.com/.
RESULTS
Predicting Week 52 Ssteroid-free Biologic Remission at Baseline
The average AuROC for the baseline model over 50 replications is 0.64. The AuROC for the baseline model under the selected training and testing split is 0.65 (95% CI: 0.53 – 0.77) as shown in Figure 1A. The variable importance plot for the baseline model is presented in Figure 1B. The 5 strongest baseline predictors of corticosteroid-free biologic remission at week 52 were: CRP , lipase, CO2, weight, and hemoglobin. Notably weak baseline predictors of this outcome were steroid use at the start of the trial (SteroidAtStart), race VDZ interval and sex. The best cutoff, number of predicted successes and failures, and the proportion of subjects with true success in the testing set (N = 142) within these 2 predicted classes are displayed in Table 3 and Supplemental Figure 2. Table 3 also provides the sensitivity of 0.64 and specificity of 0.56. The effects of the baseline predictors on the outcome of corticosteroid-free biologic remission at week 52 can be illustrated by partial dependence plots. Several of these are not linear. These are displayed in multiple panels in Supplemental Figure 3.
FIGURE 1.
Baseline model AuROC and variable importance.
1A, AuROC for the baseline model.
1B, Variable importance plot for the baseline model.
Table 3:
Model Performance
| Model | Validation Sample Size | AuROC (95% CI) | Best Cutoff | Prediction Category | Predicted Cases | True Success Rate | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|
| Baseline model | 142 | 0.65 (0.53, 0.77) | 0.17 | Predicted success (≥cutoff) | 67 | 23.9% | 0.58 | 0.64 | 0.56 |
| Predicted failure (<cutoff) | 75 | 12.0% | |||||||
| Week 6 model | 142 | 0.75 (0.64, 0.86) | 0.21 | Predicted success (≥cutoff) | 53 | 35.8% | 0.72 | 0.76 | 0.71 |
| Predicted failure (<cutoff) | 89 | 6.7% | |||||||
| Week 6 (HGB*ALB*VDZ Level)/(CRP*Weight) | 472 | 0.75 (0.70, 0.81) | 185.96 | Predicted success (≥cutoff) | 182 | 32.4% | 0.69 | 0.73 | 0.69 |
| Predicted failure (<cutoff) | 290 | 7.6% |
Number of predicted success is tp+fp; number of predicted failure is tn+fn; for predicted success, true success rate is tp/(tp+fp); for predicted failure, true success rate is fn/(tn+fn); accuracy is (tp+tn)/(tp+fp+tn+fn); sensitivity is tp/(tp+fn); specificity is tn/(tn+fp).
Predicting Week 52 Steroid-free Biologic Remission Through Week 6
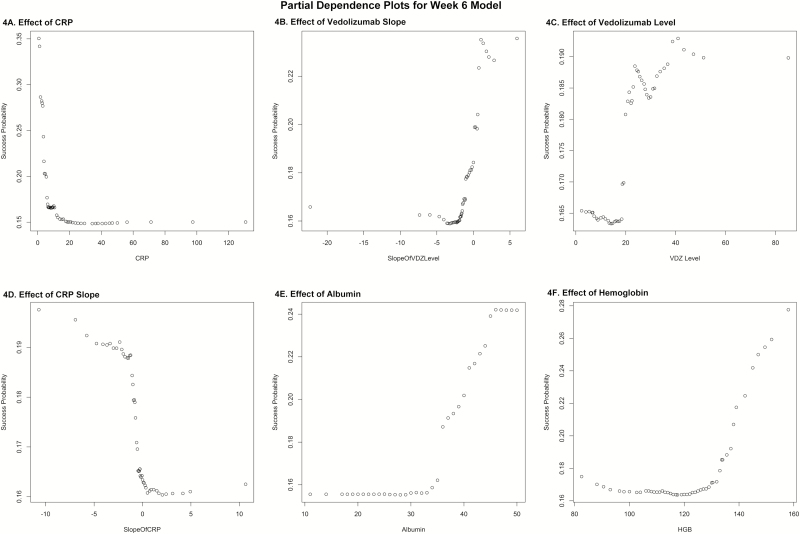
The average AuROC for the week 6 model over 50 replications is 0.75. The AuROC for the week 6 model under the selected representative training and testing split is 0.75 (95% CI: 0.64 – 0.86), as shown in Figure 2A. The P value for the AuROC of the week 6 model vs an AuROC of 0.5 is 4.46 x 10–5. The variable importance plot for the week 6 model is shown in Figure 2B. The 5 strongest predictors of corticosteroid-free biologic remission at week 52 were CRP, slope of VDZ level, hemoglobin, albumin, and VDZ level at week 6. Notably weak predictors of this outcome at week 6 were SteroidAtStart, race, VDZ interval, and sex. The best cutoff, number of predicted successes and failures, and the proportion of subjects of true success within these 2 predicted classes are displayed in Table 3 and Figure 3. Table 3 also provides the sensitivity of 0.76 and specificity of 0.71. The effects of the week 6 predictors on the outcome of corticosteroid-free biologic remission at week 52 are illustrated in partial dependence plots, displayed in multiple panels in Figure 4, and in Supplemental Figure 4.
FIGURE 2.
Week 6 model AuROC and variable importance.
2A, AuROC for the week 6 model.
2B, Variable importance plot for the week 6 model.
FIGURE 3.
Endpoint success at week 52.
FIGURE 4.
Partial dependence plots for week 6 model.
4A, Effect of CRP.
4B, Effect of VDZ slope.
4C, Effect of VDZ level.
4D, Effect of CRP slope.
4E, Effect of lbumin.
4F, Effect of hemoglobin.
Simplified Model Performance
Given the complexity of the random forest models, we considered whether a simpler model might be more pragmatic and nearly as accurate. Based on the variable importance plot at week 6, we hypothesized that a simpler model with a single ratio as a predictor would be accurate. We calculated the ratio based on week 6 values as: (HGB*ALB*VDZ level)/(CRP*weight) and tested it post hoc on the entire dataset to see if a simple model could have comparable predictive performance. Using this single ratio as a predictor on the entire dataset, we found that a cutoff of ≥185.96 predicted success well, with an AuROC of 0.75 (95% CI: 0.70 – 0.81).
DISCUSSION
These data have allowed us to develop and apply algorithms created by random forest machine learning techniques to predict corticosteroid-free biologic remission in response to VDZ in CD. Whereas the baseline model is relatively inaccurate, with a sensitivity of 64% and a specificity of 56%, our week 6 model is much more accurate, with a sensitivity of 76% and a specificity of 71%. A simple model using (HGB*ALB*VDZ level)/(CRP*weight) at week 6 with a cutoff of ≥185.96 also predicted success well, with a sensitivity of 73% and a specificity of 69%. It should be noted that the performance of the simple model is difficult to compare to the random forest model as the simple model was motivated by and tested on the entire cohort. Ideally, the simple model should be externally validated on a separate external validation cohort and should be used cautiously as VDZ levels may be obtained using different assays,7, 9 unless these have been validated against the Takeda in-house assay, which we used in this model. These models can all be implemented using HL-7 compatible laboratory information systems via a cloud-based service,10 through which one can securely submit laboratory values and accept returned calculated algorithmic results without exposing protected health information.
It should be noted that there are several limitations to this study. First, this study has been developed and validated in a clinical trial cohort and its generalizability to patients outside of clinical trials needs further exploration. Second, 228 patients of 472 patients were already corticosteroid-free at baseline but did have evidence of active disease at entry. Third, this study is limited to the data available from GEMINI II and is only as generalizable as these data that resulted in the US Food and Drug Administration approval of VDZ for use in the treatment of CD.
Given the efficacy and expense of biologic therapies,11, 12 being able to identify patients with a low rate of success after a short trial of therapy is valuable, as these patients can then move on in a timely fashion to another therapy with a different mechanism of action. This would reduce the time period of their active symptoms, likely reduce their exposure to steroids, and reduce the expense of a biologic therapy that is likely to be futile in a given patient.
Being able to identify CD patients who are at high risk of failure with VDZ at week 6 also provides an opportunity to improve the outcomes for these patients. Patients who are predicted to be likely to fail VDZ might benefit from addition of a “booster” therapy beginning at week 6, possibly including an anti-TNF therapy, an anti-IL23 therapy,13 or a JAK inhibitor,14 if these drugs are approved for this indication. These difficult-to-treat CD patients, once they achieve low CRP levels, normal albumin levels, and adequate VDZ levels, may well be able to continue on VDZ maintenance monotherapy successfully. Future randomized clinical trials in this subset of patients are needed to determine if patients in whom VDZ is likely to fail can be rescued with a timely intervention. In addition, early identification of patients at high risk of VDZ failure also may be an opportunity for physicians to start the approval process for the next drug while continuing to use VDZ to see if there is any therapeutic benefit.
CONCLUSIONS
A random forest machine learning algorithm, using laboratory data through week 6 of VDZ therapy, was able to identify which CD patients with baseline inflammation would achieve corticosteroid-free biologic remission on VDZ at week 52. This algorithm could provide value in identifying whether continuing VDZ is the right choice for a particular CD patient. These algorithms have important potential applications in clinical decision-making, guiding providers to consider options including dose escalation, adding a cotherapy, or changing to a different mechanism of action for CD as early as week 6 of VDZ therapy.
SUPPLEMENTARY DATA
Supplementary data are available at Inflammatory Bowel Diseases online.
ACKNOWLEDGEMENTS
We would like to thank Clinical Study Data Request for making this data publically available.
Glossary
Abbreviations
- VDZ
Vedolizumab
- CSDR
Clinical Study Data Request website
- CD
Crohn’s disease
- CS
Corticosteroid
- CRP
C-reactive protein
- IRB
Institutional Review Board
- ROC
Receiver Operating Characteristic
- AuROC
Area Under the Receiver Operating Characteristic curve.
Conflicts of interest: There are no conflicts of interest to report.
Supported by: Akbar K. Waljee is supported by a career development grant award (CDA 11–217) from the United States Department of Veterans Affairs (VA) Health Services Research and Development Service. The research of Peter D. R. Higgins and Akbar K. Waljee is supported by National Institutes of Health (NIH) R01 GM097117. The content is solely the responsibility of the authors and does not necessarily represent the official views of the University of Michigan, the VA, or the NIH.
REFERENCES
- 1. Loftus EV. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–17. [DOI] [PubMed] [Google Scholar]
- 2. Sandborn WJ, Feagan BG, Rutgeerts P et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2013;369:711–21. [DOI] [PubMed] [Google Scholar]
- 3. Dulai PS, Singh S, Jiang X et al. The real-world effectiveness and safety of vedolizumab for moderate-severe Crohn’s disease: results from the US VICTORY consortium. Am J Gastroenterol. 2016;111:1147–55. [DOI] [PubMed] [Google Scholar]
- 4. CSDR: Clinical Study Data Request https://clinicalstudydatarequest.com/.
- 5. Liaw A, Wiener M. Classification and regression by random Forest. R news. 2002;2:18–22. [Google Scholar]
- 6. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 7. Williet N, Boschetti G, Fovet M et al. Association between low trough levels of vedolizumab during induction therapy for inflammatory bowel diseases and need for additional doses within 6 months. Clin. Gastroenterol. Hepatol. 2017;15:1750–7. [DOI] [PubMed] [Google Scholar]
- 8. Rosario M, Dirks NL, Gastonguay MR et al. Population pharmacokinetics- pharmacodynamics of vedolizumab in patients with ulcerative colitis and Crohn’s disease. Aliment Pharmacol Ther. 2015;42:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bian S, Dreesen E, Tang HT et al. Antibodies toward vedolizumab appear from the first infusion onward and disappear over time. Inflamm Bowel Dis. 2017; 23:2202–8. [DOI] [PubMed] [Google Scholar]
- 10. Romero J, Lopez P, Noguera JLV et al. Integrated, reliable and cloud-based personal health record: a scoping review. Health Inform J. 2016;5:1–20. [Google Scholar]
- 11. Stidham RW, Lee TCH, Higgins PDR et al. Systematic review with network meta-analysis: the efficacy of anti-TNF agents for the treatment of Crohn’s disease. Aliment Pharmacol Ther. 2014;39:1349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stidham RW, Lee TCH, Higgins PDR et al. Systematic review with network meta-analysis: the efficacy of anti-tumour necrosis factor-alpha agents for the treatment of ulcerative colitis. Aliment Pharmacol Ther. 2014;39:660–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furfaro F, Gilardi D, Allocca M et al. IL-23 Blockade for Crohn’s disease: next generation of anti-cytokine therapy. Expert Rev Clin Immunol. 2017;13:457–67. [DOI] [PubMed] [Google Scholar]
- 14. Olivera P, Danese S et al. JAK inhibition in inflammatory bowel disease. Expert Rev Clin Immunol. 2017;13:693–703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.