Abstract
Background
Phosphatase and tensin homolog (Pten) is capable of mediating microbe-induced immune responses in the gut. Thus, Pten deficiency in the intestine accelerates colitis development in Il10-/- mice. As some ambient pollutants inhibit Pten function and exposure to ambient pollutants may increase inflammatory bowel disease (IBD) incidence, it is of interest to examine how Pten inhibition could affect colitis development in genetically susceptible hosts.
Methods
With human colonic mucosa biopsies from pediatric ulcerative colitis and non-IBD control subjects, we assessed the mRNA levels of the PTEN gene and the gene involved in IL10 responses. The data from the human tissues were corroborated by treating Il10-/-, Il10rb-/-, and wild-type C57BL/6 mice with Pten-specific inhibitor VO-OHpic. We evaluated the severity of mouse colitis by investigating the tissue histology and cytokine production. The gut microbiome was investigated by analyzing the 16S ribosomal RNA gene sequence with mouse fecal samples.
Results
PTEN and IL10RB mRNA levels were reduced in the human colonic mucosa of pediatric ulcerative colitis compared with non-IBD subjects. Intracolonic treatment of the Pten inhibitor induced colitis in Il10-/- mice, characterized by reduced body weight, marked colonic damage, and increased production of inflammatory cytokines, whereas Il10rb-/- and wild-type C57BL/6 mice treated with the inhibitor did not develop colitis. Pten inhibitor treatment changed the fecal microbiome, with increased abundance of colitogenic bacteria Bacteroides and Akkermansia in Il10-/- mice.
Conclusions
Loss of Pten function increases the levels of colitogenic bacteria in the gut, thereby inducing deleterious colitis in an Il10-deficient condition.
Keywords: air pollutant, early-onset colitis, interleukin 10, microbiome, Toll-like receptor
INTRODUCTION
Phosphatase and tensin homolog (Pten) is the phosphatase generating phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] from PI(3,4,5)P3.1 Downregulation of Pten activity by either genetic modifications or small molecule inhibitors leads to prolonged activation of the PI3K/Akt/mTOR signaling pathway, resulting in the promotion of cell proliferation, growth, survival, metabolism, polarization, and movement.2, 3 Loss of the Pten gene is identified in various malignancies in humans, and therefore Pten has long been considered a tumor suppressor gene.1–3 In accordance with the role of Pten in promoting various cellular functions, a downregulation of Pten activity not only provides a neuroprotective effect in central nervous system insults4, 5 but also protects the brain from ischemia/reperfusion injury.6 Thus, small molecule inhibitors of Pten may be valuable as potential therapeutics for stroke and ischemia reperfusion injury with the expectation that Pten inhibitor treatment over a limited period of time would not increase the risk of tumorigenesis.7 Indeed, it has been suggested that a Pten inhibitor, bisperoxovanadium-pic [bpV(pic)], could provide protective effects against amyloid β-peptide (25–35)–induced oxidative stress and neurotoxicity, implying that Pten inhibitors may be used therapeutically against neurodegenerative diseases, such as Alzheimer’s disease.8
Apart from the classical role for Pten as a tumor suppressor, we recently demonstrated that Pten determines the plasma membrane localization of the adaptor molecule Mal/Tirap by controlling the cellular level of PI(4,5)P2, thereby regulating the interaction between Toll-like receptor 5 (Tlr5) and Mal/Tirap. Therefore, by regulating Tlr5-mediated responses, Pten is able to regulate immune and inflammatory responses.9 This report suggested that Pten plays a role as an immune regulatory factor controlling the host-gut microbial interaction by regulating Tlr5-induced responses in the gut.
Inflammatory bowel diseases (IBDs), Crohn’s disease (CD) and ulcerative colitis (UC), are chronic inflammatory conditions in the gastrointestinal tract10 whose cause remains unclear. Studies have suggested a series of IBD-associated genetic factors such as interleukin 10 (IL10), interleukin 10 receptor (IL10R), and NOD2 that may participate in the pathogenesis of IBD. However, the involvement of a single IBD-associated gene is not sufficient to account for the development of these inflammatory intestinal disorders.11 Recently emerging evidence indicates that uncontrolled immune responses can cause IBD in a genetically susceptible individual, suggesting that a combination of multiple factors leads to IBD.12 Consistent with this hypothesis, we demonstrated that a combination of Pten gene deletion in the intestinal epithelium and global Il10 deficiency results in severe colitis in mice at very early ages, which is similar to early-onset colitis in humans.13 This study provides evidence that loss of Pten gene expression by several distinct mechanisms could be a polygenic factor interacting with an IBD-susceptible genetic factor such as Il10 deficiency in inducing intestinal inflammation.
Intriguingly, recent epidemiological studies have suggested that exposure to ambient pollutants including benzene, cadmium, and reactive oxygen species may be associated with early-onset and adult IBD.14, 15 Salim et al. demonstrated that exposure to ingested airborne pollutants can result in early-onset colitis in Il10-deficient mice.16 Benzene is one of the most common air pollutants present in both outdoor and indoor air and is released mainly from motor vehicle emissions.17 It is noteworthy that benzene suppresses Pten mRNA expression by inducing Pten gene methylation.18 Cadmium is also able to inhibit Pten.19 Reactive oxygen species can inactivate the enzymatic activity of Pten.20 These findings prompted us to hypothesize that the inhibition of Pten activity by a specific inhibitor in the colon would induce colitis in Il10-defective mice. In line with this hypothesis, in this study, we found reduced Pten gene expression in mucosal biopsies from pediatric UC patients compared with normal controls. Moreover, we also demonstrate that treatment with a Pten-specific inhibitor induces deleterious intestinal inflammation in Il10-/- mice.
Together, our findings suggest that loss of Pten function due to Pten inhibitory molecules or reduced Pten gene expression is associated with the development of colitis in the genetically susceptible host.
METHODS
Human Colonic Mucosal Tissues
Colonic mucosal tissues were obtained from pediatric UC patients (younger than age 18 years) enrolled in the Massachusetts General Hospital for Children Pediatric IBD Biorepository. Informed consent or assent was acquired from all subjects or their legal guardians. With approval from the Partners Health Care Institutional Review Board (protocol #2009P001287), sigmoid colon mucosal biopsies were collected at the time of colonoscopy, snap-frozen in liquid nitrogen, and stored at −80°C. Matched sigmoid biopsies were also obtained and processed for routine histologic evaluation. Biopsies from patients diagnosed with pediatric UC (n = 9) or non-IBD controls (n = 19) were analyzed.
RNA Isolation From Human Colonic Mucosal Tissues
Colonic biopsies were homogenized for RNA extraction using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions with minor modifications. Specifically, two additional washings of the RNA pellet with 70% ethanol were included to minimize any potential phenol contamination in the sample. The sample purity was determined by calculating the absorbance ratios of 260 nm/230 nm and 260 nm/280 nm for phenol and protein contamination, respectively, using a Synergy HTX microplate spectrophotometer (BioTek, Winooski, VT, USA). The samples having ratios ≥2.0 were utilized in this study. Furthermore, the integrity of the RNA was evaluated by Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA), and samples with an RNA integrity number (RIN) higher than 7.5 were included for evaluation.
Quantitative Real-time Polymerase Chain Reaction Analysis
Real-time polymerase chain reaction (PCR) for PTEN, IL10, IL10RA, IL10RB, and GAPDH was performed with total RNA extracted from biopsies, as described previously.21 We evaluated mRNA expression levels by quantitative real-time PCR using CFX384 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with the Exiqon PCR primer sets according to manufacturer’s instructions (Exiqon Inc. Woburn, MA, USA). The mRNA expression level was evaluated by calculating the PCR cycle number (CT) at which the exponential growth in fluorescence from the probe passes a certain threshold value (CT). Relative gene expression was determined by the difference in the CT values of the target genes after normalization to RNA input level, using the CT value of a house-keeping GAPDH gene. Relative quantification was represented by standard 2-ΔCT calculations. ΔCT = (CT-target gene − CT-GAPDH).22 Each reaction was performed in triplicate.
Primers used for PTEN were forward 5’- ATTCGACTTA GACTTGACCT-3’ and reverse 5’- GCGGTGTCATAA TGTCT-3’; IL10, forward 5’- GAAGAATGCCTTTAAT AAGCTC-3’ and reverse 5’- AGTCTATAGAGTCGCCAC-3’; IL10RA, forward 5’-TTCTCTGTGGATGAAGTGA-3’ and reverse 5’-TCATACTCTCGGAAGTGAC-3’; IL10RB, forward 5’-CAGCACCTGAAAGAGTT-3’ and reverse 5’-CTCAGA GTCTTCTGCAATG-3’; GAPDH, forward 5’- CGCTCTCTGC TCCTCCTGTT-3’ and reverse 5’-CCATGGTGTCTGAGC GATGT-3’.
Mice and Reagents
Il10-/-23 and Il10rb-/-24 on C57BL/6 background and C57BL/6 mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Mice were bred and maintained in a specific pathogen-free condition with normal drinking water ad libitum at the animal facility of UCLA with the Institutional Animal Care and Use Committee (IACUC) approval. Newborn mice were co-fostered in a breeding pair. At 3 weeks of age, male and female pups were separated from the parents, and mice were co-housed in separate cages with fewer than 4 mice per cage. Using mouse tail biopsies, the genotypes of Il10-/- and Il10rb-/- mice were determined by standard genotyping PCR protocols provided by the Jackson Laboratory. All animal experiments were approved by the Animal Research Committee of UCLA.
The vanadium complex VO-OHpic, a potent allosteric inhibitor of Pten (PTEN-i),25, 26 was purchased from Echelon Biosciences Inc. (Salt Lake City, UT, USA).
Histology
Mouse colon tissues were fixed in 10% buffered formalin, dehydrated, and embedded in paraffin. Tissue sections (5 μm) were prepared for hematoxylin and eosin (H&E) staining. Images were visualized with an Axio Imager Z1 microscope (Carl Zeiss) and captured with an AxioCam digital camera and processed with Adobe Photoshop. The histological severity of colitis was graded in a blinded fashion, as previously described.27, 28
Transmission Electron Microscopy
As described previously,13 mouse colon tissues were preserved in electron microscopy solution prepared with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M PBS, pH 7.4, for 2 hours at room temperature, followed by an incubation at 4°C overnight. Then, 0.5% tannic acid was supplemented to the tissues and incubated for 1 hour at room temperature. We washed the tissue blocks 5 times with 0.1 M PBS buffer, and they were postfixed in a solution of 1% OsO4 in PBS, pH 7.2–7.4, to increase the staining of the membranes. The samples were washed (x4) in Na acetate buffer (pH 5.5), followed by block staining in 0.5% uranyl acetate in 0.1 M Na acetate buffer (pH 5.5) for 12 hours at 4°C. The samples were dehydrated in graded ethanol (50%, 75%, 95%, 100%, 100%, 100%) 10 minutes each, passed through propylene oxide, and infiltrated in mixtures of Epon 812 and propylene oxide 1:1 and then 2:1 for 2 hours each. The tissues were then infiltrated in pure Epon 812 overnight. Then, embedding was done in pure Epon 812, and curing was performed in an oven at 60°C for 48 hours. Using a diamond knife, sections were cut on an ultramicrotome (RMC MTX) at 60-nm thickness (gray interference color). The sections were placed on single-hole grids coated with Formvar and carbon and double-stained in aqueous solutions of 8% uranyl acetate for 25 minutes at 60°C and lead citrate for 3 minutes at room temperature. We examined the thin sections with a 100CX JEOL transmission electron microscope.
Cytokine Enzyme-Linked Immunosorbent Assays
Mouse colon tissue was homogenized in RIPA lysis buffer. Total protein lysates were subjected to an enzyme-linked immunosorbent assay (ELISA) to measure the cytokines Tnfα, Cxcl1, and Il6 using appropriate kits from R&D Systems, following the manufacturer’s instructions. All assays were performed in triplicate, and data were expressed as mean ± SEM.
Intracolonic Administration of Pten Inhibitor
As previously described,27, 29 200 μL of Pten inhibitor VO-OHpic (5 mM/kg) or vehicle solution (5% DMSO) was administered via an 18G, 2.5-mm-diameter tube that was advanced through the rectum into the colon, until the tip was 3 cm proximal to the anus (midcolon). We ensured that there was no leakage after the enema injection.
Fecal Sample Collection for Microbiome Analysis
Sex- and age-matched and co-fostered C57BL/6 or Il10-/- mice were treated with intracolonic administration of the Pten inhibitor. Each mouse group was maintained in a co-housing condition in an SPF facility. The feces were harvested from the cecum and colon and snap-frozen in liquid nitrogen before sequencing.
DNA Extraction From Mouse Fecal Samples
Genomic DNA was isolated from fecal samples using the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA, USA) following the manufacturer’s instructions. Instead of the recommended 250 mg of soil, approximately 2 fecal pellets were provided to the PowerBeads tube to undergo cell lysis. The isolated DNA was eluted from the spin filter using 50 μL of solution C6 and stored at –20°C until PCR amplification.
PCR Amplification and Amplicon Sequencing Using Next-Generation Technology (bTEFAP)
Next-generation technology (bTEFAP) of amplicon sequencing has been described previously.30, 31 Next-generation technology (bTEFAP) can be used to assess a wide range of environmental and health-related microbiomes including the intestinal populations of a variety of sample types including cattle, mice, rats, and humans, and a wide array of environmental samples.30, 32, 33 A reengineered modern version of bTEFAP has adapted nonoptical sequencing technologies such as the Ion Torrent PGM and the Illumina MiSeq and HiSeq platforms and become one of the most widely published methods to evaluate microbiota.
As the primer set 27F-519R (27F AGRGTTTGATC MTGGCTCAG and 519R GTNTTACNGCGGCKGCTG) spans hypervariable regions V1 to V3 of the 16S rRNA gene,13 the 16S universal Eubacterial primer 27F-519R set was utilized for PCR amplification to evaluate the microbial ecology of each sample on the MiSeq with methods via bTEFAP DNA analysis.
A single-step 30-cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA, USA) was used under the following conditions: 94°C for 3 minutes, followed by 28 cycles of 94°C for 30 seconds; 53°C for 40 seconds and 72°C for 1 minute; after which a final elongation step at 72°C for 5 minutes was performed. Following PCR, all amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Co., Beverly, MA, USA). Samples were sequenced utilizing Illumina MiSeq chemistry following the manufacturer’s protocols for examining the microbial ecology of each sample.
Sequence Analysis
The Q25 sequence data obtained from the sequencing process was elaborated through a proprietary analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX, USA). Sequences were depleted of barcodes and primers; then short sequences <200 bp were removed, sequences with ambiguous base calls were removed, and sequences with homopolymer runs exceeding 6 bp were removed. Sequences were then denoised and chimeras removed. Operational taxonomic units (OTUs) were defined after removal of singleton sequences, clustering at 3% divergence (97% similarity).30, 31, 34–36 OTUs were then taxonomically classified using BLASTn against a curated GreenGenes/RDP/NCBI-derived database37 and compiled into each taxonomic level into both “counts” and “percentage” files. Count files contain the actual number of sequences whereas the percentage files contain the relative (proportion) percentage of sequences within each sample that map to the designated taxonomic classification. For example, if there are 1000 sequences and 100 of the sequences are classified as Staphylococcus, then we represent this as Staphylococcus being 10% of the total population.
Normalized and denoised files were then run through Quantitative Insights Into Microbial Ecology (QIIME)38 to generate alpha and beta diversity data, as described previously.30, 31, 34–36 Based upon weighted unifrac analysis in the QIIME, principal coordinates analysis (PCoA) plots were generated. Additional statistical analyses were performed with NCSS2007 (NCSS, Kaysville, UT) and XLstat2012 (Addinsoft, NY, USA). Significance reported for any analysis is defined as P < 0.05.
Alpha and Beta Diversity Analysis
Alpha diversity is essentially a means to evaluate how many different bacterial species are within the given sample or treatment group. Beta diversity allows comparison of the community of bacteria as a whole, taking into account both how many different things are in the sample and what those things are related to phylogenetically. Alpha and beta diversity analyses were conducted as described previously30, 31, 34–36 using QIIME (www.qiime.org).
Statistical Analysis
Statistical analysis was conducted with GraphPad Prism (GraphPad Software, Inc., San Diego, CA, USA). Data of body weight change were compared by 2-way analysis of variance (ANOVA), followed by the multiple-comparison Bonferroni t test. P values of less than 0.05 were considered significant. Additional information regarding statistical analysis is presented in the corresponding figure legend.
RESULTS
Colonic Mucosal Biopsies From Pediatric UC Patients Had Lower PTEN and IL10RB mRNA Levels Than Normal Tissues From Non-IBD Subjects
IL10 is the most potent anti-inflammatory cytokine contributing to maintaining immune balance. IL10 fulfills its anti-inflammatory roles through the heteromeric IL10 receptor (IL10R) complex composed of IL10RA and IL10RB subunits. A nonfunctional mutation in the gene encoding IL10 or the receptor subunit abolishes the anti-inflammatory effects driven by IL10. Recently, we identified that intestinal Pten gene deletion in Il10 -/- mice not only enhances the risk of mouse colitis but also shortens the onset and peak time of massive intestinal inflammation.13 In light of this finding, we hypothesized that a combination of intestinal Pten deficiency with defective IL10 signaling would accelerate the development of colitis. To test this hypothesis, we examined the colonic mucosa tissues from pediatric UC patients and non-IBD control pediatric subjects to evaluate the expression of PTEN and the genes involved in the IL10 signaling axis (IL10, IL10RA, IL10RB).
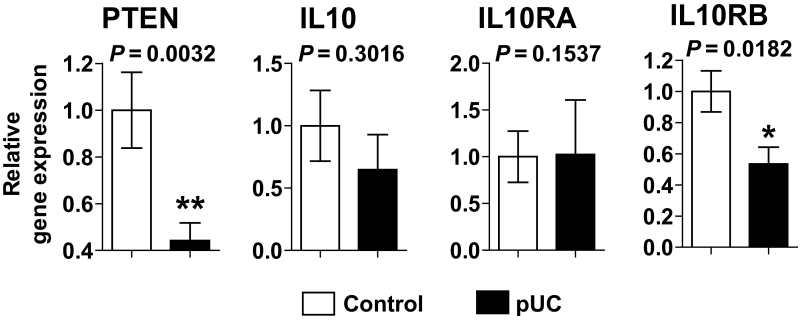
Surprisingly, we identified that PTEN mRNA expression was dramatically reduced in pediatric UC tissues compared with uninflamed control tissues (Fig. 1). Furthermore, IL10RB mRNA expression was diminished in pediatric UC biopsies compared with controls, whereas IL10 and IL10RA expression levels were preserved in these tissues. These data indicate that impaired PTEN and a defective IL10 signaling axis should be associated with deleterious colonic inflammation.
FIGURE 1.
Colonic mucosal biopsies of pediatric UC patients have lower PTEN and IL10RB mRNA expression levels than the tissues of control subjects. Quantitative real-time PCR was performed to evaluate the mRNA expression of PTEN, IL10, IL10RA, and IL10RB in colonic mucosal biopsies from pediatric UC patients (pUC; n = 9) and non-IBD control subjects (n = 19). Error bars indicate ± SD. *P < 0.05, **P < 0.01 (Mann-Whitney U test).
It is worth noting that IL10 and IL10RA expression levels are capricious, depending on cell types and the extracellular environment. IL10 expression can be induced in several immune cells such as monocytes, macrophages, dendritic cells, natural killer cells, mast cells, neutrophils, and T and B cells in response to immune stimulatory antigens such as microbe-associated molecular patterns.39 IL10RA expression identified in most types of hematopoietic cells is also upregulated by various cell-stimulatory antigens. Moreover, IL10RA expression is differentially regulated in certain types of nonhematopoietic cells such as fibroblasts in response to lipopolysaccharide (LPS).40, 41 Therefore, IL10 and IL10RA expression seem to fluctuate in an inflamed tissue, depending on the degree of inflammation, inflammatory antigen, or endogenous damage-associated molecular patterns from dying cells. Thus, it may be hard to observe consistently reduced IL10 and IL10RA expression in a chronic inflammatory condition. In contrast, IL10RB constitutively expresses in various cell types.42–44 Therefore, it is reasonable to believe that altered IL10RB expression would more directly reflect defective IL10 signaling than IL10 and IL10RA expression levels in chronically inflamed intestinal tissues. This may be the reason that reduced IL10RB, but not IL10 or IL10RA, was observed in the inflamed tissues of pediatric UC patients.
Together, our results suggest the possibility that disrupted IL10 signaling could cooperate with impaired PTEN function to induce inflammatory disorders in the colon.
Pten-Specific Inhibitor Treatment Induced Pan-Colitis in Il10-/- Mice, but not in Il10rb-/- or C57BL/6 Mice
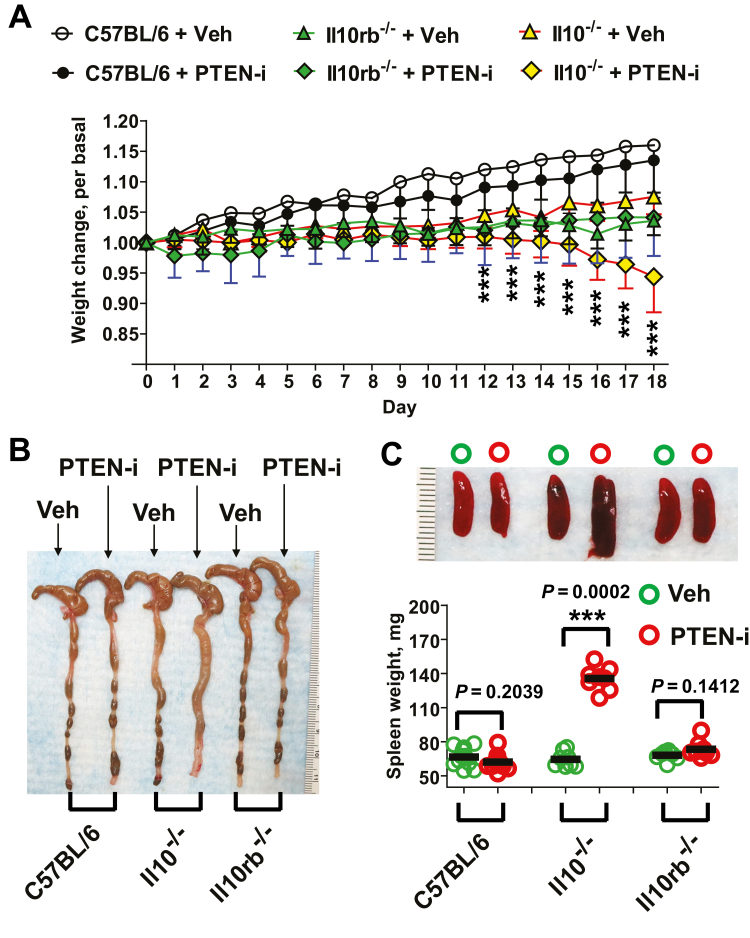
A combination of Pten gene deletion in the intestinal epithelium and global Il10 gene deletion elicits massive colitis in mice at early ages.13 As shown above, colonic mucosa tissues of pediatric UC patients were characterized by substantially reduced PTEN and IL10RB mRNA expression compared with normal subjects (Fig. 1). In light of these findings, we hypothesized that pharmacological inhibition of Pten in the colon would induce colitis in mice with disrupted IL10 signaling. To test this hypothesis, we examined whether the inhibition of Pten in the colon would elicit colitis in Il10-/- or Il10rb-/- mice. The vanadium complex VO-OHpic is a potent allosteric inhibitor of Pten,25, 26 and its inhibitory effect against Pten has been tested in mice previously.26 Therefore, Il10-/-, Il10rb-/-, and wild-type C57BL/6 mice were treated with intracolonic administration of VO-OHpic (PTEN-i, 5 mM/kg). PTEN-i administration changed body weight-gaining pattern in Il10-/- mice, compared with vehicle-treated Il10-/- mice (Fig. 2A). Il10-/- mice treated with PTEN-i exhibited pronounced weight loss in 12 days, whereas vehicle (Veh)-treated Il10-/- mice maintained a normal weight-gaining pattern throughout the experimental period. In contrast, Il10rb-/- and C57BL/6 mice treated with PTEN-i kept a consistent body weight-gaining pattern, which was comparable to control mice groups. These data indicate that intracolonic treatment of PTEN-i results in reduced body weight in Il10-/- mice, but not in Il10rb-/- or normal C57BL/6 mice.
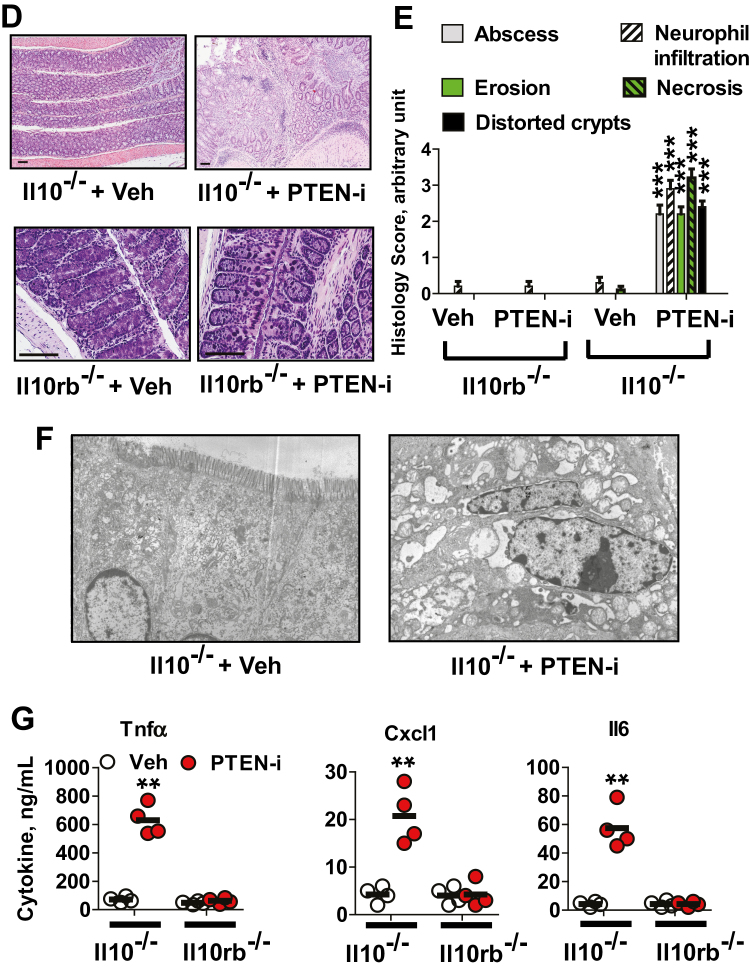
FIGURE 2.
PTEN inhibitor treatment elicits colitis in Il10-/- mice, but not in Il10rb-/- or C57BL/6 mice. A, Age- and sex-matched Il10-/-, Il10rb-/-, and C57BL/6 mice were treated with intracolonic administration of PTEN inhibitor (PTEN-i) VO-OHpic (5 mM/kg) and vehicle (Veh, 5% DMSO), given once every day during the experimental period. Body weight change was evaluated as previously described.13, 68 Results are means ± SD, C57BL/6 (PTEN-i group, n = 12; Vehicle group, n = 12), Il10rb-/- (PTEN-i, n = 7; Vehicle, n = 9), Il10-/-, (LPS, n = 8; Vehicle, n = 8). Data were compared by 2-way ANOVA (with treatment and times) followed by the multiple-comparison Bonferroni t test to assess differences between groups. ***Difference from PTEN-i-treated Il10-/- mice vs vehicle-treated Il10-/- mice at that time (P < 0.001). Data were analyzed with the results accumulated by 6 independent experiments. B, Gross appearance of the colon and cecum from these mice was compared. C, Presented is the gross appearance of the spleen obtained from the mice (upper panel). The spleen weight was measured and compared (lower panel). ***P < 0.001 (Mann-Whitney U test). D, Full length of the colon was prepared in a “swiss roll” method and subjected to H&E staining (scale bars, 100 μm). E, Histological parameters were quantified with H&E sections (n = 10/group). F, Electron micrographs of the colonic epithelium from Il10-/- mice treated with PTEN-i or vehicle. G, The total protein was extracted from the mouse colon, followed by ELISA to measure the level of Tnfα, Cxcl1, and Il6 production. Data were expressed as mean ± SEM (n = 4 mice per group, each with triplicate determinations). **P < 0.01 (Mann-Whitney U test). Representative images are presented.
Subsequently, we identified inflamed colons in PTEN-i treated Il10-/- mice, whereas vehicle-treated Il10-/- mice had normal colons. In line with preserved body weight-gaining, normal colon was observed in PTEN-i- or vehicle-treated Il10rb-/- and C57BL/6 mice (Fig. 2B). Massive inflammatory responses from the inflamed colon can affect the periphery, resulting in an enlarged spleen. In agreement with this notion, Il10-/- mice treated with PTEN-i had enlarged spleens, in contrast to vehicle-treated Il10-/- mice, which exhibited normal spleens (Fig. 2C). Just like the normal colons observed, all Il10rb-/- and C57BL/6 mouse groups exhibited normal spleens.
Subsequent microscopic examination of colon tissues demonstrated that Il10-/- mice treated with PTEN-i exhibited severe mucosal damage and massive inflammatory signs characterized by abscess, neutrophil infiltration, erosion, necrosis, and distorted crypts in the colonic mucosa, whereas no histopathological features were observed in colon tissues from vehicle-treated Il10-/- mice (Fig. 2D and E). Normal colonic mucosa was observed in Il10rb-/- mice treated with PTEN-i or vehicle. Similarly, electron micrographs of Il10-/- mice treated with the inhibitor revealed massive necrosis, damaged cell-to-cell adhesions, and effaced microvilli in the colonic epithelium, whereas normal columnar epithelia with intact microvilli and preserved cell-to-cell adhesion were observed in the colonic mucosa of vehicle-treated Il10-/- mice (Fig. 2F). Moreover, the production of inflammatory cytokines such as Tnfα, Cxcl1, and Il6 was substantially increased in the colon tissues of PTEN-i-treated Il10-/- mice compared with control Il10-/- mice (Fig. 2G). Colonic cytokine production was comparable between Il10rb-/- mice treated with PTEN-i or vehicle.
Taken together, these data demonstrate that intracolonic treatment with the Pten inhibitor elicits massive colitis in Il10-/- mice, but not in Il10rb-/- or C57BL/6 mice. In line with the reduced PTEN and IL10RB gene expression observed in human colonic mucosa tissues from pediatric UC patients, our data from Pten inhibitor–treated Il10-/- mice strongly support the cooperative involvement of impaired Pten and disrupted IL10-induced signaling in colitis development.
Pten Inhibitor Treatment Disrupted the Gut Microbiome in Mice
The colon is the primary reservoir for a large collection of commensal microbiota. Mounting evidence indicates that gut microbial dysbiosis could be associated with the development and perpetuation of intestinal inflammatory diseases in humans.45–47 Previously, we suggested that the abundance of the genus Bacteroides was greatly increased in fecal samples from double knockout (Pten∆IEC/∆IEC;Il10-/-) mice having Pten gene deletion in the intestine and global Il10 gene deletion, leading to accelerated colitis development.13 Based on these considerations, we hypothesized that intracolonic administration of PTEN-i could alter the gut microbiome to increase the risk of spontaneous colitis development in Il10-/- mice. To test this hypothesis, sex-matched 2-month-old Il10-/- and C57BL/6 mice were treated with intracolonic administration of PTEN-i or vehicle control. After 12 days, fecal samples were harvested from these mice, followed by sequencing of the 16S ribosomal RNA (rRNA) gene in the samples.
The observed operational taxonomic unit counts were evaluated for each of the groups. C57BL/6 or Il10-/- mouse groups did not show significant differences in the alpha diversity measurements with regard to the Shannon diversity index, indicating that there was no significant difference in the species diversity and richness within C57BL/6 and Il10-/- mouse groups (Table 1).
TABLE 1:
There were no significant differences in either of the alpha diversity measurements between the groups
| Mouse Group | Observed Species | Shannon Index | ||||
|---|---|---|---|---|---|---|
| Mean | Std. Dev. | Statistics | Mean | Std. Dev. | Statistics | |
| C57BL/6 + vehicle | 1039.375 | 101.871 | P = 0.529 | 4.908 | 0.448 | P = 0.059 |
| C57BL/6 + PTEN-i | 1076.875 | 153.641 | 5.326 | 0.392 | ||
| Il10-/- + vehicle | 1776.400 | 178.094 | P = 0.464 | 6.847 | 0.280 | P = 0.464 |
| Il10-/- + PTEN-i | 1847.625 | 167.177 | 6.651 | 0.469 | ||
The mean rarefaction predicted OTU data for the mouse groups were measured at a sequence depth of 26,893 (C57BL/6 group) and 26,668 (Il10-/- groups). Presented are the observed species and the shannon index showing the mean value.
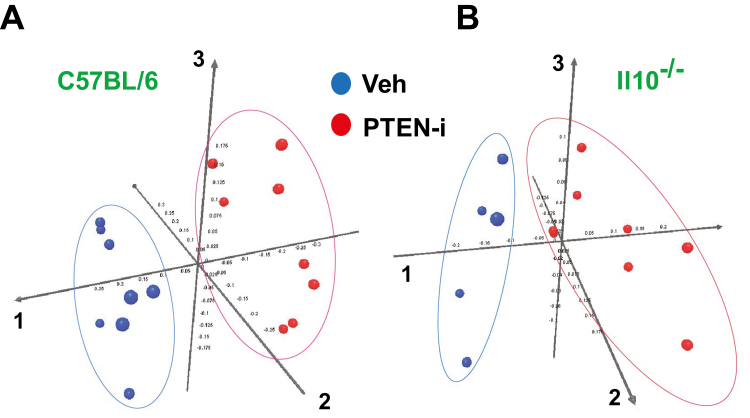
Next, the bacterial community structure was analyzed using weighted UniFrac distance matrices. Principal coordinates analysis plots were used to visualize the data in these matrices, and analysis of similarities (ANOSIM) was utilized to determine if there were any significant differences between the bacterial communities. The samples from C57BL/6 + PTEN-i and C57BL/6 + vehicle mouse groups in the weighted PCoA plot formed 2 separate clusters (Fig. 3A). The ANOSIM R value (R = 0.89, P = 0.001) indicates that there is a significant difference between C57BL/6 + PTEN-i and C57BL/6 + vehicle groups. It is noteworthy that an R value close to 1 indicates a high separation of microbial species composition between treated and control samples, whereas an R value close to 0 indicates no separation between the samples. Similarly, in Il10-/- + PTEN-i and Il10-/- + vehicle groups, microbiome phylogenetic assemblage was also formed (Fig. 3B). The ANOSIM R value (R = 0.83, P = 0.003) for these 2 Il10-/- mouse groups indicates that there is a significant difference between Il10-/- + PTEN-i and Il10-/- + vehicle groups.
FIGURE 3.
The fecal microbiome of PTEN-i treated mice is distinct from that of vehicle-treated mice. A, C57BL/6 mice treated with PTEN-i and vehicle control group. Principal coordinates analysis plot of weighted Unifrac metrics was generated based on the primary vector, which explained 49.2% of the variation between the groups. The first 3 vectors together exhibit 84.4% of the variation among the groups. B, Il10-/- + PTEN-i and Il10-/- + vehicle groups. The data were generated based on the primary vector, which explained 54.3% of the variation between the groups. The first 3 vectors together exhibit 84.3% of the variation among the groups. Each dot represents individual microbiota samples from the mouse. n = 8 per each mouse group.
Although the species diversity and species richness within the C57BL/6 and Il10-/- mouse groups were not altered by the inhibitor treatment, our data taken together demonstrate that intracolonic administration of Pten inhibitor did indeed change the species composition of the fecal microbiome.
Fecal Microbiota Communities of PTEN Inhibitor–Treated Mice Were Different From Vehicle-Treated Mice
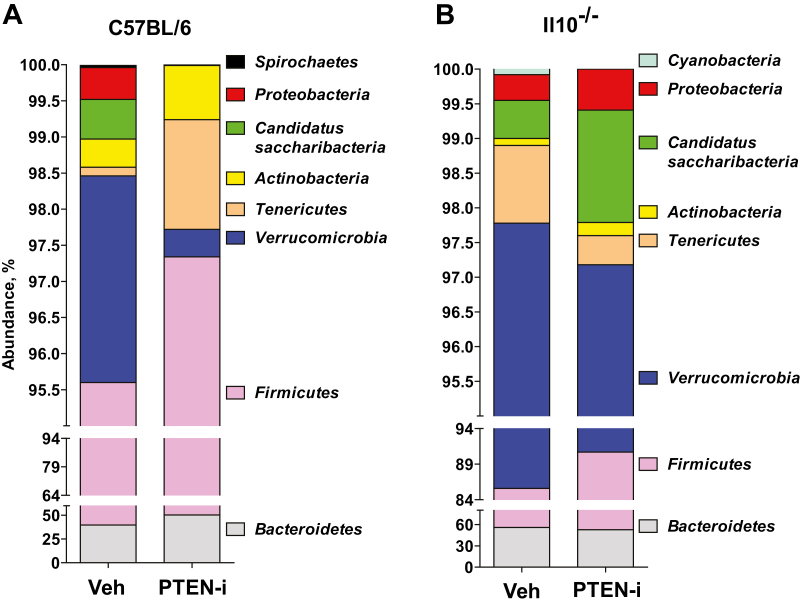
We observed that 100% of analyzed sequences from all C57BL/6 and Il10-/- mouse groups belonged to the kingdom bacteria. There was no detected archaeal or eukaryotic contamination in the sequences. The majority (>98%) of analyzed sequences from C57BL/6 mice were assigned to 4 major phyla: Firmicutes (vehicle-treated vs PTEN-i-treated, 55.83% vs 46.97%), Bacteroidetes (39.77% vs 50.37%), Verrucomicrobia (2.86% vs 0.38%), Tenericutes (0.12 vs 1.52%) (Fig. 4A). Similarly, the majority of analyzed sequences (>97%) from Il10-/- mice also had the same major phyla: Bacteroidetes (vehicle-treated vs PTEN-i-treated, 55.73% vs 52.60%), Firmicutes (29.86 vs 38.10%), Verrucomicrobia (12.18% vs 6.48%), Tenericutes (1.12% vs 0.42%) (Fig. 4B). Therefore, the major phyla constituting the fecal microbiota were not substantially different between the C57BL/6 and Il10-/- mouse groups.
FIGURE 4.
The abundance of taxonomic groups identified in the mouse fecal samples. A and B, The abundance (% of sequences) of the phyla detected in the fecal samples in C57BL/6 mice treated with PTEN-i or vehicle (A) or Il10-/- mice treated with PTEN-i or vehicle (B). In C57BL/6 mice, Firmicutes, Bacteroidetes, Verrucomicrobia, and Tenericutes are the 4 major phyla encompassing the majority of sequences (>98%). Il10-/- mice had the same major phyla with Bacteroidetes, Firmicutes, Verrucomicrobia, and Tenericutes, accounting for the majority of sequences (>97%)
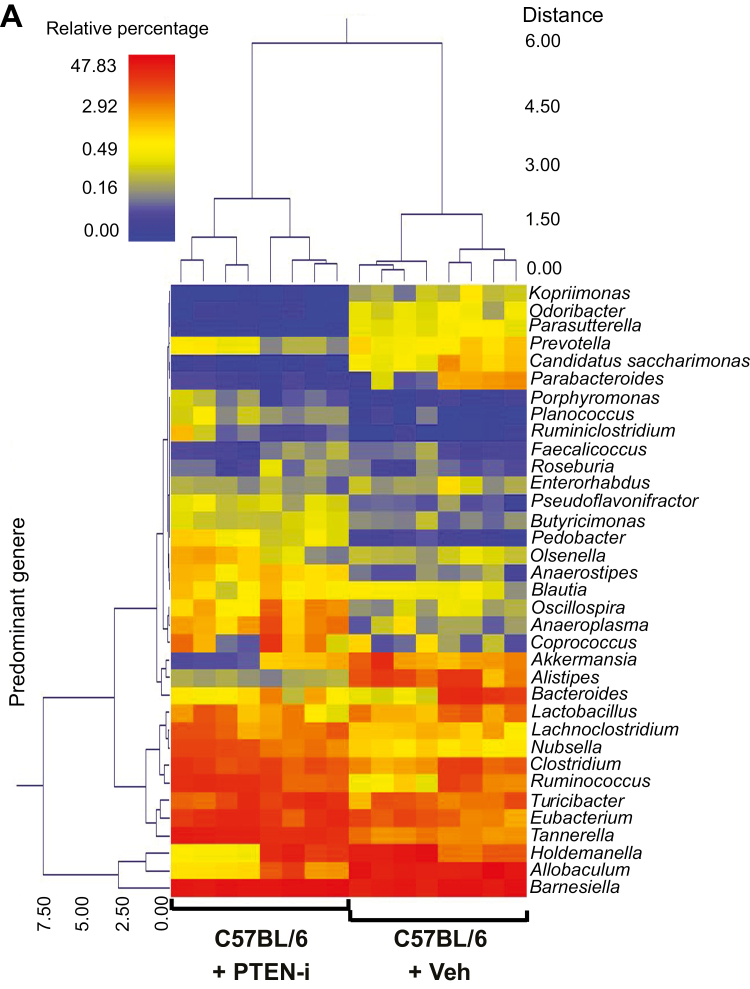
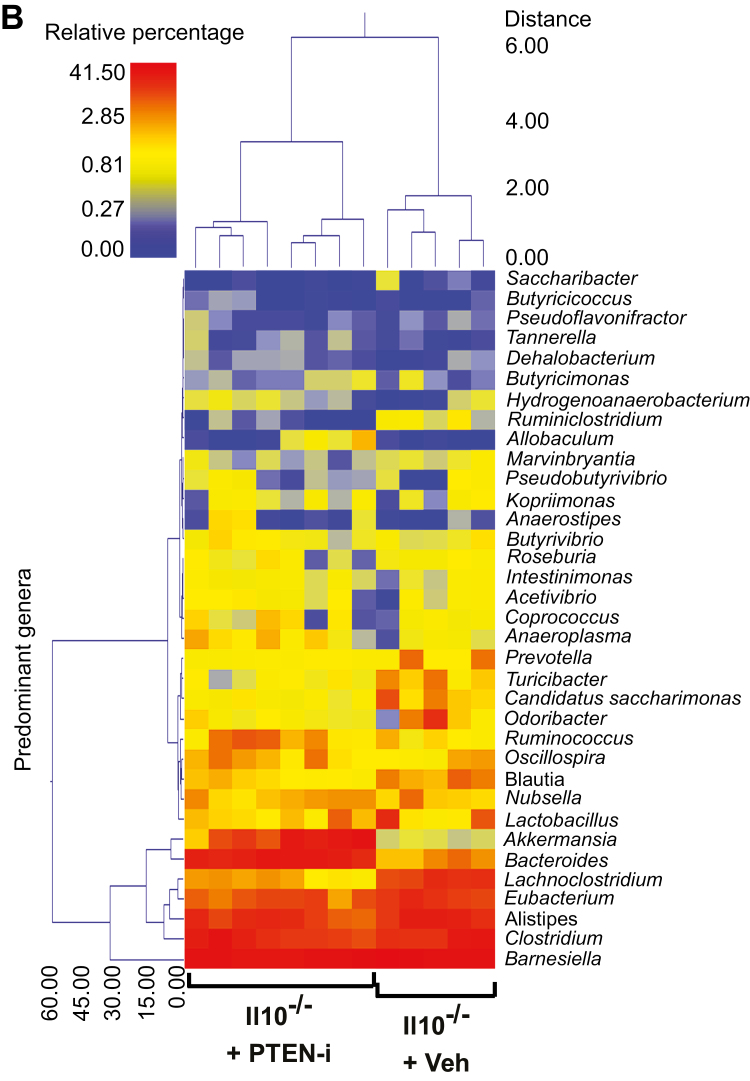
Subsequently, we evaluated whether any specific bacterial genera were significantly different between the control and treatment groups. There was a wide range of genera whose abundance was significantly different between C57BL/6 or Il10-/- mouse groups in response to inhibitor treatment. To provide a visual overview combined with analysis, we utilized a dual hierarchical dendrogram to display the data for the predominant genera with clustering related to the different groups. Based on the 2 distinct clusters seen in the data from C57BL/6 mice, a significant difference in the predominant genera was detected between the C57BL/6 + PTEN-i and C57BL/6 + control groups (Fig. 5A). The predominant genera from the Il10-/- + PTEN-i and Il10-/- + control groups formed its own single cluster (Fig. 5B).
FIGURE 5.
PTEN-i treatment altered fecal microflora in C57BL/6 and Il10-/- mice. (A and B), Based on the predominant genera using Ward’s minimum variance clustering and Manhattan distances, a dual hierarchal dendrogram was constructed. Samples were clustered along the x axis, labeled by the treatment in C57BL/6 (A) and Il10-/- mice (B). Samples with more similar microbial populations are mathematically clustered closer together. The genera (consortia) are used for clustering. Thus, the samples with more similar consortia of genera cluster closer together, with the length of connecting lines (top of heat map) related to the similarity. Shorter lines between 2 samples indicate closely matched microbial consortia. The heat map represents the relative percentages of each genus. The predominant genera are represented along the right y axis. The legend for the heat map is provided in the upper left corner.
Together with the PCoA plots, these data indicate that the fecal microbiota communities from PTEN inhibitor–treated mice are easily distinguished and indeed different from vehicle-treated mice in both C57BL/6 and Il10-/- mice, demonstrating an altered gut microbiome in Pten inhibitor–treated mice.
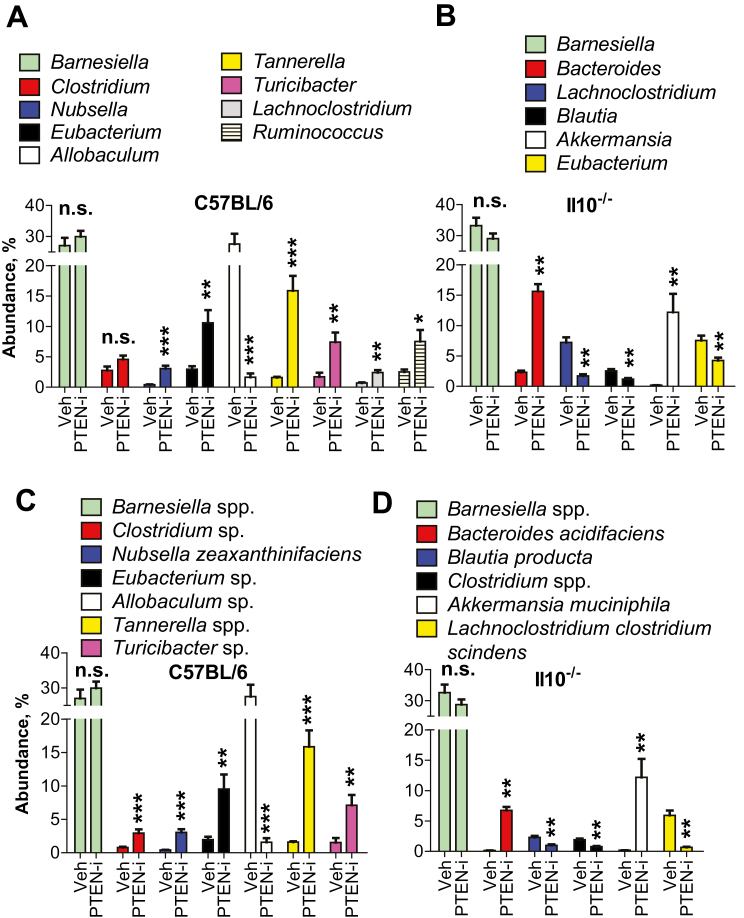
The Colitogenic Bacteria, Bacteroides and Akkermansia, Were Substantially Increased by Pten Inhibitor Treatment in Il10-/- Mice, but Not in C57BL/6 Mice
We evaluated whether the abundance of any specific bacterial genera was significantly changed in response to Pten inhibitor treatment. In C57BL/6 mice treated with PTEN-i or vehicle control (Fig. 6A), the 3 most notable differences were found in the relative abundance of Allobaculum (27.57% in C57BL/6 + vehicle → 1.62% in C57BL/6 + PTEN-i), Tannerella (1.59% → 15.87%), and Eubacterium (2.96% → 10.60%). In Il10-/- mice (Fig. 6B), the most notable difference was observed in the relative abundance of Bacteroides (2.30% in Il10-/- + vehicle → 15.61% in Il10-/- + PTEN-i) and Akkermansia (0.154% → 12.18%).
FIGURE 6.
The abundance of colitogenic bacteria, Bacteroides and Akkermansia, was elevated by PTEN inhibitor treatment in Il10-/- mice, not in C57BL/6 mice. A–D, The abundance of major bacteria at the genus (A and B) and species (C and D) levels identified in the mouse fecal samples were analyzed to compare the bacterial abundance induced by inhibitor vs vehicle treatment. Results are presented as mean ± SD. **P < 0.01; ***P < 0.001 (Mann-Whitney U test). Abbreviation: n.s., not significant (n = 8/group).
It is noteworthy that an increased abundance of Bacteroides is associated with the development of spontaneous colitis in both humans and Il10-/- mice.13, 48, 49 In addition, it was suggested that Akkermansia has great potential to promote intestinal inflammation.50 Our microbiome analysis data demonstrated that the abundance of the genera Bacteroides and Akkermansia was dramatically increased in PTEN-i-treated Il10-/- mice (Fig. 6A and B). In contrast, the inhibitor treatment did not alter the abundance of these bacteria in C57BL/6 mice, which did not develop colitis. Therefore, elevated Bacteroides and Akkermansia levels in PTEN-i-treated Il10-/- mice are not only associated with colitis development in Il10-/- mice, but may also explain why C57BL/6 mice were still able to maintain a normal colon upon inhibitor treatment.
Although we didn’t observe a change in the abundance of any species from the genus Bacteroides in the C57BL/6 mouse groups (Fig. 6C), we did observe an effect within the Il10-/- group. Within the genera of Bacteroides and Akkermansia, which were dramatically increased in the fecal sample, we identified sharp increases in the species Bacteroides acidifaciens (0.14% in Il10-/- + vehicle → 6.74% in Il10-/- + PTEN-i) and Akkermansia muciniphila (0.15% → 12.18%) in the samples of PTEN-i treated Il10-/- mice compared with control Il10-/- mice (Fig. 6D).
Taken together, these results suggest that Pten inhibitor treatment results in a disrupted gut microbiome, with greatly increased abundance of colitogenic bacteria such as Bacteroides and Akkermansia in Il10-/- mice, thereby causing the development of deleterious colitis in a genetically susceptible host.
DISCUSSION
A growing body of evidence indicates that an altered gut microbiome contributes to the initiation and perpetuation of IBD in genetically susceptible hosts.51–53 Likewise, the gut microbiome plays an essential role in the development of spontaneous colitis in mice with disrupted IL10 signaling such as Il10-/- mice, as these mice do not develop colitis under germ-free conditions.54 Further studies identified several gut bacterial species that are capable of inducing colitis. For instance, the presence of Helicobacter species in mouse intestine is essential to inducing colitis in Il10-/- mice.55, 56Bacteroides also has great potential to induce colitis. This notion is supported by separate studies, which have suggested that oral gavage of Bacteroides induces colitis in colitis-susceptible mice48 and that the abundance of Bacteroides is much higher in the colons of UC patients than healthy control subjects.49 Our recent study also suggested a colitogenic effect of Bacteroides in Il10-/- mice.13 Due to the enzymatic capacity to degrade mucin, Akkermansia exacerbates intestinal inflammation in mice.50, 57 Therefore, increased amounts of colitogenic bacteria in the intestine may contribute to the onset of colitis development in Il10-/- mice. Notably, we confirmed that mice maintained in our animal facility do not harbor any Helicobacter species.13 As the abundance of these colitogenic bacteria was dramatically increased by Pten inhibitor treatment in Il10-/- mice, it is reasonable to believe that Pten inhibitor treatment causes severe colitis in Il10-/- mice by increasing the abundance of Bacteroides and Akkermansia.
IL10, a potent anti-inflammatory cytokine, directly binds to the heteromeric receptor complex composed of IL10RA and IL10RB to mediate its anti-inflammatory effects. In contrast to IL10RA, IL10RB is shared by other cytokine receptors such as IL22, IL26, and IFNλ.44 It was suggested that IL22 could have a role in protecting the intestine from intestinal leakiness.58 Both IL22 and IL26 could induce the secretion of chemokines and antimicrobial proteins to maintain mucosal barrier function against gastrointestinal pathogens.59 IFNλ may regulate intestinal epithelial homeostasis.60 In addition to mediating anti-inflammatory responses from IL10, therefore, IL10RB can promiscuously participate in more cellular functions than IL10RA. Moreover, its specific ligand IL10 is produced by limited types of cells and is specialized for inducing anti-inflammatory responses. In line with this notion, we observed that Il10-/- mice were more susceptible to spontaneous colitis development than Il10rb-/- mice (Supplementary Fig. 1).
Despite the evidence that an impairment in IL10-induced signaling pathways is capable of inducing intestinal inflammation in both humans and mice, recent studies have indicated a disparity between these species when it comes to the involvement of IL10RB deficiency in intestinal inflammation. In humans, a deleterious mutation in the IL10, IL10RA, or IL10RB gene causes severe and extensive intestinal inflammation at very early ages.41, 61–65 In mice, Il10 gene deletion similarly causes spontaneous colitis, which is dependent on gut microbiota. Il10rb-/- mice on the mixed 129SvEv;C57BL/6 background are capable of developing colonic inflammation, but not small intestinal inflammation.24, 41 However, a recent study demonstrated that Il10rb-/- mice on the C57BL/6 background do not develop spontaneous colitis, but a combination of Il10rb gene deletion with TgfβR gene deletion elicits massive colitis.66 In agreement with this finding, our data also show that Il10rb-/- mice on the C57BL/6 background do not exhibit colitis development, whereas Il10-/- mice reveal spontaneous colitis (Supplementary Fig. 1). Moreover, Il10ra-/- mice on the C57BL/6 background do not develop spontaneous colitis.67 These findings indicate that, rather than an altered gut microbiome, an additional impairment in cytokine signaling must occur along with Il10rb or Il10ra deficiency to induce colitis in mice. We believe that this could explain why intracolonic administration of the Pten inhibitor does not induce colitis in Il10rb-/- mice (Fig. 2), whereas it does cause inflammation in Il10-/- mice, in which the development of colitis is sensitive to an altered gut microbiome.
In contrast to its classical role as a tumor suppressor gene, the immune modulatory function of Pten seems to play an important role in maintaining intestinal homeostasis. Accordingly, reduced levels of PTEN mRNA expression in the colonic mucosa biopsies of pediatric UC patients as shown here can be indicative of altered immune responses in the colon. The hypothesis that abnormal immune response to gut microbes is associated with IBD in genetically susceptible subjects is supported by our data showing reduced PTEN and IL10RB mRNA expression in human biopsies.
Intriguingly, recent studies strongly indicate that exposure to ambient pollutants is associated with the increasing incidence rate of IBD, although a pathogenic mechanism remains unknown.14, 15 It is noteworthy that ambient pollutants such as benzene,18 cadmium,19 and reactive oxygen species20 can inhibit the biological function of Pten. Therefore, our findings may provide some mechanistic insights into how ambient pollutants may be involved in the pathogenesis of IBD. Given the fact that Pten enzymatic activity is inactivated by phosphorylation at the serine and threonine residues in its C-terminal region by specific kinases,20 an inhibitory molecule specifically targeting a Pten-inhibiting kinase could be a potential therapeutic agent for Pten inactivation–associated colitis.
In summary, our study demonstrates that the inhibition of Pten in the intestine causes deleterious inflammation by increasing the abundance of colitogenic bacterial genera such as Bacteroides and Akkermansia. Due to the ability of some ambient pollutants to inhibit Pten function, our results allow us to speculate why exposure to ambient pollutants could be involved in the initiation and perpetuation of IBD.
SUPPLEMENTARY DATA
Supplementary data are available at Inflammatory Bowel Diseases online.
ACKNOWLEDGMENTS
We thank Dr. Marianne Cilluffo for supervising electron microscopy at the Electron Microscopy Core Facility of the Brain Research Institute at UCLA.
Conflicts of interest: The authors declare that they have no conflicts of interest related to the contents of this article.
Supported by: grants from Oakland University and the National Institutes of Health (DK079015, to S.H.R.), the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2016R1D1A1B03932222 to E.I.), and the Pediatric IBD Foundation (to H.S.W.); and philanthropic support from the B. Hasso Family Foundation and Martin Schlaff (to H.S.W.).
REFERENCES
- 1. Hill R, Wu H. PTEN, stem cells, and cancer stem cells. J Biol Chem. 2009;284:11755–11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta A, Leslie NR. Controlling PTEN (phosphatase and tensin homolog) stability: a dominant role for lysine 66. J Biol Chem. 2016;291:18465–18473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. [DOI] [PubMed] [Google Scholar]
- 4. Gary DS, Mattson MP. PTEN regulates Akt kinase activity in hippocampal neurons and increases their sensitivity to glutamate and apoptosis. Neuromolecular Med. 2002;2:261–269. [DOI] [PubMed] [Google Scholar]
- 5. Ning K, Pei L, Liao M et al. . Dual neuroprotective signaling mediated by downregulating two distinct phosphatase activities of PTEN. J Neurosci. 2004;24:4052–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruan H, Li J, Ren S et al. . Inducible and cardiac specific PTEN inactivation protects ischemia/reperfusion injury. J Mol Cell Cardiol. 2009;46:193–200. [DOI] [PubMed] [Google Scholar]
- 7. Spinelli L, Lindsay YE, Leslie NR. PTEN inhibitors: an evaluation of current compounds. Adv Biol Regul. 2015;57:102–111. [DOI] [PubMed] [Google Scholar]
- 8. Liu XY, Zhang LJ, Chen Z, Liu LB. The PTEN inhibitor bpV(pic) promotes neuroprotection against amyloid β-peptide (25-35)-induced oxidative stress and neurotoxicity. Neurol Res. 2017;39:758–765. [DOI] [PubMed] [Google Scholar]
- 9. Choi YJ, Jung J, Chung HK et al. . PTEN regulates TLR5-induced intestinal inflammation by controlling Mal/TIRAP recruitment. Faseb J. 2013;27:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bradford EM, Ryu SH, Singh AP et al. . Epithelial TNF receptor signaling promotes mucosal repair in inflammatory bowel disease. J Immunol. 2017;199:1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology. 2013;145:293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geremia A, Biancheri P, Allan P et al. . Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13:3–10. [DOI] [PubMed] [Google Scholar]
- 13. Im E, Jung J, Pothoulakis C et al. . Disruption of pten speeds onset and increases severity of spontaneous colitis in il10(-/-) mice. Gastroenterology. 2014;147:667–679.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ananthakrishnan AN, McGinley EL, Binion DG et al. . Ambient air pollution correlates with hospitalizations for inflammatory bowel disease: an ecologic analysis. Inflamm Bowel Dis. 2011;17:1138–1145. [DOI] [PubMed] [Google Scholar]
- 15. Kaplan GG, Hubbard J, Korzenik J et al. . The inflammatory bowel diseases and ambient air pollution: a novel association. Am J Gastroenterol. 2010;105:2412–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salim SY, Jovel J, Wine E et al. . Exposure to ingested airborne pollutant particulate matter increases mucosal exposure to bacteria and induces early onset of inflammation in neonatal IL-10-deficient mice. Inflamm Bowel Dis. 2014;20:1129–1138. [DOI] [PubMed] [Google Scholar]
- 17. Filippini T, Heck JE, Malagoli C et al. . A review and meta-analysis of outdoor air pollution and risk of childhood leukemia. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2015;33:36–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang J, Zuo X, Bai W et al. . PTEN methylation involved in benzene-induced hematotoxicity. Exp Mol Pathol. 2014;96:300–306. [DOI] [PubMed] [Google Scholar]
- 19. Chen L, Xu B, Liu L et al. . Cadmium induction of reactive oxygen species activates the mtor pathway, leading to neuronal cell death. Free Radic Biol Med. 2011;50:624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leslie NR, Downes CP. PTEN function: how normal cells control it and tumour cells lose it. Biochem J. 2004;382:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koukos G, Polytarchou C, Kaplan JL et al. . MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–852.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi YJ, Im E, Pothoulakis C et al. . TRIF modulates TLR5-dependent responses by inducing proteolytic degradation of TLR5. J Biol Chem. 2010;285:21382–21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kühn R, Löhler J, Rennick D et al. . Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. [DOI] [PubMed] [Google Scholar]
- 24. Spencer SD, Di Marco F, Hooley J et al. . The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mak LH, Woscholski R. Targeting PTEN using small molecule inhibitors. Methods. 2015;77–78:63–68. [DOI] [PubMed] [Google Scholar]
- 26. Zu L, Shen Z, Wesley J et al. . PTEN inhibitors cause a negative inotropic and chronotropic effect in mice. Eur J Pharmacol. 2011;650:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi YJ, Im E, Chung HK et al. . TRIF mediates toll-like receptor 5-induced signaling in intestinal epithelial cells. J Biol Chem. 2010;285:37570–37578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rhee SH, Ma EL, Lee Y et al. . Corticotropin releasing hormone and urocortin 3 stimulate vascular endothelial growth factor expression through the camp/CREB pathway. J Biol Chem. 2015;290:26194–26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rhee SH, Im E, Riegler M et al. . Pathophysiological role of Toll-like receptor 5 engagement by bacterial flagellin in colonic inflammation. Proc Natl Acad Sci U S A. 2005;102:13610–13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dowd SE, Callaway TR, Wolcott RD et al. . Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (btefap). BMC Microbiol. 2008;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dowd SE, Sun Y, Wolcott RD et al. . Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned salmonella-infected pigs. Foodborne Pathog Dis. 2008;5:459–472. [DOI] [PubMed] [Google Scholar]
- 32. Callaway TR, Dowd SE, Edrington TS et al. . Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J Anim Sci. 2010;88:3977–3983. [DOI] [PubMed] [Google Scholar]
- 33. Williams WL, Tedeschi LO, Kononoff PJ et al. . Evaluation of in vitro gas production and rumen bacterial populations fermenting corn milling (co)products. J Dairy Sci. 2010;93:4735–4743. [DOI] [PubMed] [Google Scholar]
- 34. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 35. Eren AM, Zozaya M, Taylor CM et al. . Exploring the diversity of gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One. 2011;6:e26732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Swanson KS, Dowd SE, Suchodolski JS et al. . Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. Isme J. 2011;5:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeSantis TZ, Hugenholtz P, Larsen N et al. . Greengenes, a chimera-checked 16S rrna gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caporaso JG, Kuczynski J, Stombaugh J et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. [DOI] [PubMed] [Google Scholar]
- 40. Weber-Nordt RM, Meraz MA, Schreiber RD. Lipopolysaccharide-dependent induction of IL-10 receptor expression on murine fibroblasts. J Immunol. 1994;153:3734–3744. [PubMed] [Google Scholar]
- 41. Shouval DS, Ouahed J, Biswas A et al. . Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv Immunol. 2014;122:177–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gibbs VC, Pennica D. CRF2-4: isolation of cDNA clones encoding the human and mouse proteins. Gene. 1997;186:97–101. [DOI] [PubMed] [Google Scholar]
- 43. Kotenko SV, Krause CD, Izotova LS et al. . Identification and functional characterization of a second chain of the interleukin-10 receptor complex. Embo J. 1997;16:5894–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. [DOI] [PubMed] [Google Scholar]
- 45. Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chu H, Khosravi A, Kusumawardhani IP et al. . Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strober W. Impact of the gut microbiome on mucosal inflammation. Trends Immunol. 2013;34:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bloom SM, Bijanki VN, Nava GM et al. . Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–624. [DOI] [PubMed] [Google Scholar]
- 50. Ganesh BP, Klopfleisch R, Loh G et al. . Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella typhimurium-infected gnotobiotic mice. PLoS One. 2013;8:e74963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garrett WS, Gallini CA, Yatsunenko T et al. . Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guo S, Nighot M, Al-Sadi R et al. . Lipopolysaccharide regulation of intestinal tight junction permeability is mediated by TLR4 signal transduction pathway activation of FAK and myd88. J Immunol. 2015;195:4999–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jacobs JP, Goudarzi M, Singh N et al. . A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol. 2016;2:750–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sellon RK, Tonkonogy S, Schultz M et al. . Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chichlowski M, Sharp JM, Vanderford DA et al. . Helicobacter typhlonius and Helicobacter rodentium differentially affect the severity of colon inflammation and inflammation-associated neoplasia in IL10-deficient mice. Comp Med. 2008;58:534–541. [PMC free article] [PubMed] [Google Scholar]
- 56. Kullberg MC, Ward JM, Gorelick PL et al. . Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–181. [DOI] [PubMed] [Google Scholar]
- 58. Hammer AM, Morris NL, Cannon AR et al. . Interleukin-22 prevents microbial dysbiosis and promotes intestinal barrier regeneration following acute injury. Shock. 2017;48:657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nava P, Koch S, Laukoetter MG et al. . Interferon-gamma regulates intestinal epithelial homeostasis through converging beta-catenin signaling pathways. Immunity. 2010;32:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Begue B, Verdier J, Rieux-Laucat F et al. . Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106:1544–1555. [DOI] [PubMed] [Google Scholar]
- 62. Glocker EO, Kotlarz D, Boztug K et al. . Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kotlarz D, Beier R, Murugan D et al. . Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. 2012;143:347–355. [DOI] [PubMed] [Google Scholar]
- 64. Engelhardt KR, Shah N, Faizura-Yeop I et al. . Clinical outcome in IL-10- and IL-10 receptor-deficient patients with or without hematopoietic stem cell transplantation. J Allergy Clin Immunol. 2013;131:825–830. [DOI] [PubMed] [Google Scholar]
- 65. Shouval DS, Biswas A, Goettel JA et al. . Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kang SS, Bloom SM, Norian LA et al. . An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pils MC, Pisano F, Fasnacht N et al. . Monocytes/macrophages and/or neutrophils are the target of IL-10 in the LPS endotoxemia model. Eur J Immunol. 2010;40:443–448. [DOI] [PubMed] [Google Scholar]
- 68. Im E, Riegler FM, Pothoulakis C et al. . Elevated lipopolysaccharide in the colon evokes intestinal inflammation, aggravated in immune modulator-impaired mice. Am J Physiol Gastrointest Liver Physiol. 2012;303:G490–G497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.