Summary
Non-human primate neuroimaging is a rapidly growing area of research that promises to transform and scale translational and cross-species comparative neuroscience. Unfortunately, the technological and methodological advances of the past two decades have outpaced the accrual of data, which is particularly challenging given the relatively few centers that have the necessary facilities and capabilities. The PRIMatE Data Exchange (PRIME-DE) addresses this challenge by aggregating independently acquired non-human primate magnetic resonance imaging (MRI) datasets and openly sharing them via the International Neuroimaging Data-sharing Initiative (INDI). Here, we present the rationale, design, and procedures for the PRIME-DE consortium, as well as the initial release, consisting of 25 independent data collections aggregated across 22 sites (total = 217 non-human primates). We also outline the unique pitfalls and challenges that should be considered in the analysis of non-human primate MRI datasets, including providing automated quality assessment of the contributed datasets.
Highlights
-
•
Openly shared, large non-human primate neuroimaging data resource
-
•
Multiple imaging modalities contributed from investigators around the world
-
•
Quality assessments of the dataset
-
•
Discussed pitfalls and challenges in analyzing the non-human primate MRI data
The PRIMatE Data Exchange (PRIME-DE) consortium is an open science resource for the neuroimaging community aiming to facilitate efforts to map the non-human primate connectome. It aggregates and shares anatomical, functional, and diffusion MRI datasets from laboratories throughout the world.
Introduction
Translational, comparative neuroscience research enables a bridging of knowledge gaps across species as well as invasive and noninvasive approaches. A growing body of research has documented the utility of magnetic resonance imaging (MRI) technologies to support in vivo examination of brain organization and function in non-human primates (Vanduffel et al., 2014, Rilling, 2014, Van Essen and Glasser, 2014, Zhang et al., 2013, Shmuel and Leopold, 2008, Schwiedrzik et al., 2015). Recent work has demonstrated the ability to recapitulate findings from gold-standard invasive methodologies (Ghahremani et al., 2017, Donahue et al., 2016, Grayson et al., 2016). This work also provides novel insights into the organizational principles of the non-human primate (NHP) connectome (Goulas et al., 2017, Hutchison and Everling, 2014, Hutchison et al., 2011, Vincent et al., 2007) and cross-species comparative connectomics (Hutchison et al., 2012, Hutchison et al., 2015, Miranda-Dominguez et al., 2014, Mars et al., 2011, Seidlitz et al., 2018a), which are possible only through in vivo studies. These advances are timely given the growing prominence of large-scale national and international initiatives focused on advancing our understanding of human brain organization and the ability to generate novel therapeutics for neurology and psychiatry (Bargmann and Newsome, 2014).
Despite the clear demonstrations of feasibility and utility, the field of non-human primate neuroimaging is still developing. Numerous unique challenges related to the acquisition and processing of non-human primate data are still being addressed (e.g., Seidlitz et al., 2018b, Hutchison and Everling, 2012), and the potential for broad reaching cross-species studies remains unexploited. Perhaps most challenging is the limited availability of data.
Here, we introduce the PRIMatE Data Exchange (PRIME-DE) to create an open science resource for the neuroimaging community that will facilitate the mapping of the non-human primate connectome. To accomplish this, we aggregate a combination of anatomical, functional, and diffusion MRI datasets from laboratories throughout the world and make these data available to the scientific community. It merits emphasis that PRIME-DE supports an ongoing process that will remain open to new contributions of data from macaques and other non-human primate species.
Results
Overview
At present, PRIME-DE contains 25 collections aggregated across 22 sites; to date, data from 217 primates are included (see Table 1 for information on each institution). Contributions will continue to be accepted and shared on a rolling basis.
Table 1.
Experimental Design
| Investigators | Speciesa | Subjects | State | Contrast Agent | Structural T1 | Structural T2 | Resting State fMRI | Naturalistic Viewing fMRI | Task fMRI | Field map | Diffusion MRI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMU | Belin, Brochier, Sein | MM | 4 | Anesthetized | No | ✔ | ✔ | – | – | – | – | ✔ |
| Caltech | Rajimehr, Tsao | MM | 2 | Awake | Yes | – | – | – | 96 min | – | – | – |
| ECNU (C) | Aihua Chen | MM | 10 | Anesthetized | No | ✔ | – | – | – | – | – | – |
| ECNU (K)b | Kwok, Zhou | MM | 4 | Anesthetized | No | ✔ | ✔ | 8 min | – | – | – | ✔ |
| Institute of Neuroscience (IoN) | Wang | MM, MF | 8 | Anesthetized | No | ✔ | – | 20–40 min | – | – | ✔ | – |
| Institut des Sciences Cognitives Marc Jeannerod | Ben Hamed, Hiba | MM | 8 | Anesthetized/Awake | Yes | ✔ | – | ✔ | – | ✔ | – | ✔ |
| Lyon Neuroscience Research Center | Hadj-Bouziane, Meunier, Guedj | MM | 1 | Anesthetized/Awake | Yes/No | ✔ | ✔ | 13 min | – | – | – | – |
| McGill University | Mok, Rudko, Shmuel | MM, MF | 3 | Anesthetized | No | ✔ | ✔ | – | – | – | – | – |
| Mount Sinai (P) | Croxson, Fleysher | MM, MF | 9 | Anesthetized | No | ✔ | ✔ | 43 min | – | – | ✔ | ✔ |
| Mount Sinai (S) | Croxson, Fleysher, Froudist-Walsh, Damatac, Nagy | MM | 5 | Anesthetized | No | ✔ | ✔ | – | – | – | – | ✔ |
| NKI | Schroeder, Milham | MM | 2 | Anesthetized/Awake | Yes/No | ✔ | 76–155 min | 55–345 min | – | – | – | |
| NIMH (L)c | Leopold, Russ | MM | 3 | Awake | Yes | ✔ | ✔ | 30–150 min | 170 min | – | – | – |
| NIMH (M)c,d | Messinger, Jung, Seidlitz, Ungerleider | MM | 3 | Anesthetized/Awake | Yes | ✔ | – | 10−15 min | – | – | – | – |
| Netherlands Institute for Neuroscience (NIN) | Klink, Roelfsema | MM | 2 | Anesthetized | No | ✔ | ✔ | 9.7 min | – | – | – | – |
| NeuroSpin | Jarraya, Dehaene | MM | 3 | Anesthetized | Yes/No | ✔ | – | ✔ | – | – | – | – |
| Newcastle | Petkov, Nacef, Thiele, Poirier, Balezeau, Griffiths, Schmid, Rios | MM | 14 | Anesthetized/Awake | No | ✔ | ✔ | 21.6 min | – | – | – | – |
| OHSU | Sullivan, Fair | MM | 2 | Anesthetized | Yes/No | ✔ | ✔ | 480 min | – | – | – | – |
| Princeton | Kastner, Pinsk | MM | 2 | Anesthetized | ✔ | ✔ | – | – | – | ✔ | ✔ | |
| Rockefeller | Schwiedrzik, Freiwald, Zarco | MM, MF | 6 | Anesthetized | Yes | ✔ | – | 80 min | – | – | ✔ | |
| SBRI | Procyk, Wilson, Amiez | MM, MF | 22 | Anesthetized | No | ✔ | ✔ | ✔ | – | – | – | – |
| UC Davis | Baxter, Croxson, Morrison | MM | 19 | Anesthetized | No | ✔ | ✔ | 13.5 min | – | – | ✔ | ✔ |
| Univ. of Minnesota (UMN) | Yacoub, Harel | M | 2 | Anesthetized | – | ✔ | – | 27 min | – | – | ✔ | ✔ |
| Univ. of Oxfordc | Sallet, Mars, Rushworth | MM | 20 | Anesthetized | No | ✔ | – | 53.43 min | – | – | – | – |
| NIN Primate Brain Bank/Utrecht University | Navarrete, Blezer, Todorov, Lindenfors, Laland, Reader | Multiplea | 51 | Post-mortem | Yes/No | ✔ | – | – | – | – | – | – |
| Univ. of Western Ontario (UWO) | Everling, Menon | MM | 12 | Anesthetized | No | ✔ | ✔ | 60 min | – | – | – | – |
General information about PRIME-DE data collections contributed prior to the time of publication. For usage agreement, CC-BY-NC-SA: Creative Commons – Attribution-NonCommercial Share Alike, Standard INDI data sharing policy, prohibits use of the data for commercial purposes; DUA: Data Usage Agreement, users must complete a DUA prior to gaining access to the data. For species information, MM: Macaca mulatta; MF: Macaca fascicularis; M: Macaca.
Detailed species information is available on the PRIME-DE site and in Navarrete et al., 2018
ECNU (K) provided magnetic resonance spectroscopy
The usage agreement is DUA for those sites, CC-BY-NC-SA for all other sites
NIMH (M) provided cortical thickness and brain template
To promote usage of a standardized data format, we organized all data using the Brain Imaging Data Structure (BIDS) format (Gorgolewski et al., 2017). All PRIME-DE datasets can be accessed through the PRIME-DE site (http://fcon_1000.projects.nitrc.org/indi/indiPRIME.html). Prior to downloading the data, users are required to establish a user account on NITRC and register with the International Neuroimaging Data-sharing Initiative (INDI; anticipated time: <1 min).
MRI Data
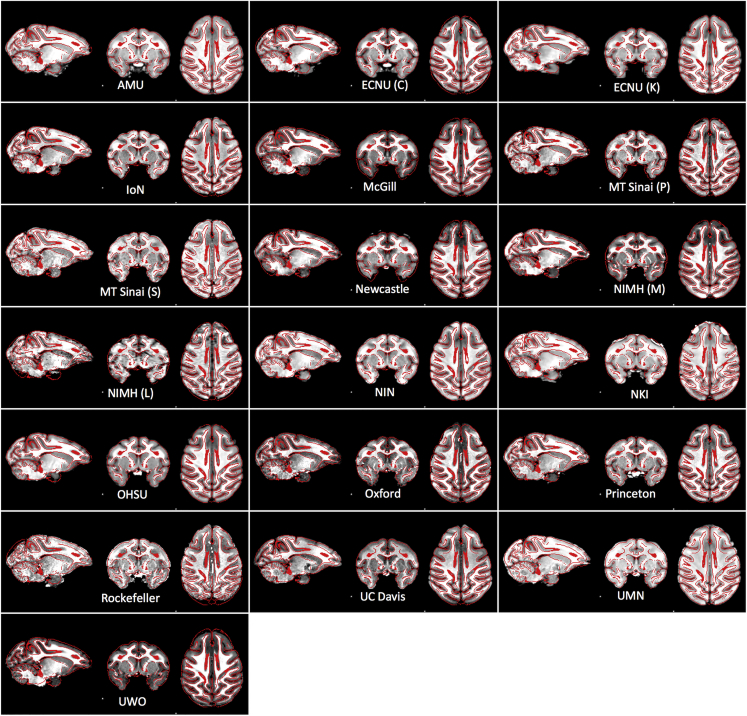
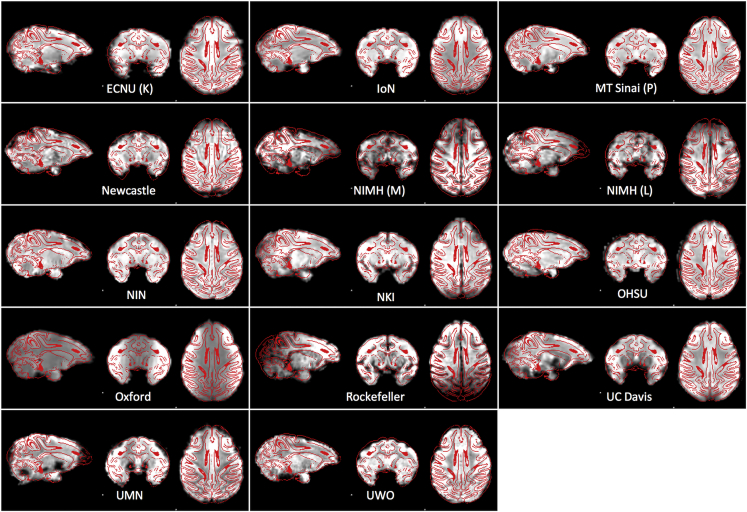
With one exception, for each of the PRIME-DE collections, at least one structural MRI (sMRI) is available for each unique ID number (see Table 1). Eighteen of the collections contain at least one corresponding resting-state functional MRI (R-fMRI) dataset, and three of the collections contain naturalistic viewing fMRI (NV-fMRI). In addition, one collection from the National Institutes of Mental Health (NIMH (M)) also provided cortical thickness data and R-fMRI data aligned to an anatomical template. Corresponding diffusion MRI (dMRI) datasets are available for nine collections. Field map images for fMRI correction are available for six collections. Consistent with its popularity in the imaging community and prior usage in INDI efforts, the NIFTI file format was selected for storage of the PRIME-DE MRI datasets. Table 2 lists the specific MRI scanners and head coils utilized for each collection. Specific MRI sequence parameters for the various data collections are summarized in Tables S1, S2, S3, and S4 and detailed on the PRIME-DE website. Across collections, R-fMRI acquisition durations varied from 8 to 155 min per subject. In two collections, subjects were in an awake state. In five collections, subjects were scanned both awake and under anesthesia. One collection scanned 51 post-mortem specimens. In the remaining 17 collections, subjects were scanned under anesthesia. For the three collections with NV-FMRI, acquisition durations varied from 55 to 375 min. See Figures 3 and 4 for example structural and functional images from the different sites aligned in a common space.
Table 2.
Scanner Information
| Site | Manufacturer | Model | Field Strength (T) | Head coil # channels |
|---|---|---|---|---|
| AMU | Siemens | Prisma | 3 | Body transmit array, 11 cm loop receiving coil |
| Caltech | Siemens | Tim Trio | 3 | 8 |
| ECNU (C) | Siemens | Tim Trio | 3 | – |
| ECNU (K) | Siemens | Tim Trio | 3 | 1-channel surface coil |
| Institute of Neuroscience (IoN) | Siemens | Tim Trio | 3 | 8-channel phased-array transceiver coils |
| Institut des Sciences Cognitives Marc Jeannerod | Siemens | Sonata/Prisma | 1.5/3 | 8-channel custom head coils/association of independent circular coils |
| Lyon Neuroscience Research Center | Siemens | Sonata/Prisma | 1.5/3 | Custom-made 10 cm loop receiving coil 2 × L11 and 1 × L7 Siemens loop-receiving coil |
| McGill University | Siemens | Tim Trio | 3 | Custom-made 8-channel phased-array receive coil |
| Mount Sinai (P) | Philips | Achieva | 3 | Single loop receive coil (T1 and T2) 4-channel phased-array receive, transmit through body coil (resting state and diffusion) |
| Mount Sinai (S) | Siemens | Skyra | 3 | 8-channel phased-array receive with a single loop transmit |
| NKI | Siemens | Tim Trio | 3 | Custom-made 8-channel phased-array receive coil (KU Leuven) with a custom 16-channel pre-amplifier (MRcoils) |
| NIMH (L) | Bruker | BiospecVertical | 4.7 | 8 |
| NIMH (M) | Bruker | BiospecVertical | 4.7 | 1–4 |
| Netherlands Institute for Neuroscience (NIN) | Philips | Ingenia | 3 | Custom-made 8-channel phased-array receive coil (KU Leuven) with a custom 16-channel pre-amplifier (MRcoils). |
| NeuroSpin | Siemens | Tim Trio/PrismaFit | 3 | 1chTxRxcoil/1Tx-8Rxchcoil |
| Newcastle | Bruker | Vertical Bruker | 4.7 | 4–8 |
| OHSU | Siemens | Tim Trio | 3 | Knee coil 15 channel |
| Princeton | Siemens | Prisma VE11C | 3 | Siemens Loop Coil, Large (11 cm) |
| Rockefeller | Siemens | TIM Trio + AC88 gradient | 3 | 8-channel phased-array receive with a single-loop transmit |
| SBRI | Siemens | Sonata/Prisma | 1.5/3 | Custom made 10 cm loop receiving coil 2 × L11 and 1 × L7 Siemens loop receiving coil |
| UC Davis | Siemens | Skyra | 3 | 4 |
| Univ. of Minnesota (UMN) | Siemens | SyngoB17 | 7 | 16-channel transmit/receive + 6 receive only |
| Univ. of Oxford | – | – | 3 | A four-channel phased-array coil |
| NIN Primate Brain Bank/Utrecht University | Varian/Siemens | Small-bore scanner/Magnetom trio | 9.4/3 | – |
| Univ. of Western Ontario (UWO) | Siemens | Magnetom | 7 | Custom-made 24-channel phased-array receive coil with an 8-channel transmit coil |
Information on scanner and head coil for PRIME-DE data collections contributed prior to the time of publication. Note that scanner information from University of Oxford is not reported due to an agreement made previously with the scanner manufacturer. For scan sequences, see also Tables S1, S2, S3, and S4.
Figure 3.
Example Structural Images
Example structural images aligned to the common space defined by the NMT template.
Figure 4.
Example Functional Images
Example functional images aligned to the common space defined by the NMT template.
Data Licensing
Contributors to PRIME-DE will be able to set the sharing policy for their data in accord with their preferences and institutional requirements. For each sample, the contributor will set the sharing permissions for their data using one or more the following three policies:
-
(1)
Creative Commons – Attribution-Non-Commercial Share Alike (CC-BY-NC-SA) (https://creativecommons.org/licenses/by-nc-sa/4.0/). Standard INDI data sharing policy. Prohibits use of the data for commercial purposes.
-
(2)
Creative Commons – Attribution (CC-BY) (https://creativecommons.org/licenses/by/4.0/). Least restrictive data sharing policy.
-
(3)
Custom Data Usage Agreement. Users must complete a data usage agreement (DUA) prior to gaining access to the data. Contributors can customize the agreement as they see fit, including determining whether or not signatures from authorized institutional official are required prior to executing the DUA. Note: this option was created to facilitate potential contributors whose institution requires completion of a formal interinstitutional agreement in order to share non-human primate data. Of note, one lesson learned from the human neuroimaging literature is that such agreements are not dissuasive, as is evidenced by the success of the Human Connectome Project (Van Essen et al., 2013) and the NKI-Rockland Sample (Nooner et al., 2012).
Automated Quality Assessment
Consistent with the established policy of INDI, all data contributed to PRIME-DE was made available to users regardless of data quality; users should check data quality before inclusion in their analyses. The rationale of this decision has been the lack of consensus on optimal quality criteria in regards to specific measures or their combinations and cutoffs—a reality that is even more pronounced in non-human primate imaging given the variation in data quality and characteristics across scan protocols. Of note, a benefit of sharing data with differing levels of quality data is also important for those working to develop methods for evaluating, and at times overcoming, such variations.
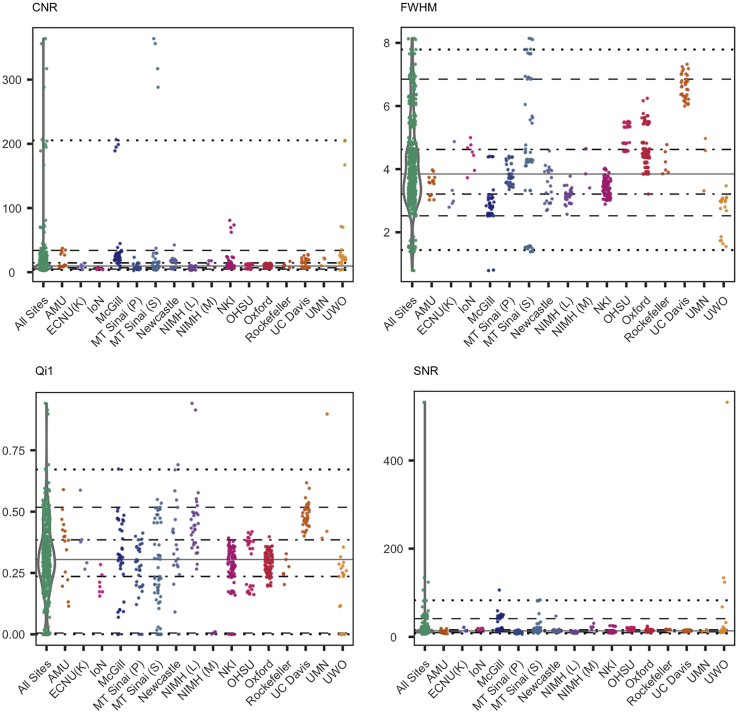
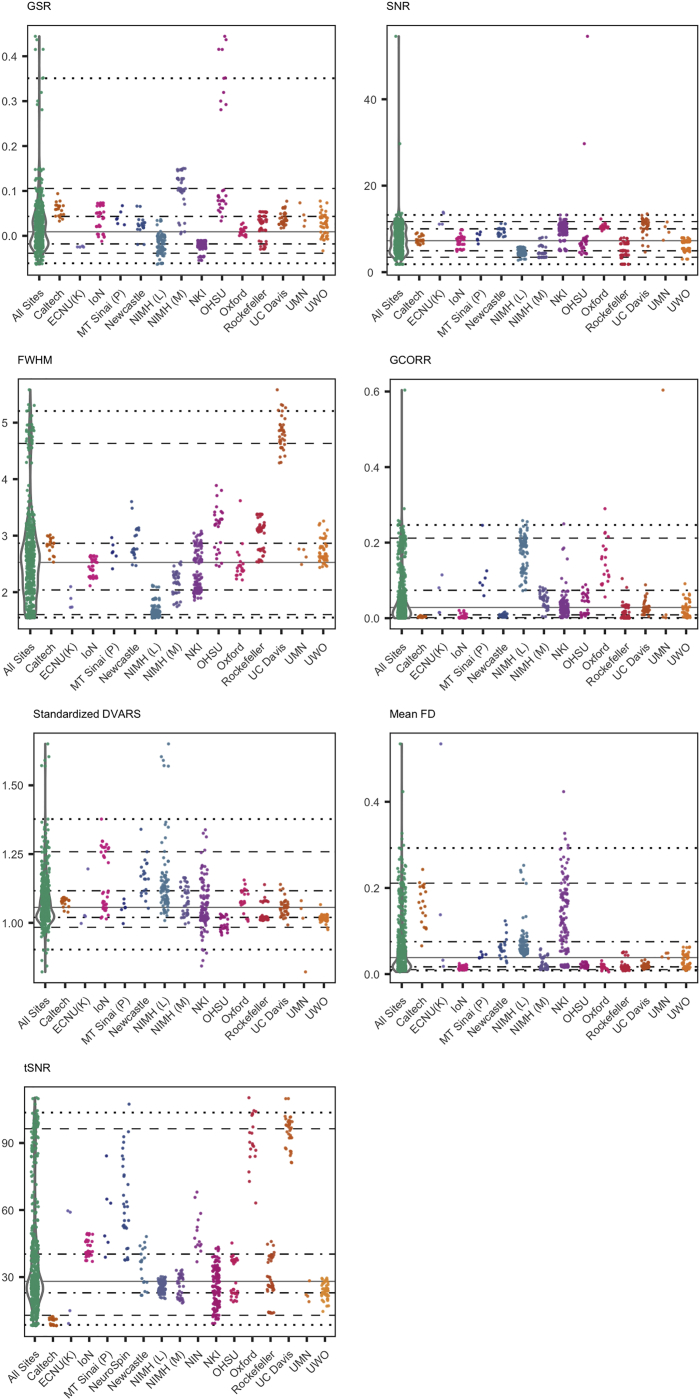
Following the tradition of recent INDI data-sharing consortia, a collection of automated, reference-free quality assurance measures, known as the Preprocessed Connectome Project Quality Assurance Protocol (PCP-QAP; Shehzad et al., 2015), is being made available with the PRIME-DE datasets. These measures focus on structural and temporal (when appropriate) aspects of the datasets. Table 3 provides a brief description of the measures included, and Figures 1 and 2 depict a subset of QAP results (Magnotta et al., 2006, Mortamet et al., 2009, Giannelli et al., 2010, Jenkinson et al., 2002, Friedman et al., 2006, Nichols, 2012). As would be expected, measures of head motion are notably smaller for sites using anesthetized scan sessions than for awake (NIMH (L), NIMH (M), NKI, Newcastle, Lyon Neuroscience Research Center). Importantly, the measures provided are not intended to be definitive for the field or all encompassing; rather, they are included to spur interest in the potential utility and further development of automated measures.
Table 3.
Description of PCP QAP Measures
| Spatial Metrics | Description | References |
|---|---|---|
| Contrast-to-noise ratio (CNR) (sMRI only) | MGM intensity—MWM intensity/SDair intensity. Larger values reflect a better distinction between WM and GM. | Magnotta et al., 2006 |
| Artifactual voxel detection (Qi1) (sMRI only) | Voxels with intensity corrupted by artifacts/voxels in the background. Larger values reflect more artifacts which likely due to motion or image instability. | Mortamet et al., 2009 |
| Smoothness of Voxels (FWHM)a | Full width at half maximum of the spatial distribution of the image intensity values. Larger values reflect more spatial smoothing perhaps due to motion or technical differences. | Friedman et al., 2006 |
| Signal-to-noise ratio (SNR) | MGM intensity/SDair intensity. Larger values reflect less noise. | Magnotta et al., 2006 |
| Temporal Metrics (fMRI and DTI only) | Description | References |
| Ghost-to-Signal Ratio (GSR)a | M signal in the “ghost” image divided by the M signal within the brain. Larger values reflect more ghosting likely due to physiological noise, motion, or technical issues. | Giannelli et al., 2010 |
| Mean frame-wise displacement- Jenkinson (meanFD)b | Sum absolute displacement changes in the x, y, and z directions and rotational changes around them. Rotational changes are given distance values based on changes across the surface of a 50 mm radius sphere. Larger values reflect more movement. | Jenkinson et al. 2002 |
| Standardized DVARSb | Spatial SD of the data temporal derivative normalized by the temporal SD and autocorrelation. Larger values reflect larger frame-to-frame differences in signal intensity due to head motion or scanner instability. | Nichols, 2012 |
| Global Correlation (GCORR)b | M correlation of all combinations of voxels in a time series. Illustrates differences between data due to motion/physiological noise. Larger values reflect a greater degree of spatial correlation between slices, which may be due to head motion or “signal leakage” in simultaneous multi-slice acquisitions. | – |
Here, we provide a brief description of the Preprocessed Connectome Project Quality Assessment Protocol. These measures have been computed for all structural MRI (sMRI) and resting-state functional MRI (R-fMRI) datasets in PRIME-DE. The table was adopted from Di Martino et al. (2017).
For R-fMRI data, these metrics are computed on mean functional data
For R-fMRI, these metrics are computed on time series data. M, mean; GM, gray matter; WM, white matter; SD, standard deviation
Figure 1.
Spatial Quality Metrics for Morphometry MRI Datasets
Spatial quality metrics include: contrast-to-noise ratio (CNR), smoothness of voxels indexed as full width at half maximum (FWHM), signal-to-noise ratio (SNR), and artifactual voxel detection (Qi1). See Table 3 for details on this and the other quality metrics released. The colored scatterplots illustrate the quality metrics distribution for each data collection. The violin plots on the left of each panel represent a kernel density estimation of the distribution across all data collections for each quality metric. Starting from the bottom: each horizontal line marks the 1st, 5th, 25th, 50th, 75th, 95th, and 99th percentiles.
Figure 2.
Spatial and Temporal Quality Metrics for Functional MRI Datasets
Spatial quality metrics include: ghost-to-single ratio (GSR), smoothness of voxels indexed as full width at half maximum (FWHM), and signal-to-noise ratio (SNR). Temporal metrics are mean frame-wise displacement (Mean FD), standardized DVARS, global correlation (GCORR), and temporal signal-to-noise ratio (tSNR). See Table 3 for details on this and the other quality metrics released. The colored scatterplots illustrate the quality metrics distribution for each data collection. The violin plots on the left of each panel represent a kernel density estimation of the distribution across all data collections for each quality metric. Starting from the bottom: each horizontal line marks the 1st, 5th, 25th, 50th, 75th, 95th, and 99th percentiles.
Discussion
Challenges in the Processing of Non-human Primate Imaging Data
We confront a variety of challenges when trying to adapt well-established methods for human neuroimaging processing to primate data. Beyond the differences between species in tissue contrast, brain shape and size, and type and amount of tissue surrounding the brain, there are significant differences in data collection equipment and acquisition protocols. Non-human primate data are often acquired at very high fields (4.7T, 7T, 9.4T, 11.7T), using some non-standardized arrangement of surface coils. These result in increased variations in image intensity due to B1 inhomogeneity and non-uniform coil coverage and in greater distortion and dephasing due to susceptibility. Another issue is that the equipment and acquisition protocols used are typically customized, resulting in substantial variation in the quality and characteristics of data collected at different sites. Consequently, there is no one-size-fits-all strategy for processing animal data, and researchers need a great deal of flexibility to optimize their pipelines for the data at hand.
Brain extraction and tissue segmentation are more challenging in non-human primate imaging data due to differences in tissue contrast and the nature of structures immediately surrounding the brain. If compromised, these steps in turn can dramatically compromise image registration and normalization procedures as well as temporal de-noising approaches. As of yet, there is no consensus for an optimal solution for each of these processing steps, in part due to the many sources of variation across studies that can differentially impact data characteristics and quality (e.g., anesthesia protocols, coil type, use of contrast agents, magnet strength, animal/rodent type). Additionally, commonly used pre-processing pipelines, used extensively with human neuroimaging datasets, often fail to work properly on non-human primate datasets. As a result, researchers commonly work to optimize individual steps for their datasets outside of traditional workflows, resulting in different pipelines and processing steps across groups. There are efforts underway to form best practices to guide this process and help researchers avoid the need to redefine pipelines themselves (e.g., Seidlitz et al., 2018b, Love et al., 2016); currently, however, it is still necessary for researchers to do so.
Resources and Solutions
Templates and Atlases
A number of macaque templates were created in the last decade, including single-animal templates, e.g., the NeuroMap macaque atlas (M.F. Dubach and D.M. Bowden, 2009, Soc. Neurosci., abstract) and the 3D Digital D99 Template (Reveley et al., 2017), and population-averaged templates based on multiple animals, e.g., 112RM-SL (McLaren et al., 2009), INIA19 (Integrative Neuroscience Initiative on Alcoholism; (Rohlfing et al., 2012), MNI (Montreal Neurological Institute; (Frey et al., 2011), CIVM MRI/DTI atlas (Calabrese et al., 2015), and the most recent NMT (National Institute of Mental Health Macaque Template; (Seidlitz et al., 2018b). In addition, there are surface-based atlases, including the macaque single-subject F99 atlas (Van Essen, 2012, Van Essen, 2002) and the group-average Yerkes19 macaque atlas (Donahue et al., 2016). Data collected in individual macaques can be aligned to these templates using affine and non-linear registration. These templates provide a common anatomical space and coordinate system for specifying specific brain locations and visualizing data collected across days, animals, and laboratories.
Of note, some templates link to volumetric digital brain atlases (Frey et al., 2011, Reveley et al., 2017, Seidlitz et al., 2018b, Saleem and Logothetis, 2012) derived from analysis of histological tissue (Saleem and Logothetis, 2012, Paxinos et al., 1999, Paxinos, 2009). These anatomical parcellations can be warped to individual subjects using standard linear and non-linear registration algorithms (e.g., AFNI’s 3dAllineate and 3dQwarp). Scripts to automate this alignment are available for the single-subject D99 template (https://afni.nimh.nih.gov/pub/dist/atlases/macaque) and the recently published National Institute of Mental Health Macaque Template (NMT; Seidlitz et al., 2018b; https://afni.nimh.nih.gov/NMT). The NMT is a high-resolution (0.25 mm isotropic) T1 template built from in vivo scans of 31 young adult macaques. This volume (and accompanying surfaces) is representative of the adult population and provides anatomical detail akin to that of ex vivo templates, which require days of scanning to acquire. The NMT is available via the PRIME-DE website as well as on GitHub (https://github.com/jms290/NMT). The database also includes resting-state data from three subjects that have been aligned to the NMT (see NIMH (M) in Table 1). A similar multi-subject template also exists for pre-pubertal rhesus monkeys (Fox et al., 2015); additionally, the publically available UNC-Wisconsin Rhesus Macaque Neurodevelopment Database features a longitudinal dataset that can be used to provide insights into age-related changes in structure (Young et al., 2017).
Other anatomical parcellations have been defined on the surface using the single-subject F99 template (available in Caret; Van Essen et al., 2012), which can be used for analysis on the cortical sheet. For example, the cortical parcellation from Markov et al. (2014) includes quantitative tract-tracing connectivity estimates for a subset of these regions.
Improving Skull Extraction, Segmentation, and Registration
A high-quality T1 image with isotropic voxels is important for skull extraction. There are a number of brain extraction algorithms and available tools, e.g., the Brain Extraction Tool (BET in FSL; Smith, 2002), 3dSkullStrip in AFNI (Cox, 1996), the Hybrid Watershed Algorithm (HWA in FreeSurfer; (Ségonne et al., 2004), BSE in BrainSuite (Shattuck and Leahy, 2002), Robust Brain Extraction (ROBEX; Iglesias et al., 2011), Primatologist toolbox (Balbastre et al., 2017), and ANTs (Avants et al., 2011). Most of these tools can be effectively applied to human data; however, the performance is suboptimal and variable in NHP due to the differences in brain structure (e.g., size, adipose tissue, olfactory bulb) and the quality of the T1 image (SNR, inhomogeneous intensity). Accordingly, the parameters and/or related atlas library need to be customized to optimize the brain extraction in NHP. For example, in AFNI, the program “3dSkullStrip” with alternative options “-monkey,” “-marmoset,” and “-surface_coil” is available for brain extraction in NHP. Population brain templates, such as the NMT, can further improve and automate the registration and brain extraction process (Seidlitz et al., 2018b).
Standard segmentation algorithms can separate gray versus white matter, but if the signal is not homogeneous, which is typically the case at higher magnetic fields, segmentation in some parts of the brain will be better than others (especially subcortically). Registration of T2 datasets to T1 structural scans also remains a challenge. Affine or non-linear registration algorithms can work well provided that intermediate scans are available. For instance, a full brain T1 structural scan from the same individual obtained along with T2 images (also with as much coverage of the brain as possible) could be crucial for registering T2 datasets to any of the freely available monkey template brains that are registered to macaque atlases.
One way to reduce or eliminate the manual intervention during brain extraction and tissue segmentation—using only the typically acquired T1 scan—is to rely on priors defined on a high-resolution and high-contrast template. The multi-subject NMT includes manually refined masks of the brain, cortical gray matter, and various tissue types (including blood vasculature; Seidlitz et al., 2018b). Applying the inverse anatomical alignment transformations to the NMT brain mask produces an approximate single-subject mask for brain extraction. A more precise individual brain mask and tissue segmentation can be obtained using the NMT’s representative brain and tissue segmentation masks as priors. The NMT distribution includes scripts that use AFNI and ANTs to perform these mask refinements (as well as morphological analysis). These improvements could be critical for later processing steps for fMRI data. Furthermore, the NMT includes surfaces for visualization of individual subject or group results in a standard coordinate space. Future work could add to these advances, such as tailoring existing surface-based processing pipelines (e.g., CIVET or FreeSurfer) to be specifically used with non-human primate MRI data.
Head Motion
Head motion in NHP imaging is an important concern, just as it is in human neuroimaging studies. For the most part, one can apply human imaging motion correction techniques to NHP data directly. However, there are a few concerns with NHP neuroimaging that will be addressed below.
Anesthesia is commonly used in NHP functional neuroimaging, in part due to the lower behavioral and technical demands compared to awake imaging. As reflected by the QAP results, another benefit is that anesthesia dramatically reduces motion artifacts during NHP scanning. However, the use of anesthesia comes with its own set of tradeoffs dealing with how the drugs used interact with neural activity. There are changes in FC patterns due to the particular set and doses of agents used and in comparison to awake imaging (Xu et al., 2018). For this reason, researchers should always assess how anesthesia may, or may not, influence the results of their study before using it. It should be noted that in some studies, anesthesia can be an experimental goal; for example, fMRI imaging in anesthetized macaques can help reveal brain mechanisms of loss of consciousness (Barttfeld et al., 2015).
In awake NHP imaging, the animals are far more likely to create motion artifacts, which need to be addressed during data preprocessing and analyses when they occur. Of note, these artifacts tend to be caused by body movements (Pfeuffer et al., 2007) rather than head movements, as the head is usually fixed and stable. Body movements can cause changes in the magnetic field, making the shimming performed at the beginning of the scan ineffective (Pfeuffer et al., 2007); the monitoring of full body position can be helpful to eliminate motion artifacts (Keliris et al., 2007). Additionally, acclimation to the chair and scanner setup and training to remain still are of great importance in reducing the amount of motion artifacts. As with human neuroimaging best practices, keeping individual scan periods to the shortest necessary for your task will help to reduce motion artifacts. Recent human studies also suggested that movie (NV-fMRI) paradigm may help to reduce head motion relative to resting conditions (e.g., Vanderwal et al., 2015, Alexander et al., 2017). This is also true in awake NHP imaging; for example, in the PRIME-DE NKI site, the mean FD for rest sessions was 0.21 (SD = 0.03), but 0.14 (SD = 0.07) during movie sessions (t = 2.82, p = 0.006, df = 128).
Regarding motion-correction algorithms, those designed for human neuroimaging data perform similarly for NHP data. As such, most groups use SPM, AFNI, ANTs, or FSL software to estimate the motion parameters and remove motion artifacts. The estimates of the movement values can be used as regressors of no interest during the analysis of functional data, if desired. The grayplot, proposed by Power (2017), can be used to illustrate the motion and the de-noising effects. However, as with all neuroimaging data, image distortions or signal drop-out caused movement correction to be suboptimal.
Next Steps
The PRIME-DE is an ongoing data-sharing consortium stewarded by INDI, which has shared more than 15,000 human imaging datasets over the past decade. As such, we invite new contributions from all investigators in the NHP imaging community, not just those involved in the consortium at the time of the initial release. It is our hope that future contributions will help to capture and promote emerging trends in the NHP community, such as the increasing ability to image during awake states and usage of high-field scanners (e.g., 7.0T), as well as the growing range of species being examined (e.g., marmosets). Additionally, we hope that other data modalities obtained in the NHP community (e.g., electrophysiology) will be shared with higher frequency. Similar to other INDI-based efforts, PRIME-DE is intended to take the first step—establishing a culture for sharing. The logical second step is building toward an optimal infrastructure for sharing. In this regard, it is our hope that open access database and computational platforms will work to increase their support for the needs of NHP imaging. Finally, it is our hope that, building upon the spirit of sharing engendered in PRIME-DE, users will share their resultant statistical maps with one another via venues such as Neurovault (Gorgolewski et al., 2015), which can now handle results from NHP studies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and Algorithms | ||
| NMT Template | Seidlitz et al., 2018b | https://github.com/TingsterX/PRIME-DE |
| Preprocessed Connectome Project Quality Assurance Protocol | Shehzad et al., 2015 | http://preprocessed-connectomes-project.org/quality-assessment-protocol/ |
| FSL | Smith et al., 2004 | https://fsl.fmrib.ox.ac.uk/fsl/fslwiki; RRID: SCR_002823 |
| AFNI | Cox, 1996 | https://afni.nimh.nih.gov/; RRID: SCR_005927 |
| FreeSurfer | Fischl, 2012 | https://surfer.nmr.mgh.harvard.edu/; RRID: SCR_001847 |
| ANTs | Avants et al., 2011 | http://stnava.github.io/ANTs/; RRID: SCR_004757 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael P. Milham (michael.milham@childmind.org).
Experimental Model and Subject Details
Ethics Approval and Consent to Participate
All experimental procedures were approved by local ethics boards prior to any data collection. UK macaque datasets were obtained with Home Office approval and abide with the European Directive on the protection of animals used in research (2010/63/EU). For the NIN Primate Brain Bank/Utrecht University dataset, post-mortem specimens were loaned from the Netherlands Institute of Neuroscience Primate Brain Bank (PBB; http://www.primatebrainbank.org/). No individuals were sacrificed for PBB brain issue. Instead, brains were collected from individuals that died from natural causes or that had to be humanely euthanized for reasons unrelated to the tissue collection.
Method Details
Criteria for Data Contributions
PRIME-DE welcomes contributions from any laboratory willing to openly share multimodal MRI datasets obtained from non-human primates, including but not limited to functional MRI, diffusion MRI and structural MRI. Contributors are responsible for ensuring that any data collected and shared were obtained in accordance with local ethical and regulatory requirements.
There are no set exclusion criteria. We encourage the sharing of all data, independent of quality. This decision is based on the realizations that: 1) there is no consensus on acceptable criteria for movement in functional MRI or diffusion MRI data, 2) high motion datasets are essential to the determination of the impact of motion on reliability, and 3) new approaches continue to be developed to account for movement artifacts. We also encourage submission of data from other modalities (e.g., ASL) or experimental paradigms (e.g., longitudinal data, pharmacologic manipulations) when available.
Metadata
Any imaging metadata (e.g., protocol parameters) provided with the data contribution are represented in the BIDS data format. In the case that data are provided in DICOM format, the metadata from the DICOM are used to population the .json file available with BIDS.
Given that this is a retrospective data collection, phenotypic data primarily focuses on basic measures that are relatively standard in the neuroimaging field, as well as those fundamental for analyses and sample characterization. Minimal phenotypic information includes: age, sex, species. The contribution of additional variables that can enhance data usage is encouraged, though not required.
When additional measurements of brain function and behavior are available (e.g., electrophysiology, eye tracking), we will share this data along with the imaging. For any data types that are not yet included in the BIDS format, we will include the relevant metadata in accompanying .csv files; a readme.txt file will facilitate any additional instructions for integration of information. In the long-run, we expect that such specifications will evolve in the BIDS format and we will adopt them accordingly.
Following the model of prior efforts, all contributions are reviewed by the INDI team following upload and corrected as needed to ensure consistent data organization within and across sites. Before open release, each contributing site reviews their reorganized phenotypic records, five random images per imaging modality and their collection-specific narrative for final approval.
Alignment to a Common Space
For the purposes of illustration, we depict sample anatomical and functional images (when available) for each contribution to PRIME-DE. Here, we provide a summary of the steps employed for alignment to the common space defined by the NMT template (Seidlitz et al., 2018b), which was essential for creation of Figures 3 and 4 (extracted brains and scripts required for generation of figure are available at: https://github.com/TingsterX/PRIME-DE).
The intensity correction was first applied to T1 images using ANTs ‘N4BiasFieldCorrection’. Then the T1 images were skull stripped using the AFNI 3dSkullstrip with ‘-monkey’ option and ANTs tools by registering the individual head image to NMT head template and then inverse transformed the NMT brain mask into the individual space. The better brain masks were selected and manually corrected if needed. The skull stripped T1 images were then registered to NMT template for the final demonstration.
The functional image was initially skull stripped using the union of the results of ‘bet2′ and ‘3dAutomask’. The T1 brain mask created from the structural processing above was then transformed back to the functional space for further refinement of the functional brain mask for a given subject; this was accomplished using the inverse transform calculated from the transformation from the space of the EPI to that of the high resolution anatomical image (i.e., rigid body transformation). Finally, the functional image was extracted again using the refined brain mask and registered to the T1 image. For the final demonstration, we combined the transformation from functional to anatomical image and the warp from anatomical to template to align functional image to the NMT template.
Data and Software Availability
Data Preparation and Aggregation
PRIME-DE data aggregation is carried out through the International Neuroimaging Data-sharing Initiative (INDI; Mennes et al., 2013); the portal is located at the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC; http://fcon_1000.projects.nitrc.org/indi/indiPRIME.html).
NMT Alignment
Extracted brains and scripts required for generation of Figures 3 and 4 are available at: https://github.com/TingsterX/PRIME-DE.
Acknowledgments
We thank Cameron Craddock for his creation and open sharing of the Preprocessed Connectome Project (PCP) Quality Assessment Project (QAP), which was used here (http://preprocessed-connectomes-project.org/quality-assessment-protocol/index.html).
Primary support for the work by M.P.M. and the INDI team was provided by gifts from Joseph P. Healy to the Child Mind Institute as well as by the BRAIN Initiative (R01MH111439). M.P.M. is a Randolph Cowen and Phyllis Green Scholar.
Primary support for the work by C.S. is provided by the BRAIN Initiative (R01MH111439) and the Sylvio O. Conte Center “Neurobiology and Dynamics of Active Sensing” (P50MH109429).
Primary support for the work by D.S.M. is provided by the Max Planck Society.
Primary support for the work by T.B. and J.S. is provided by Project PRIMAVOICE, Pascal Belin, and French Agence Nationale de la Recherche.
Primary support for the work by R.R. and D.T. is provided by NIH grant R01 EY019702.
Primary support for the work by A.C. is provided by the National Natural Science Foundation of China (No. 31371029 and No. 31571121) and the Innovation Program of Shanghai Municipal Education Commission (No. 14ZZ051, No. 20130076120021, No. 15JC1400104, and No. 16JC1400100).
Primary support for the work by S.C.K. and Y.-d.Z. is provided by the following grants: Ministry of Education of PRC Humanities and Social Sciences Research Grant 16YJC190006; STCSM Shanghai Pujiang Program 16PJ1402800l; STCSM Natural Science Foundation of Shanghai 16ZR1410200; and National Key Fundamental Research (973) Program of China Grant 2013CB329501.
Primary support for the work by Z.W. is provided by the National Natural Science Foundation of China (81571300, 31771174).
Primary support for the work by F.H.-B., M.M., and C.G. is provided by the French National Research Agency (ANR-14-CE13-0005-1 and ANR-15-CE37-0003) and the NEURODIS Foundation.
Primary support for the work by the Icahn School of Medicine at Mount Sinai was provided by a Charles H. Revson Senior Fellowship in the Biomedical Sciences to P.L.C.
Primary support for the work by M.G.B. and J.H.M. provided by NIA P01AG016765.
Primary support for the work by K.M., D.A.R., and A.S. from McGill University is provided by a grant from the Brain Canada Foundation.
Primary support for the work by the NIMH is provided by the Intramural Research Program of the NIMH (ZICMH00289).
Primary support for the work by P.R.R. and P.C.K. is provided by grants from the Netherlands Organisation for Scientific Research (NWO) and the European Union.
Primary support for the work by B. Jarraya and S.D. are provided by CEA, University Paris-Saclay, INSERM, Fondation de France, and ERC.
Primary support for the work by Newcastle University provided by Wellcome Trust, Medical Research Council, European Research Council, NC3Rs, and BBSRC.
Primary support for work by E.L.S. and D.A.F. is provided by R01MH107508.
Primary support for the work by S.K. and M.P. is provided by NIMH 1P50MH109429, NIMH R01MH064043, and NEI R01EY017699.
Primary support for the work by C.M.S. and W.F. is supported by a Human Frontier Science Program Long-Term Fellowship; the NIH; an Irma T. Hirschl/Monique Weill-Caulier Trusts Award; a Pew Scholar Award in the Biomedical Sciences; a McKnight Scholars Award; a Human Frontier Science Program Research Grant; the New York Stem Cell Foundation; the National Eye Institute; the NIMH; the NSF Science and Technology Center for Brains, Minds, and Machines; and the National Science Foundation.
Primary support for the work by E.P., C.R.E.W., and C.E. is provided by Agence National de la Recherche, Fondation Neurodis, the LabEx CORTEX ANR-11-LABX-0042, Fondation de France, Human Frontier Science Program, and Fondation pour la Recherche Médicale.
Primary support for the work by E.Y. and N.H. is provided by the following grants: R01-NS081118, R01-NS085188, P41-EB015894, P30-NS076408, and the University of Minnesota Udall Center P50NS098573.
Primary support for the work by the University of Oxford is provided by the Medical Research Council UK, the Biotechnology and Biological Sciences Research Council UK, the Royal Society, and the Wellcome Trust.
Primary support for the work by E.L.A.B., O.S.T., P.L., K.N.L., and S.M.R. was provided by the John Templeton Foundation, the Canada Foundation for Innovation, and the Anna-Greta och Holger Crafoords Stiftelse.
Primary support for the work by S.E. and R.S.M. is provided by the Canadian Institutes of Health Research and Brain Canada. Primary support for the work by C.M.S. and W.F. is supported by a Human Frontier Science Program Long-Term Fellowship (LT001118/2012).
Author Contributions
Conception and Experimental Design, D.S.M., M.P.M., and C.E.S.; Implementation and Logistics, L.A., D.S.M., M.P.M., C.E.S., B.K., and T.X.; Data Collection, M.P.M., C.A., F.B., M.G.B., S.B.H., T.B., A.C., P.L.C., C.G.D., S.E., D.A.F., L.F., W.F., S.F.-W., T.D.G., C.G., F.H.-B., N.H., B.H., B. Jarraya, B. Jung, P.C.K., S.D., S.K., S.C.K., D.A.L., R.B.M., R.S.M., A.M., M.M., K.M., J.H.M., J. Nacef, J. Nagy, M.O.R., C.I.P., M.P., C.P., E.P., R.R., P.R.R., D.A.R., M.F.S.R., B.E.R., S.M.R., E.L.A.B., O.S.T., P.L, K.N.L., J. Sallet, M.C.S., J. Seidlitz, J. Sein, C.M.S., A.S., E.L.S., L.U., A.T., D.T., Z.W., C.R.E.W., E.Y., F.Q.Y., W.Z., Y.-d.Z., D.S.M., and C.E.S.; Data Informatics, B.K. and L.A.; Data Analysis, L.A. and M.P.M.; Drafting of the Manuscript, M.P.M.; Critical Review and Editing of the Manuscript, all authors.
Declaration of Interests
The authors declare no competing interests.
Published: September 27, 2018
Footnotes
Supplemental Information includes four tables and can be found with this article online at https://doi.org/10.1016/j.neuron.2018.08.039.
Supplemental Information
FA, flip angle; TI, inversion time; TE, echo time; ES, echo spacing; BW, bandwidth per pixel; TR, repetition time; PA, parallel acquisition; PF, partial Fourier (half scan); SO, slice orientation; PE, phase encoding direction; RO, read out direction; SL, slice direction. Reconstructed resolution (RR; mm) and image dimensions (RID; px) refer to the images after they have been reconstructed from the k-space data, the matrix size, and resolution used for the acquisition may differ. For these categories, RO, read out direction; PE, phase encoding direction, and SL, slice direction. TA, acquisition time.
TE, echo time; TR, repetition time; PE, phase encoding; RO, read out direction; Reconstructed resolution (RR; mm) and image dimensions refer to the images after they have been reconstructed from the k-space data, the matrix size, and resolution used for the acquisition may differ. For these categories, RO, read out direction; PE, phase encoding direction, and SL, slice direction.
FA, flip angle; TE, echo time; TR, repetition time; BW, bandwidth per pixel; ES, echo spacing; PA, parallel acquisition; PF, partial Fourier (half scan); PE, phase encoding direction; FS, fat suppression; SO, slice orientation; SA, slice acquisition order; Gap, gap between slices; RO, read out direction; Nacq, number of volumes collected; Ndisc, number of initial volumes discarded by the scanner; TA, acquisition time. Reconstructed resolution (RR; mm) and image matrix (RIM; px) refer to the images after they have been reconstructed from the k-space data, the matrix size, and resolution used for the acquisition may differ. For these categories, RO, read out direction; PE, phase encoding direction, and SL, slice direction.
FA, flip angle; TE, echo time; TR, repetition time
References
- Alexander L.M., Escalera J., Ai L., Andreotti C., Febre K., Mangone A., Vega-Potler N., Langer N., Alexander A., Kovacs M. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci. Data. 2017;4:170181. doi: 10.1038/sdata.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbastre Y., Rivière D., Souedet N., Fischer C., Hérard A.-S., Williams S., Vandenberghe M.E., Flament J., Aron-Badin R., Hantraye P. Primatologist: a modular segmentation pipeline for macaque brain morphometry. Neuroimage. 2017;162:306–321. doi: 10.1016/j.neuroimage.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Newsome W.T. The Brain Research through Advancing Innovative Neurotechnologies (BRAIN) initiative and neurology. JAMA Neurol. 2014;71:675–676. doi: 10.1001/jamaneurol.2014.411. [DOI] [PubMed] [Google Scholar]
- Barttfeld P., Uhrig L., Sitt J.D., Sigman M., Jarraya B., Dehaene S. Signature of consciousness in the dynamics of resting-state brain activity. Proc. Natl. Acad. Sci. USA. 2015;112:887–892. doi: 10.1073/pnas.1418031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E., Badea A., Coe C.L., Lubach G.R., Shi Y., Styner M.A., Johnson G.A. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage. 2015;117:408–416. doi: 10.1016/j.neuroimage.2015.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Di Martino A., O’Connor D., Chen B., Alaerts K., Anderson J.S., Assaf M., Balsters J.H., Baxter L., Beggiato A., Bernaerts S. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci. Data. 2017 doi: 10.1038/sdata.2017.10. Published online March 14, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue C.J., Sotiropoulos S.N., Jbabdi S., Hernandez-Fernandez M., Behrens T.E., Dyrby T.B., Coalson T., Kennedy H., Knoblauch K., Van Essen D.C., Glasser M.F. Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J. Neurosci. 2016;36:6758–6770. doi: 10.1523/JNEUROSCI.0493-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.S., Oler J.A., Shackman A.J., Shelton S.E., Raveendran M., McKay D.R., Converse A.K., Alexander A., Davidson R.J., Blangero J. Intergenerational neural mediators of early-life anxious temperament. Proc. Natl. Acad. Sci. USA. 2015;112:9118–9122. doi: 10.1073/pnas.1508593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Pandya D.N., Chakravarty M.M., Bailey L., Petrides M., Collins D.L. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space) Neuroimage. 2011;55:1435–1442. doi: 10.1016/j.neuroimage.2011.01.040. [DOI] [PubMed] [Google Scholar]
- Friedman L., Glover G.H., Krenz D., Magnotta V., FIRST BIRN Reducing inter-scanner variability of activation in a multicenter fMRI study: role of smoothness equalization. Neuroimage. 2006;32:1656–1668. doi: 10.1016/j.neuroimage.2006.03.062. [DOI] [PubMed] [Google Scholar]
- Ghahremani M., Hutchison R.M., Menon R.S., Everling S. Frontoparietal functional connectivity in the common marmoset. Cereb. Cortex. 2017;27:3890–3905. doi: 10.1093/cercor/bhw198. [DOI] [PubMed] [Google Scholar]
- Giannelli M., Diciotti S., Tessa C., Mascalchi M. Characterization of Nyquist ghost in EPI-fMRI acquisition sequences implemented on two clinical 1.5 T MR scanner systems: effect of readout bandwidth and echo spacing. J. Appl. Clin. Med. Phys. 2010;11:3237. doi: 10.1120/jacmp.v11i4.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K.J., Varoquaux G., Rivera G., Schwarz Y., Ghosh S.S., Maumet C., Sochat V.V., Nichols T.E., Poldrack R.A., Poline J.-B. NeuroVault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front. Neuroinform. 2015;9:8. doi: 10.3389/fninf.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K.J., Alfaro-Almagro F., Auer T., Bellec P., Capotă M., Chakravarty M.M., Churchill N.W., Cohen A.L., Craddock R.C., Devenyi G.A. BIDS apps: improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput. Biol. 2017;13:e1005209. doi: 10.1371/journal.pcbi.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A., Stiers P., Hutchison R.M., Everling S., Petrides M., Margulies D.S. Intrinsic functional architecture of the macaque dorsal and ventral lateral frontal cortex. J. Neurophysiol. 2017;117:1084–1099. doi: 10.1152/jn.00486.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D.S., Bliss-Moreau E., Machado C.J., Bennett J., Shen K., Grant K.A., Fair D.A., Amaral D.G. The rhesus monkey connectome predicts disrupted functional networks resulting from pharmacogenetic inactivation of the amygdala. Neuron. 2016;91:453–466. doi: 10.1016/j.neuron.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison R.M., Everling S. Monkey in the middle: why non-human primates are needed to bridge the gap in resting-state investigations. Front. Neuroanat. 2012 doi: 10.3389/fnana.2012.00029. Published online July 26, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison R.M., Everling S. Broad intrinsic functional connectivity boundaries of the macaque prefrontal cortex. Neuroimage. 2014;88:202–211. doi: 10.1016/j.neuroimage.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Hutchison R.M., Leung L.S., Mirsattari S.M., Gati J.S., Menon R.S., Everling S. Resting-state networks in the macaque at 7 T. Neuroimage. 2011;56:1546–1555. doi: 10.1016/j.neuroimage.2011.02.063. [DOI] [PubMed] [Google Scholar]
- Hutchison R.M., Gallivan J.P., Culham J.C., Gati J.S., Menon R.S., Everling S. Functional connectivity of the frontal eye fields in humans and macaque monkeys investigated with resting-state fMRI. J. Neurophysiol. 2012;107:2463–2474. doi: 10.1152/jn.00891.2011. [DOI] [PubMed] [Google Scholar]
- Hutchison R.M., Culham J.C., Flanagan J.R., Everling S., Gallivan J.P. Functional subdivisions of medial parieto-occipital cortex in humans and nonhuman primates using resting-state fMRI. Neuroimage. 2015;116:10–29. doi: 10.1016/j.neuroimage.2015.04.068. [DOI] [PubMed] [Google Scholar]
- Iglesias J.E., Liu C.-Y., Thompson P.M., Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging. 2011;30:1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Keliris G.A., Shmuel A., Ku S.-P., Pfeuffer J., Oeltermann A., Steudel T., Logothetis N.K. Robust controlled functional MRI in alert monkeys at high magnetic field: effects of jaw and body movements. Neuroimage. 2007;36:550–570. doi: 10.1016/j.neuroimage.2007.02.057. [DOI] [PubMed] [Google Scholar]
- Love S.A., Marie D., Roth M., Lacoste R., Nazarian B., Bertello A., Coulon O., Anton J.-L., Meguerditchian A. The average baboon brain: MRI templates and tissue probability maps from 89 individuals. Neuroimage. 2016;132:526–533. doi: 10.1016/j.neuroimage.2016.03.018. [DOI] [PubMed] [Google Scholar]
- Magnotta V.A., Friedman L., FIRST BIRN Measurement of signal-to-noise and contrast-to-noise in the fBIRN multicenter imaging study. J. Digit. Imaging. 2006;19:140–147. doi: 10.1007/s10278-006-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov N.T., Ercsey-Ravasz M.M., Ribeiro Gomes A.R., Lamy C., Magrou L., Vezoli J., Misery P., Falchier A., Quilodran R., Gariel M.A. A weighted and directed interareal connectivity matrix for macaque cerebral cortex. Cereb. Cortex. 2014;24:17–36. doi: 10.1093/cercor/bhs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars R.B., Jbabdi S., Sallet J., O’Reilly J.X., Croxson P.L., Olivier E., Noonan M.P., Bergmann C., Mitchell A.S., Baxter M.G. Diffusion-weighted imaging tractography-based parcellation of the human parietal cortex and comparison with human and macaque resting-state functional connectivity. J. Neurosci. 2011;31:4087–4100. doi: 10.1523/JNEUROSCI.5102-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren D.G., Kosmatka K.J., Oakes T.R., Kroenke C.D., Kohama S.G., Matochik J.A., Ingram D.K., Johnson S.C. A population-average MRI-based atlas collection of the rhesus macaque. Neuroimage. 2009;45:52–59. doi: 10.1016/j.neuroimage.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennes M., Biswal B.B., Castellanos F.X., Milham M.P. Making data sharing work: the FCP/INDI experience. Neuroimage. 2013;82:683–691. doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O., Mills B.D., Grayson D., Woodall A., Grant K.A., Kroenke C.D., Fair D.A. Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology. J. Neurosci. 2014;34:5552–5563. doi: 10.1523/JNEUROSCI.4229-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortamet B., Bernstein M.A., Jack C.R., Jr., Gunter J.L., Ward C., Britson P.J., Meuli R., Thiran J.-P., Krueger G., Alzheimer’s Disease Neuroimaging Initiative Automatic quality assessment in structural brain magnetic resonance imaging. Magn. Reson. Med. 2009;62:365–372. doi: 10.1002/mrm.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete A.F., Blezer E.L.A., Pagnotta M., de Viet E.S.M., Todorov O.S., Lindenfors P., Laland K.N., Reader S.M. Primate brain anatomy: New volumetric MRI measurements for neuroanatomical studies. Brain, Behavior and Evolution. 2018;91:1–9. doi: 10.1159/000488136. [DOI] [PubMed] [Google Scholar]
- Nichols T.E. Neuroimaging Statistics Tips & Tools; 2012. Standardizing DVARS, 28/10/12. [Google Scholar]
- Nooner K.B., Colcombe S.J., Tobe R.H., Mennes M., Benedict M.M., Moreno A.L., Panek L.J., Brown S., Zavitz S.T., Li Q. The NKI-Rockland sample: a model for accelerating the pace of discovery science in psychiatry. Front. Neurosci. 2012;6:152. doi: 10.3389/fnins.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G. Academic Press; 2009. The Rhesus Monkey Brain in Stereotaxic Coordinates. [Google Scholar]
- Paxinos G., Huang X.-F., Toga A.W. Academic Press; 1999. The Rhesus Monkey Brain in Stereotaxic Coordinates. [Google Scholar]
- Pfeuffer J., Shmuel A., Keliris G.A., Steudel T., Merkle H., Logothetis N.K. Functional MR imaging in the awake monkey: effects of motion on dynamic off-resonance and processing strategies. Magn. Reson. Imaging. 2007;25:869–882. doi: 10.1016/j.mri.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Power J.D. A simple but useful way to assess fMRI scan qualities. Neuroimage. 2017;154:150–158. doi: 10.1016/j.neuroimage.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveley C., Gruslys A., Ye F.Q., Glen D., Samaha J., E Russ B., Saad Z., K Seth A., Leopold D.A., Saleem K.S. Three-dimensional digital template atlas of the macaque brain. Cereb. Cortex. 2017;27:4463–4477. doi: 10.1093/cercor/bhw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling J.K. Comparative primate neuroimaging: insights into human brain evolution. Trends Cogn. Sci. 2014;18:46–55. doi: 10.1016/j.tics.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Rohlfing T., Kroenke C.D., Sullivan E.V., Dubach M.F., Bowden D.M., Grant K.A., Pfefferbaum A. The INIA19 template and NeuroMaps atlas for primate brain image parcellation and spatial normalization. Front. Neuroinform. 2012;6:27. doi: 10.3389/fninf.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem K.S., Logothetis N.K. Academic Press; 2012. A Combined MRI and Histology Atlas of the Rhesus Monkey Brain in Stereotaxic Coordinates. [Google Scholar]
- Schwiedrzik C.M., Zarco W., Everling S., Freiwald W.A. Face patch resting state networks link face processing to social cognition. PLoS Biol. 2015;13:e1002245. doi: 10.1371/journal.pbio.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F., Dale A.M., Busa E., Glessner M., Salat D., Hahn H.K., Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Seidlitz J., Váša F., Shinn M., Romero-Garcia R., Whitaker K.J., Vértes P.E., Wagstyl K., Kirkpatrick Reardon P., Clasen L., Liu S., NSPN Consortium Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron. 2018;97:231–247.e7. doi: 10.1016/j.neuron.2017.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidlitz J., Sponheim C., Glen D., Ye F.Q., Saleem K.S., Leopold D.A., Ungerleider L., Messinger A. A population MRI brain template and analysis tools for the macaque. Neuroimage. 2018;170:121–131. doi: 10.1016/j.neuroimage.2017.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck D.W., Leahy R.M. BrainSuite: an automated cortical surface identification tool. Med. Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Shehzad Z., Giavasis S., Li Q., Benhajali Y., Yan C., Yang Z., Milham M., Bellec P., Craddock C. The Preprocessed Connectomes Project Quality Assessment Protocol - a resource for measuring the quality of MRI data. Front. Neurosci. 2015 Published online August 5, 2015. [Google Scholar]
- Shmuel A., Leopold D.A. Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: Implications for functional connectivity at rest. Hum. Brain Mapp. 2008;29:751–761. doi: 10.1002/hbm.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E.J., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C. Windows on the brain: the emerging role of atlases and databases in neuroscience. Curr. Opin. Neurobiol. 2002;12:574–579. doi: 10.1016/s0959-4388(02)00361-6. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C. Cortical cartography and Caret software. Neuroimage. 2012;62:757–764. doi: 10.1016/j.neuroimage.2011.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D.C., Glasser M.F. In vivo architectonics: a cortico-centric perspective. Neuroimage. 2014;93:157–164. doi: 10.1016/j.neuroimage.2013.04.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D.C., Glasser M.F., Dierker D.L., Harwell J. Cortical parcellations of the macaque monkey analyzed on surface-based atlases. Cereb. Cortex. 2012;22:2227–2240. doi: 10.1093/cercor/bhr290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D.C., Smith S.M., Barch D.M., Behrens T.E.J., Yacoub E., Ugurbil K., WU-Minn HCP Consortium The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwal T., Kelly C., Eilbott J., Mayes L.C., Castellanos F.X. Inscapes: a movie paradigm to improve compliance in functional magnetic resonance imaging. Neuroimage. 2015;122:222–232. doi: 10.1016/j.neuroimage.2015.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanduffel W., Zhu Q., Orban G.A. Monkey cortex through fMRI glasses. Neuron. 2014;83:533–550. doi: 10.1016/j.neuron.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J.L., Patel G.H., Fox M.D., Snyder A.Z., Baker J.T., Van Essen D.C., Zempel J.M., Snyder L.H., Corbetta M., Raichle M.E. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Xu T., Falchier A., Sullivan E.L., Linn G., Ramirez J.S.B., Ross D., Feczko E., Opitz A., Bagley J., Sturgeon D. Delineating the macroscale areal organization of the macaque cortex in vivo. Cell Rep. 2018;23:429–441. doi: 10.1016/j.celrep.2018.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.T., Shi Y., Niethammer M., Grauer M., Coe C.L., Lubach G.R., Davis B., Budin F., Knickmeyer R.C., Alexander A.L., Styner M.A. The UNC-Wisconsin Rhesus Macaque Neurodevelopment Database: a structural MRI and DTI database of early postnatal development. Front. Neurosci. 2017;11:29. doi: 10.3389/fnins.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Guo L., Zhu D., Li K., Li L., Chen H., Zhao Q., Hu X., Liu T. Diffusion tensor imaging reveals evolution of primate brain architectures. Brain Struct. Funct. 2013;218:1429–1450. doi: 10.1007/s00429-012-0468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FA, flip angle; TI, inversion time; TE, echo time; ES, echo spacing; BW, bandwidth per pixel; TR, repetition time; PA, parallel acquisition; PF, partial Fourier (half scan); SO, slice orientation; PE, phase encoding direction; RO, read out direction; SL, slice direction. Reconstructed resolution (RR; mm) and image dimensions (RID; px) refer to the images after they have been reconstructed from the k-space data, the matrix size, and resolution used for the acquisition may differ. For these categories, RO, read out direction; PE, phase encoding direction, and SL, slice direction. TA, acquisition time.
TE, echo time; TR, repetition time; PE, phase encoding; RO, read out direction; Reconstructed resolution (RR; mm) and image dimensions refer to the images after they have been reconstructed from the k-space data, the matrix size, and resolution used for the acquisition may differ. For these categories, RO, read out direction; PE, phase encoding direction, and SL, slice direction.
FA, flip angle; TE, echo time; TR, repetition time; BW, bandwidth per pixel; ES, echo spacing; PA, parallel acquisition; PF, partial Fourier (half scan); PE, phase encoding direction; FS, fat suppression; SO, slice orientation; SA, slice acquisition order; Gap, gap between slices; RO, read out direction; Nacq, number of volumes collected; Ndisc, number of initial volumes discarded by the scanner; TA, acquisition time. Reconstructed resolution (RR; mm) and image matrix (RIM; px) refer to the images after they have been reconstructed from the k-space data, the matrix size, and resolution used for the acquisition may differ. For these categories, RO, read out direction; PE, phase encoding direction, and SL, slice direction.
FA, flip angle; TE, echo time; TR, repetition time