Abstract
Representing an enormous health care and socioeconomic challenge, breast cancer is the second most common cancer in the world and the second most common cause of cancer-related death. Although many of the challenges associated with preventing, treating, and ultimately curing breast cancer are addressable in the laboratory, successful translation of groundbreaking research to clinical populations remains an important barrier. Particularly when compared with research on other types of solid tumors, breast cancer research is hampered by a lack of tractable in vivo model systems that accurately recapitulate the relevant clinical features of the disease. A primary objective of this article was to provide a generalizable overview of the types of in vivo model systems, with an emphasis primarily on murine models, that are widely deployed in preclinical breast cancer research. Major opportunities to advance precision cancer medicine facilitated by molecular imaging of preclinical breast cancer models are discussed.
Keywords: animal imaging, molecular imaging, breast cancer, coclinical trials, mouse models, precision medicine
Breast cancer is the second most common cancer in the world, with an estimated 1.67 million new cases diagnosed in 2012, and the second most common cause of cancer-related death (1). In the United States alone, the American Cancer Society estimates diagnoses of more than 231,000 new cases of invasive breast cancer among women and approximately 2,350 new cases among men in 2015 (2). Uniquely, the term “breast cancer” does not reflect a single disease; rather, breast cancer should be thought of as a repertoire of related diseases classifiable into distinct subtypes, each portending distinct prognoses and potentially actionable phenotypic, molecular, or genetic characteristics. Although targeting certain molecular vulnerabilities inherent in specific breast cancer subtypes has improved clinical outcomes in a limited number of patients, a sobering reality is that more than 40,000 individuals in the United States will die from this disease in 2015 (2); this information underscores the numerous challenges that still remain in the clinical care of individuals with this disease.
Although many of the challenges associated with preventing, treating, and ultimately curing breast cancer are addressable in the laboratory, successful translation of groundbreaking laboratory research to clinical populations remains an important barrier. Particularly when compared with research on other types of solid tumors, breast cancer research is hampered by a lack of tractable in vivo model systems that accurately recapitulate the relevant clinical features of the disease. Although certain models necessarily will be highlighted as a consequence of illuminating examples and opportunities, more exhaustive catalogs of previously described models are reviewed in several suggested references (3–7). Here, a generalizable overview of the types of in vivo model systems, with an emphasis primarily on murine models that are widely deployed in preclinical breast cancer research, is provided; this overview encompasses the specific relationship of the models with the clinical disease and how imaging within the context of the models might be exploited to maximize translational gains to combat breast cancer.
A distinguishing feature of this article is that the key attributes of various preclinical breast cancer models and their utility are developed from the perspective of noninvasive molecular imaging. Despite major successes and lessons learned from the genomic landscape of cancer, it is now widely recognized that individual cancer genomes, like individual patients, are exquisitely heterogeneous; each contains a unique spectrum of drivers accompanied by passengers of less obvious significance. Tools that illuminate the cellular and molecular underpinnings of tumors on a patient by-patient basis, such as noninvasive molecular imaging, will be essential to bringing precision cancer therapy to fruition. As such, preclinical imaging techniques relevant to mouse models of breast cancer, with an emphasis on molecular imaging, are also discussed in some detail.
MICE AS MODELS OF HUMAN CANCER
Although it might seem obvious, it is worth noting at the outset that all “models” of human disease are imperfect. Regardless of the degree of sophistication, model systems are, by definition, not humans. Rationales for late-phase clinical failures of new drugs are frequently based on a (healthy) skepticism of the translational value of certain preclinical models; much has been written about this issue already, and the value of model systems as a translational bridge to clinical applications is not debated in this article. However, in vivo modeling provides gains to the breast cancer field that complement what can be discovered at the laboratory bench. Indeed, the strongest experimental approaches will test hypotheses in multiple model systems. Therefore, it is critical to understand both the strengths and the limitations of in vivo models of breast cancer to maximize what can be learned with this approach.
The laboratory mouse (Mus musculus) represents a truly ideal model system for simulating the entire spectrum of events that lead to advanced breast cancer in humans. Mouse model systems enable elucidation of distinct facets of cancer biology that may not be frankly addressable in patients. Some of the advantages of the mouse as a model system are as follows: it is a mammal of small size, facilitating inexpensive housing and convenient handling; rapid breeding can facilitate colony expansion on convenient time scales; it has a relatively long life-span (~3 y); the complete sequence and characterization of the mouse genome are available; and manipulation of the mouse genome can be accomplished with relative ease. Additionally, mice and other rodents share many physiologic similarities with humans (8) and therefore are commonly used in drug metabolism and pharmacokinetic and toxicity studies. Ironically, for imaging studies, the small size of the mouse can be a limitation, particularly when studies aim to image tumors whose diameters approximate or are smaller than the effective resolution of the imaging modality of choice. Some notable differences between humans and mice include a higher metabolic rate in mice, an altered telomere length in inbred mouse strains, and an altered time frame for cancer onset (9).
HUMAN BREAST CANCER SUBTYPES: WHAT MODELS AIM TO RECAPITULATE
Several clinical and pathologic features of human breast cancer that allow stratification of patients on the basis of risk, prognosis, and likelihood of a response to certain types of therapy have been identified (10); in this light, for clinical breast cancer there are several impressive precision medicine–related success stories (11) and opportunities for future drug development (Table 1). Distinct molecular subtypes can be initially stratified on the basis of hormone receptor status; luminal breast cancers are typically hormone receptor–positive, whereas human epidermal growth factor receptor 2 (HER2) and basallike breast cancers (BLBCs) are hormone receptor– negative. Other potential molecular subtypes, including luminal C and normallike tumors, have been reported; at present, however, little is known about these subtypes (10).
Table 1.
Major Subtypes of Human Breast Cancer
| Molecular subtype | Gene expression features | Clinical features | Treatment and prognosis |
|---|---|---|---|
| Luminal | Elevated expression of hormone receptors and associated genes (luminal A > luminal B) | ~65% of invasive breast cancers are ER- or PR-positive | Respond to endocrine therapy (responses to tamoxifen and aromatase inhibitors may differ in luminal A and B cancers) |
| Luminal B cancers tend to be of higher histologic grade than luminal A cancers | Variable response to chemotherapy (greater in luminal B cancers than in luminal A cancers) | ||
| Some overexpress HER2 (luminal B) | Prognosis is better for luminal A cancers than for luminal B cancers | ||
| HER2 | Elevated expression of HER2 and other genes in amplicon | ~15% of invasive breast cancers are ER- or PR-negative | Respond to trastuzumab (Herceptin) |
| Respond to anthracycline-based chemotherapy | |||
| Low expression of ER, PR, and associated genes | High probability of being highgrade and node-positive | Prognosis is typically poor | |
| Basallike | Elevated expression of basal epithelial genes and basal cytokeratins | ~15% of invasive breast cancers | No response to endocrine therapy or trastuzumab (Herceptin) |
| Low expression of ER, PR, and associated genes | Most are ER-, PR-, and HER2- negative (TNBC) | Appear to be sensitive to platinum-based chemotherapy and polyadenosine ribose polymerase inhibitors | |
| Low expression of HER2 | BRCA1 dysfunction (germ line, sporadic) | Prognosis is typically poor (but not uniformly poor) | |
| Particularly common in African American women |
Adapted with permission of (10).
Luminal A and Luminal B Subtypes
Luminal breast cancers are characterized by the expression of the estrogen receptor (ER) and the progesterone receptor (PR), which are nuclear hormone receptors, and other associated genes (12). Taken together, luminal A and luminal B subtypes account for approximately 65% of all breast cancers, although there are some differences between these subtypes. Luminal A breast cancers tend to express greater quantities of hormone receptors, particularly the PR, than luminal B breast cancers. In contrast, luminal B tumors tend to exhibit characteristics associated with higher-grade disease, are frequently more proliferative, are clinically more aggressive, and have a poorer prognosis than luminal A tumors. Because of their hormone receptor expression and activity, luminal A and luminal B breast cancers are routinely treated with endocrine therapies that block ER activity, including selective ER modulators (such as tamoxifen), selective ER downregulators (such as fulvestrant), and aromatase inhibitors (such as letrozole) that block the systemic production of the native ligand (β-estradiol). Luminal A and luminal B tumors exhibit disparate responses to chemotherapy, with higher-grade luminal B tumors frequently responding more favorably to chemotherapy (10). Given the hormone receptor expression and activity of luminal A and luminal B breast cancers, PET imaging with an 18F-labeled form of estradiol (16α−18F-fluoro-17β-estradiol [18F-FES]) is often useful and may represent a suitable companion diagnostic approach for predicting a response to anti-ER therapy in selected patients (13,14).
HER2-Enriched Subtype
The HER2 gene is amplified in approximately 15% of invasive breast cancers. Some breast cancers of this subtype have been shown to express ER, but most HER2-enriched tumors lack ER or PR expression. HER2-enriched tumors are frequently highergrade tumors, with positive lymph node involvement. Precision medicine approaches to this cancer include the use of trastuzumab (Tz) (Herceptin; Genentech), a monoclonal antibody that selectively targets the HER2 gene product, a receptor tyrosine kinase, as well as small-molecule kinase inhibitors (lapatinib and everolimus) (15,16). HER2-enriched breast cancers with metastatic disease are additionally treated with anthracyclines (doxorubicin) and often display an initial response to treatment, although recurrence is seen in nearly all cases. Other strategies targeting the HER2 receptor and its pathway include novel small-molecule inhibitors and HER2 antibodies, heat shock protein 90 inhibitors, agents targeting downstream components of the HER2 signaling pathway, and antibody–drug conjugates. Certain molecular imaging strategies targeting HER2-enriched tumors have leveraged the selectivity of Tz labeled with a positron-emitting isotope (64Cu or 89Zr). Promising clinical results in patients with metastatic breast cancer have been shown for these strategies (17,18).
BLBCs
BLBCs abundantly express epithelial genes, such as those for cytokeratins 5 and 17, but the expression of ER, PR, and HER2 is notoriously absent. On the basis of their lack of ER, PR, and HER2 expression, many BLBCs are deemed “triple-negative breast cancer” (TNBC). BLBCs are especially common in African American women (10) and are generally associated with a poor prognosis. Given the typical lack of ER, PR, and HER2 expression in BLBCs, molecularly targeted agents used to treat other breast cancer subtypes are often highly ineffective for BLBCs; therefore, chemotherapy is a mainstay for treating BLBCs (19). However, recent efforts to develop increasingly effective therapies against TNBC have led to the identification of several novel TNBC subtypes distinguishable by gene expression profiles and with potential vulnerabilities (20). Provocatively, noninvasive imaging of the androgen receptor by PET with 16b-18F-fluoro-5α-dihydrotestosterone (18F-FDHT), a structural analog of 5α-dihydrotestosterone, may represent a companion diagnostic approach for this challenging subtype. At present, a study is exploring the feasibility of using 18F-FDHT PET to assess androgen receptor expression in metastatic breast cancer (ClinicalTrials.gov NCT01988324); this study is examining whether the effects of antiandrogens on tumor 18F-FDHT uptake could aid in identifying optimum dosing for blocking the androgen receptor in metastatic breast cancer.
ATTRIBUTES OF PRECLINICAL MOUSE MODELS OF CANCER
Rapidly increasing knowledge about breast cancer molecular subtypes may affect the genesis of, progression of, and therapeutic strategy for any given breast cancer and underscores the importance of mouse model selection in designing preclinical studies and coclinical trials. Astounding growth in the reported number as well as the biologic elegance of mouse models for cancer research has been witnessed in the last decade. An extensive repertoire of mouse models with which to study breast cancer progression and treatment is now available. In genetically engineered mouse models (GEMMs), the tumor develops through all stages of epithelial transformation with the native stroma, immune system, and microenvironment (21). This trend has been propelled in part by the sheer volume of laboratories developing and deploying innovative mouse models to advance basic cancer research as well as by the adoption of contemporary and comparatively inexpensive genome editing technologies, such as the clustered regularly interspersed short palindromic repeats (CRISPR)/CRISPRassociated protein 9 (Cas9) system (22) and RNA interference approaches (23). Another important contribution to the volume of mouse models recently described has come from the assembly of patient-derived xenograft (PDX) banks and, particularly for some cancer types, standardization of the infrastructure and protocols required to support these systems (24). Here we describe 4 types of mouse model systems that can be used for breast cancer research, identifying both the strengths and the limitations of each (Table 2).
Table 2.
Preclinical Murine Models of Human Cancer
| Model | Main components | Advantages | Limitations | Time and Cost* |
|---|---|---|---|---|
| Xenograft (cell line) | Immortalized human tumor cell lines transplanted into immunodeficient host (mouse) | Numerous established and well-annotated cell lines | Immunodeficient host | 2–4 wk, $ |
| Representation from various human tumor types | Subcutaneous location may not allow cultivation of key tissue-specific stromal infiltrate | |||
| Features of tumor microenvironment, including stromal and vascular cells, incorporated within tumor | Cross-species divide; stromal components are mouse, whereas tumor cells are human | |||
| Tumors are easily and precisely measured | Limited or no genetic heterogeneity present within tumor | |||
| Xenograft (patient-derived) | Human tumor explant propagated in immunodeficient host (mouse) | Heterogeneity and genetic diversity within tumors | Immunodeficient host | 8–24 wk†, $$$ |
| Representation from various human tumor types | Subcutaneous location may not allow cultivation of key tissue-specific stromal infiltrate | |||
| Features of tumor microenvironment, including stromal and vascular cells, incorporated within tumor | Surgical implantation required | |||
| Tumors are easily and precisely measured | Cross-species divide; stromal components are mouse, whereas tumor cells are human | |||
| Genetic and phenotypic drift with passage | ||||
| Syngeneic | Immortalized mouse tumor cell line allografted into immunocompetent host (mouse) | Presence of intact immune system | Limited number of established cell lines, which are poorly annotated | 2–4 wk, $ |
| Features of tumor microenvironment, including stromal and vascular cells, incorporated within tumor | Strong immunogenicity of some lines promotes spontaneous regression | |||
| All cell types within tumor are of mouse origin | Rapid growth rate of many lines limits use in longer-term studies | |||
| Tumors are easily and precisely measured | ||||
| GEMMs | Genetic modification that permits induced or spontaneous tumor development | Tumors develop in tissue of origin | Limited genetic mosaicism and heterogeneity of tumors | 12–24 wk†, $$ |
| Presence of intact immune system | Technical hurdlesfor monitoring tumor response in internal organs | |||
| All cell types within tumor are of mouse origin | Low throughput and high investment | |||
| Features of tumor microenvironment, including stromal and vascular cells, and immune system components |
=low
=low
=intermediate cost
=high cost.
Up to 1 y to observe metastases.
Adapted with permission of (21).
Cell Line Xenograft Models
Mouse models of breast cancer derived by transplanting immortalized human cancer cell lines into an immunocompromised murine host are among the simplest and most frequently deployed model systems in cancer research. Most preclinical drug treatment studies performed in vivo have involved the use of immortalized human breast cancer cell lines growing within the subcutaneous dorsal flank of immunocompromised mice. Given the vast research history accumulated for many immortalized breast cancer cell lines and the numerous, diverse cell lines that represent all breast cancer molecular subtypes, xenografting breast cancer cell lines has become a staple in preclinical breast cancer research.
Although these models are technically simple to establish and are inexpensive to maintain over the short term, they have critical weaknesses that should be considered before larger programmatic efforts are based solely on them. In particular, shortcomings inherent in cell line xenograft models are commonly cited as the Achilles’ heel of drug discovery efforts, especially when preclinical and clinical results are incongruent (25). An insightful commentary suggested that cell line xenografts are useful as a bridge between in vitro and in vivo studies (3). Objectively, cell line xenograft models have clear strengths, especially for rapid hypothesis testing, including the following: the development and extensive characterization of a panoply of human breast cancer cell lines from all molecular subtypes; the development of tumor stromal characteristics that can mimic the characteristics of human tumors (albeit incompletely); easily interrogated tumors; and quick tumor manifestation, which reduces attendant housing costs and speeds discovery. These strengths are balanced by the following limitations of cell line xenografts: the immunodeficiency of the host in which tumors arise, resulting in major contributions from the immune system to cancer development, cancer progression, and a therapeutic response being ignored; subcutaneous tumor propagation, which fails to simulate organotypic tumor microenvironments; a species disconnect between the tumor cells (human) and the stroma (mouse); and extreme homogeneity within the tumor, which poorly reflects the intratumoral heterogeneity seen in clinical breast tumors.
PDX Models
An often overlooked shortcoming of the cell line xenograft model is the fact that immortalized cell lines are developed through clonal attrition, resulting in cell populations that are propagated through multiple passages on a (typically) plastic surface. Selective pressures and genetic drift give rise to genotypic and phenotypic changes that may irreversibly distinguish daughter clones from paternal tumors (26); this scenario may poorly recapitulate the original underlying cancer biology of the patient. Models developed from patient-derived tumors, otherwise known as PDX models—in which patient tumors are surgically implanted into recipient murine hosts without being cultured—overcome this limitation. PDX models of various human tumors have been developed with great success, although breast cancer PDX models have historically been especially challenging (27). DeRose et al. reported exemplary success when multiple PDX tumor models derived from patient specimens recapitulated ER- or PR-positive, ER- or PR-negative, and HER2-positive tumors and TNBC (28).
The major strengths of the PDX approach include genetic diversity and heterogeneity that more accurately reflect human breast cancer; the ability to model various cancer subtypes; the incorporation of contextually correct human stroma within the tumor, including vascularity and inflammation; the documented ability to model metastasis; and easy interrogation of tumors, such as breast cancers, for correlative studies. This approach maintains the genetic and phenotypic integrity of the tumor cells, without the clonal selection or inadvertent genetic drift seen in immortalized breast cancer cell lines. PDX models are increasingly being used on the basis of the observation that the histologic and molecular (gene expression and copy number variations) characteristics of the PDX can be maintained through several mouse “passages.” Importantly, PDX models retain clinical responses to many drug treatments, making them ideal for coclinical trials.
Nevertheless, there are several potential drawbacks of PDX models, including the requirement to use a severely immunodeficient murine host; the fact that the surgical procedure for implanting tumors into mice is invasive and requires skill (29); a species disconnect between the implanted tumor cells and stroma (human) and subsequently infiltrating stroma (mouse); and the time required to generate the models, which can require several months simply for the establishment of engraftment (30). Technical issues aside, the fact that establishing and maintaining PDX model systems require major capital investments in supporting infrastructure and personnel must not be overlooked.
Syngeneic Models
The requirement for the use of immunocompromised mice in xenograft models fails to incorporate the impact of the immune system on the tumor response. This area of cancer research is in its early stages, with rapid progress and vast promise that underscores the need for immunocompetent models of breast cancer for more rigorous analyses. Adequately modeling cancer immunology requires a propagating tumor within an immunocompetent host. One approach is to use mouse mammary tumors or mouse mammary tumor cell lines implanted into syngeneic immunocompetent murine hosts. Devoid of the species constraints inherent in xenografts and xenotransplants, allografted mouse tumors are not typically rejected by the murine host, given the similar genetic backgrounds. Syngeneic model systems offer the distinct advantage of studying cancer biology within the context of an intact immune system and speciesspecific tumor microenvironment. However, mouse tumor cell lines are limited and annotated to various degrees, and although smallmolecule therapies may be adequately evaluated within these models, the species specificity of antibody imaging agents and therapies generally precludes their evaluation in syngeneic model systems.
GEMMs
GEMMs are the most sophisticated in vivo platforms used to simulate human cancer. These models are capable of not only accurately mimicking many relevant pathophysiologic features of human cancer but also recapitulating the sequence of molecular events that give rise to cancer. The transgenic expression of an oncogene specifically within the mouse mammary epithelium under the control of a strong mammary epithelial promoter is frequently used to induce mammary tumor formation. This is a clinically relevant model of tumor initiation and progression, enforcing the stepwise procession of cells from hyperplasia to ductal carcinoma in situ and then to invasive ductal carcinoma. Importantly, this process occurs within the context of the native stromal matrix (requiring stromal remodeling and angiogenesis) and the native immune system (requiring immune system evasion). The genetic manipulations can drive oncogene expression in a reversible or irreversible manner, in a tissue-specific manner (3) or, more broadly, throughout an entire organism. Frequently, GEMMs that harbor oncogenic driver genes (e.g., HER2) or lack tumor suppressor genes (e.g., p53), thus genetically mimicking human cancers, are developed.
The diverse array of oncogenes used to generate transgenic models of breast cancer has resulted in a multitude of models that mimic many of the specific molecular subtypes seen in clinical breast cancers, as confirmed by comparative expression analyses of mouse and human breast tumor samples (31). The advantages of GEMMs include tumor formation in the contextually appropriate tissue and potentially cell of origin through the use of tissue-specific or cellspecific promoters; an intact immune system; and a native tumor microenvironment that more accurately reflects human disease, including stromal components, vascularity, and inflammation. However, GEMMs are limited by the time, expense, and resources required to derive, establish, and maintain them; these demands can be overly burdensome given the potentially low experimental throughput of GEMMs. Few GEMMs of breast cancer truly harbor ER expression, despite commonalities in expression profiles between mouse and human luminal breast cancers. Although metastases in mouse breast cancer models are hematogenous and almost exclusively pulmonary, human breast cancer metastases occur though lymphatic spread that often precedes hematogenous metastasis to the lungs, liver, bone, brain, and elsewhere.
Molecular Imaging Applications: Biomarkers, Drug Discovery, and Coclinical Trials
Molecular imaging is an indispensable tool uniquely poised to address major challenges obstructing the delivery of personalized cancer therapy. Capable of noninvasively quantifying the cellular and molecular underpinnings of tumors on a patient-by-patient basis, molecular imaging enables the detection of tumors at early, potentially curable stages and provides a means to accurately predict the response of a tumor to therapy well before conventional means of assessment.
Numerous excellent review articles that thoroughly discuss the attributes of various molecular imaging modalities in both patients and preclinical animal models have been disseminated. Rather than recapitulate a description of specific imaging systems and methods, we simply suggest that interested readers consult specific articles that already relate directly to this topic (32–34). However, as an introduction to preclinical molecular imaging in breast cancer models, it is worth noting that a range of imaging modalities can be entirely suitable for this purpose; such modalities include optical techniques (bioluminescence and fluorescence), ultrasound, MRI, MR spectroscopy, and nuclear imaging techniques that use ionizing radiation, namely, PET and SPECT (Table 3).
Table 3.
Imaging Modalities
| Modality | Signal/contrast | Translational | Preclinical | Sensitivity* | Resolution | Depth | Quantitative | Target | Acquisition time (s) | Cost† |
|---|---|---|---|---|---|---|---|---|---|---|
| PET | 11C, 13N, 15O, 18F, 64Cu, 68Ga, 89Zr, 124I | Yes | Yes | 1 | 1–2 mm | No limit | Yes (very good) | Molecular | 10–100 | $$$ |
| SPECT | 99mTc 123/125I 201Tl 111 In, 177Lu, 67Ga, 133Xe |
Yes | Yes | 10−1-10−2 | <1 mm | No limit | Yes (good) | Molecular | 100–1,000 | $$ |
| MRI | Hydrogen, gadolinium, magnetic or paramagnetic particles | Yes | Yes | 10−5 | 10–100 μm | No limit | Yes (fair) | Anatomic, molecular |
100–1,000 | $$$ |
| MR spectroscopy Hyperpolarization (13C, 15N, 129Xe, 3He) | Yes | Yes | <10−5 | 5 mm | No limit | Yes (fair) | Molecular | 100–1,000 | $$$ | |
| Ultrasound | Echoes, microbubbles |
Potential | Yes | 10−3 | 50 μm | Centimeter Yes (poor) | Anatomic, molecular |
<1 | $$ | |
| Bioluminescence | Luciferase (reporter gene) | No | Yes | 1–102* | <10 mm | Centimeter Yes (poor to fair)† | Molecular | 1–10 | $$ | |
| Fluorescence | Fluorescent proteins, fluorochromes, quantum dots | Potential | Yes | 10−2-1* | 2–3 mm | <1 cm | Yes (poor to fair)† | Molecular | 1–10 | $ |
Relative to that of PET.
For high-resolution, small-animal imaging systems (clinical imaging systems differ). Cost is based on purchase price of imaging systems in United States.
=low
=intermediate cost
=high cost.
Depth-dependent
The modalities can be generally parsed into 2 major categories: anatomic, which centers on morphology (gross and fine), and molecular, which centers on underlying biologic function (metabolism, biochemistry, gene expression, and systems). The choice of imaging modality for addressing in vivo hypotheses depends largely on the biologic question of interest and is often guided by the strengths and limitations inherent in the modality. As highlighted in Table 3, certain modalities are better suited for molecular imaging (PET, SPECT, MR spectroscopy, and optical techniques), whereas others may serve in both capacities under some scenarios (ultrasound and MRI). Although all have been used in preclinical studies, only a select few are considered eminently translational.
Once the modality and the model have been selected, numerous clinically unmet needs can potentially be addressed in the laboratory through the marriage of noninvasive molecular imaging and preclinical mouse models of breast cancer. For example, the development of inhibitors targeting various portions of the ErbB signaling axis is an active and clinically important area of breast cancer research. Tz is a Food and Drug Administration–approved, recombinant, humanized monoclonal antibody that selectively binds to the extracellular domain of HER2, yet objective means to assess the treatment response to Tz therapy remain undeveloped. To this end, Whisenant et al. recently reported the use of 3ʹ-deoxy-3ʹ−18F-fluorothymidine (18F-FLT) PET as an early marker of the response to Tz in HER2-overexpressing xenografts (35). The researchers showed that 18F-FLT PET was sensitive to early molecular changes in Tz-sensitive, HER2-overexpressing breast cancer xenografts and that it could differentiate mouse models of HER2-overexpressing breast cancer with various Tz sensitivities.
The development of noninvasive imaging methods that could identify nonresponders earlier during therapeutic intervention is of great clinical interest because of the desire to spare patients any delay in the initiation of effective combination therapies. For example, Kramer-Marek et al. reported the feasibility of Affibody (Affibody AB)–based (18F-FBEM-HER2:342) PET for quantifying changes in ErbB2 (HER2/neu) expression and predicting the response to Tz in mouse (BT474) breast cancer xenografts (36). In addition to immunohistochemical correlation of the overall decrease in 18F-FBEM-HER2:342 Affibody uptake with a tumor response and downregulation of ErbB2 expression, their work also reaffirmed that the number of vessels in a tumor could act as a useful prognostic marker for a treatment response. Moreover, molecular imaging with PET could serve as a valuable strategy for predicting the tumor response to Tz.
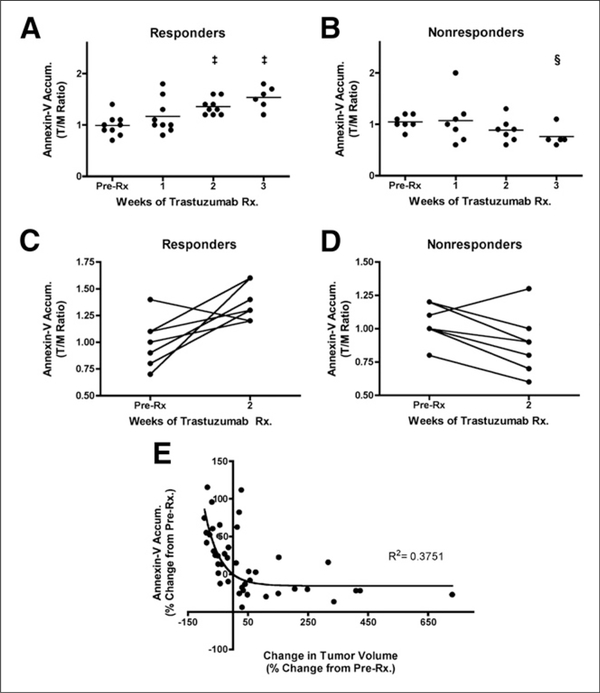
Our laboratory has also evaluated a suite of translational, noninvasive molecular imaging metrics in an attempt to predict the response to Tz in preclinical mouse models of HER2-overexpressing breast cancer (37). In that study, mammary tumors from MMTV/ HER2 transgenic female mice were transplanted into syngeneic female mice. Tumor cell apoptosis was assessed with an optical imaging tracer based on annexin V, glucose metabolism was assessed with 18F-FDG PET, and proliferation was assessed with 18F-FLT PET. The results of that study suggested that molecular imaging of apoptosis accurately predicted Tz-induced regression of HER2-positive tumors and warranted clinical exploration as a means to predict an early response to neoadjuvant Tz (Fig. 1). That noninvasive imaging of apoptosis was superior to both 18F-FDG PET and 18F-FLT PET in that setting was a factor that prompted our development of other imaging probes for apoptosis, including a caspase-targeted PET tracer (38). In an analogous study seeking the elucidation of mechanisms that affect Tz efficacy, Miller et al. used combined imaging of apoptosis and glucose metabolism to determine that the inhibition of both mammalian target of rapamycin and phosphoinositide 3-kinase was required for the growth-inhibitory effect of HER2 antagonists (39).
FIGURE 1.
Longitudinal assessment of NIR700-annexin V accumulation (Accum.) in MMTV/HER2 tumors, illustrating efficacy of Tz treatment (Rx.). (A) NIR700-annexin V accumulation ratios (tumor to normal muscle [T/M]) increased and attained statistical significance after 2 wk of Tz treatment in responding MMTV/HER2 tumor–bearing mice. PreRx5before treatment. ‡P >, 0.001, as determined with paired t test. (B) NIR700-annexin V did not accumulate in nonresponding tumors, and overall uptake of imaging probe decreased as tumors progressed. §P < 0.05. (C and D) On individual basis, NIR700-annexin V uptake (pretreatment imaging compared with 2-wk posttreatment imaging) significantly increased after treatment in most responding MMTV/HER2 tumor–bearing mice (C). However, nonresponding cohorts typically showed decreased NIR700-annexin V accumulation after 2 wk of Tz treatment (D). (E) Magnitude of tumor regression was proportional to degree of uptake of NIR700-annexin V. Nonlinear inverse correlation was observed between change in NIR700-annexin V uptake from baseline and change in tumor volume from baseline when all imaging time points and all mice were considered. (Reprinted with permission of (37).)
In addition to HER2-enriched breast cancer, BLBCs and luminal breast cancers have been the focus of other studies. BLBCs, such as the TNBC BRCA1-related subtype, often exhibit high rates of glucose uptake that can be noninvasively assessed with 18F-FDG PET. Using a GEMM (MMTV-CreBrca1f/fTrp53+/−) to study and develop polyadenosine ribose polymerase and phosphoinositide 3-kinase combination therapies, Juvekar et al. showed how the inhibition of 18F-FDG uptake may serve as an early and predictive pharmacodynamic marker of a treatment response (40). Because of de novo and acquired resistance of luminal breast cancers to endocrine therapy, there remains a need to identify which ER- or PR-positive tumors are most likely to respond to therapy (fulvestrant). Using small-animal PET and a preclinical mouse model of human luminal breast cancer (GEMM), Fowler et al. identified a profile that delineated fulvestrant-sensitive from fulvestrant-resistant ERα-and PR-positive tumors before changes in tumor size were apparent (41). Noninvasive imaging of baseline tumoral 18F-FES uptake and initial changes in 18F-fluorofuranylnorprogesterone uptake could be used as a prognostic strategy to identify responders and nonresponders to endocrine therapy at an early stage of disease. Although 18F-FDG remains the clinical standard for PET imaging in oncology, recent work by Prignon et al. with a PDX mouse model (human ZR75–1) revealed the potential of gastrin-releasing peptide receptor PET with the 68Ga-labeled bombesin analog AMBA (68Ga-DOTA-CHCO-Gly-4-aminobenzyl-Gln-Trp-Ala-Val-GlyHis-Leu-Met-NH) as a possible alternative (42). 68Ga-AMBA not only visualized but also monitored the therapeutic response (tamoxifen) better than 18F-FDG.
Another research area in which noninvasive molecular imaging and preclinical mouse models have natural synergy is the coclinical trial concept. A coclinical oncology trial can be loosely defined as coupled laboratory and clinical investigations developed around a common hypothesis that features a clinical trial in patients with cancer and analogous, complementary studies in mouse models that have been carefully selected to recapitulate elements of individual patients (43,44). Such concepts may have a stated therapeutic goal, although studies exploring cancer biomarkers or cancer risk could also be envisioned. Essentially any of the preclinical mouse models discussed here could be deployed as part of a coclinical trial, but each system has unique considerations, and some may be more suitable than others. For example, parallel clinical trials and studies in PDX models of individual patient tumors could be attractive. The predictive clinical value of these approaches, which are sometimes referred to as avatar models, is being explored (45) but currently remains unknown. A complicating factor is the rather long lead time required to generate PDX models compared with the time line of typical clinical trials, so that coclinical trials are not necessarily parallel in a temporal sense.
Nonetheless, even with the limitations inherent in cell line xenograft models, coclinical imaging trials can be instrumental in elucidating mechanisms of response and potential resistance and rationalizing clinical results in focused populations. For example, with the aim of determining whether the small-molecule mitogen-activated protein kinase kinase inhibitor selumetinib could increase the efficacy of standard-of-care chemotherapy (docetaxel), Chen et al. demonstrated in a murine lung cancer coclinical trial that the concomitant loss of either of 2 clinically relevant tumor suppressors (p53 or Lkb1) markedly impaired the response of Kras mutant cancers to docetaxel monotherapy (46). The investigators showed that the addition of selumetinib provided substantial benefit for mice with lung cancer driven by Kras or Kras and p53 mutations but not for mice with Kras and Lkb1 mutations, which possessed primary resistance to this combination therapy. Further pharmacodynamic studies with PET helped identify, in both mice and patients, biologic markers that provided a rationale for the differential efficacies of these therapies in the different genotypes. That study highlighted the rationale for synchronous coclinical trials to not only anticipate the results of ongoing human clinical trials but also generate clinically relevant hypotheses to iteratively inform the design and analysis of human studies.
With an emphasis on predicting the response to the monoclonal antibody cetuximab in a preclinical mouse model xenograft of wildtype KRAS colorectal cancer, we recently reported that 18F-FLT PET closely reflected prosurvival responses to targeted therapy mediated by the activity of phosphoinositide 3-kinase–mammalian target of rapamycin; we concluded that this imaging marker could play a novel and potentially critical role in predicting tumors that exhibit molecular features reflecting recalcitrance to mitogen-activated protein kinase–targeted therapy (47). In parallel, for a focused cohort of patients with wild-type KRAS rectal cancer, we evaluated the early molecular response with clinical 18F-FLT PET after one cycle of cetuximab (48). Interestingly, 18F-FLT uptake was reduced in 3 of 4 patients after monotherapy, yet 1 patient exhibited elevated 18FFLT uptake at this time point. Overall, we interpret these findings to suggest that 18F-FLT PET reflected mechanistic insight into tumor responses to cetuximab given that imaging accurately predicted posttreatment levels of p27, an inhibitor of the cell cycle, in agreement with the results of parallel preclinical studies (47). In fact, 18F-FLT PET results supported by Ki-67 and p27 immunoreactivity suggested that this 1 patient did not experience an antiproliferation benefit from cetuximab monotherapy. We envision that in future studies with similar regimens, 18F-FLT PET after cetuximab treatment could be incorporated as a noninvasive assay to predict the patients most likely to benefit from this drug before changes in tumor size are ascertained by anatomic imaging measures, such as CT.
CONCLUSION
Although all model systems remain an approximation of human disease, mouse models of cancer are indispensable resources that facilitate and accelerate research and ultimately have tremendous potential to affect the care of patients with breast cancer and other solid tumors. Mouse models provide a unique means to elucidate causative and contributory factors related to cancer as well as to provide useful platforms for evaluating novel therapies with translational potential. Molecular imaging is a complementary tool that allows investigators to maximize the potential utility of mouse models in laboratory and translational settings.
Acknowledgments
DISCLOSURE
This work was supported by Congressionally Directed Medical Research Programs awards W81XWH-13–1 (to Rebecca S. Cook) and W81XWH-14–1 (to Rebecca S. Cook), National Institutes of Health grants 5P50CA098131 (to Rebecca S. Cook and H. Charles Manning) and 1R01CA163806 (to H. Charles Manning and Jason R. Buck), the Lustgarten Foundation (to H. Charles Manning and Jason R. Buck), and the Kleberg Foundation (to H. Charles Manning and Jason R. Buck). No other potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortalityworldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2015. Atlanta, GA: American Cancer Society; 2015. [Google Scholar]
- 3.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–658. [DOI] [PubMed] [Google Scholar]
- 4.Cheon DJ, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95–119. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson JN, Muller WJ. Transgenic mouse models of human breast cancer. Oncogene. 2000;19:6130–6137. [DOI] [PubMed] [Google Scholar]
- 6.Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim IS, Baek SH. Mouse models for breast cancer metastasis. Biochem Biophys Res Commun. 2010;394:443–447. [DOI] [PubMed] [Google Scholar]
- 8.Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically engineered mousemodels in cancer research. Adv Cancer Res. 2010;106:113–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangarajan A, Weinberg RA. Opinion: comparative biology of mouse versushuman cells—modelling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. [DOI] [PubMed] [Google Scholar]
- 10.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(suppl 2):S60–S64. [DOI] [PubMed] [Google Scholar]
- 11.Baselga J, Norton L, Albanell J, Kim YM, Mendelsohn J. Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res. 1998;58:2825–2831. [PubMed] [Google Scholar]
- 12.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson LM, Kurland BF, Schubert EK, et al. A phase 2 study of 16a-[18F]fluoro-17b-estradiol positron emission tomography (FES-PET) as a marker of hormone sensitivity in metastatic breast cancer (MBC). Mol Imaging Biol. 2014;16: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Kruchten M, Glaudemans AW, de Vries EF, et al. PET imaging of estrogenreceptors as a diagnostic tool for breast cancer patients presenting with a clinical dilemma. J Nucl Med. 2012;53:182–190. [DOI] [PubMed] [Google Scholar]
- 15.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. [DOI] [PubMed] [Google Scholar]
- 16.Murphy CG, Modi S. HER2 breast cancer therapies: a review. Biologics. 2009;3 289–301. [PMC free article] [PubMed] [Google Scholar]
- 17.Kurihara H, Hamada A, Yoshida M, et al. 64Cu-DOTA-trastuzumab PET imaging and HER2 specificity of brain metastases in HER2-positive breast cancer patients. EJNMMI Res. 2015;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dijkers EC, Oude Munnink TH, Kosterink JG, et al. Biodistribution of 89Zrtrastuzumab and PET imaging of HER2-positive lesions in patients with metastatic breast cancer. Clin Pharmacol Ther. 2010;87:586–592. [DOI] [PubMed] [Google Scholar]
- 19.Toft DJ, Cryns VL. Minireview: basal-like breast cancer—from molecular profiles to targeted therapies. Mol Endocrinol. 2011;25:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negativebreast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gould SE, Junttila MR, de Sauvage FJ. Translational value of mouse models inoncology drug development. Nat Med. 2015;21:431–439. [DOI] [PubMed] [Google Scholar]
- 22.Doudna JA, Charpentier E. Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- 23.Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol. 2014;16:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mousemodels in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. [DOI] [PubMed] [Google Scholar]
- 26.Daniel VC, Marchionni L, Hierman JS, et al. A primary xenograft model ofsmall-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts asmodels for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeRose YS, Wang G, Lin YC, et al. Tumor grafts derived from women withbreast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeRose YS, Gligorich KM, Wang G, et al. Patient-derived models of humanbreast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr Protoc Pharmacol. 2013;60:14.23.1–14.23.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2:247–250. [DOI] [PubMed] [Google Scholar]
- 31.Pfefferle AD, Herschkowitz JI, Usary J, et al. Transcriptomic classification ofgenetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14:R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson TE, Manning HC. Molecular imaging: 18F-FDG PET and a whole lot more. J Nucl Med Technol. 2009;37:151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons SK. Imaging mouse models of cancer. Cancer J. 2015;21:152–164. [DOI] [PubMed] [Google Scholar]
- 35.Whisenant JG, McIntyre JO, Peterson TE, et al. Utility of [18 F]FLT-PET to assesstreatment response in trastuzumab-resistant and trastuzumab-sensitive HER2overexpressing human breast cancer xenografts. Mol Imaging Biol. 2015;17:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer-Marek G, Gijsen M, Kiesewetter DO, et al. Potential of PET to predictthe response to trastuzumab treatment in an ErbB2-positive human xenograft tumor model. J Nucl Med. 2012;53:629–637. [DOI] [PubMed] [Google Scholar]
- 37.Shah C, Miller TW, Wyatt SK, et al. Imaging biomarkers predict response toanti-HER2 (ErbB2) therapy in preclinical models of breast cancer. Clin Cancer Res. 2009;15:4712–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hight MR, Cheung YY, Nickels ML, et al. A peptide-based positron emissiontomography probe for in vivo detection of caspase activity in apoptotic cells. Clin Cancer Res. 2014;20:2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller TW, Forbes JT, Shah C, et al. Inhibition of mammalian target ofrapamycin is required for optimal antitumor effect of HER2 inhibitors against HER2-overexpressing cancer cells. Clin Cancer Res. 2009;15: 7266–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juvekar A, Burga LN, Hu H, et al. Combining a PI3K inhibitor with a PARPinhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2:1048–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fowler AM, Chan SR, Sharp TL, et al. Small-animal PET of steroid hormonereceptors predicts tumor response to endocrine therapy using a preclinical model of breast cancer. J Nucl Med. 2012;53:1119–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prignon A, Nataf V, Provost C, et al. 68Ga-AMBA and 18F-FDG for preclinical PET imaging of breast cancer: effect of tamoxifen treatment on tracer uptake by tumor. Nucl Med Biol. 2015;42:92–98. [DOI] [PubMed] [Google Scholar]
- 43.Lunardi A, Pandolfi PP. A co-clinical platform to accelerate cancer treatmentoptimization. Trends Mol Med. 2015;21:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nardella C, Lunardi A, Patnaik A, Cantley LC, Pandolfi PP. The APLparadigm and the “co-clinical trial” project. Cancer Discov. 2011;1:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derivedxenograft mouse models. Nat Rev Cancer. 2015;15:311–316. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trialidentifies genetic modifiers of therapeutic response. Nature. 2012;483:613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKinley ET, Zhao P, Coffey RJ, Washington MK, Manning HC. 39-deoxy39-[18F]-fluorothymidine PET imaging reflects PI3K-mTOR-mediated prosurvival response to targeted therapy in colorectal cancer. PLoS One. 2014; 9:e108193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKinley ET, Watchmaker JM, Bapsi Chakravarthy A, et al. [18F]-FLT PET to predict early response to neoadjuvant therapy in KRAS wild-type rectal cancer: a pilot study. Ann Nucl Med. 2015;29:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]