Abstract
Molluscan shells, mainly composed of calcium carbonate, also contain organic components such as proteins and polysaccharides. Shell organic matrices construct frameworks of shell structures and regulate crystallization processes during shell formation. To date, a number of shell matrix proteins (SMPs) have been identified, and their functions in shell formation have been studied. However, previous studies focused only on SMPs extracted from adult shells, secreted after metamorphosis. Using proteomic analyses combined with genomic and transcriptomic analyses, we have identified 31 SMPs from larval shells of the pearl oyster, Pinctada fucata, and 111 from the Pacific oyster, Crassostrea gigas. Larval SMPs are almost entirely different from those of adults in both species. RNA-seq data also confirm that gene expression profiles for larval and adult shell formation are nearly completely different. Therefore, bivalves have two repertoires of SMP genes to construct larval and adult shells. Despite considerable differences in larval and adult SMPs, some functional domains are shared by both SMP repertoires. Conserved domains include von Willebrand factor type A (VWA), chitin-binding (CB), carbonic anhydrase (CA), and acidic domains. These conserved domains are thought to play crucial roles in shell formation. Furthermore, a comprehensive survey of animal genomes revealed that the CA and VWA–CB domain-containing protein families expanded in molluscs after their separation from other Lophotrochozoan linages such as the Brachiopoda. After gene expansion, some family members were co-opted for molluscan SMPs that may have triggered to develop mineralized shells from ancestral, nonmineralized chitinous exoskeletons.
Keywords: biomineralization, larval shell, mollusca, proteome, shell matrix protein
Introduction
In living organisms, biomineralized tissues serve multiple functions: tissue support, storage of mineral ions, protecting the soft body from predators and from environmental factors such as UV radiation (Lowenstam 1989; Simkiss and Wilbur 2012). As in other metazoan lineages, acquisition of diverse mineralized exoskeletons is one of the reasons for the rapid establishment of shell-bearing molluscs at the dawn of Cambrian times (Kawasaki et al. 2004; Killian and Wilt 2008). Despite being a minor component in the shells by mass, organic matrices, composed mainly of proteins, glycoproteins, chitin, and acidic polysaccharides, have essential roles in numerous aspects of shell formation, such as calcium carbonate nucleation, crystal growth, and choice of calcium carbonate polymorphs (Addadi et al. 2006; Marin et al. 2008). Recent proteomic and transcriptomic studies of molluscan shell matrix proteins (SMPs) revealed that repertoires of SMPs differ substantially among molluscan species, and among shell structures (e.g., the nacreous layer and the prismatic layer) within the same species, whereas some functional domains are shared among them (Jackson et al. 2010; Marie et al. 2012, 2013, 2017; Zhang et al. 2012; Mann and Edsinger 2014; Mann and Jackson 2014; Liu et al. 2015). Even though the number of identified molluscan SMPs is rapidly increasing with the help of high-throughput DNA sequencing technology, previous research has focused only on shells formed by adult animals.
During larval development, bivalves form two types of shells, called prodissoconch I and II (Werner 1939; Gosling 2015). The first shell, prodissoconch I, begins to be secreted from the shell field during the trochophore larval stage, and forms D-shaped shells, usually about 18–24 h after fertilization (Alagarswami et al. 1983; Rose and Baker 1994; Doroudi and Southgate 2003; Kakoi et al. 2008; Wassnig and Southgate 2012; Gosling 2015). Immediately after prodissoconch I, prodissoconch II is secreted from mantle tissue of the veliger larva. After settlement and metamorphosis of veliger larvae to become juveniles, adult shells, or dissoconch, are formed (Waller 1981; Mao Che et al. 2001). Adult molluscan shells are comprised mainly of calcite or aragonite, or both, which are assembled in highly variable microstructures (Kobayashi 1969; Taylor 1973; Carter 1990). Molluscan larval shells, however, have similar microstructures and are composed only of aragonite (LaBarbera 1974; Iwata 1980; Eyster 1983, 1986; Weiss et al. 2002), implying that larval shells are evolutionarily highly conserved (Taylor 1973). With this commonality in mineralogy and microstructures, studies of larval shell proteins could help to infer the evolutionary antiquity of larval types. For instance, they could be used to test the hypothesis that “set-aside cells” in molluscan larvae provide the evolutionary and developmental ground on which adult structures are built (Peterson et al. 1997). In addition, with the shorter time required to see potential effects of in vivo manipulation experiments on shell formation genes, larval shells would arguably be a suitable model for understanding shell formation processes. However, due to technical challenges, such as isolating an adequate amount of larval shell material, proteomic analyses of molluscan larval shells have not been performed. It is still unknown how many and what kinds of proteins are required to form a larval shell.
In this study, we identified a large number of SMPs from 24-h (hour after fertilization) larval shells of two pteriomorph bivalve species, the pearl oyster, Pinctada fucata, and the Pacific oyster, Crassostrea gigas, for which whole genome sequences are available (Takeuchi et al. 2012; Zhang et al. 2012; Takeuchi, Koyanagi, et al. 2016). We found that larval SMP repertoires are almost entirely distinct from adult repertoires. Nevertheless, there are functional similarities between larval and adult SMPs, such as von Willebrand factor type A (VWA), chitin-binding (CB) domains, carbonic anhydrases (CAs), and the acidic nature of some SMPs. These common features shed light on essential components for shell formation.
Results
SMPs of Larval and Adult Shells
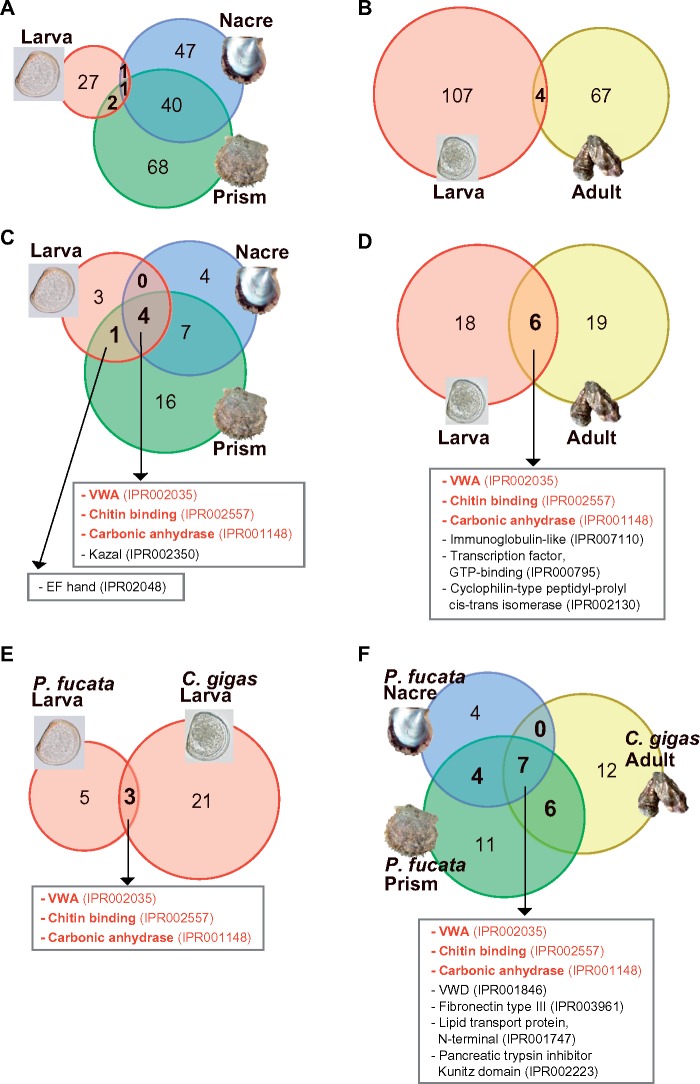
In the larval shell proteomes of P. fucata and C. gigas, 31 and 111 proteins were identified, respectively (fig. 1; supplementary data S1–S6, Supplementary Material online). In both species, the majority of larval SMPs are different from adult SMPs (fig. 1A and B). Only four SMPs are shared between larval and adult SMPs in P. fucata (fig. 1A). One SMP, VWA and CB domain-containing protein (pfu_cdna2.0_089203), is present in larval, nacre, and prism shell proteomes. A novel aspartic acid-rich protein (pfu_aug2.0_2162.1_08762.t1) is found in larval and nacre proteomes. In addition, two valine-rich proteins, for which genes are tandemly arranged in the genome (pfu_aug2.0_618.1_27594.t1 and pfu_aug2.0_618.1_27595.t1), are identified in larval and prism proteomes. Similarly, four larval SMPs of C. gigas were found among adult SMPs reported by Zhang et al. (2012). They include L-ascorbate oxidase (CGI_10008969), Elongation factor 1-alpha (CGI_10012474), ATP synthase subunit alpha (CGI_10024501), and ATP synthase subunit beta (CGI_10013347).
Fig. 1.
Repertoires of larval and adult SMPs differ, but they share some functional domains. Numbers of shared and unshared shell matrix proteins among larval, adult nacreous, and adult prismatic shells in P. fucata (A), and between larval and adult shells in C. gigas (B). Comparisons of functional domains found in larval and adult SMPs of P. fucata (C) and C. gigas (D). Functional domains in larval (E) and adult (F) SMPs of the two species were also compared. Domains shared by both larvae and adults of two species are colored in red.
Although larval and adult SMPs are largely different, these proteins share some functional domains (fig. 1C and D). Among eight domain categories identified in the larval shell proteome of P. fucata, five were found in adult SMPs (fig. 1C). In C. gigas, six functional domains were shared between larval and adult SMPs (fig. 1D). Cross-species comparison of functional domains showed that three and seven domains are shared among larval (fig. 1E) and adult (fig. 1F) SMPs, respectively. It is important to note that only three domains including VWA (IPR002035), CB (IPR002557), and CA (IPR001148) domains were commonly found in both larval and adult SMPs in P. fucata and C. gigas, implying that these conserved domains are essential for shell formation.
In order to identify larval SMPs common to these two bivalve species, amino acid sequences of P. fucata larval SMPs were searched against C. gigas predicted gene models (Zhang et al. 2012) using BLASTP. A total of 21 larval SMPs of P. fucata showed significant sequence similarity (E-value ≤1e−5) to C. gigas proteins (supplementary table S1, Supplementary Material online). Among them, 12 proteins were detected in larval SMPs of C. gigas.
Larval and Adult SMPs Show Different Expression Patterns during Development
Expression of SMP genes in larvae and the adult mantle tissues of P. fucata was analyzed using RNA-seq (fig. 2). Gene expression profiles during larval and adult shell formation are distinct. Most larval SMP genes are highly expressed during larval shell formation in trochophore and D-shaped larval stages, whereas larval SMP gene expression was not detected during adult shell formation. Some SMP genes, including pfu_aug2.0_2162.1_08762.t1, pfu_aug2.0_618.1_27594.t1, and pfu_aug2.0_618.1_27595.t1, common to both larval and adult shells of P. fucata are intensely expressed during both larval and adult stages. On the other hand, genes responsible for the nacre and prism shell layers were rarely expressed in larval stages (fig. 2; supplementary data S3, Supplementary Material online). In C. gigas, about two-thirds of SMP genes are expressed only during larval or adult stages, whereas others are highly expressed during both. The latter comprise mainly house-keeping genes such as elongation factor 1 alpha and ribosomal protein genes (supplementary fig. S1 and data S2, Supplementary Material online).
Fig. 2.
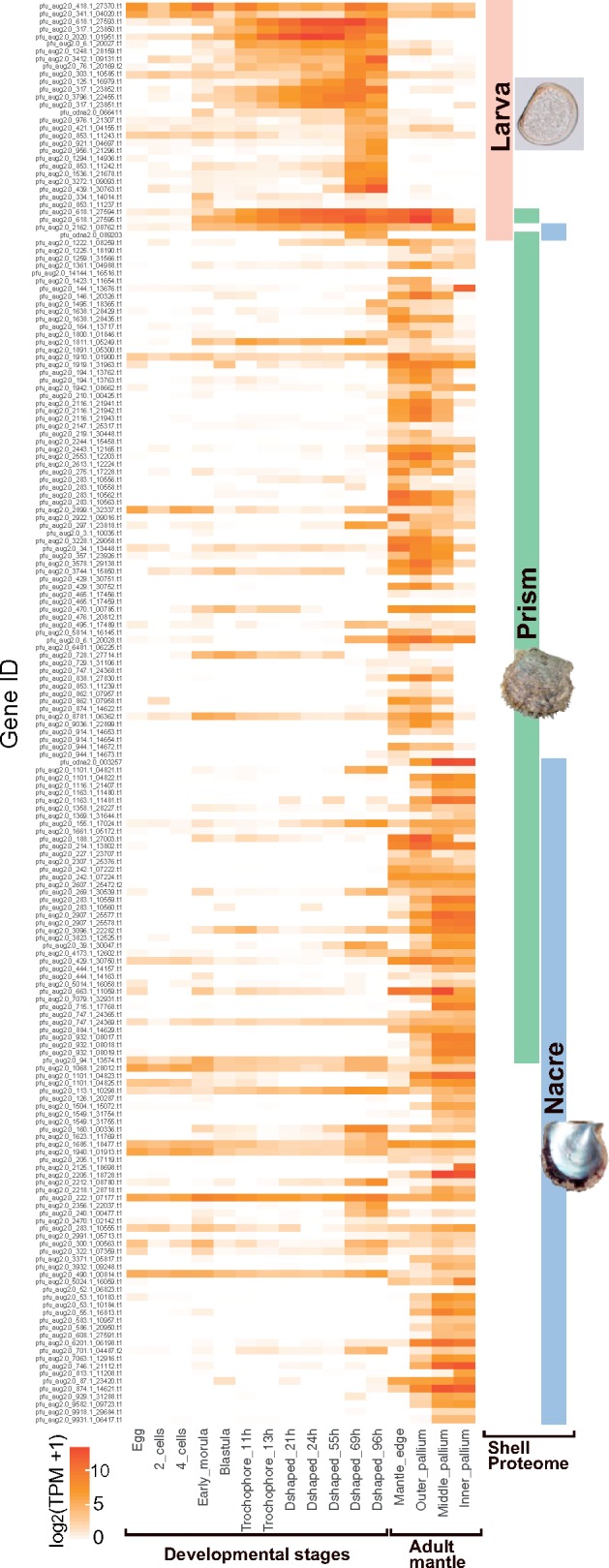
Stage-specific expression of larval and adult SMP genes in P. fucata. The heatmap shows expression levels of identified SMP genes at different developmental stages and different regions of mantle tissues. Colored bars on the right indicate the presence of the SMPs in larval, adult nacre, and adult prism shells. The gene expression pattern is congruent with localization of corresponding SMPs in the shells.
We also analyzed expression of 21 C. gigas gene models listed in supplementary table S1, Supplementary Material online, amino acid sequences of which are significantly similar to P. fucata larval SMPs (supplementary fig. S2, Supplementary Material online). Among the 21 gene models, 12 genes are larval SMP genes. All of these larval SMP genes were expressed during trochophore and/or D-shaped stages, confirming their role in larval shell formation. In addition, seven gene models (CGI_10024592, CGI_10000503, CGI_10011145, CGI_10007856, CGI_10028014, CGI_10025645, and CGI_10021720) were expressed during larval shell-forming stages.
SMPs with VWA and CB Domains
Despite major differences between larval and adult SMPs, some functional domains, including VWA, CB, and CA domains, are common to both larval and adult SMPs in both species. Therefore, these functional domains are essential for shell formation. Thus, we further investigated the evolutionary origins of proteins having these conserved domains.
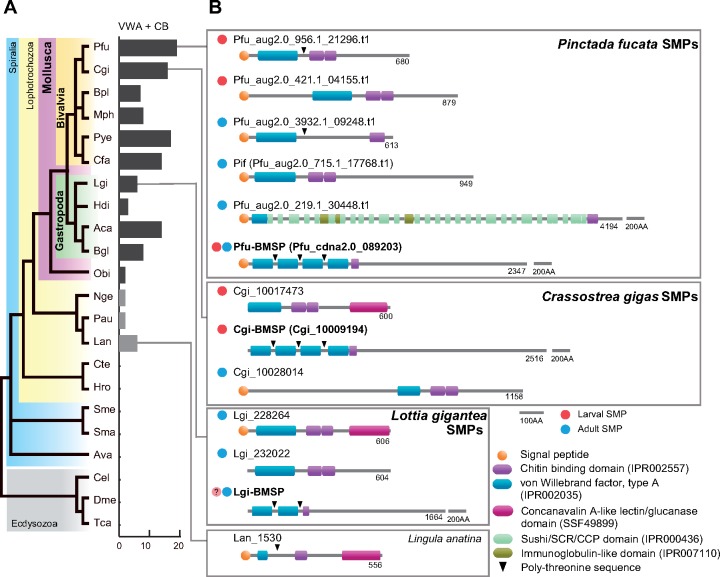
VWA and CB domains are common in diverse animal phyla (supplementary tables S2 and S3, Supplementary Material online). The VWA domain is widely present in extracellular matrix proteins such as collagen and integrin, and is thought to be involved in interactions between proteins that compose multiprotein complexes (Tuckwell 1999; Whittaker and Hynes 2002). The CB domain is well-characterized in arthropod chitinases and peritrophic matrix proteins (Elvin et al. 1996; Shen and Jacobs-Lorena 1999). Our whole genomic survey confirmed that genes encoding CB domain-containing proteins are significantly expanded in the Mollusca, as well as in the Arthropoda (supplementary table S2, Supplementary Material online). Although VWA and CB domains are widely distributed in many organisms, proteins having both domains are found almost exclusively in molluscs and other lophotrochozoans, such as the Nemertea, the Phoronida, and the Brachiopoda (Luo et al. 2015; 2018) (fig. 3A; supplementary table S2, Supplementary Material online). The VWA and CB domain-containing protein (hereafter VWA–CB dcp) family has expanded in the molluscan lineage. The P. fucata genome encodes a total of 19 VWA–CB dcp genes, and three of them (pfu_aug2.0_956.1_21296.t1, pfu_aug2.0_421.1_04155.t1, pfu_cdna2.0_089203) are found in larval SMPs (fig. 3B;supplementary data S1, Supplementary Material online). Notably, pfu_cdna2.0_089203 was also found in adult shell layers. In addition, three VWA–CB dcps including Pif (pfu_aug2.0_715.1_17768.t1), which was identified from the nacreous layer of P. fucata (Suzuki et al. 2009), were found in adult shells (fig. 3B). SMPs From the C. gigas larval shell proteome, two VWA–CB dcps (CGI_10017473 and CGI_10009194) were identified (fig. 3B;supplementary data S2, Supplementary Material online). Gene expression patterns of VWA–CB domain-containing SMPs are congruent with their localization in the shells. Larval VWA–CB SMP genes are expressed during larval stages, whereas adult VWA–CB genes are active in the adult mantle tissues (supplementary fig. S3, Supplementary Material online).
Fig. 3.
VWA and chitin-binding domain-containing protein (VWA–CB dcp) is essential for molluscan shell biomineralization. (A) Numbers of VWA–CB dcp genes in protostomian animal genomes. The phylogeny of Spiralia, with Ecdysozoa as an out group, is based on Luo et al. (2018). VWA–CB dcps are found only in Mollusca (purple) and Lophotrochozoa (yellow). (B) Conserved domain architecture of VWA–CB domain-containing SMPs in P. fucata, C. gigas, and L. gigantea. One VWA–CB dcp found in the brachiopod (L. anatina) genome is shown for comparison. Red and cyan circles on the left indicate larval and adult SMPs, respectively. BMSPs are found both in P. fucata and C. gigas larval proteomes. See supplementary table S3, Supplementary Material online for species name abbreviations.
We comprehensively searched for VWA–CB domain architecture among animal genomes and found that VWA–CB dcps with multiple VWA domains are exclusive to molluscs (supplementary figs. S4 and S5, Supplementary Material online). In bivalves, domain architecture typically includes four tandemly arranged VWA domains intercalated with poly-threonine sequences, followed by one or two CB domains (fig. 3; supplementary fig. S5, Supplementary Material online). This domain architecture was first identified in BMSP (blue mussel shell protein) from Mytilus galloprovincialis shells (Suzuki et al. 2011). Based on their highly conserved domain architecture and phylogenetic distribution, we conclude that genes encoding the BMSP domain architecture are orthologous, and denote the corresponding proteins as Pfu-BMSP (pfu_cdna2.0_089203) in P. fucata and Cgi-BMSP (CGI_10009194) in C. gigas, respectively. In addition, one SMP possessing the BMSP domain architecture was identified in Lottia shells (fig. 3; supplementary fig. S5, Supplementary Material online) (Marie et al. 2013). Notably, the Lottia BMSP transcript was expressed in the larval stage (Marie et al. 2013), implying that the BMSP is also involved in larval shell formation in gastropods.
Chitin is one of the major polysaccharides comprising the structural scaffold of larval and adult shells (Weiner and Traub 1984; Weiss and Schönitzer 2006; Suzuki et al. 2007). In the P. fucata genome, 14 genes encoding the chitin synthase domain (PF03142) were found (supplementary fig. S6, Supplementary Material online). One gene (pfu_cdna2.0_ 086716), which encodes a chitin synthase with a myosin head domain (Suzuki et al. 2007), is expressed both in D-shaped larva and in mantle tissue (supplementary fig. S6, Supplementary Material online). A chitin synthase gene (pfu_aug2.0_2.1_06699) begins to be expressed in early trochophore larvae, which form the shell field prior to shell crystallization, but is not expressed in the adult mantle. It is possible that genes responsible for chitin synthesis during larval shell formation may also be different from those of adults, as is the SMP repertoire.
Carbonic Anhydrases
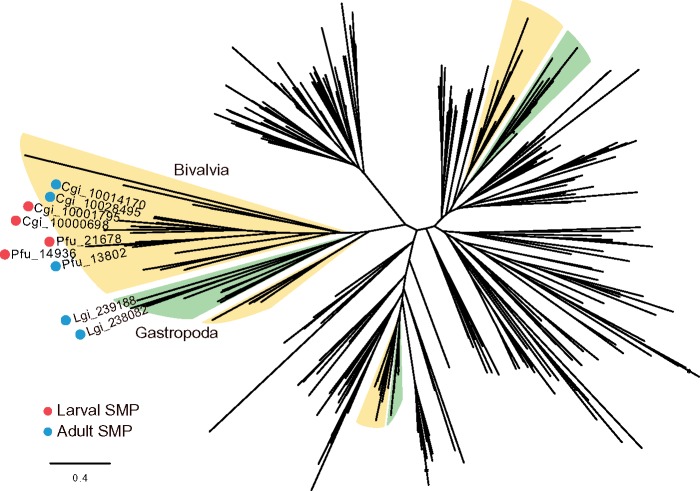
CA is considered one of the most important enzymes in CaCO3 biomineralization due to its ability to catalyze the hydration of carbon dioxide, CO2 +H2O → HCO3− + H+, which provides bicarbonate ions that react with Ca2+ to form CaCO3. In molluscs, Nacrein is an SMP containing a CA domain (Miyamoto et al. 1996). This protein has been identified in both nacreous and prismatic layers of pearl oysters (Miyamoto et al. 1996; Miyashita et al. 2002) and in various other mollusc shells (Miyamoto et al. 2003; Norizuki and Samata 2008; Marie et al. 2011; Mann et al. 2012). Two larval SMPs containing a CA domain were found in P. fucata (pfu_aug2.0_1294.1_14936.t1 and pfu_aug2.0_1536.1_21678.t1) and in C. gigas (CGI_10000698 and CGI_10001795), respectively (fig. 4; supplementary data S1 and S2, Supplementary Material online). Expression levels of genes encoding larval α-CA SMPs peak during the D-shape larval stage or before that, and are either reduced or disappear in the adult stage at the mantle, which is the main tissue in charge of the adult shell formation (fig. 2; supplementary fig. S7, Supplementary Material online). In contrast, expression of adult α-CA SMP (pfu_aug2.0_214.1_13802.t1) or Nacrein gene was not detected during larval stages, but increases abruptly during adult shell-forming tissues.
Fig. 4.
Molecular phylogeny of protostomian alpha carbonic anhydrase (α-CA) shows lineage-specific expansion of the gene family and its recruitment for shell formation in molluscs. Red and cyan circles indicate larval and adult SMPs, respectively. Colored branches indicate that the cluster is exclusively dominated by α-CA genes of specific taxonomic clades (yellow: Bivalvia, green: Gastropoda). We analyzed protein sequences containing CA domains and having more than 100 amino acids. The tree was constructed using the ML method with the LG model.
In order to test gene family expansion in molluscs, copy numbers of genes encoding α-CA in protostome animal genomes were investigated (supplementary fig. S8A and table S2, Supplementary Material online). The number of α-CA in molluscan genomes is 24.8 on average, ranging from 15 (Aplysia) to 40 (Modiolus), whereas the number in the other protostome genomes is much smaller (7.6 on average excluding some exceptions such as Helobdella, Adineta, and Drosophila) (supplementary fig. S8A and table S2, Supplementary Material online). The molecular phylogenetic tree constructed for protostomian α-CA showed that there are clusters consisting entirely of bivalve or gastropod genes (fig. 4; supplementary data S7, Supplementary Material online), indicating that lineage-specific expansions of the CA family occurred in bivalve and gastropod lineages. Larval and adult SMPs with an α-CA domain of P. fucata and C. gigas were included within a bivalve-specific cluster.
We noticed that in α-CA gene families, multiple homologs occur in larval shells in P. fucata and C. gigas (fig. 4). In addition, two α-CAs were reported from adult shell of the limpet, Lottia gigantea (Marie et al. 2013). We constructed a molecular phylogenetic tree of α-CAs from these three molluscan species and confirmed that α-CAs responsible for shell formation was independently duplicated in each species (supplementary fig. S8B, Supplementary Material online). This result is congruent with the fact that SMP genes are frequently duplicated in the P. fucata genome (Takeuchi, Koyanagi, et al. 2016).
Acidic Proteins
The existence of unusually acidic proteins has been confirmed by pioneering work on the organic matrix of molluscan shells (Hare 1963; Weiner and Hood 1975; Weiner 1979, 1983). Acidic amino acids have side chains that are negatively charged under physiological conditions. Acidic proteins, usually rich in aspartic and/or glutamic acid, bind calcium ions (Hare 1963; Weiner and Hood 1975; Takeuchi et al. 2008; Suzuki et al. 2009). EF-hand calcium-binding proteins were also considered to be involved in shell biomineralization (Huang et al. 2007). Until now, only a few acidic SMPs have been reported (Sarashina and Endo 1998, 2001; Tsukamoto et al. 2004; Gotliv et al. 2005; Suzuki et al. 2009; Marie et al. 2013), because of technical difficulties in isolating acidic proteins (Gotliv et al. 2003).
We found three novel proteins with extremely acidic natures (pI<4.5) in the larval shell of P. fucata (supplementary data S1, Supplementary Material online). SMPs encoded by gene models pfu_aug2.0_3272.1_09093 and pfu_aug2.0_2162.1_08762 are secreted proteins showing high aspartic acid content (19.0% and 18.6%, respectively) and low pIs (4.2 and 3.8, respectively). Another acidic SMP (pfu_aug2.0_3796.1_22455) is rich in aspartic acid (12.2%) and glutamic acid (18.5%) with a pI of 4.4, containing two calcium-binding EF-hand domains (IPR002048). Acidic proteins are also found in C. gigas larval shells (supplementary data S2, Supplementary Material online). The most acidic protein is CGI_10012241, with a predicted pI of 4.3, carrying calcium-binding EF-hand_5 domains (PF13202). Two acidic SMPs (CGI_10005282 and CGI_10022165) were also found. These acidic SMPs showed no sequence similarity to known protein sequences in public databases (nr and UniProt). Their actual functions in biomineralization should be tested by further experiments.
Discussion
Function of Larval SMPs
Unlike adult shells, with diverse shell CaCO3 polymorphs and microstructure, larval molluscan shells all contain aragonite and similar microstructures (Iwata 1980; Waller 1981; Eyster 1982, 1986; Weiss et al. 2002). Shell proteomes and gene expression profiles showed that the gene set for larval shell formation is totally different from that in adults.
By comparing larval SMP repertoires of two bivalves, P. fucata and C. gigas, which diverged around 240–330 Ma (Simakov et al. 2015; Sun et al. 2017; Luo et al. 2018), we found that 12 larval SMPs of the two species showed significant sequence similarity (supplementary table S1, Supplementary Material online). Conserved larval SMPs include VWA–CB domain-containing proteins, CAs, and acidic proteins (fig. 1; supplementary table S1, Supplementary Material online). Conservation of functional domains among SMPs indicates their fundamental role in shell formation. VWA–CB dcps may interact with chitin and other SMPs, such as acidic proteins to organize the organic shell matrix complex (Suzuki et al. 2009; 2011). CAs supply bicarbonate ions necessary for CaCO3 crystallization (Miyamoto et al. 1996), and acidic proteins interact with the crystal phase to regulate the crystallization processes (Addadi and Weiner 1985; Gotliv et al. 2003). Conserved larval SMPs also include 6 SMPs without specific functional domains (supplementary fig. S1, Supplementary Material online). To understand their role in larval shell formation, in vivo functional analysis, such as gene knockdown experiments, is necessary.
In contrast to the shared 12 SMPs, the other 19 larval SMPs of P. fucata do not have counterparts among C. gigas larval SMPs. One explanation of the incongruity between P. fucata and C. gigas larval SMP repertoires is a technical issue. SMP identification using proteomic analysis is not comprehensive and the present SMP list may not include all larval SMPs. For example, different extraction and preparation methods may isolate different SMPs. In our case, only proteins with trypsin cleavage sites can be detected. Notably, seven proteins of C. gigas, with sequences significantly similar to those of P. fucata larval SMPs, were not identified in the C. gigas larval shell proteome (supplementary table S1, Supplementary Material online). However, the genes were expressed during D-shaped larval stages in C. gigas (supplementary fig. S2, Supplementary Material online). This implies that the gene products are actually larval SMPs of C. gigas, but not identified in the proteome for technical reasons. In order to develop a complete list of larval SMPs, more experiments using different protein extraction protocols and broader taxonomic sampling are necessary.
A previous study demonstrated that expression of six genes responsible for adult shell formation was absent or very weak before metamorphosis in P. fucata, implying these adult SMP genes do not participate in larval shell formation (Miyazaki et al. 2010). Our RNA-seq data further confirmed that gene expression profiles during larval and adult shell formation are distinct (fig. 2). There are two expression patterns of larval SMP genes. About half of larval SMP genes begin to be expressed during early trochophore stages, and the other half of SMP genes are expressed only at later D-shaped larval stages, indicating their different roles in larval shell formation. In the early trochophore stage, the shell field appears and organic materials extend to cover the entire larva prior to crystallization of the shell (Mouëza et al. 2006; Wassnig and Southgate 2012). The SMPs provide the framework for the shell structure. SMP genes, which exhibit strong expression during D-shaped larval stages, participate in shell mineralization and accumulation processes.
In C. gigas, one-third of SMP genes are expressed not only in larvae, but also in adult mantle tissues (supplementary fig. S1, Supplementary Material online). These genes encode cellular proteins such as elongation factor alpha and ribosomal proteins (supplementary data S2, Supplementary Material online). One explanation of this result is that cells were included in the larval shells. A previous study also reported house-keeping proteins in C. gigas adult shells (Zhang et al. 2012). These cellular proteins may be derived from hemocytes, which generate CaCO3 crystals during shell regeneration (Mount et al. 2004) and the cells are eventually incorporated into the shell. However, we do not have enough evidence of cell-mediated biomineralization in larval shell formation. We cannot exclude the possibility of soft tissue contamination despite intensive cleaning of the C. gigas samples. Contamination might also explain the difference in number of larval SMPs between P. fucata and C. gigas (31 and 111).
Evolutionary Origin of Molluscan Shells
Comparative genomics of a wide range of animals revealed that VWA–CB dcp and CA gene families expanded in the common ancestor of the Mollusca, and some members of the families were utilized to form larval and adult shells in P. fucata and C. gigas. In addition, these proteins are also present in gastropod SMPs, implying their essential role in mollusc shell formation.
A genome-wide survey showed that the VWA–CB domain architecture appeared in the common ancestor of the Mollusca and Brachiopoda (fig. 3; supplementary fig. S4, Supplementary Material online). Brachiopods are other lophotrochozoans with mineralized shells. In the Lingula anatina genome, one gene (Lan_1530) encodes a VWA and a CB domain with a poly-threonine sequence between them (fig. 3; supplementary fig. S4, Supplementary Material online). This protein also carries a concanavalin A-like lectin/glucanase domain at the C-terminus. This domain or a similar sequence is also found in some SMPs in C. gigas, L. gigantea, and P. fucata (fig. 3). The conserved domain architecture with VWA, CB, and concanavalin A-like lectin/glucanase domains and a poly-threonine sequence may represent a “prototype” of VWA–CB dcps.
In the Lingula genome, six VWA–CB dcp genes were found (supplementary fig. S4 and table S2, Supplementary Material online). None of the VWA–CB dcps were found in the Lingula shell proteome (Luo et al. 2015). In addition, proteomic studies for rhynchonelliformean brachiopod shells reported no VWA–CB domain-containing SMPs (Immel et al. 2015; Isowa et al. 2015; Jackson et al. 2015). Therefore, VWA–CB dcps are not related to shell formation in brachiopods. Furthermore, gene expression profiles of all VWA–CB dcp genes in P. fucata show that half of VWA–CB dcp genes were not expressed in shell-forming tissues (supplementary fig. S3, Supplementary Material online), implying that their functions are not related to shell formation. These results are congruent with the hypothesis that the ancestral function of the VWA–CB dcp gene family was something other than shell biomineralization. In summary, the VWA–CB dcp gene family appeared before the split of the Mollusca and Brachiopoda. Then the gene family was specifically expanded and co-opted for shell formation in the molluscan lineage.
Chitin is often found in conchiferan shells, as well as in aculiferan shell plates, sclerites, and cuticles (Peters 1972; Furuhashi et al. 2009). Notably, unmineralized sclerites of Wiwaxia, a stem group of Mollusca, is likely chitinous (Vinther 2015). It is possible that a chitinous element on the body surface was a precursor of mineralized shell. VWA–CB dcps secreted from the mantle tissue can bind chitin and other SMPs to form organic matrix that provide scaffold of the shells. Particularly, BMSP homologs are found only in molluscan genomes (fig. 3; supplementary figs. S3 and S4, Supplementary Material online). It is hypothesized that the BMSP gene is crucial to shell biomineralization in the molluscan lineage. To test this hypothesis, it is essential to investigate this gene in other molluscs, such as the Monoplacophora and Polyplacophora, and to perform functional analyses of the gene in vivo.
Conclusion
We extracted molluscan larval SMPs and effectively identified a considerable number of proteins involved in biomineralization. Larval and adult SMP repertoires are unambiguously distinguishable. Substantial differences highlight conserved components between larval and adult SMP repertoires, including VWA–CB dcps, α-CAs, and acidic proteins, which may play crucial roles in shell formation. Larval and adult SMPs within the same gene family that encode the same functional domains, follow stage-specific expression patterns; that is, one SMP gene is expressed during larval stages and another in adult stages. Furthermore, a comprehensive genomic survey revealed that the VWA–CB dcp and α-CA families became expanded in the molluscan lineage and that some members of these families were co-opted for shell biomineralization. By identifying novel SMP repertoires in larval shells, we hope to better understand molluscan shell formation. Functional analyses of SMPs, for example, in vivo gene manipulation experiments, will help illuminate the process.
Materials and Methods
Protein Extraction
Twenty-four-hour D-shaped larvae of P. fucata and C. gigas were gifts from Mikomoto Pearl Research Institute and Nagasaki Prefectural Institute of Fisheries, respectively. The following shell cleaning and protein extraction method was the same for both species. Three milliliters of larvae were collected in a 50-ml tube and incubated with 1 M sodium NaOH overnight. Shells were washed with Milli-Q water five times and were observed with a stereo microscope. The cleaning process was repeated until soft tissues and contaminants were completely removed. Then cleaned shells were decalcified in 1 M acetic acid. The solution was centrifuged at 4,000 × g for 30 min to separate the supernatant and the pellet. The insoluble pellet was rinsed with Milli-Q water three times and was lyophilized as an acid-insoluble matrix (AIM). The acid-soluble matrix (ASM) dissolved in the supernatant was recovered by methanol/chloroform precipitation method as previously described (Takeuchi, Yamada, et al. 2016).
Adult shells of P. fucata were provided by Mikimoto Pearl Research Institute. Shells were incubated in 1% NaOCl for 24 h and mechanically washed to remove superficial epibionts and periostracum. The outer prismatic and inner nacreous layers were separated and finely crushed. Ten grams of each shell layer were decalcified in 1 M acetic acid overnight. Afterward, acid soluble and insoluble matrices were obtained, as mentioned above.
Liquid Chromatography–Tandem Mass Spectrometry
AIM and ASM were suspended in solubilization buffer (1% SDS, 10 mM DTT, 50 mM Tris–HCl, pH 8.0) and used for SDS-PAGE in 10–20% gradient gels. Gel portions that contained proteins were excised and used for further analysis.
For P. fucata shells, proteomic analysis was conducted as described by Yamada et al. (2009). Gels were digested with trypsin and analyzed by LC/MS/MS using a capillary liquid chromatography system (Ultima3000; DIONEX) connected online to a mass spectrometer (LTQ-XL, ThermoScientific). Raw spectra were processed using SEQUEST software to extract peak lists (Araki et al. 2012), which were analyzed using an in-house MASCOT (ver. 2.3.2) server against P. fucata gene models (Takeuchi, Koyanagi, et al. 2016). The false discovery rate was set to 0.05. Proteins supported by at least two unique peptide sequences were kept for further analyses.
For C. gigas shells, excised gel bands were subjected to reduction/alkylation with dithiothreitol and iodoacetamide, respectively, followed by trypsin digestion overnight at 37°C. Peptides were extracted from gels using 5% formic acid and 50% acetonitrile in water. After extraction, peptides were concentrated in a Genevac EZ-2 Elite speed vacuum concentrator, and then resuspended in 0.1% formic acid in water for LC/MS analysis. A 5 μl of each sample was injected into a Dionex Ultimate 3000 nano-UPLC system in tandem with a Thermo Q-Exactive Plus Mass Spectrometer, acquiring MS1 and MS2 spectra of the 10 most intense peaks. Peptides were separated on a Zorbax 300SB-C18 (0.3× 150 mm; Agilent) column at 40°C, with a flow rate of 3 µl/min using a 90-min gradient. Acquired MS/MS spectra were subjected to database searches against protein sequences of C. gigas gene models (7) and a transcriptome assembly of C. gigas D-shaped larva, described below, complemented with the common Repository of Adventitious Proteins (cRAP; http://www.thegpm.org/crap/; last accessed on May 8, 2017) database, using Proteome Discoverer software v1.4 (ThermoFisher Scientific)—SEQUEST HT algorithm. Results were filtered with a 0.1% false discovery rate cutoff at protein level. Spectra not assigned by SEQUEST HT were analyzed using PEAKs Studio 7 with de novo sequencing first and then refined with database-assisted searches for alignment.
Sequence Analysis of SMPs
BLASTP searches of the UniProtKB/Swiss-Prot database (https://blast.ncbi.nlm.nih.gov/Blast.cgi; last accessed on December 22, 2017) were performed using default settings. Functional and low-complexity domains were identified using InterProScan 5.27 platform (Jones et al. 2014) and SMART online service (http://smart.embl-heidelberg.de; last accessed on December 22, 2017). Signal peptide prediction employed SignalP 4.0 (Petersen et al. 2011), and transmembrane domains were assessed with TMHMM 2.0 (Krogh et al. 2001) software. Molecular masses and isoelectric points (pIs) of sequences were predicted using the ExPASy ProtParam tool (https://web.expasy.org/protparam/; last accessed on December 22, 2017). All of these programs were run using default settings and thresholds. Amino acid sequences of larval SMPs of P. fucata were searched against translated sequences of C. gigas gene models (Zhang et al. 2012) using BLASTP with default settings except E-value threshold as ≤1e−5.
Transcriptome Analysis
Total RNA of P. fucata was extracted from adult mantle tissues and 12 developmental stages, including eggs, two-cell embryo, four-cell embryo, early morula, blastula, 11-h (hour after fertilization) trochophore, 13-h trochophore, 21-h D-shaped larva, 24-h D-shaped larva, 55-h D-shaped larva, 69-h D-shaped larva, and 96-h D-shaped larva using Trizol reagent (Chomczynski and Sacchi 1987). RNA-seq libraries were prepared using a TruSeq RNA sample Prep Kit v2 (Illumina) and sequenced with the Illumina GAIIx platform. Raw sequences were quality filtered and trimmed with Trimmomatic 0.36 (Bolger et al. 2014), and then mapped to P. fucata gene models (Takeuchi, Koyanagi, et al. 2016) using Bowtie2 (Langmead and Salzberg 2012) with default parameters. For gene expression analysis, transcripts per kilobase million (TPM) were calculated using eXpress 1.5.1 (Roberts and Pachter 2012). By the same means, an RNA-seq library of 24-h D-shaped larvae of C. gigas was also prepared and sequenced with Illumina MiSeq. The transcriptome was assembled with Trinity 2.2.1 (Grabherr et al. 2011). For gene expression analysis of C. gigas, RNA-seq data were retrieved from GigaDB (http://gigadb.org/; last accessed on April 12, 2017) (Zhang et al. 2012) and TPM was estimated as mentioned above.
Phylogenetic Analysis
Protein sequences from diverse metazoans were retrieved from public databases as described in supplementary table S3, Supplementary Material online. Multiple sequence alignments of amino acid sequences were made using MAFFT 7.271 (Katoh and Standley 2013) with default parameters. Phylogenetic analysis was conducted using the maximum likelihood (ML) method in RAxML 8.2.4 (Stamatakis 2014). Reliability of the topology was checked by bootstrap analysis on the basis of 100 replicates. The trees were visualized with Jalview version 2.8.2 (Waterhouse et al. 2009).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Steven D. Aird for editing the manuscript. This work was supported by JSPS (Japan Society for the Promotion of Science) KAKENHI (grant number 23780209 to T.T.) and by internal funds from the Okinawa Institute of Science and Technology (OIST).
Authors’ Contributions
R.Z., T.T., and K.E. conceived and designed the experiments. R.Z., T.T., Y.J.L., A.I., T.K., R.K., A.V.B., and L.Y. performed experiments and analyzed data. H.S., I.S., K.N., N.S., and K.E. contributed reagents and materials. R.Z., T.T., and K.E. wrote the paper.
References
- Addadi L, Joester D, Nudelman F, Weiner S.. 2006. Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chem Eur J. 12(4): 980–987. [DOI] [PubMed] [Google Scholar]
- Addadi L, Weiner S.. 1985. Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc Natl Acad Sci U S A. 82(12): 4110–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagarswami K, Dharmaraj S, Velayudhan TS, Chellam A, Victor ACC, Gandhi AD.. 1983. Larval rearing and production of spat of pearl oyster Pinctada fucata (Gould). Aquaculture 34(3–4): 287–301. [Google Scholar]
- Araki Y, Shimizu HD, Saeki K, Okamoto M, Yamada L, Ishida K, Sawada H, Urushihara H.. 2012. A surface glycoprotein indispensable for gamete fusion in the social amoeba Dictyostelium discoideum. Eukaryotic Cell 11(5): 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30(15): 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JG. 1990. Skeletal biomineralization: Atlas and Index. New York: Van Nostrand Reinhold Company. [Google Scholar]
- Chomczynski P, Sacchi N.. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 162(1): 156–159. [DOI] [PubMed] [Google Scholar]
- Doroudi MS, Southgate PC.. 2003. Embryonic and larval development of Pinctada margaritifera (Linnaeus, 1758). Molluscan Res. 23(2): 101–107. [Google Scholar]
- Elvin CM, Vuocolo T, Pearson RD, East IJ, Riding GA, Eisemann CH, Tellam RL.. 1996. Characterization of a major peritrophic membrane protein, Peritrophin-44, from the larvae of Lucilia cuprina: cDNA and deduced amino acid sequences. J Biol Chem. 271(15): 8925–8935. [DOI] [PubMed] [Google Scholar]
- Eyster LS. 1982. Embryonic shell formation in the Nudibranch Aeolidia papillosa. Am Zool. 22:981. [DOI] [PubMed] [Google Scholar]
- Eyster LS. 1983. Ultrastructure of early embryonic shell formation in the opisthobranch gastropod Aeolidia papillosa. Biol Bull. 165(2): 394–408. [DOI] [PubMed] [Google Scholar]
- Eyster LS. 1986. Shell inorganic composition and onset of shell mineralization during bivalve and gastropod embryogenesis. Biol Bull. 170(2): 211–231. [Google Scholar]
- Furuhashi T, Schwarzinger C, Miksik I, Smrz M, Beran A.. 2009. Molluscan shell evolution with review of shell calcification hypothesis. Comp Biochem Physiol B: Biochem Mol Biol. 154(3): 351–371. [DOI] [PubMed] [Google Scholar]
- Gosling E. 2015. Marine bivalve molluscs. John Wiley & Sons. [Google Scholar]
- Gotliv B-A, Addadi L, Weiner S.. 2003. Mollusk shell acidic proteins: in search of individual functions. Chembiochem 4(6): 522–529. [DOI] [PubMed] [Google Scholar]
- Gotliv BA, Kessler N, Sumerel JL, Morse DE, Tuross N, Addadi L, Weiner S.. 2005. Asprich: a novel aspartic acid-rich protein family from the prismatic shell matrix of the bivalve Atrina rigida. Chembiochem 6(2): 304–314. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 29(7): 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PE. 1963. Amino acids in proteins from aragonite and calcite in shells of Mytilus californianus. Science 139(3551): 216–217. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang C, Ma Z, Xie L, Zhang R.. 2007. A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim Biophys Acta 1770(7): 1037–1044. [DOI] [PubMed] [Google Scholar]
- Immel F, Gaspard D, Marie A, Guichard N, Cusack M, Marin F.. 2015. Shell proteome of rhynchonelliform brachiopods. J Struct Biol. 190(3): 360–366. [DOI] [PubMed] [Google Scholar]
- Isowa Y, Sarashina I, Oshima K, Kito K, Hattori M, Endo K.. 2015. Proteome analysis of shell matrix proteins in the brachiopod Laqueus rubellus. Proteome Sci. 13(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata K. 1980. Mineralization and architecture of the larval shell of Haliotis discus hannai Ino (Archaeogastropoda). J Fac Sci. 19:305–320. [Google Scholar]
- Jackson DJ, Mann K, Häussermann V, Schilhabel MB, Lüter C, Griesshaber E, Schmahl W, Wörheide G.. 2015. The Magellania venosa biomineralizing proteome: a window into brachiopod shell evolution. Genome Biol Evol. 7(5): 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, McDougall C, Woodcroft B, Moase P, Rose RA, Kube M, Reinhardt R, Rokhsar DS, Montagnani C, Joubert C, et al. 2010. Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol. 27(3): 591–608. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9): 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakoi S, Kin K, Miyazaki K, Wada H.. 2008. Early Development of the Japanese Spiny Oyster (Saccostrea kegaki): characterization of Some Genetic Markers. Zool Sci. 25(5): 455–464. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in Performance and Usability. Mol Biol Evol. 30(4): 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Suzuki T, Weiss KM.. 2004. Genetic basis for the evolution of vertebrate mineralized tissue. Proc Natl Acad Sci U S A. 101(31): 11356–11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian CE, Wilt FH.. 2008. Molecular aspects of biomineralization of the echinoderm endoskeleton. Chem Rev. 108(11): 4463–4474. [DOI] [PubMed] [Google Scholar]
- Kobayashi I. 1969. Internal microstructure of the shell of bivalve molluscs. Am Zool. 9(3): 663–672. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3): 567–580. [DOI] [PubMed] [Google Scholar]
- LaBarbera M. 1974. Calcification of the first larval shell of Tridacna squamosa (Tridacnidae: bivalvia). Mar Biol. 25(3): 233–238. [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Method 9(4): 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li S, Kong J, Liu Y, Wang T, Xie L, Zhang R.. 2015. In-depth proteomic analysis of shell matrix proteins of Pinctada fucata. Sci Rep. 5:17269.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstam HAWS. 1989. On biomineralization. New York: Oxford University Press. [Google Scholar]
- Luo Y-J, Kanda M, Koyanagi R, Hisata K, Akiyama T, Sakamoto H, Sakamoto T, Satoh N.. 2018. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat Ecol Evol. 2(1): 141–151. [DOI] [PubMed] [Google Scholar]
- Luo Y-J, Takeuchi T, Koyanagi R, Yamada L, Kanda M, Khalturina M, Fujie M, Yamasaki S-i, Endo K, Satoh N.. 2015. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun. 6(1): 8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Edsinger E.. 2014. The Lottia gigantea shell matrix proteome: re-analysis including MaxQuant iBAQ quantitation and phosphoproteome analysis. Proteome Sci. 12:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Edsinger-Gonzales E, Mann M.. 2012. In-depth proteomic analysis of a mollusc shell: acid-soluble and acid-insoluble matrix of the limpet Lottia gigantea. Proteome Sci. 10(1): 28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Jackson DJ.. 2014. Characterization of the pigmented shell-forming proteome of the common grove snail Cepaea nemoralis. BMC Genomics 15:249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Che L, Golubic S, Le Campion-Alsumard T, Payri C.. 2001. Developmental aspects of biomineralisation in the Polynesian pearl oyster Pinctada margaritifera var. cumingii. Oceanol Acta 24:37–49. [Google Scholar]
- Marie B, Arivalagan J, Mathéron L, Bolbach G, Berland S, Marie A, Marin F.. 2017. Deep conservation of bivalve nacre proteins highlighted by shell matrix proteomics of the Unionoida Elliptio complanata and Villosa lienosa. J R Soc Interface 14(126): 20160846.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Jackson DJ, Ramos-Silva P, Zanella-Cléon I, Guichard N, Marin F.. 2013. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage-specific novelties. FEBS J. 280(1): 214–232. [DOI] [PubMed] [Google Scholar]
- Marie B, Joubert C, Tayalé A, Zanella-Cléon I, Belliard C, Piquemal D, Cochennec-Laureau N, Marin F, Gueguen Y, Montagnani C.. 2012. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci. 109(51): 20986–20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Le Roy N, Zanella-Cléon I, Becchi M, Marin F.. 2011. Molecular evolution of mollusc shell proteins: insights from proteomic analysis of the edible mussel Mytilus. J Mol Evol. 72(5–6): 531–546. [DOI] [PubMed] [Google Scholar]
- Marin F, Luquet G, Marie B, Medakovic D.. 2008. Molluscan shell proteins: primary structure, origin, and evolution. Curr Top Dev Biol. 80:209–276. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Miyashita T, Okushima M, Nakano S, Morita T, Matsushiro A.. 1996. A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci. 93(18): 9657–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H, Yano M, Miyashita T.. 2003. Similarities in the structure of nacrein, the shell-matrix protein, in a bivalve and a gastropod. J Molluscan Stud. 69(1): 87–89. [Google Scholar]
- Miyashita T, Takagi R, Miyamoto H, Matsushiro A.. 2002. Identical carbonic anhydrase contributes to nacreous or prismatic layer formation in Pinctada fucata (Mollusca: bivalvia). Veliger 45:250–255. [Google Scholar]
- Miyazaki Y, Nishida T, Aoki H, Samata T.. 2010. Expression of genes responsible for biomineralization of Pinctada fucata during development. Comp Biochem Physiol B: Biochem Mol Biol. 155(3): 241–248. [DOI] [PubMed] [Google Scholar]
- Mouëza M, Gros O, Frenkiel L.. 2006. Embryonic development and shell differentiation in Chione cancellata (Bivalvia, Veneridae): an ultrastructural analysis. Invertebr Biol. 125(1): 21–33. [Google Scholar]
- Mount AS, Wheeler AP, Paradkar RP, Snider D.. 2004. Hemocyte-mediated shell mineralization in the eastern oyster. Science 304(5668): 297.. [DOI] [PubMed] [Google Scholar]
- Norizuki M, Samata T.. 2008. Distribution and function of the Nacrein-related proteins inferred from structural analysis. Mar Biotechnol. 10(3): 234–241. [DOI] [PubMed] [Google Scholar]
- Peters W. 1972. Occurrence of chitin in Mollusca. Comp Biochem Physiol B: Biochem Mol Biol. 41(3): 541–550. [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10): 785–786. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Cameron RA, Davidson EH.. 1997. Set-aside cells in maximal indirect development: evolutionary and developmental significance. Bioessays 19(7): 623–631. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pachter L.. 2012. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10(1): 71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RA, Baker SB.. 1994. Larval and spat culture of the Western Australian silver- or goldlip pearl oyster, Pinctada maxima Jameson (Mollusca: pteriidae). Aquaculture 126(1–2): 35–50. [Google Scholar]
- Sarashina I, Endo K.. 1998. Primary structure of a soluble matrix protein of scallop shell; implications for calcium carbonate biomineralization. Am Mineral 83(11–12 Part 2): 1510–1515. [Google Scholar]
- Sarashina I, Endo K.. 2001. The complete primary structure of molluscan shell protein 1 (MSP-1), an acidic glycoprotein in the shell matrix of the scallop Patinopecten yessoensis. Mar Biotechnol. 3(4): 362–369. [DOI] [PubMed] [Google Scholar]
- Shen Z, Jacobs-Lorena M.. 1999. Evolution of chitin-binding proteins in invertebrates. J Mol Evol. 48(3): 341–347. [DOI] [PubMed] [Google Scholar]
- Simakov O, Kawashima T, Marlétaz F, Jenkins J, Koyanagi R, Mitros T, Hisata K, Bredeson J, Shoguchi E, Gyoja F, et al. 2015. Hemichordate genomes and deuterostome origins. Nature 527(7579): 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkiss K, Wilbur KM.. 2012. Biomineralization. New York: Elsevier. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang Y, Xu T, Zhang Y, Mu H, Zhang Y, Lan Y, Fields CJ, Hui JHL, Zhang W, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 1(5): 0121. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Iwashima A, Tsutsui N, Ohira T, Kogure T, Nagasawa H.. 2011. Identification and characterisation of a calcium carbonate-binding protein, blue mussel shell protein (BMSP), from the nacreous layer. Chembiochem 12(16): 2478–2487. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Sakuda S, Nagasawa H.. 2007. Identification of chitin in the prismatic layer of the shell and a chitin synthase gene from the Japanese pearl oyster, Pinctada fucata. Biosci Biotechnol Biochem. 71(7): 1735–1744. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Saruwatari K, Kogure T, Yamamoto Y, Nishimura T, Kato T, Nagasawa H.. 2009. An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 325(5946): 1388–1390. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, Ikuta T, Shoguchi E, Fujiwara M, Shinzato C, Hisata K, et al. 2012. Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res. 19(2): 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Koyanagi R, Gyoja F, Kanda M, Hisata K, Fujie M, Goto H, Yamasaki S, Nagai K, Morino Y, et al. 2016. Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zool Lett. 2(1): 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Sarashina I, Iijima M, Endo K.. 2008. In vitro regulation of CaCO(3) crystal polymorphism by the highly acidic molluscan shell protein Aspein. FEBS Lett. 582(5): 591–596. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N.. 2016. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS One 11(6): e0156424.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JD. 1973. The structural evolution of the bivalve shell. Palaeontology 16:519–534. [Google Scholar]
- Tsukamoto D, Sarashina I, Endo K.. 2004. Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem Biophys Res Commun. 320(4): 1175–1180. [DOI] [PubMed] [Google Scholar]
- Tuckwell D. 1999. Evolution of von Willebrand factor A (VWA) domains. Biochem Soc Trans. 27(6): 835–840. [DOI] [PubMed] [Google Scholar]
- Vinther J. 2015. The origins of molluscs. Palaeontology 58:19–34. [Google Scholar]
- Waller TR. 1981. Functional morphology and development of veliger larvae of the European oyster, Ostrea edulis Linné. Smithsonian Contrib Zool. 328(328): 1–70. [Google Scholar]
- Wassnig M, Southgate PC.. 2012. Embryonic and larval development of Pteria penguin (Röding, 1798) (Bivalvia: pteriidae). J Molluscan Stud. 78(1): 134–141. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9): 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner S. 1979. Aspartic acid-rich proteins: major components of the soluble organic matrix of mollusk shells. Calcif Tissue Int. 29(2): 163–167. [DOI] [PubMed] [Google Scholar]
- Weiner S. 1983. Mollusk shell formation: isolation of two organic matrix proteins associated with calcite deposition in the bivalve Mytilus californianus. Biochemistry 22(17): 4139–4145. [Google Scholar]
- Weiner S, Hood L.. 1975. Soluble protein of the organic matrix of mollusk shells: a potential template for shell formation. Science 190(4218): 987–989. [DOI] [PubMed] [Google Scholar]
- Weiner S, Traub W.. 1984. Macromolecules in mollusc shells and their functions in biomineralization. Philos Trans R Soc Lond B: Biol Sci. 304:425–434. [Google Scholar]
- Weiss IM, Schönitzer V.. 2006. The distribution of chitin in larval shells of the bivalve mollusk Mytilus galloprovincialis. J Struct Biol. 153(3): 264–277. [DOI] [PubMed] [Google Scholar]
- Weiss IM, Tuross N, Addadi L, Weiner S.. 2002. Mollusc larval shell formation: amorphous calcium carbonate is a precursor phase for aragonite. J Exp Zool. 293(5): 478–491. [DOI] [PubMed] [Google Scholar]
- Werner B. 1939. Über die Entwicklung und Artunterscheidung von Muschellarven des Nordseeplanktons, unter besuderer Berucksichtigung der Schalen-entwicklung. Zool Jahrbucher 66:1–54. [Google Scholar]
- Whittaker CA, Hynes RO.. 2002. Distribution and evolution of von Willebrand/integrin a domains: widely dispersed adhesion and elsewhere. Mol Biol Cell 13(10): 3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada L, Saito T, Taniguchi H, Sawada H, Harada Y.. 2009. Comprehensive egg coat proteome of the ascidian Ciona intestinalis reveals gamete recognition molecules involved in self-sterility. J Biol Chem. 284(14): 9402–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418): 49–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.