Abstract
The oxymonad Monocercomonoides exilis was recently reported to be the first eukaryote that has completely lost the mitochondrial compartment. It was proposed that an important prerequisite for such a radical evolutionary step was the acquisition of the SUF Fe–S cluster assembly pathway from prokaryotes, making the mitochondrial ISC pathway dispensable. We have investigated genomic and transcriptomic data from six oxymonad species and their relatives, composing the group Preaxostyla (Metamonada, Excavata), for the presence and absence of enzymes involved in Fe–S cluster biosynthesis. None possesses enzymes of mitochondrial ISC pathway and all apparently possess the SUF pathway, composed of SufB, C, D, S, and U proteins, altogether suggesting that the transition from ISC to SUF preceded their last common ancestor. Interestingly, we observed that SufDSU were fused in all three oxymonad genomes, and in the genome of Paratrimastix pyriformis. The donor of the SUF genes is not clear from phylogenetic analyses, but the enzyme composition of the pathway and the presence of SufDSU fusion suggests Firmicutes, Thermotogae, Spirochaetes, Proteobacteria, or Chloroflexi as donors. The inventory of the downstream CIA pathway enzymes is consistent with that of closely related species that retain ISC, indicating that the switch from ISC to SUF did not markedly affect the downstream process of maturation of cytosolic and nuclear Fe–S proteins.
Keywords: Preaxostyla, SUF, amitochondriate, CIA, oxymonads
Iron–sulfur clusters are small inorganic prosthetic groups, which are among the most ancient and versatile cofactors. Their main function is mediating electron transport, which makes them a key part of many important processes such as photosynthesis, respiration, DNA replication and repair, and regulation of gene expression (Rudolf et al. 2006; Fuss et al. 2015; Paul and Lill 2015).
There are three pathways for the Fe–S clusters synthesis known in prokaryotes—NIF (nitrogen fixation), ISC (iron sulfur cluster), and SUF (sulfur utilization factor). The basic process of the Fe–S cluster biogenesis is similar in all three (Roche et al. 2013·). Sulfur (S2–) is provided by cysteine desulfurase (NifS, IscS, SufS). The source of iron (Fe2+) is unclear, however, for the mitochondrial ISC pathway frataxin is expected to be the provider (Pastore and Puccio 2013; Yoon et al. 2015). The sulfur and iron ions are first combined into a cluster on a scaffold protein (NifU, IscU, SufB–SufD complex), from which the cluster is transferred onto an apoprotein.
In eukaryotic cells, three compartments have distinct pathways for Fe–S cluster synthesis. Mitochondria typically use the ISC pathway, which was inherited from the alphaproteobacterial endosymbiont (Tachezy et al. 2001; Braymer and Lill 2017). This holds also for most mitochondrion-related organelles including the mitosomes of Giardia intestinalis (Tovar et al. 2003) and microsporidia (Katinka et al. 2001; Goldberg et al 2008), and hydrogenosomes of Trichomonas vaginalis (Sutak et al. 2004). Exceptions to this rule are found in mitochondrion-related organelles of Pygsuia biforma, Mastigamoeba balamuthi, and Entamoeba histolytica (Ali et al. 2004; van der Giezen et al. 2004; Mi-ichi et al. 2009; Nyvltova et al. 2013; Stairs et al. 2014), which contain SUF, NIF, or possibly none of these pathways, respectively. Eukaryotic plastids contain the SUF pathway, which was inherited from the cyanobacterial ancestor (Balk and Pilon 2011).
In the eukaryotic cytosol, the Fe–S cluster-containing proteins are formed by a cytosolic iron–sulfur cluster assembly (CIA), which is also responsible for maturation of nuclear Fe–S proteins. In yeast and human, the pathway contains at least eleven essential proteins (Sharma et al. 2010; Netz et al. 2014; Lill et al. 2015). CIA is unique to eukaryotes and most of its components do not have prokaryotic homologs, apart from Nbp35 (Boyd et al. 2009) and Cia2 (Tsaousis et al. 2014). It has been experimentally shown that the mitochondrial ISC pathway is necessary for the function of the CIA, probably because it synthesizes and transports an uncharacterized sulfur containing precursor to the cytosol (Kispal et al. 1999; Gerber et al. 2004; Biederbick et al. 2006; Pondarré et al. 2006). Dependency on ISC is interpreted as a major reason for the retention of mitochondrion-related organelles in anaerobic eukaryotes (Williams et al. 2002). Maturation of the cytosolic Fe–S proteins by the CIA pathway starts with the formation of [4Fe–4S] cluster on the Cfd1-Nbp35 scaffold (Hausmann et al. 2005; Netz et al. 2012), transfer of electrons from NADPH is mediated by Dre2 and diflavin reductase Tah18 is required for this process (Zhang et al. 2008; Netz et al. 2010). The [4Fe–4S] cluster from Cfd1-Nbp35 is transferred to a target apoprotein by Nar1 and the late-acting CIA components Cia1, Cia2, and Met18, which form the so-called CIA-targeting complex (Lill et al. 2015).
A unique combination of the Fe–S cluster assembly enzymes has been found in a flagellate Monocercomonoides exilis (strains PA203; Treitli et al. 2018) from the group of Oxymonadida (Karnkowska et al. 2016). Monocercomonoides exilis contains the CIA pathway, however, the ISC pathway is absent together along with all other mitochondrial proteins. As there is no microscopic evidence for the existence of a mitochondrion, this is interpreted as showing the mitochondrion has been lost altogether (Karnkowska et al. 2016), which makes this oxymonad unique among eukaryotes. Instead of ISC, M. exilis contains SUF pathway, which is slightly reduced compared with bacterial pathways, containing only three proteins—SufB, SufC, and SufSU. SufSU represents a fusion of SufS (cysteine desulfurase) and SufU (an enhancer of SufS in prokaryotes; Albrecht et al. 2010, 2011; Riboldi et al. 2011; Karnkowska et al. 2016).
The M. exilis SUF proteins are not specifically related to plastid homologues, or homologues from any other microbial eukaryotes, but rather to enzymes found in eubacteria, and to homologues in the transcriptome of Paratrimastix pyriformis, a sister taxon of oxymonads (Zhang et al. 2015; Karnkowska et al. 2016). It has been proposed that the pathway was acquired by horizontal gene transfer (HGT) from a eubacterium in the common ancestor of Monocercomonoides and Paratrimastix. This apparently preceded the loss of mitochondria in M. exilis, because P. pyriformis retains a mitochondrion-related organelle (Hampl et al. 2008; Zubáčová et al. 2013). Localization of the SUF pathway in P. pyriformis is unknown and in M. exilis heterologous localizations in Saccharomyces cerevisiae and T. vaginalis suggest cytosolic localization (Karnkowska et al. 2016).
Oxymonads and P. pyriformis are classified into the group Preaxostyla within phylum Metamonada (supergroup Excavata; Hampl et al. 2009; Adl et al. 2012). All representatives of two other Metamonada lineages (Parabasalia and Fornicata) contain the ancestral ISC pathway and no genes for enzymes in the SUF pathway have been observed in these lineages, indicating that this modification of Fe–S cluster assembly is specific to Preaxostyla. This may have served as a precondition for the mitochondrial loss in oxymonads. To further reveal the evolutionary history of this unique evolutionary switch, we investigated genomes and transcriptomes of 16 members of Preaxostyla for presence of genes and/or transcripts involved in Fe–S cluster synthesis.
Results and Discussion
The following Preaxostyla data sets were investigated in this study: genomic assemblies of M. exilis strain PA203 (Karnkowska et al. 2016), Blattamonas nauphoetae strain NAU3, and P. pyriformis strain ATCC 50935, single cell genome assembly of Streblomastix strix, three single cell transcriptome assemblies of Saccinobaculus doroaxostylus (SD1, SD2, SDN), three single cell transcriptome assemblies of Saccinobaculus ambloaxostylus (Amblo-1, Amblo-5, Amblo-5), one single cell transcriptome assembly of Oxymonas sp., two single cell transcriptome assemblies of Streblomastix sp. (Streblo-1, Streblo-4), one single cell transcriptome assembly of Pyrsonympha sp., transcriptome assembly of Trimastix marina strain PCT (Leger et al. 2017), and transcriptome assembly of trimastigid “MORAITIKA”. Quality and coverage varied extremely between data sets, which was probably the main reason for the lack of some genes/transcripts especially in the single cell genomes and transcriptomes.
The data were searched for all known components of ISC, SUF, CIA, and NIF pathways. In all examined data sets, we were unable to identify any gene involved in the ISC and NIF pathways, but we identified genes or gene fragments for proteins involved in the SUF and CIA pathways in all examined organisms with exception of “Streblo-4” (figs. 1 and 2). Mitochondrial targeting peptides were not predicted in any complete SUF proteins (supplementary table S1, Supplementary Material online), indicating cytosolic localization of these proteins.
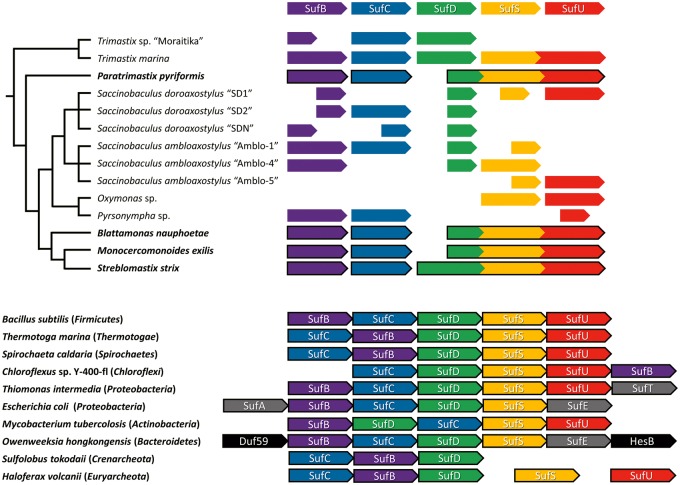
Fig. 1.
Inventory of SUF proteins in Preaxostyla. The scheme shows SUF genes/transcripts identified in the members of Preaxostyla. The relationship within this groups is indicated by the tree. For organisms in bold, genomic data were investigated, in others transcriptomic or single cell transcriptomic data sets were used. Completeness of a gene/transcript is indicated by the length of the arrow. The order of Preaxostyla genes does not reflect their order in the genome. Gene fusions are marked by fused arrows. At the bottom are given schemes of typical SUF gene operons in representatives of prokaryotic groups, see supplementary figure S5, Supplementary Material online for broader prokaryotic representation.
Fig. 2.
Inventory of CIA proteins in Preaxostyla. Scheme shows the presence (black) or absence (white/gray) of CIA genes/transcripts in Preaxostyla with reference to Metamonada represented by G. intestinalis and T. vaginalis. White/grey shading indicates that the gene was not identified in available genome/transcriptome, respectively. The gene inventory of T. vaginalis and G. intestinalis was taken from Pyrih et al. 2016. Question marks indicate uncertain orthology to Tah18.
Contaminants among the CIA proteins were not expected, because they have no close prokaryotic homologues. To filter prokaryotic contamination of SUF genes/transcripts and to exclude those that were severely truncated, sequences with >70% nucleotide similarity to prokaryotic sequences in NCBI and also sequences shorter than 65 aa were not included in further analyses. Phylogenetic trees were then constructed for individual SUF components (supplementary figs. S1–S4, Supplementary Material online). Every protein tree resolved a major Preaxostyla clade (shown in green in supplementary figs. S1–S4, Supplementary Material online). Sequences derived from the genomic assemblies were present only in this major clade, suggesting that this clade represents bona fide Preaxostyla genes. Species composition of this clade in every protein tree suggests that it was acquired prior to the last common ancestor of Preaxostyla.
Some sequences branched robustly (ML bootstrap >90%) outside this major clade (shown in red in supplementary figs. S1–S4, Supplementary Material online). Origin of these sequences is unclear, they may result from prokaryotic contaminations or more recent lineage specific HGT. For this reason, these sequences were not included in the concatenation analysis. Such outlying SUF proteins were especially common in the data set of Oxymonas sp. (supplementary figs. S1–S4, Supplementary Material online). These were all unique to Oxymonas and no closely related orthologues were found in any other oxymonad, suggesting that this data set contains high level of prokaryotic contamination. In contrast, we identified six closely related SufS genes from S. doroaxostylus isolate SDN, Streblomastix sp. (Streblo-1), S. ambloaxostylus (Amblo-5), and trimastigid “MORAITIKA.” They formed a well-supported clade (ML bootstrap 100) that was deeply nested in a clade containing mixture of Archaea and Eubacteria (supplementary fig. S4, Supplementary Material online). These sequences may represent contaminants from closely related prokaryotes, but it is also possible given the fact that these genes do not share a high level of identity and were found in a larger number of species, that they represent a second and independent acquisition of SufS by a subset of Preaxostyla. It should be noted, however, that these homologues were identified only in transcriptomes with overall low quality and high level of contamination, and most critically that the sequences putatively ascribed to Streblomastix do not contain evidence of the alternative genetic code typical for this oxymonad (Keeling and Leander 2003).
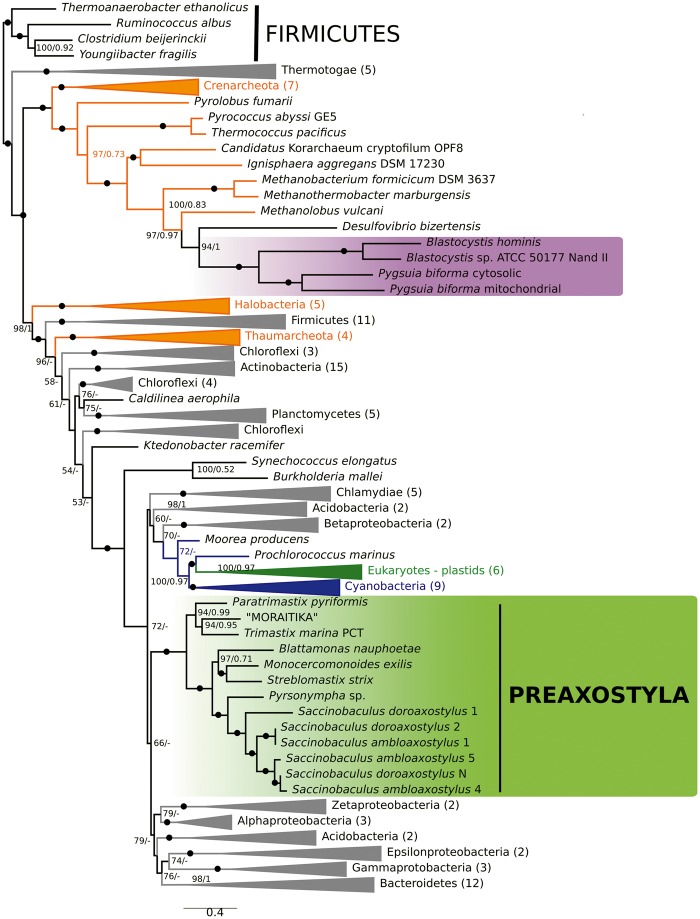
A concatenated tree of SufB, SufC, SufD, and SufS was constructed including sequences from “green clades” with reliable Preaxostyla origin (fig. 3). In this tree, all Preaxostyla formed a single and well-supported clade (RaxML/MrBayes support 100/1) with an internal topology consistent with the relationships among Preaxostyla (Treitli et al. 2018). This result strongly suggests that the whole pathway originated in their common ancestor and was inherited vertically since then.
Fig. 3.
Phylogenetic analysis of concatenated SufB, C, D, and S proteins. The topology of the tree was calculated by ML in IQ-TREE using partition-specific models. Numbers at nodes represent statistical support in regular ML bootstraps/Bayesian posterior probabilities. The support 99/0.99 and higher is indicated by filled circles, values <50 and 0.5 are not shown.
Three proteins of the pathway (SufD, S, and U) are fused (SufDSU) in M. exilis, B. nauphoetae, S. strix, and P. pyriformis. The homology of the N-terminal part of this protein with SufD has not been recognized previously (Karnkowska et al. 2016). Similar fusion is also present in the transcriptome of T. marina, where we identified a fusion of SufSU but SufD is on an independent contig. No fusions were evident in other data sets, but this may be a result of the fragmented nature of the data, making fusions hard to detect. A scheme of the detected SUF genes/transcripts and their fusions is shown in figure 1 (see supplementary fig. S5, Supplementary Material online for broader representation of prokaryotes). The phylogeny of SUFs (fig. 3) was unable to resolve the position of Preaxostyla sequences within prokaryotes. However, the arrangement of SUF operons in sequenced prokaryotic genomes suggests Firmicutes, Thermotogae, Spirochaetes, Proteobacteria, or Chloroflexi as probable donors. Operons in these groups are consistent with the SUF pathway composition as well as the SufDSU gene order found in Preaxostyla fusion genes.
The inventory of the CIA genes in Preaxostyla is in general very similar to that of other metamonads, including Nbp35, Cia1, Nar1, and Cia2 (Pyrih et al. 2016; fig. 2). Preaxostyla lack proteins associated with the mitochondrion (Erv1 and Atm1), and proteins Dre2 and Tah18. Their absence is not surprising as the Dre2 is often missing in anaerobes (Basu et al. 2014; Tsaousis et al. 2014) and neither Erv1 nor Atm1 was found in other metamonads (Pyrih et al. 2016). The primary function of Tah18 is to provide electrons for Dre2 (Netz et al. 2010), so in the absence of Dre2 it is reasonable that Tah18 was probably lost as well. We were not able to identify MMS19 in any of the studied oxymonads, but protein containing N-terminal MMS19 domain was present in P. pyriformis (fig. 2). The conserved inventory of CIA proteins in Preaxostyla contrasts with the major switch of the upstream Fe–S cluster assembly pathway in this group and indicates functional robustness of the CIA pathway.
Materials and Methods
For single cell transcriptomes (S. doroaxostylus, S. ambloaxostylus, Oxymonas sp., Streblomastix spp., and Pyrsonympha), cells were manually picked by micropipette, washed 1–2 times, and then deposited directly into single cell lysis buffer and frozen in –80 °C freezer. Single cell cDNA was then amplified following Picelli et al. (2014) and Kolisko et al. (2014) protocols. Illumina Nextera XT protocol was used for sequencing library construction. Transcriptomes were assembled by Trinity 2.0.6 (Grabherr et al. 2011) and for quality trimming trimmomatic0.32 (Bolger et al. 2014) with default settings was used.
The trimastigid “MORAITIKA” was maintained as a monoeukaryotic polyxenic culture in the ATCC medium 1525 at room temperature. Total RNA was isolated from 300 ml of culture using TRI Reagent (Sigma). Isolated RNA was purified by Qiagen RNeasy Mini Kit (Qiagen) and RNase-Free DNase Set (Qiagen) according to the manufacturer's protocol. Total RNA was sent to EMBL where the libraries were prepared. Contigs were assembled by Trinity 2014-04-13p1 (Grabherr et al. 2011), quality trimming was done by fastx version 0.0.13 (fastq_quality_filter -Q33 -q 20 -p 70), contigs shorter than 200 nt were discarded.
The single cell of S. strix was manually picked by micropipette from gut content of the termite Zootermopsis angusticolis, three times washed in Trager U media, then DNA was isolated and the whole genome was amplified using illustra Single Cell GenomiPhi DNA Amplification Kit (GE Healthcare) according to the manufacturer's protocol. The amplified DNA was purified using Agencourt AMPure XP (Beckman Coulter), and sequencing libraries were prepared using Illumina TruSeq DNA PCR-Free (Illumina) for HiSeq 2500 or with Ligation Sequencing Kit 1D (Oxford Nanopore Technologies) for Oxford Nanopore sequencing. Draft genome was assembled as a hybrid assembly using SPAdes 3.10.0 (Bankevich et al. 2012; Antipov et al. 2016). Binning of the assembled data and separation of the eukaryotic genome from bacterial sequences was done using tetranucleotide frequencies using tetraESOM method (Dick et al. 2009), together with blast analysis of the assembled data.
Blattamonas nauphoetae strain NAU3 was grown as a monoeukaryotic polyxenic culture in modified TYSGM media (Diamond 1982) without gastric mucin. The genomic DNA was sequenced using Illumina MiSeq (coverage 62x) and Oxford Nanopore MinIon (coverage 2x) technology and assembled using the SPAdes 3.7.1 (Bankevich et al. 2012; Antipov et al. 2016) followed by scaffolding with SSPACE basic V2 (Boetzer et al. 2011) using the Illumina mate-pair reads.
Paratrimastix pyriformis was grown in a monoeukaryotic polyxenic culture on rye grass cerophyll infusion (Sonneborn's Paramecium medium, ATCC #802) at room temperature. The genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen). The P. pyriformis draft genome sequence was sequenced and assembled from raw genomic reads produced by 454, Illumina HiSeq (coverage 894x), and PacBio (coverage 11x) sequencing technologies using SPAdes 3.11.1 (Bankevich et al. 2012; Antipov et al. 2016) assembly toolkit. Automatic gene prediction for the S. strix, B. nauphoetae and P. pyriformis draft genomes was done using Augustus 3.2.3 (Stanke and Waack 2003).
Nucleotide data sets predicted proteins were searched by TBLASTN algorithms; six-frame translations of the transcriptomes and predicted proteomes were searched by BLASTP. Proteins not identified by BLAST were searched for by HMMER. In the BLAST searches, we have used full gene inventories of M. exilis, Escherichia coli, and Bacillus subtilis for the SUF pathway, S. cerevisiae, T. vaginalis, G. intestinalis, and E. coli for the ISC pathway, Azotobacter vinelandii, E. histolytica, and M. balamuthii for the NIF pathway and S. cerevisiae and human for the CIA pathway. HMMER searches were performed by HMMER 3.1b2 (Eddy 2011) using curated Pfam and custom created models for the aforementioned genes. Sequences of SUF and CIA pathway genes/proteins retrieved from unpublished data sets were deposited in GenBank under accession numbers MH608120–MH608208.
Sequences were aligned by MAFFT v. 7.222 (Katoh et al. 2002) and trimmed with BMGE 1.12 software (Criscuolo and Gribaldo 2010) using blosum30 matrix. Gene fragments originating from different assemblies of the same species (namely SufC—SDN_lcl|TR24651|c0_g1_i1 and SDN_lcl|TR10532|c0_g1_i1, SufS—SD1_lcl|TR11348|c0_g2_i1 and SD1_lcl|TR17340|c0_g1_i1) were concatenated to increase phylogenetic resolution. Alignments are available upon request. Phylogenetic trees were constructed by IQ-TREE v 1.6.1 (Nguyen et al. 2015) using the best fitting models according to Bayesian information criterion predicted by ModelFinder (Kalyaanamoorthy et al. 2017)—LG4M for SufS and SufD, C20 for SufC, and EX_EHO for SufD. For analysis of the concatenated alignment, gene-partition-specific models given above were used. Bayesian analysis was performed for concatenated data set using MrBayes 3.2 (Ronquist et al. 2012) with two runs, each of four chains of 10 mil. generations, sampling frequency 500 generations and uniform WAG+gamma+covarion model for the whole concatenate. The value of the average standard deviation of split frequencies did not drop below the 0.01; however, both chains have shown stable plateau of likelihood values after 1 × 106 generations. Tree from first 1 × 106 generations were discarded as burn-in before consensus tree calculation.
Supplementary Material
Acknowledgments
This work was supported by the Czech Science Foundation (project 15-16406S to V.H.), by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 771592 to VH) the Grant Agency of Charles University (Project 1584314 to V.V.), the Ministry of Education, Youth and Sports of CR within the National Sustainability Program II (Project BIOCEV-FAR) LQ1604, by the project “BIOCEV” (CZ.1.05/1.1.00/02.0109), by the Centre for research of pathogenicity and virulence of parasites reg. nr.: CZ.02.1.01/0.0/0.0/16_019/0000759, by the Natural Sciences and Engineering Council of Canada (project RGPIN-2014-03994 to P.J.K.), by a grant from the Tula Foundation to the UBC Centre for Microbial Diversity and Evolution, by Purkyne Fellowship to M.K. (Czech Academy of Sciences). Computational resources were provided by the CESNET LM2015042 and the CERIT Scientific Cloud LM2015085, provided under the programme “Projects of Large Research, Development, and Innovations Infrastructures.”
References
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59(5):429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht AG, Netz DJA, Miethke M, Pierik AJ, Burghaus O, Peuckert F, Lill R, Marahiel MA.. 2010. SufU is an essential iron–sulfur cluster scaffold protein in Bacillus subtilis. J Bacteriol. 192(6):1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht AG, Peuckert F, Landmann H, Miethke M, Seubert A, Marahiel MA.. 2011. Mechanistic characterization of sulfur transfer from cysteine desulfurase SufS to the iron–sulfur scaffold SufU in Bacillus subtilis. FEBS Lett. 585(3):465–470. [DOI] [PubMed] [Google Scholar]
- Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T.. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron–sulfur cluster assembly under anaerobic conditions. J Biol Chem. 279(16):16863–16874. [DOI] [PubMed] [Google Scholar]
- Antipov D, Korobeynikov A, McLean JS, Pevzner PA.. 2016. HybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32(7):1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Pilon M.. 2011. Ancient and essential: the assembly of iron–sulfur clusters in plants. Trends Plant Sci. 16(4):218–226. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Netz DJ, Haindrich AC, Herlerth N, Lagny TJ, Pierik AJ, Lill R, Lukeš J.. 2014. Cytosolic iron–sulphur protein assembly is functionally conserved and essential in procyclic and bloodstream Trypanosoma brucei. Mol Microbiol. 93(5):897–910. [DOI] [PubMed] [Google Scholar]
- Biederbick A, Stehling O, Rosser R, Niggemeyer B, Nakai Y, Elsasser H-P, Lill R.. 2006. Role of human mitochondrial Nfs1 in cytosolic iron–sulfur protein biogenesis and iron regulation. Mol Cell Biol. 26(15):5675–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Drevland RM, Downs DM, Graham DE.. 2009. Archaeal ApbC/Nbp35 homologs function as iron–sulfur cluster carrier proteins. J Bacteriol. 191(5):1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braymer JJ, Lill R.. 2017. Iron–sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem. 292(31):12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10(1):210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LS. 1982. A new liquid medium for xenic cultivation of Entamoeba histolytica and other lumen-dwelling protozoa. J Parasitol. 68(5):958–959. [PubMed] [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, Banfield JF.. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 10(8):R85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. Pearson WR, editor. PLoS Comput Biol. 7(10):e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss JO, Tsai CL, Ishida JP, Tainer JA.. 2015. Emerging critical roles of Fe–S clusters in DNA replication and repair. Biochim Biophys Acta-Mol Cell Res. 1853(6):1253–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Neumann K, Prohl C, Muhlenhoff U, Lill R.. 2004. The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol Cell Biol. 24(11):4848–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M, Cox S, Tovar J.. 2004. The iron–sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol Biol. 4:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, Delbac F, Vivares CP, Hirt RP, Lill R, Embley TM.. 2008. Localization and functionality of microsporidian iron–sulphur cluster assembly proteins. Nature 452(7187):624–628. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ.. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 106(10):3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Silberman JD, Stechmann A, Diaz-Triviño S, Johnson PJ, Roger AJ.. 2008. Genetic evidence for a mitochondriate ancestry in the “amitochondriate” flagellate Trimastix pyriformis. PLoS One 3(1):e1383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann A, Netz DJA, Balk J, Pierik AJ, Muhlenhoff U, Lill R.. 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron–sulfur protein assembly machinery. Proc Natl Acad Sci USA. 102(9):3266–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, Novák L, Žárský V, Barlow LD, Herman EK, et al. 2016. A eukaryote without a mitochondrial organelle. Curr Biol. 26(10):1274–1284. [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414(6862):450–453. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Leander BS.. 2003. Characterisation of a Non-canonical genetic code in the oxymonad Streblomastix strix. J Mol Biol. 326(5):1337–1349. [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R.. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18(14):3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolisko M, Boscaro V, Burki F, Lynn DH, Keeling PJ.. 2014. Single-cell transcriptomics for microbial eukaryotes. Curr Biol. 24(22):R1081–R1082. [DOI] [PubMed] [Google Scholar]
- Leger MM, Kolisko M, Kamikawa R, Stairs CW, Kume K, Čepička I, Silberman JD, Andersson JO, Xu F, Yabuki A, et al. 2017. Organelles that illuminate the origins of Trichomonas hydrogenosomes and Giardia mitosomes. Nat Ecol Evol. 1(4):0092.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Dutkiewicz R, Freibert SA, Heidenreich T, Mascarenhas J, Netz DJ, Paul VD, Pierik AJ, Richter N, Stümpfig M, et al. 2015. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron–sulfur proteins. Eur J Cell Biol. 94(7–9):280–291. [DOI] [PubMed] [Google Scholar]
- Mi-ichi F, Yousuf MA, Nakada-Tsukui K, Nozaki T.. 2009. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci USA. 106(51):21731–21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJA, Mascarenhas J, Stehling O, Pierik AJ, Lill R.. 2014. Maturation of cytosolic and nuclear iron–sulfur proteins. Trends Cell Biol. 24(5):303–312. [DOI] [PubMed] [Google Scholar]
- Netz DJA, Pierik AJ, Stümpfig M, Bill E, Sharma AK, Pallesen LJ, Walden WE, Lill R.. 2012. A bridging [4Fe–4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron–sulfur protein maturation. J Biol Chem. 287(15):12365–12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJA, Stümpfig M, Doré C, Mühlenhoff U, Pierik AJ, Lill R.. 2010. Tah18 transfers electrons to Dre2 in cytosolic iron–sulfur protein biogenesis. Nat Chem Biol. 6(10):758–765. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvltova E, Sutak R, Harant K, Sedinova M, Hrdy I, Paces J, Vlcek C, Tachezy J.. 2013. NIF-type iron–sulfur cluster assembly system is duplicated and distributed in the mitochondria and cytosol of Mastigamoeba balamuthi. Proc Natl Acad Sci USA. 110(18):7371–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Puccio H.. 2013. Frataxin: a protein in search for a function. J Neurochem. 126:43–52. [DOI] [PubMed] [Google Scholar]
- Paul VD, Lill R.. 2015. Biogenesis of cytosolic and nuclear iron–sulfur proteins and their role in genome stability. Biochim Biophys Acta-Mol Cell Res. 1853(6):1528–1539. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R.. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 9(1):171–181. [DOI] [PubMed] [Google Scholar]
- Pondarré C, Antiochos BB, Campagna DR, Clarke SL, Greer EL, Deck KM, McDonald A, Han AP, Medlock A, Kutok JL, et al. 2006. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron–sulfur cluster biogenesis. Hum Mol Genet. 15(6):953–964. [DOI] [PubMed] [Google Scholar]
- Pyrih J, Pyrihová E, Kolísko M, Stojanovová D, Basu S, Harant K, Haindrich AC, Doležal P, Lukeš J, Roger A, et al. 2016. Minimal cytosolic iron–sulfur cluster assembly machinery of Giardia intestinalis is partially associated with mitosomes. Mol Microbiol. 102(4):701–714. [DOI] [PubMed] [Google Scholar]
- Riboldi GP, Larson TJ, Frazzon J.. 2011. Enterococcus faecalis sufCDSUB complements Escherichia coli sufABCDSE. FEMS Microbiol Lett. 320(1):15–24. [DOI] [PubMed] [Google Scholar]
- Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F.. 2013. Reprint of: iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta-Bioenergy. 1827(8–9):923–937. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J, Makrantoni V, Ingledew WJ, Stark MJR, White MF.. 2006. The DNA repair helicases XPD and FancJ have essential iron–sulfur domains. Mol Cell. 23(6):801–808. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Pallesen LJ, Spang RJ, Walden WE.. 2010. Cytosolic iron–sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J Biol Chem. 285(35):26745–26751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs CW, Eme L, Brown MW, Mutsaers C, Susko E, Dellaire G, Soanes DM, Van Der Giezen M, Roger AJ.. 2014. A SUF Fe–S cluster biogenesis system in the mitochondrion-related organelles of the anaerobic protist Pygsuia. Curr Biol. 24(11):1176–1186. [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S.. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl. 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Sutak R, Dolezal P, Fiumera HL, Hrdy I, Dancis A, Delgadillo-Correa M, Johnson PJ, Muller M, Tachezy J.. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc Natl Acad Sci USA. 101(28):10368–10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy J, Sánchez LB, Müller M.. 2001. Mitochondrial type iron–sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol. 18(10):1919–1928. [DOI] [PubMed] [Google Scholar]
- Tovar J, León-Avila G, Sánchez LB, Sutak R, Tachezy J, Van Der Giezen M, Hernández M, Müller M, Lucocq JM.. 2003. Mitochondrial remnant organelles of Giardia function in iron–sulphur protein maturation. Nature 426(6963):172–176. [DOI] [PubMed] [Google Scholar]
- Treitli SC, Kotyk M, Yubuki N, Jirounková E, Vlasáková J, Smejkalová P, Šípek P, Čepička I, Hampl V.. 2018. Molecular and morphological diversity of the Oxymonad Genera Monocercomonoides and Blattamonas gen. nov. Protist 169(5):744–783. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Gentekaki E, Eme L, Gaston D, Roger AJ.. 2014. Evolution of the cytosolic iron–sulfur cluster assembly machinery in Blastocystis species and other microbial eukaryotes. Eukaryot Cell. 13(1):143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BAP, Hirt RP, Lucocq JM, Embley TM.. 2002. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418(6900):865–869. [DOI] [PubMed] [Google Scholar]
- Yoon H, Knight SAB, Pandey A, Pain J, Turkarslan S, Pain D, Dancis A.. 2015. Turning Saccharomyces cerevisiae into a frataxin-independent organism. PLoS Genet. 11(5):e1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Táborský P, Silberman JD, Pánek T, Čepička I, Simpson AGB.. 2015. Marine isolates of Trimastix marina form a plesiomorphic deep-branching lineage within preaxostyla, separate from other known trimastigids (Paratrimastix n. gen.). Protist 166(4):468–491. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee D-W, Bi E, Ohnishi T, Daldal F, Pain D, et al. 2008. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol. 28(18):5569–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubáčová Z, Novák L, Bublíková J, Vacek V, Fousek J, Rídl J, Tachezy J, Doležal P, Vlček Č, Hampl V.. 2013. The mitochondrion-like organelle of Trimastix pyriformis contains the complete glycine cleavage system. PLoS One 8(3):e55417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.