Abstract
Human flap endonuclease 1 (FEN1) is a structure-specific, multi-functional endonuclease essential for DNA replication and repair. We and others have shown that during DNA replication, FEN1 processes Okazaki fragments via its interaction with the proliferating cell nuclear antigen (PCNA). Alternatively, in response to DNA damage, FEN1 interacts with the PCNA-like Rad9–Rad1–Hus1 complex instead of PCNA to engage in DNA repair activities, such as homology-directed repair of stalled DNA replication forks. However, it is unclear how FEN1 is able to switch between these interactions and its roles in DNA replication and DNA repair. Here, we report that FEN1 undergoes SUMOylation by SUMO-1 in response to DNA replication fork-stalling agents, such as UV irradiation, hydroxyurea, and mitomycin C. This DNA damage-induced SUMO-1 modification promotes the interaction of FEN1 with the Rad9–Rad1–Hus1 complex. Furthermore, we found that FEN1 mutations that prevent its SUMO-1 modification also impair its ability to interact with HUS1 and to rescue stalled replication forks. These impairments lead to the accumulation of DNA damage and heightened sensitivity to fork-stalling agents. Altogether, our findings suggest an important role of the SUMO-1 modification of FEN1 in regulating its roles in DNA replication and repair.
Keywords: flap endonuclease 1, Rad9–Rad1–Hus1 complex, replication stress, SUMOylation
Introduction
Flap endonuclease-1 (FEN1) is a structure-specific, multi-functional nuclease. FEN1 possesses flap endonuclease (FEN), 5′ exonuclease (EXO), and gap endonuclease (GEN) activities (Liu et al., 2004; Zheng and Shen, 2011; Zheng et al., 2011b). Its FEN activity is required for Okazaki fragment maturation (Tishkoff et al., 1997; Bae et al., 2001; Ayyagari et al., 2003) and long-patch base excision repair (Klungland and Lindahl, 1997; Kim et al., 1998). Its EXO activity, as we have recently demonstrated (Liu et al., 2015), is critical for editing DNA polymerase α errors during Okazaki fragment maturation. In addition, its concerted EXO and GEN activities play an important role in resolving DNA secondary structures (Singh et al., 2007; Yang and Freudenreich, 2007), processing stalled replication forks (Zheng et al., 2005; Chung et al., 2015), and facilitating DNA replication at difficult-to-replicate regions, including rDNA and telomeres (Guo et al., 2008; Saharia et al., 2008; Sampathi and Chai, 2011). FEN1 mutations that eliminate its EXO activity increase the rate of base-substitution mutations and favor the incidence of cancer (Zheng et al., 2007b; Liu et al., 2015). Mutations that impair its EXO or/and GEN, but not FEN, activities result in trinucleotide repeat expansion, instabilities in telomeric and rDNA regions, and increased sensitivity to replication fork-stalling agents (Zheng et al., 2005; Singh et al., 2007; Guo et al., 2008; Saharia et al., 2008; Sampathi and Chai, 2011). Not surprisingly, yeast and mammalian cells deficient in FEN1 display defects in cell proliferation and are sensitive to DNA alkylating agents (Gary et al., 1999).
FEN1 interacts with more than 30 proteins, including proliferating cell nuclear antigen (PCNA), replication protein A (RPA), WRN protein, and the Rad9–Rad1–Hus1 heterotrimeric complex (Zheng et al., 2011b). It has been suggested that the protein–protein interactions between FEN1 and its partners dictates its role in various DNA replication and repair pathways. By interacting with the DNA clamp PCNA, FEN1 is recruited to DNA replication sites where it uses its FEN activity to cleave the RNA–DNA flap during Okazaki fragment maturation (Gary et al., 1999; Zheng et al., 2007a). F343A/F344A (FFAA) mutations to FEN1 completely disrupt its interaction with PCNA. Thus, homozygous FFAA mutations impair DNA replication and cause growth retardation and death in newborn mice (Zheng et al., 2007a), and heterozygous FFAA mutations result in aneuploidy-associated cancer in adult mice (Zheng et al., 2011a, 2012). FEN1 also binds to the Rad9–Rad1–Hus1 checkpoint clamp protein complex, which is structurally similar to PCNA (Wang et al., 2004; Querol-Audi et al., 2012) but specifically envelops regions of damaged DNA (Aravind et al., 1999; Parrilla-Castellar et al., 2004; Dore et al., 2009; Bai et al., 2010). The binding of FEN1 to the Rad9–Rad1–Hus1 complex enhances its nuclease activities and its coordination with DNA ligase 1, which is essential for DNA repair.
How FEN1 is directed to switch from its interaction with PCNA to interact with the Rad9–Rad1–Hus1 complex in response to DNA damage is a critical question that remains unanswered. Previous studies have revealed that FEN1 undergoes cell cycle- and DNA damage-dependent post-translational modifications, such as methylation, acetylation, and phosphorylation (Hasan et al., 2001; Henneke et al., 2003; Guo et al., 2008, 2010, 2012), which may play a role in its functional regulation. We found that methylation and phosphorylation of FEN1 are critical for regulating its interactions with its partners, including PCNA. For example, we observed that FEN1 is normally methylated by PRMT5 during the G1 phase, and this methylation blocks FEN1 phosphorylation by CDK1/cyclin A. After FEN1 completes RNA primer removal, however, it is demethylated, allowing phosphorylation (Guo et al., 2010). Phosphorylated FEN1 triggers its dissociation from PCNA, enabling DNA ligase 1 to access PCNA to join Okazaki fragments (Guo et al., 2010). We also found that FEN1 phosphorylation is also induced by ultraviolet (UV) irradiation and oxidative DNA damage (Zhou et al., 2017). Thus, phosphorylation of FEN1 likely serves as a switch to trigger its dissociation from PCNA and the replication site. Additional post-translational modifications to the dissociated FEN1 may stimulate its binding to DNA repair proteins, such as the Rad9–Rad1–Hus1 complex, to participate in DNA repair.
We previously reported that FEN1 can be modified by SUMO family proteins, including SUMO-1, SUMO-2, and SUMO-3, in vitro. The modification of FEN1 by SUMO-2/3 is important for its cell cycle-dependent degradation (Guo et al., 2012), but the biological role of its modification by SUMO-1 has remained unclear. Here, we report that UV irradiation and exposure to the fork-stalling agents hydroxyurea (HU) and mitomycin C (MMC) induce sequential FEN1 phosphorylation and SUMOylation by SUMO-1. We also define FEN1 phosphorylation stimulates its SUMO-1 modification. Furthermore, we demonstrate that SUMO-1 modification of FEN1 enhances its interaction with the HUS1 subunit of the Rad9–Rad1–Hus1 complex, and FEN1 mutations disrupting this modification impair UV- and HU-induced interactions of FEN1 with HUS1. As a consequence, cells carrying mutant FEN1 that cannot be modified by SUMO-1 are hypersensitive to UV, HU, and MMC treatment and accumulate UV- and HU-induced DNA double-strand breaks.
Results
DNA damage induces modification of FEN1 by SUMO-1
UV irradiation (120 J/m2) induced the emergence of a modified form of FEN1 with a molecular weight slightly greater than 50 kDa in HeLa cells harvested 3 h after treatment (Figure 1A). siRNA knockdown of the SUMO-conjugating enzyme Ubc9 reduced levels of this UV-induced modified FEN1, suggesting that it was the product of FEN1 SUMOylation (SUMO-FEN1; Figure 1A). We used immunoprecipitation (IP) to pull down FEN1 from HeLa cells and analyzed levels of SUMO-1 modified FEN1 (SUMO-1-FEN1) using an antibody specifically against SUMO-1. We observed that SUMO-1-FEN1 levels were elevated considerably in response to UV irradiation (Figure 1B). These findings suggest that UV irradiation induces SUMO-1 modification of FEN1. To confirm that UV irradiation induced FEN1 SUMO-1 modification, we expressed 3×FLAG-tagged FEN1 in HeLa cells, which we treated with UV irradiation. Three hours after treatment, we harvested the cells and performed IP using anti-FLAG M2 beads to pull down the 3×FLAG-tagged FEN1. UV irradiation stimulated SUMO-1 modification of exogenous FEN1, similar to that of endogenous FEN1, in HeLa cells (left two lanes, Figure 1C). siRNA knockdown of Ubc9 led to the reduction of SUMO-1 modification of 3×FLAG-tagged FEN1 in response to UV irradiation, but overexpression of exogenous Myc-tagged Ubc9 restored it (right three lanes, Figure 1C). Consistent with these findings, incubation of recombinant FEN1 and SUMO-1 with Ubc9 and the E1 SUMO-activating enzyme SAE1/UBA2 in an ATP-containing SUMOylation buffer produced SUMO-1-modified FEN1 (SUMO-1-FEN1; Figure 1D). We then evaluated the dynamic levels of SUMO-1-FEN1 in HeLa cells post-UV irradiation. We found that immediately (0 h) post-UV irradiation, there was no obvious elevation in SUMO-1-FEN1 levels compared to levels in untreated control cells (Figure 1E). At 2 or 4 h post-UV irradiation, however, there was significantly more SUMO-1-FEN1 in UV-exposed HeLa cells than in untreated controls (Figure 1E). By 6 h post-UV irradiation, SUMO-1-FEN1 levels were no longer significantly different between groups (Figure 1E). In addition to UV irradiation, other DNA damaging agents, including HU (1 mM, 3 h), camptothecin (CPT, 5 μM, 3 h), and MMC (18 μM, 3 h) also induced SUMO-1 modification of FEN1 (Figure 1F).
Figure 1.
Identification and validation of FEN1 SUMO-1 modification. (A) Western blot analysis was used to detect SUMOylated and unmodified FEN1 in UV-treated (120 J/m2, 3-h recovery) HeLa cells treated with siRNA targeting the coding region of Ubc9 gene (si-Ubc9) or a scrambled siRNA control (–). Cells were harvested 3 h post-UV irradiation. FEN1 proteins in whole cell extracts were detected using an antibody against human FEN1. Relative protein levels of Ubc9 and β-actin (loading control) were detected using anti-Ubc9 and anti-β-actin antibodies. (B) FEN1 is SUMO-1 modified in HeLa cells in response to UV treatment. UV treatment was conducted as described for Panel 1A, and cells were harvested at 3 h post-UV irradiation. FEN1 was isolated from whole cell extracts via IP, and FEN1 and SUMO-1-FEN1 were detected by western blot analysis using antibodies against FEN1 and SUMO-1, respectively. β-actin was used as a control to ensure equal input for each IP reaction. To accurately reflect the amount of FEN1 in different samples, we could not overexpose the immunoblot. Therefore, the SUMO-1-FEN1 was not shown here. (C) Exogenously expressed 3×FLAG-tagged FEN1 (FLAG-FEN1) is SUMO-1-modified after UV treatment. HeLa cells stably expressing FLAG-FEN1 were treated with siRNA targeting the 5′UTR of Ubc9 gene si-Ubc9 or negative scrambled siRNA control. To restore Ubc9 expression, HeLa cells were co-transfected with si-Ubc9 and a vector encoding human Ubc9. FLAG-FEN1 was immunoprecipitated using anti-FLAG M2 beads. SUMO-1-FLAG-FEN1 and FLAG-FEN1 were detected by western blot using anti-SUMO-1 or FEN1 antibodies. Ubc9 levels were verified using an anti-Ubc9 antibody. β-actin was used as a loading control. (D) SUMO-1 modification of recombinant FEN1 in vitro is Ubc9-mediated. Purified recombinant FEN1 as incubated with Ubc9 and SUMO-1 for 60 min at 37°C. Unmodified FEN1 and SUMO-1-FEN1 were visualized using Coomassie Brilliant Blue staining and western blot analysis using antibodies against FEN1 and SUMO-1. (E) HeLa cells stably expressing 3×FLAG-tagged FEN1 were exposed to UV irradiation and allowed to recover for 0, 2, 4, or 6 h. Cells not exposed to UV irradiation were used as controls (CON). Cells were harvested and total 3×FLAG-FEN1 was isolated via IP. 3×FLAG-FEN1 and SUMO-1-3×FLAG-FEN1 were detected by western blot using anti-FEN1 or anti-SUMO-1 antibodies. The top panel shows the representative western blot images, and the bottom panel shows the quantification of SUMO-1-FEN1 relative to levels in UV-unexposed control cells at 0 h. The intensity of SUMO-1-3×FLAG-FEN1 bands in the SUMO-1 blot was normalized to the corresponding 3×FLAG-FEN1 band in the FLAG blot. Values shown are mean ± SD of three independent assays. P-values were calculated using Student’s t-test for each time point. ns, not significant, *P < 0.05. (F) HeLa cells stably expressing 3×FLAG-tagged FEN1 were exposed to UV irradiation (120 J/m2, 3-h recovery) or treated with HU (1 mM, 3 h) or MMC (18 μM, 3 h). FEN1 was purified from treated cells and untreated controls using anti-FLAG M2 magnetic beads, and 3×FLAG-FEN1 and SUMO-1-3×FLAG-FEN1 were detected by western blot analysis using anti-FEN1 and anti-SUMO-1 antibodies.
Determination of SUMO-1 modification sites of FEN1
To identify the sites of FEN1 that are modified by SUMO-1, we conducted SUMO-1 modification of FEN1 using a recombinant SUMO-1 mutant (T95K), Ubc9, and FEN1, using methods similar to those used in our previous study (Guo et al., 2012). SUMOylation with the T95K SUMO-1 mutant tags modified lysines with a diglycine (GG) remnant, which can be detected using mass spectrometry (Knuesel et al., 2005). Thus, we subjected T95K SUMO-1-modified FEN1 to liquid chromatography–electrospray ionization–tandem mass spectrometry (LC–ESI–MS/MS) analyses after GluC and Trypsin endoproteinase digestion. We identified Lys366, Lys367, Lys369, and Lys375 as potential SUMO-1 modification sites of FEN1 (Supplementary Figure S1). To validate that these four lysine residues are indeed SUMO-1 modification sites of FEN1, we constructed, expressed, and purified 6×His-tagged FEN1 harboring the point mutations K366R, K367R, K369R, or K375R, or all four mutations (4KR). The K367R single point mutation did not significantly alter Ubc9-mediated SUMO-1 modification of FEN1. The point mutations at K366, K369, and K375, however, reduced Ubc9-mediated FEN1 SUMO-1 modification by approximately 40% relative to that of wild-type (WT) FEN1, and the 4KR mutation nearly abolished FEN1 SUMO-1 modification to <10% that of WT FEN1 (Figure 2A and B). We then stably overexpressed 3×FLAG-tagged WT and 4KR mutant FEN1 in HeLa cells (Supplementary Figure S2). Co-IP and western blot analysis showed that the 4KR FEN1 mutation reduced SUMO-1-FEN1 levels in the cells under normal culture conditions (Figure 2C), as well as under exposure to UV irradiation and other DNA damaging agents, as described above (Figure 2D). In addition, we used the Duolink®in situ proximity ligation assay (PLA), which has been used to detect and quantify in situ protein interactions (Soderberg et al., 2006), to directly visualize co-localization of SUMO-1 and FEN1 in HeLa cells. When PLA probes are in close proximity (<40 nm), a fluorescent signal is emitted. The PLA signal for SUMO-1-FEN1 was significantly higher in the UV-treated WT FEN1-expressing cells than that in untreated WT FEN1-expressing cells (Figure 2E and Supplementary Figure S3), whereas low PLA signals were detected in the 4KR cell line both with and without UV treatment (Figure 2E). These findings demonstrate that Lys366, Lys367, Lys369, and Lys375 residues are the primary modification sites for the SUMO-1 modification of FEN1.
Figure 2.
K366, K367, K369, and K375 residues are the primary SUMO-1 modification sites of FEN1. (A) Purified FLAG-tagged WT or mutant (K366R, K367R, K369R, K375R, or 4KR) FEN1 proteins were incubated with SUMO-1 and SUMO-1 modification reaction components. FEN1 and SUMO-1-FEN1 were detected by western blot analysis using anti-FLAG and anti-SUMO-1 antibodies. The quantified intensities of SUMO-1 modification of the mutant FEN1 proteins, normalized to corresponding 3×FLAG FEN1 levels and relative to that of WT FEN1, are shown. (B) WT or 4KR FEN1 were incubated with SUMO-1 modification reaction components, with or without SUMO-1. FEN1 and SUMO-1-FEN1 levels were detected in a single blot using an anti-FEN1 antibody. (C) HeLa cells stably expressing 3×FLAG-tagged WT or 4KR mutant FEN1 were exposed to UV (120 J/m2, 3-h recovery) and 3×FLAG-tagged WT or 4KR FEN1 was purified with anti-FLAG M2 magnetic beads. 3×FLAG-tagged FEN1 and SUMO-1-3×FLAG-FEN1 were detected by western blot analysis. (D) HeLa cells stably expressing 3×FLAG-tagged WT or 4KR mutant FEN1 were exposed to UV irradiation (120 J/m2, 3-h recovery) or treated with HU (1 mM, 3 h), CPT (5 μM, 3 h), or MMC (18 μM, 3 h). 3×FLAG-tagged WT and 4KR mutant FEN1 were purified with anti-FLAG M2 beads, and 3×FLAG-FEN1 and SUMO-1-3×FLAG-FEN1 were detected by western blot analysis. The intensities of SUMO-1-FEN1, normalized to corresponding 3×FLAG FEN1 levels and relative to that of untreated WT FEN1, are shown. (E) SUMO-1 modification of WT and 4KR FEN1 was visualized in HeLa cells using the Duolink®in situ PLA with anti-SUMO-1 and anti-FLAG antibodies (PLA-SUMO-1/3×FLAG). Nuclei were stained with DAPI. Scale bars: 10 μm.
Phosphorylation of FEN1 is required to stimulate its SUMO-1 modification
We have previously demonstrated that UV damage induces FEN1 phosphorylation (Guo et al., 2008). Subsequently, we sought to determine if UV-induced phosphorylation is involved in UV-induced SUMO-1 modification of FEN1. We found that FEN1 phosphorylation and FEN1 SUMO-1 modification were higher 3 h after UV irradiation (120 J/m2 UV) than in untreated cells, whereas FEN1 methylation was lower than that in untreated cells (Figure 3A). These results are consistent with our previous finding that FEN1 methylation blocks its phosphorylation. To define the relationship between FEN1 phosphorylation and SUMO-1 modification, we treated HeLa cells with a CDK inhibitor, olomoucine, to suppress FEN1 phosphorylation. The CDK inhibitor reduced UV-induced SUMO-1-FEN1 levels (Figure 3B), indicating a positive correlation between FEN1 phosphorylation and SUMO-1 modification. To further elucidate this relationship, we overexpressed WT FEN1 or phosphorylation-deficient (S187A) or phosphorylation-mimicking (S187D) FEN1 mutants in HeLa cells (Supplementary Figure S5) and measured SUMO-1-FEN1 levels. The level of SUMO-1-S187D FEN1 was higher than that of SUMO-1-WT FEN1, whereas SUMO-1-S187A FEN1 was barely detected (Figure 3C). We then measured in vitro SUMOylation of recombinant WT, S187A, and S187D FEN1 proteins. Recombinant WT FEN1 was phosphorylated (Supplementary Figure S4) and SUMO-1-modified in the presence of ATP (Figure 3D) in vitro. S187D, which mimics phosphorylated FEN1 displayed a higher level of SUMO-1 modification than WT FEN1 (Figure 3D). Conversely, no SUMO-1 modification was observed for S187A FEN1 (Figure 3D). These data suggest that FEN1 phosphorylation is required for its SUMO-1 modification.
Figure 3.
Phosphorylation of FEN1 stimulates its SUMO-1 modification. (A) Various FEN1 modifications were analyzed in HeLa cells expressing 3×FLAG-FEN1 with or without UV irradiation (120 J/m2, 3-h recovery). 3×FLAG-FEN1 was purified using M2 beads. Methylated (Me-FEN1), phosphorylated (Phos-FEN1), and SUMO-1-modified FEN1 were detected by western blot analysis using anti-FLAG, anti-methylarginine, anti-phosphoserine, and anti-SUMO-1 antibodies. β-actin served as a control to ensure equal input for each IP reaction. (B) HeLa cells expressing 3×FLAG-FEN1 were treated with the CDK inhibitor olomoucine (40 μM, 24 h) or a vehicle control (DMSO) and exposed to UV irradiation. FLAG-FEN1, SUMO-FEN1, and Phos-FEN1 were detected, as in Panel A. GAPDH served as a control to ensure equal input for each IP reaction. Left panel: representative western blot images. Right panel: Quantification of phosphorylated and SUMO-1 modified FEN1. The intensities of Phos-FEN1 or SUMO-1-FEN1 bands, normalized to the corresponding 3×FLAG-FEN1 band and relative to that of UV-unexposed cells, are shown. Values shown are mean ± SD of three independent assays. P-values were calculated using Student’s t-test. *P < 0.05, **P < 0.01. (C) HeLa cells were transfected with myc-tagged WT, S187A phosphorylation-defective, or S187D phosphorylation-mimic FEN1. Myc-tagged WT, S187A, or S187D FEN1 was isolated using myc beads. SUMO-1-myc-FEN1 (WT or mutant) was detected using western blot analysis with an anti-SUMO-1 antibody. FEN1 levels were determined using anti-myc and anti-FEN1 antibodies. GAPDH served as a control to ensure equal input for each IP reaction. (D) SUMO-1 modification of purified WT, S187A, and S187D FEN1 was conducted in vitro. ATP was or was not added to initiate the reaction. Anti-FEN1 and anti-SUMO-1 antibodies were used to detect FEN1 and SUMO-1-FEN1.
SUMO-1 modification of FEN1 facilitates its interaction with RAD1 and HUS1
We evaluated the impact of the SUMO-1 modification of FEN1 on its interaction with PCNA and the Rad9–Rad1–Hus1 complex. We measured binding of MBP-tagged PCNA and GST-tagged HUS1 (a subunit of the Rad9–Rad1–Hus1 complex) to FEN1 and SUMO-1-FEN1 in vitro. Unmodified 6×His-tagged FEN1 (first two lanes in each blot, Figure 4A) bound to MBP-tagged PCNA and GST-tagged HUS1, but not MBP or GST protein tags alone (Figure 4A). We also found that SUMO-1 modification (lane 3 in each blot, Figure 4A) enhanced the amount of GST-HUS1 that was co-pulled down with 6×His-tagged FEN1, but reduced the amount of MBP-PCNA that was co-pulled down with 6×His-tagged FEN1, suggesting that SUMO-1 modification of FEN1 stimulates its interaction with HUS1 and inhibits its interaction with PCNA. We then tested FEN1 interactions with PCNA and the Rad9–Rad1–Hus1 complex in HeLa cells without or with UV irradiation, which induced SUMO-1 modified FEN1 (Figure 1A and B). Under normal culture conditions, FEN1 interacted with both PCNA and the HUS1 and RAD1 subunits of the Rad9–Rad1–Hus1 complex (Figure 4B and C). In response to UV irradiation, the FEN1/PCNA interaction was reduced, but FEN1/HUS1 and FEN1/RAD1 interactions increased (Figure 4B). Under both normal and UV-treated conditions, 4KR mutant FEN1, which had significantly lower SUMO-1 modification levels than WT FEN1, also had significantly reduced interactions with HUS1 but increased interactions with PCNA (Figure 4C and D). Interactions with RAD1 were similar for WT and 4KR mutants under normal conditions, but lower for 4KR FEN1 after UV treatment (Figure 4C). In addition, we noticed that UV irradiation induced the interaction of 4KR mutant FEN1 with HUS1 (Figure 4C and D), suggesting that the FEN1/HUS1 interaction can be stimulated through a K366/K367/K369/K375 SUMO-1 modification-independent mechanism. Notably, in situ PLA revealed that after UV treatment, the WT FEN1/HUS1 complex was enriched in the nucleus including the nucleolus (Figure 4D), where replication forks are frequently stalled by secondary structures resulting from repetitive DNA sequences.
Figure 4.
SUMOylation of FEN1 facilitates its interaction with RAD1 and HUS1. (A) In vitro binding assays were conducted for FEN1 or SUMO-1-FEN1 with PCNA or HUS1. Purified recombinant 6×His-tagged FEN1 was incubated with Ubc9, SUMO-1, and other components of the SUMOylation kit, with or without ATP, at 37°C for 60 min. Unmodified FEN1 or SUMO-1-FEN1 was incubated with Ni-NTA beads. After extensive washing, the beads were incubated with a mixture of MBP-tagged PCNA and GST-tagged HUS1 proteins. Incubation of PCNA and HUS1 with beads only (no FEN1) and incubation of FEN1-Ni-NTA beads with non-conjugated MBP and GST proteins without ATP served as negative controls for non-specific binding of the proteins or tags to the beads. FEN1-bound MBP-PCNA and GST-HUS1 were analyzed by western blot analysis using anti-PCNA, anti-HUS1, anti-MBP, and anti-GST antibodies. The intensities of PCNA and HUS1 in the pull-down were quantified and normalized to their input levels. The relative levels of PCNA and HUS1 binding to FEN1, with versus without SUMO-1 modification (ATP), are shown. (B) WT HeLa cells were treated with 120 J/m2 UV irradiation and allowed to recover for 3 h. FEN1 complexes were immunoprecipitated using an anti-FEN1 antibody. FEN1 and SUMO-1-FEN1, as well as PCNA, HUS1, RAD1 that were co-IPed with FEN1, were detected by western blot analysis. A bead-only (no anti-FEN1) control was used as a negative control for IP. The relative levels of PCNA, HUS1, and RAD1 binding to FEN1, normalized to the loading control IgG and relative to that of untreated WT cells, are shown. (C) HeLa cells stably expressing 3×FLAG-tagged WT or 4KR mutant FEN1 were treated with 120 J/m2 UV irradiation and allowed to recover for 3 h. 3×FLAG-FEN1 complexes were immunoprecipitated using anti-FLAG M2 beads. 3×FLAG-FEN1 and SUMO-1-FEN1, as well as PCNA, HUS1, and RAD1 that were co-immunoprecipitated with FEN1, were detected by western blot analysis. (D) The interactions of WT or 4KR mutant FEN1 with PCNA, HUS1, and RAD1 with and without UV treatment were analyzed using the Duolink®in situ PLA with antibody mixtures containing anti-FLAG/anti-PCNA, anti-FLAG/anti-HUS1, and anti-FLAG/anti-RAD-1. Nuclei were stained with DAPI. Upper panels show representative PLA assay images (scale bars: 10 μm). and the bottom panels show PLA intensities, relative to that of control WT cells. Values shown are mean ± SD of three independent assays. P-values were calculated using Student’s t-test. ns, not significant, *P < 0.05, **P < 0.01.
We then investigated the impact of the SUMO-1 modification of FEN1 on its FEN and GEN activities, which are important for processing DNA intermediates to restart stalled replication forks (Sharma et al., 2004; Zheng et al., 2005). Using FAM-labeled synthetic DNA substrates, we found that SUMO-1-FEN1 had considerably higher FEN and GEN activities than the unmodified FEN1 (Figure 5A and B). On the other hand, the 4KR mutation impaired the FEN and GEN activities of FEN1 (Supplementary Figure S6). These findings are consistent with a previous study showing that the C-terminus of FEN1 mediates DNA substrate binding (Stucki et al., 2001) and suggest a new, SUMO-1-mediated mechanism for regulating FEN1 nuclease activities.
Figure 5.
Nuclease activities of unmodified- and SUMO-1-modified FEN1. (A and B) FEN activity on double flap substrates (A) and GEN activity on gapped duplex substrates (B) were assessed in vitro. Purified FEN1 was incubated for 60 min with the SUMO-1 modification kit reaction mixture at 37°C in the absence or presence of ATP. The resultant unmodified (no ATP control) FEN1 or SUMO-1-FEN1 was then incubated with FAM-labeled DNA substrates in reaction buffer at 37°C for 5, 10, 15, 20, or 25 min (duration indicated by the wedge above the gel). The reactions were resolved in a 15% denaturing PAGE gel and visualized with a Typhoon FLA 9500 imager.
SUMO-1 modification of FEN1 affects its replication and repair responses under replication stress
Because FEN1 SUMO-1 modification is induced by UV irradiation, HU, CPT, and MMC, which cause replication forks to stall, we hypothesized that a major function of FEN1 SUMO-1 modification is to stimulate FEN1 to rescue stalled replication forks. To test this hypothesis, we used a DNA fiber assay—labeling thymidine analogs IdU and CIdU—to track newly synthesized DNA (Figure 6A). We assessed the ability of WT or SUMO-1-FEN1-deficient 4KR mutant HeLa cells to restart spontaneous and HU-induced stalled replication forks and found that 4KR mutant cells had a much higher percentage of stalled forks that failed to restart (non-restarted forks) under normal culture conditions, as well as in response to HU treatment (Figure 6B). Under normal culture conditions, there were approximate 27% stalled forks in 4KR cells, compared to 8% in WT cells. Upon HU treatment, there were 34% stalled forks in 4KR cells, compared to 14% in WT cells. In addition, 4KR mutant cells had a lower percentage of newly firing forks (Figure 6B). These results suggest that defective replication fork processing may result in the accumulation of DNA damage, which may suppress origin firing. Consistent with this notion, using an EdU incorporation assay, we found that treated and untreated 4KR cells exhibited more defects in overall DNA replication (Figure 6C and D). Flow cytometric analyses of cell cycle progression of the WT and the 4KR mutant cells revealed that more 4KR cells than WT cells accumulated at the late S/G2 stage (Supplementary Figure S7A). In addition, we observed that the 4KR cells had a slower cell proliferation rate than the WT cells (Supplementary Figure S7B). These findings suggest that the 4KR FEN1 mutation results in late S/G2 cell cycle arrest.
Figure 6.
SUMOylation of FEN1 affects its DNA damage repair activity in vivo and in vitro. (A) WT or 4KR mutant HeLa cells were pulse-labeled with the thymidine analog IdU (25 μM) for 30 min, treated with HU (4 mM) or DMSO for 4 h, and then pulse-labeled with the thymidine analog CIdU (125 μM) for 40 min. The cells were lysed and the DNA fibers were spread, and stained using anti-IdU and anti-CIdU antibodies. Representative images show stalled DNA forks that failed to restart (green), newly firing forks (red), and restarted forks (green/red). (B) Percentages of each type of replication fork (restarted, non-restarted, and newly firing) in WT and 4KR cells with or without HU treatment. Values are mean ± SD of three independent assays. (C) DNA replication and proliferation of WT and 4KR mutant HeLa cells were analyzed by EdU incorporation after cells were synchronized to S phase. Representative microimages show EdU-positive cells in WT and 4KR cells with or without UV irradiation (120 J/m2 UV, 3-h recovery). Nuclei were stained with DAPI (Hoechst blue), EdU incorporation (green) marks cells undergoing proliferation. Scale bars: 60 μm for the images with low magnification and 10 μm for the enlarged images. (D) Percentages of EdU-positive WT and 4KR cells with or without UV irradiation. Values are mean ± SD of three assays. P-values were calculated using Student’s t-test. *P < 0.05.
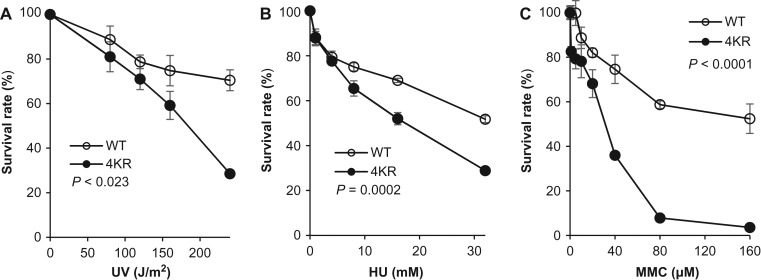
To determine if the 4KR FEN1 mutation results in the accumulation of DNA damage, we stained γH2AX and 53BP1, two typical readouts for DNA strand breaks (Rogakou et al., 1998; Anderson et al., 2001), in WT and 4KR cells. We found that 4KR cells had significantly more γH2AX- and 53BP1-positive nuclei than WT cells, spontaneously and in response to UV irradiation or HU or MMC treatment (Figure 7A–D). In order to determine if the SUMO-1-FEN1-deficient 4KR cells are sensitive to fork-stalling agents, we treated cells with different doses of UV irradiation, HU, or MMC and measured cell proliferation using an MTS assay. We observed that 4KR cells were considerably more sensitive to UV, HU, and MMC than WT cells (Figure 8A–C). We also noticed that 4KR mutant cells were more sensitive to MMC than to UV or HU treatment. One possible reason is that MMC forms inter-strand cross-links (Noll et al., 2006), which induce the homology-directed repair pathway to rescue the stalled replication fork. We previously showed that the GEN activity of FEN1 cleaves stalled replication forks to initiate homology-directed repair (Zheng et al., 2005). This likely explains why 4KR cells show greater sensitivity to MMC, especially compared to HU, which stalls replication forks via depletion of the dNTP pool but not DNA lesions (Petermann et al., 2010).
Figure 7.
FEN1 SUMOylation-defective cells accumulate DNA damage. (A and B) Representative immunofluorescence images of DNA damage markers γH2AX and 53BP1 in the nuclei of WT and 4KR mutant HeLa cells. Cells were stained using anti-γH2AX and anti-53BP1 antibodies. Scale bars: 20 μm. (C) Representative immunofluorescence images of γH2AX and 53BP1 in the nuclei of WT of 4KR cells after UV irradiation (120 J/m2 UV, 3-h recovery) or treated with HU (1 mM, 3 h) or MMC (18 μM, 3 h). Scale bars: 30 μm. (D) Quantification of γH2AX- and 53BP1-positive nuclei under each DNA damaging condition. Values are mean ± SD of three independent assays. P-values were calculated using Student’s t-test. *P < 0.05, **P < 0.01. (E) Western blot analysis using anti-γH2AX and anti-53BP1 antibodies in WT and 4KR cells with or without exposure to UV irradiation. β-actin was used as a loading control.
Figure 8.
Defective SUMO-1 modification of FEN1 impairs cell cycle progression and sensitizes cells to replication fork-stalling agents. Survival rates of WT and 4KR mutant HeLa cells, determined using MTS cell proliferation assays after treatment with varying levels of UV irradiation (A) or HU (B) or MMC (C) treatment, are shown. Survival rates were calculated as the absorbance 490 (A490) of a sample divided by the A490 of the corresponding untreated control, multiplied by 100%. Values are mean ± SD of three independent assays. P-values, indicated in each panel, were calculated using ANOVA test.
Discussion
We have demonstrated SUMO-1 modification of FEN1 in vitro and have determined that, like other SUMOylation reactions, FEN1 SUMO-1 modification is mediated by the SUMO-conjugating enzyme Ubc9, as knockdown of Ubc9 reduces SUMO-1-modified FEN1 in HeLa cells. We also revealed that, similar to SUMO-3 modification, SUMO-1 modification of FEN1 is stimulated by its phosphorylation. However, whereas SUMO-3 modification of FEN1 plays an important role in controlling FEN1 protein levels in a cell cycle-dependent manner (Guo et al., 2012), SUMO-1 modification of FEN1 is induced by DNA damage or replication stresses. FEN1 is demethylated in response to DNA insults, which allows it to be phosphorylated and subsequently linked to the SUMO-1 protein. Unlike SUMO-3 modification, which induces ubiquitination and degradation of FEN1 (Guo et al., 2012), SUMO-1 modification does not trigger FEN1 ubiquitination. Instead, FEN1 SUMO-1 modification promotes the interaction of FEN1 with the Rad9–Rad1–Hus1 complex, a PCNA-like DNA clamp heterotrimer (Wang et al., 2004; Querol-Audi et al., 2012).
The Rad9–Rad1–Hus1 complex is recruited early in response to DNA damage and serves as a platform to recruit other DNA damage response and repair proteins to damaged DNA sites (Aravind et al., 1999; Parrilla-Castellar et al., 2004; Dore et al., 2009; Bai et al., 2010). Previous studies found that targeted deletion of the HUS1 subunit of the Rad9–Rad1–Hus1 complex in mice resulted in spontaneous chromosomal abnormalities and embryonic lethality, indicating that HUS1, a cell cycle checkpoint protein, plays a central role in genome maintenance by mediating cellular responses to DNA damage and replication stress (Levitt et al., 2005). Our current study reveals a new function of the Rad9–Rad1–Hus1 complex, together with PCNA, in mediating the function of in DNA replication and repair. Under physiological conditions, FEN1 binds to PCNA during S phase to cleave RNA–DNA flaps, generating ligatable DNA ends to join two Okazaki fragments during lagging-strand DNA synthesis. However, UV irradiation or exposure to DNA damaging agents induces FEN1 phosphorylation and subsequent SUMO-1 modification. FEN1 phosphorylation triggers the nuclease to dissociate from PCNA and SUMO-1 modification may induce conformational changes that favor the binding of FEN1 to the Rad9–Rad1–Hus1 complex rather than to PCNA. Thus, FEN1 switches from RNA primer removal to processing of stalled replication forks.
DNA replication stress is a feature of the initiation and progression of cancers (Zheng et al., 2012; Macheret and Halazonetis, 2015). Thus, DNA replication stress has been proposed as a new target for cancer prevention (Zheng et al., 2012; O’Connor, 2015; Liu et al., 2016). Meanwhile, conventional chemotherapy agents, including cisplatin and MMC, have been found to kill cancer cells by producing DNA lesions that block replication forks (Cheung-Ong et al., 2013). To counteract endogenous and exogenous replication stresses, cancer cells have hijacked various cellular mechanisms, including the expression of DNA repair genes and the induction of protein modifications, to facilitate DNA replication and DNA damage responses and repair (Peng et al., 2012; Zheng et al., 2012; Wang et al., 2015). Therefore, inhibition of these mechanisms may sensitize cancer cells to chemotherapy or radiotherapy. We previously showed that inhibiting YY1-mediated FEN1 expression sensitizes cancer cells to MMC (Wang et al., 2015). Here, we demonstrated that blocking SUMO-1 modification of FEN1 renders HeLa cells more sensitive to HU and MMC. Therefore, our current study has provided a new target for increasing cancer cell sensitivity to chemotherapy and radiotherapy.
Materials and methods
Reagents
Mouse monoclonal antibodies against FEN1, CDK2, and CIdU were from Abcam. Anti-IdU was from BD Biosciences; anti-FLAG was from Santa Cruz Biotechnology; antibodies against PCNA, FEN1, SUMO-1, HUS1, RAD9, RAD1, and β-actin were from Proteintech; anti-γH2AX and secondary antibodies were from Cell Signaling Technology. Mouse/rabbit Duolink® PLA and anti-FLAG M2 magnetic beads were from Sigma-Aldrich. Dulbecco’s modified Eagle’s medium (DMEM) with high glucose, fetal bovine serum (FBS), penicillin and streptomycin (PS), and phosphate buffer solution (PBS) were obtained from HyClone, GE Healthcare.
Cell culture and transfection
Cells were cultured in DMEM medium supplemented with 10% FBS and 1% PS. All cells were cultured in a humidified incubator containing 5% CO2 at 37°C. HeLa cells were transfected with plasmids carrying pEZ-3×FLAG-FEN1-WT (or 4KR)-neo using TG-transfection (Top Gene Technologies). HeLa cells stably expressing WT or 4KR FEN1 were selected using G418 (Sangon) and verified by DNA sequencing. For Ubc9 knockdown, cells were transfected with siRNA against Ubc9 (si-Ubc9) or a scrambled siRNA control. Knockdown efficiency was evaluated by western blot analysis. To suppress phosphorylation, cells were treated with the CDK inhibitor olomoucine (ab120938) before harvest (Henneke et al., 2003).
Cell synchronization
HeLa cells were seeded until they reached the logarithmic growth phase and then were synchronized by incubation in 2.5 mmol/L radiolabeled thymidine (TdR) medium for 16 h, followed by release in TdR-free medium for 9 h. Cells were treated again with TdR for 16 h, followed by TdR-free medium, in which they were released to enter S phase after 3 h (Harper, 2005). To synchronize cells at the G2/M phase, HeLa cells were treated with nocodazole (400 ng/ml) for 16 h as we previously described (Guo et al., 2012). Synchronized cells were washed with PBS buffer and cultured in regular DMEM to release them to cell cycle progression. The cell cycle was monitored by flow cytometry analysis.
Protein expression and purification
Plasmids encoding 6×His-tagged FEN1 (pET28b-FEN1), MBP-tagged PCNA (pET28a-MBPTEV-PCNA, no 6His tag), GST-tagged HUS1 (pQE30-HUS1), was transformed into Escherichia coli BL21 (DE3). The transformants were grown at 37°C overnight in LB medium in the presence of Kanamycin (50 μg/ml) or ampicillin (50 μg/ml). Bacterial cultures were diluted with fresh LB medium and incubated for 3 h at 30°C. The cells were induced by 0.4 mM isopropyl-thio-β-D-galactopyroanoside (Sigma) for 4 h, and then collected by centrifugation. Cell lysates were prepared by ultrasonication. 6×His-tagged FEN1 proteins were purified using the Ni2+ NTA affinity chromatograph as previously described (Frank et al., 2001). MBP-tagged PCNA was purified using the amylose affinity chromatograph following the previously described procedure (Lebendiker and Danieli, 2011). GST-tagged HUS1 was expressed and purified according to a GST-based affinity purification protocol (Harper and Speicher, 2011). Protein concentrations were determined using the Bradford protein assay, and protein purity was evaluated by SDS-PAGE (Bio-Rad).
In vitro SUMOylation analysis
A commercial SUMO-1 modification kit (R&D Systems) was used for in vitro SUMOylation. Purified His-tagged FEN1 (5 μg) was incubated with the reaction mixture containing 1 μg of SUMO-activating enzyme E1 (SAE1/SAE2), 1 μg of the conjugating enzyme E2 (Ubc9), 5 μg of SUMO-1 in reaction buffer (50 mM Tris pH 8.0, 5 mM or no ATP, and 5 mM MgCl2) in a total volume of 20 μl. The reaction was carried out at 37°C for 60 min. To detect the unmodified FEN1 and SUMO-1-FEN1 in the same blot, western blot analysis using the anti-FEN1 antibody was conducted. To specifically detect SUMO-1-FEN1, western blot analysis using the anti-SUMO-1 antibody was conducted.
Immunoprecipitation and western blot analysis
Cell pellets were suspended in whole cell extraction buffer containing 50 mM Tris–HCl (pH 7.5), 5 mM EDTA, 150 mM NaCl, 0.1% NP-40 lysis buffer, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Sangon). To isolate endogenous FEN1 complexes, the whole cell lysates were incubated with anti-FEN1 at 4°C overnight and incubated with Protein A/G PLUS Agarose (Santa Cruz Biotechnology). After washing five times with RIPA buffer (Sangon) and PBS. To purify the 3×FLAG-tagged FEN1 complex proteins, the cell extracts were incubated with anti-FLAG M2 magnetic beads. After extensively washing, 3×FLAG-FEN1 proteins were eluted by directly boiling the beads in SDS loading buffer. The proteins were resolved in 4%–15% SDS-PAGE and transferred onto a PVDF membrane for western blot analysis. To detect SUMO-1-FEN1, immunoblot analysis was conducted using the anti-SUMO-1 antibody, which specifically detect SUMO-1 but not SUMO-2/3 modified FEN1. The total FEN1 or 3×FLAG-FEN1 was detected using the anti-FEN1 or anti-FLAG antibody. To detect PCNA, RAD1, or HUS1 that is co-pulled down with FEN1, immunoblot analysis was conducted using the anti-PCNA, anti-RAD1, or anti-HUS1, respectively. The β-actin or GAPDH in the input was used as a control to ensure equal loading. Protein bands were developed using Pierce ECL substrate from Thermo Fisher Scientific and visualized on X-ray film. The band intensity was quantified using the Image J program.
In vitro pull-down assay
Purified recombinant 6×His-tagged FEN1 was incubated with Ubc9, SUMO-1, and other components of the SUMOylation kit, with or without ATP, at 37°C for 60 min. Unmodified FEN1 or SUMO-1-FEN1 was incubated with Ni-NTA beads. After extensive washing with PBS buffer, the beads were incubated with a mixture of purified MBP-tagged PCNA and GST-tagged HUS1 proteins at room temperature for 30 min. Incubation of FEN1-Ni-NTA beads with non-conjugated MBP and GST proteins without ATP served as negative controls for non-specific binding of the tag proteins to FEN1. In addition, to exclude the non-specific binding to beads, Ni-NTA beads only (no FEN1) were incubated with the mixture of MBP-PCNA and GST-HUS1. Unmodified FEN1 or SUMO-1 FEN1 was detected by western blot analysis using the anti-FEN1 antibody. FEN1-bound MBP-PCNA and GST-HUS1 were analyzed by western blot analysis using anti-PCNA, anti-HUS1, anti-MBP, and anti-GST antibodies.
Duolink® PLA
Duolink® PLA was used to identify the SUMO-1 modification of FEN1, as well as its interactions with other proteins. After treatment with 120 J/m2 UV irradiation or DNA damaging agents, freshly dissociated cells stably expressing 3×FLAG-WT-neo and 3×FLAG-4KR-neo were plated at a density of 2 × 104 per cm2 on glass cover slips (25 mm diameter) in a 6-well plate. Cells were then fixed, permeabilized, blocked, and incubated with primary antibodies against 3×FLAG and the other protein (SUMO-1, PCNA, HUS1, or RAD1), as well as the PLA probe anti-mouse MINUS (DUO92004) and PLA probe anti-rabbit PLUS (DUO92002). Proximity ligation was then conducted in situ according to the manufacturer’s instructions. HeLa cells not expressing 3×FLAG-tagged FEN1 served as negative controls and HeLa cells stably expressing 3×FLAG-tagged FEN1 were positive controls. When the PLA probes are in close proximity (<40 nm), bright fluorescent emissions can be detected and quantified by a fluorescence microscope. Fluorescent emissions were observed using confocal laser scanning microscopy (Nikon).
Immunofluorescence staining
DNA damage was determined using γH2AX and 53BP1 as readouts. After UV irradiation or treatment with DNA damaging agents, cells were allowed to recover for 1 h at 37°C before fixation with 4% formaldehyde in PBS for 20 min at room temperature. The fixed cells were washed twice with PBS and permeabilized in 0.2% Triton X-100 in PBS for 5 min. After washing twice with PBS, the slides were blocked with blocking buffer (0.2% Triton X-100, 5% BSA in PBS) for 60 min and incubated with anti-γH2AX (EMD Millipore) or anti-53BP1 (Abcam) antibody. After washing with PBS, the slide was incubated with the anti-rabbit secondary antibody Alexa 594 (Proteintech). The slides were visualized using Leica DM400 and quantified by ImagePro.
DNA replication and cell proliferation
DNA synthesis and cell proliferation were analyzed using the EdU incorporation assay (Cell-Light EdU Apollo 488 In Vitro Imaging Kit; RiboBio), as previously described (Salic and Mitchison, 2008). Cells were synchronized to S phase and pre-incubated with EdU for 30 min, washed three times with PBS, and treated with 120 J/m2 UV. Immunofluorescence imaging of EdU was performed as described by the manufacturer. Images were taken with a Zeiss AXIO Observer microscope.
MTS cell proliferation assay
Sensitivity to various DNA damage reagents was determined using an MTS cell proliferation assay. HeLa cells were seeded in 96-well plates (1500/well), incubated (overnight, 37°C), synchronized to S phase, and treated with varying concentrations of HU and MMC (for 18 h, 37°C) or exposed to different doses of UV irradiation. The cells were then washed in a fresh medium (DMEM containing 10% FBS), incubated under normal growth conditions (37°C, 5% CO2) for 1.5 h. The number of viable cells was determined using the CellTiter 96® AQueous One Solution Reagent (Promega). At least four replications were averaged for each treatment.
DNA fiber assay
DNA replication progression was analyzed using the DNA fiber assay (Xu et al., 2011; Liu et al., 2016). Cells were labeled with IdU (25 μM) for 30 min, followed by exposure to HU (4 mM) or DMSO for 4 h, and chased with CldU (125 μM) for 40 min. Cells were lysed, DNA fibers were spread, and the IdU and CldU tracts were stained and detected, as previously described (Xu et al., 2011; Liu et al., 2016). DNA fibers were imaged using a Leica DM4000 fluorescent microscope.
Cell cycle analysis
HeLa cells stably expressing WT or 4KR FEN1 were synchronized at G2/M phase by nocodazole treatment (0.4 μg/ml, 18 h). The cells were released into cell cycle progression by replacing the medium with fresh DMEM containing no nocodazole. Cells were harvested at 4, 5, 6, 16, 17, 21, 22, 23, and 24 h, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle phases (Late S/G2, S, and G1) were determined based on DNA contents using MultiCycle AV DNA Analysis software.
Mass spectrometry analysis of SUMOylation sites
SUMOylation of FEN1 was performed with the T95K SUMO-1 mutant tags modified lysines with a diglycine (GG) remnant, which can be detected using mass spectrometry (Knuesel et al., 2005). SUMOylated FEN1 bands were excised from SDS-PAGE gels, followed by in-gel reduction, alkylation, and endoproteinase digestion with GluC (Roche), Trypsin (Promega), or a mixture of GluC and Trypsin. LC-MS/MS data were acquired using an Eksigent nanoLC-2D equipped with a self-packed C18 column connected to a hybrid linear ion trap (LTQ-FT) mass spectrometer (Thermo Electron). MS/MS spectra were matched to a database of generated FASTA files created by ChopNSpice (chopnspice.gwdg.de) using the Global Proteome Machine Database (GPMDB) X!Tandem search engine. All identified SUMOylated MS spectra were reconfirmed manually. In the figures, the b and y ion series members are numbered from the N-terminus.
Supplementary Material
Acknowledgements
We thank Dr Kerin K. Higa of City of Hope for her critical reading and editing of this manuscript. We are grateful to the flow cytometry and microscopy core facilities of Zhejiang University School of Medicine and Institute of Translational Medicine for their contributions to this work. We also thank Dr David Waugh (National Cancer Institute) for the generous gift of the protein expression vector pET28a-MBPTEV.
Funding
This work was supported by grants from the National Basic Research Program of China (2015CB910600), the National Natural Science Foundation of China (31700688), the National Key Research and Development Program of China (2017YFA0503900), and the Natural Science Foundation of Zhejiang Province (LY16C050003) to Y.J.H. and H.X. A part of the work presented in the current article was supported by the National Institutes of Health grants R01CA073764 to B.H.S and R50CA211397 to L.Z.
Conflict of interest
none declared.
References
- Anderson L., Henderson C., and Adachi Y. (2001). Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21, 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Walker D.R., and Koonin E.V. (1999). Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R., Gomes X.V., Gordenin D.A., et al. (2003). Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278, 1618–1625. [DOI] [PubMed] [Google Scholar]
- Bae S.H., Bae K.H., Kim J.A., et al. (2001). RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461. [DOI] [PubMed] [Google Scholar]
- Bai H., Madabushi A., Guan X., et al. (2010). Interaction between human mismatch repair recognition proteins and checkpoint sensor Rad9-Rad1-Hus1. DNA Repair (Amst) 9, 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Ong K., Giaever G., and Nislow C. (2013). DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem. Biol. 20, 648–659. [DOI] [PubMed] [Google Scholar]
- Chung L., Onyango D., Guo Z., et al. (2015). The FEN1 E359K germline mutation disrupts the FEN1-WRN interaction and FEN1 GEN activity, causing aneuploidy-associated cancers. Oncogene 34, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore A.S., Kilkenny M.L., Rzechorzek N.J., et al. (2009). Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex—implications for clamp loading and regulation. Mol. Cell 34, 735–745. [DOI] [PubMed] [Google Scholar]
- Frank G., Qiu J., Zheng L., et al. (2001). Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J. Biol. Chem. 276, 36295–36302. [DOI] [PubMed] [Google Scholar]
- Gary R., Park M.S., Nolan J.P., et al. (1999). A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19, 5373–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kanjanapangka J., Liu N., et al. (2012). Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol. Cell 47, 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Qian L., Liu R., et al. (2008). Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol. Cell. Biol. 28, 4310–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zheng L., Xu H., et al. (2010). Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 6, 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.V. (2005). Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol. Biol. 296, 157–166. [DOI] [PubMed] [Google Scholar]
- Harper S., and Speicher D.W. (2011). Purification of proteins fused to glutathione S-transferase. Methods Mol. Biol. 681, 259–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S., Stucki M., Hassa P.O., et al. (2001). Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell 7, 1221–1231. [DOI] [PubMed] [Google Scholar]
- Henneke G., Koundrioukoff S., and Hubscher U. (2003). Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene 22, 4301–4313. [DOI] [PubMed] [Google Scholar]
- Kim K., Biade S., and Matsumoto Y. (1998). Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 273, 8842–8848. [DOI] [PubMed] [Google Scholar]
- Klungland A., and Lindahl T. (1997). Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M., Cheung H.T., Hamady M., et al. (2005). A method of mapping protein sumoylation sites by mass spectrometry using a modified small ubiquitin-like modifier 1 (SUMO-1) and a computational program. Mol. Cell. Proteomics 4, 1626–1636. [DOI] [PubMed] [Google Scholar]
- Lebendiker M., and Danieli T. (2011). Purification of proteins fused to maltose-binding protein. Methods Mol. Biol. 681, 281–293. [DOI] [PubMed] [Google Scholar]
- Levitt P.S., Liu H., Manning C., et al. (2005). Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics 86, 212–224. [DOI] [PubMed] [Google Scholar]
- Liu S., Lu G., Ali S., et al. (2015). Okazaki fragment maturation involves α-segment error editing by the mammalian FEN1/MutSα functional complex. EMBO J. 34, 1829–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhou M., Li Z., et al. (2016). A selective small molecule DNA2 inhibitor for sensitization of human cancer cells to chemotherapy. EBioMedicine 6, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kao H.I., and Bambara R.A. (2004). Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73, 589–615. [DOI] [PubMed] [Google Scholar]
- Macheret M., and Halazonetis T.D. (2015). DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 10, 425–448. [DOI] [PubMed] [Google Scholar]
- Noll D.M., Mason T.M., and Miller P.S. (2006). Formation and repair of interstrand cross-links in DNA. Chem. Rev. 106, 277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor M.J. (2015). Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar E.R., Arlander S.J., and Karnitz L. (2004). Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair 3, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Peng G., Dai H., Zhang W., et al. (2012). Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA end resection. Cancer Res. 72, 2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Orta M.L., Issaeva N., et al. (2010). Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol-Audi J., Yan C.L., Xu X.J., et al. (2012). Repair complexes of FEN1 endonuclease, DNA, and Rad9-Hus1-Rad1 are distinguished from their PCNA counterparts by functionally important stability. Proc. Natl Acad. Sci. USA 109, 8528–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., et al. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868. [DOI] [PubMed] [Google Scholar]
- Saharia A., Guittat L., Crocker S., et al. (2008). Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 18, 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., and Mitchison T.J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA 105, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathi S., and Chai W. (2011). Mapping the FEN1 interaction domain with hTERT. Biochem. Biophys. Res. Commun. 407, 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Otterlei M., Sommers J.A., et al. (2004). WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol. Biol. Cell 15, 734–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Zheng L., Chavez V., et al. (2007). Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 282, 3465–3477. [DOI] [PubMed] [Google Scholar]
- Soderberg O., Gullberg M., Jarvius M., et al. (2006). Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000. [DOI] [PubMed] [Google Scholar]
- Stucki M., Jonsson Z.O., and Hubscher U. (2001). In eukaryotic flap endonuclease 1, the C terminus is essential for substrate binding. J. Biol. Chem. 276, 7843–7849. [DOI] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi N., Gaida G.M., et al. (1997). A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88, 253–263. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhou L., Li Z., et al. (2015). YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC Cancer 15, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Brandt P., Rossi M.L., et al. (2004). The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl Acad. Sci. USA 101, 16762–16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zheng L., Dai H., et al. (2011). Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice. Oncogene 30, 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., and Freudenreich C.H. (2007). Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene 393, 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Hegde M.L., et al. (2011. a). Fen1 mutations that specifically disrupt its interaction with PCNA cause aneuploidy-associated cancer. Cell Res. 21, 1052–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Qiu J., et al. (2007. a). Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol. Cell. Biol. 27, 3176–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Zhou M., et al. (2007. b). Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat. Med. 13, 812–819. [DOI] [PubMed] [Google Scholar]
- Zheng L., Dai H., Zhou M., et al. (2012). Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat. Commun. 3, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Jia J., Finger L.D., et al. (2011. b). Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 39, 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., and Shen B. (2011). Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 3, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Zhou M., Chai Q., et al. (2005). Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 6, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Dai H., Wu J., et al. (2017). Role of FEN1 S187 phosphorylation in counteracting oxygen-induced stress and regulating postnatal heart development. FASEB J. 31, 132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.