Abstract
Phylogenomics and genome scale positive selection analyses were performed on 29 Corynebacterium pseudotuberculosis genomes that were isolated from different hosts, including representatives of the Ovis and Equi biovars. A total of 27 genes were identified as undergoing adaptive changes. An analysis of the clades within this species and these biovars, the genes specific to each branch, and the genes responding to selective pressure show clear differences, indicating that adaptation and specialization is occurring in different clades. These changes are often correlated with the isolation host but could indicate responses to some undetermined factor in the respective niches. The fact that some of these more-rapidly evolving genes have homology to known virulence factors, antimicrobial resistance genes and drug targets shows that this type of analysis could be used to identify novel targets, and that these could be used as a way to control this pathogen.
Introduction
Population genetics and genomic approaches increase our understanding of both natural selection and molecular evolution. Alleles with adaptive mutations increase in frequency in what is known as positive selection, and these mutations have been identified by comparing nucleotide sequences between different populations [1–3]. Codon substitution models, which compare a non-synonymous (dN) to synonymous (dS) substitution rate (as ω = dN / dS), can be used to determine if the mutations that change the amino acid (dN) in a specific position are adaptive (ω > 1, positive selection), deleterious (ω < 1, negative selection) or neutral (ω = 1, neutral evolution) [4]. Research has shifted from looking at selective pressures on individual genes to a broad examination that looks for genes under selective pressure across entire genomes [5–7], and the pipelines developed to examine this often involve orthologous group identification, codon based alignments, phylogenetic tree reconstruction, and models of codon evolution [3,8].
The interactions between a host and its infecting pathogen have been of particular interest to those interested in positive selection, particularly in the interactions that involve the immune and defense mechanisms deployed by the host. Pathogen genes that have been identified as being under positive selection have been found to be involved in regulation, modulation and modification of the host immune response, membrane lipid metabolism, certain cell wall processes, and receptor mediated binding [6,9], all of which could play a role in host-pathogen interactions. Several studies have examined selective pressures and the response in many important pathogenic bacteria, including Escherichia coli [9,10], Salmonella [10], Staphylococcus aureus [11], Mycobacterium tuberculosis [12–14], Shigella flexneri [9], and members of the Streptoccocus [15], Campylobacter [16] and Leptospira [17,18] genera.
Corynebacterium pseudotuberculosis is a Gram-positive, pleomorphic and facultative intracellular bacterium of veterinary and medical relevance. It has a global distribution [19], and it causes economic losses in animal production. Control methods, such as diagnosis, vaccines and antibiotics remain elusive [19]. It is separated into two biovars based on host preference and nitrate reduction. Biovar Ovis (nitrate negative) is the causative agent of Caseous Lymphadenitis (CLA), a chronic disease in goats and sheep [20,21]. Ovis has also been isolated from cattle [22], camels [23], and humans [21,24,25], causing skin lesions or lymphadenitis. Isolates from the Equi biovar (nitrate positive) are known for causing Oedematous Skin Disease (OSD) in buffaloes [26]. Equi isolates have also been found in horses [27,28], cattle [22,29] and camels [30], with different manifestation in each host species. Cattle and camels are the only cross-over hosts in that both Ovis and Equi strains have been isolated from them, but each biovars present a different disease phenotype. Ovis has never been found in horses or buffalo, and no sheep or goats have been found to be infected by any strain belonging to the Equi biovar. However, an experimental infection of a strain isolated from a buffalo and part of the Equi biovar caused CLA in sheep [31].
While previous work has identified changes specific to each of the C. pseudotuberculosis biovars [32,33], no one has been able to identify any genes that are involved in the interactions between pathogen and host species. A single exception is probably the presence of a particular prophage that harbors the diphtheria toxin (DT) and is found only in strains isolated from buffalo (Equi biovar) [33]. Phylogeny of the species show that the two biovars are clearly distinct. In this work, we examined nucleotide changes in genes shared by both biovars in order to identify differences in selective pressure as a means to explore the evolution of this pathogen, and to distinguish genes that might be involved in host-pathogen interactions and host preference.
Materials and methods
Genomes and reannotation
Positive selection analysis that includes all of the genes in a single genome, and then compares a group genomes, is computationally expensive [5,6]. We limited the sampling to 29 complete genomes of C. pseudotuberculosis that were retrieved from GenBank. These genomes represent isolates from both biovars, and from each type of host that has been found to be infected with C. pseudotuberculosis. A maximum of five genomes were included from each type of host, depending upon availability (Table 1). All genomes were all consistently annotated using the RASTtk (Rapid Annotation Using Subsystem Technology) [34] annotation service in the Pathosystem Resource Integration Center (PATRIC) [35].
Table 1. Corynebacterium pseudotuberculosis genomes used in positive selection analysis.
| Strain | Biovar | Host | Country | Access no |
|---|---|---|---|---|
| E56 | Ovis | Sheep | Egypt | CP013699.1 |
| PA01 | Ovis | Sheep | Brazil | CP013327.1 |
| C231 | Ovis | Sheep | Australia | CP001829.1 |
| MEX25 | Ovis | Sheep | Mexico | CP013697.1 |
| N1 | Ovis | Sheep | Equatorial Guinea | CP013146.1 |
| 1002B | Ovis | Goat | Brazil | CP012837.1 |
| VD57 | Ovis | Goat | Brazil | CP009927.1 |
| PO222/4-1 | Ovis | Goat | Portugal | CP013698.1 |
| MEX1 | Ovis | Goat | Mexico | CP017711.1 |
| MEX9 | Ovis | Goat | Mexico | CP014543.1 |
| P54B96 | Ovis | Wildebeest | South Africa | CP003385.1 |
| 267 | Ovis | Llama | USA | CP003407.1 |
| 48252 | Ovis | Human | Norway | CP008922.1 |
| FRC41 | Ovis | Human | France | CP002097.1 |
| I19 | Ovis | Cow | Israel | CP002251.1 |
| 29156 | Ovis | Cow | Israel | CP010795.1 |
| 262 | Equi | Cow | Belgium | CP012022.1 |
| I37 | Equi | Cow | Israel | CP017384.1 |
| 162 | Equi | Camel | UK | CP013260.1 |
| 258 | Equi | Horse | Belgium | CP003540.2 |
| MB14 | Equi | Horse | USA | CP013261.1 |
| E19 | Equi | Horse | Chile | CP003540.2 |
| MEX30 | Equi | Horse | Mexico | CP017291.1 |
| CIP52.97 | Equi | Horse | Kenya | CP003061.2 |
| 31 | Equi | Buffalo | Egypt | CP003421.3 |
| 32 | Equi | Buffalo | Egypt | CP015183.1 |
| 33 | Equi | Buffalo | Egypt | CP015184.1 |
| 36 | Equi | Buffalo | Egypt | CP015186.1 |
| 48 | Equi | Buffalo | Egypt | CP015191.1 |
Genome scale positive selection analysis
Positive selection analysis using branch-site models has been used to identify genes and specific codons (sites) that are under positive selection in specific phylogenetic lineages (also called directional selection. When doing this type of comparison, the lineage to be tested for positive selection is identified as the “foreground”, and the genomes compared to that foreground lineage are labeled as “background”. This comparison will identify specific sites that are under positive selection (ω > 1) only in the foreground lineage, evidencing its adaptive mutations [36,37]. Once identified, the functional roles of these genes can be explored, and they can play in part in future hypothesis generation [38].
The PosiGene pipeline [7] was used to perform genome-scale positive selection in this analysis using branch-site models. Multifasta files containing the protein-coding sequences of each gene of the 29 genomes were generated, with the RASTtk sequence IDs modified to a format suitable for PosiGene (RASTtk-based IDs) using a modified version of the script extract_aa_nt_from_gb.pl (S1 File) [6]. The input files for each genome are provided in S2 File.
Ortholog group assignment
The PosiGene module “create_catalog”, which uses a BLASTp best-bidirectional hit analysis [39,40], was used to assign ortholog groups. Each group was named after the ID from the sequence of a reference genome and only ortholog groups that have a sequence from an anchor genome were analyzed. A reference or anchor genome was selected according to the biovar of the foreground genomes, to avoid missing genes that are more common in a specific biovar. Strain 31, a buffalo isolate, was selected as the reference and anchor genome for Equi biovar, and strain 1002B (goat) was selected for Ovis.
Alignments, gene trees and species tree
The PosiGene module “alignments” was used to generate multiple sequence alignments. This module also created a phylogenetic tree for each ortholog group, and a species tree (consensus tree). The species tree was used for realignment of the nucleotide sequences by codon and posterior identification of the target groups.
A sequence filter based on similarity, with a minimal sequence identity of 50%, was performed to ensure the analysis of one sequence per genome on each ortholog group [7]. For each gene sequence from the reference genome, the orthologs from all genomes were assigned by progressive protein alignments using CLUSTALW [41,42].
A phylogenetic tree of each ortholog group was generated by alignment filtering using GBLOCKS [43] and phylogenetic reconstruction by the parsimony method and jackknifing using DNAPARS from the PHYLIP package [44]. For the species tree, a consensus tree was calculated using PHYLIP’s CONSENSE program. Codon level alignments were generated using PRANK [45] for each ortholog group that had at least three sequences, and also for the species tree.
Target groups
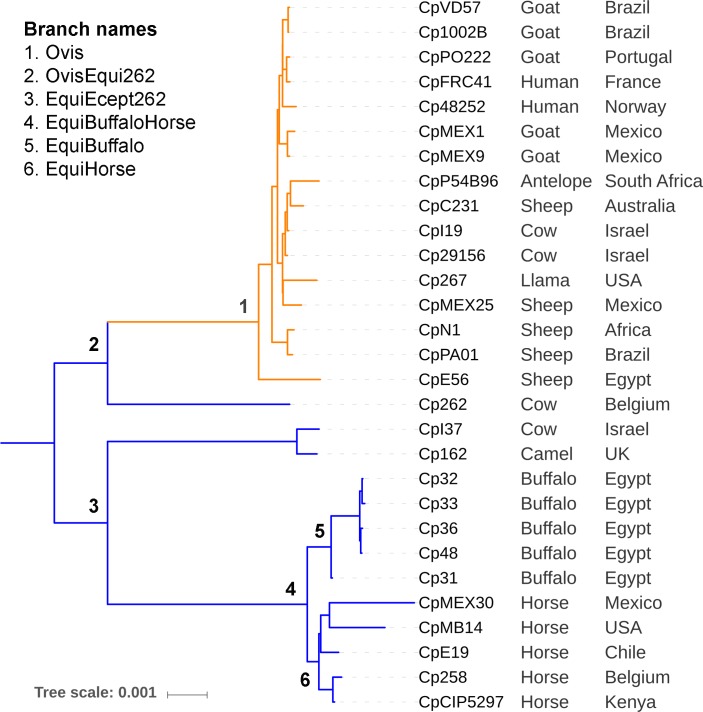
The species tree had to be manually rooted prior to the selection target groups. To identify the most ancestral branch of C. pseudotuberculosis, a second tree was generated that included C. ulcerans strain 210932 (CP009500.1) [46] to root the tree (S1 Fig) and identify the most ancestral C. pseudotuberculosis clade. The first C. pseudotuberculosis species tree (without C. ulcerans) was then manually rooted using MEGA 7 [47] and visualized with iTOL 4.2.3 (itol.embl.de) (Fig 1) to identify foreground groups and to be used in the next step of the PosiGene pipeline. We compared this tree (Fig 1) with trees generated by other two methods to compare and confirm phylogenetic placement. One of these comparison trees was built using the PEPR (https://github.com/enordber/pepr.git) (S2 Fig), a pipeline that uses the core proteome and builds an alignment of all the genes shared across all genomes. Another comparison tree was built using MEGA 7 and the Maximum Likelihood method [48]. This tree was generated based on the alignment the rpoB gene (S3 Fig), which has been described a good discriminator for differentiating between Corynebacterium species [49].
Fig 1. Target groups (foreground branches) 1 to 6 of a Corynebacterium pseudotuberculosis phylogeny.
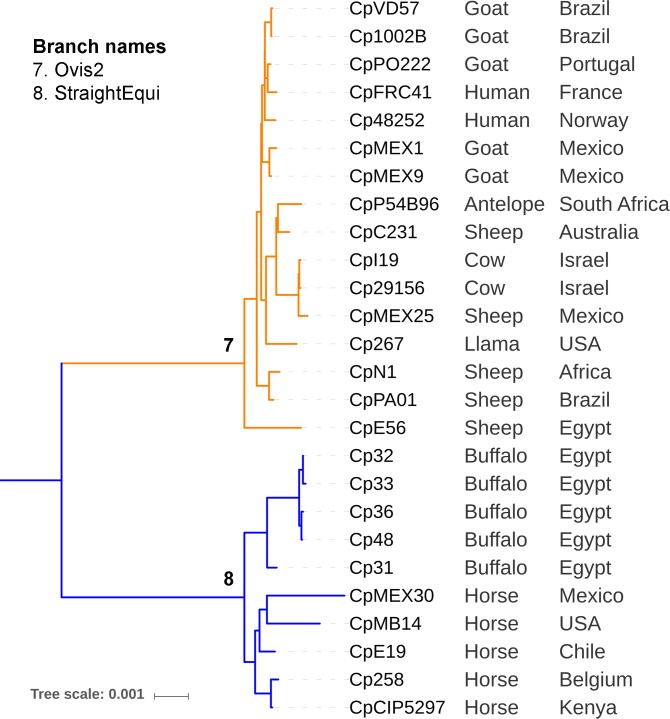
Eight separate foreground groups were used as input for PosiGene. These were selected based on the clades that were identified by the phylogenetic trees (Figs 1 and 2). This resulted in eight separate analyses, each one comparing a foreground group with the remaining groups in the tree (background), to identify adaptive mutations that occurred only in the last common ancestor of the foreground group. The target groups are listed in Table 2 and are represented in the phylogenomic trees of Figs 1 and 2.
Fig 2. Target groups (foreground branches) 7 and 8 of a Corynebacterium pseudotuberculosis phylogeny excluding the Equi strains 262, I37 and 162.
Table 2. Groups of foreground and background lineages of Corynebacterium pseudotuberculosis analyzed by branch-site models.
| Group number | Group name | Foreground (genomes) | Background (genomes) | Reference/anchor genome |
|---|---|---|---|---|
| 1 | Ovis | All Ovis genomes (16) | All Equi genomes (13) | Cp1002B (Ovis) |
| 2 | OvisEqui262 | All Ovis genomes and Equi 262 (17) | All Equi genomes except 262 (12) | Cp1002B (Ovis) |
| 3 | EquiExcept262 | All Equi genomes except 262 (12) | All Ovis genomes and Equi 262 (17) | Cp31 (Equi) |
| 4 | EquiBuffaloHorse | Equi genomes from buffalo and horse only (10) | All other Ovis and Equi genomes (19) | Cp31 (Equi) |
| 5 | EquiBuffalo | Equi genomes from buffalo only (5) | All other Equi and Ovis genomes (24) | Cp31 (Equi) |
| 6 | EquiHorse | Equi genomes from horse only (5) | All other Equi and Ovis genomes (25) | Cp31 (Equi) |
| 7 | Ovis2 | All Ovis genomes (16) | Equi genomes from buffalo and horse only (10) | Cp1002B (Ovis) |
| 8 | StraightEqui | Equi genomes from buffalo and horse only (10) | All Ovis genomes (16) | Cp31 (Equi) |
Positive selection module
The codeml program of the PAML package [8] was used to identify sites under positive selection by a branch-site test [36,37], which uses each gene sequence alignment and its phylogenetic gene tree as input. The likelihood ratio test (LRT) calculates and compares the likelihood of a null model, where all sites are considered to evolve under neutral (ω = 1) or negative selection (ω < 1), and an alternative model that assumes that the same sites are under positive selection (ω > 1) on the foreground branch only. The p-value for the LRT is calculated via a χ2 distribution, with one degree of freedom. For each site with a significant p-value, the Bayes empirical Bayes (BEB) method was used to calculate the posterior probability [50]. In addition to the p-value, the PosiGene pipeline provides the significance value for the Bonferroni correction and Benjamini–Hochberg false discovery rate (FDR) [51]. We considered positive selection when p < 0.05 for FDR only, as Bonferroni is too conservative and can lead to many false negatives [52]. For each gene that was identified as being under positive selected, the sequence alignment was tested for evidence of intragenic recombination, as it can lead to an alignment of non-homologous codons and possible false positive results [53,54]. As no single method performs optimally under all scenarios, our strategy involved a combination of all of them [55]. We used PhiPack [56] to test for evidence of recombination using the methods Pairwise Homoplasy Index (PHI) [56], Neighbor Similarity Score (NSS) [57] and Maximum Chi-Square [58]. We considered recombination when q < 0.05 for PHI and at least one another test [6].
Gene functional characterization and location
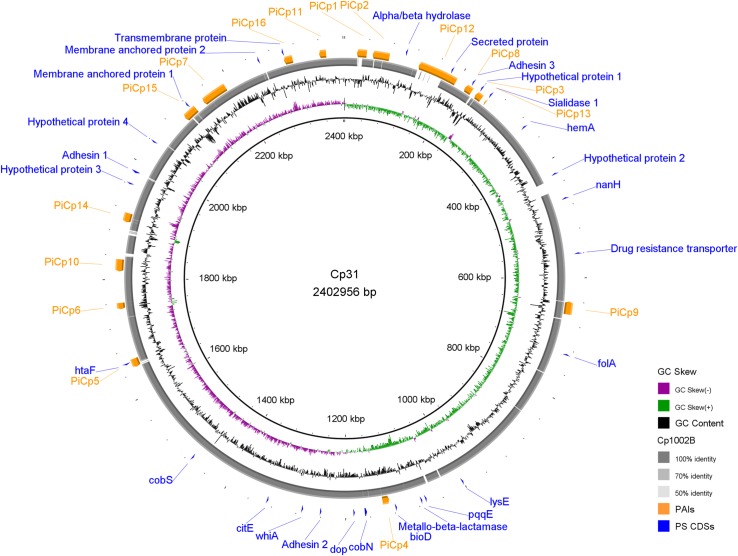
For each gene that the pipeline identified as being under positive selection, the sequence from the anchor genome was checked for the presence of functional domains using the InterProScan Database (https://www.ebi.ac.uk/interpro/search/sequence-search), and for metabolic activity using PATRIC’s Pathway Summary [35]. PATRIC’s Protein Family Sorter was used to verify the distribution of specific genes across the genomes. GIPSy [59] was used to verify the location of positively selected genes in relation to 16 pathogenicity islands that have been previously described [32], using C. glutamicum ATCC1302 (NC_006958.1) as the non-pathogenic reference. The positions of the positively selected genes were plotted in a circular map generated using BRIG Fig 3 [60].
Fig 3. Circular map showing the position of pathogenicity islands and positively selected genes in relation to Corynebacterium pseudotuberculosis strain 31 genome.
PAI–Pathogenicity Island, PS–positively selected, CDS–coding sequences.
Results and discussion
We used genome-scale positive selection analyses to identify adaptive mutations in specific lineages (branches or foregrounds) of C. pseudotuberculosis, and explored differences that could be correlated with biovar and isolation host.
Positively selected genes
The complete results for positive selection analysis for each foreground are provided (S3 File), as are the GenBank and RASTtk locus tags for each gene (S1 Table). Twenty-seven genes were identified as being under positive selection (Table 3) and the number of positively selected sites for each foreground is given in Table 4. Seven of the eight foreground groups had genes that were identified as being under positive selection, with the sole exception being Branch 6 (EquiHorse, Table 2). None of these 27 genes were significant for the recombination detection method (S2 Table).
Table 3. List of positively selected genes in Corynebacterium pseudotuberculosis in different branches (FDR < 0.05).
| GenBank ID (Equi/ Ovis) |
1 | 2 | 3 | 4 | 5 | 7 | 8 | Product (Gene) | Function | PAI | Reference (Drug target or Vaccine) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cp31_0488/ Cp1002B_0499 | X | Drug resistance transporter | Resistance | - | [62,63] | ||||||
| Cp31_1168/ Cp1002B_1500 | X | Citrate lyase beta chain (citE) | Metabolism | - | [64] | ||||||
| Cp31_1468/ Cp1002B_1186 | X | Cell-surface hemin receptor (hatF) | Transport | PiCp5 | - | ||||||
| Cp31_2169/ Cp1002B_0189 | X | Hypothetical protein 1 (no domains) | Unknown | PiCp3 | - | ||||||
| Cp31_0206/ Cp1002B_0207 | X | X | Sialidase 1 | Virulence | PiCp13 | [65,66] | |||||
| Cp31_0638/ Cp1002B_2037 | X | Dihydrofolate reductase (folA) | Metabolism | - | [67] | ||||||
| Cp31_0945/ Cp1002B_1731 | X | Coenzyme PQQ biosynthesis protein E (pqqE) | Metabolism | - | - | ||||||
| Cp31_0950/ Cp1002B_1726 | X | Metallo-beta-lactamase | Resistance | - | [68] | ||||||
| Cp31_0985/ Cp1002B_1689 | X | Dethiobiotin synthetase (bioD) | Metabolism | - | [69,70] | ||||||
| Cp31_1044/ Cp1002B_1624 | X | Pup deaminase (dop) | Metabolism | - | [71,72] | ||||||
| Cp31_1309/ Cp1002B_1363 | X | X | X | X | Cobalt chelatase subunit CobS (cobS) | Metabolism | - | - | |||
| Cp31_1724/ Cp1002B_0908 | X | X | X | X | Hypothetical protein 3 (no domains) | Unknown | - | - | |||
| Cp31_1868/ Cp1002B_0763 | X | Membrane anchored protein 1 | Unknown | PiCp13 | - | ||||||
| Cp31_0109/ Cp1002B_0104 | X | Alpha / beta hydrolase | Unknown | - | [73] | ||||||
| Cp31_2015/ Cp1002B_2083 | X | Transmembrane protein | Unknown | PiCp16 | - | ||||||
| Cp31_2279/ - | X | X | Adhesin 1 (membrane anchored) | Adhesion | - | - | |||||
| Cp31_0366/ Cp1002B_0381 | X | Hypothetical protein 2 (no domains) | Unknown | - | - | ||||||
| Cp31_1094/ Cp1002B_1575 | X | Adhesin 2 (membrane anchored) | Adhesion | - | - | ||||||
| Cp31_1977/ Cp1002B_0655 | X | Membrane anchored protein 2 | Unknown | - | - | ||||||
| Cp31_0279/ Cp1002B_0289 | X | Glutamyl-tRNA reductase (hemA) | Metabolism | - | [74] | ||||||
| Cp31_1028/ Cp1002B_1640 | X | Cobaltochelatase subunit CobN (cobN) | Metabolism | - | - | ||||||
| Cp31_1117/ Cp1002B_1551 | X | Sporulation regulator WhiA-like (whiA) | Cell division | - | - | ||||||
| Cp31_0142/ Cp1002B_0139 | X | X | Secreted protein | Unknown | PiCp12 | - | |||||
| Cp31_0180/ Cp1002B_0178 | X | X | Adhesin 3 (thioester domain) | Adhesion | PiCp8 | - | |||||
| Cp31_0399/ Cp1002B_0408 | X | X | Sialidase 2 (nanH) | Metabolism | - | [65,66] | |||||
| Cp31_0893/ Cp1002B_1784 | X | X | Lysine exporter protein (lysE) | Transport | - | [75] | |||||
| Cp31_2281/ Cp1002B_0835 | X | Hypothetical protein 4 (no domains) | Unknown | - |
PAI–Pathogenicity island
Table 4. Number and percentage of positively selected sites in Corynebacterium pseudotuberculosis.
| GenBank ID (Equi/ Ovis) | Alignment | Positively selected sizes per foreground (%) | Product (Gene) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 7 | 8 | |||
| Cp31_0488/ Cp1002B_0499 | 473 | 1 (0.21) | - | - | - | - | - | - | Drug resistance transporter |
| Cp31_1168/ Cp1002B_1500 | 300 | 2 (0.67) | - | - | - | - | - | - | Citrate lyase beta chain (citE) |
| Cp31_1468/ Cp1002B_1186 | 721 | 1 (0.14) | - | - | - | - | - | - | Cell-surface hemin receptor (hatF) |
| Cp31_2169/ Cp1002B_0189 | 208 | 13 (6.25) | - | - | - | - | - | - | Hypothetical protein 1 (no domains) |
| Cp31_0206/ Cp1002B_0207 | 465 | - | 5 (1.08) | 5 (1.08) | - | - | - | - | Sialidase 1 |
| Cp31_0638/ Cp1002B_2037 | 175 | - | 1 (0.57) | - | - | - | - | - | Dihydrofolate reductase (folA) |
| Cp31_0945/ Cp1002B_1731 | 412 | - | 1 (0.24) | - | - | - | - | - | Coenzyme PQQ biosynthesis protein E (pqqE) |
| Cp31_0950/ Cp1002B_1726 | 201 | - | 2 (1.00) | - | - | - | - | - | Metallo-beta-lactamase |
| Cp31_0985/ Cp1002B_1689 | 229 | - | 1 (0.44) | - | - | - | - | - | Dethiobiotin synthetase (bioD) |
| Cp31_1044/ Cp1002B_1624 | 510 | - | 3 (0.59) | - | - | - | - | - | Pup deaminase (dop) |
| Cp31_1309/ Cp1002B_1363 | 360 | - | 3 (0.83) | 3 (0.83) | - | - | 4 (1.11) | 4 (1.11) | Cobalt chelatase subunit CobS (cobS) |
| Cp31_1724/ Cp1002B_0908 | 42 | - | 1 (2.38) | 1 (2.38) | - | - | 1 (2.38) | 1 (2.38) | Hypothetical protein 3 (no domains) |
| Cp31_1868/ Cp1002B_0763 | 297 | - | 10 (3.37) | - | - | - | - | - | Membrane anchored protein 1 |
| Cp31_0109/ Cp1002B_0104 | 286 | - | - | 2 (0.7) | - | - | - | - | Alpha / beta hydrolase |
| Cp31_2015/ Cp1002B_2083 | 347 | - | - | 3 (0.86) | - | - | - | - | Transmembrane protein |
| Cp31_2279/ - | 868 | - | - | 20 (2.3) | 23 (2.65) | - | - | - | Adhesin 1 (membrane anchored) |
| Cp31_0366/ Cp1002B_0381 | 44 | - | - | - | 2 (4.55) | - | - | - | Hypothetical protein 2 (no domains) |
| Cp31_1094/ Cp1002B_1575 | 604 | - | - | - | 14 (2.32) | - | - | - | Adhesin 2 (membrane anchored) |
| Cp31_1977/ Cp1002B_0655 | 298 | - | - | - | 6 (2.01) | - | - | - | Membrane anchored protein 2 |
| Cp31_0279/ Cp1002B_0289 | 432 | - | - | - | - | 1 (0.23) | - | - | Glutamyl-tRNA reductase (hemA) |
| Cp31_1028/ Cp1002B_1640 | 1201 | - | - | - | - | 2 (0.17) | - | - | Cobaltochelatase subunit CobN (cobN) |
| Cp31_1117/ Cp1002B_1551 | 329 | - | - | - | - | 1 (0.30) | - | - | Sporulation regulator WhiA-like (whiA) |
| Cp31_0142/ Cp1002B_0139 | 213 | - | - | - | - | - | 15 (7.04) | 15 (7.04) | Secreted protein |
| Cp31_0180/ Cp1002B_0178 | 518 | - | - | - | - | - | 31 (5.98) | 31 (5.98) | Adhesin 3 (thioester domain) |
| Cp31_0399/ Cp1002B_0408 | 680 | - | - | - | - | - | 92 (13.53) | 92 (13.53) | Sialidase 2 (nanH) |
| Cp31_0893/ Cp1002B_1784 | 243 | - | - | - | - | - | 3 (1.23) | 3 (1.23) | Lysine exporter protein (lysE) |
| Cp31_2281/ Cp1002B_0835 | 60 | - | - | - | - | - | 10 (16.67) | - | Hypothetical protein 4 (no domains) |
The branch-site models used in the analysis identify sites under positive selection only in the foreground group (branch). To confirm our results, we checked to see if the same sites identified as being under positive selected sites are also identified when a subset of the genomes that had been previously tested as foreground were used as the new foreground genomes (S4 Fig). In this case, the previously identified sites would not be expected to be identified with the new foreground genomes. The results show that none of the previously identified genes were positively selected within these genome subsets (S4 File), thus confirming the previous results (Table 3).
An analysis of the 27 genes identified as being under positive selection showed that they played a variety of functional roles that includes activity in metabolism, cell division, resistance, transport, adhesion, or were identified as hypothetical proteins with unknown functions. Many of these genes have previously been suggested as drug or vaccine targets (Tables 3 and 4). Seven of them are located in areas that have been previously identified as pathogenicity islands [32] (Fig 3, Table 3). The functional categories assigned to the genes in these islands have previously been included in a list of niche/virulence factors involved in pathogenesis for the Corynebacterium genus [61]. Some of the genes appeared to be exposed on the cell surface. Proteins located at the interface between bacteria and the environment are more likely to undergo positive selection [9], so it would not be surprising if some of the genes we detected (Table 3) play a role in the dynamics of the host-pathogen interaction. Some of the processes that had genes identified as being under positive selection include nutrient uptake, modulation of the host immune response, resistance and receptor-mediated binding [6,9] (Table 3). In those proteins, positive selection could act as a protective measure to avoid attachment by antibodies or phages, instead of a response related to the protein function [9].
Positive selection in each target group
Adaptations in Ovis biovar (Foreground 1: Ovis)
Several studies have identified phenotypic and genotypic changes that differentiate the Ovis and Equi biovars. These include differences in nitrate reduction [76], changes in serotype and disease manifestation in the guinea pig model host [77], and pathogenicity islands that are biovar specific [32]. In addition, the Ovis clade has been documented as having a higher genomic similarity across its members than what is seen in Equi [32,33].
Our examination of the Ovis clade (Foreground 1, Fig 1) compared the genomes from 16 Ovis isolates to 13 from Equi, with Cp1002B selected as the anchor (Table 2). This comparison revealed adaptive mutations in four genes (Cp31_1168, Cp31_0488, Cp31_1468 and Cp31_2169) that have occurred in Ovis since it separated from Equi (Table 3), providing an indication of specific selective pressures imposed upon this group. Three of these specific genes (Cp31_1168, Cp31_0488, Cp31_1468) have defined functions, while the fourth (Cp31_2169) is annotated as a hypothetical protein. Two of the genes with described functions are involved in the use of carbon and iron sources (citE, Cp31_1168 and htaF, Cp31_1468), and the third is a drug transporter that is used in competition with other microorganisms (Drug transporter, Cp31_0488) (Table 3). Two of these genes, citE (Cp31_1168) [64] and the drug resistance transporter Cp13_0488 [62], are homologs to previously identified drug targets [62,64], and the hypothetical protein (Cp31_2169) is located in a pathogenicity island.
Adaptations shared by Ovis and Equi strain 262 (Foreground 2: OvisEqui262)
Phylogenetic analysis showed that Equi strain 262 is closer to the Ovis biovar than it is to the Equi (Fig 1). To identify probable adaptive mutations that Equi 262 and genomes in the Ovis biovar share that differentiates them from the broader Equi clade, we compared these 17 genomes to the remaining 12 Equi genomes, with Cp1002B once again used as the anchor (Table 2). The 262 genome and all of the 17 belonging to Ovis share nine genes that were identified as being under adaptive selection (Table 3). Two play a role in virulence or antimicrobial resistance, and five have well-established roles in metabolism (Table 3). Sialidases have been associated with virulence in Corynebacterium [65,78], and Cp31_0206 is the one of two genes in this group that is located in a known pathogenicity island. The role of beta lactamases in drug resistance is well known, and the gene with this functional description (Cp31_0950) appears to be experiencing selective pressure within this group. Other genes indicated in making adaptive changes play an important metabolic role (Cp31_0638, Cp31_0638, Cp31_0945, Cp31_0985, Cp31_1044 and Cp31_1309), while the functions of the membrane anchored protein (Cp31_1868) in PiCp13 and a hypothetical protein (Cp31_1724) are not yet known.
Several of these genes identified in this group have homology to genes that have previously been suggested as possible drug targets in Mycobacterium tuberculosis, which is part of the CNMR group that includes Corynebacterium. These include the sialidase [66], dethiobiotin synthetase (bioD) [69], dihydrofolate reductase (folA) [67], pup deamidase (dop) [71,72] and the metallo-beta-lactamase [68].
Adaptations in the monophyletic Equi clade (Foreground 3: EquiExcept262)
In this group, we searched for positive selection only within the monophyletic lineage of Equi, which includes twelve genomes that were isolated from a variety of large mammals (Table 1). Although Cp262 is part of the Equi biovar, it was not included in this particular analysis because our phylogenetic analysis showed that it is more closely aligned with the Ovis clade than with the other Equi genomes (Fig 1). This comparison is a reverse of the previous one, as it looks for adaptive changes in the 12 Equi genomes compared to the 17 genomes that include the single 262 Equi and the 16 Ovis isolates. Strain Cp31 was used as the anchor (Table 2). This comparison revealed six genes under positive, adaptive selection in the 12 Equi genomes, and the fact that they do not occur in the other genomes show that the changes occurred after divergence with the common ancestor these Equi genomes share with 262. These include genes related to nutrition and evasion of the host immune response (Sialidase 1, Cp31_0206), acetyl-CoA and DNA synthesis, fermentation (cobS, Cp31_1309), an adhesion (Adhesin 1, Cp31_2279), and three genes of undetermined function (Cp31_1724, Cp31_0109 and Cp31_2015). Several of these genes (Cp31_0109, Cp31_2015 and Cp31_2279) were only identified in this particular comparison, with Adhesin 1 (Cp31_2279) being perhaps the most interesting as these types of genes are known virulence factors. It has 20 sites under positive selection (Table 4 and S3 File). Other genes found to be under positive selection in this group include an alpha/beta hydrolase (Cp31_0109), a transmembrane protein (Cp31_2015), and a hypothetical protein (Cp31_1724), but the roles that these genes have in the interaction with the hosts they infect has yet to be determined.
Adaptations shared by strains isolated from buffalo and horse (Foreground 4: EquiBuffaloHorse)
An examination of the Equi clade (Foreground 3, Fig 1) shows two distinct sub-branches that separate Equi genomes isolated from a cow (CpI37) and a camel (Cp162) from those isolated from horses and buffalo (Foreground 4, Fig 1). To identify genes under positive selection in the genomes united by Foreground 4, we compared the 10 buffalo and horse isolates to all the other 19 genomes in the analysis, using Cp31 as the anchor (Table 2). This comparison revealed four genes under positive selection within these genomes isolated from horses and buffalo, which included known surface exposed proteins and a hypothetical protein. Positive selection was found in Adhesin 2 (Cp31_1094) and in the Equi exclusive Adhesin 1 (Cp31_2279, 23 sites). Seeing the adhesin genes responding to selective pressure in the Equi biovar indicates that these proteins play an important role in the particular niche these organisms inhabit. These differences could help the Equi isolates adapt to the different hosts that they are able to utilize, which presumably includes adhesion to specific cell receptors. Moreover, one of these adhesins (Cp31_2279) was also identified in the Branch 3 comparison mentioned above, indicating that this particular gene is responding uniquely to different selective pressures that are imposed on each of these clades.
Adaptations in strains isolated from buffalo (Foreground 5: EquiBuffalo) and horse (Branch 6: EquiHorse)
In a previous analysis, buffalo strains were shown to be clonal, with 94.7% shared genes in the core genome [33]. They compose a monophyletic cluster and they were seen to differ from the horse isolates mainly by an exclusive tox+ prophage [33]. Isolates from buffalo were the only C. pseudotuberculosis strains shown to produce diphtheria toxin [31,79–83]. This information supports the hypothesis where the presence of the prophage, specifically its diphtheria toxin (tox), is required for C. pseudotuberculosis to infect this buffalo, and this has been suggested as a potential vaccine target [33]. In contrast, the genomes isolated from horses only share 42.5% of their genes in a prior study and no genes related to the different disease phenotypes were found [84]. It is clear that one of the main differences between the horse and buffalo isolates are the presence of the prophage and the diphtheria toxin [33], which fits the “stable ecotype” model where adaptive genes allowed expansion into a new niche (the buffalo host), and then the founder mutant reproduces clonally [85].
We searched for positive selection in the Equi clades isolated from buffalo and horses separately (Foreground 5, Fig 1). We compared the 5 genomes isolated from buffalo to all other C. psedotuberculosis genomes used in the analysis, using the Cp31 genome as the anchor (Table 2). Three genes were found to be under positive selection only in these buffalo isolates, and they include genes hemA (Cp31_0279), cobN (Cp31_1028 are related to biosynthesis of cofactors used in important biological process, while whiA (Cp31_1117) is involved in cell division regulation (Table 3), suggesting adaptations across a wide range of cellular processes. Among the three genes, hemA has been previously suggested as a dug target in Vibrio cholerae [74].
We did not find any genes identified as experiencing positive selection when we compared the five isolates from horses (Foreground 6, Fig 1) to the rest of the genomes used in the analysis, making it unique across all of our comparisons.
Adaptation in Ovis (Foreground 7: Ovis2) and the monophyletic Equi clade (Foreground 8: StraightEqui)
In order to identify genes that under selection in the Ovis and Equi biovars, we compared the genomes from the Ovis clade (Foreground 7, Fig 2) to what we consider to be “Straight Equi” (isolates from buffalo and horses in Foreground 8, Fig 2). We excluded Equi I37 and 162 as they were closer to the Ovis biovars than the other Equi genomes (Fig 1). In this comparison, the Cp1002B genome was used as the anchor for Ovis2 (Foreground 7), and Cp31 for StraightEqui (Foreground 8) (Table 2). Surprisingly, both of these branches shared the same genes undergoing positive selection, the sole exception being a hypothetical protein (Cp31_2281) that was only found to be changing within the Ovis clade (Foreground 7). The fact that both of the clades share the six remaining genes identified as undergoing positive selection indicates that these genes are responding differently to selective pressures that they are experiencing in these environments that these clades are exposed to. These pressures could be different hosts, or something else that we do not yet understand.
Positive selection was identified in sialidase 2 (nanH, Cp31_0399), cobaltochelatase subunit CobS (cobS, Cp31_1309), lysine exporter protein (lysE, Cp31_0893), adhesin 3 (Cp31_0180), and a secreted protein (Cp31_0142). Only Ovis2 had positive selection in Hypothetical protein 4 (Cp31_2281) (Table 3). Sialidase 2 (nanH) is also found in C. diphtheria and C. ulcerans [86]. Different sialidases in a bacterium can have differences in their substrate specificities and could play important roles in the interaction with other organisms or in the infection of a specific tissue [66]. In C. pseudotuberculosis, we detected positive selection in 92 sites of sialidase nanH and 31 sites in Adhesin 3 (Table 4), suggesting a very active response to whatever the selective pressures are imposing.
Phylogeny and ecological adaptation
The phylogenetic trees separate biovar Ovis from Equi with at least 90% of confidence value, clearly showing it as a monophyletic group (S2 and S3 Figs). This confirms what has been seen in previous studies [32,33,87]. In addition, the Equi from buffalo and horse formed a clade with two different clusters representing each host. In the phylogenomic trees (S1 and S2 Figs), Equi strain 262 was found to be a sister group of Ovis, as was found in a previous phylogenetic tree using 44 genomes [33]. The rpoB gene tree (S3 Fig) shows Equi 262 as the most primitive, but have a similar topology regarding to the other groups. The rpoB gene is more efficient at differentiating Corynebacterium species than 16S gene [49] and was shown to have power to differentiate biovars and Equi hosts. This tree topologies suggests that Ovis originated from an Equi ancestor, and that the last one is a paraphyletic group [88].
In a previous study, C. pseudotuberculosis was suggested to be under anagenesis and that Ovis would replace Equi [87]. However, Equi has horse and buffalo as exclusive hosts [19,31] and infections of horses are increasing in frequency in North America [28]. This implies that at least Equi has exclusive hosts in which it would not be outcompeted and replaced by Ovis, and that both biovars (lineages) will probably continue to coexist. Newly divergent lineages can coexist indefinitely when they have exclusive resources [89,90].
Based on our analysis, we feel that C. pseudotuberculosis evolution fits the “stable ecotype” model of ecological diversification, in which the acquisition of adaptive genes and mutations allows an exploration of a new resource, in this case a new host, creating a new “ecotype” [85,89]. This results in unique selective pressures during the initial expansion by the new clonal population, decreases genetic diversity within the new population by periodic positive selection and genetic drift, and decreases the fitness for the ancestral niche [85,89]. Both populations coexist long enough to accumulate neutral sequence divergence at every locus, being distinguished as multilocus sequence clusters [85,89]. Indeed, Ovis was shown to be i) derived from Equi (this study), ii) more clonal its ancestral biovar [32,33], probably due to decrease in genetic diversity by periodic selection and genetic drift, and iii) to have decreased the fitness for the ancestral niche by losing its capacity to infect horse. The results of our positive selection analysis identified genes under different selective pressures across lineages of C. pseudotuberculosis that are probably related to changes in ecological niches, which could be represented by expansion into new host ranges.
False positives for positive selection
The codon models of positive selection analysis are sensitive to data quality. Errors in sequencing, assembly, annotation, alignment and ortholog assignment can lead to false polymorphisms and alignments of non-homologous sites resulting in a statistical signal that is misinterpreted as positive selection [7,91–93]. In this work, five of the total results were identified as false positives (Table 5).
Table 5. False positive for positive selection in Corynebacterium pseudotuberculosis.
| Product | Artifact | Branch | GenBank ID |
|---|---|---|---|
| Sodium/alanine symporter family protein | Frameshifts in Ovis and Equi strain 262 (cow) | 1: Ovis | Cp1002B_0653 |
| Zinc ABC transporter, permease protein (znuB1) | Frameshift in Equi strains I37 (cow) and 162 (camel) | 1. Ovis | Cp1002B_0053 |
| HNH endonuclease | Frameshift in Ovis and Equi MEX30 | 1. Ovis, 7. Ovis2 | Cp1002B_1784 |
| Lysine exporter protein | Two frameshifts in Ovis | 1. Ovis | Cp1002B_1784 |
Frameshifts causing alignment of non-homologous codons were identified in proteins mainly related to transport. The false positive found in the Sodium/alanine symporter (Cp1002B_0653) is due to different frameshifts in Ovis and Equi 262, suggesting an independent loss of function, presumably because neither needs this gene for survival.
Frameshift mutations were found in znuB1 from Equi strains I37 and 262. In fact, the entire znuB1C1A1 operon of zinc transporter is frameshifted in all the other Equi strains. This operon is in pathogenicity island PiCp2, but another zinc transport operon (znuB2C2A2) is found in all C. pseudotuberculosis strains, which is not located in a pathogenicity island. The loss of function in the zinc transport operon znuB1C1A1 only in Equi suggests a different selective pressure on this group, with the sequence changes helping it adapt to its particular niche. The loss of specific functionality in specific branches or clades have been suggested as adaptation to different selective pressures in particular niches [94,95]. In bacteria, there is a strong mutational bias toward deleting superfluous sequences by mutation, drift, and selective pressure to reduce the size and redundancy in a genome [90,94].
Genome variation and the evolution of C. pseudotuberculosis
Different genome changes involved in host adaptation have been described in bacteria [95,96]. First, the already existent genes can be fine-tuned by positive selection. Second, new genes can be acquired by functional divergence, gene duplication, intragenic recombination or horizontal gene transfer. Third, the genome size can be reduced by loss of sequences due to redundant functions provided by the host, or negative selection [95,96]. Here, we analyzed the positive selection and gene acquisition/loss that could be related to the host preferences of C. pseudotuberculosis.
In the circular map (Fig 3), there is a gap between PiCp3 and PiCp8 of Cp1002B genome. We examined this region and found an adhesin containing the “Fibrogen-binding domain 1” (RASTtk Cp31_247, GenBank Cp31_2168), flanked by the genes that encode Aspartokinase (lysC, Cp31_0184) and Aspartate-semialdehyde dehydrogenase (asd, Cp31_0185). Both biovars have this adhesin, but the difference in nucleotide sequence (> 50%) was high enough to be considered a non-homologous sequence by BRIG. The identity between the sequences of the protein in Cp31 and Cp1002B (RASTtk Cp1002B_180, GenBank Cp1002B_184) is 39% with a coverage of 98%. This variation is probably related to adhesion to tissues from different hosts, within the range of each biovar.
Previous studies identified an exclusive sigma factor in PiCp5 of Ovis strains [32,97]. Also, two additional characteristics that differentiate the biovars were recently identified in two other genomic regions using comparative genomics, a Type III restriction-modification system found only in Ovis and a CRISPR-Cas system found only in Equi (Parise et al., accepted). Assuming Ovis as a monophyletic clade derived from Equi (S1 to S3 Figs), we checked whether these features are primitive or derived by checking their presence across Equi strains using PATRIC’s Protein Family Sorter [35] and their position in relation to the pathogenicity islands, using GIPSy. The Type III restriction-modification system is in the pathogenicity island PiCp15, which is found only in genomes belonging to Ovis and is absent in all Equi strains. This indicates that PiCp15 was acquired after the separation of Ovis and Equi, presumably by the last common ancestor of all the Ovis isolates.
The CRISPR-Cas genes are in PiCp1 and present in all Equi strains, including strain 262, and one gene is reminiscent in Ovis. This suggests that the CRISPR-Cas genes were acquired by the common ancestor of C. pseudotuberculosis strains and were lost from the Ovis biovar.
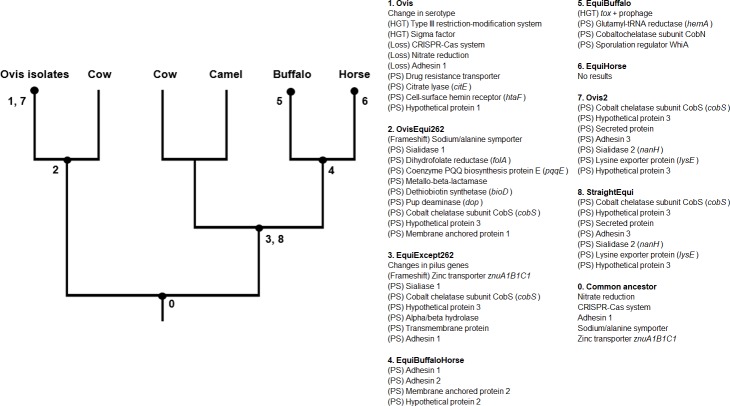
Various comparative genomics studies have been done in C. pseudotuberculosis [32,33,84,97–99]. We mapped our data and differences described in previous studies to our phylogenetic tree to clarify the specific changes that have occurred during the evolution and host expansion of this pathogen (Fig 4). In Ovis, previous analyses documented the loss of nitrate reduction related genes [33,76,100], changes in serotype [77], an exclusive Type III restriction-modification system (this study), and a sigma factor in PiCp5 [32,97]. In Equi, previous studies have described frameshifts in pilus genes [32,33] and acquisition of a tox+ prophage in PiCp12 [33,101]. Previously, variations in the presence of pathogenicity islands were said to explain most of the phenotypic differences seen between the Ovis and Equi biovars [32]. Here, for the first time, we can see that selective pressures are also occurring, and that they play a likely role in the adaptation of C. pseudotuberculosis to selective pressures that correspond to the observed differences in phylogeny.
Fig 4. Genome variations in different branches of Corynebacterium pseudotuberculosis.
HGT–horizontal gene transfer, PS–positive selection.
Conclusions
By performing genome scale positive selection analysis, we have identified what appear to be adaptive mutations in specific genes found in defined phylogenetic clades of C. pseudotuberculosis. These differences can be seen to correlate with the different hosts that the genomes were isolated from, and with the two biovars described for this species. Many of the proteins identified as being under selection are involved in important processes that are known to increase of survival, including metabolism, cell division, resistance, transport, adhesion. Some of the genes that are under positive selection have previously been identified as potential drug targets in other bacteria, which could indicate a possible future role in treatment or infection prevention. In addition, we have combined a phylogenomic analysis with previously documented changes, and this analysis of positive selection, to show specific changes that have occurred during the evolution of this species. These changes are correlated with both ecological diversification as an expanding host range in this pathogen.
Supporting information
Equi branches are in blue and Ovis branches are in Orange.
(TIF)
Equi branches are in blue and Ovis branches are in Orange. The blue circles represent jackknife values above 90%.
(TIF)
Equi branches are in blue and Ovis branches are in Orange. The blue circles represent bootstrap values above 90%.
(TIF)
The target groups 9 to 12 are subsets of genomes used in target group 1 (Ovis). Target group 13 is a subset of genomes used in target group 5 (EquiBuffalo).
(TIF)
(XLSX)
(XLSX)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by: Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (www.capes.gov.br), Conselho Nacional de Desenvolvimento Científico e Tecnológico (cnpq.br), and Pró-Reitoria de Pesquisa e Extensão of Universidade Federal de Minas Gerais (www.ufmg.br/prpq). A.R. Wattam was supported in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (www.nih.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stephan W. Detecting strong positive selection in the genome. Mol Ecol Resour. 2010;10: 863–872. 10.1111/j.1755-0998.2010.02869.x [DOI] [PubMed] [Google Scholar]
- 2.Casillas S, Barbadilla A. Molecular population genetics. Genetics. 2017;205: 1003–1035. 10.1534/genetics.116.196493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedge J, Wilson DJ. Practical Approaches for Detecting Selection in Microbial Genomes. PLoS Comput Biol. 2016;12: 1–12. 10.1371/journal.pcbi.1004739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11: 725–36. 10.1093/oxfordjournals.molbev.a040153 [DOI] [PubMed] [Google Scholar]
- 5.Moretti S, Murri R, Maffioletti S, Kuzniar A, Castella B, Salamin N, et al. Gcodeml: A grid-enabled tool for detecting positive selection in biological evolution. Stud Health Technol Inform. 2012;175: 59–68. 10.3233/978-1-61499-054-3-59 [DOI] [PubMed] [Google Scholar]
- 6.Hongo JA, de Castro GM, Cintra LC, Zerlotini A, Lobo FP. POTION: an end-to-end pipeline for positive Darwinian selection detection in genome-scale data through phylogenetic comparison of protein-coding genes. BMC Genomics. 2015;16: 567 10.1186/s12864-015-1765-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahm A, Bens M, Platzer M, Szafranski K. PosiGene: automated and easy-to-use pipeline for genome-wide detection of positively selected genes. Nucleic Acids Res. 2017;45: 1–11. 10.1093/nar/gkw1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 9.Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17: 1336–1343. 10.1101/gr.6254707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay S, Paul S, Kisiela DI, Linardopoulou E V., Sokurenko E V. Convergent molecular evolution of genomic cores in Salmonella enterica and Escherichia coli. J Bacteriol. 2012;194: 5002–5011. 10.1128/JB.00552-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guinane CM, Ben Zakour NL, Tormo-Mas MA, Weinert LA, Lowder B V, Cartwright RA, et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol Evol. 2010;2: 454–66. 10.1093/gbe/evq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osório NS, Rodrigues F, Gagneux S, Pedrosa J, Pinto-Carbó M, Castro AG, et al. Evidence for diversifying selection in a set of Mycobacterium tuberculosis genes in response to antibiotic- and nonantibiotic-related pressure. Mol Biol Evol. 2013;30: 1326–1336. 10.1093/molbev/mst038 [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Xu Y, Gu Z, Liu N, Jin K, Li Y, et al. Identification of new antibacterial targets in RNA polymerase of Mycobacterium tuberculosis by detecting positive selection sites. Comput Biol Chem. 2017; 10.1016/j.compbiolchem.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 14.Tan JL, Ng KP, Ong CS, Ngeow YF. Genomic Comparisons Reveal Microevolutionary Differences in Mycobacterium abscessus Subspecies. Front Microbiol. 2017;8: 1–10. 10.3389/fmicb.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefébure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8: R71 10.1186/gb-2007-8-5-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefébure T, Stanhope MJ. Pervasive, genome-wide positive selection leading to functional divergence in the bacterial genus Campylobacter. Genome Res. 2009;19: 1224–1232. 10.1101/gr.089250.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann JS, Corey VC, Ricaldi JN, Vinetz JM, Winzeler EA, Matthias MA. Whole Genome Shotgun Sequencing Shows Selection on Leptospira Regulatory Proteins During in vitro Culture Attenuation. Am J Trop Med Hyg. 2016;94: 302–313. 10.4269/ajtmh.15-0401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Zhu YY, Wang Y, Chang Y-F, Zhang Y, Jiang X, et al. Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira. Sci Rep. Nature Publishing Group; 2016;6: 20020 10.1038/srep20020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorella FA, Carvalho Pacheco L, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res. 2006;37: 201–218. 10.1051/vetres:2005056 [DOI] [PubMed] [Google Scholar]
- 20.Williamson LH. Caseous Lymphadenitis in Small Ruminants. Vet Clin North Am Food Anim Pract. 2001;17: 359–371. 10.1016/S0749-0720(15)30033-5 [DOI] [PubMed] [Google Scholar]
- 21.Baird GJ, Fontaine MC. Corynebacterium pseudotuberculosis and its Role in Ovine Caseous Lymphadenitis. J Comp Pathol. 2007;137: 179–210. 10.1016/j.jcpa.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 22.Yeruham I, Friedman S, Perl S, Elad D, Berkovich Y, Kalgard Y. A herd level analysis of a Corynebacterium pseudotuberculosis outbreak in a dairy cattle herd. Vet Dermatol. 2004;15: 315–320. 10.1111/j.1365-3164.2004.00388.x [DOI] [PubMed] [Google Scholar]
- 23.Hawari AD. Corynebacterium pseudotuberculosis Infection (Caseous Lymphadenitis) in Camels (Camelus dromedarius) in Jordan. Am J Anim Vet Sci. 2008;3: 68–72. 10.3844/ajavsp.2008.68.72 [DOI] [Google Scholar]
- 24.Trost E, Ott L, Schneider J, Schröder J, Jaenicke S, Goesmann A, et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics. BioMed Central Ltd; 2010;11: 728 10.1186/1471-2164-11-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heggelund L, Gaustad P, Havelsrud OE, Blom J, Borgen L, Sundset A, et al. Corynebacterium pseudotuberculosis Pneumonia in a Veterinary Student Infected During Laboratory Work. Open Forum Infect Dis. 2015;2: ofv053–ofv053. 10.1093/ofid/ofv053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selim SA. Oedematous Skin Disease of Buffalo in Egypt. J Vet Med Ser B. 2001;48: 241–258. 10.1046/j.1439-0450.2001.00451.x [DOI] [PubMed] [Google Scholar]
- 27.Foley JE, Spier SJ, Mihalyi J, Drazenovich N, Leutenegger CM. Molecular epidemiologic features of Corynebacterium pseudotuberculosis isolated from horses. Am J Vet Res. 2004;65: 1734–1737. 10.2460/ajvr.2004.65.1734 [DOI] [PubMed] [Google Scholar]
- 28.Spier SJ, Azevedo V. Corynebacterium pseudotuberculosis infection in horses: Increasing frequency and spread to new regions of North America. Equine Vet Educ. 2016; 10.1111/eve.12566 [DOI] [Google Scholar]
- 29.Yeruham I, Braverman Y, Shpigel NY, Chizov‐Ginzburg A, Saran A, Winkler M. Mastitis in Dairy Cattle Caused by Corynebacterium pseudotuberculosis and the Feasibility Of Transmission by Houseflies I. Vet Q. 1996;18: 87–89. 10.1080/01652176.1996.9694623 [DOI] [PubMed] [Google Scholar]
- 30.Tejedor-Junco MT, Lupiola P, Schulz U, Gutierrez C. Isolation of nitrate-reductase positive Corynebacterium pseudotuberculosis from dromedary camels. Trop Anim Health Prod. 2008;40: 165–167. 10.1007/s11250-007-9077-2 [DOI] [PubMed] [Google Scholar]
- 31.Moussa IM, Ali MS, Hessain AM, Kabli SA, Hemeg HA, Selim SA. Vaccination against Corynebacterium pseudotuberculosis infections controlling caseous lymphadenitis (CLA) and oedematousskin disease. Saudi J Biol Sci. 2016;23: 718–723. 10.1016/j.sjbs.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soares SC, Silva A, Trost E, Blom J, Ramos R, Carneiro A, et al. The Pan-Genome of the Animal Pathogen Corynebacterium pseudotuberculosis Reveals Differences in Genome Plasticity between the Biovar ovis and equi Strains. PLoS One. 2013;8 10.1371/journal.pone.0053818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viana MVC, Figueiredo H, Ramos R, Guimarães LC, Pereira FL, Dorella FA, et al. Comparative genomic analysis between Corynebacterium pseudotuberculosis strains isolated from buffalo. Lin B, editor. PLoS One. 2017;12: e0176347 10.1371/journal.pone.0176347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brettin T, Davis JJ, Disz T, Edwards R a, Gerdes S, Olsen GJ, et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5: 8365 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45: D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Nielsen R. Codon-Substitution Models for Detecting Molecular Adaptation at Individual Sites Along Specific Lineages. Mol Biol Evol. 2002;19: 908–917. 10.1093/oxfordjournals.molbev.a004148 [DOI] [PubMed] [Google Scholar]
- 37.Zhang J. Evaluation of an Improved Branch-Site Likelihood Method for Detecting Positive Selection at the Molecular Level. Mol Biol Evol. 2005;22: 2472–2479. 10.1093/molbev/msi237 [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Dos Reis M. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28: 1217–1228. 10.1093/molbev/msq303 [DOI] [PubMed] [Google Scholar]
- 39.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altenhoff AM, Dessimoz C. Phylogenetic and Functional Assessment of Orthologs Inference Projects and Methods. Eisen JA, editor. PLoS Comput Biol. 2009;5: e1000262 10.1371/journal.pcbi.1000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 42.Liu K, Linder CR, Warnow T. Multiple sequence alignment: a major challenge to large-scale phylogenetics. PLoS Curr. 2011;2: RRN1198 10.1371/currents.RRN1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castresana J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol Biol Evol. 2000;17: 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6 Department of Genome Sciences, University of Washington, Seattle; 2005. [Google Scholar]
- 45.Loytynoja A, Goldman N. Phylogeny-Aware Gap Placement Prevents Errors in Sequence Alignment and Evolutionary Analysis. Science (80-). 2008;320: 1632–1635. 10.1126/science.1158395 [DOI] [PubMed] [Google Scholar]
- 46.Viana MVC, de Jesus Benevides L, Batista Mariano DC, de Souza Rocha F, Bagano Vilas Boas PC, Folador EL, et al. Genome Sequence of Corynebacterium ulcerans Strain 210932. Genome Announc. 2014;2: 1–2. 10.1128/genomeA.01233-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10: 512–26. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- 49.Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol. 2005;43: 1934–1936. 10.1128/JCM.43.4.1934-1936.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z. Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection. Mol Biol Evol. 2005;22: 1107–1118. 10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- 51.Noble WS. How does multiple testing correction work? Nat Biotechnol. 2009;27: 1135–1137. 10.1038/nbt1209-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100: 9440–5. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shriner D, Nickle DC, Jensen MA, Mullins JI. Potential impact of recombination on sitewise approaches for detecting positive natural selection. Genet Res. 2003;81: 115–121. 10.1017/S0016672303006128 [DOI] [PubMed] [Google Scholar]
- 54.Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164: 1229–1236. 10.1093/bioinformatics/btn086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Posada D, Crandall KA, Holmes EC. Recombination in Evolutionary Genomics. Annu Rev Genet. 2002;36: 75–97. 10.1146/annurev.genet.36.040202.111115 [DOI] [PubMed] [Google Scholar]
- 56.Bruen TC, Philippe HH, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172: 2665–81. 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12: 291–295. 10.1093/bioinformatics/12.4.291 [DOI] [PubMed] [Google Scholar]
- 58.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34: 126–129. 10.1007/BF00182389 [DOI] [PubMed] [Google Scholar]
- 59.Soares SC, Geyik H, Ramos RTJ, de Sá PHCG, Barbosa EGV, Baumbach J, et al. GIPSy: Genomic island prediction software. J Biotechnol. 2016;232: 2–11. 10.1016/j.jbiotec.2015.09.008 [DOI] [PubMed] [Google Scholar]
- 60.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12: 402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tauch A, Burkovski A. Molecular armory or niche factors: virulence determinants of Corynebacterium species. FEMS Microbiol Lett. 2015;67: fnv185 10.1093/femsle/fnv185 [DOI] [PubMed] [Google Scholar]
- 62.Li X, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2009;69: 1555–1623. 10.2165/11317030-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schroeder M, Brooks B, Brooks A. The Complex Relationship between Virulence and Antibiotic Resistance. Genes (Basel). 2017;8: 39 10.3390/genes8010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goulding CW, Bowers PM, Segelke B, Lekin T, Kim CY, Terwilliger TC, et al. The Structure and Computational Analysis of Mycobacterium tuberculosis Protein CitE Suggest a Novel Enzymatic Function. J Mol Biol. 2007;365: 275–283. 10.1016/j.jmb.2006.09.086 [DOI] [PubMed] [Google Scholar]
- 65.Kim S, Oh D-B, Kang HA, Kwon O. Features and applications of bacterial sialidases. Appl Microbiol Biotechnol. 2011;91: 1–15. 10.1007/s00253-011-3307-2 [DOI] [PubMed] [Google Scholar]
- 66.Kim S, Oh D-B, Kwon O. Sialidases of Corynebacteria and their Biotechnological Applications Corynebacterium diphtheriae and Related Toxigenic Species. Dordrecht: Springer; Netherlands; 2014. pp. 247–262. 10.1007/978-94-007-7624-1_13 [DOI] [Google Scholar]
- 67.Santa Maria JP, Park Y, Yang L, Murgolo N, Altman MD, Zuck P, et al. Linking High-Throughput Screens to Identify MoAs and Novel Inhibitors of Mycobacterium tuberculosis Dihydrofolate Reductase. ACS Chem Biol. 2017;12: 2448–2456. 10.1021/acschembio.7b00468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palzkill T. Metallo-β-lactamase structure and function. Ann N Y Acad Sci. 2013;1277: 91–104. 10.1111/j.1749-6632.2012.06796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salaemae W, Azhar A, Booker GW, Polyak SW. Biotin biosynthesis in Mycobacterium tuberculosis: physiology, biochemistry and molecular intervention. Protein Cell. 2011;2: 691–695. 10.1007/s13238-011-1100-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salaemae W, Yap MY, Wegener KL, Booker GW, Wilce MCJ, Polyak SW. Nucleotide triphosphate promiscuity in Mycobacterium tuberculosis dethiobiotin synthetase. Tuberculosis. Elsevier Ltd; 2015;95: 259–266. 10.1016/j.tube.2015.02.046 [DOI] [PubMed] [Google Scholar]
- 71.Zhang S, Burns-Huang KE, Janssen G V, Li H, Ovaa H, Hedstrom L, et al. Mycobacterium tuberculosis Proteasome Accessory Factor A (PafA) Can Transfer Prokaryotic Ubiquitin-Like Protein (Pup) between Substrates. Rubin EJ, editor. MBio. 2017;8: e00122–17. 10.1128/mBio.00122-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delley CL, Müller AU, Ziemski M, Weber-Ban E. Prokaryotic Ubiquitin-Like Protein and Its Ligase/Deligase Enyzmes. J Mol Biol. Elsevier Ltd; 2017;429: 3486–3499. 10.1016/j.jmb.2017.04.020 [DOI] [PubMed] [Google Scholar]
- 73.Carr PD, Ollis DL. Alpha/beta hydrolase fold: an update. Protein Pept Lett. 2009;16: 1137–1148. 10.2174/092986609789071298 [DOI] [PubMed] [Google Scholar]
- 74.Ravichandran M, Ali SA, Rashid NHA, Kurunathan S, Yean CY, Ting LC, et al. Construction and evaluation of a O139 Vibrio cholerae vaccine candidate based on a hemA gene mutation. Vaccine. 2006;24: 3750–3761. 10.1016/j.vaccine.2005.07.016 [DOI] [PubMed] [Google Scholar]
- 75.Gideon HP, Wilkinson KA, Rustad TR, Oni T, Guio H, Kozak RA, et al. Hypoxia Induces an Immunodominant Target of Tuberculosis Specific T Cells Absent from Common BCG Vaccines. Deretic V, editor. PLoS Pathog. 2010;6: e1001237 10.1371/journal.ppat.1001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biberstein EL, Knight HD, Jang S. Two biotypes of Corynebacterium pseudotuberculosis. Vet Rec. 1971;89: 691–692. 10.1136/vr.89.26.691 [DOI] [PubMed] [Google Scholar]
- 77.Barakat A., Selim SA, Atef A, Saber MS, Nafie EK, El-Ebeedy AA. Two serotypes of Corynebacterium pseudotuberculosis isolated from different animal species. Rev Sci Tech Off Int Epiz. 1984;3: 151–163. [DOI] [PubMed] [Google Scholar]
- 78.Kim S, Oh DB, Kwon O, Kang HA. Identification and functional characterization of the NanH extracellular sialidase from Corynebacterium diphtheriae. J Biochem. 2010;147: 523–533. 10.1093/jb/mvp198 [DOI] [PubMed] [Google Scholar]
- 79.Maximescu P, Oprişan A, Pop A, Potorac E. Further studies on Corynebacterium species capable of producing diphtheria toxin (C. diphtheriae, C. ulcerans, C. ovis). J Gen Microbiol. 1974;82: 49–56. 10.1099/00221287-82-1-49 [DOI] [PubMed] [Google Scholar]
- 80.Wong TP, Groman N. Production of diphtheria toxin by selected isolates of Corynebacterium ulcerans and Corynebacterium pseudotuberculosis. Infect Immun. 1984;43: 1114–6. Available: http://www.ncbi.nlm.nih.gov/pubmed/6321350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Groman N, Schiller J, Russell J. Corynebacterium ulcerans and Corynebacterium pseudotuberculosis responses to DNA probes derived from corynephage beta and Corynebacterium diphtheriae. Infect Immun. 1984;45: 511–7. 10.1159/000114687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Syame SM, Hakim AS, Hedia RH, Marie HSH, Selim SA. Characterization of Virulence Genes Present in Corynebacterium pseudotuberculosis Strains Isolated From Buffaloes. 2013;10: 585–591. 10.5829/idosi.gv.2013.10.5.7388 [DOI] [Google Scholar]
- 83.Selim SA, Mohamed FH, Hessain AM, Moussa IM. Immunological characterization of diphtheria toxin recovered from Corynebacterium pseudotuberculosis. Saudi J Biol Sci. King Saud University; 2015; 0–5. 10.1016/j.sjbs.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baraúna RA, Ramos RTJ, Veras AAO, Pinheiro KC, Benevides LJ, Viana MVC, et al. Assessing the Genotypic Differences between Strains of Corynebacterium pseudotuberculosis biovar equi through Comparative Genomics. Munderloh UG, editor. PLoS One. 2017;12: e0170676 10.1371/journal.pone.0170676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Curr Biol. Elsevier Ltd; 2008;18: R1024–34. 10.1016/j.cub.2008.09.014 [DOI] [PubMed] [Google Scholar]
- 86.Ott L, Burkovski A. Toxigenic Corynebacteria: Adhesion, Invasion and Host Response In: Burkovski A, editor. Corynebacterium diphtheriae and Related Toxigenic Species. 1st ed Dordrecht: Springer; Netherlands; 2014. pp. 143–170. 10.1007/978-94-007-7624-1_8 [DOI] [Google Scholar]
- 87.Oliveira A, Teixeira P, Azevedo M, Jamal SB, Tiwari S, Almeida S, et al. Corynebacterium pseudotuberculosis may be under anagenesis and biovar Equi forms biovar Ovis: a phylogenic inference from sequence and structural analysis. BMC Microbiol. 2016;16: 100 10.1186/s12866-016-0717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vandamme A-M. Basic concepts of molecular evolution In: Lemey P, Salemi M, Vandamme A-M, editors. The Phylogenetic Handbook—A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2nd ed Cambridge: Cambridge University Press; 2009. pp. 3–29. [Google Scholar]
- 89.Cohan FM. Transmission in the Origins of Bacterial Diversity, From Ecotypes to Phyla. Microbiol Spectr. 2017;5: 1–26. 10.1128/microbiolspec.MTBP-0014-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lassalle F, Muller D, Nesme X. Ecological speciation in bacteria: reverse ecology approaches reveal the adaptive part of bacterial cladogenesis. Res Microbiol. 2015;166: 729–41. 10.1016/j.resmic.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 91.Schneider A, Souvorov A, Sabath N, Landan G, Gonnet GH, Graur D. Estimates of Positive Darwinian Selection Are Inflated by Errors in Sequencing, Annotation, and Alignment. Genome Biol Evol. 2009;1: 114–118. 10.1093/gbe/evp012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mallick S, Gnerre S, Muller P, Reich D. The difficulty of avoiding false positives in genome scans for natural selection. Genome Res. 2009;19: 922–33. 10.1101/gr.086512.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Markova-Raina P, Petrov D. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res. 2011;21: 863–74. 10.1101/gr.115949.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bobay L-M, Ochman H. The Evolution of Bacterial Genome Architecture. Front Genet. 2017;8: 1–6. 10.3389/fgene.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Toft C, Andersson SGE. Evolutionary microbial genomics: Insights into bacterial host adaptation. Nat Rev Genet. Nature Publishing Group; 2010;11: 465–475. 10.1038/nrg2798 [DOI] [PubMed] [Google Scholar]
- 96.Sheppard SK, Guttman DS, Fitzgerald JR. Population genomics of bacterial host adaptation. Nat Rev Genet. Springer US; 2018; 1 10.1038/s41576-018-0032-z [DOI] [PubMed] [Google Scholar]
- 97.Ruiz JC, D’Afonseca V, Silva A, Ali A, Pinto AC, Santos AR, et al. Evidence for reductive genome evolution and lateral acquisition of virulence functions in two corynebacterium pseudotuberculosis strains. PLoS One. 2011;6 10.1371/journal.pone.0018551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soares SC, Abreu VAC, Ramos RTJ, Cerdeira L, Silva A, Baumbach J, et al. PIPS: Pathogenicity Island Prediction Software. Mokrousov I, editor. PLoS One. 2012;7: e30848 10.1371/journal.pone.0030848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soares SC, Trost E, Ramos RTJ, Carneiro AR, Santos AR, Pinto AC, et al. Genome sequence of Corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. J Biotechnol. 2013;167: 135–141. 10.1016/j.jbiotec.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 100.Almeida S, Sousa C, Abreu V, Diniz C, Dorneles EMS, Lage AP, et al. Exploration of Nitrate Reductase Metabolic Pathway in Corynebacterium pseudotuberculosis. Int J Genomics. 2017;2017: 1–12. 10.1155/2017/9481756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramos RTJ, Carneiro AR, de Castro Soares S, Barbosa S, Varuzza L, Orabona G, et al. High efficiency application of a mate-paired library from next-generation sequencing to postlight sequencing: Corynebacterium pseudotuberculosis as a case study for microbial de novo genome assembly. J Microbiol Methods. 2013;95: 441–447. 10.1016/j.mimet.2013.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Equi branches are in blue and Ovis branches are in Orange.
(TIF)
Equi branches are in blue and Ovis branches are in Orange. The blue circles represent jackknife values above 90%.
(TIF)
Equi branches are in blue and Ovis branches are in Orange. The blue circles represent bootstrap values above 90%.
(TIF)
The target groups 9 to 12 are subsets of genomes used in target group 1 (Ovis). Target group 13 is a subset of genomes used in target group 5 (EquiBuffalo).
(TIF)
(XLSX)
(XLSX)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.