Abstract
Hematopoietic stem cells (HSCs) are rare cells, with the mouse bone marrow containing only ~25,000 phenotypic long term repopulating HSCs. A Western blotting protocol was optimized and suitable for the analysis of small numbers of HSCs (500 - 15,000 cells). Phenotypic HSCs were purified, accurately counted, and directly lysed in Laemmli sample buffer. Lysates containing equal numbers of cells were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the blot was prepared and processed following standard Western blotting protocols. Using this protocol, 2,000 - 5,000 HSCs can be routinely analyzed, and in some cases data can be obtained from as few as 500 cells, compared to the 20,000 to 40,000 cells reported in most publications. This protocol should be generally applicable to other hematopoietic cells, and enables the routine analysis of small numbers of cells using standard laboratory procedures.
Keywords: Biochemistry, Issue 138, Western blotting, hematopoietic stem cells, small cell numbers, SDS-PAGE, animal testing reduce, bone marrow
Introduction
Hematopoietic stem cells (HSCs) are self-renewing cells that can give rise to all blood lineages. They are relatively rare cells in the bone marrow, rendering biochemical analyses difficult. Approaches suitable for analyzing rare cells, such as flow cytometry, have been extremely useful for quantifying relative amounts of cell surface markers and intracellular proteins. However, the analysis of intracellular proteins necessitates the use of cell permeabilization procedures to enable antibody access, and not all cell surface epitopes survive these procedures1,2. In addition, antibodies that discriminate between different protein isoforms or cleavage products are not often available for flow cytometry, and therefore investigators still rely on Western blots for certain types of analyses.
Western blot analysis of cell lysates is a routine procedure in most laboratories. Cells can be purified under native conditions that preserve the epitopes of cell surface molecules, and cell lysates can subsequently be prepared and analyzed. However, the analysis of proteins in rare primary cell populations by Western blot can require euthanizing large numbers of animals to obtain enough cells. By making small adjustments to several steps, a conventional Western blotting protocol was able to detect proteins in relatively small numbers of HSCs (500 - 15,000, depending on the protein of interest). The adjustments include accurately counting the cells, carefully handling the cell pellet, reducing transfers of cells between tubes to minimize cell loss, and lysing a defined number of cells with a concentrated loading buffer containing proteasome and phosphatase inhibitors. Many published reports include Western blots obtained with 20,000 or more HSCs3,4,5,6,7; this simple procedure will reduce the number of cells and experimental animals required to produce equivalent data by between 4 and 40 fold. The protocol is designed to normalize results on a per cell basis, rather than to an internal control. This enables detection of overall reductions in protein levels that can be overlooked if data are normalized to an internal control. The importance of normalizing on a per cell basis was described for the analysis of gene expression data8, and the same principle applies to quantifying proteins by Western blot. This optimized protocol should be useful for anyone needing to analyze small numbers of cells.
Protocol
All procedures must be performed in accordance with institutional animal use and care guidelines. The procedure was developed for the analysis of murine hematopoietic stem and progenitor cells (HSCs and HPs), but can be adapted for the analysis of other cell populations.
1. Flow Cytometry Isolation of Murine HSCs and HPs
Harvest murine bone marrow cells as described in the literature6. NOTE: In data presented in Figure 1 and Figure 2, bone marrow was harvested from C57BL/6J mice.
Perform lineage depletion of bone marrow cells using a mouse lineage depletion kit, following the manufacturer's instructions.
Incubate the cells with antibodies against the cell surface markers of interest as described7. In the experiments shown in Figure 1 and Figure 2, antibodies to Sca-1, Kit, Flt3, and lineage (Lin) markers Mac1, Gr1, CD3e, B220, and Ter119 are used.
Sort 2,000 - 20,000 Lin- Sca1+Kit+ Flt3- cells (HSCs) or Lin-Sca1-Kit+ cells (HPs) into 1.5 mL Eppendorf tubes containing 0.1 mL of phosphate buffered saline (PBS) with 2% fetal bovine serum (FBS). NOTE: For the data shown in Figure 1 and Figure 2, cells were sorted using a Flow Cytometer with a 70 µM nozzle. The maximum collection volume is 1.4 mL.
2. Sample Preparation
NOTE: This step is critical. Process the sample very carefully. When removing the supernatant, be very careful not to disturb the cell pellet.
- Centrifuge the 1.5 mL tubes containing sorted cells in a swinging bucket rotor at 4 °C, 500 x g, for 5 min. Remove all but approximately 100 µL of the supernatant from the cell pellet very gently using a pipette and a 20 µL pipet tip. Do not disturb the pellet.
- If the sample contains fewer than 5,000 cells, and the total volume of the sorted sample is less than 0.2 mL, transfer the cells first to low binding 0.2 mL PCR tubes before centrifugation.
- If sample contains fewer than 5,000 cells and volume is more than 0.2 mL, spin the cells down with a centrifuge at 4 °C, 500 x g, for 5 min, and remove all but 200 µL of the supernatant. Re-suspend the pelleted cells in the remaining 200 µL of supernatant, then transfer the cells to the 0.2 mL PCR tubes and spin down again. Remove all but 20 - 30 µL from the pellet after the second spin and re-suspend cells.
Resuspend the cells in the supernatant left in the tube. If necessary,add additional PBS in 2% FBS/PBS (you can also use 2% bovine serum albumin (BSA)/PBS, or PBS alone) to obtain a concentration 5 x104 to 5 x 105 cells/mL. NOTE: Use the cell counts collected by cytometry to estimate the volume used to suspend cells.
Use 2 - 5 µL of the re-suspended cells to determine an accurate cell concentration using an automated cell counter (following the manufacturer's instructions) or a hemocytometer9.
Transfer a volume of re-suspended cells containing the desired number of cells to new 1.5 mL tubes. For the experiment in Figure 1, 2,000, 1,000, or 500 HSCs or HPs were transferred. The desired number of cells depends on the strength of the signal obtained by Western blot, and is determined empirically. Perform the transfer using a 20 µL or 100 µL Eppendorf pipette tip.
Centrifuge the cells in a swinging bucket rotor at 4 °C, 500 x g, for 5 min.
To generate 100x stock solutions, dissolve proteasome and phosphatase inhibitors in DMSO according to the manufacturer's instructions.
Prepare 2x Laemmli sample buffer by diluting the 4x Laemmli sample buffer provided by the manufacturer with an equivalent amount of distilled water. Add an appropriate amount of the 100x stock of proteasome and phosphatase inhibitors to obtain a final concentration of 2x inhibitors in 2x Laemmli sample buffer. NOTE: The 2x Laemmli sample buffer can be made in advance, but the proteasome and phosphatase inhibitors should be added immediately before adding the 2x Laemmli sample buffer (plus inhibitors) to the cell pellet to lyse the cells, as described in the following step.
Carefully remove a portion of the supernatant from the cell pellet, and to the supernatant remaining in the tube with the cell pellet, add an equal volume of 2x Laemmli sample buffer plus proteasome and phosphatase inhibitors to achieve a final concentration of 500 cells/µL in 1x Laemmli sample buffer. NOTE: For example, if you transfer 2,000 cells in a total volume of 20 µL to a tube in Step 2.5, after centrifuging the cells (Step 2.6) you should remove 18 µL of the supernatant from the pellet, and to the 2 µL of supernatant that is remaining add an equal volume (2 µL) of 2x Laemmli sample buffer to achieve a final concentration of 500 cells/µL in 1x Laemmli sample buffer.
Resuspend the pellet to generate the lysate as soon as possible. At this point, the samples can be immediately electrophoresed through SDS-PAGE gels, or can be snap frozen in liquid N2 and stored at -80 °C for future use.
3. Electrophoresis
Heat the lysates in a heat block at 95 °C or in boiling water for 5 min.
Electrophorese 1 - 40 µL of the lysates (containing from 500 up to 20,000 cell equivalents) through SDS-PAGE gels at 100V using standard protocols10. NOTE: The volume of lysate loaded into the gel is determined by the number of cells needed to generate the desirable signal, and will depend on the abundance of the protein of interest, and the quality of the antibody. The data in Figure 1 were obtained with 2,000, 1,000, and 500 cell equivalents of lysate. The width of the well should be in the range of 3 mm to 1.5 mm. 1.5 mm teeth can be cut from a commercially available comb with wider teeth using scissors.
4. Transfer and Block
NOTE: Perform a Western blot following standard protocols11. The steps are briefly outlined here:
Pretreat a polyvinylidene difluoride (PVDF) membrane with methanol for 10 s, then wash the membrane briefly with distilled water.
Transfer the protein from the SDS-PAGE gel to the membrane following the instructions in the manual provided by the manufacturer of the transfer apparatus.
Following transfer, block the membrane with 2 mL of 5% bovine serum albumin (BSA) in PBST (PBS with 0.1% Tween) overnight at 4 °C following standard protocols11.
5. Antibody Labeling
Incubate the membrane with antibodies on a shaker overnight at 4 °C using antibody dilutions recommended by the vendor. NOTE: In the Table of Materials, antibodies, which we have validated for HSCs and HPs and the corresponding antibody dilutions, are shown. Perform the antibody labeling using standard procedures8.
6. Detection
Wash the membrane 3 times, 5 min for each wash in PBST at room temperature.
Incubate the membrane with 2 mL of enhanced chemiluminescence buffer for 1 min at room temperature. Detect the signals using an imaging system following the manufacturer's instructions, or autoradiography film and film processor.
Representative Results
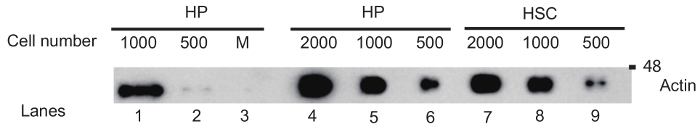
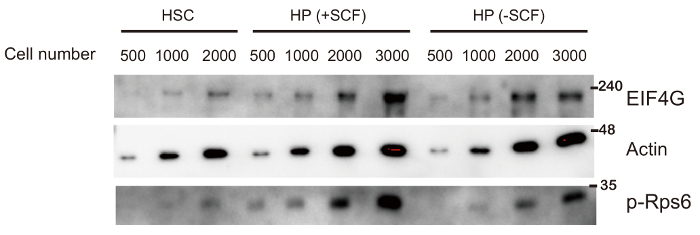
Representative results from 500 - 2,000 purified HSCs and HPs are shown in Figure 1 and Figure 2. The β-actin signal in Figure 1 can be detected from as few as 500 HSCs and HPs purified from the bone marrow of one mouse. Note that loading the lysates into 1.5 mm wells produced a much stronger signal from 500 HPs than loading into 3.0 mm wells. Figure 2 is a Western blot of EIF4G and the phosphorylation of Rps6 (p-Rps6), both of which are involved in the regulation of protein translation12, in HSCs, and in HPs with and without stimulation by stem cell factor (SCF) in vitro.
Figure 1: Western Blot for β-actin performed with lysates from murine HSCs and HPs. HSCs were sorted from lineage depleted murine bone marrow cells as Lin-Sca1+Kit+ Flt3- cells, and HPs were Lin-Sca1-Kit+. Lysates prepared from different numbers of cells were electrophoresed through a 12% SDS-PAGE gel prepared using mini glass plates with 1 mm spacers. The blot was probed with antibody to β-actin. The blot shows the comparative β-actin signals from 500 to 2,000 HPs when the lysates were loaded into 1.5 mm wells (lanes 4 - 6) as compared to 3 mm wells (lanes 1 - 2). Lysates from 2,000 to 500 cells were loaded onto lanes 7 to 9. M, molecular weight markers. Please click here to view a larger version of this figure.
Figure 2: Western Blot analysis of freshly isolated HSCs, and HPs stimulated in vitro with the cytokine Stem Cell Factor (SCF). HPs were purified by fluorescence activated cell sorting (FACS), centrifuged, resuspended in 2% FBS/PBS, counted, then split into two tubes. The HPs in the first tube were stimulated with SCF (10 ng/mL) for 5 min at 37 °C (+SCF), and HPs in the second tube were cultured for 5 min in the absence of SCF (-SCF). The cells were then centrifuged and the cell pellet lysed with Laemmli sample buffer. HSCs were purified and directly lysed with Laemmli sample buffer without culturing. The lysates were loaded into 1.5 mm wells and electrophoresed through 12% SDS-PAGE gels. The blot was developed with antibodies to elongation initiating factor 4G (EIF4G), β-actin, and phosphorylated small ribosome subunit S6 (p-Rps6). Please click here to view a larger version of this figure.
Discussion
Western Blotting is a common technique for detecting specific proteins and the activation of signaling pathways in tissues or cells. By introducing small adjustments to a commonly used procedure, we were able to routinely detect 15 different proteins (Table of Materials) in 15,000 HSCs, and in some cases in as few as 500 HSCs. The most critical steps in this protocol are: 1) accurately counting the cells, 2) minimizing the number of transfers between tubes, and 3) lysing the cells directly with Laemmli sample buffer. When lysing the cell pellet with Laemmli sample buffer, we found that rather than removing all of the supernatant from the cell pellet and re-suspending it in Laemmli Sample Buffer, if we instead left a small volume of supernatant in the tube and added an equivalent volume of 2x Laemmli Sample Buffer, we could avoid cell loss. Centrifuging the cells in a swinging bucket rotor also decreased the risk of cell loss. Reducing the width of the well by trimming the comb teeth improved the sensitivity of detection.
With these simple adjustments to the procedure, we were able to obtain reproducible results equivalent to those in published papers that had used 10 - 20 times more cells3,4,5,6,7. Further, the blots can be stripped for re-blotting following standard procedures13, increasing the amount of information that can be obtained from a small number of cells. The ability to detect proteins of interest will be limited by the quality of the antibody and the protein abundance. This modified technique should greatly reduce the number of animals necessary to obtain protein data from rare cell populations.
Disclosures
The authors have no conflict of interests to declare.
Acknowledgments
National Institutes of Health grant R01 CA149976 (N.A.S) supported this work.
References
- Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. J Immunol. 2005;175(4):2357–2365. doi: 10.4049/jimmunol.175.4.2357. [DOI] [PubMed] [Google Scholar]
- Du J, et al. Signaling profiling at the single-cell level identifies a distinct signaling signature in murine hematopoietic stem cells. Stem Cells. 2012;30(7):1447–1454. doi: 10.1002/stem.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509(7498):49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148(5):1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Hoshii T, et al. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J Clin Invest. 2012;122(6):2114–2129. doi: 10.1172/JCI62279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, et al. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 2016;30(15):1698–1703. doi: 10.1101/gad.282756.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell. 2015. [DOI] [PMC free article] [PubMed]
- Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JoVE Science Education Database. Using a Hemoacytometer to Count Cells. Basic Methods in Cellular and Molecular Biology. 2017.
- JoVE Science Education Database. Separating Protein with SDS-PAGE. Basic Methods in Cellular and Molecular Biology. 2017.
- JoVE Science Education Database. The Western Blot. Basic Methods in Cellular and Molecular Biology. 2017.
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton SL, et al. A guide to modern quantitative fluorescent western blotting with troubleshooting strategies. J Vis Exp. 2014. p. e52099. [DOI] [PMC free article] [PubMed]


