Abstract
The immune system is an essential component of a healthy heart. The myocardium is home to a rich population of different immune cell subsets with functional compartmentalization both during steady state and during different forms of inflammation. Until recently, the study of immune cells in the heart required the use of microscopy or poorly developed digestion protocols, which provided enough sensitivity during severe inflammation but were unable to confidently identify small — but key — populations of cells during steady state. Here, we discuss a simple method combining enzymatic (collagenase, hyaluronidase and DNAse) and mechanical digestion of murine hearts preceded by intravascular administration of fluorescently-labelled antibodies to differentiate small but unavoidable intravascular cell contaminants. This method generates a suspension of isolated viable cells that can be analyzed by flow cytometry for identification, phenotyping and quantification, or further purified with fluorescence-activated cell sorting or magnetic bead separation for transcriptional analysis or in vitro studies. We include an example of a step-by-step flow cytometric analysis to differentiate the key macrophage and dendritic cell populations of the heart. For a medium sized experiment (10 hearts) the completion of the procedure requires 2–3 h.
Keywords: Immunology and Infection, Issue 138, Myocardium, immune cells, macrophages, dendritic cells, cell isolation, tissue digestion
Introduction
Different forms of myocardial stress or injury, including ischemic (ischemia reperfusion or myocardial infarction) and non-ischemic (hypertension or myocarditis), promote the recruitment of inflammatory cells with reparative and protective, but also pathogenic properties. As early as 1891, Romberg first described the presence of cellular infiltrates in the myocardium of patients infected with typhus and scarlet fever1. However, the detailed study of cardiac immune cells required the development of more advanced immunophenotyping techniques. As a consequence, only recently have we started to understand that a diverse population of immune cells with essential maintenance roles during steady state resides within the myocardium.
Histology has been, and still is, the most common method to characterize cardiac immune cells during inflammation. However, while histology represents a valuable diagnostic tool, its use in the study of cardiac immune cells has important limitations. The immune cells residing in the myocardium represent a very small proportion of the total cells and are more or less evenly distributed in a proportionally immense space. Similarly, several forms of cardiac inflammation, such as viral myocarditis, show focal patterns of inflammation. This means that accurate analysis of the immune system of the heart often require higher degrees of histological sampling, increasing the cost to reduce bias. Moreover, histology provides a very limited amount of usable information in cell identification (e.g. number of parameters). With an ever increasing number of cell subsets that require the use of multiple expression markers (including surface markers, transcription factors or secreted molecules), flow cytometry has established itself as the most powerful tool for immunophenotyping, due to low-cost and high-throughput.
The use of immunophenotyping techniques is closely dependent on the development of efficient digestion protocols that allowed for single-cells analysis of large numbers of cells. The development of exhaustive protocols of cell extraction opened a new window of opportunities in the study of cardiac immunology. Through the combination of digestion, flow cytometry and transcriptomics, the major populations of macrophages and dendritic cells residing in the heart have been characterized2,3. The most abundant population of cardiac immune cells during steady state are CD64+MerTK+ macrophages, which can be further divided based on their expression of CCR2 or CD11c2. CCR2+ macrophages originate from adult bone marrow hematopoiesis, whereas the majority of CCR2- macrophages, which can express high or low levels of MHC-II, are mainly of prenatal origin. Macrophages perform key functions such as repair4 and removal of debris5 during injury or supporting electrical connectivity6 in steady state. During different forms of inflammation an important influx of Ly6Chi MHC-IIlo CD64int monocytes occur, which later differentiate into Ly6Chi CCR2+ macrophages2,7. A significantly smaller, but important, population of cells residing in the myocardium of healthy individuals is composed of dendritic cells8,9. The two major subsets of cardiac conventional DCs (cDCs) have been recently characterized: cDC1 (CD103+ DCs) and cDC2 (CD11b+ DCs). DCs play important roles in defense against infection8 but can also promote self-injury during inflammation, particularly during myocardial infarction9.
Here, we describe a simple method for isolation of viable cardiac immune cells from mouse myocardium. The method combines enzymatic and mechanical digestion with a cell-strainer filtration in order to obtain a single cell suspension that can be analyzed, or further purified, by flow cytometry sorting or magnetic bead enrichment. Accurate measurements of extravascular myocardial cells require cardiac perfusion in order to remove possible contaminants from the bloodstream of the cardiac microvasculature. In addition, we present an optional step of intravascular labeling of immune cells that can be used to further differentiate myocardial cells from intravascular contaminants, based on the Galkina et al. protocol10. Finally, we present a basic flow cytometry analysis to identify the major macrophage and cDC subpopulations.
Protocol
Ethics statement: This protocol has been reviewed and approved by the Animal Care Committee at the University Health Network (Toronto, Canada) and is in compliance with the Canadian Council on Animal Care.
1. Buffer Preparation
Prepare HBB buffer (Hank's Balanced Salt Solution (HBSS), 2% heat inactivated bovine serum, 0.2% bovine serum albumin). Add 10 mL of heat inactivated bovine serum and 1 g of bovine serum albumin to 500 mL of HBSS. Filter sterilize through a 0.2 µm filter and store at 4 °C.
Prepare FACS buffer (Phosphate buffered saline (PBS), 2% heat inactivated bovine serum, 1 mM Ethylenediaminetetraacetic acid (EDTA, pH 8.0)). Add 10 mL heat inactivated bovine serum and 1 mL of 0.5 M EDTA to 500 mL of PBS. Filter sterilize through a 0.2 µm filter and store at 4 °C. NOTE: Solutions can be stored at 4 °C for up to 1 month.
2. Labelling of Circulating Leukocytes
Aseptically prepare anti-mouse CD45 injection with 1 µg of antibody diluted in sterile phosphate buffered saline (PBS) for a final injection volume of 200 µL/mouse. Load volume into a 28.5G insulin syringe and keep away from light. NOTE: Use two different fluorochrome-labelled anti-CD45 antibodies in order to differentiate the intravascular (stained through i.v. administration) from the intracardiac staining (stained after digestion; see step 4).
- Pre-warm mice (8–12 weeks old) by exposing the tail to a red heat lamp for 5 min. CAUTION: Do not overheat the animal. Keep the lamp at a safe distance (>30 cm).
- Restrain the animal in the sternal position using an appropriate device. Expose and stabilize the tail with the non-dominant hand by immobilizing the base of the tail with the index and middle finger and the mid-distal region of the tail with the thumb and ring finger. CAUTION: Do not apply too much force at the tip of the tail as the skin can be pulled off. Insert the needle with the bevel facing up into the vein. Keep the needle parallel to the vein at all times as it is easy to puncture the vein. NOTE: Blood entering the syringe may occur and is a sign of a good injection. Bending the needle to a 90° angle can facilitate i.v. administration. Confirm with your safety office as bending needles may represent a hazard.
- Inject 200 µL intravenously into the tail vein. The fluid should flow easily and temporarily clear the vein. CAUTION: Do not force fluid in if resistance is met. This is sign of misplacement of the needle.
- Allow antibody to circulate for 5 min before humanely euthanizing mouse.
3. Heart Isolating and Digestion
- Euthanize mice using CO2 or by other institutionally accepted euthanasia procedure.
- For CO2 euthanasia, place the mouse in the CO2 chamber and set the fill rate to 20% of the chamber volume per minute (i.e., if the chamber is 10 L, fill at 2 L/min). Run for 2 min and monitor mouse until breathing has ceased.
- Shut off the CO2 and leave the mouse for an additional 1-2 min. Ensure breathing has ceased before opening the chamber and perform a toe pinch to confirm the mouse is dead before proceeding to process the mouse.
Spray the mouse with 70% ethanol. Lift the skin at the abdomen and dissect the mouse by cutting along the ventral midline beginning at the level of the hips up to the sternum. Open the abdomen and move the liver to expose the diaphragm.
- Cut the diaphragm, then the ribs on both sides of the midline, being careful to avoid puncturing the lungs and heart.
- Expose the heart by peeling back the ribs, then cut first the left atria followed by the right atria. CAUTION: Use caution when cutting the atria, ensuring no arteries or veins are injured, as this can decrease the efficiency of flushing the blood contained in the organs.
- Perfuse the heart with 15–20 mL of cold PBS using a 60 mL syringe fitted with a 21 G needle and cold PBS. Gently grasp the base of the heart with forceps and insert the needle into the left ventricle near the apex. Repeat the procedure in the right ventricle. A proper injection results in white lungs and pale liver. NOTE: Perfusion should be performed within 5 minutes after euthanasia in order to avoid blood coagulation, which can contaminate the organs of interest with circulating cells.
- Hold the base of the heart with forceps and gently pull the heart up, then detach the heart by cutting the major pulmonary arteries and veins, aorta and the pericardial sack.
- Clean the heart by gently holding the heart between the thumb and index fingers in a clean lint-free paper towel and pull off the atria and remnants of major arteries and veins and any other contaminants with forceps. NOTE: Ensure the entirety of the atria is removed and no other tissues, such as the lung or blood vessels, are present as these tissues have their own resident immune cells. The lung in particular has a large immune cell population that can mask the smaller populations of those within the myocardium.
- Place the heart in a 5 cm plastic dish with 50 µL of cold PBS.
- Mince the heart using curved dissecting scissors until there are no visible pieces and it has a smooth consistency. NOTE: Try to keep the heart in a contained area of the dish during chopping to minimize tissue loss.
- Transfer thetrituratedheart tissue using curved forceps into 2 mL microcentrifuge tube with 800 µL cold Dulbecco's Modified Eagle Medium (DMEM).
- Add 450 U/mL ofcollagenase I, 60 U/mL of hyaluronidase type I-S, and 60 U/mL of DNase-I to each sample.
- Incubate samples at 37 °C on a rocking shaker at 50 rpm for 45–60 min.
- After digestion, briefly vortex samples (20 s) and immediately place on ice to keep samples at 4 °C.
- Using a 1 mL pipettor, pipette samples up and down until tissue samples are homogeneous and do not clog the pipette tip. NOTE: Defining a specific number of times to pipette may increase reproducibility. If the hearts are not optimally digested, pieces of myocardium may obstruct the pipette. Gently grinding the pipette tip against the microcentrifuge tube will help homogenize bigger pieces of tissue.
- Pre-wet a 40 µm cell strainer placed on a 50 mL conical tube with 1 mL of cold HBB.
- Add homogenized sample to the filter and wash through by slowly pouring 12–14 mL of cold HBB.
- Transfer filtered samples to a labelled 15 mL conical tube and keep at 4 °C.
- Centrifuge samples at 400 x g for 5 min at 4 °C. Aspirate off supernatant.
- Lyse red blood cells by adding 1 mL of Ammonium-Chloride-Potassium (ACK) lysis buffer to each sample. Resuspend samples by vortexing and incubate at room temperature for 5 min.
- Once lysis has completed, add 5–8 mL of cold Hank's Balanced Salt Solution (HBSS) to stop hemolysis.
Centrifuge samples at 400 x gfor 5 min at 4 °C. Aspirate off supernatant.
Add 1 mL of FACS buffer to each sample and transfer to a labelled 1.5 mL microcentrifuge tube. NOTE: The protocol can be paused here for up to 2 h with samples stored at 4 °C.
4. Flow Cytometry Staining and Analysis
- Centrifuge samples at 400 x g for 5 min at 4 °C.
- To reduce non-specific antibody binding, dilute 1:100 anti-CD16/CD32 and Monocyte Blocker (1:20) (see Table of Materials) accounting for 50 µL FACS per sample. Incubate at room temperature for 15 min.
- Prepare the antibody dilution (1:250 for each antibody) accounting for 50 µL of FACS per sample. NOTE: A viability dye can be included in the antibody cocktail in order to exclude dead cells. Alternatively, small size exclusion by flow analysis can be used (Figure 1C and D).
- Without washing, add 50 µL of antibody master mix to each sample and mix.
- Incubate samples at 4 °C for 30 min in the dark.
Add 600 µL FACS to each sample to wash and centrifuge samples at 400 x g for 5 min at 4 °C.
Aspirate off supernatant and resuspend pellet in 300 µL FACS. Keep samples at 4 °C in the dark. NOTE: Some viability dyes, such as DAPI, require staining within 10 min prior to running flow cytometry.
Analyze by flow cytometry (see Figures 1 and 2) within the next 3 h. If a longer time is required, short term fixation using a low concentration paraformaldehyde (0.5–1% in PBS) is recommended to preserve staining. NOTE: It is highly recommended to filter samples through a 40 µm cell strainer prior to initiate flow cytometry analysis in order to prevent obstruction of the fluidics of the flow cytometer.
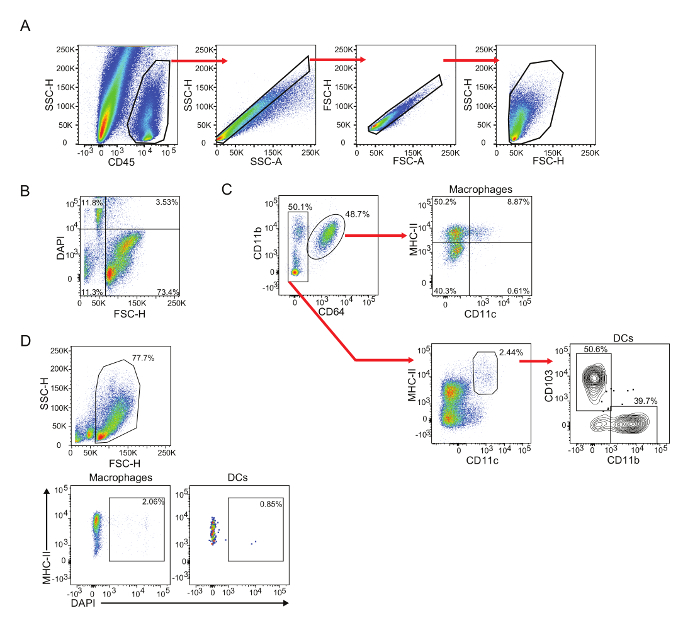
Representative Results
To date, no good method has been designed to isolate immune cells from other cardiac cell components, such as cardiomyocytes. Hence, analysis of the cardiac single cell suspension by flow cytometry requires a pre-gating with CD45 to identify the immune cell populations, followed by single cell and small size exclusion gating (Figure 1A). Alternatively, a viability staining can be performed to exclude dead cells. Small size exclusion and viability dye staining represent nearly equivalent methods to exclude dead cells (Figure 1B and 1D). It is highly recommended to proceed with the antibody staining soon after the digestion procedure and always keep the cell suspension at 4 °C, as the viability of the cells may decay. This procedure typically yields about 80–90% of viable immune cells.
The myocardium houses a diverse colony of immune cells, many of which are still pending identification and characterization. Macrophages, identified as CD64+ cells, are the major immune population of the heart, representing 60–70% of all immune cells (Figure 1C, top). The digestion of an adult murine heart yields about 3–5 x 104 macrophages, depending of the heart's size. Cardiac macrophages can be further subdivided based on their expression of CD11c or CCR2 and their levels of MHC-II2.
The other cardiac immune subpopulation that has been well characterized are conventional dendritic cells. Conventional DCs are CD64- cells that express intermediate to high levels of MHC-II and high levels of CD11c (Figure 1C, bottom). These cells can be further divided based on their expression of either CD103 or CD11b8,9. DCs represent a much smaller cardiac subpopulation compared to macrophages. The digestion of an adult murine heart yields between 150 and 500 DCs of each subset.
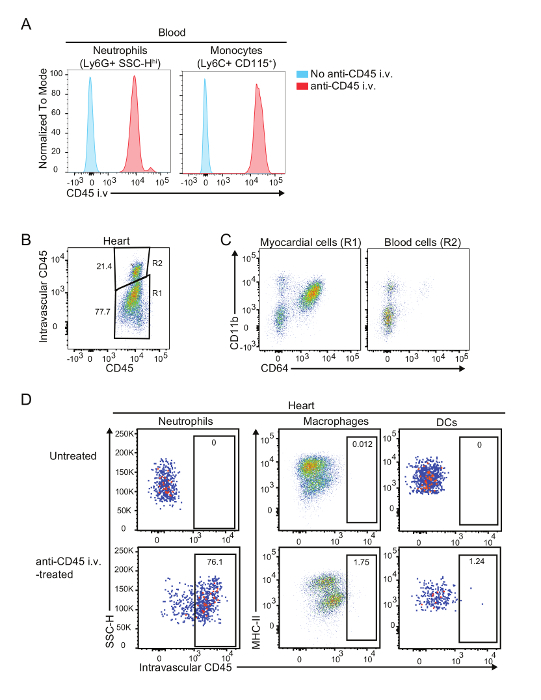
Perfusion of the heart with cold PBS is essential when identifying immune cell populations within the myocardium. However, imperfect perfusion may increase the amounts of intravascular contaminants in the samples, leading to important bias that can be based on peripheral changes (blood) rather than changes in the cardiac infiltrates. In these cases, intravascular administration of fluorescent-labelled anti-CD45 antibodies, which labels all immune cells circulating through macro- and microvasculature (Figure 2A), can help differentiate myocardial cells from intravascular contaminants. Figure 2B shows an example of an imperfectly perfused heart in which 20% of immune cells are intravascular contaminants. Importantly, this level of contamination has little effect on macrophage and DC analysis, since these cells are rarely found in blood (Figure 2D).
The use of flow cytometry of a single cell suspension of myocardium allows for the study of the small subpopulations of the heart. For example, the deficiency of the transcription factor BATF3 has been shown to prevent the development of mature CD103+ DCs in several tissues11. In order to study if this is also true in the case of the myocardium, single cell suspensions of cardiac cells from 8–15-week-old BATF3-deficient or control littermates (in C57BL/6 background) were analyzed using flow cytometry. Due to the small population size and number of markers needed to differentiate the DC subsets, histology could not have provided enough power for analysis to reach a definitive answer. However, using flow cytometry analysis to compare the immune cells of the myocardium of wild-type versus a Batf3-/- mice, a lack of CD103+ DCs can be confirmed in the latter (Figure 3A and B). In addition, we observed that BATF3-deficiency did not significantly alter the composition of other cardiac populations (Figure 3C and D). These results exemplify the degree of sensitivity that can be achieved by using this combined method of enzymatic and mechanical digestion to achieve a single cell suspension.
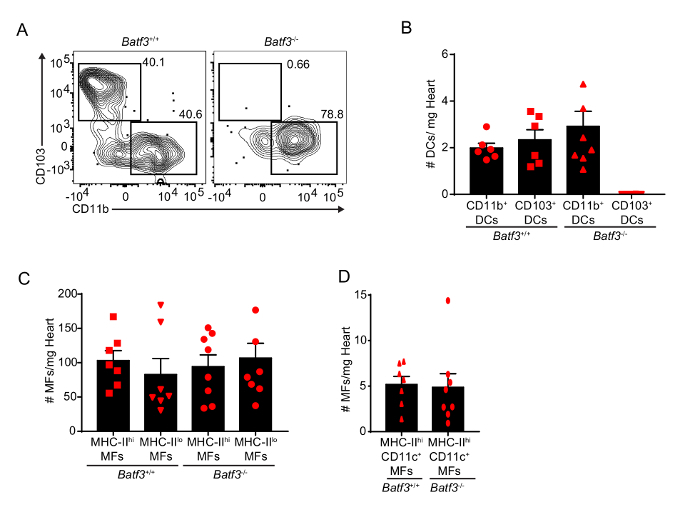
Figure 1: Gating strategy for cardiac macrophages and DCs. Isolated hearts from C57BL/6 mice were digested as described, and single cell suspension was analyzed by flow cytometry. (A) Gating of CD45+ live singlet cardiac cells. Use CD45 labeling to identify immune cells from the cardiac single cell homogenate, which includes very high levels of non-immune cells (i.e., cardiomyocytes, endothelial cells and fibroblasts). Exclude doublets by comparing SSC-H vs SSC-A followed by FSC-H vs FSC-A. Note: Any other comparison between SSC-A, SSC-H and SSC-W or FSC-A, FSC-H and FSC-W are similarly useful. Selection of live cells can be performed by excluding very small cells (SSC-H vs FSC-H gate). (B) Comparison of dead cell exclusion using size vs viability staining (DAPI). (C) Gating strategy for identification of cardiac macrophages (CD11b+ CD64+ cells) and DCs (CD64- MHC-IIhi CD11c+). Cardiac macrophages can be further classified based on their expression of MHC-II and CD11c. Cardiac DCs can be subdivided based on their expression of CD103 or CD11b. (D) Representative plots of DAPI+ dead cells within the macrophage and DC compartments after small size exclusion. Please click here to view a larger version of this figure.
Figure 2: Labelling of intravascular cells contaminants. (A) Comparison of intravascular CD45 labelling of blood neutrophils and Ly6Chi monocytes in anti-CD45 antibody treated (red) or non-treated (blue) mice. (B) Identification of intravascular contaminants in a cardiac preparation by intravascular CD45 labelling. (C) Representative flow plots of myocardial and intravascular cells. (D) Representative plots of labelled cells within the macrophage and DC compartments after anti-CD45 i.v. administration. Note that macrophages and DCs are exclusively extravascular. Intravascular contaminants are typically lymphocytes, monocytes and neutrophils2. Please click here to view a larger version of this figure.
Figure 3: Quantification of cardiac macrophages and DCs in BATF3-deficient mice. (A) Representative flow plots of cardiac DCs in Batf3+/+ and Batf3-/- mice. (B) Quantification of cardiac DCs in Batf3+/+ and Batf3-/- mice. Note the absence of CD103+ DCs in Batf3-/- mice. (C and D) Quantification of cardiac macrophages (MF) in Batf3+/+ and Batf3-/- mice. Error bars represent SEM. Red symbols represent individual animals. Please click here to view a larger version of this figure.
Discussion
Myocardial inflammation, or myocarditis, is a feature of most cardiovascular diseases. However, the myocardium is not devoid of its own immune components in non-disease states. During steady state, many immune cells reside in the myocardium and play the essential roles of maintenance and protection. The characterization of these diverse populations of cells would not have been possible without methods such as the one presented in this protocol.
A combination of mechanical and enzymatic digestion of the myocardium permits the isolation of a single cell suspension that can be used in combination with other methods of analysis, such as flow cytometry or immunomagnetic bead isolation. Several considerations regarding the present method must be taken into account. First, all of our analyses refer to the populations within the myocardium; we exclude the atria from our analysis as these are specialized tissues with its own idiosyncrasies. Second, the amounts of enzymes used are expressed as activity units to help in the set-up of digestions using enzymes from other sources. However, empiric optimization of the final concentration of enzymes and the time of digestion is highly recommended, as limiting enzyme can significantly reduce the cell yield and too much digestion can drastically affect viability. Third, the optional step involving intravascular administration of labelled antibodies is especially recommended when studying uncommon populations in the normal myocardium (such as lymphocytes, monocytes or neutrophils)2 or under circumstances when numbers of populations in the blood may change (monocytosis, granulocytosis or lymphocytosis) which may give a false impression of changes within the myocardium that need to be confirmed using this method or, alternatively, microscopy. However, this step is of limited utility when analyzing macrophages or mature DCs, as these cells are rarely found in significant quantities in the blood (Figure 2D). It is important to optimize the quantity of antibody administered and the incubation time before sacrificing the mouse, as long term incubation can allow for some antibody diffusion into the heart, especially in an inflamed myocardium. Finally, it is very important to be meticulous in the isolation of the heart, eliminating remnants of major arteries, veins and or any other tissue, such as the lung. These contaminant tissues have highly elevated numbers of immune cells that can easily mask the myocardial indigenous populations.
It is very important to perform exclusion of dead cells, whether using viability dyes or through small size exclusion during flow cytometry analysis, as digestion (or tissue damage in disease models) can cause a relatively large amount of dead cells. We have compared the efficiency of dead cell exclusion by size and using a viability label (Figure 1B and D), demonstrating their near equivalence. Importantly, size exclusion cannot be used after fixation and permeabilization as this procedure significantly reduces cell size.
Although autofluorescence is rarely a problem to identify cardiac macrophages using flow cytometry, this represents a more important problem when trying to characterize the expression of some markers, especially with fluorochromes excited by high energy (shorter) light wavelengths (350–500 nm). In these experiments, it is highly recommended to use isotype controls, avoid the blue and violet lasers (fluorescein isothiocyanate, phycoerythrin, Pacific Blue/Brilliant Violet 421) and use fluorochromes with emissions above 600 nm, such as allophycocyanin.
The use of protocols as the one presented herein are becoming more important as an increasing amount of literature is identifying the role of intensity and type of inflammation in evaluating and characterizing the outcomes of cardiovascular disease. This method permits higher levels of cell characterization than histology. For instance, the use of lineage tracing allowed for the identification of ontogenically different subsets of macrophages with their programmed different functions2. Similarly, this method can be used to study T cell responses during myocardial inflammation and combined with ex vivo stimulation to study T cell polarization8.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (148808 and 148792), SE was supported by a Heart and Stroke Foundation, personnel award from the Ontario Provincial Office, March of Dimes, Ted Rogers Centre for Heart Research and the Peter Munk Cardiac Centre. XCC holds a CIHR Banting Fellowship. LA holds a Heart & Stroke/Richard Lewar Studentship Award.
References
- Marboe CC, Fenoglio JJ. Pathology and natural history of human myocarditis. Pathology and Immunopathology Research. 1988;7(4):226–239. doi: 10.1159/000157119. [DOI] [PubMed] [Google Scholar]
- Epelman S, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40(1):91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AR, et al. An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS One. 2012;7(5):36814. doi: 10.1371/journal.pone.0036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(45):16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Wang J, Nagai R. Cardioprotective function of cardiac macrophages. Cardiovascular Research. 2014;102(2):232–239. doi: 10.1093/cvr/cvu059. [DOI] [PubMed] [Google Scholar]
- Hulsmans M, et al. Macrophages Facilitate Electrical Conduction in the Heart. Cell. 2017;169(3):510–522. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahrendorf M, et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. Journal of Experimental Medicine. 2007;204(12):3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Casares X, et al. A CD103(+) Conventional Dendritic Cell Surveillance System Prevents Development of Overt Heart Failure during Subclinical Viral Myocarditis. Immunity. 2017;47(5):974–989. doi: 10.1016/j.immuni.2017.10.011. [DOI] [PubMed] [Google Scholar]
- Van der Borght K, et al. Myocardial Infarction Primes Autoreactive T Cells through Activation of Dendritic Cells. Cell Reports. 2017;18(12):3005–3017. doi: 10.1016/j.celrep.2017.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkina E, et al. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. Journal of Clinical Investigation. 2005;115(12):3473–3483. doi: 10.1172/JCI24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. Journal of Experimental Medicine. 2010;207(4):823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]


