Abstract
The mammalian epididymis generates one of the most complex intraluminal fluids of any endocrine gland in order to support the post-testicular maturation and storage of spermatozoa. Such complexity arises due to the combined secretory and absorptive activity of the lining epithelial cells. Here, we describe the techniques for the analysis of epididymal protein synthesis and secretion by focusing on the model protein family of dynamin (DNM) mechanoenzymes; large GTPases that have the potential to regulate bi-directional membrane trafficking events. For the study of protein expression in epididymal tissue, we describe robust methodology for immunofluorescence labeling of target proteins in paraffin-embedded sections and the subsequent detection of the spatial distribution of these proteins via immunofluorescence microscopy. We also describe optimized methodology for the isolation and characterization of exosome like vesicles, known as epididymosomes, which are secreted into the epididymal lumen to participate in intercellular communication with maturing sperm cells. As a complementary approach, we also describe the immunofluorescence detection of target proteins in an SV40-immortalized mouse caput epididymal epithelial (mECap18) cell line. Moreover, we discuss the utility of the mECap18 cell line as a suitable in vitro model with which to explore the regulation of epididymal secretory activity. For this purpose, we describe the culturing requirements for the maintenance of the mECap18 cell line and the use of selective pharmacological inhibition regimens that are capable of influencing their secretory protein profile. The latter are readily assessed via harvesting of conditioned culture medium, concentration of secreted proteins via trichloroacetic acid/acetone precipitation and their subsequent analysis via SDS-PAGE and immunoblotting. We contend that these combined methods are suitable for the analysis of alternative epididymal protein targets as a prelude to determining their functional role in sperm maturation and/or storage.
Keywords: Developmental Biology, Issue 138, Dynamin, epididymis, epididymosome, exosome, immunofluorescence, protein secretion, sperm, sperm maturation
Introduction
The spermatozoa of all mammalian species acquire the potential to display forward progressive motility and to fertilize an ovum during their prolonged descent through the epididymis, a highly specialized region of the male extra-testicular duct system, which may take 7 - 14 days to navigate (depending on the species)1. Due to the extreme condensation of the paternal chromatin and the shedding of the majority of cytoplasm that accompanies the cytodifferentiation of spermatozoa within the testes, their subsequent functional maturation is driven exclusively by their interaction with the epididymal microenvironment. This milieu is, in turn, created by the secretory and absorptive activity of the lining epididymal soma and displays an exceptional level of segment-segment variation1. Thus, the most active segments in terms of protein synthesis and secretion are those located in the proximal portion of the epididymis (namely, the caput and corpus)2. This activity mirrors the functional profile of spermatozoa, with the cells first beginning to display hallmarks of functional competence (i.e., progressive motility and the ability to bind to acid-solubilized zona glycoproteins) following their passage through the caput epididymis3. These functional attributes continue to develop before reaching optimal levels as the sperm reach the distal epididymal segment (cauda), wherein they are stored in a quiescent state in readiness for ejaculation. The formation and maintenance of this sperm storage reservoir is also intimately tied to the lining epithelium, which in the cauda is dominated by strong absorptive activity4,5. Although anatomical differences have been reported6,7,8, such regionalized division of labor appears to be a characteristic of the epididymis that is shared among the majority of mammalian species studied to date, including our own9,10. Indeed, from a clinical perspective, it is known that epididymal dysfunction makes an important contribution to the etiology of male factor infertility11, thus highlighting the importance of understanding the regulation of this specialized tissue.
It is therefore regrettable that our understanding of epididymal physiology, and the mechanisms that regulate the sequential phases of sperm maturation and storage within this tissue, remain to be fully resolved. Among the contributing factors, limiting advances in epididymal research are the overall complexity of this tissue and knowledge of the mechanisms that exert regulatory control over its luminal microenvironment. Anatomically, we know that beyond the distinction of caput, corpus and cauda segments, the epididymis can be further subdivided into several zones (Figure 1A), each separated by septa12 and characterized by discrete profiles of gene/protein expression13,14,15,16,17,18. Indeed, on the basis of detailed transcriptional profiling of segmental gene expression in the epididymis, as many as 6 and 9 distinct epididymal zones have been reported in the mouse and rat models, respectively19,20. Such complexity presumably reflects the composition of the epididymal soma, a pseudostratified epithelium comprising numerous different cell types; each differing with respect to their abundance, distribution and secretory/absorptive activities along the length of the tract. Thus, principal cells are by far the most abundant epididymal cell type constituting upwards of 80% of all epithelial cells. Accordingly, principal cells are responsible for the bulk of epididymal protein biosynthesis and secretion5. In contrast, the clear cell population, which rank as the second most abundant cell type within the epididymal soma, are primarily involved in selective absorption of luminal components and the acidification of this microenvironment5. Adding another tier of complexity, androgens and other lumicrine factors of testicular origin exert differential control over each of these epididymal cell types depending on their positioning along the tract.
Despite the limitations imposed by such complexity, significant inroads continue to be made into resolving the mechanistic basis of epididymal function. A key to these studies has been the application of advanced mass spectrometry strategies to establish broad scale inventories of the epididymal proteome, in tandem with detailed analyses of individual proteins selected from among these initial surveys. An illustration of this approach is our recent characterization of the DNM family of mechanoenzymes in the mouse model21. Our initial interest in DNM was fueled by its dual action in the coupling of exo- and endocytotic processes. Building on these observations, we were able to demonstrate that the three canonical isoforms of DNM (DNM1 - DNM3) are highly expressed in the mouse epididymis and appropriately positioned to fulfill regulatory roles in protein secretion and absorption21. Moreover, we were able to clearly differentiate each DNM isoform on the basis of their cellular and sub-cellular localization, thus suggesting that they possess complementary, as opposed to redundant, activity within the epididymal epithelium21.
Here, we describe the experimental methodology employed for the study of DNM expression in the mouse epididymis with the hope that this information will find wider application in the characterization of alternative epididymal proteins and thus contribute to our understanding of the function of this important element of the male reproductive tract. Specifically, we describe the development of robust methodology for immunofluorescence labeling of target proteins in paraffin-embedded epididymal sections and the subsequent detection of the spatial distribution of these proteins via immunofluorescence microscopy. We further document our recently optimized protocols22 for the isolation and characterization of epididymosomes; small exosome-like vesicles that constitute key elements of the epididymal secretory profile and appear to hold a prominent role in promoting sperm maturation23. As a complementary approach, we also describe the immunofluorescence detection of target proteins in an immortalized mouse caput epididymal epithelial (mECap18) cell line and the use of this resource as a model with which to explore the regulation of epididymal secretory activity in vitro.
Protocol
All experimental procedures involving animal tissue collection were approved by the University of Newcastle's Animal Care and Ethics Committee.
1. Immunofluorescence Staining of the Paraffin-embedded Epididymal Sections (Figures 1 and 2)
Immediately after the euthanasia of adult mice via CO2 inhalation (Swiss mice, over 8 weeks old), carefully dissect the epididymis (using surgical scissors and tweezers) free of overlying connective tissue and fat and immerse in Bouin’s fixative solution (> ten times volume/tissue weight) for overnight fixation.
Wash the tissue with 70% ethanol with 2× changes daily for 2 days and then dehydrate through graded ethanol (70%, 95% and 100%) in preparation for infiltration and embedding into a paraffin block.
Section the paraffin blocks at a thickness of 4-6 µm and mount on the slides in preparation for immunofluorescence staining.
In a fume hood, dewax the epididymal paraffin sections by adding a sufficient amount of xylene to the slide jar to completely immerse the tissue section (3× 5 min each time).
Rehydrate the tissue sections by the immersion in graded ethanol solutions diluted in purified H2O (100% ethanol 5 min, 100% ethanol 5 min, 90% ethanol 1 min, 80% ethanol 1 min, 70% ethanol 1 min, and 50% ethanol 1 min).
Wash the sections in a slide jar once for 5 min with sufficient phosphate buffered saline (PBS) to completely immerse the entire tissue section (follow these directions for all subsequent washes).
Decant appropriate antigen retrieval solution (i.e., 10 mmol/L sodium citrate, 50 mmol/L Tris pH 10.5 or alternative antigen retrieval solution(s), depending on the antigen to be detected) into a slide rack and microwave until boiling. Immerse the slides into this solution and subject the tissue sections to heat-induced antigen retrieval conditions optimized for individual antibodies (see Table 1). Caution: Ensure that the slides are fully immersed in antigen retrieval solution during the antigen retrieval process.
Remove the slide container from the microwave and cool to room temperature.
Rinse the slides with PBS and use a liquid-repellent slide marker pen to trace around the tissue section.
Place the slides in a humidified container (created by a moistened tissue at the base of the container), and apply blocking solution (3% BSA/PBS, previously filtered through a 0.45 µm filter) for 1 h at 37 °C.
Rinse the slides once with PBS.
Incubate the sections with appropriate primary antibody diluted to an experimentally optimized concentration in filtered 1% BSA/PBS at 4 °C overnight (1:60 for anti-DNM1, DNM2 and DNM3 antibodies; 1:100 for anti-ATP6V1B1 antibody, see Table of Materials for antibody details). Note: To distinguish specific from non-specific antibody binding, it is necessary to include stringent negative (i.e., secondary antibody only, primary antibody preabsorbed against immunizing peptide) and positive controls24.
Rewarm the slides by placing at room temperature for 30 min.
Wash the slides 3× with PBS on a shaking platform (60 rpm) for 10 min each.
Incubate the sections with appropriate secondary antibody diluted in 1% BSA/PBS (filtered through a 0.45 µm filter) at 37 °C for 1 h (1:400 dilution for all secondary antibodies, see Table of Materials for antibody details). CAUTION: Keep the slide container in the dark from this step onwards. For dual labeling, choose a compatible combination of secondary antibodies (i.e., secondary antibodies must have been raised in different species).
Wash the slides 3× with PBS on a shaking platform (60 rpm) for 10 min each.
Counterstain the sections with propidium iodide (PI, 7.48 μmol/L) or 4΄,6-diamidino-2-phenylindole (DAPI, 4.37 μmol/L) for 2 min at room temperature to label the cell nucleus.
Wash the slides twice with PBS on a shaking platform (60 rpm) for 5 min each.
Mount the sections with 10% Mowiol 4-88 prepared in a solution of 30% glycerol in 0.2 mol/L Tris (pH 8.5) and 2.5% 1, 4-diazabicyclo-(2.2.2)-octane.
Seal the coverslip with nail varnish and store the slides at 4 °C for future observation. CAUTION: It is recommended to perform the imaging of the slides as soon as practical after the preparation to avoid excessive loss of fluorescence.
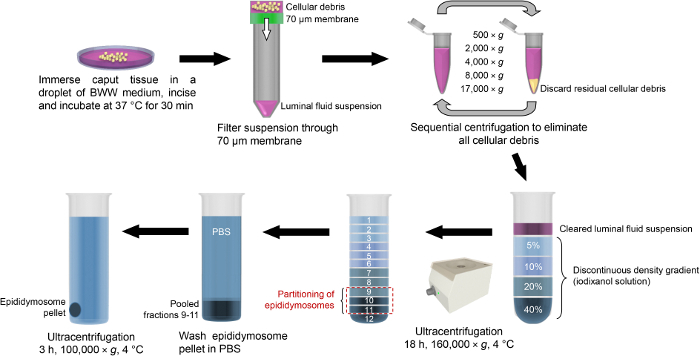
2 Isolation of Epididymosomes from the Mouse Caput Epididymis (Figure 3)
Immediately after the euthanasia of adult mice via CO2 inhalation (Swiss mice over 8 weeks old), perfuse their vasculature with PBS (pre-warmed to 37 °C) to minimize the blood contamination of epididymal tissue. CAUTION: Blood plasma contains diverse populations of exosomes, which are of similar size to epididymosomes25. The efficacy of blood clearance from epididymal tissue can be accessed via the inspection of the initial segment, a highly vascularized epididymal segment located proximal to the caput segment (i.e., zone 1 in Figure 1A)
Carefully dissect the epididymis free of overlying fat and connective tissue, and rinse with modified Biggers, Whitten, and Whittingham medium (BWW; pH 7.4, osmolality of 300 mmol/kg water26,27) to reduce any potential for surface blood contamination.
Blot the epididymal tissue to remove excess media, dissect the caput epididymis (i.e., zones 2-5 in Figure 1A) and transfer to a fresh Petri dish (35 × 10 mm) containing BWW medium. Ensure that the amount of medium is sufficient for the final recovery. Note: For 6 caput epididymides, it is recommended to use 1.1 mL of the medium to allow for a recovery of ~900 µL, which is then evenly split and applied atop of 2 pre-prepared gradients (see step 2.9).
Make a number of small incisions into the caput tissue with a razor blade. Do not mince the tissue and thus avoid contaminating the sample with excessive cytosolic contents. Incubate the plate containing the tissue with mild agitation at 37 °C for 30 min to release the luminal contents.
Filter the resultant suspension through 70 µm membranes to remove the cellular debris.
Collect the filtrate and subject this to successive centrifugation steps at 4 °C with increasing velocity in order to eliminate cellular debris (i.e., 500 × g, 2,000 × g, 4,000 × g, 8,000 × g, 5 min each; 17,000 × g for 20 min, and finally 17,000 × g for an additional 10 min or until no pellet is formed after centrifugation). CAUTION: It is important to assess the color of the pellet after the initial 500 × g centrifugation step to ensure minimal blood contamination is present. Discard any samples in which this pellet displays pink coloration.
Prepare discontinuous iodixanol gradients (comprising 40%, 20%, 10%, 5% layers) by diluting a density gradient medium (comprising 60% (w/v) aqueous iodixanol) with a solution of 0.25 mol/L sucrose and 10 mmol/L Tris (pH 7.5).
Prepare the gradient in an ultracentrifuge tube (11 × 35 mm), with each fraction of 450 µL (Figure 3). Visually inspect the gradient after the application of each fraction to ensure that the interfaces are successfully formed between each layer prior to loading the epididymal fluid sample. Prepare each gradient fresh on the day of use, however, the epididymal luminal fluid sample can be preserved at 4 °C for up to 2 h prior to loading.
Carefully add 450 µL of epididymal luminal fluid suspension (corresponding to the material collected from the caput of 3 epididymides) atop of a single gradient.
Ultracentrifuge the gradients at 160,000 × g at 4 °C for 18 h. CAUTION: Since this centrifugation is conducted at very high speed, all ultracentrifuge tubes must be paired and balanced precisely. Check the tubes to ensure that they are free of any visible damage that could compromise their integrity.
Gently remove 12 equal fractions (each consisting of 185 µL) starting from the uppermost layer and progressing toward the bottom of the gradient. Pool the equivalent fractions recovered from each gradient if applicable (up to two gradients). Note: Mouse epididymosomes are most highly enriched in fractions 9 - 1122, see Figure 4 and Discussion.
After the recovery and pooling of fractions 9 – 11, dilute into 2 mL of PBS, and ultracentrifuge the samples at 100,000 × g at 4 °C for 3 h (13 × 56 mm tube) to pellet the epididymosomes. CAUTION: Since the epididymosome pellet can be difficult to see, ensure that the orientation of the tubes is noted as they are placed into the rotor and mark the tube to indicate the expectant position of the epididymosome pellet. Ensure that each tube contains a sufficient volume (i.e., exceeding 50% of its total capacity) to preclude the risk of tube collapse.
Carefully aspirate and discard the supernatant without disturbing the epididymosome pellet.
Assess the epididymosome purity (Figure 4).
Resuspend the epididymosome pellet into desired medium according to the downstream application(s). For instance, BWW medium is generally used for the experiments involving co-incubation with spermatozoa or alternatively an appropriate lysis buffer in preparation for the resolution of the epididymal proteome via SDS-PAGE.
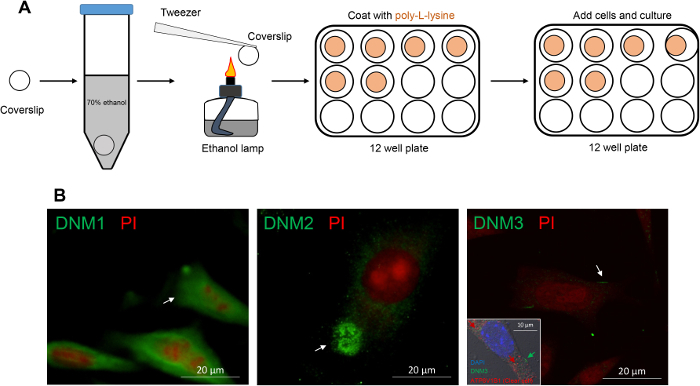
3. Immunofluorescence Staining of mECap18 Cells
- Preparation of sterile coverslips (to be conducted in a cell culture hood)
- Soak the coverslips (12 × 12 mm) in 70% ethanol for 10 min and disinfect by drying under high temperature above an ethanol lamp.
- Cool the coverslip for 10 s before transferring to a 12 well plate.
- Apply sterile poly-L-lysine solution to cover the coverslip and settle for 10 min at room temperature.
- Discard the poly-L-lysine solution and rinse the coverslip with sterile H2O or appropriate medium.
- Preparation mECap18 cells
- Passage the aliquots of 2 × 105 mECap18 cells in each well of the 12 well plate containing the coverslips.
- Culture the cells with mECap18 cell medium (DMEM supplemented with 1% L-glutamine, 1% sodium pyruvate, 1% penicillin/streptomycin, and 50 μmol/L 5α-androstan-17β-ol-3-oneC-IIIN) containing 10% fetal calf serum (FBS) in a 37 °C incubator under an atmosphere 5% CO2 overnight.
- Once the cells adhere to the coverslip, discard the medium and rinse the cells twice with PBS.
- Add a sufficient amount of 4% paraformaldehyde (PFA) diluted in PBS to immerse the entire coverslip and fix the cells at room temperature for 15 min.
- Discard the PFA solution and rinse the coverslips twice in PBS.
- Immunofluorescence staining
- Permeabilize mECap18 cells by immersion in 0.1% Triton X-100 in PBS for 10 min.
- Rinse the coverslips with PBS.
- Block mECap18 cells with 3% BSA and proceed with immunolabeling of cells utilizing equivalent protocols to those described for epididymal tissue sections.
4. Isolation of Proteins from Conditioned Cell Culture Medium
- Collection of conditioned cell culture medium
- Passage aliquots of 4 × 105 mECap18 cells in each well of 6 well plate with mECap18 cell medium supplemented with 10% FBS for 24 h.
- Wash mECap18 cells three times with mECap18 cell medium (prepared without FBS) to remove residual FBS and any associated protein contaminants.
- Add 1.5 mL of mECap18 cell medium (prepared without FBS) to each well and incubate with mECap18 cells for 12 h in a 37°C incubator under 5% CO2. Note: mECap18 cells at this step can be assessed for different target antigens according to experimental design.
- After 12 h incubation, collect the cell medium and centrifuge at 2,000 × g for 10 min to remove all cellular debris. Note: The duration of incubation is able to be altered in accordance with experimental design/endpoint assessment and in consideration of the cell’s tolerance to applied treatment(s). It is recommended to tailor the timing of incubation based on specific experimental regimens to ensure that optimal results are achieved.
- Assess mECap18 cell viability via the application of a standard trypan blue exclusion assay28. Discard all material in which cell viability has declined below 90% to eliminate bias introduced by proteins released from dead or moribund cells.
- Isolate proteins from cell medium as follows or preserve medium at -80 °C.
- Protein isolation (to be conducted in a fume hood)
- Add 20% volume of chilled 100% trichloroacetic acid to 80% volume of conditioned cell medium to precipitate the proteins released from the cultured mECap18 cells. Incubate at 4 °C overnight with constant mixing.
- After the incubation, pellet precipitated protein by centrifugation (17, 000 × g, 4 °C for 10 min). Note: Due to the limited quantity of protein expected to be secreted into the medium, it is possible that the pellet will not be easily visualized after centrifugation. It is therefore imperative to correctly orientate the tube prior to the centrifugation and take care not to disturb the expectant pellet location during removal of the supernatant.
- Discard the supernatant and wash the pellet twice with chilled acetone prior to the re-centrifugation (17, 000 × g, 4 °C for 10 min).
- Carefully remove and discard the supernatant before air-drying any residual acetone within a fume hood.
- Resuspend the protein pellet in an appropriate extraction buffer in preparation for endpoint analysis to detect complete secretory protein profiles and/or individual target proteins (e.g., SDS-PAGE, immunoblotting).
Representative Results
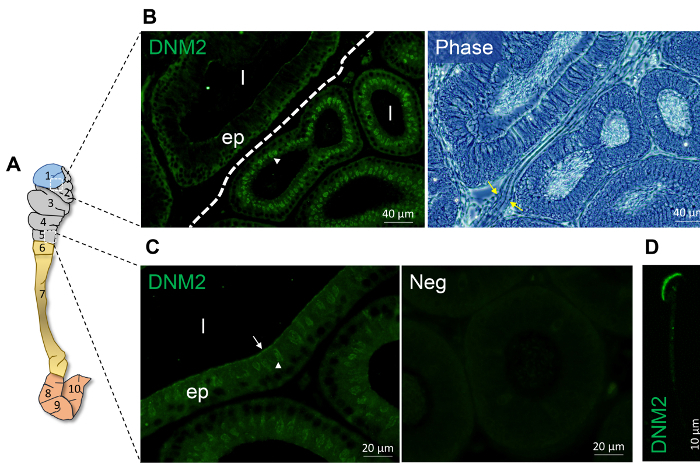
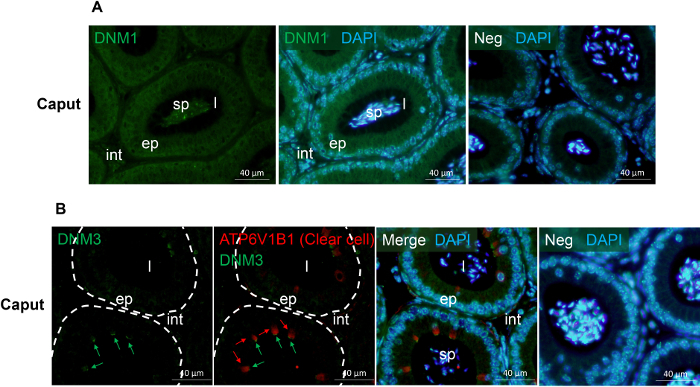
Figure 1 and Figure 2 show representative results of immunofluorescence localization of DNM in the mouse caput epididymis. Each of the three DNM isoforms investigated display distinct localization profiles. Thus, DNM1 is characterized by relatively modest diffuse labeling of the epididymal cells throughout the initial segment and caput epididymis (Figure 2A). By contrast, the DNM2 isoform was first detected in the vicinity of the opposing basal and apical border of cells in the initial segment, before being repositioned to the supranuclear domain in cells within the adjacent downstream caput segment (i.e., zones 2 - 5) (Figure 1B, C). Notably, however, the intensity of DNM2 labeling gradually decreased between zones 2 to 5 of the caput epididymis, a result that essentially mirrors the secretory activity of these epididymal segments21 (Figure 1B, C). Accordingly, the supranuclear labeling of DNM2 was subsequently shown to correspond to the distribution of the Golgi apparatus within caput principal cells21. Spermatozoa isolated from the same epididymal region showed intense acrosomal labeling for DNM2 (Figure 1D). As a caveat, however, equivalent DNM2 labeling was not routinely detected on luminal spermatozoa within our tissue sections. This phenomenon is the one we have encountered on several occasions when applying a range of antibodies targeting different epididymal/sperm antigens and presumably arises due to issues associated with antigen presentation and/or masking in the paraffin-embedded tissue sections. In any case, such differences emphasize the importance of conducting parallel immunofluorescent labeling of isolated spermatozoa alongside that of the epididymal tissue itself. Differing from both DNM1 and DNM2, the DNM3 isoform was mainly detected in the apical domain of a small number of caput epithelial cells (Figure 2B, green arrows), which were shown to correspond to the clear cell sub-population by co-labeling with the recognized clear cell marker, ATP6V1B1 (Figure 2B, red arrows). In a similar manner, representative markers that have proven suitable for differentiating the different epididymal epithelial cell types are summarized in Table 229,30,31,32,33,34.
In addition to the description of the techniques for the subcellular localization of proteins residing within the epididymal epithelium, here, we also report our recently optimized protocols for the study of secretory proteins encapsulated within epididymosomes, small extracellular vesicles that represent an important component of the luminal milieu responsible for supporting sperm maturation and storage22. Combined, step 2 and Figure 3 provide a detailed step by step account of the methodology used for the isolation of highly enriched populations of epididymosomes from mouse caput epididymal tissue. Notably, however, these methods are readily applicable for the isolation of alternate populations of epididymosomes originating from more distal epididymal segments. Owing to the potential for contamination of these samples, we also described the stringent characterization protocols that we routinely employ for each epididymosome preparation. These include the assessment of the size and heterogeneity of the epididymosome populations using both high-resolution electron microscopy and dynamic light scattering techniques. In tandem, we also utilize immunoblotting strategies to assess the enrichment of recognized extracellular vesicle markers and the corresponding absence of proteins that are characteristic of potential contaminants (i.e., anti-hemoglobin (HBB) as a marker of blood contamination and anti-arachidonate 15-lipoxygenase (ALOX15) and anti-IZUMO1 antibodies as markers of cytoplasmic droplet and sperm contamination, respectively)22. Although we have found that the contaminants are rare, if they are encountered, we immediately discard the epididymosome preparation.
The non-overlapping localization of DNM isoforms in the caput epididymis prompted a further investigation of their potential roles in regulating the epididymal microenvironment. For this purpose, an immortalized mECap18 cell line was utilized as an in vitro model to study epididymal cell secretory activity. Previous characterization of this cell line has shown that it harbors a mixed cell population, which stain positive for either principal or clear cell markers. Moreover, mECap18 cells have also proven suitable for reporting physiological profiles of epididymal gene and protein expression under different in vitro treatment regimens35. Prior to use, DNM localization was assessed in cultured mECap18 cells by settling these onto poly-L-lysine treated coverslips (Figure 5A) and subjecting them to immunofluorescence detection. Consistent with the distribution patterns recorded in caput epididymal tissue sections, DNM1 was detected throughout the cytoplasm of mECap18 cells, while DNM2 was concentrated within the supranuclear domain of these cells and DNM3 was characterized by discrete foci of membrane staining within a small sub-population number (i.e., 11%) of the mECap18 cells which were ATP6V1B1 positive (Figure 5B). These data affirm the utility of the mECap18 cell line as a valuable resource for investigating the role of DNM in regulating epididymal cell secretory/absorptive activity.
Accordingly, step 4 describes the methodology for the analysis of mECap18 cell secretory activity; the techniques which are broadly amenable for assessing the impact of a range of different experimental conditions. In our study, we applied selective pharmacological interventions to suppress the activity of DNM1 and DNM2 prior to the visualization and quantification of the profile of proteins released from mECap18 cells into conditioned medium21. An important feature of this analysis, however, was to ensure that mECap18 cells were thoroughly washed and cultured in the absence of FBS supplementation. Whilst such a step was essential to preclude the contamination of conditioned medium with FBS derived proteins, it nevertheless carries the attendant risk of negatively impacting mECap18 cell growth and/or viability. In controlling for this possibility, we noted that the mECap18 cell line tolerated FBS free culture and the introduction of DNM inhibitors for the duration of our incubation window (i.e., 12 h). Indeed, over this time course, cell viability remained above 90% in all experimental replicates. This approach could therefore serve as a useful proof-of-concept strategy to identify the function of specific epididymal proteins before committing to investment into gene manipulation strategies.
| Heat induced epitope retrieval solution | 10 mmol/L sodium citrate | 50 mmol/L Tris (pH 10.5) |
| Time | 3 min | 3 min |
| 6 min | 6 min | |
| 9 min | 9 min | |
| 12 min | 12 min |
Table 1: General conditions for the optimization of heat-induced antigen retrieval for the use with paraffin-embedded epididymal sections. The fixation process can be problematic as different epitopes often require the use of different fixation techniques, thereby necessitating that the methodology is optimized for each antigen.
| Epithelial cell type | Distribution | Marker | References (PMID) |
| Principal cell | Whole epididymis | AQP9 | 11027599, 17360690 |
| Clear cell | Caput, corpus and cauda | V-ATPases, CIC-5 | 19448084, 12475763 |
| Basal cell | Whole epididymis | CLDN1 | 11159859, 21441423 |
| Narrow cell | Initial segment | V-ATPases, CIC-5 | 19448084, 12475763 |
Table 2 : Representative markers suitable for the detection of different primary epididymal epithelial cell types.
Figure 1: Spatial expression of DNM 2 within the proximal mouse epididymis. (A) Schematic model of epididymis depicting the partitioning on the mouse epididymis into 10 zones physically separated septa as reported by Turner and collegues20. In this model, zone 1 corresponds to the initial segment, zones 2-5 correspond to the caput epididymis, zones 6-7 correspond to the corpus epididymis and zones 8-10 represent the cauda epididymis. (B-C) Immunofluorescence localization of DNM2 revealed zone-specific distribution patterns (indicated by white arrowhead and arrow). The border between zone 1 and 2 is demarcated by a dotted line or denoted by yellow arrows. (D) DNM2 is also expressed in the peri-acrosomal domain of spermatozoa isolated from the caput epididymis. However, no such staining was routinely detected in luminal spermatozoa within the corresponding epididymal sections. ep, epithelial cells; l, lumen; Neg, secondary antibody only control. Experiments were replicated on material from three animals and representative immunofluorescence images are presented. Please click here to view a larger version of this figure.
Figure 2: Immunofluorescence detection of DNM 1 and DNM 3 in the mouse caput epididymis. (A) The localization of DNM136 was examined in the mouse caput epididymis. (B) Co-localization of DNM336 and the clear cell marker, ATP6V1B137 in the mouse caput epididymis. This analysis confirmed that both DNM3 (green arrows) and ATP6V1B1 (red arrows) reside in the clear cell sub-population but display minimal sub-cellular overlap. ep, epithelial cells; int, interstitium; l, lumen; sp, sperm; Neg, secondary antibody only control. Cell nuclei were counterstained with DAPI (blue). Experiments were replicated on material from three animals and representative immunofluorescence images are presented. Please click here to view a larger version of this figure.
Figure 3: Schematic of isolation protocols used for enrichment of mouse caput epididymosomes. After the dissection, caput epididymal tissue is immersed into a droplet of BWW medium and incised to release the luminal contents. The luminal fluid is then filtered through a 70 µm membrane and the resultant suspension is centrifuged at increasing velocity in order to pellet any residual cell debris. The cleared suspension is then loaded atop of a discontinuous density gradient (iodixanol solution) and subjected to overnight ultracentrifugation. Epididymosomes partition into fractions 9 - 11, which are pooled, washed via dilution into PBS and returned to the ultracentrifuge to pellet the epididymosomes. Please click here to view a larger version of this figure.
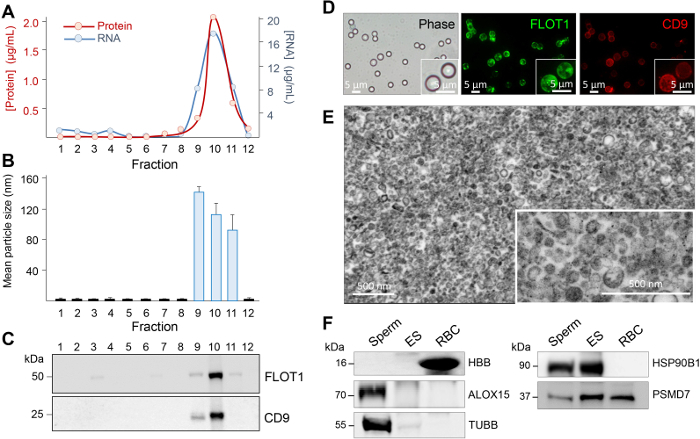
Figure 4: Assessment of epididymosome purity. Twelve equal fractions were recovered after the ultracentrifugation of the gradient and an aliquot of each prepared for (A) protein and RNA quantification, (B) size heterogeneity assessment by using dynamic light scattering, and (C) immunoblot analysis of epididymosome marker distribution. Additional characterization steps included (D) dual-labeling of epididymosomes concentrated onto aldehyde/sulphate latex beads, (E) transmission electron microscopy assessment, and (F) immunoblot assessment of spermatozoa (Sperm) and red blood cell (RBC) contamination by using either anti-arachidonate 15-lipoxygenase (ALOX15, cytoplasmic droplet/sperm contamination) or anti-hemoglobin (HBB, RBC contamination). Immunoblots were also probed with known epididymosome cargo (26S proteasome non-ATPase regulatory subunit 7, PSMD7; heat shock protein 90kDa beta member 1, HSP90B1; and beta tubulin, TUBB). These data were originally published in Scientific Reports (PMID: 27549865) and have been reproduced here with the permission of the publisher, Springer Nature. Please click here to view a larger version of this figure.
Figure 5: Immunofluorescence detection of DNM isoforms in mECap18 cells reveal distribution patterns that accord with those detected in caput epididymal tissue. (A) Schematic of coverslip preparation for sterile mECap18 cell culture. (B) Representative immunofluorescence images of DNM staining revealed cellular distribution patterns (arrows and inset (dual labeling of DNM3 and clear cell marker ATP6V1B1)) that mirrored those detected within epididymal tissue sections. Cell nuclei were counterstained with either propidium iodide (PI; red) or DAPI (blue). Experiments were replicated on material from three animals and representative immunofluorescence images are presented. Please click here to view a larger version of this figure.
Discussion
These studies incorporated the use of Bouin's fixed epididymal tissue that had been subjected to paraffin embedding and standard sectioning protocols. Bouin's fixative solution comprises a mixture of formaldehyde, picric acid and acetic acid, with each component having a specific and complementary function. Thus, formaldehyde reacts with primary amines to form protein cross-links, picric acid slowly penetrates the tissue forming salts and hence coagulation of basic proteins and conversely, acetic acid rapidly penetrates the tissue and causes the coagulation of nucleic acids. These combined properties have engendered Bouin's as a fixative of choice for the preservation of morphological detail and its use is widely reported in the epididymal literature. However, Bouin's solution is not without its limitations, which include the propensity for fixative induced fluorescence and for formaldehyde induced cross-linking which may mask target antigens.
The potential for background fluorescence necessitates the use stringent negative controls, which in our studies include the omission of the primary antibody, omission of the secondary antibody and, where the reagents are available, the use of primary antibodies preabsorbed against the immunizing peptide from which they were generated. Details of the application of such controls are exemplified in our previous study of dynamin DNM expression in the mouse epididymis21. Ideally, such results should also be validated through the use of tissue derived from knockout animals, however, this material is not always readily available. In seeking to counter the secondary problem of cross-linked or chemically modified target antigens, it is frequently necessary to perform some form of antigen retrieval in order to unmask epitopes altered by fixation and thus restore their potential for antibody binding. The methodology used for retrieval depends on many variables, including the target antigen, antibody, tissue type, and the method of fixation. However, the most widely adopted techniques feature the application of either heat-mediated or proteolytic induced antigen retrieval. The former features as our favored approach owing to a higher success rate for restoring immunoreactivity, with the details of the heat regimens and retrieval solutions we commonly utilize being documented in Table 1.We caution however, that this is by no means an exhaustive list and ultimately the optimization of antigen retrieval for each protein target/antibody combination requires preliminary studies using a matrix of time, temperature, and pH combinations. Additional considerations include the potential for heat retrieval to elicit tissue damage and/or cause artefactual labeling. Thus, in addition to the application of the negative controls documented above, we also routinely incorporate positive controls featuring antibodies, such as anti-Golgin-97, which recognize distinct cellular organelles.
In seeking to establish whether proteins such as those belonging to the DNM family fulfill redundant, as opposed to complementary, functions in epididymal tissue, we have found it particularly informative to perform dual labeling experiments such as those illustrated in Figure 2B. This strategy involves sequential labeling of tissue sections with pairs of primary antibodies (raised in different species) followed appropriate secondary antibodies conjugated to different fluorophores. However, a confounding feature that occasionally arises in seeking to perform these dual labeling studies is the incompatibility of the antigen retrieval protocols needed for optimal labeling with each primary antibody. This limitation was encountered in the case of co-labeling of DNM 2 and Golgin-97 in the mouse caput epididymis, leading us to use consecutive serial sections (as opposed to the same section)21. Nevertheless, either of these approaches are extremely useful in the context of ascribing protein expression to a particular cell type among those represented in the pseudostratified epididymal epithelium. With this goal in mind, we have included a list of representative cell type markers and their reported distribution patterns along the length of the epididymal tubule (Table 2). When one wishes to go beyond cell type and begin to explore the subcellular distribution of target proteins, the use of dual labeling with recognized organelle markers, such as Golgin-97, offers distinct advantages. Alternatively, the application of high-resolution electron microscopy in tandem with immunogold labeling remains the method of choice for detailed ultrastructural localization and validation of staining patterns achieved using immunofluorescence21.
Among the limitations posed by the study of epididymosomes are their small size and the difficulty of obtaining sufficient quantities for detailed end point analyses, particularly in commonly used laboratory species such as the mouse. However, by capitalizing on the pioneering studies of Sullivan and colleagues38,39,40, we have been able to optimize robust methodology for epididymosome isolation from the mouse model (see step 2). We do stress, however, the need to impose stringent controls to assess and physical characteristics of the enriched epididymosome populations41 due to the potential contamination from spermatozoa, cytoplasmic droplets and/or blood-borne exosomes (see Figure 4). For this purpose, we routinely use a combination of: (i) high resolution electron microscopy to visualize the size and heterogeneity of the epididymosome preparation, (ii) calculation of the mean particle size and heterogeneity (iii) concentration of the epididymosomes onto 4 µm aldehyde/sulphate latex beads and fluorescent labeling of recognized exosome surface markers, including CD9 and FLOT1, and (iv) immunoblotting of isolated epididymosomes with a suite of antibodies recommended for experimental validation of exosomes (e.g., anti-CD9, anti-FLOT1), as well as negative controls corresponding to antigens that should be restricted to spermatozoa (anti-IZUMO1), sperm cytoplasmic droplets (anti-ALOX15), or blood (anti-HBB)22. If these standards are met, then the epididymosome preparations isolated are readily amenable for use in downstream applications including co-incubation with spermatozoa and/or cargo profiling analyses22,42, both of which are powerful approaches for enhancing our understanding of the role of epididymosomes in regulating epididymal sperm maturation1.
In this study, we describe the application of an SV40-immortalized mouse caput epididymal epithelial (mECap18) cell line, which we have utilized to study the involvement of DNM in the regulation of epididymal secretory activity21 as well as the impact of environmental toxicants on epididymal physiology43. An important feature of the mECap18 cell line is that it displays phenotypic stability between passages and features a representative population of both principal and clear cells21,35,44. Compared to primary epididymal cell cultures, the mECap18 cell line also displays tolerance to culturing in fetal calf serum free medium, which extends the duration and nature of experimental interventions these cells can be exposed to, whilst also being permissive of recovering a higher abundance of secreted proteins from the conditioned medium. A limitation of the mECap18 cell line, however, is that it has been immortalized and may thus respond differently to stress and / or immune-related stimuli compared to that of primary cell cultures or those cells present in vivo. With this limitation in mind, it is recommended to compare the results obtained using mECap18 cells to in vivo responses whenever possible. In summary, the protocols we describe highlight the utility of this cell line as a tool with which to begin to study the functionality of target proteins within the caput epididymis. Indeed, in combination with the use of commercial protein inhibitors and/or genome-editing tools (such as CRISPR-Cas9), the mECap18 cell line holds considerable potential to help resolve the mechanistic basis of epididymal function.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors would like to acknowledge the National Health and Medical Research Council of Australia Project Grant APP1103176 for the support of this work.
References
- Zhou W, De Iuliis GN, Dun MD, Nixon B. Characteristics of the Epididymal Luminal Environment Responsible for Sperm Maturation and Storage. Frontiers in Endocrinology. 2018;9:59. doi: 10.3389/fendo.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux JL, Gatti JL, Dacheux F. Contribution of epididymal secretory proteins for spermatozoa maturation. Microscopy research and technique. 2003;61(1):7–17. doi: 10.1002/jemt.10312. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, et al. Proteomic changes in mammalian spermatozoa during epididymal maturation. Asian journal of andrology. 2007;9(4):554–564. doi: 10.1111/j.1745-7262.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- Hermo L, Dworkin J, Oko R. Role of epithelial clear cells of the rat epididymis in the disposal of the contents of cytoplasmic droplets detached from spermatozoa. The American journal of anatomy. 1988;183(2):107–124. doi: 10.1002/aja.1001830202. [DOI] [PubMed] [Google Scholar]
- Robaire B, Hinton B, Orgebin-Crist M. The epididymis. Vol. 3. Knobil and Neill's Physiology of Reproduction; 2006. [Google Scholar]
- Nixon B, et al. Formation and dissociation of sperm bundles in monotremes. Biology of Reproduction. 2016;95(4) doi: 10.1095/biolreprod.116.140491. [DOI] [PubMed] [Google Scholar]
- Cleland K. The structure and fuction of the Epididymis. 1. The histology of the Rat Epididymis. Australian Journal of Zoology. 1957;5(3):223–246. [Google Scholar]
- Belleannée C, et al. Identification of luminal and secreted proteins in bull epididymis. Journal of proteomics. 2011;74(1):59–78. doi: 10.1016/j.jprot.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Turner TT. De Graaf's thread: the human epididymis. Journal of andrology. 2008;29(3):237–250. doi: 10.2164/jandrol.107.004119. [DOI] [PubMed] [Google Scholar]
- Holland MK, Nixon B. The specificity of epididymal secretory proteins. Journal of reproduction and fertility. 1998;53:197–210. [PubMed] [Google Scholar]
- Cooper TG, Hing-Heiyeung C, Nashan D, Nieschlag E. Epididymal markers in human infertility. Journal of andrology. 1988;9(2):91–101. doi: 10.1002/j.1939-4640.1988.tb01016.x. [DOI] [PubMed] [Google Scholar]
- Turner TT, Johnston DS, Jelinsky SA, Tomsig JL, Finger JN. Segment boundaries of the adult rat epididymis limit interstitial signaling by potential paracrine factors and segments lose differential gene expression after efferent duct ligation. Asian journal of andrology. 2007;9(4):565–573. doi: 10.1111/j.1745-7262.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- Garrett SH, Garrett JE, Douglass J. In situ histochemical analysis of region-specific gene expression in the adult rat epididymis. Molecular reproduction and development. 1991;30(1):1–17. doi: 10.1002/mrd.1080300102. [DOI] [PubMed] [Google Scholar]
- Lareyre JJ, et al. A 5-kilobase pair promoter fragment of the murine epididymal retinoic acid-binding protein gene drives the tissue-specific, cell-specific, and androgen-regulated expression of a foreign gene in the epididymis of transgenic mice. Journal of Biological Chemistry. 1999;274(12):8282–8290. doi: 10.1074/jbc.274.12.8282. [DOI] [PubMed] [Google Scholar]
- Cornwall GA, Orgebin-Crist MC, Hann SR. The CRES gene: a unique testis-regulated gene related to the cystatin family is highly restricted in its expression to the proximal region of the mouse epididymis. Molecular Endocrinology. 1992;6(10):1653–1664. doi: 10.1210/mend.6.10.1280328. [DOI] [PubMed] [Google Scholar]
- Nixon B, Jones RC, Hansen LA, Holland MK. Rabbit epididymal secretory proteins. I. Characterization and hormonal regulation. Biology of Reproduction. 2002;67(1):133–139. doi: 10.1095/biolreprod67.1.133. [DOI] [PubMed] [Google Scholar]
- Nixon B, Jones RC, Clarke HG, Holland MK. Rabbit epididymal secretory proteins. II. Immunolocalization and sperm association of REP38. Biology of Reproduction. 2002;67(1):140–146. doi: 10.1095/biolreprod67.1.140. [DOI] [PubMed] [Google Scholar]
- Nixon B, Hardy CM, Jones RC, Andrews JB, Holland MK. Rabbit epididymal secretory proteins. III. Molecular cloning and characterization of the complementary DNA for REP38. Biology of Reproduction. 2002;67(1):147–153. doi: 10.1095/biolreprod67.1.147. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, et al. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biology of Reproduction. 2007;76(4):561–570. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- Johnston DS, et al. The Mouse Epididymal Transcriptome: Transcriptional Profiling of Segmental Gene Expression in the Epididymis 1. Biology of Reproduction. 2005;73(3):404–413. doi: 10.1095/biolreprod.105.039719. [DOI] [PubMed] [Google Scholar]
- Zhou W, et al. Developmental expression of the dynamin family of mechanoenzymes in the mouse epididymis. Biology of Reproduction. 2017;96(1):159–173. doi: 10.1095/biolreprod.116.145433. [DOI] [PubMed] [Google Scholar]
- Reilly JN, et al. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Scientific reports. 2016;6 doi: 10.1038/srep31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R. Epididymosomes: a heterogeneous population of microvesicles with multiple functions in sperm maturation and storage. Asian journal of andrology. 2015;17(5):726–729. doi: 10.4103/1008-682X.155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AT, et al. Glycogen synthase kinase 3 regulates acrosomal exocytosis in mouse spermatozoa via dynamin phosphorylation. The FASEB Journal. 2015;29(7):2872–2882. doi: 10.1096/fj.14-265553. [DOI] [PubMed] [Google Scholar]
- Danesh A, et al. Exosomes from red blood cell units bind to monocytes and induce proinflammatory cytokines, boosting T-cell responses in vitro. Blood. 2014;123(5):687–696. doi: 10.1182/blood-2013-10-530469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AL, et al. Assessment of microRNA expression in mouse epididymal epithelial cells and spermatozoa by next generation sequencing. Genomics data. 2015;6:208–211. doi: 10.1016/j.gdata.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggers J, Whitten W, Whittingham D. The culture of mouse embryos in vitro. In: Daniels J, editor. Methods in mammalian embryology. San Francisco: Freeman; 1971. pp. 86–116. [Google Scholar]
- Strober W. Trypan blue exclusion test of cell viability. Current protocols in immunology. 2001. A3. B. 1-A3. B. 3. [DOI] [PubMed]
- Elkjær M-L, et al. Immunolocalization of AQP9 in liver, epididymis, testis, spleen, and brain. Biochemical and biophysical research communications. 2000;276(3):1118–1128. doi: 10.1006/bbrc.2000.3505. [DOI] [PubMed] [Google Scholar]
- Gregory M, Dufresne J, Hermo L, Cyr DG. Claudin-1 is not restricted to tight junctions in the rat epididymis. Endocrinology. 2001;142(2):854–863. doi: 10.1210/endo.142.2.7975. [DOI] [PubMed] [Google Scholar]
- Isnard-Bagnis C, et al. Detection of ClC-3 and ClC-5 in epididymal epithelium: immunofluorescence and RT-PCR after LCM. American Journal of Physiology-Cell Physiology. 2003;284(1):C220–C232. doi: 10.1152/ajpcell.00374.2001. [DOI] [PubMed] [Google Scholar]
- Rojek AM, et al. Defective glycerol metabolism in aquaporin 9 (AQP9) knockout mice. Proceedings of the National Academy of Sciences. 2007;104(9):3609–3614. doi: 10.1073/pnas.0610894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum WW, Da Silva N, Brown D, Breton S. Regulation of luminal acidification in the male reproductive tract via cell-cell crosstalk. Journal of Experimental Biology. 2009;212(11):1753–1761. doi: 10.1242/jeb.027284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum WW, Ruan YC, Silva N, Breton S. Establishment of cell-cell cross talk in the epididymis: Control of luminal acidification. Journal of andrology. 2011;32(6):576–586. doi: 10.2164/jandrol.111.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipilä P, Shariatmadari R, Huhtaniemi IT, Poutanen M. Immortalization of epididymal epithelium in transgenic mice expressing simian virus 40 T antigen: characterization of cell lines and regulation of the polyoma enhancer activator 3. Endocrinology. 2004;145(1):437–446. doi: 10.1210/en.2003-0831. [DOI] [PubMed] [Google Scholar]
- Feugang JM, et al. Profiling of relaxin and its receptor proteins in boar reproductive tissues and spermatozoa. Reproductive Biology and Endocrinology. 2015;13(1):46. doi: 10.1186/s12958-015-0043-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullberg M, et al. Cytokine detection by antibody-based proximity ligation. Proceedings of the National Academy of Sciences. 2004;101(22):8420–8424. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette G, Girouard J, Sullivan R. Comparison between epididymosomes collected in the intraluminal compartment of the bovine caput and cauda epididymidis. Biology of Reproduction. 2006;75(6):885–890. doi: 10.1095/biolreprod.106.054692. [DOI] [PubMed] [Google Scholar]
- Fornes M, Barbieri A, Sosa M, Bertini F. First observations on enzymatic activity and protein content of vesicles separated from rat epididymal fluid. Andrologia. 1991;23(5):347–351. doi: 10.1111/j.1439-0272.1991.tb02578.x. [DOI] [PubMed] [Google Scholar]
- Eickhoff R, et al. Influence of macrophage migration inhibitory factor (MIF) on the zinc content and redox state of protein-bound sulphydryl groups in rat sperm: indications for a new role of MIF in sperm maturation. Molecular human reproduction. 2004;10(8):605–611. doi: 10.1093/molehr/gah075. [DOI] [PubMed] [Google Scholar]
- Lötvall J, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheon K, et al. Analysis of the small non-protein-coding RNA profile of mouse spermatozoa reveals specific enrichment of piRNAs within mature spermatozoa. RNA biology. 2017;14(12):1776–1790. doi: 10.1080/15476286.2017.1356569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. Genetic basis of the spread of antibiotic resistance genes. Annali dell'Istituto superiore di sanita. 1987;23(4) [PubMed] [Google Scholar]
- Nixon B, et al. Next generation sequencing analysis reveals segmental patterns of microRNA expression in mouse epididymal epithelial cells. PloS one. 2015;10(8):e0135605. doi: 10.1371/journal.pone.0135605. [DOI] [PMC free article] [PubMed] [Google Scholar]


