Abstract
Microbial biofilms are a leading cause of chronic infections in humans and persistent biofouling in industries due to extremely high-level tolerance of biofilm cells to antimicrobial agents. Eradicating mature biofilms is especially challenging because of the protection of the extracellular matrix and slow growth of biofilm cells. Recently, we reported that established biofilms can be effectively removed (e.g. 99.9% dispersion of 48 h Pseudomonas aeruginosa PAO1 biofilms) by shape memory polymer-based dynamic changes in surface topography. Here, we demonstrate that such biofilm dispersion also sensitizes biofilm cells to conventional antibiotics. For example, shape recovery in the presence of 50 µg/mL tobramycin reduced biofilm cell counts by more than 3 logs (2,479-fold) compared to the static flat control. The observed effects were attributed to the disruption of biofilm structure and increase in cellular activities as evidenced by an 11.8-fold increase in intracellular level of adenosine triphosphate (ATP), and 4.1-fold increase in expression of the rrnB gene in detached cells. These results can help guide the design of new control methods to better combat biofilm associated antibiotic-resistant infections.
Keywords: Biofilm, removal, antifouling, shape memory polymer, antibiotic susceptibility, ATP
Graphical Abstract
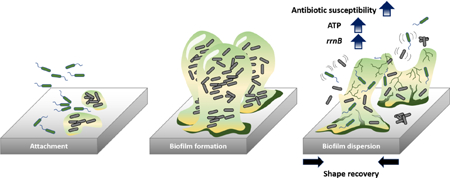
Introduction
Bacteria can survive in challenging environments by attaching to a surface and developing a biofilm that consists of sessile bacterial cells and an extracellular matrix [1]. Cells in mature biofilms are also associated with slow growth, which renders most antibiotics ineffective [2,3]. Consequently, biofilms are up to 1,000 times more tolerant to antibiotics compared to planktonic cells; and biofilms are involved in more than 65% of nosocomial infections according to the Centers for Disease Control and Prevention (CDC) [4–6].
The economical and clinical significance of biofilm-related problems has stimulated intensive research to design more effective anti-fouling strategies [7–10]. To prevent bacteria from colonizing a surface, different approaches have been explored to alter the properties of the substrate materials such as surface chemistry [11–24], topography [8,25–35], and stiffness [36– 39]. Strategies for modifying surface chemistry include coating with antibacterial agents [11– 14,17,20,21,24] or other compounds that can change the charge [15] or hydrophobicity [19,22,23]. Surface hydrophobicity can also be changed by altering surface topography [27]. Inspired by natural anti-fouling surfaces such as shark skin [30], lotus leaves [19], taro leaves [23], and cicada wings [28], static micron- and nano-scale patterns and roughness have been created and demonstrated to prevent biofilm formation without using antimicrobial agents that can potentially promote resistance [8,25,26,28–35]. These chemical and physical approaches have been demonstrated to inhibit bacterial adhesion for up to 14 days; however, challenges such as the sustaining efficacy of agents, adverse effects of environmental and host factors (e.g., covering by body fluid or metabolic products during bacterial growth), and the remarkable capabilities of bacteria to adapt to challenging environments can allow bacteria to overcome unfavorable surface properties and eventually form biofilms over time [7]. Thus, it is important to develop new technologies that can effectively remove mature biofilms.
Previous studies showed that, by altering the surface features using pneumatic actuation [40], electrical voltage [41], and air-pressure or water inflation generated strain [42,43], up to 90% of mature biofilm could be removed. Recently, we demonstrated strong activities of biofilm removal by dynamic changes in surface topography using shape memory polymer (SMP). Using tert-butyl acrylate (tBA) based SMP, on-demand shape recovery of the substrate material (both flat SMP and that with microns-scale topographic patterns) can be triggered with gentle heating (10 min at 40°C), which led to effective removal of 48 h Pseudomonas aeruginosa biofilms by 99.9% [44]. The observed biofilm removal was attributed to physical disruption of biofilm structure and cell-surface interactions. Because biofilm and planktonic cells have major differences in physiology and antibiotic susceptibility [45], we hypothesize that shape recovery triggered biofilm dispersion can also alter the antibiotic susceptibility of biofilm cells. To test this hypothesis, we followed the antibiotic susceptibility of biofilm cells before and after shape recovery and compared it with the control surfaces that were not programmed to have shape change (henceforth “static flat control”). We also tracked the changes in intracellular ATP level and gene expression profiles to understand the mechanism of observed results. The findings of this study may help design the next generation smart anti-fouling materials by combining dynamic surface topography with antimicrobials.
2. Materials and methods
2.1. Bacterial strains and medium
P. aeruginosa PAO1 [46] was grown in Lysogeny Broth (henceforth LB medium) [47] consisting of 10 g/L tryptone, 10 g/L NaCl, and 5 g/L yeast extract (Thermo Fisher Scientific, Waltham, MA, USA). The reporter strain PAO1::rrnBP1-gfp(AGA) was constructed by integrating rrnBP1-gfp(AGA) into the genome of P. aeruginosa PAO1 using miniTn5 system to monitor the expression of rrnB gene with the signal from unstable GFP(AGA).
2.2. SMP substrate fabrication
The shape memory polymer was synthesized by following the protocols reported previously [44,48]. Briefly, the shape memory polymer (SMP) was synthesized using t-butyl acrylate (tBA), poly (ethylene glycol)n dimethacrylate (PEGDMA) with Mn=750 molecular weight, and photo initiator 2,2-dimethoxy-2-phenylacetophenone (DMPA) (Sigma Aldrich, St. Louis, MO, USA). The weight ratio between tBA and PEGDMA was set as 9:1; and a photo initiator, DMPA, was added as 0.4wt% to synthesize the tBA-co-PEGDMA polymer networks with a transition temperature slightly above the body temperature (37°C). In our previous study [44], this tBA based SMP exhibited a recovery ratio of 98.9% with glass transition temperature of 44.3°C. To make flat SMP, a sandwich structure was assembled with two glass slides as frames and a 1 mm thick PDMS spacer in-between. To minimize the adhesion of SMP to the glass slides, the surfaces of both glass slides were modified with RainX. A mixture of tBA, PEGDMA, and DMPA was injected between two glass slides. The mixture spread uniformly into the gap between two glass slides (created by the PDMS spacer) due to capillary effect. To cure the mixture for pre-polymerization, 365 nm UV radiation was applied for 10 min. Post-curing was conducted at 90°C for 1 h to finish the synthesis of SMP networks. To ensure complete crosslinking, we compared the swelling ratios after 1, 3, 5, and 10 min of UV exposure and different amounts of post-curing time. As shown in Supplementary Figs. S1a&b, extending UV exposure time beyond 3 min did not further change the swelling ratio, indicating that 3 min is sufficient. The results also show that increasing post-curing time beyond 1 h did not change the swelling ratio. Thus, we chose 10 min UV exposure with 1 h post-curing under 90°C to ensure complete crosslinking, and keep consistency with the protocol that we followed [48] and our previous study [44]. If further developed for real applications, it will be important to test other sterilization methods that are easier to scale up, e.g. gamma radiation. This is beyond the scope of this study. However, because we have achieved complete crosslinking, we do not expect significant changes in biofilm control activities if gamma were used for sterilization.
2.3. Programmable SMP substrate preparation
To obtain the stretched temporary shape, flat SMPs were cut into a dog bone shape using a manual stretcher. The manual stretcher with the dog bone shape SMP was incubated at 50°C for 8 min and stretched gently by 50% elongation. After the deformation, SMP was cooled to room temperature for 5 min. To recover the SMP with temporary shape, it was incubated in 0.85wt.% NaCl solution at 40°C for 10 min. In our previous study [44], we have tested the recovery ratio of this SMP and found it is 98.9%.
2.4. Biofilm formation
To grow biofilms, SMPs were cut into 0.5 cm by 1.5 cm coupons and then sterilized by exposing to UV for 1 h for each side. Overnight cultures of P. aeruginosa PAO1 grown in LB medium were used to inoculate the biofilm cultures in petri dishes containing SMPs to an optical density at 600 nm (OD600) of 0.05. Each petri dish held three biological replicates of SMP coupons. Biofilms were cultured for 48 h at room temperature.
2.5. Antibiotic susceptibility test
After 48 h incubation, SMPs with attached biofilms were washed with 0.85wt.% NaCl solution three times to remove non-specifically attached planktonic cells. After washing, each SMP was transferred to a pre-warmed test tube containing 2 mL of 0.85wt.% NaCl solution and incubated for 10 min at 40°C to trigger shape change. During this process, the programmed SMP recovered to its permanent shape, while the static flat control maintained its own shape. After the 10 min incubation, shape recovery dispersed biofilm cells were harvested for analysis. For the static flat control samples (biofilms on surfaces without stretching), biofilm cells were harvested by 25 Hz bead beating for 30 s using 0.1 g of 0.1 mm zirconia/silica bead (BioSpec Products, Inc., Bartlesville, OK, USA). This approach was found effective to detach PAO1 biofilm cells without affecting PAO1 cell viability (Supplementary Figs. S2a&b). To avoid any possible confounding effect of bead beating, cells detached by shape recovery were also processed with bead beating for 30 s before further analysis. The harvested biofilm cells were transferred to a 96-well plate and tested for susceptibility to six antibiotics including tobramycin (Tokyo Chemical Industry Co., Tokyo, Japan), ofloxacin (Sigma Aldrich, St. Louis, MO, USA), tetracycline (Sigma Aldrich), minocycline (Sigma Aldrich), ciprofloxacin hydrochloride (Sigma Aldrich), and chloramphenicol (Sigma Aldrich) added at different concentrations. After 1 h incubation at 37°C, samples were washed three times with 0.85wt.% NaCl solution before plating on LB agar plates to count colony forming units (CFU) by following a published protocol [49] after a series dilution.
2.6. Biomass quantification and cell viability test
The 48 h P. aeruginosa PAO1 biofilm cells on SMP were stained with SYTO®9 and propidium iodine from the Live/Dead® Backlight™ bacterial viability kit (Life technologies, Carlsbad, CA, USA) after three times of washing with 0.85wt% NaCl solution. Imaging analysis was conducted using an upright fluorescence microscope (Axio Imager M1, Carl Zeiss Inc., Berlin, Germany). To quantify the biomass, z stack images with 3D information were obtained followed by quantification analysis using software COMSTAT [50]. Three biological replicates were tested for each condition and five images were randomly obtained for each surface.
2.7. Scanning electron microscope (SEM) analysis
The biofilm cells on SMP substrates with different conditions were analyzed including 48 h biofilms without treatment, biofilm cells detached by bead beating/shape recovery, and SMP substrate surfaces after bead beating/shape recovery. The samples were immersed in a fixing agent containing 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) for 1 h after three times of washing with 0.85wt% NaCl solution. Then, the substrates were transferred into 1% Osmium tetroxide (OsO4, Sigma Aldrich) solution for post-fixation for 1 h followed by further washing steps with 15, 30, 50, 70, 95, and 100% ethanol for 15 min each. The 100% ethanol washing step was conducted three times. The samples were coated using a platinum sputter (Edwards S150A, Edwards, Burgess Hill, England) under 30 mV with 75 sec deposition time. SEM images were obtained using JEOL JSM-IT100LA (JEOL Ltd., Tokyo, Japan). Three biological replicates were imaged with five positions randomly selected from each sample.
2.8. Intracellular level of adenosine triphosphate (ATP)
The ENLITEN ATP Assay system (Promega, Madison, WI, USA) was used for ATP test by following the manufacture’s protocol. Briefly, the biofilm cells of both stretched and static flat control samples were obtained as described above. The luminescence of each sample was measured using a microplate reader (BioTek Synergy 2, BioTek, Winooski, VT, USA). We first established a standard curve using samples with known concentrations of ATP. The amount of ATP in actual samples was determined by fitting the ATP standard curve and normalized by the number of cells in each sample. Three replicates were tested for each condition.
2.9. Expression level of rrnB
To monitor the growth activity of biofilm cells released by shape recovery and those of the static flat control, an engineered reporter strain, PAO1::rrnBP1-gfp, was used to determine the rrnB expression level as indicated by the GFP signal intensity. Biofilm cells were harvested as described above in antibiotic susceptibility test. The intensity of GFP signal was measured using a BioTek Synergy2 microplate reader and normalized by cell number. Each condition was tested with three replicates.
2.10. RNA extraction and cDNA synthesis
Total RNA of detached biofilm cells was extracted using RNeasy mini kit (Qiagen, Hilden, Germany). Biofilm cells were cultured in the same way as described above except that more and bigger SMP coupons were used to obtain 9 times more cells per sample to ensure the abundance of RNA needed for RNA-seq and quantitative PCR (qPCR) analyses. The cells were collected by centrifugation for 3 min at 8,000 rpm at 4°C. RNA was isolated by following the protocol of RNeasy mini kit. The purity of RNA samples was evaluated using a Nanodrop tool of microplate reader EPOCH 2 (BioTek, Winooski, VT, USA). The quality of extracted RNA samples was quantified using an Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and the RNA samples with an RNA integrity number (RIN) > 9 were chosen for rRNA depletion using Ribo-zero rRNA removal kit (Illumina, San Diego, CA, USA) prior to RNA-seq analysis. For qPCR analysis, the extracted RNA samples were used to synthesize cDNA using iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). The quality of the cDNA samples was checked using the microplate reader as mentioned above.
2.11. RNA-seq library construction
RNA-seq libraries were constructed using the NEBNext Ultra RNA Library Prep kit (New England Biolabs, Ipswich, MA, USA). Each library was quantified with Qubit 2.0 (dsDNA HS kit; Thermo Fisher Scientific, Waltham, MA, USA) and the size distribution was determined using a Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA, USA) prior to pooling. Libraries were sequenced on a NextSeq 500 instrument (Illumina, San Diego, CA, USA) at the RNA Sequencing Core (RSC) Facility at Cornell University. At least 20 M single-end 75 bps reads were generated per library.
2.12. Validation of RNA-seq results using quantitative PCR (qPCR) analysis
qPCR analysis was conducted to validate the RNA-seq results. The synthesized cDNA template, DNA primer templates of interest (Table 1), and SYBR Green PCR master mix (Thermo Fisher Scientific, Waltham, MA, USA) were well mixed. The qPCR reactions were conducted using an Eppendorf Mastercycler Realplex Thermal Cycler (Eppendorf, Hamburg, Germany) with the following condition: initial denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 1 min. The melting curve was conducted at 95°C for 20 min. The fluorescent signals were measured at the end of each cycle. The expression ratios of the genes of interest were analyzed by the LinReg PCR program (Heart Failure Research Center, Amsterdam, Netherlands). Five representative genes were tested including proC, cynT, hirQ, hdhA, phnW, oprB, rrnB, and kdpB (Table 1). proC was chosen as a housekeeping gene as used in previous studies both by us and other groups [51,52].
Table 1.
Primers used in this study
| Selected genes | Forward primer sequence 5′ → 3′ | Reverse primer sequence 5′ → 3′ |
|---|---|---|
| proC (housekeeping gene) | ACCCCGCATAGCGTTCATC | GGAGACGATCAGTTGCTCCG |
| cynT | GCTCGCAACTGTTCAAGTCC | GCCGCTTTCGATGTCGTAGA |
| kdpB | ATGCTGGTGGTCGAACTGAC | CAGGAAGATCAGGGTCAGGC |
| nirQ | GCGGTATCTGCTACCTGGAC | GGGTTGTAGGACACCACCAG |
| hdhA | TACTTCACCAACACCTCGCC | AAGCCCTGGACGACATTGAG |
| phnW | TGGGACAGCGATTTCAACGA | TCATGGCATCGACGATCAGG |
| rrnB | TGCCTGGTAGTGGGGGATAA | GGACCGTGTCTCAGTTCCAG |
2.13. Analysis of RNA-seq results
RNA-seq reads were processed with Cutadapt (version 1.8) to trim low quality and adaptor sequences [53]. The mapping process to align the paired end reads against P. aeruginosa PAO1 reference genome was performed using Tophat (version 2.1). Cufflinks (version 2.2) was used to generate fragments per kilobase of transcript per million (FPKM) values and statistical analysis of differential gene expression [54,55]. RNA-seq analysis was conducted with two biological replicates. The results with absolute value of fold change > 2, p < 0.05, and q < 0.05 were considered significant using Cufflinks (version 2.2) as mentioned above.
2.14. Statistics
SAS 9.1.3, Windows version (SAS, Cary, NC, USA), was used for all statistical analyses. Results with p < 0.05 were considered as statistically significant.
3. Results
3.1. Shape recovery sensitized biofilm cells to bactericidal antibiotics.
To understand if better biofilm control can be obtained by concurrent treatment of biofilms with antibiotic during shape recovery, we first tested shape recovery with 48 h P. aeruginosa PAO1 biofilms in the presence of selected conventional antibiotics (including both bactericidal and bacteriostatic agents). The unstretched samples were used as static flat control. As shown in Fig. 1, after such concurrent treatment with 50 µg/mL tobramycin, 5 µg/mL ofloxacin, 500 µg/mL tetracycline, or 200 µg/mL minocycline, the number of viable cells attached on the surface was reduced by 4.4 ± 0.3 logs, 2.9 ± 0.06 logs, 2.1 ± 0.1 logs, and 3.1 ± 0.05 logs of the original biofilm cell numbers, respectively. These correspond to 2,480, 710, 116, and 962 folds of reduction by tobramycin, ofloxacin, tetracycline, and minocycline, respectively (p values < 0.001, one-way ANOVA adjusted by Tukey test) compared to the static flat control biofilm cells, which went through the same treatment except that the cells were not detached (the SMP was not stretched and thus no shape change) during incubation with antibiotic.
Fig. 1.
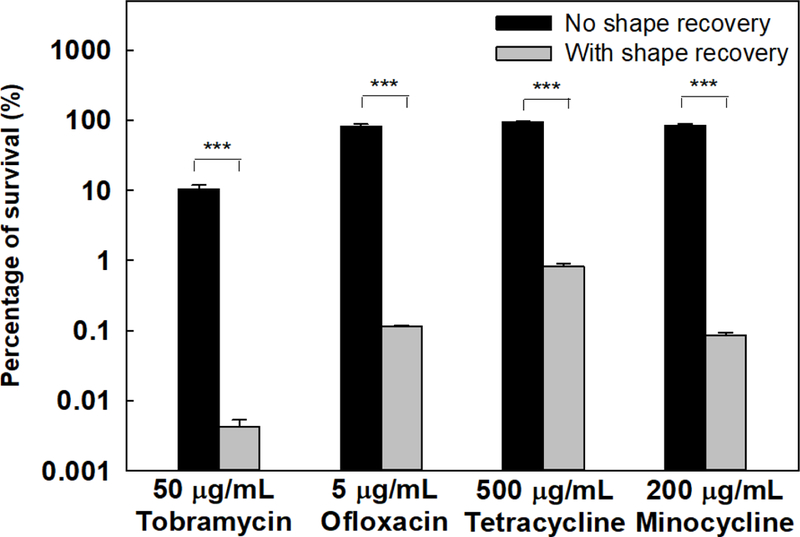
Concurrent treatment of P. aeruginosa PAO1 biofilm cells. Shape recovery (10 min at 40°C) was triggered in the presence of an antibiotic. Four antibiotics were tested including tobramycin, ofloxacin, tetracycline, and minocycline. *** p<0.001.
The above results demonstrate potent activities in biofilm control. However, the data do not reveal if the effects were due to dispersion, killing by antibiotics, or both. To more specifically evaluate the antibiotic susceptibility of detached cells, we also conducted a sequential treatment with shape recovery followed by antibiotic treatment. After growing P. aeruginosa PAO1 for biofilm formation on stretched SMP and static flat controls for 48 h, two types of biofilm cells were harvested including (1) cells dispersed by shape recovery during 10 min incubation of stretched SMPs at 40°C and (2) biofilms cells on static flat controls that went through the same 10 min incubation and detached by bead beating (no effects on cell viability, Supplementary Fig. S2) prior to antibiotic treatment. To specifically study the effects of shape recovery on bacterial antibiotic susceptibility, biofilm cells detached by shape recovery were also treated with the same bead beating step as the control samples (the method to harvest biofilm cells of the control samples) before antibiotic treatment. The bead beating process was verified effective for biofilm removal. As shown in Fig. 2a, compared to the 9.1 ± 0.8 µm3/µm2 biomass of 48 h P. aeruginosa PAO1 biofilms, it was dramatically reduced to 0.04 ± 0.004 µm3/µm2 and 0.04 ± 0.03 µm3/µm2 after bead beating or shape recovery, respectively (p = 0.001 for both; one-way ANOVA adjusted by Turkey test). These results were corroborated by the SEM images shown in Fig. 2. To verify that the bead beating condition is safe to cells, we further examined the cells using Live/Dead staining and SEM analysis. No cell death was noted based on Live/Dead staining (Supplementary Fig. S3a) and the cell integrity was verified by SEM results (Supplementary Fig. S3b).
Fig. 2.
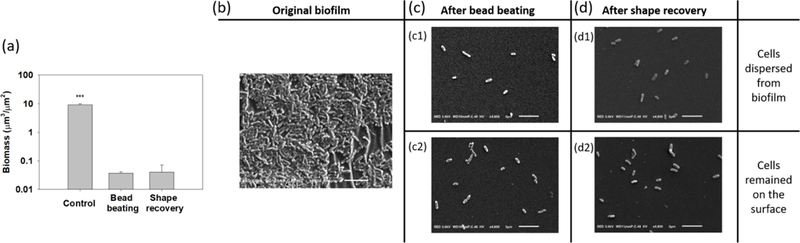
SEM analyysis of biofilm removal by shape recovery and bead beating. (a) Biomass of 48 h P. aeruginosa PAO1 biofilm cells before and after shape recovery or bead beating. (b) Image of 48 h biofilm cells prior to treatment. (c) Biofilm cells after bead beating (c1: detached biofilm cells. c2: Biofilm cells remained on surface). (d) Biofilm cells after shape recovery (d1: detached biofilm cells. c2: Biofilm cells remained on surface). *** p<0.001. Bar = 5 µm.
After harvesting the biofilm cells, tobramycin was added to treat both the static flat control and shape recovery-dispersed biofilm cells for 1 h. As shown in Fig. 3a, the log reduction after treatment with 2, 10, and 50 μg/ml tobramycin was 0.7 ± 0.1, 1.2 ± 0.1, and 1.7 ± 0.1, respectively, for static flat control biofilm cells. In comparison, 1.6 ± 0.2, 2.1 ± 0.1, and 2.4 ± 0.1 logs of shape recovery-dispersed biofilm cells were killed, indicating a 0.9 ± 0.2, 0.9 ± 0.01, and 0.7 ± 0.02 log increase in antibiotic susceptibility compared to static flat control (p = 0.01, 0.01, and 0.002, one-way ANOVA adjusted by Turkey test). This suggests that shape recovery triggered dispersion did not simply detach biofilm cells via physical forces but affected the physiological stage of biofilm cells.
Fig. 3.
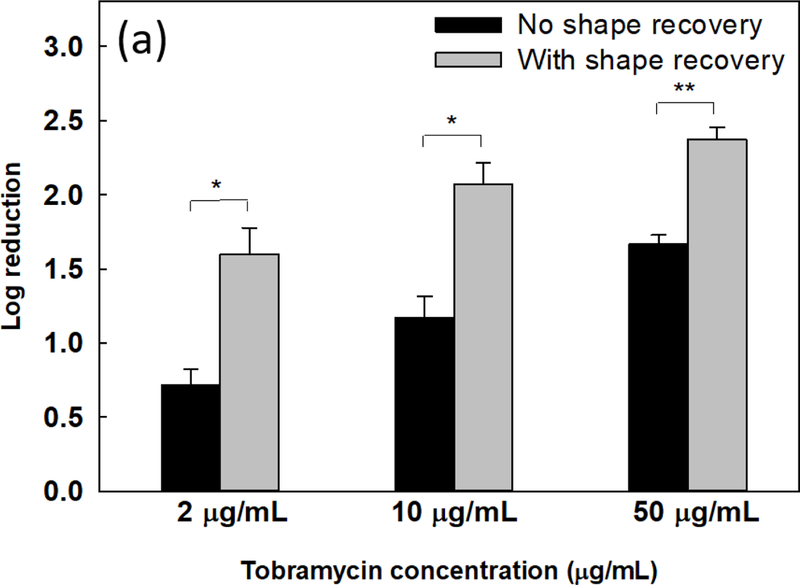
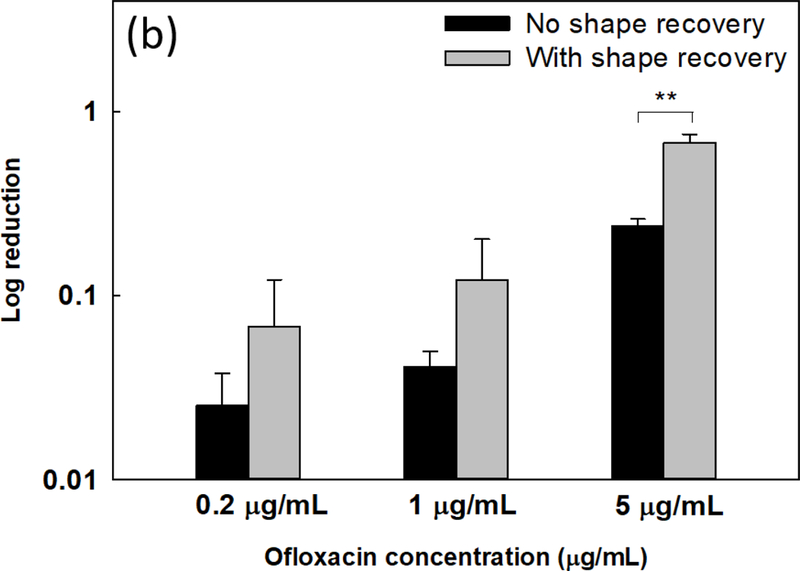
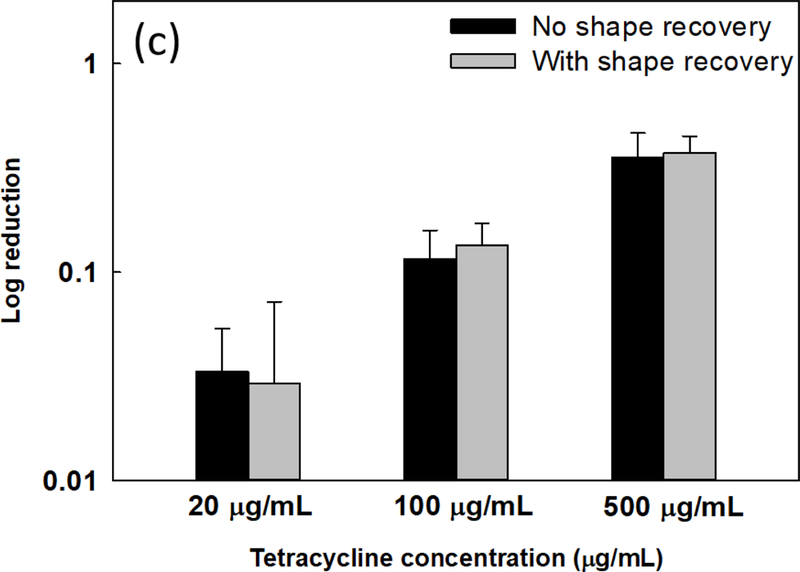
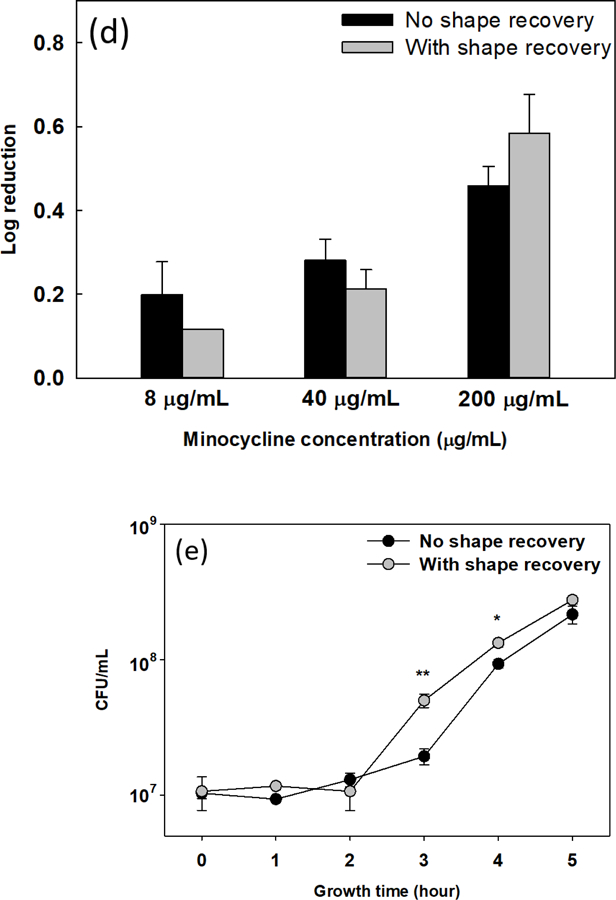
Sequential antibiotic susceptibility test on P. aeruginosa PAO1 biofilm cells. Four antibiotics were tested by adding to the biofilm cells dispersed by shape receovery including tobramycin (a), ofloxacin (b), tetracycline (c), and minocycline (d). (e) Growth curves of collected biofilm cells. The biofilm cells of static flat control were detached by bead beating. The biofilm cells released by shape recovery were also processed with bead beating to avoid any confounding effects. * p<0.05** p<0.01.
Consistent with the result of tobramycin, shape recovery triggered dispersion also sensitized the biofilm cells to ofloxacin. As shown in Fig. 3b, shape recovery released biofilm cells were 0.4 ± 0.1 log (p = 0.001, one-way ANOVA adjusted by Turkey test) more sensitive to 5 μg/mL ofloxacin than the static flat control biofilm cells. Similar results were also obtained for ciprofloxacin (Supplementary Fig. S4a). Compared to these three bactericidal antibiotics, biofilms were not sensitized to bacteriostatic antibiotics tested including tetracycline, minocycline, and chloramphenicol (Figs. 3c, d, and Supplementary Fig. S4b). This is likely due to the static nature of these agents and indicates that the detached cells were not actively growing.
3.2. Effects of shape recovery triggered biofilm dispersion on physiology of P. aeruginosa cells.
Increase in antibiotic susceptibility of dispersed cells led to our speculation that shape recovery may change the physiological stage of biofilm cells. To answer this question, we first tested if dispersion affected the growth of these cells by incubating detached cells in LB medium. After 2 h of inoculation, there was no difference in cell number between shape recovery released cells and the static flat control sample released by bead beating (both were in lag phase; Fig. 3e). The cells released by shape recovery were also processed by bead beating to avoid any confounding effects. After the lag phase, cells in both samples started growing but at different growth rates. The CFU number of shape recovery released biofilm cells after 3 h and 4 h of incubation was 2.7 ± 0.6 and 1.5 ± 0.2 times higher than the static flat control biofilm cells, respectively (p = 0.008 and 0.02, respectively, one-way ANOVA adjusted by Turkey test). This result indicates that the shape recovery released biofilm cells were at a relatively more active stage, which is consistent with their enhanced antibiotic susceptibility.
To understand if shape recovery released cells were more active metabolically, we compared the intracellular level of ATP in P. aeruginosa PAO1 biofilm cells between shape recovery samples and static flat controls. ATP level is an indicator of cellular activities and known to be associated with bacterial antibiotic susceptibility [56]. As shown in Fig. 4a, the ATP level in biofilm cells dispersed by shape recovery was 11.8 ± 2.7 times of the static flat control cells (p = 0.003, one-way ANOVA adjusted by Tukey test). This result indicates higher metabolic activities in shape recovery-dispersed cells and corroborates the increase in antibiotic susceptibility of these cells.
Fig. 4.
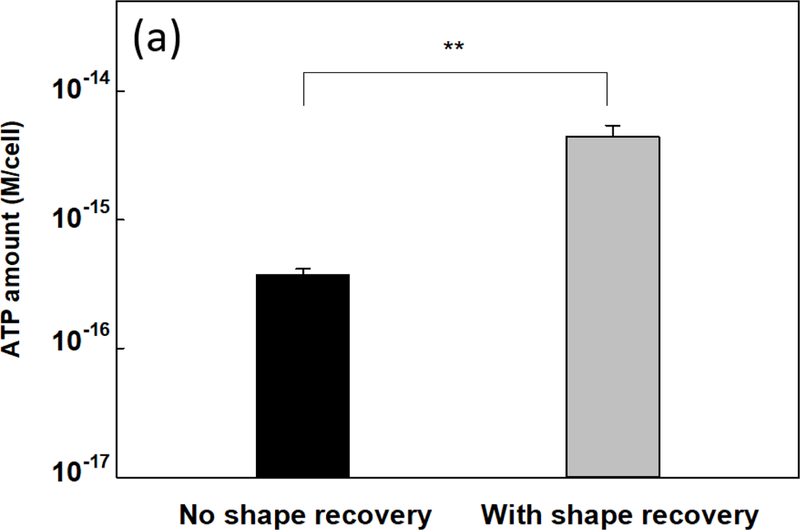
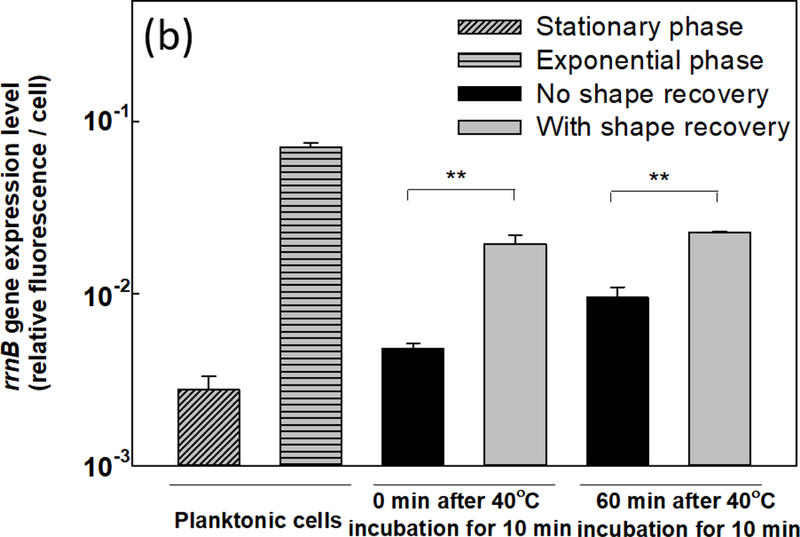
(a) Intracellular ATP level in shape recovery released P. aeruginosa PAO1 biofilm cells and static flat control. (b) Expression level of rrnB gene in P. aeruginosa PAO1::rrnBP1gfp(AGA) including planktonic cells, shape recovery released cells, and static flat control. ** p<0.01.
Intracellular ATP level is also known to affect the expression of rrnB gene, which encodes 16s rRNA for cell growth [57,58]. Thus, we measured the expression level of rrnB gene using a reporter strain PAO1::rrnBP1-gfp(AGA). Consistent with the increase in ATP level, shape recovery triggered dispersion led to a 4.1 ± 0.4-fold increase in rrnB expression compared to the static flat control (Fig. 4b) (p = 0.007, one-way ANOVA adjusted by Tukey test). The higher expression level of rrnB gene in dispersed cells was also verified using qPCR (2.0 ± 0.2-fold increase compared to static flat control; p = 0.002, one-way ANOVA adjusted by Tukey test). The rrnB expression results are consistent with the increase in ATP level and higher antibiotic susceptibility in dispersed biofilm cells.
3.3. Effects of shape recovery on P. aeruginosa gene expression
To further understand the effects of shape recovery triggered biofilm dispersion at the genome-wide scale, RNA-seq analysis was used to compare the gene expression profiles between biofilm cells dispersed by shape recovery and the static flat control. The RNA-seq results indicate that 70 genes were differentially expressed between dispersed cells and the control, including 47 up-regulated genes and 23 down-regulated genes (Supplementary Tables S1&S2). Eight up-regulated genes and 6 down-regulated genes are related to ATP or metabolic activities (Fig. 5a). Among these genes, cynT, PA2843, mdlC, katB, phnW, hisD, and PA5312 were up-regulated and PA2550, acsA, hdhA, and glpK were down-regulated. For ATP-related genes, nirQ was up-regulated by 3.1-fold, while kdpB was down-regulated by 3.6-fold. nirQ encodes denitrification regulatory protein (nitric oxide reductase), also known as ATP-related protein NirQ, which reduces nitric oxide (NO) to nitrous oxide (N2O) to avoid the accumulation of toxic NO in the cell [59]. During the denitrification process, NirQ induces a concentration gradient of hydrogen ion through cell membrane which leads the synthesis of ATP [60]. kdpB is associated with potassium ion (K+) transport, which requires ATP as energy source [61]. Thus, the induction of nirQ and repression of kapB are consistent with the increase in ATP level in dispersed cells. To validate the RNA-seq data especially the genes related to metabolic activities, qPCR was conducted for 5 representative genes, including cynT, nirQ, phnW, hdhA, and kdpB, plus rrnB discussed above. The rrnB gene was not shown in the RNA-seq results because rRNA was depleted during the pretreatment step before sequencing. All 5 representative genes showed consistent results between RNA-seq and qPCR (Fig. 5b). Thus, the qPCR data validated the RNA-seq results and provided additional evidence that the shape recovery triggered dispersion rendered P. aeruginosa biofilm cells to leave the physiological stage of biofilm growth, becoming more active metabolically and consequently more sensitive to antibiotics.
Fig. 5.
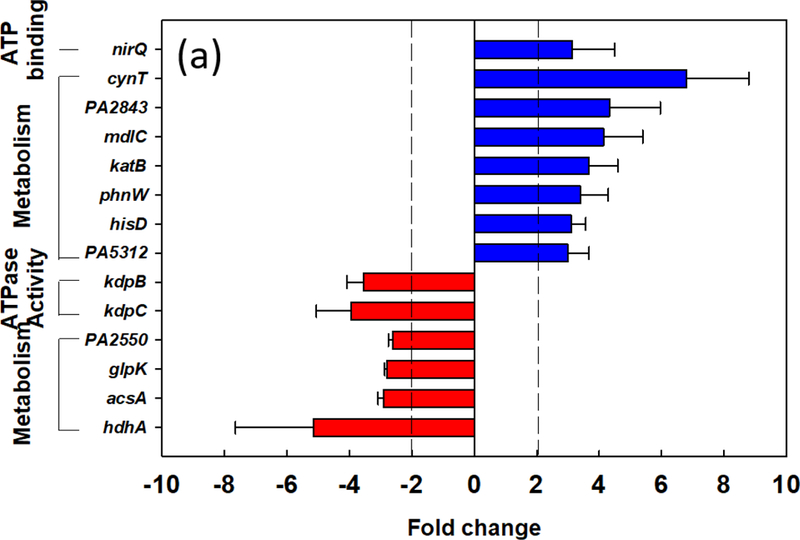
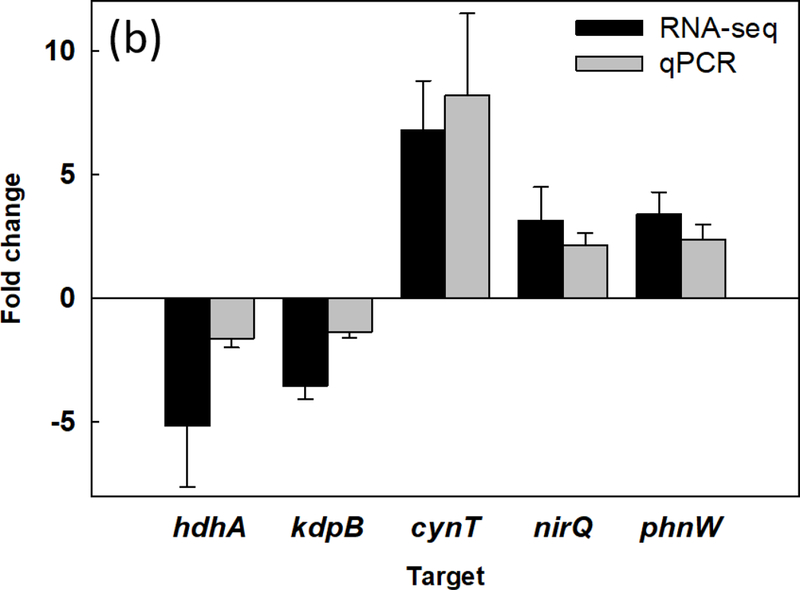
Effects of shape recovery triggered dispersion on P. aeruginosa PAO1 gene expression. (a) RNA-seq results of induced/repressed genes. (b) qPCR results of representative genes.
4. Discussion
Despite the well-recognized significance, biofilm control strategies have been largely limited to biofilm prevention and direct killing of biofilm cells. Eradicating established biofilms remains challenging. Previous research on biofilm removal has been largely based on the use of forces generated by air bubble [62,63], shock wave [64,65], water jet [66], acoustic energy [67], and magnetically rotating microrods [68,69]. These conditions can be harsh and require additional equipment, which may hinder in vivo applications. In comparison, SMP enabled shape recovery can be achieved under rather gentle conditions such as moderate temperature change in this study, or by electrical current [70–72] and light [73].
Recently, we demonstrated that mature biofilms can be effectively removed by using on-demand changes in substrate configuration of SMP [44]. In the present study, we further demonstrate that such on-demand dispersion can also sensitize biofilm cells to conventional antibiotics. Up to 9-fold increase in antibiotic susceptibility was observed when antibiotics were added after dispersion and more than 3 logs (2,479 times) reduction of biofilm cells was obtained by adding antibiotics during shape recovery. While bactericidal antibiotic showed significant differences between shape recovery conditions and control biofilm cells during sequential treatment, there was no significant difference for bacteriostatic antibiotics tested. This is not unexpected because what we did was a killing test and thus static agents would not show the same effects. It will be interesting to further test different classes of bactericidal compounds.
Synergy between physical factors and antibiotics in biofilm control has been reported. For example, using ultrasound [74,75] or ultrasound targeted microbubble destruction [76] in combination with antibiotics such as gentamicin and vancomycin can enhance the killing of biofilm cells due to the disruption of cell membranes [45]. However, the condition of shape recovery in this study alone did not cause direct killing of biofilm cells as evidenced by Live/Dead staining and SEM analysis. Also, the released cells were able to grow faster than the static flat control that was detached by bead beating (verified not to affect viability). This result suggests that the effects were through a different mechanism and the cells were not just passively dispersed by shape recovery. Instead, it might be through physiological changes in these cells.
Consistent with the results of antibiotic susceptibility, shape recovery triggered biofilm dispersion led to a higher level of intracellular ATP, slightly faster growth, and significant changes in gene expression in the dispersed cells. No change in the expression of biofilm matrix genes was observed. This is not unexpected because shape recovery happened in minutes; and thus, biofilm dispersion can be largely attributed to physical factors. Nevertheless, the results do indicate that dispersion caused physiological changes to the dispersed cells, which rendered these cells to enter a more active stage and thus more susceptible to bactericidal antibiotics.
Increasing evidence indicates that bacteria have complex systems to sense environmental cues when deciding biofilm formation vs. planktonic growth [37,77–80]. Biofilm cells are also known to disperse naturally when the environment changes to be unfavorable for bacteria to stay [81,82]. Some cell signaling systems have been shown to trigger biofilm dispersion [82–84]. Based on the results of this study, we speculate that biofilm cells may also be able to sense and respond to physical factors and adjust their physiological status for dispersion, which alters antibiotic susceptibility of these cells. Further study on such sensing mechanism may shed new light on the fundamental understanding of biofilm life cycle.
Different technologies have been developed for biofilm removal, biofilm killing, or both. However, the options for biofilm removal with gentle conditions are limited. In a recent study, we reported effective (up to 99.9%) biofilm removal using shape memory polymers [44]. Here we demonstrate that such removal also sensitizes biofilm cells to bactericidal antibiotics. It is encouraging to us since effective eradication of biofilm cells with lower doses of antibiotics can help reduce the risk of resistance development.
We chose room temperature incubation for biofilm growth and 40°C for triggering shape change to be consistent with our previous report [44], and allow us to study the effects on antibiotic susceptibility of dispersed cells specifically. To further develop this technology for in vivo applications, the polymer needs to be tested for antifouling activities at human body temperature and evaluated for cytotoxicity to mammalian cells. The temperature for triggering shape change can be adjusted by altering the ratio of tBA and PEGDMA. Alternatively, some shape memory polymers allow shape recovery to be triggered by other means such as electric signal [70–72] and light [73], which may ease medical applications. With further development, this technology has potential applications in medical devices that have major polymer components, e.g. catheters. This is part of our ongoing work.
5. Conclusions
The results of this study revealed that dynamic topography by shape recovery can sensitize the detached biofilm cells to conventional antibiotics. Specifically, the biofilm cells released by shape recovery were up to 9-fold more susceptible to antibiotics than the static flat control in sequential treatments; and more than 3 logs of biofilm reduction was achieved by concurrent treatment (shape recovery in the presence of antibiotics). Consistent with the increase in susceptibility to antibiotics, 11.8 times more ATP production and 4.1 times higher rrnB expression level were observed in biofilm cells dispersed by shape recovery compared to the static flat control. These findings were corroborated by RNA-seq and qPCR results and indicate that shape recovery triggered dispersion rendered bacterial cells to leave the physiological stage of biofilm growth and entered a more active and drug susceptible stage. The graphical abstract summarizes the main findings of this study. Collectively, the findings from this study suggest that effective controls can be developed to eradicate biofilm cells with combined physical (dynamic surface topography) and chemical (antibiotics) factors.
Supplementary Material
Statement of Significance.
Microbial infections are challenging due to high-level antibiotic resistance of biofilm cells. The protection of an extracellular matrix and slow growth of biofilm cells render conventional antibiotics ineffective. Thus, it is important to develop new technologies that can remove mature biofilms and sensitize biofilm cells to antibiotics. Recently, we demonstrated that dynamic change in surface topography can remove 48 h Pseudomonas aeruginosa PAO1 biofilms by 99.9%. In this study, we investigated how shape recovery triggered dispersion affect the physiology of biofilm cells and associated antibiotic susceptibility. These results are helpful for understanding biofilm dispersion and developing more effective control methods.
Acknowledgments.
We thank the U.S. National Institutes of Health (1R21EY0257 50-01A1) and U.S. National Science Foundation (CBET-1706061) for partial support of this work. We are grateful to former group member Dr. Xianyu Yao for helping construct the rrnB reporter stain and Dr. Karin Sauer at Binghamton University for sharing P. aeruginosa PAO1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Flemming Hans-Curt, Jost Wingender Abstract, The Biofilm Matrix, Biofouling 19 (2003) 139–150. doi: 10.1080/0892701031000072190. [DOI] [Google Scholar]
- [2].Olsen I, Biofilm-specific antibiotic tolerance and resistance, Eur. J. Clin. Microbiol. Infect. Dis 34 (2015) 877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- [3].Hall CW, Mah T-F, Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria, FEMS Microbiol. Rev 41 (2017) 276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- [4].Hall-Stoodley L, Costerton JW, Stoodley P, Bacterial biofilms: from the Natural environment to infectious diseases, Nat. Rev. Microbiol 2 (2004) 95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- [5].Sreeramoju P, Preventing Healthcare-Associated Infections, Am. J. Med. Sci (2012) 1. doi: 10.1097/MAJ.0b013e31824435e6. [DOI] [PubMed] [Google Scholar]
- [6].Wolcott RD, Ehrlich GD, Biofilms and chronic infections, JAMA - J. Am. Med. Assoc 299 (2008) 2682–2684. doi: 10.1001/jama.299.22.2682. [DOI] [PubMed] [Google Scholar]
- [7].Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L, Targeting microbial biofilms: Current and prospective therapeutic strategies, Nat. Rev. Microbiol 15 (2017) 740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gu H, Ren D, Materials and surface engineering to control bacterial adhesion and biofilm formation: A review of recent advances, Front. Chem. Sci. Eng 8 (2014) 20–33. doi: 10.1007/s11705-014-1412-3. [DOI] [Google Scholar]
- [9].Song F, Koo H, Ren D, Effects of Material Properties on Bacterial Adhesion and Biofilm Formation, J. Dent. Res 94 (2015) 1027–1034. doi: 10.1177/0022034515587690. [DOI] [PubMed] [Google Scholar]
- [10].Liu K, Jiang L, Bio-inspired design of multiscale structures for function integration, Nano Today 6 (2011) 155–175. doi: 10.1016/j.nantod.2011.02.002. [DOI] [Google Scholar]
- [11].Zhao L, Chu PK, Zhang Y, Wu Z, Antibacterial coatings on titanium implants, J. Biomed. Mater. Res. - Part B Appl. Biomater 91 (2009) 470–480. doi: 10.1002/jbm.b.31463. [DOI] [PubMed] [Google Scholar]
- [12].Hume EBH, Baveja J, Muir B, Schubert TL, Kumar N, Kjelleberg S, Griesser HJ, Thissen H, Read R, Poole-Warren LA, Schindhelm K, Willcox MDP, The control of Staphylococcus epidermidis biofilm formation and in vivo infection rates by covalently bound furanones, Biomaterials 25 (2004) 5023–5030. doi: 10.1016/j.biomaterials.2004.01.048. [DOI] [PubMed] [Google Scholar]
- [13].Kim WH, Lee SB, Oh KT, Moon SK, Kim KM, Kim KN, The release behavior of CHX from polymer-coated titanium surfaces, Surf. Interface Anal 40 (2008) 202–204. doi: 10.1002/sia.2809. [DOI] [Google Scholar]
- [14].Russell AD, Hugo WB, Antimicrobial Activity and Action of Silver, Prog. Med. Chem 31 (1994) 351–370. doi: 10.1016/S0079-6468(08)70024-9. [DOI] [PubMed] [Google Scholar]
- [15].Campoccia D, Montanaro L, Arciola CR, A review of the biomaterials technologies for infection-resistant surfaces, Biomaterials 34 (2013) 8533–8554. doi: 10.1016/j.biomaterials.2013.07.089. [DOI] [PubMed] [Google Scholar]
- [16].Mabboux F, Ponsonnet L, Morrier JJ, Jaffrezic N, Barsotti O, Surface free energy and bacterial retention to saliva-coated dental implant materials - An in vitro study, Colloids Surfaces B Biointerfaces 39 (2004) 199–205. doi: 10.1016/j.colsurfb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- [17].Price JS, Tencer AF, Arm DM, Bohach GA, Controlled release of antibiotics from coated orthopedic implants, J. Biomed. Mater. Res 30 (1996) 281–286. doi:. [DOI] [PubMed] [Google Scholar]
- [18].Hasan J, Crawford RJ, Ivanova EP, Antibacterial surfaces: The quest for a new generation of biomaterials, Trends Biotechnol 31 (2013) 295–304. doi: 10.1016/j.tibtech.2013.01.017. [DOI] [PubMed] [Google Scholar]
- [19].Zhang X, Wang L, Levänen E, Superhydrophobic surfaces for the reduction of bacterial adhesion, RSC Adv 3 (2013) 12003. doi: 10.1039/c3ra40497h. [DOI] [Google Scholar]
- [20].Ewald A, Glückermann SK, Thull R, Gbureck U, Lew D, Antimicrobial titanium/silver PVD coatings on titanium, Biomed. Eng. Online 5 (2006) 22. doi: 10.1186/1475-925X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Popat KC, Eltgroth M, LaTempa TJ, Grimes CA, Desai TA, Decreased Staphylococcus epidermis adhesion and increased osteoblast functionality on antibiotic-loaded titania nanotubes, Biomaterials 28 (2007) 4880–4888. doi: 10.1016/j.biomaterials.2007.07.037. [DOI] [PubMed] [Google Scholar]
- [22].Fadeeva E, Truong VK, Stiesch M, Chichkov BN, Crawford RJ, Wang J, Ivanova EP, Bacterial retention on superhydrophobic titanium surfaces fabricated by femtosecond laser ablation, Langmuir 27 (2011) 3012–3019. doi: 10.1021/la104607g. [DOI] [PubMed] [Google Scholar]
- [23].Ma J, Sun Y, Gleichauf K, Lou J, Li Q, Nanostructure on taro leaves resists fouling by colloids and bacteria under submerged conditions, Langmuir 27 (2011) 10035–10040. doi: 10.1021/la2010024. [DOI] [PubMed] [Google Scholar]
- [24].Chambers JR, Cherny KE, Sauer K, Susceptibility of Pseudomonas aeruginosa dispersed cells to antimicrobial agents is dependent on the dispersion cue and class of the antimicrobial agent used, Antimicrob. Agents Chemother 61 (2017) 1–18. doi: 10.1128/AAC.00846-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Anselme K, Davidson P, Popa AM, Giazzon M, Liley M, Ploux L, The interaction of cells and bacteria with surfaces structured at the nanometre scale, Acta Biomater 6 (2010) 3824–3846. doi: 10.1016/j.actbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- [26].Yan Lin H, Liu Y, Wismeijer D, Crielaard W, Mei Deng D, Effects of Oral Implant Surface Roughness on Bacterial Biofilm Formation and Treatment Efficacy, Int. J. Oral Maxillofac. Implants 28 (2013) 1226–1231. doi: 10.11607/jomi.3099. [DOI] [PubMed] [Google Scholar]
- [27].Perera-Costa D, Bruque JM, González-Martín ML, Gómez-García AC, Vadillo-Rodríguez V, Studying the influence of surface topography on bacterial adhesion using spatially organized microtopographic surface patterns, Langmuir 30 (2014) 4633–4641. doi: 10.1021/la5001057. [DOI] [PubMed] [Google Scholar]
- [28].Dickson MN, Liang EI, Rodriguez LA, Vollereaux N, Yee AF, Nanopatterned polymer surfaces with bactericidal properties, Biointerphases 10 (2015) 021010. doi: 10.1116/1.4922157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hochbaum AI, Aizenberg J, Bacteria pattern spontaneously on periodic nanostructure arrays, Nano Lett 10 (2010) 3717–3721. doi: 10.1021/nl102290k. [DOI] [PubMed] [Google Scholar]
- [30].Ball P, Shark skin and other solutions, Nat. Eng 400 (1999) 507–508. doi: 10.1038/22883. [DOI] [Google Scholar]
- [31].Gu H, Chen A, Song X, Brasch ME, Henderson JH, Ren D, How Escherichia coli lands and forms cell clusters on a surface: A new role of surface topography, Sci. Rep 6 (2016) 1–14. doi: 10.1038/srep29516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Siegismund D, Undisz A, Germerodt S, Schuster S, Rettenmayr M, Quantification of the interaction between biomaterial surfaces and bacteria by 3-D modeling, Acta Biomater 10 (2014) 267–275. doi: 10.1016/j.actbio.2013.09.016. [DOI] [PubMed] [Google Scholar]
- [33].Gu H, Kolewe KW, Ren D, Conjugation in Escherichia coli Biofilms on Poly(dimethylsiloxane) Surfaces with Microtopographic Patterns, Langmuir 33 (2017) 3142–3150. doi: 10.1021/acs.langmuir.6b04679. [DOI] [PubMed] [Google Scholar]
- [34].Xing R, Lyngstadaas SP, Ellingsen JE, Taxt-Lamolle S, Haugen HJ, The influence of surface nanoroughness, texture and chemistry of TiZr implant abutment on oral biofilm accumulation, Clin. Oral Implants Res 26 (2015) 649–656. doi: 10.1111/clr.12354. [DOI] [PubMed] [Google Scholar]
- [35].Ionescu A, Wutscher E, Brambilla E, Schneider-Feyrer S, Giessibl FJ, Hahnel S, Influence of surface properties of resin-based composites on in vitro Streptococcus mutans biofilm development, Eur. J. Oral Sci 120 (2012) 458–465. doi: 10.1111/j.1600-0722.2012.00983.x. [DOI] [PubMed] [Google Scholar]
- [36].Song F, Wang H, Sauer K, Ren D, Cyclic-di-GMP and oprF are involved in the response of Pseudomonas aeruginosa to substrate material stiffness during attachment on polydimethylsiloxane (PDMS), Front. Microbiol 9 (2018) 1–13. doi: 10.3389/fmicb.2018.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Song F, Brasch ME, Wang H, Henderson JH, Sauer K, Ren D, How Bacteria Respond to Material Stiffness during Attachment: A Role of Escherichia coli Flagellar Motility, ACS Appl. Mater. Interfaces 9 (2017) 22176–22184. doi: 10.1021/acsami.7b04757. [DOI] [PubMed] [Google Scholar]
- [38].Zhao Y, Song F, Wang H, Zhou J, Ren D, Phagocytosis of Escherichia coli biofilm cells with different aspect ratios: a role of substratum material stiffness, Appl. Microbiol. Biotechnol 101 (2017) 6473–6481. doi: 10.1007/s00253-017-8394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Song F, Ren D, Stiffness of cross-linked poly(dimethylsiloxane) affects bacterial adhesion and antibiotic susceptibility of attached cells, Langmuir 30 (2014) 10354–10362. doi: 10.1021/la502029f. [DOI] [PubMed] [Google Scholar]
- [40].Shivapooja P, Wang Q, Szott LM, Orihuela B, Rittschof D, Zhao X, López GP, Dynamic surface deformation of silicone elastomers for management of marine biofouling: laboratory and field studies using pneumatic actuation, Biofouling 31 (2015) 265–274. doi: 10.1080/08927014.2015.1035651. [DOI] [PubMed] [Google Scholar]
- [41].Shivapooja P, Wang Q, Orihuela B, Rittschof D, Lõpez GP, Zhao X, Bioinspired surfaces with dynamic topography for active control of biofouling, Adv. Mater 25 (2013) 1430–1434. doi: 10.1002/adma.201203374. [DOI] [PubMed] [Google Scholar]
- [42].Leveri V, Wang Q, Shivapooja P, Zhao X, Lõpez GP, Soft Robotic Concepts in Catheter Design: an On-demand Fouling-release Urinary Catheter, Adv Heal. Mater 3 (2014) 1588–1596. doi: 10.1002/ana.22528.Toll-like. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shivapooja P, Yu Q, Orihuela B, Mays R, Rittschof D, Genzer J, López GP, Modification of Silicone Elastomer Surfaces with Zwitterionic Polymers: Short-Term Fouling Resistance and Triggered Biofouling Release, ACS Appl. Mater. Interfaces 7 (2015) 25586–25591. doi: 10.1021/acsami.5b09199. [DOI] [PubMed] [Google Scholar]
- [44].Gu H, Lee SW, Buffington SL, Henderson JH, Ren D, On-Demand Removal of Bacterial Biofilms via Shape Memory Activation, ACS Appl. Mater. Interfaces 8 (2016) 21140–21144. doi: 10.1021/acsami.6b06900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Runyan CM, Carmen JC, Beckstead BL, Nelson JL, a Robison R, Pitt WG, Low-frequency ultrasound increases outer membrane permeability of Pseudomonas aeruginosa., J. Gen. Appl. Microbiol 52 (2006) 295–301. doi: 10.2323/jgam.52.295. [DOI] [PubMed] [Google Scholar]
- [46].Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR, The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin, Environ. Microbiol 86 (2013) n/a-n/a. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bertani G, Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli., J. Bacteriol 62 (1951) 293–300. doi:citeulike-article-id:149214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yakacki CM, Willis S, Luders C, Gall K, Deformation limits in shape-memory polymers, Adv. Eng. Mater 10 (2008) 112–119. doi: 10.1002/adem.200700184. [DOI] [Google Scholar]
- [49].Sieuwerts S, De Bok FAM, Mols E, De Vos WM, Van Hylckama Vlieg JET, A simple and fast method for determining colony forming units, Lett. Appl. Microbiol 47 (2008) 275–278. doi: 10.1111/j.1472-765X.2008.02417.x. [DOI] [PubMed] [Google Scholar]
- [50].Heydorn A, Heydorn A, Nielsen AT, Nielsen AT, Hentzer M, Hentzer M, Quantication of biofilm structures by the novel computer program, Image Process 146 (2000) 2395– 2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- [51].Pan J, Bahar AA, Syed H, Ren D, Reverting Antibiotic Tolerance of Pseudomonas aeruginosa PAO1 Persister Cells by (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one, PLoS One 7 (2012). doi: 10.1371/journal.pone.0045778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H, Expression stability of six housekeeping genes: A proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR, J. Med. Microbiol 52 (2003) 403–408. doi: 10.1099/jmm.0.05132-0. [DOI] [PubMed] [Google Scholar]
- [53].Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet.Journal 17 (2011) 10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- [54].Dobin A, Gingeras TR, Comment on “ TopHat2 : accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions “ by Kim et al, (2013) 0–9. doi: 10.1101/000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L, Differential analysis of gene regulation at transcript resolution with RNA-seq, Nat. Biotechnol 31 (2013) 46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yue S, Gandt AB, Rowe SE, Deisinger JP, Conlon BP, Lewis K, ATP-Dependent Persister Formation in Escherichia coli, 8 (2017) 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schneider DA, Gaal T, Gourse RL, NTP-sensing by rRNA promoters in Escherichia coli is direct, Proc. Natl. Acad. Sci 99 (2002) 8602–8607. doi: 10.1073/pnas.132285199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Paul BJ, Ross W, Gaal T, Gourse RL, rRNA Transcription in Escherichia coli, Annu. Rev. Genet 38 (2004) 749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- [59].Arai H, Kodama T, Igarashi Y, Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa, FEMS Microbiol. Lett 170 (1999) 19–24. doi: 10.1016/S0378-1097(98)00517-5. [DOI] [PubMed] [Google Scholar]
- [60].Thauer RK, Jungermann K, Decker K, Energy conservation in chemotrophic anaerobic bacteria., Bacteriol. Rev 41 (1977) 100–180. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kühlbrandt W, Biology, structure and mechanism of P-type ATPases, Nat. Rev. Mol. Cell Biol 5 (2004) 282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- [62].Jang H, Rusconi R, Stocker R, Biofilm disruption by an air bubble reveals heterogeneous age-dependent detachment patterns dictated by initial extracellular matrix distribution, Npj Biofilms Microbiomes 3 (2017) 0–1. doi: 10.1038/s41522-017-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Crusz SA, Popat R, Rybtke MT, Cámara M, Givskov M, Tolker-Nielsen T, Diggle SP, Williams P, Bursting the bubble on bacterial biofilms: A flow cell methodology, Biofouling 28 (2012) 835–842. doi: 10.1080/08927014.2012.716044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Francis NC, Yao W, Grundfest WS, Taylor ZD, Laser-Generated Shockwaves as a Treatment to Reduce Bacterial Load and Disrupt Biofilm, IEEE Trans. Biomed. Eng 64 (2017) 882–889. doi: 10.1109/TBME.2016.2581778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gnanadhas DP, Elango M, Janardhanraj S, Srinandan CS, Datey A, Strugnell RA, Gopalan J, Chakravortty D, Successful treatment of biofilm infections using shock waves combined with antibiotic therapy, Sci. Rep 5 (2015) 1–13. doi: 10.1038/srep17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Abramzon N, Joaquin JC, Bray J, Brelles-Mariño G, Biofilm destruction by RF high-pressure cold plasma jet, IEEE Trans. Plasma Sci 34 (2006) 1304–1309. doi: 10.1109/TPS.2006.877515. [DOI] [Google Scholar]
- [67].Dror N, Mandel M, Hazan Z, Lavie G, Advances in Microbial Biofilm Prevention on Indwelling Medical Devices with Emphasis on Usage of Acoustic Energy, Sensors 9 (2009) 2538–2554. doi: 10.3390/s90402538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mair LO, Nacev A, Hilaman R, Stepanov PY, Chowdhury S, Jafari S, Hausfeld J, Karlsson AJ, Shirtliff ME, Shapiro B, Weinberg IN, Biofilm disruption with rotating microrods enhances antimicrobial efficacy, J. Magn. Magn. Mater 427 (2017) 81–84. doi: 10.1016/j.jmmm.2016.10.100. [DOI] [Google Scholar]
- [69].Gomes IB, Lemos M, Mathieu L, Simões M, Simões LC, The action of chemical and mechanical stresses on single and dual species biofilm removal of drinking water bacteria, Sci. Total Environ 631–632 (2018) 987–993. doi: 10.1016/j.scitotenv.2018.03.042. [DOI] [PubMed] [Google Scholar]
- [70].Mahapatra SS, Yadav SK, Yoo HJ, Ramasamy MS, Cho JW, Tailored and strong electro-responsive shape memory actuation in carbon nanotube-reinforced hyperbranched polyurethane composites, Sensors Actuators, B Chem 193 (2014) 384–390. doi: 10.1016/j.snb.2013.12.006. [DOI] [Google Scholar]
- [71].Qi X, Dong P, Liu Z, Liu T, Fu Q, Selective localization of multi-walled carbon nanotubes in bi-component biodegradable polyester blend for rapid electroactive shape memory performance, Compos. Sci. Technol 125 (2016) 38–46. doi: 10.1016/j.compscitech.2016.01.023. [DOI] [Google Scholar]
- [72].Cho JW, Kim JW, Jung YC, Goo NS, Electroactive shape-memory polyurethane composites incorporating carbon nanotubes, Macromol. Rapid Commun 26 (2005) 412–416 doi: 10.1002/marc.200400492. [DOI] [Google Scholar]
- [73].Lendlein A, Jiang H, Jünger O, Langer R, Light-induced shape-memory polymers, Nature 434 (2005) 879–882. doi: 10.1038/nature03496. [DOI] [PubMed] [Google Scholar]
- [74].Ensing GT, Roeder BL, Nelson JL, Van Horn JR, Van Der Mei HC, Busscher HJ, Pitt WG, Effect of pulsed ultrasound in combination with gentamicin on bacterial viability in biofilms on bone cements in vivo, J. Appl. Microbiol 99 (2005) 443–448. doi: 10.1111/j.1365-2672.2005.02643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ensing GT, Neut D, van Horn JR, van der Mei HC, Busscher HJ, The combination of ultrasound with antibiotics released from bone cement decreases the viability of planktonic and biofilm bacteria: An in vitro study with clinical strains, J. Antimicrob. Chemother 58 (2006) 1287–1290. doi: 10.1093/jac/dkl402. [DOI] [PubMed] [Google Scholar]
- [76].He N, Hu J, Liu H, Zhu T, Huang B, Wang X, Wu Y, Wang W, Qu D, Enhancement of vancomycin activity against biofilms by using ultrasound-targeted microbubble destruction, Antimicrob. Agents Chemother 55 (2011) 5331–5337. doi: 10.1128/AAC.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Song F, Koo H, Ren D, Effects of material properties on bacterial adhesion and biofilm formation, J. Dent. Res 94 (2015) 1027–1034. doi: 10.1177/0022034515587690. [DOI] [PubMed] [Google Scholar]
- [78].Renner LD, Weibel DB, Physiochemical regulation of biofilm formation, MRS Bull 36 (2011) 347–355. doi: 10.1557/mrs.2011.65.Physicochemical. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Belas R, Biofilms, flagella, and mechanosensing of surfaces by bacteria, Trends Microbiol 22 (2014) 517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- [80].Lele PP, Hosu BG, Berg HC, Dynamics of mechanosensing in the bacterial flagellar motor, Proc. Natl. Acad. Sci 110 (2013) 11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kim SK, Lee JH, Biofilm dispersion in Pseudomonas aeruginosa, J. Microbiol 54 (2016) 71–85. doi: 10.1007/s12275-016-5528-7. [DOI] [PubMed] [Google Scholar]
- [82].Kaplan JB, Biofilm Dispersal: Mechanisms, Clinical Implications, and Potential Therapeutic Uses, J. Dent. Res 89 (2010) 205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Solano C, Echeverz M, Lasa I, Biofilm dispersion and quorum sensing, Curr. Opin. Microbiol 18 (2014) 96–104. doi: 10.1016/j.mib.2014.02.008. [DOI] [PubMed] [Google Scholar]
- [84].Marques CNH, Davies DG, Sauer K, Control of biofilms with the fatty acid signaling molecule cis-2-Decenoic acid, Pharmaceuticals 8 (2015) 816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


