Abstract
Mammalian sociality is regulated in part by the neuropeptide oxytocin. In prairie voles, subtle variation in early life experience changes oxytocin receptor-mediated social behaviors. We report that low levels of early care in voles leads to de novo DNA methylation at specific regulatory sites in the oxytocin receptor gene (Oxtr), impacting gene expression and protein distribution in the nucleus accumbens. DNA methylation state of the blood predicts expression in the brain indicating the utility of the blood as a biomarker for the transcription state of the brain. These experience-sensitive CpG sites are conserved in humans, are related to gene expression in the brain, and have been associated with psychiatric disorders and individual differences in neural response to social stimuli. These results identify a mechanism by which early care regulates later displays of typical prairie vole social behavior and suggest the potential for nurture driven epigenetic tuning of OXTR in humans.
Keywords: OXTR, DNA methylation, oxytocin, social behavior, prairie vole
1. Introduction
Experiences early in life have the potential to alter social behavior and emotion regulation across the lifespan (Levine, 1957; Harlow et al., 1965; Bowlby, 1969). One of the most critical relationships in early life is the parent-offspring dyad, where variation in experience can permanently alter the developmental trajectory of the offspring. Variability in early care may be adaptive, serving to prepare offspring for their likely future environment, and is also a key mechanism allowing for the development of individual differences in behavior (Denenberg et al., 1962; Levine et al., 1967; Francis et al., 1999).
In the prairie vole (Microtus ochrogaster), variability in the early life environment is linked to later differences in the expression of species-typical social behavior. Naturally occurring high levels of early biparental care lead to offspring who engage in high levels of alloparental behavior towards unrelated infants and an increase in the propensity to form a selective partner preference with an opposite sex mate (Perkeybile et al., 2013; Perkeybile et al., 2015; del Razo and Bales, 2016). Likewise, a single episode of handling on the first day of life results in later increases in both alloparental care and partner preference formation (Bales et al., 2007). Both alloparenting and pair bonding behaviors are regulated by oxytocin receptor (OXTR) expression in the nucleus accumbens. Prairie voles with higher levels of OXTR binding in this region engage in a greater amount of both of these social behaviors (Liu and Wang, 2003; Olazabal and Young, 2006a; Ross et al., 2009), and artificially upregulating or downregulating OXTR expression acts to increase or decrease these behaviors, respectively (Ross et al., 2009; Keebaugh and Young, 2011; Keebaugh et al., 2015).
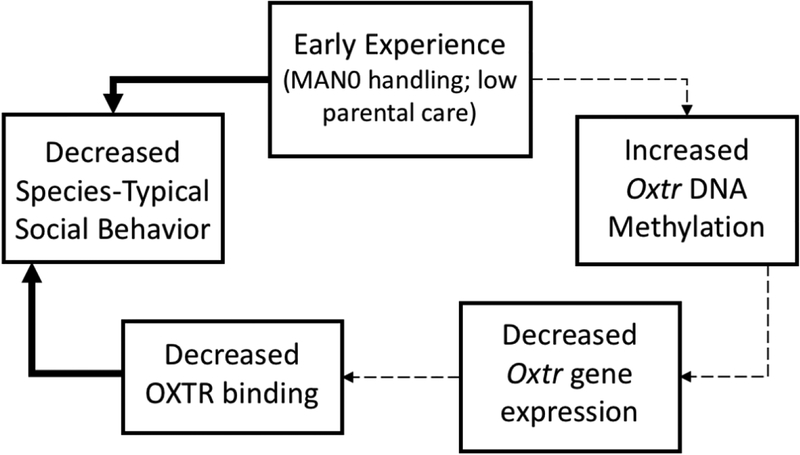
The expression of OXTR in the nucleus accumbens is remarkably variable both between and within species, although levels of the oxytocin peptide are less variable (Insel and Shapiro, 1992; Young, 1999; Gimpl and Fahrenholz, 2001; Olazabal and Young, 2006b). Mechanisms regulating expression of the receptor, rather than oxytocin itself, may provide an improved explanation for the variability in social behavior seen after differing early experiences. Here we explore one such mechanism by which early experience results in changes to OXTR-dependent social behaviors in the prairie vole. Previous work has demonstrated that both early parenting and early handling alter alloparental behavior and pair bonding behavior, both of which are dependent on OXTR expression in the nucleus accumbens. Using a candidate gene approach, we propose an epigenetic mechanism where early experience regulates DNA methylation of the oxytocin receptor gene, Oxtr, leading to downstream consequences for Oxtr gene expression and eventually for OXTR protein levels. We hypothesize that low levels of early handling and early parental care lead to increased Oxtr DNA methylation, decreased Oxtr gene expression, and low levels of OXTR binding in the nucleus accumbens, which is known to predict decreases in the social behaviors impacted by our early experience models (see Figure 1).
Figure 1. Proposed epigenetic mechanism for early experiences to impact oxytocin receptor-dependent social behavior.
Early experiences act to regulate Oxtr DNA methylation in the nucleus accumbens, which alters Oxtr gene expression and eventually OXTR protein distribution in this region. This change in OXTR protein is responsible for the changes in social behaviors seen after varying early experiences. Known relationships are indicated by solid lines. Proposed mechanisms are indicated by dashed lines.
2. Materials and Methods
2.1. Animal model
Subjects were laboratory-bred prairie voles (Microtus ochrogaster), descendants of a wild-caught stock captured near Champaign, Illinois. Breeding pairs were housed in large polycarbonate cages (44cmx22cmx16cm) and same sex offspring pairs were housed in smaller polycarbonate cages (27cmx16cmx16cm) after weaning on postnatal day (PND) 20 (date of birth: PND0). Animals were given food (high-fiber Purina rabbit chow) and water ad libitum, cotton nestlets for nesting material in breeding cages, and were maintained on a 14:10 light:dark cycle.
Procedures involved in measuring the effects of early handling on parental behavior were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois, Chicago. Procedures involved in generating tissue for the analysis of DNA methylation, gene expression, and OXTR protein following handling and naturally occurring parental care variation were reviewed and approved by the IACUC at the University of California, Davis. Procedures involved in collection of embryonic tissue were reviewed and approved by the IACUC at Northeastern University.
In all cases, animals were euthanized via cervical dislocation and rapid decapitation under deep isoflurane anesthesia. Brains were extracted, flash frozen on dry ice, and stored at −80 °C until analysis. Trunk blood samples were immediately frozen and stored at −80 °C until analysis.
2.2. Early handling manipulation (the MAN paradigm)
Within 24 hours of giving birth, breeding pairs underwent a single treatment experience, herein termed MAN0 (no direct handling manipulation) or MAN1 (direct handling manipulation) handling. MAN0 litters were picked up for 30 seconds in a clear plastic cup (no direct handling). If animals were sitting, they were scooped up in the cup. If they were walking around the cage, the cup was positioned in front so that they would walk into it. In this condition, pups were supported by the cup. Pairs in the MAN1 condition were picked up by the scruff of the neck by a gloved hand for 30 seconds (direct handling). Prairie vole pups have milk teeth to attach to the mother’s nipple, so in this condition, pups hung unsupported from the mother and were therefore not touched by the experimenter (if pups were unattached, researchers waited to perform the handling). Both the mother and father of each pair were handled in the same manner. Pups were only manipulated during the handling of the mother because they were attached to her.
2.3. Quantification of parental behavior following early handling
Maternal and paternal pup directed care were characterized and scored for 60 minutes immediately after handling on PND1, and again on PND8 and PND15 (with no further manipulation) between 17:00 and 20:00. Parental pup-directed care was video recorded and later scored by two trained observers using Observer 5.0 behavior tracking software (Noldus Inc., Wageningen, Netherlands). Each observation period was divided into 15-second segments (240 segments total) and behaviors were recorded in a one-zero fashion for each segment. Observations were conducted while the animals were in their home cage and they were not disturbed throughout the observation period. Behaviors observed included active nursing (maternal only), huddling, lateral contact with pups, hunched contact with pups, sniffing and licking/grooming of pups, anogenital licking/grooming of pups, rearranging pups, and retrieval or removal of pups from the dam. Behavioral score reflects the percentage of segments active pup-directed behaviors were observed during the observation period (MAN0, n = 7; MAN1, n = 6). During each observation, parents were distinguished from one another by distinct fur color and markings, body size, and the presence of pups visibly attached to the nipple. Inter-rater reliability was 0.89. Intra-rater reliabilities were 0.95 for the primary coder and 0.96 for the secondary coder using the kappa statistic.
2.4. Identifying conserved MT2 region in prairie vole
The Multiz Alignment of 100 Vertebrates track in the UCSC genome database (Blanchette et al., 2004) was used to identify a 71 base pair alignment block that contained similar sequence in rats, mice, and prairie voles. To assess the significance of this similarity between human and prairie vole we used the UVa FASTA server (http://fasta.bioch.virginia.edu/) and PRSS (DNA:DNA) to shuffle the prairie vole sequence 200 times and estimate the statistical significance of the shuffled scores. This analysis identified a conserved 62 base pair region between human and prairie vole with 72.6% identity. PRSS output is provided in Supplemental Figure 2C.
2.5. Tissue collection for epigenetic analyses following early handling
An additional cohort of offspring underwent early handling procedures as described above (13 male/female sibling pairs; MAN0: 7 females, 7 male; MAN1: 6 females, 6 male). On PND24, brain and blood tissues were collected, immediately frozen on dry ice, and then stored at −80 °C until DNA and RNA isolation. Brains were equilibrated to −20°C for two hours prior to sectioning. Brain samples were dissected by 1) a coronal cut to remove the olfactory bulbs, 2) a coronal cut at the nerve chiasma just rostral to the hypothalamus and bregma and, 3) bilateral punches (1mm in diameter, 2mm in depth) to isolate nucleus accumbens tissue. Following sectioning, nucleus accumbens tissue was placed in a DNAse/RNAse free microcentrifuge tube and flash frozen with liquid nitrogen. Brain tissue was then crushed using a mortar and pestle in preparation for DNA/RNA isolation.
2.6. Prenatal pup brain collection
Adult males and females were mated using a timed mating paradigm to accurately predict birth. Males and females were initially paired for 24 hours in a large polycarbonate cage. Prairie vole females experience estrus induction 48–72 hours after exposure to a male and will typically not allow mating within the first 24 hours of pairing. Therefore, a divider was placed in the cage 24 hours after pairing to allow for olfactory and auditory interaction but minimal physical contact and no mating. The divider was removed after 72 hours, at which point pairs were observed for mating. Pairs were then left undisturbed throughout gestation except for routine cage changes. On the expected day of birth, pregnant females (n = 9) were euthanized and pups were immediately removed from the uterine horns via caesarean section. Male/female sibling pairs (n=18; 9 females, 9 male) were used for forebrain (which includes the nucleus accumbens) Oxtr DNA methylation analysis, and only pups that weighed 1.8 g or greater were included in analysis.
2.7. Quantification of natural variation in parental care
Following methods previously established by this laboratory (Perkeybile et al., 2013; Perkeybile and Bales, 2015), the type and amount of naturally occurring parental care directed toward offspring was observed between PND 1–3 in real time by a trained observer using Behavior Tracker software (www.behaviortracker.com) for 30 established breeding pairs. Behavior observations were conducted in the same manner for two separate litters for each breeding pair. Each parent was observed for 20 minutes in the morning and 20 minutes in the afternoon two days between PND 1–3 for a total of 4 maternal care and 4 paternal care observations per litter. Parents were distinguished from one another based on individual characteristics such as body size, fur color and markings, or the presence of pups visibly attached to the nipple.
When ranking breeder pairs in relation to one another, total contact times for pup-directed behaviors were summed across each of the 4 observations for the mother and the father. A mean was then calculated for pup-directed behavior for these 4 summed scores for both the mother and the father, and these two means were then summed to produce an average total contact score for the breeder pair for a single litter. These scores for all 30 breeding pairs were then rank-ordered and split into approximate quartiles. Parental care of a second litter was characterized in the same way to determine if breeder pairs ranked in the same quartile for a subsequent litter. In all cases, pairs fell into the same quartile for both litters and were normally distributed, replicating previous findings (Perkeybile et al., 2013). The top quartile breeder pairs (n = 8) produced high contact offspring (n = 16), the middle two quartiles (n = 14) produced medium contact offspring (n = 90), and the bottom quartile breeder pairs (n = 8) produced low contact offspring (n = 49). The number of offspring born to breeder pairs in each contact group does not vary significantly across groups, nor does offspring mortality. Rather, the variation in number of offspring used here for each contact group is a result of animal availability due to other ongoing research at the time of this study.
Observations were done with the animals in their home cage; animals were not disturbed during the observations. Behaviors quantified included maternal and paternal huddling, non-huddling contact, licking/grooming, retrievals, and non-pup directed behaviors, such as nest building, eating and drinking. Maternal nursing postures were also recorded, including neutral, lateral, and active nursing. Behaviors recorded were based on an ethogram presented in Stone and Bales (Stone and Bales, 2010). Following weaning, offspring were housed with a same-sex sibling until euthanasia. Trunk blood samples were taken between PND 48–52 for DNA isolation.
In a separate cohort of 10 breeding pairs, naturally occurring early parental care was observed using the same behavioral ethogram. In this case, pairs were not ranked; instead care was used as a continuous variable. Offspring were euthanized between PND 24–26 and brains were collected for oxytocin receptor autoradiography (n = 32).
2.8. Oxtr DNA methylation analysis
Extraction of DNA was done using the Qiagen AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA) following manufacturer instructions. Samples included DNA and RNA isolated from the nucleus accumbens and DNA from whole blood from animals that experienced early handling (i.e. MAN1, MAN0); and DNA from whole blood from animals experiencing naturally varying amounts of early parental care. One hundred nanograms (ng) of DNA from early manipulation subjects (due to low DNA extraction volumes for 2 subjects) or 200 ng from the natural variation in parenting subjects were subject to bisulfite treatment (Kit MECOV50, Invitrogen, Carlsbad, CA) per manufacturer instructions. Bisulfite conversion allows for the detection of methylated cytosines by sequencing. Twelve nanograms of bisulfite converted DNA was used as a template for PCR using a Pyromark PCR kit (Qiagen, Valencia, CA) and 0.2 uM of primers TSL201F 5’-GGGGATAGGATGGTTAGTTAGTATT-3’ and TSL201R 5’-CCAACAACCTCAAAACTCTACT-3’. Samples were amplified in triplicate on three identical PCR machines (S1000 Thermal Cycler, Bio-Rad, Hercules, CA.) The following cycling conditions were used for amplification of the target Oxtr fragment, which included CpG sites −934_1, −934_2, −924 and −901: [Step 1: (95°C/15 min)/1 cycle, Step 2: (94°C/30 s, 58°C/30 s, 72°C/30 s)/50 cycles, Step 3: (72°C/10 min)/1 cycle, Step 4: 4°C hold]. Standard controls of 0% and 100% methylated DNA, as well as a no DNA control and a positive control vole standard were included for each PCR plate. Pyrosequencing was performed using two primers: TSL201S 5’-GAGGGAAGGTTTTGGAGTTTTTTATAT-3’ and TSL201S2 5’-AGGGATTGAAAAGTGA-3’ on a Pyromark Q24 using PyroMark Gold Q24 Reagents (Qiagen, Valencia, CA) per the manufacturer protocol. Epigenotypes reported are an average of the three replicates. On average, nucleus accumbens samples deviated from the mean by, −934_1: 1.12%; −934_2: 1.37%; −924: 1.13%, −901: 1.63%. On average, whole blood samples deviated from the mean by, −934_1: 0.95%; −934_2: 1.02%; −924: 0.95%, −901: 0.92%.
2.9. Oxtr gene expression analysis
Extraction of RNA was done using the Qiagen AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA) following manufacturer instructions. RNA was processed for cDNA synthesis following the protocol provided in the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). Real-time PCR was conducted using a 7500 Fast Real-Time PCR System (Applied Biosystems) using Power SYBR Green (Applied Biosystems No. 4367659). The cycling conditions are as follows: for Oxtr [Step 1: (95°C/10 min) 1 cycle, Step 2: (95°C/15 s, 63.4°C/60 s) 35 cycles], for Pgk1 [Step 1: (95°C/10 min) 1 cycle, Step 2: (95°C/15 s, 65.3°C/60 s) 35 cycles]. All reactions were run in triplicate (replicate standard deviation was <0.05) and their specificity verified by melting curve analysis and separation on a 2% agarose gel. Primers performance was evaluated using standard serial dilution and both primer sets performed within acceptable range for efficiency >90% (Oxtr efficiency = 99.7%, R2=0.993; Pgk1 efficiency = 93.5%, R2=0.998). The primer sequences used for Oxtr are TSL401_F 5’-GCCTTTCTTCTTCGTGCAGATG-3’ (Fwd) and TSL401_R 5’-ATGTAGATCCAGGGGTTGCAG-3’ (Rev); for Pgk1 TSL402_F 5’-TTGCCCGTTGACTTTGTCAC-3’ (Fwd) and TSL402_R 5’-GCCACAGCCTCAGCATATTTC-3’ (Rev). Relative gene expression is presented using the comparative Ct method, 2−Δ Ct. Pgk1 was chosen as a reference based on data in mouse brain showing its reliability across brain regions and developmental time points (Boda et al., 2009).
2.10. Oxtr SNP analysis
Ten nanograms of DNA was used as a template for PCR using a Pyromark PCR kit (Qiagen, Valencia, CA) and 0.2 uM of primers TSL202F 5’- CAGGGACGTTCACGTTACATG-3’ and TSL202R 5’- GACAGAGTCTCCAGCCAAGAAG-3’. The following cycling conditions were used for amplification of the target Oxtr fragment, which included SNP NT213739: [Step 1: (95°C/15 min)/1 cycle, Step 2: (94°C/30 s, 56°C/30 s, 72°C/30 s)/45 cycles, Step 3: (72°C/10 min)/1 cycle, Step 4: 4°C hold]. A single product corresponding to 99 base pairs was identified via gel electrophoresis. Pyrosequencing was performed to detect the C/T SNP using primer: TSL202S 5’- GAATCATCCCACCGT-3’ on a Pyromark Q24 with PyroMark Gold Q24 Reagents (Qiagen, Valencia, CA) per the manufacturer protocol.
2.11. Oxytocin receptor autoradiography
Oxytocin receptor autoradiography was performed following previously established methods (Bales et al., 2007; Perkeybile et al., 2015). Brains were sectioned at 20 μm into six series, mounted onto Super-frost slides, and stored at −80°C until assayed. Slides were allowed to thaw at room temperature and were then fixed in 0.1% paraformaldehyde (7.4 pH) for 2 minutes. Slides were washed two times for 10 minutes in 50mM Tris-HCl buffer solution (7.4 pH), then incubated at room temperature for 60 minutes in tracer buffer (50mM Tris-HCl buffer with 10mM MgCl2, 0.1% bovine serum albumin, and 50pM of radiotracer [125I]-ornithine vasotocin analog [(125I)OVTA] [vasotocin, d(CH2)5 [Tyr(Me)2, Thr4, Orn8, (125I)Tyr9−NH2]; 2200 Ci/mmol]). Following incubation, slides were rinsed in 50 mM Tris-HCl buffer with 10mM MgCl2 at 4° C four times for 5 minutes each, followed by a 30 minute wash in the same solution at room temperature while agitating. Sections were briefly dipped in 4°C dH2O and then rapidly dried with a stream of cool air. The following day slides were exposed to Kodak Bio Max MR film (Kodak, Rochester, NY, USA) with 125I microscale standards (American Radiolabeled Chemicals, Inc., St. Louis, MO, USA) for 168 hours. Receptor binding was quantified from film using NIH ImageJ. The 125I microscale standards were used to convert uncalibrated optical density to disintegrations per minute (DPM). Nucleus accumbens was quantified in both hemispheres and sides were compared for any differences, of which none were found. A measure of non-specific binding (NSB) was taken for each section by quantifying binding in an area of cortex with no oxytocin receptors. The NSB value was subtracted from the binding value for each section and a mean was then calculated for each section, followed by a mean for the entire area for each subject. Area means were used in data analysis and are referred to as normalized DPM.
2.12. Experimental design and statistical analysis
Statistical analyses were conducted using GraphPad Prism 6.0. For each analysis, p < 0.05 was regarded as statistically significant. Parental care following early handling manipulation was analyzed using a 2-way parent x handling condition analysis of variance (ANOVA). Oxtr methylation in nucleus accumbens and whole blood in the prairie vole was analyzed using a 2-way CpG site x handling condition ANOVA with exploratory post-hoc testing with Bonferroni multiple comparisons correction. One set of male/female sibling pairs per unique parenting pair were included for analysis. Offspring from three MAN0 pairs and four MAN1 pairs were excluded from analyses because the litters had only female offspring. Levels of de novo Oxtr DNA methylation following an early handling manipulation were analyzed using unpaired two-tailed t tests. Oxtr expression in the nucleus accumbens was assessed with an unpaired two-tailed t test. A Partial Least Squares (PLS) regression was used to explore how Oxtr methylation within each of the four assayed CpG sites in each tissue correlated with Oxtr expression. Given the high correlations in methylation values between these sites (see Supplemental Table 1), ordinary least squares regression is not appropriate due to multicollinearity. We therefore used PLS regression which operates on the entire data structure at once to find latent components which maximize the variability of predictors and have maximum correlation with the response. Model fitting and selection was performed with leave-oneout cross-validation using the PLS package (Mevik et al., 2016) in R (Team, 2017). The optimal number of components to retain for each analysis was determined by selecting the first local minimum in the root mean squared error of prediction curve. For both tissue types, a 1-component model emerged as optimal, indicating that all four CpG sites show a similar relationship to Oxtr expression and can be represented by a single latent methylation component. For visualization purposes only, we plotted trend lines from linear models predicting methylation from expression. Oxtr polymorphism impact on gene expression was analyzed using a 2-way handling condition x SNP ANOVA. Correlation was used to examine the relationship between Oxtr DNA methylation in central and peripheral prairie vole tissue. Normality was assessed by Kolmogorov-Smirnov (K-S) test. Spearman’s rank correlation was used when data were not normally distributed, whereas Pearson’s correlation was used when data were normally distributed. Oxtr methylation after experiencing naturally varying early parental care was analyzed with a nested ANOVA with a post-hoc Bonferroni multiple comparisons correction to account for the use of multiple offspring of each sex from a single breeding pair in the DNA methylation analysis. One animal from the medium contact group was removed due to pyrosequencing failure. The relationship between early care and oxytocin receptor protein levels in offspring was determined using Spearman’s rank correlation.
3. Results
3.1. Early handling increases parental care
Prairie voles reared under conditions in which they are left relatively undisturbed, with no direct handling, for the first week of life (MAN0), display marked disruptions later in life in alloparenting, pair bonding, and parenting, likely as a result of increased anxiety (Bales et al., 2007; Bales et al., 2011). In contrast, offspring that experience a brief direct handling manipulation (MAN1) on postnatal day (PND) 1, in which mother, father, and pups are lifted by an investigator’s gloved hand for 30 seconds, show later adult social behaviors typical of this species. In rats, brief early handling leads to similar changes in adult offspring behavior (Levine, 1957; Denenberg et al., 1962), likely because dams increase pup licking and grooming immediately after the handling episode (Smotherman et al., 1977; Boccia and Pedersen, 2001). Alternately, rats receiving little to no early life handling disturbance had increased anxiety-like behavior in adulthood (Levine, 2002). We observed maternal and paternal behaviors immediately following the MAN1 handling or in unmanipulated (MAN0) voles at comparable times on PND1. Total pup-directed behavior rather than a single behavior (as typically done in rats) was quantified to more accurately characterize the total biparental stimulation experienced by prairie vole pups in early life. MAN1 parents were more attentive to their pups (F (1, 20) = 7.24, p = 0.014) with higher amounts of pup directed care being provided by the mother (F (1, 20) = 13.76, p = 0.0014; Figure 2A). No differences in parental behavior were found on PND8 or PND15 (Supplemental Figure 1) indicating that the effects of manipulation are transient. The increase in care after direct (MAN1) handling may reflect conditions experienced by offspring in the field. During maternal foraging bouts, offspring are briefly separated from the mother. Upon her return to the nest, she typically licks and grooms offspring for a short time. MAN1 handling appears to mimic this condition. We hypothesized that, as in other models (Weaver et al., 2004), this transient increase in early care would lead to experience-based differences in DNA methylation impacting both gene and protein expression. We focused our investigation on Oxtr because the social behaviors influenced by early handling are heavily oxytocin-dependent.
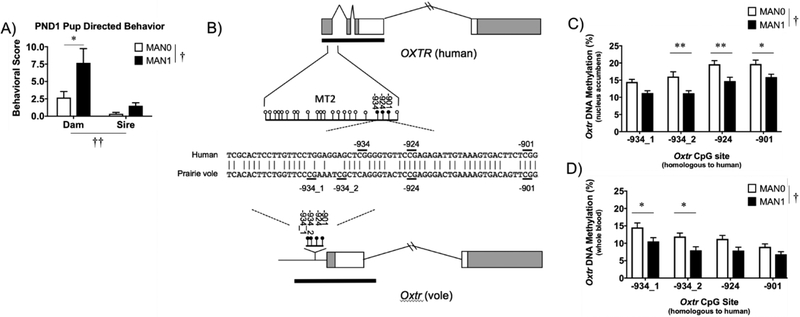
Figure 2. Early handling increases parental behavior directed toward offspring and alters DNA methylation in the brain and blood at conserved CpG sites in the promoter region of Oxtr in the prairie vole.
(A) Direct handling manipulation (MAN1) increases parental care of offspring on PND 1, specifically increasing maternal pup-directed behavior (2-way parent x handling condition ANOVA; MAN0, n=7; MAN1, n=6; † main effect of handling; †† main effect of parent; *p<0.05; error bars indicate standard error). (B) DNA methylation at CpG sites −934, −924, and −901 in OXTR MT2 region of the human OXTR are conserved in prairie vole Oxtr. Exons are displayed as boxes (coding, white; untranslated, grey) and introns are solid lines. CpG islands are indicated with black bars. Specific CpG dinucleotides analyzed here are indicated with sticks and circles, with closed circles indicating sites that exhibit significant association with expression. Partial alignment of conserved human and prairie vole gene sequence is indicated, conserved nucleotides are represented with a vertical line (|) and CpG sites are highlighted with a horizontal line (−). (C) Offspring receiving less maternal attention (MAN0) have higher levels of Oxtr DNA methylation in the nucleus accumbens and (D) in whole blood at the CpG sites conserved from the human OXTR (2-way CpG site x handling condition ANOVA; 13 male/female sibling pairs; MAN0: n=7 females, 7 males; MAN1: n=6 females, 6 males; † main effect of handling; Bonferroni correction, *p<0.05, **p<0.01; error bars indicate standard error).
3.2. Early handling alters DNA methylation of the oxytocin receptor in both brain and blood
In order to identify CpG sites in the prairie vole Oxtr that may be sensitive to early life experience and impact the expression of the gene, we used a region of the human OXTR promoter that contains a DNA methylation specific regulatory region (termed MT2, hg38: chr3:8,769,033–8,769,438) to probe the prairie vole genome (Kusui et al., 2001). First, we identified a 71 base pair alignment block in the UCSC genome browser (Kent et al., 2002) that contains a portion of the MT2 region in several rodent species including the prairie vole (Supplemental Figure 2). Next, we estimated statistical significance of the human and vole alignment by shuffling and identified significant homology (200 shuffles, z=239.9; bits=48.3; E(10000)=1.5X10−11) (Pearson, 2013). Remarkably, this region in the human contains four CpG sites (−959, −934, −924, −901) which when methylated are associated with decreased transcription in the human brain (Gregory et al., 2009) and variability in DNA methylation of one of these sites (−934) has been associated with several psychiatric disorders (24–28) as well as individual variability in the brain’s response to social perception (22, 23). Two of these CpG sites, −924 and −901 were conserved in the prairie vole and the third site, −934, may be functionally related to −934_1 and −934_2, (Figure 2B). For ease in highlighting CpG site conservation with the human sequence, we named the prairie vole CpG sites after the sites that have been previously reported in the human. Though conservation does not imply function, it is interesting to note that this CpG site conservation was not present in the reference genome of the laboratory rat and only one site, −924, was conserved in the mouse (Supplemental Figure 2). Identification of these conserved sites specifically in the prairie vole provides a starting point for DNA methylation analysis.
To examine the hypothesis that differential early care alters DNA methylation of Oxtr in the brain, we subjected prairie vole pups to the early handling manipulation paradigm described above. On PND24, brain tissue was obtained from the nucleus accumbens, a high OXTR expressing region that has been implicated in several social behaviors including pair bonding and alloparenting (13 male/female siblings; MAN0, n = 7 females, 7 males; MAN1, n = 6 females, 6 males). DNA methylation analysis was targeted to the conserved region containing CpG sites −934_1, −934_2, −924, and −901. Confirming our prediction, offspring receiving less early care (MAN0) had higher levels of Oxtr DNA methylation in the nucleus accumbens (main effect of handling, F (1, 96) =34.22, p = <0.0001; main effect of CpG site, F (3, 96) =34.22, p = <0.0001; no interaction; Bonferroni post hoc analysis, −934_1: p = 0.108; −934_2: p = 0.004; −924: p= 0.004; −901: p = 0.037; Figure 2C). A higher level of Oxtr DNA methylation in MAN0 compared to MAN1 offspring was also observed in whole blood of the same animals (main effect of handling, F (1, 96) =22.03, p<0.0001; main effect of CpG site, F (3, 96) =7.16, p=0.0002; no interaction; Bonferroni post hoc analysis, −934_1: p = 0.025; −934_2: p = 0.027; −924: p= 0.092; −901: p = 0.546; Figure 2D; Supplemental Table 1). Sex differences were not observed for either methylation analysis. The higher levels of DNA methylation found in the nucleus accumbens of MAN0 offspring indicate experience sensitive differences in the epigenetic state of Oxtr in a brain region important in the control of social attachment. That the same pattern of DNA methylation after early handling was found in the whole blood of offspring suggests that blood DNA may be used as a biomarker of this change.
3.3. Heightened early care prevents de novo methylation of Oxtr
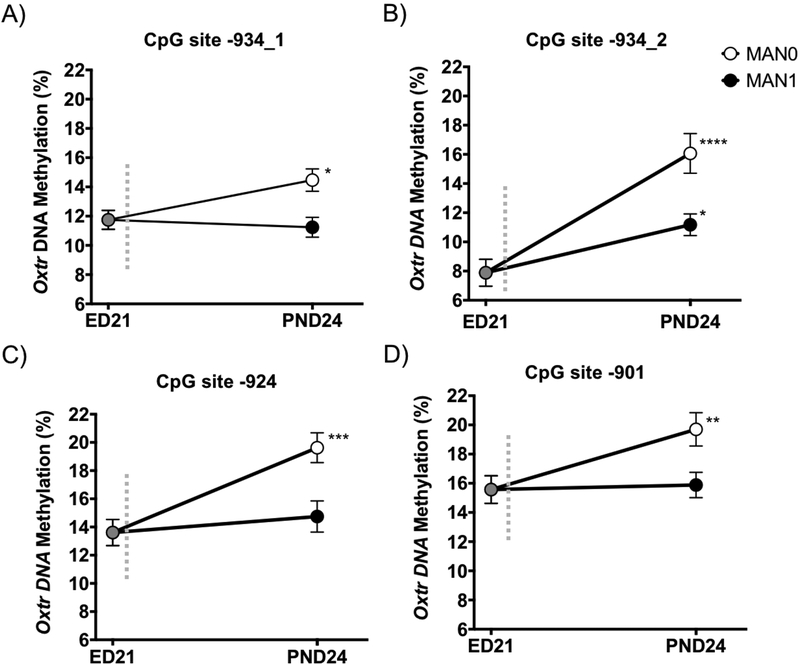
It was not clear if high amounts of early care, induced here by postnatal handling, act to decrease methylation or if less early care leads to de novo DNA methylation. To determine this, we collected the forebrain (which contains the nucleus accumbens) from embryonic day (ED) 21 offspring dissected from the uterus on the expected day of birth and prior to the receipt of any parental care to compare Oxtr DNA methylation levels in these ED21 offspring to levels observed in PND24 offspring in the above handling experiment. Compared to ED21 offspring, animals that received less early care (MAN0) had significant increases in DNA methylation across all of the conserved CpG sites (−934_1: t(30)=2.72, p=0.011; −934_2: t(30)=5.14, p<0.0001; −924: t(30)=4.27, p=0.0002; −901: t(30)=2.80, p=0.009), while methylation levels of MAN1 offspring that received high early care tended to not differ from ED21 offspring (−934_1: t(28)=0.524, p=0.605; −934_2: t(28)=2.56, p=0.016; −924: t(28)=0.78, p=0.444; −901: t(28)=0.23, p=0.821, Figure 3). High amounts of early care, then, prevent de novo DNA methylation of Oxtr.
Figure 3. Early handling prevents de novo DNA methylation of Oxtr.
Oxtr DNA methylation levels at CpG sites (A) −934_1, (B) −934_2, (C) −924, and (D) −901 in the forebrain of ED21 offspring (n=18, 9 male/female sibling pairs) are similar to MAN1 offspring that received increased early parental care, whereas MAN0 offspring that received low amounts of early care have a significant increase in postnatal de novo DNA methylation in the nucleus accumbens (unpaired two-tailed t test; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; error bars indicate standard error).
3.4. Heightened early care increases Oxtr gene expression
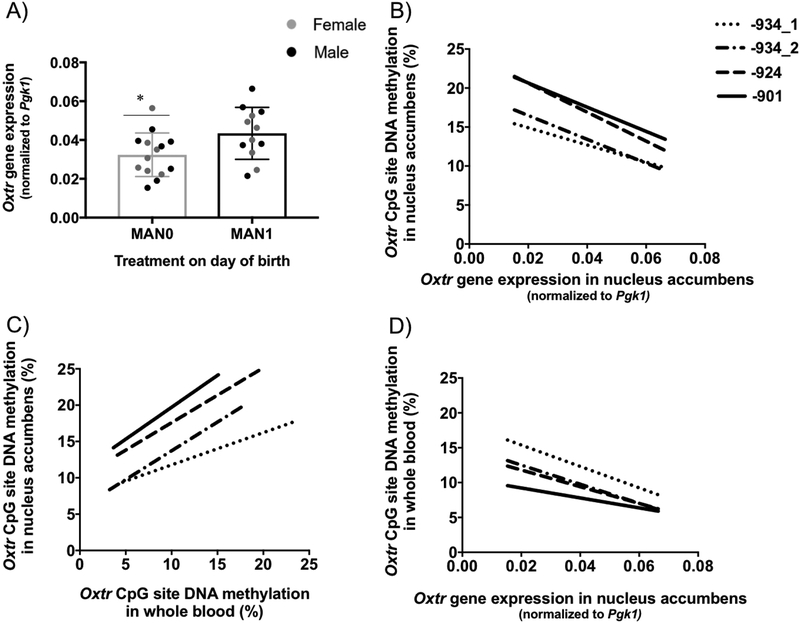
To establish that differences in the amount of early care lead to changes in gene expression in our model, we measured Oxtr gene expression in the nucleus accumbens of the same PND 24 animals. Offspring receiving less early care and also displaying increased levels of Oxtr DNA methylation (MAN0) exhibited decreased Oxtr gene expression compared to MAN1 animals (t(24)=2.28, p=0.032, Figure 4A). We then explored how Oxtr DNA methylation within the four assayed CpG sites correlated with Oxtr expression. Using a Partial Least Squares (PLS) regression, a 1-component model emerged as optimal, indicating that all four CpG sites show a similar relationship to Oxtr expression and can be represented by a single latent methylation component. This 1-component model was capable of explaining 79.07% of the variance in Oxtr DNA methylation and 26.27% of the variance in Oxtr expression in the nucleus accumbens. All methylation sites significantly negatively loaded onto the component, indicating a significant negative association between Oxtr DNA methylation and Oxtr expression in this region (Table 1). Trend lines plotted to visualize the data support conclusions of a negative relationship between Oxtr DNA methylation and Oxtr expression in the nucleus accumbens (Figure 4B). Sex differences were not observed, nor was there a relationship with a previously associated Oxtr polymorphism (King et al., 2016) (Supplemental Figure 3). These data suggest that methylation at these CpG sites may be relevant in regulating transcription of Oxtr and also provide a mechanism through which individual differences in social behavior may develop following differences in early care.
Figure 4. Experience sensitive de novo DNA methylation of Oxtr decreases gene expression in the brain, is reflected in the blood and can be used as a biomarker of the transcription state of the nucleus accumbens.
Offspring receiving less maternal attention (MAN0) (A) have decreased Oxtr gene expression in the nucleus accumbens (unpaired two-tailed t test). (B) Oxtr DNA methylation is negatively correlated with Oxtr gene expression in the nucleus accumbens (Pearson’s r). (C) DNA methylation in whole blood is positively correlated with DNA methylation levels in the nucleus accumbens and (D) negatively correlated to Oxtr gene expression in the nucleus accumbens (Pearson’s r). Trend lines are plotted predicting methylation from expression for data visualization purposes only (n=26, 13 male/female sibling pairs; MAN0=14, MAN1=12; unpaired, two tailed t-test, *p<0.05; error bars indicate standard error).
Table 1. Oxtr DNA methylation in the nucleus accumbens and in whole blood is negatively associated with Oxtr gene expression in the nucleus accumbens.
All methylation sites significantly negatively load onto the single CpG site component, indicating a significant negative association between Oxtr methylation and Oxtr expression in Nacc. In a separate analysis, all methylation sites significantly negatively load onto the single CpG site component, indicating a significant negative association between Oxtr methylation in whole blood and Oxtr expression in Nacc. Loadings, regression coefficients, and jackknife approximate t tests of regression coefficients for each model are listed in Table 1.
| Model | Site | Loading | Estimate | Std. Error | Df | t value | p value |
|---|---|---|---|---|---|---|---|
| Nucleus Accumbens | 934_1 | −0.364 | −3.10E-04 | 1.09E-04 | 25 | −2.84 | 0.009 |
| 934_2 | −0.567 | −4.29E-04 | 1.67E-04 | 25 | −2.57 | 0.017 | |
| 924 | −0.56 | −5.22E-04 | 1.62E-04 | 25 | −3.22 | 0.004 | |
| 901 | −0.489 | −4.36E-04 | 8.89E-05 | 25 | −4.91 | <.0001 | |
| Whole Blood | 934_1 | −0.599 | −4.52E-04 | 1.71E-04 | 25 | −2.64 | 0.014 |
| 934_2 | −0.515 | −4.04E-04 | 1.61E-04 | 25 | −2.52 | 0.019 | |
| 924 | −0.499 | −3.53E-04 | 1.45E-04 | 25 | −2.43 | 0.022 | |
| 901 | −0.367 | −2.10E-04 | 1.04E-04 | 25 | −2.03 | 0.053 |
3.5. Peripheral measures of DNA methylation predict central levels
Data in humans suggest a positive association between levels of OXTR DNA methylation in central and peripheral tissues (Gregory et al., 2009). Here we found a strong positive correlation between Oxtr DNA methylation in whole blood and the nucleus accumbens at PND 24 at all four conserved CpG sites (−934_1: r(26) = 0.684, p= 0.0001; −934_2: r(26) = 0.677, p= 0.0001; −924: r(26) = 0.650, p= 0.0003; −901: r(26) = 0.654, p= 0.0003, Figure 4C). The methylation state of the blood was also associated with the level of transcription in the brain at three of the four CpG sites. Using PLS regression, the 1-component model for whole blood was capable of explaining 94.92% of the variance in Oxtr DNA methylation and 18.20% of the variance in Oxtr expression. All methylation sites significantly negatively loaded onto the component, indicating a significant negative association between whole blood Oxtr DNA methylation and Oxtr expression in the nucleus accumbens (Table 1). Trend lines plotted to visualize the data support conclusions of a negative relationship between Oxtr DNA methylation in whole blood and Oxtr expression in the nucleus accumbens (Figure 4D). Peripheral measures of Oxtr DNA methylation, therefore, provide useful information on central epigenetic markers, at least in the nucleus accumbens.
3.6. Naturally varying early care alters DNA methylation
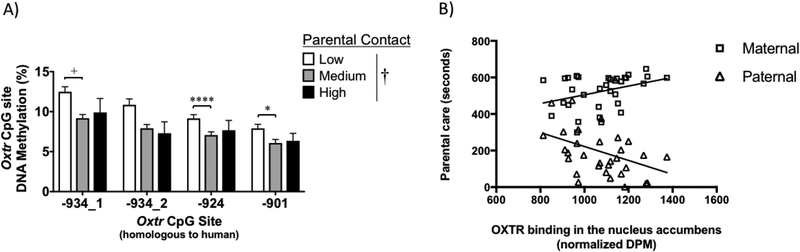
To validate findings from the handling model, we assessed naturally occurring variation in early care in prairie voles and Oxtr DNA methylation in the blood since variation in early care impacts adult social behavior (Perkeybile et al., 2013; Perkeybile et al., 2015; del Razo and Bales, 2016). Whole blood was collected from low contact (LC; n = 49), medium contact (MC; n = 90), and high contact (HC; n = 16) offspring in young adulthood (PND48–52) and assessed for DNA methylation at the conserved CpG sites. LC offspring who received the lowest levels of early care had increased levels of Oxtr DNA methylation at two of the four conserved CpG sites and trended toward increased DNA methylation at a third CpG site (main effect of parental contact, F (2, 604) =22.05, p <0.0001; main effect of CpG site, F (3, 604) =12.34, p <0.0001; no interaction; Bonferroni post hoc analysis, −934_1: p = 0.0605; −934_2: p = 0.1227; −924: p < 0.0001; −901: p = 0.0376; Figure 5A) compared to MC offspring. These results support findings of the impact of early handling, where MAN0 offspring experienced lower amounts of early care and had increased Oxtr DNA methylation levels. This offers further support for the hypothesis that the epigenetic modifications seen here are a result of the amount of total care received in the early postnatal period and that the blood can be used as a biomarker of this change.
Figure 5. Offspring reared by parents who exhibit naturally low levels of parental care exhibit increased Oxtr DNA methylation in the blood and decreased OXTR distribution in the nucleus accumbens.
(A) Offspring of low contact parents (n = 49) have higher levels of Oxtr DNA methylation in whole blood compared to offspring of medium (n = 89) or high (n = 16) contact parents (2-way CpG site x parenting ANOVA; † main effect of handling; *p<0.05, ****p<0.0001, +p = 0.06). (B) High levels of early maternal care and low amounts of early paternal care are associated with increased OXTR binding in the nucleus accumbens of adolescent offspring (Spearman’s rho; n = 32).
3.7. Heightened early care increases OXTR protein
For variation in early care to impact later life social behavior and attachment in offspring through pathways involving oxytocin, the changes seen in Oxtr DNA methylation and gene expression need to correspond to changes in receptor protein in the same region. We tested this using autoradiography methods to visualize OXTR in the nucleus accumbens in an additional cohort of PND 24–26 (n = 32) animals following naturally varying early care. Increased binding of OXTR in this region was associated with increased early maternal care (p(32) = 0.388, p = 0.028) and with decreased early paternal care (p(32) = −0.4263, p = 0.015, Figure 5B). Maternal care was also inversely related to paternal care (Supplemental Figure 4), which matches previous data from our lab (Perkeybile et al., 2013) as well as data from parental care after early handling presented above. That lower early maternal care is associated with low OXTR protein binding in the nucleus accumbens provides additional support that differences in these epigenetic markers are altering the function of oxytocin pathways via changes at the receptor level.
4. Discussion
Our findings provide strong evidence that early parental care directly impacts functioning of oxytocin pathways in the prairie vole through altering DNA methylation of Oxtr at four CpG sites conserved from the human sequence. This highlights for the first time a mechanism through which early experience can create individual differences in oxytocin-dependent behaviors. Whether naturally occurring or experimentally induced via reduced early handling, decreased early care in the prairie vole leads to decreased alloparenting in adolescence and adulthood (Bales et al., 2007; Bales et al., 2011; Perkeybile et al., 2013; del Razo and Bales, 2016) and these effects are passed to a subsequent generation through nongenomic transmission (Stone and Bales, 2010; Perkeybile et al., 2015). Alloparental behavior in prairie voles is regulated by oxytocin receptors in the nucleus accumbens (Olazabal and Young, 2006b, a; Kenkel et al., 2017); increased oxytocin receptor density in this region is positively correlated with alloparenting within individual prairie voles and between vole species. Similarly, formation of pair bonds, a behavior that is also linked to distribution and density of oxytocin receptors (Insel and Shapiro, 1992; Young, 1999; Ross et al., 2009; Ophir et al., 2012; Keebaugh et al., 2015; Johnson et al., 2016), is reduced following decreased early care (Bales et al., 2007; del Razo and Bales, 2016). That we find increased Oxtr DNA methylation in two models of decreased early parental care, then, helps to explain previously seen decreases in alloparenting and pair bonding. Higher levels of Oxtr DNA methylation work to decrease Oxtr gene expression, theoretically resulting in decreased oxytocin receptor density. Our results support this, with low levels of early parenting being associated with both increased Oxtr DNA methylation and decreased OXTR receptor density in the nucleus accumbens. We hypothesize that early care alters OXTR in this region by altering DNA methylation patterns. This lower receptor density would then work to decrease expression of species-typical alloparenting and pair bonding. This provides a mechanism by which early experiences can alter an organism’s developmental trajectory and create individual differences in social behavior and adds to the growing literature on the role epigenetic markers play in shaping variation in behavior.
Site-specific analyses in humans have revealed associations between CpG DNA methylation levels and both psychiatric diagnosis and neural functioning in neurotypical populations. Increased levels of OXTR methylation at CpG site −934 measured in blood have been linked to postpartum depression (Bell et al., 2015), autism (Gregory et al., 2009), increased neural activity in emotion processing regions (Puglia et al., 2015), and increased activity in brain regions responsible for social perception in response to socially ambiguous stimuli (Jack et al., 2012). Our work here demonstrates the need for further investigation of the impacts of the early life environment on OXTR DNA methylation in both neurotypical and atypical individuals. This focus will allow us to gain a better understanding of the consequences of early life experience on oxytocin system regulation and its link to psychiatric health and well-being.
Early life experiences, particularly those occurring within the parent-infant dyad, have long been known to alter the developmental trajectory of offspring. This has now been demonstrated with regard to epigenetic regulation of OXTR. In humans, low amounts of childhood maternal care are associated with increased levels of OXTR DNA methylation in adults (Unternaehrer et al., 2015) and childhood abuse interacts with OXTR DNA methylation to predict depression and anxiety (Smearman et al., 2016). Our results support these findings, where we see higher levels of CpG site-specific DNA methylation at sites that are associated with psychiatric disorder in humans. Therefore, future work investigating these specific CpG sites in humans following varying early life experience would be useful to support our and others’ previous findings. This may help to uncover new pathways via which early life neglect influences late mental health outcomes and allow for the development of new interventions to counteract these outcomes.
The strong relationship shown here between central and peripheral Oxtr DNA methylation levels verifies the usefulness of peripherally obtained DNA as we seek to understand epigenetic mechanisms regulating social behavior. As more research aims to understand the role of epigenetic markers on a range of human outcomes, it will be necessary to have access to biological samples that can serve as a useful proxy for central tissue. Our results indicate easily accessible blood samples may be a useful tool for this. The ability to use peripheral sampling techniques also opens the door for lifespan development animal studies with multiple sampling time points in a single animal that previously were not possible.
Our findings provide strong evidence for a mechanism by which early experiences can alter an organism’s developmental trajectory and create individual differences in social behavior and adds to the growing literature on the role epigenetic markers play in shaping variation in behavior. Future work should combine detailed behavioral measures with analyses of epigenetic markers to establish a direct link between the two outcomes following varying early experiences. As more research aims to understand the role of epigenetic markers on a range of human outcomes, it will be vital to have a comparable animal model. Our findings suggest the prairie vole will be useful in this regard, allowing for an expanded understanding of the role of epigenetic markers in controlling oxytocin pathways and impacting complex social behavior that can then inform and guide work on human conditions.
Supplementary Material
Highlights:
4 CpG sites in the Oxtr promoter region are homologous in humans and prairie voles
Low levels of early parenting lead to high Oxtr DNA methylation in brain and blood
High Oxtr methylation is associated with low Oxtr gene expression
Low levels of early care lead to low OXTR density, likely via high Oxtr methylation
Peripheral measures of Oxtr DNA methylation predict central levels
Acknowledgements
The authors thank Amber Tyler for data collection assistance, Joshua Danoff and Drs. Jason Yee and Ben Ragen for helpful comments on drafts of this manuscript, and the animal facility staffs at University of Illinois, Chicago; University of California, Davis; and Northeastern University for animal care. This research was supported by Autism Speaks grant #7110 to J.J.C. and C.S.C., NIH grant HD075750 to C.S.C. and J.J.C., National Alliance for Autism Research grant to C.S.C., NIH grant MH073022 to C.S.C. and K.L.B., NIH grant HD060117 to K.L.B., and NSF grant 0437523 to K.L.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no competing financial interests.
References Cited
- 1.Bales KL, Lewis-Reese AD, Pfeifer LA, Kramer KM, Carter CS (2007) Early experience affects the traits of monogamy in a sexually dimorphic manner. Developmental Psychobiology 49:335–342. [DOI] [PubMed] [Google Scholar]
- 2.Bales KL, Boone E, Epperson P, Hoffman G, Carter CS (2011) Are behavioral effects of early experience mediated by oxytocin? Front Psychiatry 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell AF, Carter CS, Steer CD, Golding J, Davis JM, Steffen AD, Rubin LH, Lillard TS, Gregory SP, Harris JC, Connelly JJ (2015) Interaction between oxytocin and receptor DNA methylation and genotype is associated with risk of postpartum depression in women without depression in pregnancy. Frontiers in Genetics 6:243-Article No.: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, al e (2004) Aligning multiple genomic sequences with the threaded blockset aligner. Genome Research 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boccia ML, Pedersen CA (2001) Brief vs. long maternal separations in infancy: contrasting relationships with adult maternal behavior and lactation levels of aggression and anxiety. Psychoneuroendocrinology 26:657–672. [DOI] [PubMed] [Google Scholar]
- 6.Boda E, Pini A, Hoxha E, Parolisi R, Tempia F (2009) Selection of Reference Genes for Quantitative Real-time RT-PCR Studies in Mouse Brain. Journal of Molecular Neuroscience 37:238–253. [DOI] [PubMed] [Google Scholar]
- 7.Bowlby J (1969) Attachment and Loss. New York: Basic Books, Inc. [Google Scholar]
- 8.del Razo RA, Bales KL (2016) Exploration in a dispersal task: Effects of early experience and correlation with other behaviors in prairie voles (Microtus ochrogaster). Behavioural Processes 132:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denenberg VH, Ottinger DR, Stephens MW (1962) Effects of maternal factors upon growth and behavior of the rat. Child Development 33:65–71. [DOI] [PubMed] [Google Scholar]
- 10.Francis D, Diorio J, Liu D, Meaney MJ (1999) Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science 286:1155–1158. [DOI] [PubMed] [Google Scholar]
- 11.Gimpl G, Fahrenholz F (2001) The Oxytocin Receptor System: Structure, function, and regulation. Physiol Rev 81:629–683. [DOI] [PubMed] [Google Scholar]
- 12.Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, Lintas C, Abramson RK, Wright HH, Ellis P, Langford CF, Worley G, Delong GR, Murphy SK, Cuccaro ML, Persico A, Pericak-Vance MA (2009) Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. Bmc Medicine 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow HF, Dodsworth RO, Harlow MK (1965) Total social isolation in monkeys. Proc Natl Acad Sci U S A 54:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Insel TR, Shapiro LE (1992) Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Proceedings of the National Academy of Sciences of the United States of America 89:5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack A, Connelly JJ, Morris JP (2012) DNA methylation of the oxytocin receptor gene predicts neural response to ambiguous social stimuli. Frontiers in Human Neuroscience 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson ZV, Walum H, Jamal YA, Xiao Y, Keebaugh AC, Inoue K, Young LJ (2016) Central oxytocin receptors mediate mating-induced partner preferences and enhance correlated activation across forebrain nuclei in male prairie voles. Hormones and Behavior 79:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keebaugh AC, Young LJ (2011) Increasing oxytocin receptor expression in the nucleus accumbens of pre-pubertal female prairie voles enhances alloparental responsiveness and partner preference formation as adults. Hormones and Behavior 60:498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, Young LJ (2015) RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibits social attachment and parental care in monogamous female prairie voles. Soc Neurosci 10:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenkel WM, Perkeybile AM, Carter CS (2017) The neurobiological causes and effects of alloparenting. Developmental Neurobiology 77:214–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Research 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King LB, Walum H, Inoue K, Eyrich NW, Young LJ (2016) Variation in the Oxytocin Receptor Gene Predicts Brain Region-Specific Expression and Social Attachment. Biological Psychiatry 80:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusui C, Kimura T, Ogita K, Nakamura H, Matsumura Y, Koyama M, Azuma C, Murata Y (2001) DNA methylation of the human oxytocin receptor gene promoter regulates tissue-specific gene suppression. Biochemical and biophysical research communications 289:681–686. [DOI] [PubMed] [Google Scholar]
- 23.Levine S (1957) Infantile experience and resistance to physiological stress. Science 126:405–405. [DOI] [PubMed] [Google Scholar]
- 24.Levine S (2002) Enduring effects of early experience on adult behavior In: Hormones, Brain, and Behavior (Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, eds), pp 535–542. New York: Academic Press. [Google Scholar]
- 25.Levine S, Haltmeyer GC, Kargs GG, Denenberg VH (1967) Physiological and behavioral effects of infantile stimulation. Physiol Behav 2:5. [Google Scholar]
- 26.Liu Y, Wang ZX (2003) Nucleus accumbens oxytocin and dopamine interact to regulate pair bond formation in female prairie voles. Neuroscience 121:537–544. [DOI] [PubMed] [Google Scholar]
- 27.Mevik BH, Wehrens R, Liland KH (2016) PLS: Partial Least Squares and Principal Component Regression. In: R package, 2.6–0 Edition. [Google Scholar]
- 28.Olazabal DE, Young LJ (2006a) Oxytocin receptors in the nucleus accumbens facilitate “spontaneous” maternal behavior in adult female prairie voles. Neuroscience 141:559–568. [DOI] [PubMed] [Google Scholar]
- 29.Olazabal DE, Young LJ (2006b) Species and individual differences in juvenile female alloparental care are associated with oxytocin receptor density in the striatum and the lateral septum. Hormones and Behavior 49:681–687. [DOI] [PubMed] [Google Scholar]
- 30.Ophir AG, Gessel A, Zheng DJ, Phelps SM (2012) Oxytocin receptor density is associated with male mating tactics and social monogamy. Hormones and Behavior 61:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson WR (2013) An introduction to sequence similarity (“homology”) searching. Current Protocols in Bioinformatics 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkeybile AM, Bales KL (2015) Early rearing experience is related to altered aggression and vasopressin production following chronic social isolation in the prairie vole. Behav Brain Res 283:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkeybile AM, Griffin LL, Bales KL (2013) Natural variation in early parental care correlates with social behaviors in adolescent prairie voles (Microtus ochrogaster). Front Behav Neurosci 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkeybile AM, Delaney-Busch N, Hartman S, Grimm KJ, Bales KL (2015) Intergenerational transmission of alloparental behavior and oxytocin and vasopressin receptor distribution in the prairie vole. Front Behav Neurosci 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puglia MH, Lillard TS, Morris JP, Connelly JJ (2015) Epigenetic modification of the oxytocin receptor gene influences the perception of anger and fear in the human brain. Proceedings of the National Academy of Sciences of the United States of America 112:3308–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross HE, Freeman SM, Spiegel LL, Ren XH, Terwilliger EF, Young LJ (2009) Variation in oxytocin receptor density in the nucleus accumbens has differential effects on affiliative behaviors in monogamous and polygamous voles. Journal of Neuroscience 29:1312–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smearman EL, Almli LM, Conneely KN, Brody GH, Sales JM, Bradley B, Ressler KJ, Smith AK (2016) Oxytocin Receptor Genetic and Epigenetic Variations: Association With Child Abuse and Adult Psychiatric Symptoms. Child Development 87:122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smotherman WP, Brown CP, Levine S (1977) Maternal responsiveness following differential pup treatment and mother-pup interactions. Horm Behav 8:242–253. [DOI] [PubMed] [Google Scholar]
- 39.Stone AI, Bales KL (2010) Intergenerational transmission of the behavioral consequences of early experience in prairie voles. Behavioural Processes 84:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Team RC (2017) R: A language and environment for statistical computing. In. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 41.Unternaehrer E, Meyer AH, Burkhardt SCA, Dempster E, Staehli S, Theill N, Lieb R, Meinlschmidt G (2015) Childhood maternal care is associated with DNA methylation of the genes for brain-derived neurotrophic factor (BDNF) and oxytocin receptor (OXTR) in peripheral blood cells in adult men and women. Stress-the International Journal on the Biology of Stress 18:451–461. [DOI] [PubMed] [Google Scholar]
- 42.Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl Jr., Dymov S, Szyf M, Meaney MJ (2004) Epigenetic programming by maternal behavior. Nat Neurosci 7:847–854. [DOI] [PubMed] [Google Scholar]
- 43.Young LJ (1999) Frank A. Beach Award. Oxytocin and vasopressin receptors and species-typical social behaviors. Horm Behav 36:212–221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.