Abstract
Background
Numerous articles have reported that abnormal expression levels of microRNAs (miRNAs) are related to the survival times of esophageal carcinoma (EC) patients, which contains esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). Nevertheless, there has not been a comprehensive meta-analysis to assess the accurate prognostic value of miRNAs in EC.
Methods
Studies published in English up to April 12, 2018 that evaluated the correlation of the expression levels of miRNAs with overall survival (OS) in EC were identified by online searches in PubMed, EMBASE, Web of Science, and the Cochrane Database of Systematic Reviews performed by two independent authors. The pooled hazard ratios (HRs) with 95% confidence intervals (CIs) were used to estimate the correlation between OS and miRNA expression. HR ≥ 2 was considered cutoff for considering the miRNA as prognostic candidate.
Results
Forty-four pertinent articles with 22 miRNAs and 4310 EC patients were ultimately included. EC patients with tissue expression levels of high miR-21 or low miR-133a (HR = 2.48, 95% CI = 1.50–4.12), miR-133b (HR = 2.15, 95% CI = 1.27–3.62), miR-138 (HR = 2.27, 95% CI = 1.68–3.08), miR-203 (HR = 2.83, 95% CI = 1.35–5.95), miR-375 and miR-655 (HR = 2.66, 95% CI = 1.16–6.12) had significantly poorer OS (P < 0.05). In addition, EC patients with blood expression levels of high miR-21 (HR = 2.19, 95% CI = 1.31–3.68) and miR-223 had significantly shorter OS (P < 0.05).
Conclusions
In conclusion, tissue expression levels of miR-21, miR-133a, miR-133b, miR-138, miR-203, miR-375, and miR-655 and blood expression levels of miR-21 and miR-223 demonstrate significant prognostic value. Among them, the expression levels of miR-133a, miR-133b, miR-138, miR-203, and miR-655 in tissue and the expression level of miR-21 in blood are potential prognostic candidates for predicting OS in EC.
Introduction
During the past 10 years, a substantial number of articles have reported the survival of esophageal carcinoma (EC) patients with dysregulated microRNA (miRNA) expression1–96. As the twelfth origin of incident cases and the seventh major cause of cancer-related death all over the world97, it contains two main types: esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). In the United States, EAC nowadays occupies about 7% of all EC cases. ESCC is the main subtype of EC among Asian patients. Although the treatment and prognosis of EC have been improved by multimodal therapies, the rate of 5-year overall survival (OS) remains poor98.
It is well known that EC is a complex inherited disease that is characterized by altered expression levels of certain coding or non-coding genes. As the high-throughput analysis develops, an increasing number of cancer-related non-coding RNAs have been recognized99. miRNAs, a class of small non-coding RNAs <25 nucleotides in length, act as negative regulatory factors of gene expression by depressing translation or causing deadenylation-dependent degradation of target messenger RNAs (mRNAs)100. They have been shown to be involved in various processes of tumor progression, including proliferation and metastasis of cancer cells101. In particular, EC-related miRNAs have been proved to exert functional diversity via multiple biological processes.
Despite comprehensive research aimed at illuminating the molecular mechanisms in EC, there are still challenges facing the identification of prognostic biomarkers that are minimally invasive and sensitive. Therefore, it is crucial to develop prognostic cancer biomarkers that can be expediently and reliably applied in the clinical setting, improving the survival of EC patients.
Recently, an increasing amount of evidence indicates that miRNAs can act as possible biomarkers for cancer prognosis in clinical practice that is fairly encouraging and exciting1–96. However, there has not been a systematic review and meta-analysis to estimate the associations between miRNA expression and the survival of EC patients. Therefore, the current study aimed to identify that correlation by searching the recently published evidence regarding miRNAs as prognostic tools for EC in cancer tissues and in blood.
Materials and methods
Search strategy
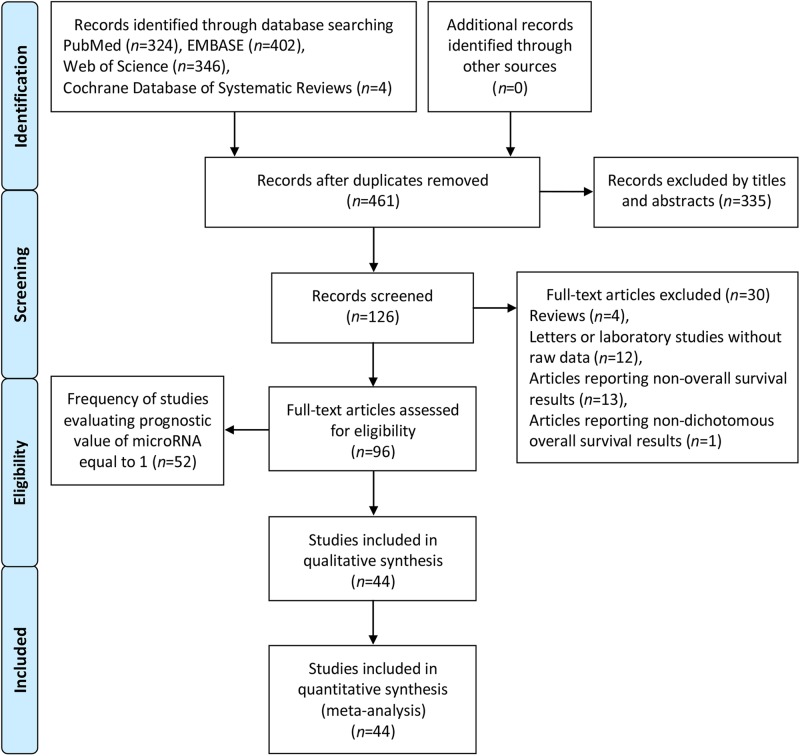
The comprehensive online search about articles from four databases, PubMed, Web of Science, Embase, and Cochrane Database of Systematic Reviews, was performed by two independent authors (S.G. and Z.-Y.Z.). Subsequently, Y.Z. re-evaluated uncertain data. A comprehensive search was conducted employing the subject terms: “microRNA,” “miRNA,” “miR”, and “esophageal carcinoma,” “esophagus carcinoma,” “oesophageal carcinoma.” Of the four databases, there were 461 records after duplicates were removed. Subsequently, we excluded 335 records by titles and abstracts. For the remaining 126 records, 30 full-text articles were excluded. The details are shown in Fig. 1. The search deadline was April 12, 2018.
Fig. 1.
Flow diagram of the literature search and selection
Inclusion criteria
We came up with inclusion criteria for qualified articles that were analyzed by our full-text estimation: (1) articles concerning the pertinence between miRNA level and prognosis of EC patients; (2) the survival results that were estimated by OS; (3) full-text articles published in English.
Exclusion criteria
Articles that were not satisfied with the aforementioned inclusion criteria, reviews, letters, or laboratory studies without raw data (Kaplan–Meier survival curves or HR with 95% CI) were excluded. Articles of non-dichotomous miRNA expression levels and frequency of studies assessing prognostic value of miRNAs equal to 1 were also excluded. If more than one paper had been published on the identical study cohort, only the most well-rounded investigation was selected for the current study. Besides, if both of the univariate and multivariate outcomes were reported, only the latter were chosen, since they were adjusted for confounding factors.
Quality assessment
S.G. and Z.-Y.Z. confirmed all eligible investigations that analyzed the prognostic value of miRNAs in EC, and Y.Z. reassessed uncertain data. Quality assessment for each study was done using modified Newcastle–Ottawa Scale (NOS)102. NOS scores were calculated on the basis of selection, comparability, and outcome. Papers with NOS scores ≥6 were considered high-quality articles103.
Study selection
A flow diagram with details of the study selection process was presented in Fig. 1.
Study frequency
The frequency of studies estimating the prognostic value of miRNAs in EC was shown in Supplementary Tables 1 (tissue) and 2 (blood), including the miRNA names, the frequency of studied miRNAs, and the references. In addition, frequency of strong miRNAs is shown in Table 1.
Table 1.
Frequency of strong microRNAs studied in esophageal carcinoma
| Tissue | Blood | ||||
|---|---|---|---|---|---|
| miR | F | R | miR | F | R |
| 133a | 2 | 32.33 | 21 | 2 | 87.89 |
| 133b | 2 | 13.34 | |||
| 138 | 2 | 35.36 | |||
| 203 | 2 | 49.50 | |||
| 655 | 2 | 75.76 | |||
F frequency of the studied microRNAs, R reference
Study characteristics
The basic information of the included articles is comprehensively detailed in Supplementary Table 3. If the data were not provided in the text but only as Kaplan–Meier survival curves, the data were extracted from the graphical survival plots, and estimations of the HR with 95% CI were then performed using a previously described method104 with the software Engauge Digitizer version 4.1.
Statistical analysis
All analyses were conducted using Stata version 13.0 (StataCorp, College Station, TX, USA). OS was the main and only reference standard for prognostic value of miRNAs. The HR was considered significant at the P < 0.05 level if the 95% CI did not include the value 1. In addition, a single miRNA was regarded as the strong candidate if its HR was ≥2. Owing to different types of samples (formalin-fixed, paraffin-embedded, frozen tissue, plasma, and serum) from EC patients at different stages, cutoff values, and miRNA methods in individual studies, random-effects models were more appropriate than fixed-effects models for most of the analyses. Accordingly, the former were employed in the current meta-analysis. Publication bias was estimated using Begg’s funnel plot. A two-tailed P value <0.05 was considered significant. A sensitivity analysis (influence analysis) was carried out to test how sensitive the combined effect size was to the removal of individual investigations. If the point assessment was outside of the 95% CI of the pooled effect size after it was removed from the analysis, an individual study was considered to have excessive influence.
Results
Meta-analysis
A summary of the HR with 95% CI evaluated from the whole combined analysis for all the miRNAs is shown in Table 2. The forest plots, Begg’s funnel plots, and sensitivity analyses are shown in Supplementary Figures 1–8 according to the logic sequencing of miRNA names. For the included 96 studies, 52 were excluded because the frequency of them evaluating prognostic value of miRNA was equal to 13,4,16,18–23,26,30,31,39–46,48,53,54,56,60,61,64–74,77–86,92–96. In addition, although one article reported OS results about miR-193a-5p, it was excluded because it had non-dichotomous miRNA expression value105. The mean NOS score of the included researches was 6.5 (4.0–8.0), indicating that the quality of them was adequate (Supplementary Table 4).
Table 2.
HR with 95% CI of microRNA expression in esophageal carcinoma
| Sample | MicroRNA | N | Included articles | HR | 95% CI | Figure | P value | Heterogeneity (Higgins I2 statistic) | Total patients |
|---|---|---|---|---|---|---|---|---|---|
| Tissue | Low let-7g | 2 | 1, 2 | 1.27 | 0.66–2.45 | Supplementary Figure 1 | 0.47 | I2 = 58.6%, P = 0.12 | 197 |
| Tissue | High miR-9 | 2 | 2, 5 | 1.07 | 0.45–2.57 | Supplementary Figure 1 | 0.88 | I2 = 72.8%, P = 0.06 | 342 |
| Tissue | High miR-21 | 10 | 1, 2, 7– 14 | 1.63 | 1.26–2.11 | Supplementary Figure 1 | <0.01 | I2 = 23.8%, P = 0.22 | 1071 |
| Tissue | High miR-26a | 2 | 15, 17 | 1.09 | 0.19–6.39 | Supplementary Figure 2 | 0.92 | I2 = 47.5%, P = 0.17 | 116 |
| Tissue | Low miR-34a | 2 | 2, 24 | 1.87 | 0.88–3.99 | Supplementary Figure 2 | 0.11 | I2 = 45.4%, P = 0.18 | 210 |
| Tissue | High miR-92a | 2 | 6, 25 | 1.47 | 0.64–3.34 | Supplementary Figure 2 | 0.36 | I2 = 54.4%, P = 0.14 | 170 |
| Tissue | Low miR-100 | 4 | 13, 27– 29 | 2.12 | 0.86–5.21 | Supplementary Figure 2 | 0.10 | I2 = 73.2%, P = 0.01 | 410 |
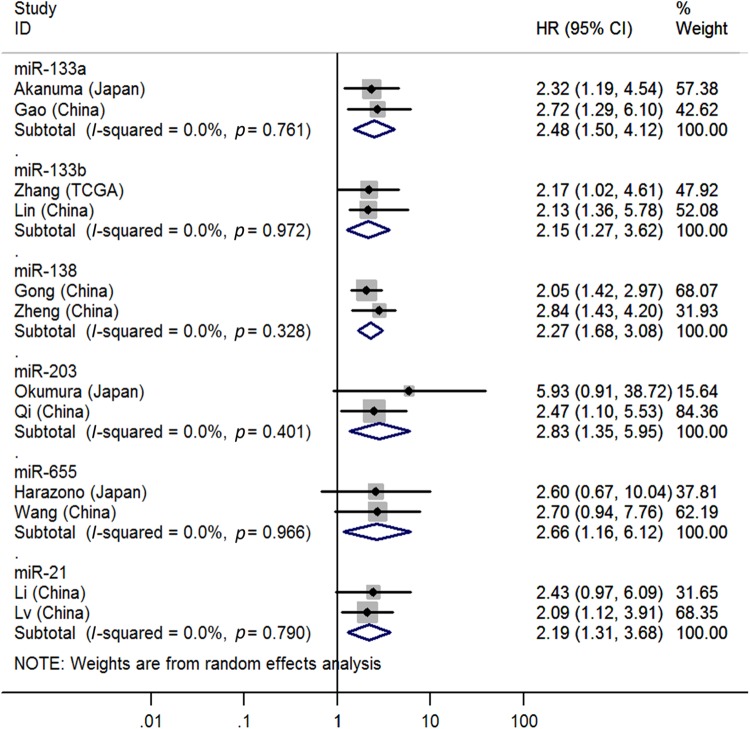
| Tissue | Low miR-133a | 2 | 32, 33 | 2.48 | 1.50–4.12 | Fig. 2 | <0.01 | I2 = 0.0%, P = 0.76 | 210 |
| Tissue | Low miR-133b | 2 | 13, 34 | 2.15 | 1.27–3.62 | Fig. 2 | <0.01 | I2 = 0.0%, P = 0.97 | 265 |
| Tissue | Low miR-138 | 2 | 35, 36 | 2.27 | 1.68–3.08 | Fig. 2 | <0.01 | I2 = 0.0%, P = 0.33 | 333 |
| Tissue | High miR-143–3p | 2 | 37, 38 | 1.12 | 0.13–9.33 | Supplementary Figure 2 | 0.92 | I2 = 95.4%, P < 0.01 | 199 |
| Tissue | High miR-145 | 2 | 1, 29 | 0.85 | 0.27–2.66 | Supplementary Figure 2 | 0.79 | I2 = 73.1%, P = 0.05 | 143 |
| Tissue | High miR-155 | 2 | 1, 13 | 1.17 | 0.64–2.14 | Supplementary Figure 3 | 0.61 | I2 = 47.6%, P = 0.17 | 283 |
| Tissue | High miR-200a | 2 | 2, 47 | 0.71 | 0.19–2.60 | Supplementary Figure 3 | 0.60 | I2 = 78.6%, P = 0.03 | 187 |
| Tissue | Low miR-203 | 2 | 49, 50 | 2.83 | 1.35–5.95 | Fig. 2 | <0.01 | I2 = 0.0%, P = 0.40 | 70 |
| Tissue | High miR-205 | 2 | 51, 52 | 0.75 | 0.09–6.45 | Supplementary Figure 3 | 0.79 | I2 = 72.4%, P = 0.06 | 57 |
| Tissue | High miR-223 | 2 | 13, 55 | 1.13 | 0.25–5.03 | Supplementary Figure 3 | 0.87 | I2 = 89.5%, P < 0.01 | 294 |
| Tissue | Low miR-375 | 6 | 7, 10, 11, 57– 59 | 1.64 | 1.05–2.58 | Supplementary Figure 3 | 0.03 | I2 = 64.8%, P < 0.01 | 729 |
| Tissue | High miR-455–3p | 2 | 62, 63 | 0.67 | 0.10–4.48 | Supplementary Figure 3 | 0.68 | I2 = 93.6%, P < 0.01 | 326 |
| Tissue | Low miR-655 | 2 | 75, 76 | 2.66 | 1.16–6.12 | Fig. 2 | 0.02 | I2 = 0.0%, P = 0.97 | 63 |
| Blood | Low miR-16 | 2 | 87, 88 | 1.23 | 0.14–10.86 | Supplementary Figure 4 | 0.86 | I2 = 90.3%, P < 0.01 | 62 |
| Blood | High miR-21 | 2 | 87, 89 | 2.19 | 1.31–3.68 | Fig. 2 | <0.01 | I2 = 0.0%, P = 0.79 | 164 |
| Blood | High miR-25 | 2 | 90, 91 | 1.75 | 0.56–5.54 | Supplementary Figure 4 | 0.34 | I2 = 67.2%, P = 0.08 | 257 |
| Blood | High miR-223 | 2 | 90, 91 | 1.62 | 1.12–2.34 | Supplementary Figure 4 | 0.01 | I2 = 0.0%, P = 0.50 | 257 |
| Blood | Low miR-375 | 3 | 87, 89, 91 | 1.44 | 0.93–2.22 | Supplementary Figure 4 | 0.10 | I2 = 29.1%, P = 0.24 | 358 |
N number of the included articles, HR hazard ratio, CI confidence interval
Tissue-based high miR-21 and low miR-133a, miR-133b, miR-138, miR-203, miR-375, and miR-655 levels predict poor OS
Ten studies1,2,7–14 analyzed the connections between high tissue miR-21 levels and OS, suggesting that EC patients with high tissue miR-21 levels had significantly worse OS than those with low levels (HR = 1.63, 95% CI = 1.26–2.11, P < 0.01, Supplementary Figure 1).
Two studies32,33 reported the associations between low tissue miR-133a levels and OS, indicating that EC patients with low tissue miR-133a levels had significantly shorter OS than those with high levels (HR = 2.48, 95% CI = 1.50–4.12, P < 0.01, Fig. 2).
Fig. 2.
Forest plot of pooled analyses of OS in association with tissue expression levels of low miR-133a, miR-133b, miR-138, miR-203, and miR-605 and blood expression levels of high miR-21
Two studies13,34 covered the relationship between low tissue miR-133b levels and OS, showing that EC patients with low tissue miR-133b levels had significantly poorer OS than those with high levels (HR = 2.15, 95% CI = 1.27–3.62, P < 0.01, Fig. 2).
Two studies35,36 focused on the pertinence between low tissue miR-138 levels and OS, suggesting that EC patients with low tissue miR-138 levels had significantly worse OS than those with high levels (HR = 2.27, 95% CI = 1.68–3.08, P < 0.01, Fig. 2).
Two studies49,50 stressed the correlations between low tissue miR-203 levels and OS, indicating that EC patients with high tissue miR-203 levels had significantly shorter OS than those with low levels (HR = 2.83, 95% CI = 1.35–5.95, P < 0.01, Fig. 2).
Six studies7,10,11,57–59 emphasized the relevance between low tissue miR-375 levels and OS, showing that EC patients with low tissue miR-375 levels had significantly poorer OS than those with high levels (HR = 1.64, 95% CI = 1.05–2.58, P = 0.03, Supplementary Figure 3).
Two studies75,76 paid attention to the relation between low tissue miR-655 levels and OS, suggesting that EC patients with low tissue miR-655 levels had significantly worse OS than those with high levels (HR = 2.66, 95% CI = 1.16–6.12, P = 0.02, Fig. 2).
Blood-based high miR-21 and miR-223 levels predict poor OS
Two studies87,89 analyzed the connections between high blood miR-21 levels and OS, suggesting that EC patients with high blood miR-21 levels had significantly worse OS than those with low levels (HR = 2.19, 95% CI = 1.31–3.68, P < 0.01, Fig. 2).
Two studies90,91 reported the associations between low blood miR-223 levels and OS, indicating that EC patients with low blood miR-223 levels had significantly shorter OS than those with high levels (HR = 1.62, 95% CI = 1.12–2.34, P = 0.01, Supplementary Figure 4).
Publication bias
Begg’s funnel plot was used to evaluate publication bias in the OS of EC patients with high tissue miR-21 levels (Supplementary Figure 5). The results showed that the P value was 0.33, indicating the absence of a publication bias.
Begg’s funnel plot was used to evaluate publication bias in the OS of EC patients with low tissue miR-375 levels (Supplementary Figure 6). The results showed that the P value was 0.73, indicating the absence of a publication bias.
Sensitivity analysis
The sensitivity analysis was applied to evaluate whether any individual study had excessive influence in the OS of EC patients with high tissue miR-21 levels (Supplementary Figure 7). The outcomes showed that no single investigation significantly influenced the merged HR and 95% CI.
The sensitivity analysis was applied to evaluate whether any individual study had excessive influence in the OS of EC patients with low tissue miR-375 levels (Supplementary Figure 8). The outcomes showed that no single investigation significantly influenced the merged HR and 95% CI.
Discussion
Primary discoveries
The present meta-analysis included 44 articles published in English that included 22 miRNAs and 4310 patients. miR-21 is the most studied miRNA, and EC patients with high tissue miR-21 levels have significantly shorter OS times than those with low levels. Similarly, high blood miR-21 levels have a significantly prognostic value for OS. In addition, some other miRNAs have significantly prognostic value for EC, including tissue miR-133a, miR-133b, miR-138, miR-203, miR-375, and miR-655 and blood miR-223. Among them, the tissue miR-133a, miR-133b, miR-138, miR-203, and miR-655 levels and the blood miR-21 level are strong biomarkers of prognosis in EC.
Molecular mechanisms of the studied miRNAs
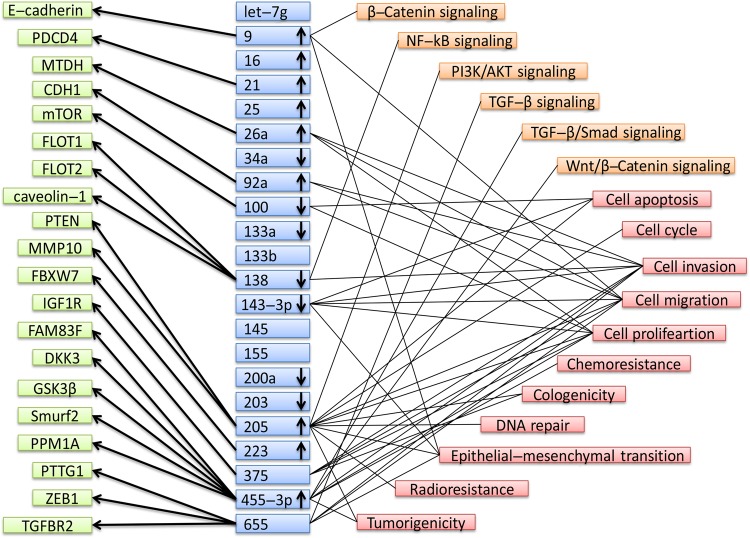
Furthermore, a summary of the 22 miRNAs with altered levels, including their potential targets and pathways, is presented in Fig. 3. Several miRNAs were not marked with up or down arrows since either they were not reported in the original articles or inconsistent expression levels of them were shown in the papers about the single miRNA. In general, Fig. 3 can help us better understand the functions of miRNAs in EC. As the strong candidate biomarkers of EC, tissue miR-133a, miR-138, and miR-203 were with downregulated expression and blood miR-21 was with upregulated expression. In addition, metadherin was the target of miR-21. miR-138 downregulation caused lipid raft formation by upregulating flotillin 1, flotillin 2, and caveolin-1 and promoted invasion of ESCC cells as well as sustained nuclear factor-κB activity. Furthermore, pituitary tumor-transforming 1, zinc finger E-box binding homeobox 1, and transforming growth factor beta receptor 2 were identified as direct targets of miR-655, overexpression of which could suppress migration and invasion of EC9706 and KYSE150 cells.
Fig. 3.
Summary of microRNAs with altered expression, potential targets, and pathways entered in this study. E-cadherin cadherin 1, type 1, E-cadherin (epithelial), PDCD4 programmed cell death 4, MTDH metadherin, CDH1 cadherin 1, mTOR mechanistic target of rapamycin kinase, FLOT1 flotillin 1, FLOT2 flotillin 2, PTEN phosphatase and tensin homolog, MMP10 matrix metallopeptidase 10, FBXW7 F-box and WD repeat domain containing 7, IGF1R insulin like growth factor 1 receptor, FAM83F family with sequence similarity 83 member F, DKK3 dickkopf WNT signaling pathway inhibitor 3, GSK3β glycogen synthase kinase 3 beta, Smurf2 SMAD specific E3 ubiquitin protein ligase 2, PPM1A protein phosphatase, Mg2+/Mn2+ dependent 1A, PTTG1 pituitary tumor-transforming 1, ZEB1 zinc finger E-box binding homeobox 1, TGFBR2 transforming growth factor beta receptor 2, NF-kB nuclear factor-kappaB, PI3K phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta, AKT AKT serine/threonine kinase 1, TGF-β transforming growth factor-β
Strengths of the meta-analysis
This work had certain strengths: (1) we searched for and identified almost all articles with survival outcomes in EC patients with altered miRNA levels. In addition, the current expression profiles of miRNAs were distinctly listed in Supplementary Tables 1 and 2 by distinguishing miRNA names and the kinds of detected samples; (2) most of our included articles had large sample sizes (≥30, except 4 studies [refs. 49,51,75,88]), strengthening and broadening the applicability of the prognostic results to EC patients; (3) combined analyses for most of miRNAs with significantly prognostic value indicated low heterogeneity (I2 ≤ 50, except tissue miR-375).
Limitations
The following limitations of our meta-analysis should be noted: (1) there were multiple variables in the present study, including different types of samples (formalin-fixed, paraffin-embedded, frozen tissue, plasma, and serum) from EC patients at different stages, cutoff values, and miRNA methods, among which the sample type and cutoffs were major limitations; (2) we only included articles published in English, probably excluding potential studies published in other languages about miRNA expression and prognosis in EC patients; (3) we only included studies estimating OS, possibly excluding potential researches with prognosis with other survival outcomes, such as cause-specific survival, disease-free survival, recurrence-free survival, progression-free survival, and metastasis-free survival; (4) although the mean NOS score of the included researches was 6.5, which indicated that the quality of them was adequate, we still could not ignore the low scores among them (the NOS scores were 4–5).
Implications for future clinical and basic research
It was worth noting that this meta-analysis was the first systematic estimation of the associations between dysregulated miRNA levels and the prognosis of EC patients. There were implications for future clinical and basic investigations: (1) combined detection of multiple miRNA levels could be used by clinical workers and other health-care providers, which might greatly augment the ability to estimate survival time of EC patients so that timely treatment could be provided; (2) the present research advances and trends regarding miRNA levels and the prognosis of EC patients could be clearly obtained by basic researchers in Tables 1 and 2. Meanwhile, the molecular mechanisms of miRNAs could be seen in Fig. 3, which could be referred to at the same time as Tables 1 and 2; (3) some conflicting results regarding the prognostic value of miRNAs might be resolved based on this work.
Conclusions
In conclusion, the tissue expression levels of miR-21, miR-133a, miR-133b, miR-138, miR-203, miR-375, and miR-655 and the blood expression levels of miR-21 and miR-223 demonstrate significant prognostic value. Among them, the expression levels of miR-133a, miR-133b, miR-138, miR-203, and miR-655 in tissue, and the expression level of miR-21 in blood are potential prognostic candidates for predicting OS in EC.
Study Highlights
What is current knowledge
Increasing evidence indicates that microRNAs can act as possible biomarkers for cancer prognosis in clinical practice.
However, there has not been a systematic review and meta-analysis to estimate the associations between microRNA expression and the survival of esophageal carcinoma patients.
What is new here
This work is the first systematic review and meta-analysis about prognostic value of microRNAs in esophageal carcinoma.
Several microRNAs suggest significantly prognostic value and are potential prognostic candidates for predicting overall survival for esophageal carcinoma.
Translational impact
Combined detection of multiple microRNA levels could be used by clinical workers and other healthcare providers, which might greatly augment the ability to estimate survival time of esophageal carcinoma patients so that timely treatment could be provided.
Electronic supplementary material
Competing interests
Guarantor of article: Yue Zhang
Specific author contributions: Study concept and design: Y.Z. Acquisition of data: S.G. and Z.-Y. Zhao. Analysis and interpretation of data: S.G., Z.-Y. Zhao and Z.-Y. Zhang. Drafting of the manuscript: Y.Z. Revision of manuscript: S.G., Z.-Y. Zhao, Z.-Y. Zhang, Y.Z. and R.W. Supervision of work: Y.Z. and R.W. All authors read and approved the final manuscript.
Financial support: None.
Potential competing interests: None.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yue Zhang, Phone: +86-531-6861-6426, Email: zhangyue0811@hotmail.com.
Rong Wu, Phone: +86-18940257577, Email: wur@sj-hospital.org.
Electronic supplementary material
The online version of this article (10.1038/s41424-018-0070-z) contains supplementary material, which is available to authorized users.
References
- 1.Hamano R, et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin. Cancer Res. 2011;17:3029–3038. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 2.Hu Y, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int. J. Cancer. 2011;128:132–143. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui D, et al. Primary tumor microRNA signature predicts recurrence and survival in patients with locally advanced esophageal adenocarcinoma. Oncotarget. 2016;7:81281–81291. doi: 10.18632/oncotarget.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara K, et al. Significance and function of microRNA-7 in oesophageal squamous cell carcinoma. Anticancer Res. 2017;37:1043–1048. doi: 10.21873/anticanres.11415. [DOI] [PubMed] [Google Scholar]
- 5.Song Y, et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu XL, et al. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann. Thorac. Surg. 2014;97:1037–1045. doi: 10.1016/j.athoracsur.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 7.Mathé EA, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin. Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, et al. microRNA and inflammatory gene expression as prognostic marker for overall survival in esophageal squamous cell carcinoma. Int. J. Cancer. 2013;132:2901–2909. doi: 10.1002/ijc.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, et al. MicroRNA-21 promotes cell growth and migration by targeting programmed cell death 4 gene in Kazakh’s esophageal squamous cell carcinoma. Dis. Markers. 2014;2014:232837. doi: 10.1155/2014/232837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng XR, Lu P, Mei JZ, Liu GJ, Fan QX. Expression analysis of miRNA and target mRNAs in esophageal cancer. Braz. J. Med. Biol. Res. 2014;47:811–817. doi: 10.1590/1414-431X20143906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winther M, et al. Evaluation of miR-21 and miR-375 as prognostic biomarkers in esophageal cancer. Acta Oncol. 2015;54:1582–1591. doi: 10.3109/0284186X.2015.1064161. [DOI] [PubMed] [Google Scholar]
- 12.Klimczak-Bitner AA, Kordek R, Bitner J, Musiał J, Szemraj J. Expression of MMP9, SERPINE1 and miR-134 as prognostic factors in esophageal cancer. Oncol. Lett. 2016;12:4133–4138. doi: 10.3892/ol.2016.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HC, Tang KF. Clinical value of integrated-signature miRNAs in esophageal cancer. Cancer Med. 2017;6:1893–1903. doi: 10.1002/cam4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Jiandong, Ma Lei, Shi Daimeng, Zhang Zhen, Yao Chuanshan, Zhao Xulin, Xu Quanxiao, Wen Penghao, He Limin. Prognostic significance of miR-21 and PDCD4 in patients with stage II esophageal carcinoma after surgical resection. Journal of Cellular Biochemistry. 2018;119(6):4783–4791. doi: 10.1002/jcb.26672. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa R, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med. Mol. Morphol. 2009;42:102–109. doi: 10.1007/s00795-009-0443-1. [DOI] [PubMed] [Google Scholar]
- 16.Hua Y, Zhao K, Tao G, Dai C, Su Y. miR-25 promotes metastasis via targeting FBXW7 in esophageal squamous cell carcinoma. Oncol. Rep. 2017;38:3030–3038. doi: 10.3892/or.2017.5995. [DOI] [PubMed] [Google Scholar]
- 17.Yang C, et al. Down-regulated miR-26a promotes proliferation, migration, and invasion via negative regulation of MTDH in esophageal squamous cell carcinoma. FASEB J. 2017;31:2114–2122. doi: 10.1096/fj.201601237. [DOI] [PubMed] [Google Scholar]
- 18.Yang S, et al. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget. 2016;7:85728–85740. doi: 10.18632/oncotarget.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi B, et al. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World J. Gastroenterol. 2017;23:7965–7977. doi: 10.3748/wjg.v23.i45.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, Zhang X, Li N, Liu Q, Chen D. miR-30b inhibits cancer cell growth, migration, and invasion by targeting homeobox A1 in esophageal cancer. Biochem. Biophys. Res. Commun. 2017;485:506–512. doi: 10.1016/j.bbrc.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Ma T, et al. MicroRNA-30c functions as a tumor suppressor via targeting SNAI1 in esophageal squamous cell carcinoma. Biomed. Pharmacother. 2018;98:680–686. doi: 10.1016/j.biopha.2017.12.095. [DOI] [PubMed] [Google Scholar]
- 22.Lin RJ, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J. Surg. Oncol. 2012;105:175–182. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 23.Cui XB, et al. MicroRNA-34a functions as a tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh esophageal squamous cell carcinoma. Oncotarget. 2017;8:92454–92469. doi: 10.18632/oncotarget.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin X, Xu XY, Chen QS, Huang C. Clinical significance of microRNA-34a in esophageal squamous cell carcinoma. Genet. Mol. Res. 2015;14:17684–17691. doi: 10.4238/2015.December.21.41. [DOI] [PubMed] [Google Scholar]
- 25.Chen ZL, et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J. Biol. Chem. 2011;286:10725–10734. doi: 10.1074/jbc.M110.165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma G, et al. MicroRNA-92b represses invasion-metastasis cascade of esophageal squamous cell carcinoma. Oncotarget. 2016;7:20209–20222. doi: 10.18632/oncotarget.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, et al. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med. Oncol. 2013;30:411. doi: 10.1007/s12032-012-0411-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhou S, et al. Prognostic value of microRNA-100 in esophageal squamous cell carcinoma. J. Surg. Res. 2014;192:515–520. doi: 10.1016/j.jss.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Feber A, et al. MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann. Thorac. Surg. 2011;91:1523–1530. doi: 10.1016/j.athoracsur.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okumura T, et al. The expression of microRNA 574-3p as a predictor of postoperative outcome in patients with esophageal squamous cell carcinoma. World J. Surg. Oncol. 2016;14:228. doi: 10.1186/s12957-016-0985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L, et al. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim. Biophys. Sin. (Shanghai). 2018;50:171–180. doi: 10.1093/abbs/gmx132. [DOI] [PubMed] [Google Scholar]
- 32.Akanuma N, et al. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br. J. Cancer. 2014;110:189–198. doi: 10.1038/bjc.2013.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao SH, Liu J, Zhang HJ, Zhao N, Zhang J. Low miR-133a expression is a predictor of outcome in patients with esophageal squamous cell cancer. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3788–3792. [PubMed] [Google Scholar]
- 34.Lin C, et al. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin. Cancer Res. 2018;24:486–498. doi: 10.1158/1078-0432.CCR-17-1851. [DOI] [PubMed] [Google Scholar]
- 35.Gong H, et al. Downregulation of miR-138 sustains NF-κB activation and promotes lipid raft formation in esophageal squamous cell carcinoma. Clin. Cancer Res. 2013;19:1083–1093. doi: 10.1158/1078-0432.CCR-12-3169. [DOI] [PubMed] [Google Scholar]
- 36.Zheng S, Zhang X, Wang X, Li J. Downregulation of miR-138 predicts poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Biomark. 2017;20:49–54. doi: 10.3233/CBM-170079. [DOI] [PubMed] [Google Scholar]
- 37.He Z, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma. Mol. Cancer. 2016;15:51. doi: 10.1186/s12943-016-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Wang Y, et al. MicroRNA expression in esophageal squamous cell carcinoma: Novel diagnostic and prognostic biomarkers. Mol. Med. Rep. 2017;15:3833–3839. doi: 10.3892/mmr.2017.6479. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, et al. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br. J. Cancer. 2016;114:290–297. doi: 10.1038/bjc.2015.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hezova R, et al. Diagnostic and prognostic potential of miR-21, miR-29c, miR-148 and miR-203 in adenocarcinoma and squamous cell carcinoma of esophagus. Diagn. Pathol. 2015;10:42. doi: 10.1186/s13000-015-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokobori T, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang Z, Dong X, Sun Q, Li X, Yan B. Clinical significance of up-regulated miR-181a in prognosis and progression of esophageal cancer. Acta Biochim. Biophys. Sin. (Shanghai). 2014;46:1007–1010. doi: 10.1093/abbs/gmu083. [DOI] [PubMed] [Google Scholar]
- 43.Gao X, et al. Overexpression of miR-191 predicts poor prognosis and promotes proliferation and invasion in esophageal squamous cell carcinoma. Yonsei Med. J. 2017;58:1101–1110. doi: 10.3349/ymj.2017.58.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun N, Ye L, Chang T, Li X, Li X. microRNA-195-Cdc42 axis acts as a prognostic factor of esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:6871–6879. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang TY, et al. Implications of microRNA-197 downregulated expression in esophageal cancer with poor prognosis. Genet. Mol. Res. 2014;13:5574–5581. doi: 10.4238/2014.July.25.12. [DOI] [PubMed] [Google Scholar]
- 46.Qi B, et al. Involvement of microRNA-198 overexpression in the poor prognosis of esophageal cancer. Asian Pac. J. Cancer Prev. 2013;14:5073–5076. doi: 10.7314/APJCP.2013.14.9.5073. [DOI] [PubMed] [Google Scholar]
- 47.Zhang HF, et al. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting Kindlin-2. Carcinogenesis. 2014;35:292–301. doi: 10.1093/carcin/bgt320. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, et al. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour Biol. 2016;37:5621–5632. doi: 10.1007/s13277-015-4432-9. [DOI] [PubMed] [Google Scholar]
- 49.Okumura T, et al. MicroRNA-203 inhibits the progression of esophageal squamous cell carcinoma with restored epithelial tissue architecture in vivo. Int. J. Oncol. 2014;44:1923–1932. doi: 10.3892/ijo.2014.2365. [DOI] [PubMed] [Google Scholar]
- 50.Qi Q, et al. Hypermethylation and low expression of miR-203 in patients with esophageal cancer in Chinese population. Int. J. Clin. Exp. Pathol. 2016;9:6245–6251. [Google Scholar]
- 51.Hezova R, et al. MiR-205 functions as a tumor suppressor in adenocarcinoma and an oncogene in squamous cell carcinoma of esophagus. Tumour Biol. 2016;37:8007–8018. doi: 10.1007/s13277-015-4656-8. [DOI] [PubMed] [Google Scholar]
- 52.Pan F, et al. Sp1-mediated transcriptional activation of miR-205 promotes radioresistance in esophageal squamous cell carcinoma. Oncotarget. 2017;8:5735–5752. doi: 10.18632/oncotarget.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi B, et al. Overregulation of microRNA-212 in the poor prognosis of esophageal cancer patients. Genet. Mol. Res. 2014;13:7800–7807. doi: 10.4238/2014.September.26.18. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Y, Hong L. Prediction value of miR-483 and miR-214 in prognosis and multidrug resistance of esophageal squamous cell carcinoma. Genet. Test. Mol. Biomark. 2013;17:470–474. doi: 10.1089/gtmb.2012.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurashige J, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br. J. Cancer. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang BJ, Gong HY, Zheng F, Liu DJ, Liu HX. Up-regulation of miR-335 predicts a favorable prognosis in esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:6213–6218. [PMC free article] [PubMed] [Google Scholar]
- 57.Kong KL, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 58.Li J, et al. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS ONE. 2013;8:e53582. doi: 10.1371/journal.pone.0053582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isozaki Y, et al. Usefulness of microRNA-375 as a prognostic and therapeutic tool in esophageal squamous cell carcinoma. Int. J. Oncol. 2015;46:1059–1066. doi: 10.3892/ijo.2014.2789. [DOI] [PubMed] [Google Scholar]
- 60.Li B, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. doi: 10.1038/onc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi B, et al. Downregulation of microRNA-382 is associated with poor outcome of esophageal squamous cell carcinoma. World J. Gastroenterol. 2015;21:6884–6891. doi: 10.3748/wjg.v21.i22.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu A, et al. Antagonizing miR-455-3p inhibits chemoresistance and aggressiveness in esophageal squamous cell carcinoma. Mol. Cancer. 2017;16:106. doi: 10.1186/s12943-017-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H, Wei YN, Zhou J, Hao TT, Liu XL. MiR-455-3p acts as a prognostic marker and inhibits the proliferation and invasion of esophageal squamous cell carcinoma by targeting FAM83F. Eur. Rev. Med. Pharmacol. Sci. 2017;21:3200–3206. [PubMed] [Google Scholar]
- 64.Xue L, et al. Upregulated miR-483-5p expression as a prognostic biomarker for esophageal squamous cell carcinoma. Cancer Biomark. 2017;19:193–197. doi: 10.3233/CBM-160506. [DOI] [PubMed] [Google Scholar]
- 65.Ren C, et al. miR-486-5p expression pattern in esophageal squamous cell carcinoma, gastric cancer and its prognostic value. Oncotarget. 2016;7:15840–15853. doi: 10.18632/oncotarget.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao Y, Li L, Liu J, Wang L, Zhou Y. MiR-495 inhibits esophageal squamous cell carcinoma progression by targeting Akt1. Oncotarget. 2016;7:51223–51236. doi: 10.18632/oncotarget.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Islam F, et al. MiR-498 in esophageal squamous cell carcinoma: clinicopathological impacts and functional interactions. Hum. Pathol. 2017;62:141–151. doi: 10.1016/j.humpath.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 68.Lin C, et al. miR-508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nat. Commun. 2014;5:4620. doi: 10.1038/ncomms5620. [DOI] [PubMed] [Google Scholar]
- 69.Chen Z, et al. Up-regulated miR-548k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp. Cell Res. 2018;362:90–101. doi: 10.1016/j.yexcr.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Guan S, et al. MiR-613: a novel diagnostic and prognostic biomarker for patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37:4383–4391. doi: 10.1007/s13277-015-4271-8. [DOI] [PubMed] [Google Scholar]
- 71.Song C, et al. MiR-622 functions as a tumor suppressor and directly targets E2F1 in human esophageal squamous cell carcinoma. Biomed. Pharmacother. 2016;83:843–849. doi: 10.1016/j.biopha.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 72.Li C, et al. The decreased expression of miR-625 predicts poor prognosis of esophageal squamous cell carcinoma. Int. J. Clin. Exp. Med. 2015;8:9560–9564. [PMC free article] [PubMed] [Google Scholar]
- 73.Jin L, et al. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim. Biophys. Sin. (Shanghai). 2016;48:810–819. doi: 10.1093/abbs/gmw073. [DOI] [PubMed] [Google Scholar]
- 74.Zhang JX, et al. Downregulation of MicroRNA-644a promotes esophageal squamous cell carcinoma aggressiveness and stem cell-like phenotype via dysregulation of PITX2. Clin. Cancer Res. 2017;23:298–310. doi: 10.1158/1078-0432.CCR-16-0414. [DOI] [PubMed] [Google Scholar]
- 75.Harazono Y, et al. miR-655 is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS ONE. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, et al. Mir-655 up-regulation suppresses cell invasion by targeting pituitary tumor-transforming gene-1 in esophageal squamous cell carcinoma. J. Transl. Med. 2013;11:301. doi: 10.1186/1479-5876-11-301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Zhou YW, et al. miR-675-5p enhances tumorigenesis and metastasis of esophageal squamous cell carcinoma by targeting REPS2. Oncotarget. 2016;7:30730–30747. doi: 10.18632/oncotarget.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge C, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6:10964–10977. doi: 10.18632/oncotarget.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gopalan V, et al. Overexpression of microRNA-1288 in oesophageal squamous cell carcinoma. Exp. Cell Res. 2016;348:146–154. doi: 10.1016/j.yexcr.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 80.Xie R, et al. Prognostic value of combined and individual expression of microRNA-1290 and its target gene nuclear factor I/X in human esophageal squamous cell carcinoma. Cancer Biomark. 2017;20:325–331. doi: 10.3233/CBM-170029. [DOI] [PubMed] [Google Scholar]
- 81.Liu K, Li L, Rusidanmu A, Wang Y, Lv X. Down-regulation of miR-1294 is related to dismal prognosis of patients with esophageal squamous cell carcinoma through elevating C-MYC expression. Cell. Physiol. Biochem. 2015;36:100–110. doi: 10.1159/000374056. [DOI] [PubMed] [Google Scholar]
- 82.Liu J, et al. Low expression of miR-1469 predicts disease progression and unfavorable post-surgical clinical outcomes in patients with esophageal squamous cell cancer. Oncol. Lett. 2017;13:4469–4474. doi: 10.3892/ol.2017.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang C, et al. Clinical potential of miR-3651 as a novel prognostic biomarker for esophageal squamous cell cancer. Biochem. Biophys. Res. Commun. 2015;465:30–34. doi: 10.1016/j.bbrc.2015.07.109. [DOI] [PubMed] [Google Scholar]
- 84.Qin HD, et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am. J. Hum. Genet. 2016;98:709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cui Y, Xue Y, Dong S, Zhang P. Plasma microRNA-9 as a diagnostic and prognostic biomarker in patients with esophageal squamous cell carcinoma. J. Int. Med. Res. 2017;45:1310–1317. doi: 10.1177/0300060517709370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, Li M, Gao F, Ge X. Serum microRNA-15a level acts as a potential diagnostic and prognostic biomarker for human esophageal squamous cell carcinoma. Cancer Biomark. 2017;18:11–17. doi: 10.3233/CBM-160667. [DOI] [PubMed] [Google Scholar]
- 87.Li BX, Yu Q, Shi ZL, Li P, Fu S. Circulating microRNAs in esophageal squamous cell carcinoma: association with locoregional staging and survival. Int. J. Clin. Exp. Med. 2015;8:7241–7250. [PMC free article] [PubMed] [Google Scholar]
- 88.Yu Q, et al. Plasma microRNAs to predict the response of radiotherapy in esophageal squamous cell carcinoma patients. Am. J. Transl. Res. 2015;7:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 89.Lv H, He Z, Wang H, Du T, Pang Z. Differential expression of miR-21 and miR-75 in esophageal carcinoma patients and its clinical implication. Am. J. Transl. Res. 2016;8:3288–3298. [PMC free article] [PubMed] [Google Scholar]
- 90.Wu C, et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS ONE. 2014;9:e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol. Biol. Rep. 2014;41:1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 92.Zhang B, et al. Pemetrexed plus dendritic cells as third-line therapy for metastatic esophageal squamous cell carcinoma. Onco Targets Ther. 2016;9:3901–3906. doi: 10.2147/OTT.S107319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan CM, et al. Serum microRNA-193b as a promising biomarker for prediction of chemoradiation sensitivity in esophageal squamous cell carcinoma patients. Oncol. Lett. 2018;15:3273–3280. doi: 10.3892/ol.2017.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu H, et al. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am. J. Transl. Res. 2013;6:71–77. [PMC free article] [PubMed] [Google Scholar]
- 95.Sun J, Song K, Feng X, Gao S. MicroRNA-367 is a potential diagnostic biomarker for patients with esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2016;473:363–369. doi: 10.1016/j.bbrc.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 96.Takeshita N, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Global Burden of Disease Cancer Collaboration et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 10.1001/jamaoncol.2018.2706 2018. [DOI] [PMC free article] [PubMed]
- 98.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J. Natl. Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 99.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lopez-Camarillo C, et al. MetastamiRs: non-coding microRNAs driving cancer invasion and metastasis. Int. J. Mol. Sci. 2012;13:1347–1379. doi: 10.3390/ijms13021347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 103.Wong WC, Cheung CS, Hart GJ. Development of a quality assessment tool for systematic reviews of observational studies (QATSO) of HIV prevalence in men having sex with men and associated risk behaviours. Emerg. Themes Epidemiol. 2008;5:23. doi: 10.1186/1742-7622-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin CH, et al. MiR-193a-5p/ERBB2 act as concurrent chemoradiation therapy response indicator of esophageal squamous cell carcinoma. Oncotarget. 2016;7:39680–39693. doi: 10.18632/oncotarget.9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.