Our study revealed a previously unappreciated role of HBSP in Fas-mediated apoptosis. The antiapoptotic activity of HBSP is important for understanding hepatitis B virus pathogenesis. In particular, HBV variants associated with hepatoma carcinoma may downregulate apoptosis of hepatocytes through enhanced HBSP expression. Our study also found that Akt is centrally involved in Fas-induced hepatocyte apoptosis and revealed that interventions directed at inhibiting the activation or functional activity of Akt may be of therapeutic value in this process.
KEYWORDS: Akt, Fas, HBSP, apoptosis, viral persistence
ABSTRACT
Hepatitis B spliced protein (HBSP) is known to associate with viral persistence and pathogenesis; however, its biological and clinical significance remains poorly defined. Acquired resistance to Fas-mediated apoptosis is thought to be one of the major promotors for hepatitis B virus (HBV) chronicity and malignancy. The purpose of this study was to investigate whether HBSP could protect hepatocytes against Fas-initiated apoptosis. We showed here that HBSP mediated resistance of hepatoma cells or primary human hepatocytes (PHH) to agonistic anti-Fas antibody (CH11)- or FasL-induced apoptosis. Under Fas signaling stimulation, expression of HBSP inhibited Fas aggregation and prevented recruitment of the adaptor molecule Fas-associated death domain (FADD) and procaspase-8 (or FADD-like interleukin-1β-converting enzyme [FLICE]) into the death-inducing signaling complex (DISC) while increasing recruitment of cellular FLICE-inhibitory protein L (FLIPL) into the DISC. Those effects may be mediated through activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway as evidenced by increased cellular phosphatidylinositol (3,4,5)-trisphosphate (PIP3) content and PI3K activity and enhanced phosphorylation of mTORC2 and PDPK1 as well as Akt itself. Confirmedly, inhibition of PI3K by LY294002 reversed the effect of HBSP on Fas aggregation, FLIPL expression, and cellular apoptosis. These results indicate that HBSP functions to prevent hepatocytes from Fas-induced apoptosis by enhancing PI3K/Akt activity, which may contribute to the survival and persistence of infected hepatocytes during chronic infection.
IMPORTANCE Our study revealed a previously unappreciated role of HBSP in Fas-mediated apoptosis. The antiapoptotic activity of HBSP is important for understanding hepatitis B virus pathogenesis. In particular, HBV variants associated with hepatoma carcinoma may downregulate apoptosis of hepatocytes through enhanced HBSP expression. Our study also found that Akt is centrally involved in Fas-induced hepatocyte apoptosis and revealed that interventions directed at inhibiting the activation or functional activity of Akt may be of therapeutic value in this process.
INTRODUCTION
Approximately 240 million people worldwide are chronically infected with hepatitis B virus (HBV), placing them at a high risk of developing liver cirrhosis and hepatocellular carcinoma (HCC) (1). HBV is a DNA virus having a 3.2-kb partially double-stranded, relaxed circular genome organized into overlapping open reading frames from which viral genes are transcribed and translated into the envelope protein (S/Pre-S), the core protein (C/pre-C), the polymerase (Pol), and the X protein (HBx). In addition to the unspliced mRNAs, a series of spliced HBV RNAs have also been discovered in HBV genome-transfected hepatoma cells and in the liver of chronically HBV infected patients (2, 3). Over a dozen splicing-derived HBV variants have been identified; the most common one is the 2.2-kb singly spliced variant that lacks intron 2447/489, thus preventing the expression of full-length DNA polymerase and precore/core protein as well as major S protein (4). Hepatitis B spliced protein (HBSP), encoded by this type of spliced variant, was first identified by Soussan et al. in the livers of patients with chronic HBV infection (5), and since then, an increasing body of evidence has suggested a relationship between HBSP and the progression of liver disease toward advanced stages (6–9). Furthermore, HBSP was found capable of hacking the tumor necrosis factor alpha (TNF-α)-stimulated signaling pathways and therefore limiting the extent of liver inflammation in a transgenic mouse model (10). More recently, it was shown that alteration in spliceosome machinery in HBV-expressing hepatocytes due to HBV infection-related liver injury enabled a remarkable reduction in liver monocyte/macrophage recruitment through HBSP expression, suggesting that alternative splicing can generate a viral product to inhibit immune-mediated inflammation, thereby alleviating the organ damage (11). However, although these findings indicate that HBV splicing is a common event during HBV infection and may be involved in the pathogenicity or persistence of HBV, the biological and clinical significance of HBV splicing is far from clear.
It has been well established that HBV infection could interfere with the apoptosis signaling to promote viral proliferation and HCC progression. The HBV-mediated alteration of apoptosis is accomplished mainly through interference with cellular signaling pathways and/or regulation of epigenetics (12). External stimuli such as Fas ligand (FasL), tumor necrosis factor alpha (TNF-α), and TRAIL (TNF-related apoptosis-inducing ligand) trigger the extrinsic pathway by binding to their respective receptors on the cell surface, resulting in the recruitment of adaptor proteins to transmit the intracellular signals via the caspase cascades (13). Fas, a 45-kDa cell surface glycoprotein, is expressed in the liver and has been shown to transduce apoptotic signals to the liver cells when agonistic anti-Fas antibody or Fas ligand (FasL) binds with it (14). Fas expression in liver tissues of patients with chronic hepatitis B (CHB) infection was closely correlated with the activity of the viral hepatitis (15). Moreover, an in situ investigation of Fas/FasL expression in CHB infection and related liver diseases revealed that Fas/FasL expression level was closely associated with the inflammatory activity for liver disease initiation and progression as a result of apoptosis following Fas-FasL interaction (16). While the induction of apoptosis is a hallmark of many viruses infecting humans, studies that aim to determine the relationship of HBV infection and apoptosis have shown contradictory results. The majority of the studies documented that HBV or, more specifically, hepatitis B virus X protein (HBx) could inhibit cellular apoptosis to facilitate virus proliferation and promote HCC progression (12). However, it has also been reported that HBx was able to activate apoptosis or sensitize hepatocytes to apoptotic inducers in vitro (17–20). HBx is the protein most frequently reported to associate with the inhibition of Fas-induced apoptosis and the activation of HCC progression (21–23). It could block apoptosis by means of sequestration of p53 signaling, activation of the phosphoinositide 3-kinase (PI3K) pathway, inhibition of the death receptor-mediated apoptotic pathway, activation of the NF-κB signaling pathway, and inhibition of the mitochondrion-mediated apoptotic pathway (21, 24–28). However, the role and significance of HBSP in Fas-mediated apoptosis are not yet known.
The serine-threonine kinase protein kinase B (PKB/Akt), one of the major downstream targets of the phosphoinositide 3-kinase (PI3K) signaling pathway, is a crucial prosurviving factor of the cell, and its deactivation is implicated in various forms of stress-induced pathological cell death, including pathogenesis of hepatocyte injury (29–33). Cross talk between inhibition of the PI3K/Akt pathway and activation of Fas-mediated apoptosis has been observed in several cancers, including gastric, prostate, colon, and pancreatic cancers (34–37). In contrast, our recent work demonstrated that activation of Akt by a novel Akt-specific activator, SC79, could effectively protect human hepatoblastoma HepG2 cells and primary mouse hepatocytes from Fas-induced apoptosis (38).
Considering the fact that hepatic infection of HBV could influence the Fas/FasL system, whose activities correlate well with disease severity, and based on our prior observation that expression of HBSP could lead to a more aggressive phenotype of hepatoma cells by activation of the Akt signaling pathway (9), we hypothesized that HBSP may be a major connector for establishing the molecular cross talk between PI3K/Akt and Fas-mediated hepatocyte apoptosis. We report here that HBSP expression suppresses Fas-mediated hepatocyte apoptosis by enhancing the activity of PI3K/Akt signaling.
RESULTS
HBSP inhibits Fas-mediated hepatocyte apoptosis.
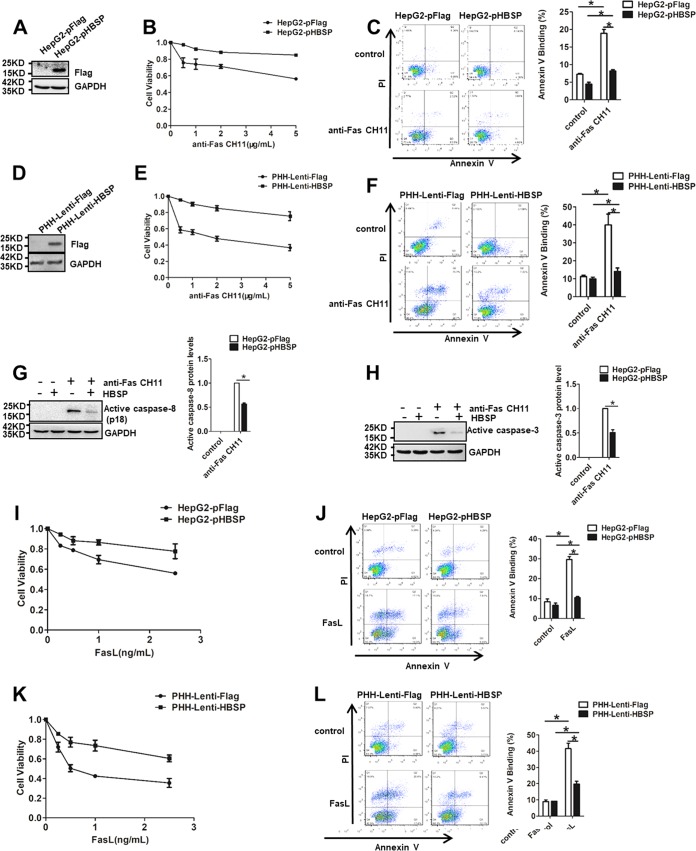
To investigate whether HBSP plays a role as a regulator of apoptosis, we generated HepG2 sublines stably transfected with empty vector pFlag or HBSP-expressing pHBSP and tested their sensitivity to agonistic anti-Fas antibody CH11, known to kill cells by the process of apoptosis. The successful expression of HBSP was confirmed by Western blot analysis (Fig. 1A). The CH11 concentration-survival curves showed that HBSP-expressing cells (HepG2-pHBSP) significantly increased the cell viability compared to the empty vector-transfected cells (HepG2-pFlag) across all the CH11 concentrations tested (Fig. 1B). To determine whether the relatively high proliferation was due to less apoptosis, we assessed the apoptotic rate of the cells by annexin V-propidium iodide (PI) assay. As shown in Fig. 1C, annexin positivity was 8.17% ± 0.43% in the CH11-treated HepG2-pHBSP cells compared to 18.91% ± 1.09% in the control HepG2-pFlag cells with the same treatment, suggesting that HBSP expression suppressed Fas-induced apoptosis in HepG2 cells, although the magnitude of this effect was relatively small. To corroborate and extend this finding, primary human hepatocytes (PHH) were infected with the recombinant lentiviruses expressing HBSP (Fig. 1D) and then tested for their viability and apoptosis after CH11 challenge. As shown in Fig. 1E and F, expression of HBSP also protected PHH from Fas-induced growth inhibition and apoptosis. Consistent with these data, CH11-stimulated HBSP-expressing HepG2-pHBSP cells exhibited less activation of both caspase-8 (Fig. 1G) and its effector caspase-3 (Fig. 1H) than the control cells. In addition, when Fas ligand (FasL) was used as the direct stimulus, similar trends of cell viability and apoptosis were observed in both HepG2-pHBSP (Fig. 1I and J) and PHH-lenti-HBSP (Fig. 1K and L) cells that express HBSP.
FIG 1.
Expression of HBSP in hepatocytes inhibits Fas-mediated apoptosis. (A) Expression of HBSP in HepG2-pHBSP and HepG2-pFlag cells assessed by Western blot analysis. (B) Cell viability determined by CCK-8 assay 72 h after HepG2-pHBSP or HepG2-pFlag cells were treated with increasing concentrations of anti-Fas CH11. (C) Quantification of apoptotic cells by PI-annexin V dual staining 12 h after HepG2-pHBSP or control HepG2-pFlag cells were treated with 2 μg/ml anti-Fas CH11. (D) Expression of HBSP in PHH cells infected with HBSP-expressing or control lentiviruses assessed by Western blot analysis. (E) Cell viability determined by CCK-8 assay 12 h after PHH-Lenti-HBSP or control PHH-Lenti-Flag cells were treated with increasing concentrations of anti-Fas CH11. (F) Quantification of apoptotic cells by PI-annexin V dual staining 12 h after PHH-Lenti-HBSP or control PHH-Lenti-Flag cells were treated with 2 μg/ml anti-Fas CH11. (G) Fas-induced activation of caspase-8 attenuated by HBSP expression. HepG2-pHBSP or HepG2-pFlag cells were treated with 2 μg/ml anti-Fas CH11 for 4 h and subjected to Western blot analysis using anti-caspase-8 antibody. (H) Fas-induced caspase-3 cleavage attenuated by HBSP expression. HepG2-pHBSP or HepG2-pFlag cells were treated with 2 μg/ml anti-Fas CH11 for 10 h and subjected to Western blot analysis using anti-caspase-3 antibody. Data from three repeat experiments were included in the statistical analysis. (I) Cell viability determined by CCK-8 assay 72 h after HepG2-pHBSP or HepG2-pFlag cells were treated with increasing concentrations of FasL. (J) Quantification of apoptotic cells by PI-annexin V dual staining 12 h after HepG2-pHBSP or control HepG2-pFlag cells were treated with 1 ng/ml FasL. (K) Cell viability determined by CCK-8 assay 12 h after PHH-Lenti-Flag or PHH-Lenti-HBSP cells were treated with increasing concentrations of FasL. (L) Quantification of apoptotic cells by PI-annexin V dual staining 12 h after PHH-Lenti-Flag or PHH-Lenti-HBSP cells were treated with 1 ng/ml FasL. Values are means ± standard deviations. *, P < 0.05.
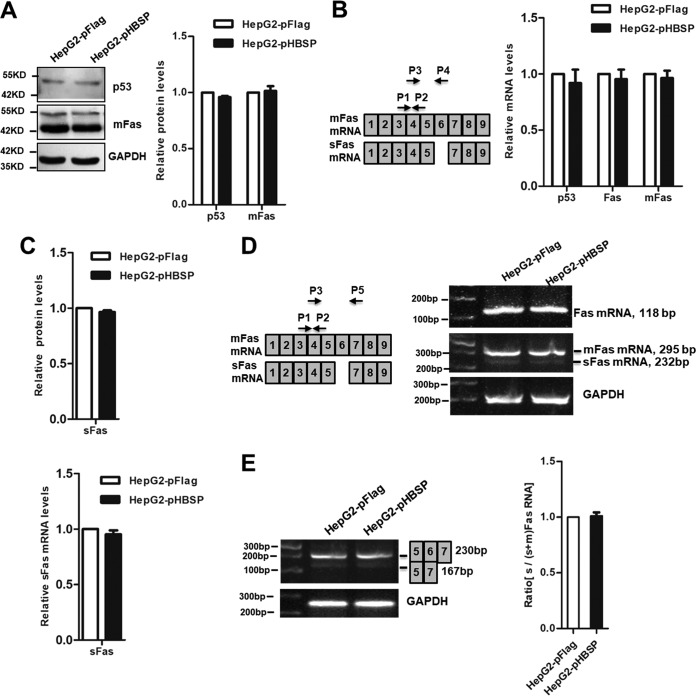
Effect of HBSP on the expression of p53, mFas, and sFas.
Fas/FasL-induced apoptosis is mediated through a number of mechanisms that include the expression levels of the ligand and receptor and efficiency of submembrane localization for receptor signaling complex assembly and activation (39), as well as p53 status in certain circumstances (40). Fas is present not only in a membrane-associated form (mFas) but also in a soluble form (sFas). Therefore, we were tempted to measure the relative expression of p53, mFas, and sFas in HepG2-pFlag and HepG2-pHBSP cells. The results showed that the protein levels of p53 and mFas did not differ at all between HepG2-pHBSP and HepG2-pFlag cells as determined by Western blot analysis (Fig. 2A), nor did the mRNA levels of p53, total Fas, and mFas as measured by quantitative real-time PCR (qRT-PCR) (Fig. 2B). We then evaluated the effects of HBSP on the expression of sFas, a soluble Fas molecule lacking exon 6 that can lead to proteins with deletions or disruptions of the single membrane-spanning domain and act as an inhibitor of Fas-mediated apoptosis signaling (41). Enzyme-linked immunosorbent assay (ELISA) and semiquantitative PCR analysis confirmed that HepG2-pHBSP cells exhibited similar expression of sFas at both the protein (Fig. 2C) and mRNA (Fig. 2D) levels as that in HepG2-pFlag cells. A Fas splicing assay was also performed in the cells transfected with Fas splicing reporter construct pCMV56 containing the human Fas receptor sequences spanning from exon 5 to position 44 of exon 7 (42). The results showed that relative mRNA levels of Fas exon 5/7 (representative of sFas) to Fas exons 5 to 7 in HepG2-pHBSP cells did not change from those in HepG2-pFlag cells (Fig. 2E).
FIG 2.
Effect of HBSP on the expression of p53, mFas, and sFas. (A) Western blot analysis showing that HBSP did not influence the expression of p53 and mFas at protein levels. (B) qRT-PCR analysis showing that HBSP did not affect the expression of p53, total Fas, and mFas at mRNA levels. (C) ELISA showing that HBSP did not alter sFas protein level. (D) Semiquantitative PCR analysis showing that HBSP did not change the expression of total Fas, mFas, and sFas at mRNA levels. The PCR product using P1 and P2 represents total Fas whereas the PCR products of P3 and P5 represent mFas and sFas varied by size. The histogram summarizes the results of three semiquantitative RT-PCR analyses of sFas mRNA levels expressed as the fold change relative to that in the control HepG2-pFlag cells. (E) Semiquantitative PCR analysis of the accumulation of alternatively spliced transcripts in HepG2-pHBSP and HepG2-pFlag cells transfected with pCMV56-Fas vector containing human Fas genomic sequences from exons 5 to 7. The products were analyzed on agarose gels with an expected size of 230 bp for exon 6 inclusion and 167 bp for exon 6 skipping. The right panel shows the ratio of exon 5 and 7 to exon 5, 6, and 7 amplification products, equivalent to the ratio of sFas to sFas-plus-mFas RNA, determined densitometrically on the band intensity from 3 independent experiments.
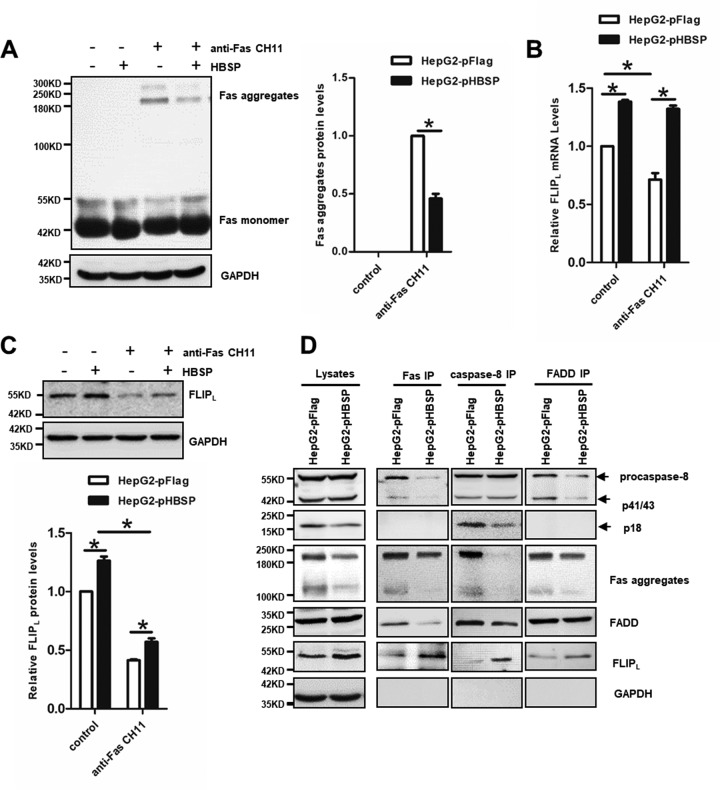
HBSP interrupts DISC assembly and function.
Upon binding of FasL or agonistic anti-Fas antibody to Fas, proapoptotic adaptor protein Fas-associated death domain (FADD) and procaspase-8 are recruited to form the death-inducing signaling complex (DISC). Meanwhile, procaspase-8 (also termed FADD-like interleukin-1β-converting enzyme [FLICE]) undergoes autocleavage and converts to active caspase-8, which activates the downstream effectors caspase-3/-7, leading to hepatocyte apoptosis (43). To examine whether HBSP could affect Fas aggregation, CH11-stimulated or unstimulated HepG2-pFlag and HepG2-pHBSP cells were subjected to Western blot analysis using an anti-Fas antibody. As shown in Fig. 3A, in the absence of CH11 stimulation only monomeric Fas was present in both HepG2-pFlag and HepG2-pHBSP cells. As expected, an SDS-stable high molecular mass of Fas aggregates was observed in both cell lines treated with CH11. However, expression of HBSP in the HepG2-pHBSP cells significantly reduced the CH11-induced formation of Fas aggregates compared to HepG2-pFlag cells. FLICE-inhibitory protein L (FLIPL) is an antiapoptotic protein with sequence homology to procaspase-8, thus functioning as a dominant negative inhibitor of caspase-8 to prevent Fas-induced apoptosis (44). Indeed, HBSP expression counteracted the reduction of FLIPL at both mRNA and protein levels caused by CH11 treatment (Fig. 3B and C). The reduction of Fas aggregates and elevation of FLIPL in CH11-treated HepG2-pHBSP cells raised the possibility that HBSP may inhibit aggregation and formation of functional DISC. Therefore, we analyzed the DISC components by immunoprecipitation and Western blot analysis, which revealed that, compared to CH11-stimulated HepG2-pFlag cells, HepG2-pHBSP cells under the same CH11 treatment recruited less FADD and procaspase-8, thus leading to incomplete cleavage of caspase-8 and an absence of active p18 prodomain at the DISC (Fig. 3D). These results indicate that HBSP blocks the recruitment and activation of procaspase-8 at the DISC probably through inhibiting the formation of higher-order Fas aggregates and enhancing the recruitment of the procaspase-8 competitor FLIPL to the DISC.
FIG 3.
HBSP disrupts DISC formation by blocking Fas aggregation and upregulating FLIPL expression. (A) The formation of higher-order Fas aggregates inhibited by HBSP. HepG2-pHBSP and HepG2-pFlag cells with or without the treatment of anti-Fas CH11 at 2 μg/ml for 4 h were subjected to Western blot analysis using anti-Fas antibody. (B) qRT-PCR analysis of FLIPL mRNA expression. The histogram shows that HBSP counteracted CH11-induced reduction in FLIPL mRNA expression. (C) The expression of FLIPL upregulated by HBSP. HepG2-pHBSP and HepG2-pFlag cells with or without the treatment of anti-Fas CH11 at 2 μg/ml for 4 h were subjected to Western blot analysis using anti-FLIPL antibody. (D) Analysis of DISC components. Lysates of CH11-stimulated HepG2-pHBSP and HepG2-pFlag cells were subjected to immunoprecipitation with anti-Fas, anti-procaspase-8, or anti-FADD antibodies and analyzed for association with the other DISC components. Data from three repeat experiments were included in the statistical analysis. Values are mean ± standard deviation. *, P < 0.05.
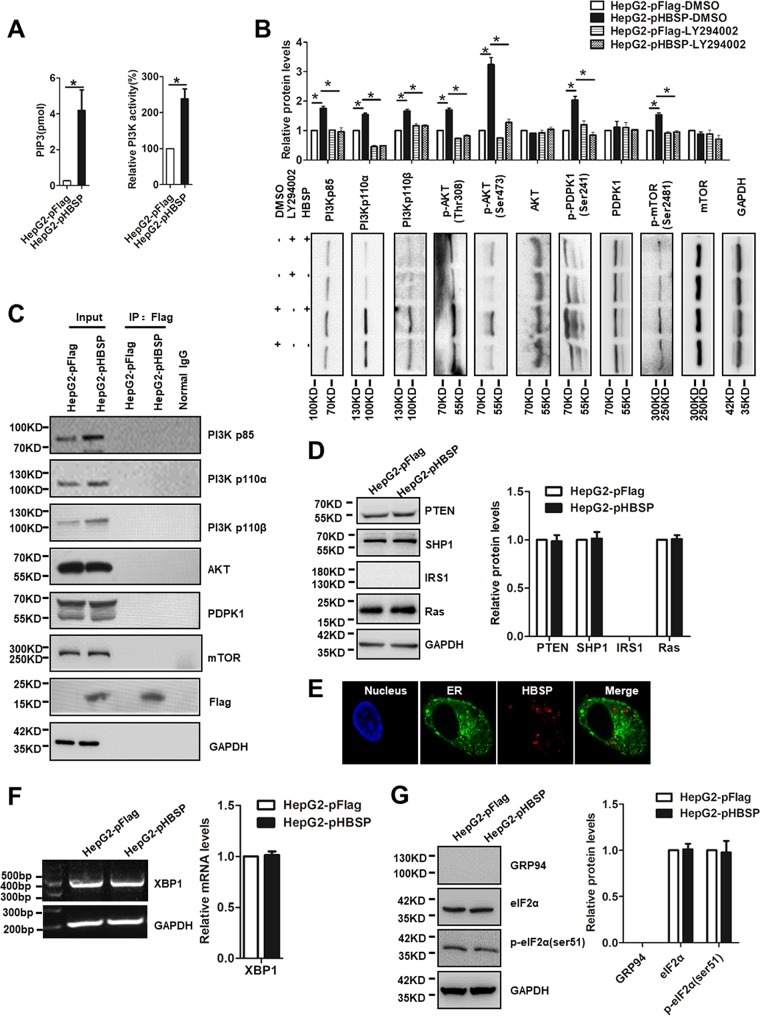
HBSP activates PI3K/Akt signaling.
So far, we have demonstrated that HBSP inhibits Fas-mediated hepatocytic apoptosis through interrupting DISC formation and its function. Activation of the PI3K signaling pathway is known to protect cancer cells from Fas-mediated apoptotic signal (31), and our previous study has documented that HBSP increases the phosphorylation of Akt, which may contribute to enhanced migration and invasion in hepatoma cells (9). Therefore, in this study we turned our attention to the upstream molecules known to be involved in Akt activation in the context of HBSP expression. Figure 4A showed that the steady-state level of phosphatidylinositol (3,4,5)-trisphosphate (PIP3), the product generated by PI3K, or PI3K activity itself was significantly higher in HepG2-pHBSP cells than the control HepG2-pFlag cells. We went further to examine the expression or phosphorylated levels of Akt upstream signaling proteins such as PI3K, mTOR, and PDPK1. As shown in Fig. 4B, both PI3K regulatory subunit p85 and the two major PI3K catalytic isoforms p110α and p110β were increased in the HBSP-expressing HepG2-pHBSP cells compared with the control HepG2-pFlag cells. HBSP expression also increased the phosphorylation of PDPK1 at Ser241 and mTOR at Ser2481 (mTORC2 specific). Consequently, increased expression of phospho-Akt at Thr308 and Ser473, which are selective substrates of PDPK1 and mTORC2, respectively, was observed in HepG2-pHBSP cells, confirming the activation of Akt upstream signaling. Notably, all of these effects were specific to the PI3K activity, as application of the PI3K inhibitor LY294002 rescued the altered expression. To assess whether HBSP could physically interact with endogenous PI3K, Akt, PDPK1, or mTOR, we performed a coimmunoprecipitation (Co-IP) study with HepG2-pHBSP cells. As shown in Fig. 4C, Flag-tagged HBSP was not immunoprecipitated with any of these signaling proteins involved in the PI3K pathway. Likewise, HBSP did not affect the expression of either PI3K upstream regulators IRS1 and Ras or PI3K negative regulators SHP1 and PTEN, the inositol phosphatases which hydrolyze PIP3 back to PIP2 (Fig. 4D). Endoplasmic reticulum (ER) stress has been implicated in regulation of Akt phosphorylation and activation through several mechanisms (44, 45). To exclude the possibility that HBSP overexpression may lead to its retention in ER and thus cause ER stress, possible localization of HBSP in ER was examined by confocal microscopy, and ER stress markers XBP1, p-eIF2α (phosphorylated α subunit of eukaryotic initiation factor 2), and GRP94 were evaluated by semiquantitative PCR or Western blot analysis. Figure 4E showed that HBSP did not localize in the ER as evidenced by lack of colocalization with ER-targeted recombinant green fluorescent protein (GFP), nor did HBSP alter the expression of XBP1, eIF2α, and GRP94 (Fig. 4F and G). Taken together, these results suggested that HBSP had an important role in boosting PI3K activity, which consequently controls its downstream signaling molecules PDPK1 and mTORC2, leading to phosphorylation and activation of their substrate Akt.
FIG 4.
Activation of PI3K/Akt signaling by HBSP. (A) PIP3 levels and activity of PI3K in HepG2-pFlag and HepG2-pHBSP cells. The cellular content of PIP3 was quantified using a competitive PIP3 ELISA; also shown is the activity of PI3K immunoprecipitated with antibody to p85 regulatory subunit of class IA PI3K. PI3K activity was expressed as percentage of control. (B) Effect of HBSP on the expression or phosphorylation of Akt upstream signaling molecules and reversal by PI3K inhibitor LY294002. Total protein extracts from HepG2-pFlag or HepG2-pHBSP cells were blotted and probed with the respective antibodies. (C) Co-IP assay showing no significant interaction between HBSP and PI3Kp85, PI3Kp110α, PI3Kp110β, Akt, PDPK1, and mTOR. The immunoprecipitated complexes with anti-Flag antibody were subjected to immunoblotting with the respective antibodies. (D) Expression of PTEN, SHP1, IRS1, and Ras showing no appreciable changes between HepG2-pFlag and HepG2-pHBSP cells determined by Western blot analysis. (E) Confocal microscopic analysis on possible localization of HBSP in the ER using ER-targeted recombinant GFP as a tracker. (F) Semiquantitative PCR analysis of XBP1 expression. (G) Western blot analysis of expression of ER stress markers eIF2α and GRP94 in HepG2-pFlag or HepG2-pHBSP cells. Data from three repeat experiments were included in the statistical analysis. Values are mean ± standard deviation. *, P < 0.05.
PI3K inhibition reverses the biological effects of HBSP.
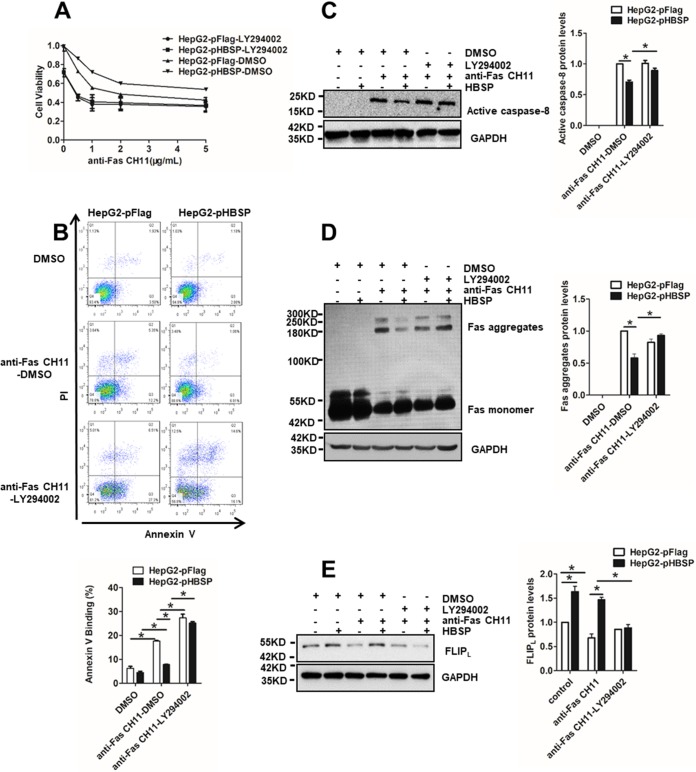
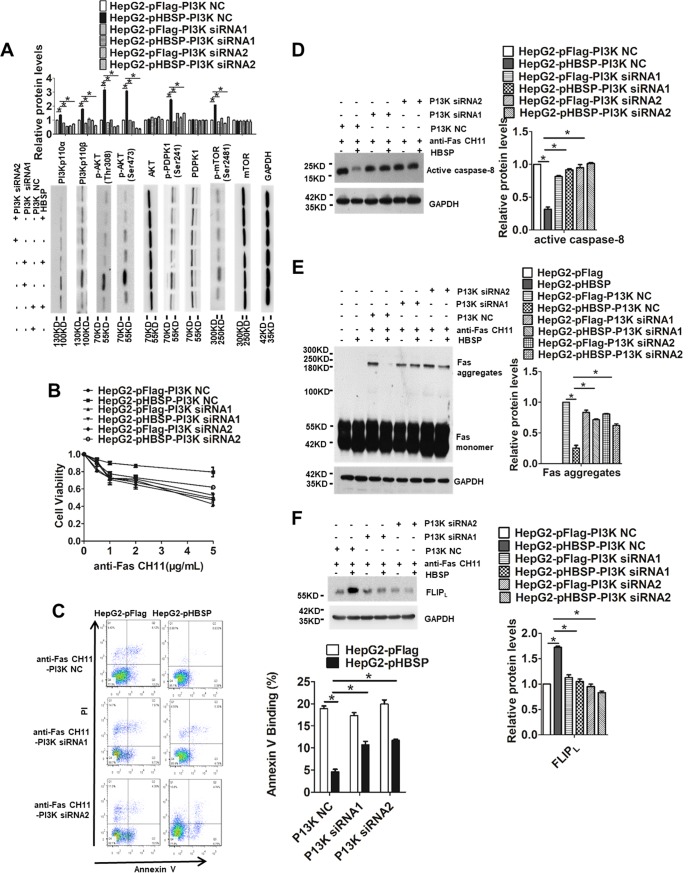
To determine whether the observed changes in HBSP-regulated and Fas-triggered apoptosis were specifically attributable to the enhancement of PI3K activity, we treated HepG2-pFlag and HepG2-pHBSP cells with CH11 in the presence or absence of the PI3K inhibitor LY294002 and compared cell viability, apoptosis rate, caspase-8 activation, and Fas aggregation as well as FLIPL expression. As shown in Fig. 5A, HBSP expression increased cellular viability under CH11 stimulation; however, the addition of LY294002 dramatically reduced the cell proliferative capability and substantially increased the apoptotic rate (Fig. 5B) and the level of active p18 prodomain as a result of caspase-8 activation (Fig. 5C). Notably, the proportion of necrotic cells (annexin V−/PI+) reflected in the upper left square of the fluorescence-activated cell sorting (FACS) profile was also increased with LY294002 treatment. To determine whether LY294002 also reversed the effect of HBSP on Fas aggregation and FLIPL expression, the levels of Fas and FLIPL were assessed by Western blot analysis in the HBSP-expressing or the control cells with or without LY294002 treatment. Consistently, inhibition of PI3K by LY294002 restored the formation of the higher-order Fas aggregates initially suppressed by HBSP (Fig. 5D) and abrogated HBSP-induced elevation of FLIPL (Fig. 5E). To further confirm the results obtained with the PI3K inhibitor, another approach to block PI3K activity was taken by using the small interfering RNA (siRNA) knockdown strategy. Successful knockdown of PI3K by the two siRNA sequences and consequent deactivation of its downstream effectors were confirmed by Western blot analysis using the respective antibodies (Fig. 6A). As anticipated, siRNA-mediated knockdown of PI3K also produced similar patterns with respect to cell proliferation (Fig. 6B) and apoptosis (Fig. 6C), caspase-8 activation (Fig. 6D), Fas aggregation (Fig. 6E), and FLIPL expression (Fig. 6F). Taken together, these results may provide more supporting evidence that the inhibitory effect of HBSP on Fas-initiated apoptosis is mainly mediated through activation of the PI3K/Akt pathway.
FIG 5.
Pharmacological inhibition of PI3K/Akt reverses the effect of HBSP on Fas-mediated hepatocyte apoptosis. (A) Cell viability determined by CCK-8 assay 72 h after HepG2-pHBSP or HepG2-pFlag cells were treated with increasing concentrations of anti-Fas CH11 in the presence or absence of 20 μM LY294002. (B) Quantification of apoptotic cells by PI-annexin V dual staining 12 h after HepG2-pHBSP or control HepG2-pFlag cells were treated with 2 μg/ml anti-Fas CH11 in the presence or absence of 20 μM LY294002. (C) LY294002 restoration of caspase-8 activation reduced by HBSP. HepG2-pHBSP or HepG2-pFlag cells were treated with 2 μg/ml anti-Fas CH11 for 4 h in the presence or absence of 20 μM LY294002 and subjected to Western blot analysis using anti-caspase-8 antibody. (D) LY294002 restoration of the formation of higher-order Fas aggregates inhibited by HBSP. HepG2-pHBSP and HepG2-pFlag cells were treated with anti-Fas CH11 at 2 μg/ml for 4 h in the presence or absence of 20 μM LY294002 and subjected to Western blot analysis using anti-Fas antibody. (E) LY294002 inhibition of FLIPL expression upregulated by HBSP. HepG2-pHBSP and HepG2-pFlag cells were treated with anti-Fas CH11 at 2 μg/ml for 4 h in the presence or absence of 20 μM LY294002 and subjected to Western blot analysis using anti-FLIPL antibody. Data from three repeat experiments were included in the statistical analysis. Values are mean ± standard deviation. *, P < 0.05.
FIG 6.
siRNA-mediated knockdown of PI3K reverses the effect of HBSP on Fas-mediated hepatocyte apoptosis. (A) Western blot analysis to confirm knockdown of PI3K and consequent deactivation of its downstream effectors in HepG2 cells. Total protein extracts from HepG2-pFlag or HepG2-pHBSP cells were blotted and probed with the respective antibodies. (B) Cell viability determined by CCK-8 assay 72 h after HepG2-pHBSP or HepG2-pFlag cells were transfected with the PI3K siRNA and treated with increasing concentrations of anti-Fas CH11. (C) Quantification of apoptotic cells by PI-annexin V dual staining 12 h after HepG2-pHBSP or control HepG2-pFlag cells were transfected with the PI3K siRNA and treated with 2 μg/ml anti-Fas CH11. (D) Knockdown of PI3K restores caspase-8 activation reduced by HBSP. HepG2-pHBSP or HepG2-pFlag cells were transfected with the PI3K siRNA, treated with 2 μg/ml anti-Fas CH11 for 4 h, and subjected to Western blot analysis using anti-caspase-8 antibody. (E) PI3K knockdown-induced restoration of the formation of higher-order Fas aggregates inhibited by HBSP. HepG2-pHBSP and HepG2-pFlag cells were transfected with the PI3K siRNA, treated with anti-Fas CH11 at 2 μg/ml for 4 h, and subjected to Western blot analysis using anti-Fas antibody. (F) PI3K knockdown-induced inhibition of FLIPL expression upregulated by HBSP. HepG2-pHBSP and HepG2-pFlag cells were transfected with the PI3K siRNA, treated with anti-Fas CH11 at 2 μg/ml for 4 h, and subjected to Western blot analysis using anti-FLIPL antibody. Data from three repeat experiments were included in the statistical analysis. Values are mean ± standard deviation. *, P < 0.05.
DISCUSSION
HBV infection is one of the most common viral diseases, accounting for most of human chronic liver diseases globally. Over 1 million people die from chronic HBV infection-related cirrhosis and liver cancer each year (46). Hepatocyte apoptosis is frequently altered in liver cancer development and correlated with HCC progression (12). A large body of studies have been performed by the investigators in the field trying to determine the relationship of HBV infection and apoptosis; however, the results are still in controversy. HBV or HBx is generally believed to inhibit cellular apoptosis, thereby facilitating virus proliferation and promoting HCC progression (12, 47–49). However, the fact that HBV infection could induce apoptosis has also been reported in several studies (17, 20, 50). In addition to the most pathogenic HBx that could interfere with apoptosis, the other HBV proteins might also regulate liver cell apoptosis either positively or negatively. For instance, HBsAg could interact with jumping translocation breakpoint protein to suppress hepatocyte apoptosis (51). HBV large surface antigen was capable of activating the Src/PI3K/Akt pathway by activation of Src kinase to inhibit apoptosis (52). Our previous study showed that HBV core protein inhibited Fas-mediated apoptosis of hepatoma cells via downregulation of Fas, p53, and FasL (53), whereas another study reported that the core protein impaired the phosphorylation of mitogen-activated protein kinase kinase 7, resulting in downregulation of the Jun N-terminal protein kinase (JNK) pathway and sensitization of HepG2 cells to TNF-α-induced apoptosis (54). As for HBSP, the study of HBSP function in full HBV genome context remains a challenge in view of the fact that the liver disease dependence of its expression is complicated with a variation of HBV alternative splicing regulation. HBSP was shown to contribute to limiting hepatic inflammation during chronic liver disease through activation of JNK and NF-κB signaling cascades (10). In concert with this, we found that HBSP functions to prevent hepatocytes from Fas-induced apoptosis via enhancing PI3K/Akt activity. However, earlier studies also indicate that HBSP could induce apoptosis when overexpressed in liver cells (5, 55). The reason for such a discrepancy is not yet known but is probably due to the different experimental conditions or the HBV genotypes used in the different laboratories. Considering that the Fas ligand receptor system plays a central role in the pathogenesis of chronic HBV infection in humans (56), it is our hypothesis that HBSP expression may contribute to HBV chronicity by increasing the resistance of infected hepatocytes to Fas-mediated cell killing. In this study, we found that HBSP remarkably protected both HepG2 and PHH cells from agonistic anti-Fas CH11- or FasL-induced hepatocytotoxicity, which was not related to the expression levels of p53, mFas, sFas, and FasL.
Fas signaling is derived from original aggregation of Fas and subsequent formation of the DISC (43). The DISC is composed of oligomerized receptors, FADD, procaspase-8, procaspase-10, and FLIPL. Activation of procaspase-8 with DISC results in the induction of Fas-mediated apoptosis (57), whereas FLIPL functions to inhibit procaspase-8 activation (58). Our observation that HBSP reduces the oligomerization of higher-order Fas and enhances the recruitment of FLIPL at the DISC may explain, at least in part, why HBSP could attenuate Fas-mediated apoptosis.
The PI3K/Akt signaling pathway is best known for its pivotal role in prosurvival of the cell. Several reports have shown that PI3K/Akt negatively regulated Fas apoptotic signaling through modulation of the proapoptotic molecules such as Bad, Bax, and Forkhead family members (59, 60) or the antiapoptotic factor NF-κB (61). Aside from these late events, early molecular modification of Fas signaling appears to play an important role as well in the antiapoptotic effect of PI3K/Akt pathway on Fas-mediated cell death. We and others have demonstrated that Akt abrogates the activation of caspase-8 by preventing procaspase-8 recruitment to the DISC in a FLIPL-dependent or -independent way (38, 61), and PI3K/Akt activation blocks the redistribution of DISC into lipid rafts (38, 62). Some cellular growth factors and cytokines are found to protect human hepatocytes from Fas-induced apoptosis in patients with active HBV infection through activation of the Akt pathway (29, 31), suggesting that activation of survival programs by Akt signaling contributes to an apoptosis-resistant phenotype in hepatocytes in the infected liver. All those observations comprised the basis of our study to evaluate the effect of HBSP on regulation of PI3K/Akt pathway in the context of Fas-mediated hepatocyte apoptosis. We have shown here that HBSP significantly increased the activity of the PI3K/Akt signaling pathway to protect hepatocytes from Fas-initiated apoptosis. Although it is not clear how HBSP activated PI3K/Akt signaling in view of its lack of physical interaction with PI3K signaling molecules and the absence of ER stress, the ability of pharmacological inhibition of PI3K to reverse the effect of HBSP on cell proliferation, apoptosis, Fas aggregation, and FLIPL expression implies that HBSP acts as an essential modulator of Fas-induced apoptosis in hepatocytes via a pathway involving PI3K/Akt activities.
PI3K/Akt/mTOR signaling is a master regulator of aerobic glycolytic metabolism (63). The active PI3K/Akt pathway has been demonstrated to enhance glucose uptake through upregulation of the expression of both cell surface glucose transporters and several glycolytic enzymes such as hexokinase (63). Moreover, Akt was found to promote activation of ATP citrate lyase, thereby enhancing the conversion of citrate to acetyl coenzyme A (acetyl-CoA) both in vitro and in vivo (64). One of the mechanisms by which PI3K/Akt signaling controls the transcription of glucose transporter is to activate mTORC1, which indirectly increases the expression of hypoxia-inducible factor 1 (HIF1) to promote glycolysis by diverting pyruvate into lactate, which is further enhanced by mTORC2-mediated activation of Akt via S473 phosphorylation (65). Although the exact molecular mechanism underlying HBSP attenuation of Fas-mediated hepatocyte apoptosis has not yet been established, the ability of HBSP to activate PI3K/Akt/mTOR signaling highlights the possibility that HBSP may participate in the modulation of cellular metabolism for hepatocyte growth and proliferation.
Although HBx is the best-studied HBV viral protein capable of inhibition of apoptosis and activation of HCC progression, the outcome of hepatocyte apoptosis is a result of complex biological processes, and there exist different regulations of multiple signaling pathways within the cell by the various HBV proteins. Our prior studies have documented that a 2.2-kb singly spliced HBV variant was present in all tumor and peritumor samples from 12 HCC patients tested (66) and that HBSP could enhance the migration and invasion of hepatoma cells and promote in vitro angiogenesis via activation of the pathways involving Akt signaling (9). Here, we demonstrated further that HBSP also contributed to the inhibition of Fas-induced hepatocyte apoptosis through strengthening the activity of the PI3K/Akt pathway. This finding may shed new light on the mechanism of HBV interference with the apoptosis signaling and HCC formation. In addition, the observation from this study that pharmacological inhibition of PI3K/Akt resensitized the HBSP-expressing hepatoma cells to Fas-induced apoptosis may serve as a steppingstone for restoring hepatoma cell death through favorable combinatory treatment using both PI3K inhibitors and extrinsic Fas agonists.
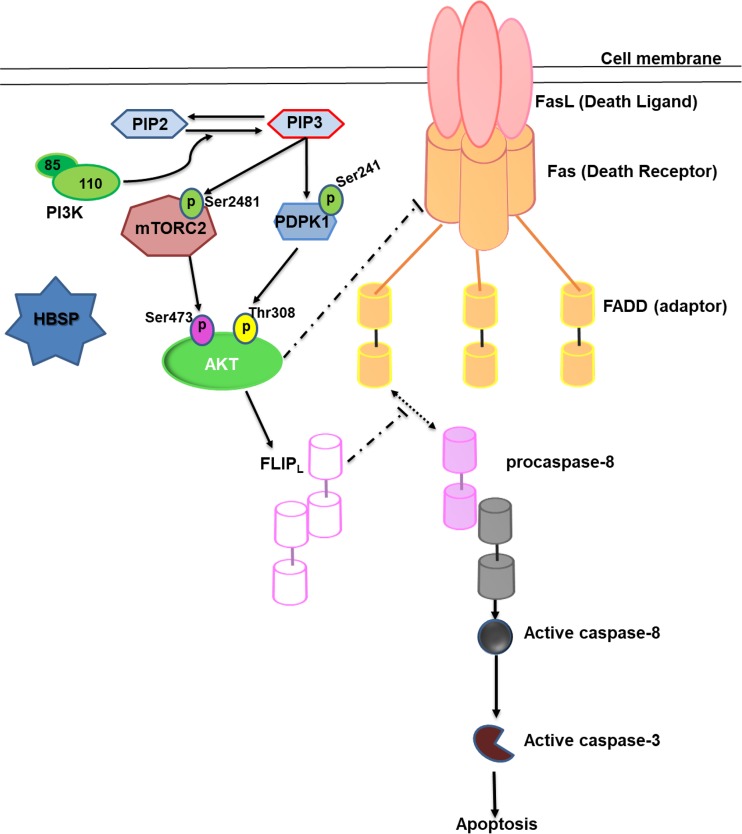
In summary, as proposed pictorially in Fig. 7, HBSP enhances PI3K activity via unknown intermediate steps resulting in the phosphorylation of mTORC2 and PDPK1 with consequent phosphorylation and activation of Akt. Upon activation, Akt inhibits Fas-mediated hepatocyte apoptosis through upregulation of FLIPL and disruption of Fas aggregation. To the best of our knowledge, this is the first evidence supporting the notion of involvement of the HBV viral protein HBSP in establishing cross talk between Fas and the PI3K/Akt pathway which may influence the outcome of HBV-related liver diseases.
FIG 7.
Schematic representation of HBSP regulation on PI3K/Akt signaling to affect Fas-apoptotic pathway.
MATERIALS AND METHODS
Plasmid construction.
A 2.2-kb spliced defective HBV DNA was previously isolated from a patient with chronic HBV infection (67) and served as a template for PCR amplification of the HBSP gene. The empty vector plasmid pFlag was constructed by inserting Flag tag sequence GATTACAAGGATGACGACGATAAG into the KpnI and XhoI sites of the plasmid pcDNA3.1/Hygro(+) (Invitrogen, Carlsbad, CA, USA). pHBSP was constructed by inserting the PCR-generated HBSP gene fused with Flag tag sequence with the forward primer 5′-CGGGGTACCGCCACCATGCCCCTATCTTATCAAC-3′ and reverse primer 5′-CCGCTCGAGCTACTTGTCGTCATCGTCTTTGTAGTCGTAAACTGAGCCA-3′ into the KpnI and XhoI sites of the plasmid pcDNA3.1/Hygro(+). pCDH-HBSP-Flag, used for establishing a stable HBSP-expressing cell line, was constructed by insertion of a PCR-generated HBSP gene into pCDH-CMV-MCS-EF1-Puro cDNA (System Biosciences, Palo Alto, CA, USA) between the XbaI and BamHI sites. The primers used were as follows: forward, 5′-CTAGTCTAGAGCCACCATGCCCCTATCTATCAACAC-3′; reverse, 5′-CGCGGATCCCTACTTGTCGTCATCGTCTTTGTAGTCGTAAACTGAGCCAGGAGAAACG-3′. pHBSP-Red expressing the fusion HBSP and Ds-Red fluorescent protein was constructed by inserting a PCR-generated HBSP gene into the XhoI and BamHI sites of the plasmid pDsRed-Monomer-Hyg-N1 vector (Clontech, Palo Alto, CA, USA). The primers used were as follows: forward, 5′-CGGAATTCATGCCCCTATCTTATCAACAC-3′; reverse, 5′-AACTGCAGCTAGTAAACTGAGCAGGAG-3′.
Cell line and culture.
The human hepatoma cell line HepG2 was maintained in minimum essential medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone Laboratories Inc., Logan, UT, USA) in a 5% CO2 humidified incubator at 37°C. Transfection was performed using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. Cryopreserved primary human hepatocytes (PHH) were purchased from Bioreclamation IVT (Brussels, Belgium) and cultured in William’s medium E (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin.
Reagents and antibodies.
Agonistic human monoclonal antibody anti-Fas CH11 was supplied by MBL (SY-001; Nagoya, Japan). Actinomycin D (ActD) was obtained from Sigma (A1410; Sigma-Aldrich), and hygromycin was supplied by Merck KGaA (400049-5MU; Darmstadt, Germany). Anti-caspase-8 antibody was purchased from Cell Signaling Technology (catalog no. 9746; Danvers, MA, USA); it detects endogenous levels of full length caspase-8 (57 kDa), the cleaved intermediate p43/p41, or the caspase-8 active fragment p18. FasL, anti-Fas, anti-P53, anti-FLIPL, anti-caspase-3, anti-FADD, anti-Akt, anti-p-Akt (Ser473 or Thr308), anti-PDPK1 and anti-mTOR, anti-p-PDPK1 (Ser241), anti-p-mTOR (Ser2481), anti-Flag, anti-PI3Kp85, anti-PI3Kp110α, anti-PI3Kp110β, anti-PTEN, anti-SHP1, anti-IRS1, anti-Ras, anti-GRP94, anti-eIF2α, anti-p-eIF2α (Ser51), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) antibodies and PI3K inhibitor LY294002 were purchased from Cell Signaling Technology. Horseradish peroxidase (HRP)-coupled secondary antibodies were from Beyotime Biotechnology (Shanghai, China).
RNA interference.
For transient knockdown of PI3K, HepG2 cells were transfected with small interfering RNA (siRNA) oligonucleotides using Lipofectamine 3000 according to the manufacturer’s instructions. The sequences of two siRNAs each targeting both PIK3CA and PI3KCB genes were as follows: 5′-GGACCUCAAUUCACCUCAUTT-3′ and 5′-GCAACAGCUUUGCAUGUUATT-3′ (PI3K siRNA1) and 5′-GCAACCUACGUGAAUGUAATT-3′ and 5′-GCUGUCAAUCAAGUGGAAUTT-3′ (PI3K siRNA2). The sequence of nontargeting scrambled control was 5′-UUCUCCGAACGUGUCACGUTT-3′ (PI3K NC). The gene silencing effect was confirmed by Western blot analysis 48 h posttransfection.
Generation of stable HBSP-expressing hepatoma cell lines.
The HepG2 cells were transfected with pHBSP or the empty vector control pFlag and then selected in the presence of 400 μg/ml hygromycin for 4 weeks. The hygromycin-resistant clones were combined to create a multiclonal cell population and screened for the extent of HBSP protein expression by Western blot analysis.
Generation of HBSP-expressing recombinant lentivirus.
Thirty percent confluent 293T cell monolayers in 10-cm culture dishes were cotransfected with pCDH-HBSP-Flag and the packaging vectors of pMDL, p-VSV-G, and pREV (Invitrogen) and cultured for 48 h. The culture medium was collected and clarified by low-speed centrifugation. The supernatant after filtration through 0.45-μm filters was added into the culture of PHH cells grown in 6-cm culture dishes. The expression of HBSP was confirmed by Western blot analysis.
CCK-8 assay.
Cells were seeded into 96-well plates with 5 × 103 cells per well and cultured for 16 h. Different concentrations of anti-Fas CH11 or FasL plus 0.5 μg/ml ActD were added to the medium, and cells were incubated for another 72 h (HepG2) or 12 h (PHH). Cell counting kit 8 (CCK-8; Donjindo, Japan) was used to detect the viability of different cell lines. The absorbance (A) at the wavelength of 450 nm was measured using a microplate reader (Bio-Tek, Winooski, VT, USA). Cell viability was expressed according to the formula mean A value of the experimental group/mean A value of the control group.
Flow cytometry.
Cells were cultured in standard cell culture medium and incubated for 12 h with 2 μg/ml of anti-Fas CH11 plus 0.5 μg/ml ActD. Cells were washed twice and mixed with 5 μl propidium iodide (PI) and 5 μl annexin V. After a 15-min incubation at room temperature in the dark, fluorescence was analyzed by flow cytometry (FACSVerse; BD Biosciences, NJ, USA) using FACSuite software (BD Biosciences).
Western blot analysis.
Cells were lysed for 30 min at 4°C in Western and immunoprecipitation (IP) lysis buffer (Beyotime Biotechnology). Protein concentration was determined using the bicinchoninic acid (BCA) protein assay kit (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Proteins were separated by SDS-PAGE on a 12% or 8% gel under reducing conditions and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA, USA). The membrane was blocked for 1 h with Tris-buffered saline (TBS)–Tween 20 (50 mmol/liter Tris, 160 mmol/liter NaCl, 0.1% Tween 20, pH 7.8) containing 5% bovine serum albumin (BSA), and all subsequent steps were done in this buffer. Specific primary antibody was incubated overnight at 4°C. After intensive washes, the peroxidase-labeled HRP-coupled secondary antibody was added for 1 h, and the proteins were visualized with the enhanced BeyoECL Star (Beyotime Biotechnology).
qRT-PCR analysis.
Total RNA was extracted using the TRIzol reagent (Invitrogen) and reverse transcribed to cDNA by using the ExScript reverse transcription-PCR (RT-PCR) kit (TaKaRa, Shiga, Japan). Quantitative real-time PCR (qRT-PCR) was performed in the Mx3000P real-time PCR system (Agilent Technologies, Santa Clara, CA, USA) with the SYBR Premix Ex Taq kit (TaKaRa) according to the manufacturer’s instructions. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a reference gene, and relative mRNA levels were calculated using the threshold cycle (2−ΔΔCT) method. The paired forward and reverse primers were as follows: P1 and P2 for total Fas amplification, P3 and P4 for mFas amplification, 5′-TAACAGTTCCTGCATGGGCGGC-3′ and 5′-AGGACAGGCACAAACACGCACC-3′ for P53, 5′-GAAGTTGACTGCCTGCTGGCTTTCT-3′ and 5′-TGGGGCAACCAGATTTAGTTTCTCC-3′ for FLIPL, and 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-AGCTCAGGGATGACCTTGCC-3′ for GAPDH.
Semiquantitative RT-PCR analysis of mFas, sFas, and XBP1.
Transcribed cDNA was used as a template for PCR amplification. The paired primers P1 and P2 were used for total Fas amplification, and P3 and P5 were used for mFas/sFas amplification; primer sequences were reported previously (53). The paired forward and reverse primers for amplification of XBP1 were 5′-CTGGAAAGCAAGTGGTAGA-3′ and 5′-CTGGGTCCTTCTGGGTAGAC-3′. PCR products were analyzed on a 3% agarose gel by electrophoresis and ethidium bromide staining. A digital image of the gel was obtained using a Syngene apparatus (Syngene, San Diego, CA) and the Syngene GeneSnap software (version 4.00.00). Individual band intensities were quantitated using the densitometric software Quantity One (Bio-Rad Laboratories). sFas or mFas mRNA level was expressed as a ratio of sFas or mFas PCR product signal to that of GAPDH.
Fas splicing assay.
The Fas splicing assay was performed as we previously reported (53). Briefly, cells were transfected with Fas splicing reporter construct pCMV56-Fas, and total RNA was extracted 48 h after transfection for reverse transcription and subsequent PCR amplification. The PCR products were run on 3% agarose gels and analyzed by a digital imaging system.
ELISA.
sFas in cell culture supernatant was detected using a human sFas enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. The PI3K activity assay was performed with an ELISA K-1000s-PI3K activity kit (ELISA: Pico; Echelon Biosciences Inc., Salt Lake City, UT, USA) according to the manufacturer’s instructions. The amount of PIP3 was measured with the competitive PIP3 mass ELISA kit (K-2500s; Echelon Biosciences, Inc.) per the manufacturer’s recommendations. The absorbance of each well was measured at 450 nm using a microplate reader (Bio-Rad Laboratories). The concentration of sFas was calibrated from a dose-response curve based on reference standards.
Co-IP assay.
For the in vivo coimmunoprecipitation (Co-IP) experiments, transiently transfected HepG2 cells were lysed and the soluble proteins were precleared with 100 μl of a 50% slurry of protein A agarose (Invitrogen). The clear lysates were then mixed with 4 μg of antibodies and 100 μl of a 50% slurry of protein A agarose. The immunoprecipitated complexes were analyzed by Western blot analysis.
Immunofluorescent confocal microscopy.
HepG2 cells were transfected with pHBSP-Red and pDSRed-Monomer-Hyg-N1 vector (Clontech). Forty-eight hours after incubation, cells were fixed with 4% paraformaldehyde for 10 min followed by washing three times with phosphate-buffered saline (PBS) and staining with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology) and then examined under a confocal microscope (TCS SP8; Leica Microsystems Inc., Buffalo Grove, IL, USA).
Statistical analysis.
Statistical analysis was performed using SPSS software (SPSS22.0; SPSS Inc., Chicago, IL, USA). The statistical significance in the difference between mRNA and protein levels was analyzed by analysis of variance (ANOVA).
ACKNOWLEDGMENTS
This work was supported by grants from the Fujian Provincial Medical Innovation Project (no. 2012-CX-14), the Joint Research Program of Health and Planning Committee and Education Department of Fujian (no. WKJ-FJ-29), the State Key Project Specialized for Infectious Diseases (2017ZX10202203-005-002), and the Joint Funds for the Innovation of Science and Technology, Fujian Province (2016Y91030022).
REFERENCES
- 1.Trepo C, Chan HL, Lok A. 2014. Hepatitis B virus infection. Lancet 384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki T, Masui N, Kajino K, Saito I, Miyamura T. 1989. Detection and mapping of spliced RNA from a human hepatoma cell line transfected with the hepatitis B virus genome. Proc Natl Acad Sci U S A 86:8422–8426. doi: 10.1073/pnas.86.21.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu HL, Chen PJ, Tu SJ, Lin MH, Lai MY, Chen DS. 1991. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J Virol 65:1680–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunther S, Sommer G, Iwanska A, Will H. 1997. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 5.Soussan P, Garreau F, Zylberberg H, Ferray C, Brechot C, Kremsdorf D. 2000. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J Clin Invest 105:55–60. doi: 10.1172/JCI8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soussan P, Pol J, Garreau F, Schneider V, Le Pendeven C, Nalpas B, Lacombe K, Bonnard P, Pol S, Kremsdorf D. 2008. Expression of defective hepatitis B virus particles derived from singly spliced RNA is related to liver disease. J Infect Dis 198:218–225. doi: 10.1086/589623. [DOI] [PubMed] [Google Scholar]
- 7.Soussan P, Tuveri R, Nalpas B, Garreau F, Zavala F, Masson A, Pol S, Brechot C, Kremsdorf D. 2003. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J Hepatol 38:343–348. doi: 10.1016/S0168-8278(02)00422-1. [DOI] [PubMed] [Google Scholar]
- 8.Bayliss J, Lim L, Thompson AJ, Desmond P, Angus P, Locarnini S, Revill PA. 2013. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol 59:1022–1028. doi: 10.1016/j.jhep.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Chen WN, Chen JY, Jiao BY, Lin WS, Wu YL, Liu LL, Lin X. 2012. Interaction of the hepatitis B spliced protein with cathepsin B promotes hepatoma cell migration and invasion. J Virol 86:13533–13541. doi: 10.1128/JVI.02095-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pol JG, Lekbaby B, Redelsperger F, Klamer S, Mandouri Y, Ahodantin J, Bieche I, Lefevre M, Souque P, Charneau P, Gadessaud N, Kremsdorf D, Soussan P. 2015. Alternative splicing-regulated protein of hepatitis B virus hacks the TNF-alpha-stimulated signaling pathways and limits the extent of liver inflammation. FASEB J 29:1879–1889. doi: 10.1096/fj.14-258715. [DOI] [PubMed] [Google Scholar]
- 11.Duriez M, Mandouri Y, Lekbaby B, Wang H, Schnuriger A, Redelsperger F, Guerrera CI, Lefevre M, Fauveau V, Ahodantin J, Quetier I, Chhuon C, Gourari S, Boissonnas A, Gill U, Kennedy P, Debzi N, Sitterlin D, Maini MK, Kremsdorf D, Soussan P. 2017. Alternative splicing of hepatitis B virus: a novel virus/host interaction altering liver immunity. J Hepatol 67:687–699. doi: 10.1016/j.jhep.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S, Zhang YJ. 2017. Interference of apoptosis by hepatitis B virus. Viruses 9:230. doi: 10.3390/v9080230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol Pathol 35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagata S. 1997. Apoptosis by death factor. Cell 88:355–365. doi: 10.1016/S0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 15.Mochizuki K, Hayashi N, Hiramatsu N, Katayama K, Kawanishi Y, Kasahara A, Fusamoto H, Kamada T. 1996. Fas antigen expression in liver tissues of patients with chronic hepatitis B. J Hepatol 24:1–7. doi: 10.1016/S0168-8278(96)80178-4. [DOI] [PubMed] [Google Scholar]
- 16.Luo KX, Zhu YF, Zhang LX, He HT, Wang XS, Zhang L. 1997. In situ investigation of Fas/FasL expression in chronic hepatitis B infection and related liver diseases. J Viral Hepat 4:303–307. doi: 10.1046/j.1365-2893.1997.00053.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, Lee H, Yun Y. 1998. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem 273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 18.Terradillos O, de La Coste A, Pollicino T, Neuveut C, Sitterlin D, Lecoeur H, Gougeon ML, Kahn A, Buendia MA. 2002. The hepatitis B virus X protein abrogates Bcl-2-mediated protection against Fas apoptosis in the liver. Oncogene 21:377–386. doi: 10.1038/sj.onc.1205110. [DOI] [PubMed] [Google Scholar]
- 19.Kim HJ, Kim SY, Kim J, Lee H, Choi M, Kim JK, Ahn JK. 2008. Hepatitis B virus X protein induces apoptosis by enhancing translocation of Bax to mitochondria. IUBMB Life 60:473–480. doi: 10.1002/iub.68. [DOI] [PubMed] [Google Scholar]
- 20.Liang X, Liu Y, Zhang Q, Gao L, Han L, Ma C, Zhang L, Chen YH, Sun W. 2007. Hepatitis B virus sensitizes hepatocytes to TRAIL-induced apoptosis through Bax. J Immunol 178:503–510. doi: 10.4049/jimmunol.178.1.503. [DOI] [PubMed] [Google Scholar]
- 21.Diao J, Khine AA, Sarangi F, Hsu E, Iorio C, Tibbles LA, Woodgett JR, Penninger J, Richardson CD. 2001. X protein of hepatitis B virus inhibits Fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J Biol Chem 276:8328–8340. doi: 10.1074/jbc.M006026200. [DOI] [PubMed] [Google Scholar]
- 22.Gottlob K, Fulco M, Levrero M, Graessmann A. 1998. The hepatitis B virus HBx protein inhibits caspase 3 activity. J Biol Chem 273:33347–33353. doi: 10.1074/jbc.273.50.33347. [DOI] [PubMed] [Google Scholar]
- 23.Pan J, Duan L-X, Sun BS, Feitelson MA. 2001. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-κB. J Gen Virol 82:171–182. doi: 10.1099/0022-1317-82-1-171. [DOI] [PubMed] [Google Scholar]
- 24.Wang XW, Gibson MK, Vermeulen W, Yeh H, Forrester K, Sturzbecher HW, Hoeijmakers JH, Harris CC. 1995. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res 55:6012–6016. [PubMed] [Google Scholar]
- 25.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. 2000. Hepatitis B virus X protein inhibits transforming growth factor-beta-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem 275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 26.Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. 2013. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog 9:e1003722. doi: 10.1371/annotation/19deb814-0602-4909-8605-61ab6d79f85f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X, Lee M, Tran T, Block T. 2005. High level expression of apoptosis inhibitor in hepatoma cell line expressing hepatitis B virus. Int J Med Sci 2:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH. 2011. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 29.Schulze-Bergkamen H, Brenner D, Krueger A, Suess D, Fas SC, Frey CR, Dax A, Zink D, Büchler P, Müller M, Krammer PH. 2004. Hepatocyte growth factor induces Mcl-1 in primary human hepatocytes and inhibits CD95-mediated apoptosis via Akt. Hepatology 39:645–654. doi: 10.1002/hep.20138. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki A, Hayashida M, Kawano H, Sugimoto K, Nakano T, Shiraki K. 2000. Hepatocyte growth factor promotes cell survival from Fas-mediated cell death in hepatocellular carcinoma cells via Akt activation and Fas-death-inducing signaling complex suppression. Hepatology 32:796–802. doi: 10.1053/jhep.2000.17738. [DOI] [PubMed] [Google Scholar]
- 31.Moumen A, Ieraci A, Patane S, Sole C, Comella JX, Dono R, Maina F. 2007. Met signals hepatocyte survival by preventing Fas-triggered FLIP degradation in a PI3k-Akt-dependent manner. Hepatology 45:1210–1217. doi: 10.1002/hep.21604. [DOI] [PubMed] [Google Scholar]
- 32.Xiao GH, Jeffers M, Bellacosa A, Mitsuuchi Y, Vande Woude GF, Testa JR. 2001. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A 98:247–252. doi: 10.1073/pnas.98.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osawa Y, Banno Y, Nagaki M, Brenner DA, Naiki T, Nozawa Y, Nakashima S, Moriwaki H. 2001. TNF-alpha-induced sphingosine 1-phosphate inhibits apoptosis through a phosphatidylinositol 3-kinase/Akt pathway in human hepatocytes. J Immunol 167:173–180. doi: 10.4049/jimmunol.167.1.173. [DOI] [PubMed] [Google Scholar]
- 34.Osaki M, Kase S, Adachi K, Takeda A, Hashimoto K, Ito H. 2004. Inhibition of the PI3K-Akt signaling pathway enhances the sensitivity of Fas-mediated apoptosis in human gastric carcinoma cell line, MKN-45. J Cancer Res Clin Oncol 130:8–14. doi: 10.1007/s00432-003-0505-z. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L, Derijard B, Chakrabandhu K, Wang BS, Chen HZ, Hueber AO. 2014. Synergism of PI3K/Akt inhibition and Fas activation on colon cancer cell death. Cancer Lett 354:355–364. doi: 10.1016/j.canlet.2014.08.038. [DOI] [PubMed] [Google Scholar]
- 36.Bertram J, Peacock JW, Tan C, Mui AL, Chung SW, Gleave ME, Dedhar S, Cox ME, Ong CJ. 2006. Inhibition of the phosphatidylinositol 3'-kinase pathway promotes autocrine Fas-induced death of phosphatase and tensin homologue-deficient prostate cancer cells. Cancer Res 66:4781–4788. doi: 10.1158/0008-5472.CAN-05-3173. [DOI] [PubMed] [Google Scholar]
- 37.Nath S, Mandal C, Chatterjee U, Mandal C. 2018. Association of cytosolic sialidase Neu2 with plasma membrane enhances Fas-mediated apoptosis by impairing PI3K-Akt/mTOR-mediated pathway in pancreatic cancer cells. Cell Death Dis 9:210. doi: 10.1038/s41419-017-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W, Jing ZT, Wu SX, He Y, Lin YT, Chen WN, Lin XJ, Lin X. 2018. A novel AKT activator, SC79, prevents acute hepatic failure induced by Fas-mediated apoptosis of hepatocytes. Am J Pathol 188:1171–1182. doi: 10.1016/j.ajpath.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Ramaswamy M, Cleland SY, Cruz AC, Siegel RM. 2009. Many checkpoints on the road to cell death: regulation of Fas-FasL interactions and Fas signaling in peripheral immune responses. Results Probl Cell Differ 49:17–47. doi: 10.1007/400_2008_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen-Schaub LB, Zhang W, Cusack JC, Angelo LS, Santee SM, Fujiwara T, Roth JA, Deisseroth AB, Zhang WW, Kruzel E. 1995. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol 15:3032–3040. doi: 10.1128/MCB.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD. 1994. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 42.Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6:1089–1098. doi: 10.1016/S1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 43.Algeciras-Schimnich A, Shen L, Barnhart BC, Murmann AE, Burkhardt JK, Peter ME. 2002. Molecular ordering of the initial signaling events of CD95. Mol Cell Biol 22:207–220. doi: 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin L, Wang Z, Tao L, Wang Y. 2010. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 45.Chen CH, Shaikenov T, Peterson TR, Aimbetov R, Bissenbaev AK, Lee SW, Wu J, Lin HK, Sarbassov DD. 2011. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Sci Signal 4:ra10. doi: 10.1126/scisignal.2001731. [DOI] [PubMed] [Google Scholar]
- 46.Revill P, Testoni B, Locarnini S, Zoulim F. 2016. Global strategies are required to cure and eliminate HBV infection. Nat Rev Gastroenterol Hepatol 13:239–248. doi: 10.1038/nrgastro.2016.7. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y, Wang J, Wang Y, Wang A, Guo H, Wei F, Mehta SR, Espitia S, Smith DM, Liu L, Zhang Y, Chen D. 2016. A novel mutant 10Ala/Arg together with mutant 144Ser/Arg of hepatitis B virus X protein involved in hepatitis B virus-related hepatocarcinogenesis in HepG2 cell lines. Cancer Lett 371:285–291. doi: 10.1016/j.canlet.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao CC. 2016. Inhibition of apoptosis by oncogenic hepatitis B virus X protein: implications for the treatment of hepatocellular carcinoma. World J Hepatol 8:1061–1066. doi: 10.4254/wjh.v8.i25.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yun C, Um HR, Jin YH, Wang JH, Lee MO, Park S, Lee JH, Cho H. 2002. NF-kappaB activation by hepatitis B virus X (HBx) protein shifts the cellular fate toward survival. Cancer Lett 184:97–104. doi: 10.1016/S0304-3835(02)00187-8. [DOI] [PubMed] [Google Scholar]
- 50.Su F, Schneider RJ. 1997. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci U S A 94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu YP, Yang XN, Jazag A, Pan JS, Hu TH, Liu JJ, Guleng B, Ren JL. 2012. HBsAg inhibits the translocation of JTB into mitochondria in HepG2 cells and potentially plays a role in HCC progression. PLoS One 7:e36914. doi: 10.1371/journal.pone.0036914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Liu H, Xu J, Zhou L, Yun X, Chen L, Wang S, Sun L, Wen Y, Gu J. 2011. Hepatitis B virus large surface antigen promotes liver carcinogenesis by activating the Src/PI3K/Akt pathway. Cancer Res 71:7547–7557. doi: 10.1158/0008-5472.CAN-11-2260. [DOI] [PubMed] [Google Scholar]
- 53.Liu W, Lin YT, Yan XL, Ding YL, Wu YL, Chen WN, Lin X. 2015. Hepatitis B virus core protein inhibits Fas-mediated apoptosis of hepatoma cells via regulation of mFas/FasL and sFas expression. FASEB J 29:1113–1123. doi: 10.1096/fj.14-263822. [DOI] [PubMed] [Google Scholar]
- 54.Jia B, Guo M, Li G, Yu D, Zhang X, Lan K, Deng Q. 2015. Hepatitis B virus core protein sensitizes hepatocytes to tumor necrosis factor-induced apoptosis by suppression of the phosphorylation of mitogen-activated protein kinase kinase 7. J Virol 89:2041–2051. doi: 10.1128/JVI.03106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu YW, Tan TL, Chan V, Chen WN. 2006. The HBSP gene is expressed during HBV replication, and its coded BH3-containing spliced viral protein induces apoptosis in HepG2 cells. Biochem Biophys Res Commun 351:64–70. doi: 10.1016/j.bbrc.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Galle PR, Hofmann WJ, Walczak H, Schaller H, Otto G, Stremmel W, Krammer PH, Runkel L. 1995. Involvement of the CD95 (APO-1/Fas) receptor and ligand in liver damage. J Exp Med 182:1223–1230. doi: 10.1084/jem.182.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Mann M, Krammer PH, Peter ME. 1997. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J 16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scaffidi C, Schmitz I, Krammer PH, Peter ME. 1999. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem 274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 59.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231–241. doi: 10.1016/S0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 60.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 61.Ozes ON, Mayo LD, Gustin JA, Pfeffer SR, Pfeffer LM, Donner DB. 1999. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature 401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 62.Pizon M, Rampanarivo H, Tauzin S, Chaigne-Delalande B, Daburon S, Castroviejo M, Moreau P, Moreau JF, Legembre P. 2011. Actin-independent exclusion of CD95 by PI3K/AKT signalling: implications for apoptosis. Eur J Immunol 41:2368–2378. doi: 10.1002/eji.201041078. [DOI] [PubMed] [Google Scholar]
- 63.Ward PS, Thompson CB. 2012. Signaling in control of cell growth and metabolism. Cold Spring Harb Perspect Biol 4:a006783. doi: 10.1101/cshperspect.a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, Jackson E, Aiello NM, Haas NB, Rebbeck TR, Judkins A, Won KJ, Chodosh LA, Garcia BA, Stanger BZ, Feldman MD, Blair IA, Wellen KE. 2014. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab 20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 66.Lin X, Wen Y, Wan D, Qian G, Gu J. 2002. Structural and functional analysis of 2.2 kb spliced variant of hepatitis B virus genomes isolated from liver tissues from hepatocellular carcinoma patients. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 16:11–15. [PubMed] [Google Scholar]
- 67.Chen JY, Chen WN, Liu LL, Lin WS, Jiao BY, Wu YL, Lin JY, Lin X. 2010. Hepatitis B spliced protein (HBSP) generated by a spliced hepatitis B virus RNA participates in abnormality of fibrin formation and functions by binding to fibrinogen gamma chain. J Med Virol 82:2019–2026. doi: 10.1002/jmv.21918. [DOI] [PubMed] [Google Scholar]