Genital infections by HSV-2 represent one of the most common sexually transmitted viral infections. The virus causes painful lesions and sores around the genitals or rectum. Intermittent release of the virus from infected tissues during sexual activities is the most common cause of transmission. At the molecular level, cell surface heparan sulfate (HS) is known to provide attachment sites for HSV-2. While the removal of HS during HSV-1 release has been shown, not much is known about the host factors and their regulators that contribute to HSV-2 release from natural target cell types. Here we suggest a role for the host enzyme heparanase in HSV-2 release. Our work reveals that in addition to the regulation of transcription by NF-κB, HPSE is also regulated posttranslationally by cathepsin L and that inhibition of heparanase activity directly affects HSV-2 release. We provide unique insights into the host mechanisms controlling HSV-2 egress and spread.
KEYWORDS: HSV-2, heparanase, viral egress
ABSTRACT
Herpes simplex virus 2 (HSV-2) can productively infect many different cell types of human and nonhuman origin. Here we demonstrate interconnected roles for two host enzymes, heparanase (HPSE) and cathepsin L, in HSV-2 release from cells. In vaginal epithelial cells, HSV-2 causes heparan sulfate shedding and upregulation in HPSE levels during the productive phase of infection. We also noted increased levels of cathepsin L and show that regulation of HPSE by cathepsin L via cleavage of HPSE proenzyme is important for infection. Furthermore, inhibition of HPSE by a specific inhibitor, OGT 2115, dramatically reduces HSV-2 release from vaginal epithelial cells. Likewise, we show evidence that the inhibition of cathepsin L is detrimental to the infection. The HPSE increase after infection is mediated by an increased NF-κB nuclear localization and a resultant activation of HPSE transcription. Together these mechanisms contribute to the removal of heparan sulfate from the cell surface and thus facilitate virus release from cells.
IMPORTANCE Genital infections by HSV-2 represent one of the most common sexually transmitted viral infections. The virus causes painful lesions and sores around the genitals or rectum. Intermittent release of the virus from infected tissues during sexual activities is the most common cause of transmission. At the molecular level, cell surface heparan sulfate (HS) is known to provide attachment sites for HSV-2. While the removal of HS during HSV-1 release has been shown, not much is known about the host factors and their regulators that contribute to HSV-2 release from natural target cell types. Here we suggest a role for the host enzyme heparanase in HSV-2 release. Our work reveals that in addition to the regulation of transcription by NF-κB, HPSE is also regulated posttranslationally by cathepsin L and that inhibition of heparanase activity directly affects HSV-2 release. We provide unique insights into the host mechanisms controlling HSV-2 egress and spread.
INTRODUCTION
Genital herpes is one of the most common, persistent, and highly infectious sexually transmitted diseases; it is caused by herpes simplex virus 2 (HSV-2) and, in many emerging first-time cases, by herpes simplex virus 1 (HSV-1) (1–4). Primarily, the sites of infection include the vulva and the vagina, with some cases involving the cervix and perianal region in women and typically the glans or the shaft of the penis in heterosexual men, whereas anal infection has also been reported for homosexual men (5–7). Primary and recurrent genital herpes infections result in lesions and inflammation around the genital area which are painful and cause distress (4). While there is no vaccination or cure against HSV-2, resistance against current therapies, such as acyclovir, has been reported (8). Furthermore, these therapies are more than a decade old and work on a single aspect of the viral life cycle, viral DNA replication. Novel therapeutic interventions that target different stages of viral infection, including viral entry, viral protein translation, and viral egress, need to be addressed to successfully curb this distressing disease. One method to generate novel antiviral drugs that target these viral pathways is to understand host factors that help facilitate the viral life cycle. In this study, we focused on the host enzyme heparanase (HPSE) and its regulators that help facilitate egress of the HSV-2 virions.
Human HPSE is an endoglycosidase with the distinction of being the only enzyme capable of degrading heparan sulfate (HS) (9–11), an evolutionarily conserved glycosaminoglycan that is present ubiquitously at the cell surface. HPSE is initially translated as a preproenzyme. Cleavage of a signal sequence by a signal peptidase leaves an inactive 65-kDa proHPSE, which undergoes further processing in the lysosomal compartment (10, 12). Proteolytic removal of an N-terminal 8-kDa linker by a lysosomal cysteine endopeptidase, cathepsin L, cleaves the C-terminal 50-kDa subunit, which remains associated as a noncovalent heterodimer in active HPSE (13, 14). Active HPSE is responsible for the degradation of cell surface HS, which is found covalently attached to a small set of extracellular matrix and plasma membrane proteins forming heparan sulfate proteoglycans (HSPG) (15, 16). Clearance of HS via HPSE modulates cell division and differentiation, tissue morphogenesis and architecture, and organismal physiology (16). HSV-2 encodes two envelope glycoproteins, gB and gC, which bind HS at the cell surface and initiate viral entry (17–19). We first reported that host-encoded HPSE is upregulated and required for the release of viral progeny from parent cells after HSV-1 infection, and subsequently, similar findings were reported for porcine reproductive and respiratory syndrome virus (PRRSV) infection (20, 21).
The premise of this study was to understand the role of HPSE in the egress of HSV-2 from its natural target cells. In this report, we show that HPSE is upregulated by the virus upon infection and serves to aid viral egress by preventing the newly released viral progeny from reattaching to cell surface HS. We also studied transcriptional and posttranslational regulators of HPSE and for the first time implicate cathepsin L in HSV release. We demonstrate that inhibition of HPSE and cathepsin L via commercially available inhibitors negatively impacts viral egress.
(This article was submitted to an online preprint archive [22].)
RESULTS
Loss of cell surface HS during infection.
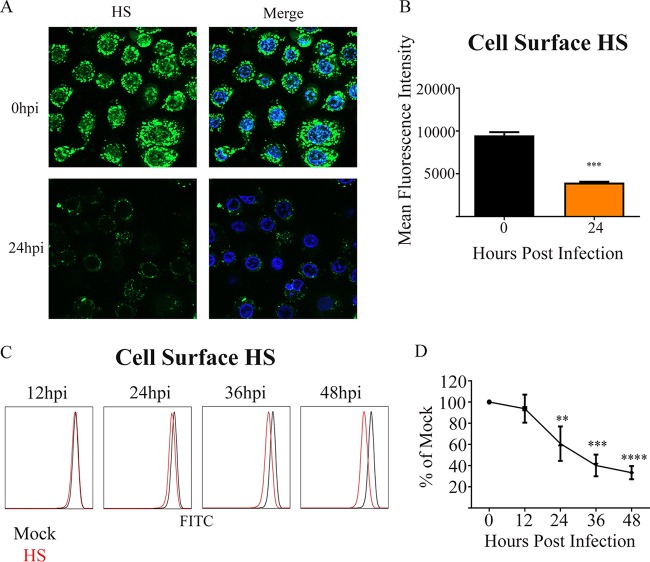
To understand how HSV-2 infection modulates cell surface HS levels, we infected a natural target cell type, human vaginal epithelial cells (VK2), with HSV-2 at a multiplicity of infection (MOI) of 1 for a period of 48 h. We observed that while mock-infected cells consistently showed large amounts of cell surface HS, most HS was cleared in HSV-2-infected cells by 24 h postinfection (hpi) (Fig. 1A and B). We also observed that there was a progressive loss of HS on the cell surface with time during infection using flow cytometric analysis (Fig. 1C and D).
FIG 1.
Loss of cell surface HS during infection. (A) Representative immunofluorescent images of HS stain. HSV-2 333 was used to infect cells at an MOI of 1 for 24 h. The upper left shows HS stain only in an uninfected sample, the upper right shows Hoechst and HS stains merged for an uninfected sample, the lower left shows HS stain only for an infected sample at 24 hpi, and the lower right shows Hoechst and HS stains merged for an infected sample at 24 hpi. (B) Quantification of HS cell surface expression. (C) Representative flow cytometry histogram showing change in cell surface HS expression, with red representing infected samples and black representing the uninfected control. (D) Quantification of cell surface HS flow cytometry experiments.
HPSE is upregulated after HSV-2 infection.
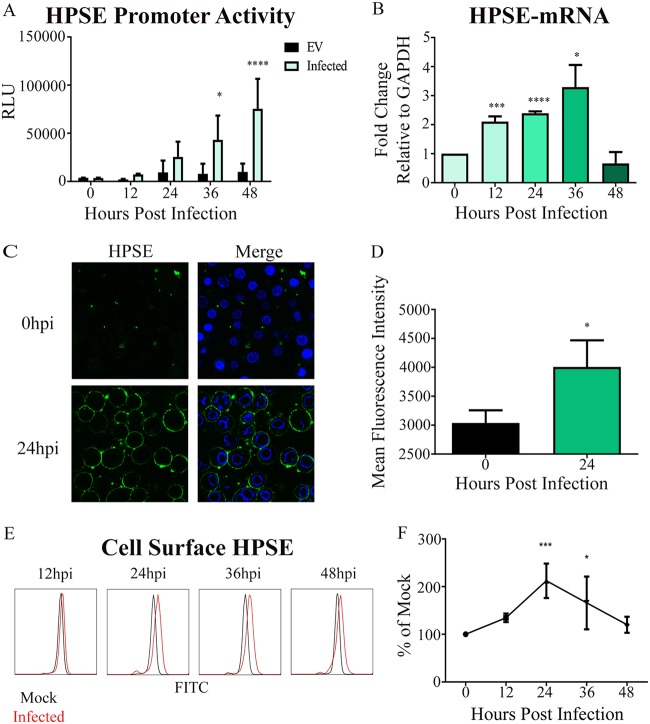
Given that clearance of HS correlated with duration of HSV-2 infection, we hypothesized an upregulation of HPSE expression. HPSE is the only mammalian enzyme known to cleave HS (9–11). In order to test our hypothesis, we analyzed HPSE promoter activity at 12, 24, 36, and 48 hpi using a luciferase reporter assay. As expected, we observed a significant increase in the HPSE promoter activity during the duration of infection (Fig. 2A). To understand this further, we then looked at HPSE mRNA transcript levels after HSV-2 infection. We observed by quantitative real-time PCR (qRT-PCR) analysis that HPSE mRNA was significantly elevated at 12, 24, and 36 hpi (Fig. 2B). Since HPSE expression was clearly upregulated inside the cells, we decided to look at HPSE translocation to the surface, which is the primary site for HS removal (16). Complementary to our HPSE mRNA results, after infection at an MOI of 1, there was a significant increase in cell surface HPSE protein levels, observed first by immunofluorescence microscopy, that showed a significant increase at 24 hpi (Fig. 2C and D). This increase in cell surface levels of HPSE was subsequently verified by flow cytometry (Fig. 2E and F). Taken together, our results confirmed an upregulation in HPSE levels upon infection.
FIG 2.
HPSE is upregulated after HSV-2 infection. (A) Increase in promoter activity of HPSE gene upon infection in HCE cells. HSV-2 333 was used to infect cells at an MOI of 1 for 12, 24, 36, and 48 h. Shown is the average fold increase over the uninfected control. Experimental values are normalized to those obtained with pGL3 as a control for transfection efficiency. (B) Increase in HPSE mRNA levels. Shown is the average fold increase over that at 0 h postinfection. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Representative immunofluorescence microscopy images of cell surface HPSE stain. HSV-2 333 was used to infect cells at an MOI of 1 for 24 h. The upper left shows HPSE stain only in an uninfected sample, the upper right shows Hoechst and HPSE stains merged for an uninfected sample, the lower left shows HPSE stain only for an infected sample at 24 hpi, and the lower right shows Hoechst and HPSE stains merged for an infected sample at 24 hpi. (D) Quantification of HPSE cell surface expression from multiple immunofluorescent images. (E) Representative flow cytometry histogram showing change in cell surface HPSE expression, with red representing infected samples and black representing the uninfected control. (F) Quantification of cell surface HPSE flow cytometry experiments compared to that at 0 h postinfection.
Mechanism of HPSE upregulation and activation upon infection.
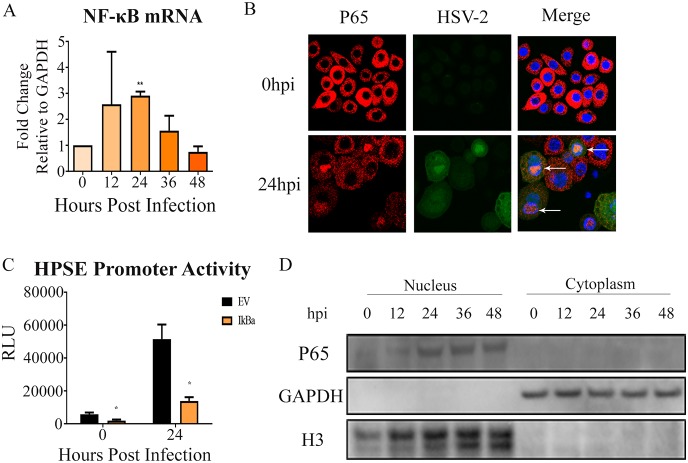
To understand the mechanism of HPSE regulation during HSV-2 infection, we studied transcriptional and posttranslational regulators of HPSE. It is reported that during HSV-1 infection, nuclear factor NF-κB (p65) activation and translocation to the nucleus could transcriptionally increase HPSE expression (23, 24). To understand if HSV-2 used a similar mode of action, we analyzed p65 activity during HSV-2 infection. As expected, there was a consistent increase in p65 mRNA expression in HSV-2-infected cells through 36 hpi (Fig. 3A). We also observed the nuclear translocation of p65 (Fig. 3B) at 24 hpi using immunofluorescence microscopy. These results corroborated our Western blot data, which showed significantly increased p65 in the nuclear fraction after HSV-2 infection (Fig. 3D). Next we wanted to understand if the inhibition of p65 activation and nuclear localization would affect HPSE promoter activity. We overexpressed a plasmid encoding mutant IkBa (S32A/S36A) in VK2 cells. This mutant IkBa is incapable of being phosphorylated and degraded, and as a result, it acts as a dominant negative protein that inhibits NF-κB activation and nuclear translocation (25). We did observe that expression of the IkBa mutant led to a decrease in HPSE promoter activity (Fig. 3C). Taken together, our findings suggest that virus-induced activation of NF-κB regulates HPSE expression.
FIG 3.
NF-κB as a mechanism for HPSE upregulation. (A) Increase in NF-κB (p65) mRNA levels. Shown is the average fold increase over the uninfected control. (B) Representative immunofluorescence microscopy images of nuclear translocation of p65 upon infection with HSV-2 333 GFP, with NF-κB P65 labeled red, HSV-2 green, and the nucleus blue. (C) Inhibition of NF-κB activation results in decreased HPSE promoter activity. VK2 cells were transfected with mutant IkBa incapable of degradation (S32A/S36A), thereby specifically inhibiting NF-κB activation and nuclear translocation. Twenty-four hours after transfection, cells were infected with HSV-2 333 for 24 h at an MOI of 1. Cell lysates were isolated, and a luciferase assay was performed. Results are normalized to those with empty pGL3 vector (EV) as a control for transfection efficiency. (D) Representative Western blot of nuclear translocation of p65 upon HSV-2 infection with HSV-2 333.
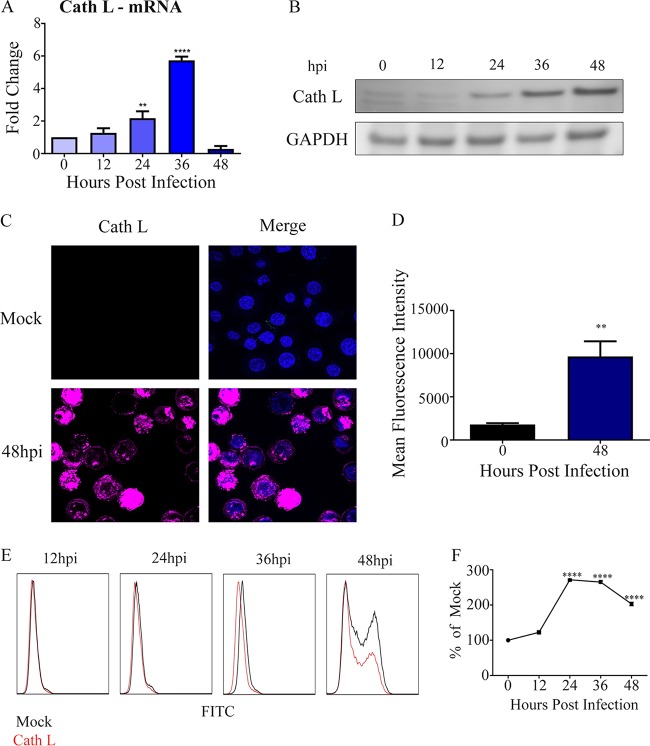
As described by others in the field, cathepsin L is the only known posttranslational activator of HPSE in mammalian cells (13, 26). Given that HPSE is upregulated upon HSV-2 infection, we wanted to assess whether its lysosomal activator, cathepsin L, also increased with HSV-2 infection. As observed, cathepsin L mRNA levels were upregulated at 12 hpi compared to those at 0 hpi and continued to increase at 24 and 36 hpi (Fig. 4A). We observed a similar trend in mature cathepsin L, which is known to be markedly stable (27); protein levels were highest at 48 hpi by Western blot analysis (Fig. 4B). These important findings were confirmed using immunofluorescence microscopy (Fig. 4C and D) and flow cytometry analysis (Fig. 4E and F). Collectively, our data suggest a connection between HSV-2 infection and HPSE/cathepsin L upregulation.
FIG 4.
Cathepsin L (Cath L) as a mechanism of HPSE activation (A) Increase in cathepsin L mRNA levels. Shown is the average fold increase over that at 0 h. (B) Representative Western blot showing increase of cathepsin L over time after infection with HSV-2 333 at an MOI of 1. (C) Representative immunofluorescence microscopy images of cathepsin L stain. HSV-2 333 was used to infect cells at an MOI of 1 for 24 h. The upper left shows cathepsin L stain only in an uninfected sample, the upper right shows Hoechst and cathepsin L stains merged for an uninfected sample, the lower left shows cathepsin L stain only for an infected sample at 24 hpi, and the lower right shows Hoechst and cathepsin L stains merged for an infected sample at 48 hpi. (D) Quantification of total cathepsin L expression from multiple immunofluorescent images. (E) Representative flow cytometry histogram showing change in cathepsin L expression, with red representing infected samples and black representing an uninfected control. (F) Quantification of cathepsin L flow cytometry experiments.
Effect of inhibition of cathepsin L and HPSE on infection.
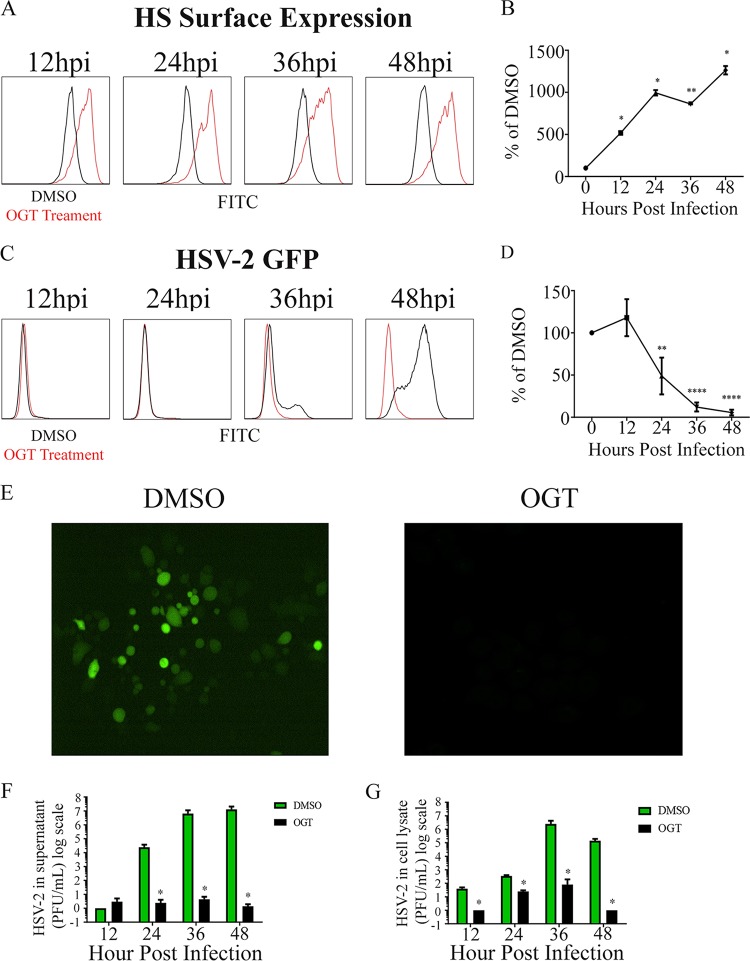
Having established a mechanism through which HPSE is upregulated and activated, we wanted to assess whether the inhibition of HPSE and cathepsin L would affect the HSV-2 viral life cycle. To ascertain the role of HPSE we used a well-characterized and commercially available small-molecule HPSE activity inhibitor, OGT 2115. This compound functionally blocks HPSE activity and does not significantly affect its expression (28). To verify the inhibition of HPSE activity during infection, we analyzed the cell surface HS expression. While cell surface HS expression during HSV-2 infection usually decreases, our results after pharmacological inhibition of HPSE showed a drastic increase in HS expression in cells treated with OGT cells versus those treated with dimethyl sulfoxide (DMSO) (Fig. 5A and B). Furthermore, we also observed a decrease in HSV-2 infection as measured by green fluorescent protein (GFP) reporter activity in the presence of OGT 2115 at 10 μM (Fig. 5C and D). Immunofluorescence microscopy data were in accordance with these results when we used the same HSV-2 GFP reporter virus and the same OGT 2115 concentration (Fig. 5E). We also observed a significant decrease in virus production and release using cell culture supernatant plaque assays through 48 hpi (Fig. 5F). These data combined with observations showing higher levels of virus in lysates of OGT 2115-treated cells (Fig. 6F) suggest that pharmacological inhibition of HPSE using OGT 2115 significantly increases HS expression on the cell surface while reducing the overall viral egress and spread.
FIG 5.
Effect of inhibition of HPSE on HSV-2 infection. (A) Representative flow cytometry histogram showing change in cell surface HS expression, with red representing OGT 2115 treatment and black representing DMSO treatment. Both samples were infected with HSV-2 333 at an MOI of 1. (B) Quantification of results of cell surface HS OGT flow cytometry experiments. (C) Representative flow cytometry histogram showing change in infection following OGT treatment, with red representing OGT 2115 treatment and black representing DMSO treatment. Both samples were infected with HSV-2 333 GFP. (D) Quantification of infection after treatment with OGT flow cytometry experiments. (E) Representative immunofluorescence microscopy images taken 24 h after infection with HSV-2 333 GFP after treatment with OGT or DMSO, with green representing infected cells. (F) Average change in virus released from cells with and without OGT treatment every 12 h up to 48 h. (G) Average change in virus from cell lysate with and without OGT treatment every 12 h up to 48 h.
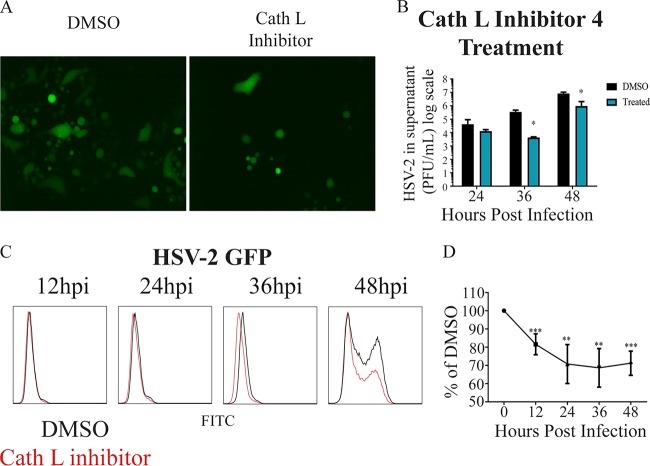
FIG 6.
Effect of inhibition of cathepsin L on HSV-2 infection. (A) Representative immunofluorescence microscopy images taken 24 h after infection with HSV-2 333 GFP after treatment with cathepsin L inhibitor IV or DMSO, with green representing infected cells. (B) Average change in virus released from cells with and without cathepsin L inhibitor IV treatment every 12 h up to 48 h. (C) Representative flow cytometry histogram showing change in infection after cathepsin L inhibitor IV treatment, with red representing cathepsin L inhibitor IV treatment and black representing DMSO treatment. Both samples were infected with HSV-2 333 GFP. (D) Quantification of infection after treatment with cathepsin L inhibitor IV flow cytometry experiments.
Next we wanted to understand if the inhibition of cathepsin L, the lysosomal activator of HPSE, would affect HSV-2 viral egress and spread. We used a well-characterized and commercially available small-molecule inhibitor of cathepsin L, cathepsin L inhibitor IV (29). Given that active HPSE remains in the lysosomal compartment for a period of 48 h before it is degraded (30), we hypothesized that cathepsin L would need to be inhibited for a period up to 48 h prior to and during infection to make sure no active HPSE is present throughout. As hypothesized, we saw a loss of infection in cathepsin L inhibitor IV-treated cells compared to the DMSO-treated control. These results were consistent when analyzed by immunofluorescence microscopy (Fig. 6A) and flow cytometry (Fig. 6C and D). We also observed through plaque assay that the amount of egressed virus found in the infected cell supernatant also decreased in cathepsin L inhibitor IV-treated cells compared to mock (DMSO)-treated samples, reaching significance at 48 hpi (Fig. 6B).
Effect of overexpression of HPSE during HSV-2 infection.
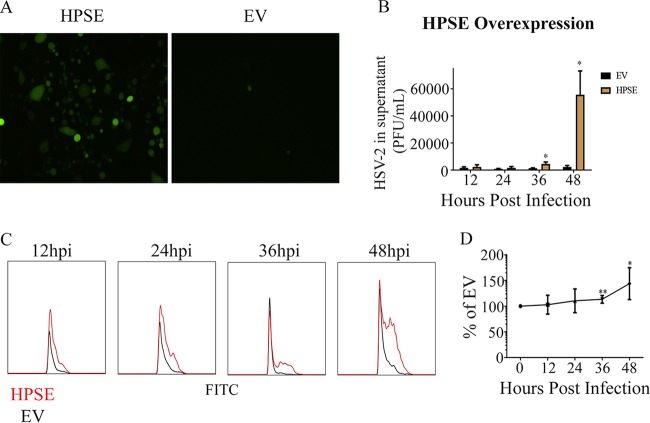
Through the experiments described in previous sections, we were able to establish that HPSE is important for HSV-2 release. Lastly, we wanted to understand if the overexpression of HPSE during a HSV-2 infection would be beneficial during viral proliferation. To study this, we overexpressed HPSE in VK2 cells for a period of 24 h followed by infection with HSV-2 (333) GFP at an MOI of 1. The cells were plated without any methylcellulose to allow release and spread of the extracellular virions. We observed via immunofluorescence microscopy a clear increase in virally infected cell clusters in the HPSE-transfected cells compared to the empty-vector control (Fig. 7A). It is important to note that although we saw less infection than expected, we normally find less infection in samples that have been previously transfected. Our results were in accordance with flow cytometry results (Fig. 7C), which showed that overexpression of HPSE led to a faster spread of infection, reaching significance at 36 and 48 hpi (Fig. 7D). We also observed an increase in released virus after HPSE overexpression (Fig. 7B) at 36 hpi and 48 hpi. Taken together, our results suggest a direct connection between higher levels of HPSE and HSV-2 release in the culture supernatant.
FIG 7.
Effect of overexpression of HPSE during HSV-2 infection. (A) Representative immunofluorescence microscopy images taken 24 h after infection with HSV-2 333 GFP after overexpression of HPSE, with green representing infected cells. (B) Average change in virus released from cells overexpressing HPSE and infected with HSV-2 333 every 12 h up to 48 h. (C) Representative flow cytometry histogram showing change in newly infected cells after overexpression of HPSE, with red representing HPSE overexpression and black representing EV. Both samples were infected with HSV-2 333 GFP. (D) Quantification of infection after overexpression of HPSE flow cytometry experiments.
DISCUSSION
Active HPSE is responsible for the degradation of cell surface HS. HS is found covalently attached to a small set of extracellular matrix and plasma membrane proteins forming heparan sulfate proteoglycans (HSPG) (15, 16). Clearance of HS via HPSE modulates cell division and differentiation, tissue morphogenesis and architecture, and organismal physiology (16). HSV-2 encodes two envelope glycoproteins, gB and gC, which bind HS at the cell surface and initiate viral entry (17–19). However, the new virions released can face a significant challenge if they bind with HS during viral egress. This phenomenon is commonly seen with other viruses, including influenza virus, in which the viral hemagglutinin binds to host sialic acid chains for entry and if host sialic acid is not removed, it can restrict viral release (31, 32). To eliminate the possibility of new virions binding upon release, the surface sialic acid residues are cleaved by a virus-encoded sialidase, neuraminidase, to prevent this problem for influenza virus (33, 34). In this regard, HSV-2 does not have any known viral proteins that can cleave cell surface HS. HSV-2 may have found the ability to cleave HS nevertheless from a host enzyme (20, 21). Given that HPSE is the only known mammalian enzyme capable of cleaving HS chains (9–11), we hypothesized and then demonstrated that its upregulation during infection may be beneficial for HSV-2 release from host cells.
Our evidence of HPSE upregulation during infection may suggest an additional role for HPSE in the exacerbation of the genital disease caused by HSV-2. During genital infection, distressing, painful lesions and sores around the genitals or rectum can result from a productive HSV-2 infection. These lesions are accompanied by inflammation and localized loss to tissue architecture (6). Recent studies have implicated HPSE overexpression in multiple pathological processes, including inflammation (35), angiogenesis (36), tumor metastasis (37), and atherosclerosis (38). It has been shown how heparanase contributes to HS cleavage and how this specifically results in rearrangement of the extracellular matrix as well as in controlling the release of many cell surface HS-linked molecules, such as growth factors, cytokines, and enzymes, in larger tissue-wide changes (9, 39).
To understand potential contributions of HPSE during HSV-2 infection, we first looked at HS chain expression on the cell surface during HSV-2 infection (41, 42). It became very clear to us that HS expression decreases over time during infection. This information also strengthened our goal to see how HPSE is modulated during infection. As suspected, HPSE was upregulated both transcriptionally and translationally, showing higher protein levels on the cell surface at 24, 36, and 48 hpi. Together, these results suggest that HSV-2-infected cells upregulate HPSE, which is then translocated to the cell surface, decreasing HS on the cell surface after infection.
Next we wanted to understand the mechanism behind the upregulation and activation of HPSE during infection. It was reported that nuclear factor NF-κB is upregulated upon HSV infection and that NF-κB upregulation has been linked to the transcriptional regulation of HPSE (20). Also, it is well known that cathepsin L, a lysosomal endopeptidase, is the only known activator of HPSE in mammalian cells. Hence, we looked at the transcriptional and protein levels of these two factors and how they change with infection. Through this study, we were able to show that HSV-2 infection of VK2 cells upregulated NF-κB and cathepsin L, which, in turn, orchestrated the expression and activation of HPSE.
Finally, we asked whether the inhibition of HPSE and cathepsin L would downregulate viral egress and spread. To study this, we used two well-known inhibitors, OGT 2115 and cathepsin L inhibitor IV, on VK2 cells during HSV-2 infection. We saw a significant loss of infection using both these inhibitors compared to vehicle controls, suggesting an alternate therapeutic modality against HSV-2 infections. Additionally, when we upregulated HPSE expression using an HPSE expression plasmid, we saw a significant increase in viral progeny and spread.
For our model, we propose that during the productive phase of HSV-2 infection of VK2 cells, HSV-2 causes an upregulation in HPSE levels that is mediated by an increased NF-κB (p65) nuclear localization and a resultant activation of HPSE transcription. In parallel, increased levels of cathepsin L contribute to HPSE proenzyme cleavage and activation. Together they contribute to the removal of heparan sulfate from the cell surface and thus facilitate virus release from cells. Higher levels of HPSE may also be a trigger for the breakdown of extracellular matrix and eventually a trigger for local inflammation. While the latter part is yet to be demonstrated, our studies discussed here directly implicate HPSE and cathepsin L in HSV-2 release.
MATERIALS AND METHODS
Cells and viruses.
The human vaginal epithelial cell VK2/E6E7 was obtained from the ATCC. VK2/E6E7 cells were passaged in keratinocyte serum-free medium (KSFM) (Gibco/BRL, Carlsbad, CA) supplemented with epidermal growth factor (EGF), bovine pituitary extract (BPE), and 1% penicillin-streptomycin. For convenience this cell line is referred to as VK2 throughout. All infections were done with HSV-2 333 at an MOI of 0.1 or 1 on VK2 cells unless mentioned otherwise. The Vero cell line (African green monkey kidney) was generously given by Patricia G. Spear (Northwestern University, Chicago, IL) and cultured in Dulbecco modified Eagle medium (DMEM; Gibco) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cathepsin L inhibitor IV (Santa Cruz Biotechnology) was used as a cathepsin L activity inhibitor (29). The cathepsin L inhibitor was used at a concentration of 10 μg/ml in DMSO unless otherwise specified. OGT 2115 (Tocris Biosciences) was used for heparanase activity inhibition and has been previously described as an HPSE inhibitor (28). OGT 2115 was used at 10 μM unless otherwise specified. Viruses used were wild-type (WT) HSV-2 333 and HSV-2 333 GFP (40). Virus stocks were grown and titered on Vero cells and stored at −80°C.
Antibodies and plasmids.
HPSE antibody H-80 (Santa Cruz Biotechnology) was used for imaging (1:100) and flow cytometry studies (1:100). Cathepsin L antibody ab58991 (Abcam, Cambridge, MA) was used for Western blot analysis (1:2,000), imaging (1:100), and flow cytometry studies (1:200). Histone H3 antibody (Cell Signaling Technology, Danvers, MA) was used at a dilution of 1:500 for Western blot analysis. Anti-human HS monoclonal antibody 10E4 (U.S. Biological, Salem, MA) was used for flow cytometry (1:100) and cell imaging (1:100). NF-κB (p65) antibody C-20 (Santa Cruz Biotechnology) was used for imaging (1:500) and Western blot analysis (1:2,000). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology) was used for Western blot analysis at a dilution of 1:2,000. The IkBa (S32/36A) plasmid was provided by Michael Karin (University of California, San Diego, La Jolla, CA). All transfections were performed using Lipofectamine 2000 (Life Technologies).
Western blot analysis.
Proteins from samples in this study were collected using radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s protocol. After gel electrophoresis, membranes were blocked in 5% bovine serum albumin (BSA) for 1 h, followed by incubation with primary antibody for 1 h and then incubation with respective secondary antibodies (anti-mouse [1:10,000] and anti-rabbit [1:10,000]) for 1 h. Protein bands were visualized using the SuperSignal West Femto maximum-sensitivity substrate (Thermo Scientific, Waltham, MA) with an ImageQuant LAS 4000 biomolecular imager (GE Healthcare Life Sciences, Pittsburgh, PA). The densities of the bands were quantified using ImageJ 1.52a image analysis software (NIH, USA).
PCR.
TRIzol (Life Technologies) was used to obtain RNA by following the protocol from the manufacturer, and then a cDNA reverse transcription kit from Applied Biosystems was used to transcribe to DNA by following the protocol from the manufacturer. Fast SYBR green master mix (Applied Biosystems) was used to perform real-time quantitative PCR, using QuantStudio 7 Flex (Applied Biosystems). The primers used in this study are as follows: HPSE forward primer 5ʹ-CTCGAAGAAAGACGGCTA-3ʹ and reverse primer 5ʹ-GTAGCAGTCCGTCCATTC-3ʹ, NF-κB (p65) forward primer 5ʹ-TGGGGACTACGACCTGAATG-3ʹ and reverse primer 5ʹ-GGGGGCACGATTGTCAAAGA-3ʹ, GAPDH forward primer 5ʹ-TCCACTGGCGTCTTCACC-3ʹ and reverse primer 5ʹ-GGCAGAGATGATGACCCTTTT-3ʹ, and cathepsin L forward primer 5ʹ- ATCTGGGCATGAACCACCTG-3ʹ and reverse primer 5ʹ-CAGCAAGCACCACAAGAACC-3ʹ.
Luciferase assay.
An empty vector, pGL3-Basic (Promega, Madison, WI), was used to control for transfection efficiency. The pHep1(0.7 kb)-luc plasmid, which expresses firefly luciferase that is driven by the 0.7-kb upstream promoter for transcription of human HPSE, was provided by Xiulong Xu (Rush University). Luciferase activity was measured using a Berthold Detection Systems luminometer.
Flow cytometry.
Measurement of HS and HPSE cell surface expression was performed after infection with WT HSV-2 333 infection. Monolayers of VK2 cells were infected at an MOI of 0.1 or 1 and then harvested at different times postinfection (12 hpi, 24 hpi, 36 hpi, and 48 hpi). Cells were harvested, fixed with 4% paraformaldehyde (PFA) for 10 min, and then incubated with 5% BSA for 1 h, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated anti-HS diluted 1:100 in phosphate-buffered saline (PBS) with 1% BSA for 1 h. Cells were then suspended in fluorescence-activated cell sorting (FACS) buffer (PBS, 5% FBS, 0.1% sodium azide). For detection of HPSE on the cell surface, cells were harvested, fixed with 4% PFA for 4 min, incubated with 5% BSA for 1 h, and then incubated with primary antibody diluted in PBS with 5% BSA for 1 h, followed by incubation with FITC-conjugated secondary antibody. Cells were then resuspended in FACS buffer. Cells stained with respective FITC-conjugated secondary antibodies only were used as background controls. Entire cell populations were used for the mean fluorescence intensity calculations.
Immunofluorescence microscopy.
VK2 cells were cultured in glass bottom dishes (MatTek Corporation, Ashland, MA). Cells were fixed in 4% PFA for 10 min, permeabilized with 0.1% Triton X-100 for intracellular labeling, incubated with 5% BSA for 1 h, and diluted with primary antibody in PBS with 5% BSA for 1 h and then with FITC-conjugated secondary. For HPSE surface staining, the protocol was 5% BSA for 1 h and dilution with primary antibody in PBS with 5% BSA for 1 h and then with FITC-conjugated secondary antibody. Imaging was done with a confocal microscope (710; Zeiss, Germany). The pinhole was set to 1 Airy unit. The fluorescence intensity of images was calculated using Zen software.
Plaque assay.
Viral egress titers were measured using a plaque assay. Monolayers of VK2 cells were plated in 12-well plates and infected with HSV-2 333 at an MOI of 0.1 or 1. Media were collected at different time points postinfection, and titers were determined on Vero cells. Primary incubation of collected media was performed with DPBS+/+ (Life Technologies) with 1% glucose and 1% heat-inactivated serum for 2 h. Vero cells were then incubated with growth media containing 5% methylcellulose for 72 h, followed by fixation with 100% methanol and staining with crystal violet solution.
Statistics.
Error bars in all figures represent standard errors from three independent experiments, unless otherwise specified. In figures, significant difference as determined by Student’s t test is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
ACKNOWLEDGMENTS
We acknowledge Ruth Zhelka for help with using the departmental imaging facilities.
This work was supported by grants from the NIH (AI128171, EY029426, and AI139768) to D.S. J.H. was supported by ISPB grant G3108. T.Y. was supported by ISPB grant G3126. A.M.A. was supported by fellowship F30EY025981.
REFERENCES
- 1.Xu F, Sternberg MR, Gottlieb SL, Berman SM, Markowitz LE, Forhan SE, Taylor LD. 2010. Seroprevalence of herpes simplex virus type 2 among persons aged 14–49 years—United States, 2005–2008. MMWR Morb Mortal Wkly Rep 59:456–459. [PubMed] [Google Scholar]
- 2.Kinghorn GR. 1993. Genital herpes: natural history and treatment of acute episodes. J Med Virol 1:33–38. [DOI] [PubMed] [Google Scholar]
- 3.Scoular A, Norrie J, Gillespie G, Mir N, Carman WF. 2002. Longitudinal study of genital infection by herpes simplex virus type 1 in Western Scotland over 15 years. BMJ 324:1366–1367. doi: 10.1136/bmj.324.7350.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wald A. 2006. Genital HSV-1 infections. Sex Transm Infect 82:189–190. doi: 10.1136/sti.2006.019935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, Berman SM, Markowitz LE. 2006. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296:964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- 6.Jaishankar D, Shukla D. 2016. Genital herpes: insights into sexually transmitted infectious disease. Microb Cell 3:438–450. doi: 10.15698/mic2016.09.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halpern-Felsher BL, Cornell JL, Kropp RY, Tschann JM. 2005. Oral versus vaginal sex among adolescents: perceptions, attitudes, and behavior. Pediatrics 115:845–851. doi: 10.1542/peds.2004-2108. [DOI] [PubMed] [Google Scholar]
- 8.Strick LB, Wald A, Celum C. 2006. Management of herpes simplex virus type 2 infection in HIV type 1-infected persons. Clin Infect Dis 43:347–356. doi: 10.1086/505496. [DOI] [PubMed] [Google Scholar]
- 9.Vlodavsky I, Ilan N, Naggi A, Casu B. 2007. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des 13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 10.Fairbanks MB, Mildner AM, Leone JW, Cavey GS, Mathews WR, Drong RF, Slightom JL, Bienkowski MJ, Smith CW, Bannow CA, Heinrikson RL. 1999. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem 274:29587–29590. doi: 10.1074/jbc.274.42.29587. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Viola CM, Brzozowski AM, Davies GJ. 2015. Structural characterization of human heparanase reveals insights into substrate recognition. Nat Struct Mol Biol 22:1016–1022. doi: 10.1038/nsmb.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fux L, Feibish N, Cohen-Kaplan V, Gingis-Velitski S, Feld S, Geffen C, Vlodavsky I, Ilan N. 2009. Structure-function approach identifies a C-terminal domain that mediates heparanase signaling. Cancer Res 69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulombe R, Grochulski P, Sivaraman J, Ménard R, Mort JS, Cygler M. 1996. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J 15:5492–5503. doi: 10.1002/j.1460-2075.1996.tb00934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishidoh K, Towatari T, Imajoh S, Kawasaki H, Kominami E, Katunuma N, Suzuki K. 1987. Molecular cloning and sequencing of cDNA for rat cathepsin L. FEBS Lett 223:69–73. doi: 10.1016/0014-5793(87)80511-2. [DOI] [PubMed] [Google Scholar]
- 15.Bishop JR, Schuksz M, Esko JD. 2007. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 16.Esko JD, Selleck SB. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 17.Spear PG, Eisenberg RJ, Cohen GH. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 18.Gruenheid S, Gatzke L, Meadows H, Tufaro F. 1993. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J Virol 67:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spear PG, Shieh MT, Herold BC, WuDunn D, Koshy TI. 1992. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol 313:341–353. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- 20.Hadigal SR, Agelidis AM, Karasneh GA, Antoine TE, Yakoub AM, Ramani VC, Djalilian AR, Sanderson RD, Shukla D. 2015. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat Commun 6:6985. doi: 10.1038/ncomms7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo C, Zhu Z, Guo Y, Wang X, Yu P, Xiao S, Chen Y, Cao Y, Liu X. 2017. Heparanase upregulation contributes to porcine reproductive and respiratory syndrome virus release. J Virol 91:e00625-17. doi: 10.1128/JVI.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopkins J, Yadavalli T, Agelidis AM, Shukla D. 2018. Host enzymes heparanase and cathepsin L promote herpes simplex virus 2 release from cells. bioRxiv doi: 10.1101/364562. [DOI] [PMC free article] [PubMed]
- 23.Kasperczyk H, La Ferla-Bruhl K, Westhoff MA, Behrend L, Zwacka RM, Debatin KM, Fulda S. 2005. Betulinic acid as new activator of NF-kappaB: molecular mechanisms and implications for cancer therapy. Oncogene 24:6945–6956. doi: 10.1038/sj.onc.1208842. [DOI] [PubMed] [Google Scholar]
- 24.Goldshmidt O, Zcharia E, Abramovitch R, Metzger S, Aingorn H, Friedmann Y, Schirrmacher V, Mitrani E, Vlodavsky I. 2002. Cell surface expression and secretion of heparanase markedly promote tumor angiogenesis and metastasis. Proc Natl Acad Sci U S A 99:10031–10036. doi: 10.1073/pnas.152070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. 1997. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature 388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 26.Bakhshi R, Goel A, Seth P, Chhikara P, Chauhan SS. 2001. Cloning and characterization of human cathepsin L promoter. Gene 275:93–101. doi: 10.1016/S0378-1119(01)00650-3. [DOI] [PubMed] [Google Scholar]
- 27.Jordans S, Jenko-Kokalj S, Kühl NM, Tedelind S, Sendt W, Brömme D, Turk D, Brix K. 2009. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem 10:23. doi: 10.1186/1471-2091-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Courtney SM, Hay PA, Buck RT, Colville CS, Phillips DJ, Scopes DI, Pollard FC, Page MJ, Bennett JM, Hircock ML, McKenzie EA, Bhaman M, Felix R, Stubberfield CR, Turner PR. 2005. Furanyl-1,3-thiazol-2-yl and benzoxazol-5-yl acetic acid derivatives: novel classes of heparanase inhibitor. Bioorg Med Chem Lett 15:2295–2299. doi: 10.1016/j.bmcl.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Yasuma T, Oi S, Choh N, Nomura T, Furuyama N, Nishimura A, Fujisawa Y, Sohda T. 1998. Synthesis of peptide aldehyde derivatives as selective inhibitors of human cathepsin L and their inhibitory effect on bone resorption. J Med Chem 41:4301–4308. doi: 10.1021/jm9803065. [DOI] [PubMed] [Google Scholar]
- 30.Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea BB, Joyce JA, Vlodavsky I. 2008. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem 283:18167–18176. doi: 10.1074/jbc.M801327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki Y, Ito T, Suzuki T, Holland RE, Chambers TM, Kiso M, Ishida H, Kawaoka Y. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 74:11825–11831. doi: 10.1128/JVI.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, Cox NJ, Paulson JC, Donis RO. 2012. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baum LG, Paulson JC. 1991. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology 180:10–15. doi: 10.1016/0042-6822(91)90003-T. [DOI] [PubMed] [Google Scholar]
- 34.Schulman JL. 1969. The role of antineuraminidase antibody in immunity to influenza virus infection. Bull World Health Organ 41:647–650. [PMC free article] [PubMed] [Google Scholar]
- 35.Changyaleket B, Chong ZZ, Dull RO, Nanegrungsunk D, Xu H. 2017. Heparanase promotes neuroinflammatory response during subarachnoid hemorrhage in rats. J Neuroinflammation 14:137. doi: 10.1186/s12974-017-0912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poupard N, Badarou P, Fasani F, Groult H, Bridiau N, Sannier F, Bordenave-Juchereau S, Kieda C, Piot JM, Grillon C, Fruitier-Arnaudin I, Maugard T. 2017. Assessment of heparanase-mediated angiogenesis using microvascular endothelial cells: identification of lambda-carrageenan derivative as a potent anti angiogenic agent. Mar Drugs 15:134. doi: 10.3390/md15050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiao F, Bai S, Ma Y, Yan Z, Yue Z, Yu Y, Wang X, Wang J. 2014. DNA methylation of heparanase promoter influences its expression and associated with the progression of human breast cancer. PLoS One 9:e92190. doi: 10.1371/journal.pone.0092190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Planer D, Metzger S, Zcharia E, Wexler ID, Vlodavsky I, Chajek-Shaul T. 2011. Role of heparanase on hepatic uptake of intestinal derived lipoprotein and fatty streak formation in mice. PLoS One 6:e18370. doi: 10.1371/journal.pone.0018370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderson RD, Elkin M, Rapraeger AC, Ilan N, Vlodavsky I. 2017. Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J 284:42–55. doi: 10.1111/febs.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez WM, Spear PG. 2002. Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex virus types 1 and 2 but not for pseudorabies virus or bovine herpesvirus 1. J Virol 76:7255–7262. doi: 10.1128/JVI.76.14.7255-7262.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla D, Spear PG. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest 108:503–510. doi: 10.1172/JCI200113799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agelidis AM, Shukla D. 2015. Cell entry mechanisms of HSV: what we have learned in recent years. Future Virol 10:1145–1154. doi: 10.2217/fvl.15.85. [DOI] [PMC free article] [PubMed] [Google Scholar]