Our finding that HCMV IE1 protected hPML from degradation by HSV ICP0 is important, because the PML body (aka ND10) is believed to be the first line of host intrinsic defense against herpesviral infection. How the infected viruses overcome the nuclear defensive structure (PML body) has not been fully understood. Herpesviral proteins, ICP0 of HSV and IE1 of CMV, have been identified to interact with PML. Here, we report that HCMV IE1 incompletely deSUMOylated PML, resulting in the mono-SUMOylated PML, which is consistent with the report of Schilling et al. (J Virol 91:e02049-16, 2017, https://doi.org/10.1128/JVI.02049-16). The mono-SUMOylated PML was subjected to degradation by HSV ICP0. However, it was protected by IE1 from degradation by ICP0 or HSV-1 infection. In contrast, IE1 with L174P mutation lost the function of deSUMOylating PML and failed to protect the degradation of the mono-SUMOylated PML. Whether the mono-SUMOylated PML has any defensive function against viral infection will be further investigated.
KEYWORDS: cytomegalovirus (CMV), immediate-early protein 1 (IE1), herpes simplex virus (HSV), infected cellular protein (ICP0), promyelocytic leukemia protein (PML), nuclear domain 10 (ND10), SUMOylation, HSV-1
ABSTRACT
To countermeasure the host cellular intrinsic defense, cytomegalovirus (CMV) and herpes simplex viruses (HSV) have evolved the ability to disperse nuclear domain 10 (ND10, aka PML body). However, mechanisms underlying their action on ND10 differ. HSV infection produces ICP0, which degrades the ND10-forming protein PML. Human CMV (HCMV) infection expresses IE1 that deSUMOylates PML to result in dispersion of ND10. It has been demonstrated that HSV ICP0 degraded only the SUMOylated PML, so we hypothesized that HCMV IE1 can protect PML from degradation by ICP0. HCMV IE1-expressing cell lines (U-251 MG-IE1 and HELF-IE1) were used for infection of HSV-1 or transfection of ICP0-expressing plasmid. Multilabeling by immunocytochemistry assay and protein examination by Western blot assay were performed to determine the resultant fate of PML caused by ICP0 in the presence or absence of HCMV IE1. Here, we report that deSUMOylation of human PML (hPML) by HCMV IE1 was incomplete, as mono-SUMOylated PML remained in the IE1-expressing cells, which is consistent with the report by E. M. Schilling, M. Scherer, N. Reuter, J. Schweininger, et al. (J Virol 91:e02049-16, 2017, https://doi.org/10.1128/JVI.02049-16). As expected, we found that IE1 protected PML from degradation by ICP0 or HSV-1 infection. An in vitro study found that IE1 with mutation of L174P failed to deSUMOylate PML and did not protect PML from degradation by ICP0; hence, we conclude that the deSUMOylation of PML is important for IE1 to protect PML from degradation by ICP0. In addition, we revealed that murine CMV failed to deSUMOylate and to protect the HSV-mediated degradation of hPML, and that HCMV failed to deSUMOylate and protect the HSV-mediated degradation of mouse PML. However, IE1-expressing cells did not enhance wild-type HSV-1 replication but significantly increased ICP0-defective HSV-1 replication at a low multiplicity of infection. Therefore, our results uncovered a host-virus functional interaction at the posttranslational level.
IMPORTANCE Our finding that HCMV IE1 protected hPML from degradation by HSV ICP0 is important, because the PML body (aka ND10) is believed to be the first line of host intrinsic defense against herpesviral infection. How the infected viruses overcome the nuclear defensive structure (PML body) has not been fully understood. Herpesviral proteins, ICP0 of HSV and IE1 of CMV, have been identified to interact with PML. Here, we report that HCMV IE1 incompletely deSUMOylated PML, resulting in the mono-SUMOylated PML, which is consistent with the report of Schilling et al. (J Virol 91:e02049-16, 2017, https://doi.org/10.1128/JVI.02049-16). The mono-SUMOylated PML was subjected to degradation by HSV ICP0. However, it was protected by IE1 from degradation by ICP0 or HSV-1 infection. In contrast, IE1 with L174P mutation lost the function of deSUMOylating PML and failed to protect the degradation of the mono-SUMOylated PML. Whether the mono-SUMOylated PML has any defensive function against viral infection will be further investigated.
INTRODUCTION
The promyelocytic leukemia gene, also known as MYL, RNF71, TRIM19, and PP8675, is located in chromosome 15 and encodes a tumor suppressor protein, PML (1). Aberrant PML protein, resulting from the breakpoint translocation between chromosomes 15 and 17, is related to acute promyelocytic leukemia (APL) (2–5). The translocation results in fusion of PML with the retinoic acid receptor (RAR), generating PML-RARα. PML-RARα is seen in more than 98% of APL cases. The PML gene overrides more than 50 kb, contains 9 exons, and potentially produces 7 isoforms through alternative splicing (1, 6, 7). All PML isoforms share the N-terminal 1 to 418 amino acids (aa). PML I is the longest, with 882 aa, and PML VII the shortest, with only 435 aa. The N-terminal 418 aa contains RING (R), two B-box domains, and coiled coil (CC) domains. Therefore, PML is an RBCC protein, or a protein with tripartite motifs (TRIM). PML I to VI are the nuclear PML isoforms that share the N-terminal 560 aa, which, in addition to RBCC, contains a nuclear localization signal (NLS) and a SUMO-interacting motif (SIM) (8). PML VIIb (also called PML VII) is a cytoplasmic isoform.
The nuclear isoforms of PML are essential for forming a nuclear structure, designated nuclear domain 10 (ND10, or PML body) (9, 10). ND10 isoforms are spherical bodies distributed throughout the nucleoplasm and measure around 0.2 to 1.0 μm. The molecular mechanism of the biogenesis of ND10 was a complete mystery until PML was identified as forming the matrix of ND10. Phosphorylation is required for the high SUMOylation of PML; SUMOylation of PML is required to form ND10, and SUMOylated PML localizes to ND10, where it functions as a transcription factor and tumor suppressor (11). Its expression is cell cycle related; therefore, ND10 morphology and number in the nucleus are dependent on the cell cycle (12). It regulates BCL-2 and the p53 response to oncogenic signals and therefore may explain how the translocation of PML with RARα causes APL (13, 14).
As nuclear structures, ND10’s biological function remains largely unclear. The interaction between virus and ND10 has been a focus of study in the field of host-virus interaction. The first virus found to be associated with ND10 was herpes simplex virus 1 (HSV-1); it was found that Vmw110, also called ICP0 (infected cell protein 0), localizes to ND10 (15, 16). Interestingly, the C-terminal portion of ICP0 was shown to be sufficient to disrupt the normal distribution of PML (16). It has been determined (with certainty) that ICP0 disrupts ND10 by mediating the loss of the SUMO-1-modified forms of PML and the subsequent proteasome-mediated degradation of the PML protein (17–20). The results were consistent with the findings that PML residue lysine 160 is the SUMOylation site, and the mutation of this residue makes PML resistant to degradation by ICP0 (21, 22). However, a recent study suggested that ICP0 also targets and degrades unSUMOylated PML (23). Therefore, the mechanisms that ICP0 uses to degrade PML are likely more intricate than previously thought.
Cytomegalovirus (CMV) infection can also disrupt ND10, but the mechanism underlying the dispersion is different from that by HSV-1. IE1 protein has been identified for CMV to disperse ND10, but PML is not degraded (24–26). It has been demonstrated that human CMV (HCMV) IE1 induces deSUMOylation of PML and Sp100, and one amino acid mutation (L174P) abolishes its function of deSUMOylation of PML and Sp100 (27). This L174P IE1 cannot disperse ND10 either (27–29). Therefore, the mechanisms of dispersing ND10 by HCMV is IE1-mediated deSUMOylation, rather than degradation, of ND10 proteins (30).
We asked whether the PML deSUMOylated by IE1 can be degraded by HSV-1. We hypothesized that HCMV IE1 can protect PML from degradation by HSV ICP0. Here, we found that PML in the HCMV IE1-expressing stable cell line cannot be degraded by HSV-1 infection or ICP0 transfection. More detailed investigation revealed that deSUMOylation of PML is required for IE1 to prevent ICP0 from degrading PML. In addition, we found that wild-type (WT) HSV-1 does not replicate significantly better in IE1-expressing cells than in non-IE1-expressing cells, but the ICP0-mutated HSV-1 that lost the ability to degrade PML grew much better in HCMV IE1-expressing cells than in non-IE1 cells.
RESULTS
HCMV IE1 protects deSUMOylated or mono-SUMOylated PML from being degraded by HSV-1 in U-251 MG-IE1 cells.
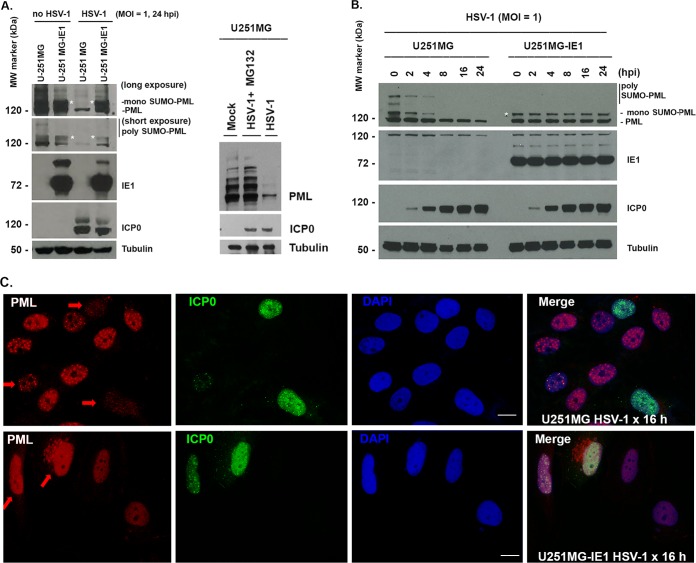
It has been well demonstrated that HSV-1 infection causes degradation of PML and SP100, which is mediated by the viral protein ICP0 (18). Interestingly, SUMOylation of PML is important for ICP0 to undertake the degradation of PML, because PML with mutation of lysine 160 failed to be SUMOylated and is resistant to proteasome-mediated degradation by ICP0 (21). HCMV IE1 is able to deSUMOylate PML (24, 27, 31, 32), so we were curious about whether HCMV IE1 is able to protect PML from being degraded by HSV-1. To investigate, we utilized a cell line, U-251MG-IE1, that stably expresses HCMV IE1. The U-251MG and U-251MG-IE1 cells were infected with HSV-1 at a multiplicity of infection (MOI) of 1 for 24 h; uninfected cells were used as a control. The whole-cell lysates were collected as samples for running Western blot assays using antibodies recognizing PML, ICP0, IE1, and tubulin. As shown in the first two lanes of Fig. 1A, left (no HSV-1 infection), PML from U-251MG presents several bands. The ∼120-kDa band is the unSUMOylated PML, above the ∼120-kDa band is the mono-SUMOylated PML, as reported by Schilling et al. (33), and the upper bands are the poly-SUMOylated PML (34). The poly-SUMOylated PML is not detectable in U-251MG-IE1 cells, because IE1 prevents the poly-SUMOylation of PML (compare the bands between the first lane and the second lane). When the cell lines were infected with HSV-1 for 24 h, as shown in the 3rd and 4th lanes of Fig. 1A, left, PML degradation occurred in the U-251MG cell line but not in the U-251MG-IE1 cell line. The band immediately above the unSUMOylated PML, indicated by an asterisk, is not seen in HSV-1-infected U-251MG due to HSV-1-mediated degradation but can be seen in HSV-1-infected U-251MG-IE1 cells. Western blotting also showed ICP0 in HSV-1-infected cells and IE1 in U-251MG-IE1 cells, which confirmed the HSV-1 infection or IE1 expression. The results suggested that HCMV IE1 protects PML from degradation by HSV-1 infection. To exclude the possibility that the PML reduction was caused by reduced protein production instead of degradation, we treated the cells with MG132 in the HSV-1-infected U-251 MG cells. As can be seen in Fig. 1A, right, treatment with MG132 can prevent PML degradation by HSV-1 infection, which is consistent with the report of Parkinson and Everett (35).
FIG 1.
HCMV IE1 deSUMOylated hPML and protected hPML from degradation by ICP0. (A, right) U-251 MG or U-251 MG-IE1 cells were mock infected or infected with WT HSV-1 (strain 17) for 24 h at an MOI of 1. The whole-cell lysates were collected for Western blotting to detect the proteins PML, IE1, ICP0, and tubulin. (Left) U-251 MG cells were mock infected (lane 1), HSV-1 infected plus MG132 treated (lane 2), or HSV-1 infected (lane 3) for 12 h. The whole-cell lysates were used for Western blotting to detect the proteins. MW, molecular weight. (B) U-251 MG or U-251 MG-IE1 cells were infected with WT HSV-1 (strain 17) for the indicated times at an MOI of 1. The whole-cell lysates were collected for Western blotting to detect the proteins PML, IE1, ICP0, and tubulin. (C) U-251 MG or U-251 MG-IE1 cells grown on coverslips were infected with WT HSV-1 (strain 17) for 16 h, and the cells were then fixed and permeabilized for IFA to detect PML, ICP0, and DAPI. The arrows show the PML in ICP0-positive cells. Bar, 5 μm.
To further demonstrate the assumption that HCMV IE1 protects PML from degradation by HSV-1, we infected U-251MG or U-251MG-IE1 with HSV-1 at an MOI of 1. The whole-cell lysate samples were collected at different times, as indicated in Fig. 1B. Western blot assay was performed to detect PML, ICP0, IE1, and tubulin. As can be seen, SUMOylated PML of U-251MG was degraded by HSV-1 following infection and cannot be detected at and after 8 h postinfection (hpi). However, the SUMOylated PML (as shown by the asterisk) are not degraded in U-251MG-IE1 cells. This experimental result further demonstrated that IE1 protects PML from degradation by HSV-1.
Two observations cannot be explained by our current knowledge. (i) PML from U-251MG-IE1 cells is still SUMOylated; does this imply that IE1 can only partially deSUMOylate PML? (ii) The unSUMOylated PML can still be degraded by HSV-1 (Fig. 1A), as seen by comparing the PML density in HSV-1-infected U-251MG with that in HSV-1-infected U-251MG-IE1 cells. These questions will be investigated in the future.
We next wondered if we could visualize the PML degradation that is protected by HCMV IE1 using the immunofluorescence assay (IFA) method. To that end, we infected U-251MG or U-251MG-IE1 cells with HSV-1 at an MOI of 0.5 for 16 h. The cells were fixed and permeabilized for IFA to show PML in red, ICP0 in green, and DAPI in blue. The images were then merged to ensure that the split images colocalize. As can be seen in Fig. 1C, PML density decreased sharply in the HSV-1-infected U-251MG cells, as shown by the arrows, suggesting that PML degradation occurred in these cells. The PML density has not been changed in HSV-1-infected U-251MG-IE1 cells. The IFA results demonstrated that HCMV IE1 protected PML from degradation by HSV-1.
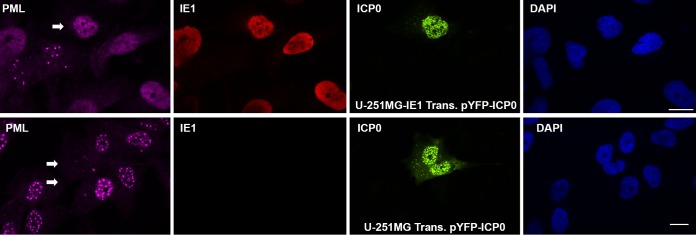
Since ICP0 was demonstrated to be responsible for HSV-1-mediated PML degradation (27), we wondered whether the same results could be seen in an ICP0 transfection system. To that end, we transfected U-251-MG or U-251MG-IE1 cells with pYFP-ICP0 that expresses ICP0 fused with yellow fluorescent protein (YFP). We fixed the cells at 24 h after transfection to perform IFA to show PML in pink, IE1 in red, ICP0 in green, and nuclei in blue, as shown in Fig. 2. PML in the IE1-positive U-251MG-IE1 cells exists as a diffuse pattern. We need to point out here that, in the U-251MG-IE1 cells, we saw two cells with PML in a dot pattern and no IE1. This is because the cell line loses IE1 during propagation. As indicated by the arrow in the PML image of the upper panel (U-251MG-IE1 cells), ICP0 does not affect the density of PML compared to that of other cells. However, in the U-251MG cells (lower), as indicated by the 2 arrows, ICP0 degraded PML such that PML can barely be detected. Therefore, we demonstrated that HCMV IE1 is able to protect PML from HSV-1 ICP0-mediated degradation.
FIG 2.
IFA to show that HCMV IE1 protected hPML from degradation by ICP0. (Upper) U-251 MG-IE1 cells grown on coverslips were transfected with pYFPICP0 for 24 h, and the cells were then fixed and permeabilized for IFA to detect PML, IE1, ICP0, and DAPI. The arrows show the PML in ICP0-positive cells. (Lower) The same experiments were performed in U-251 MG cells. Bar, 10 μm.
HCMV, not MCMV, infection protects PML from degradation.
CMV-infected cells produce IE1 protein immediately after infection. We previously demonstrated that murine CMV (MCMV) infection in human cells can process to DNA replication stage, and MCMV IE1 does not disperse human ND10 (36, 37). We were curious about whether HCMV or MCMV infection is able to play a protective role on human PML from degradation by HSV-1. We first mock infected or infected MRC-5 cells with HCMV or MCMV at an MOI of 1 for 24 h. The cells then were washed twice with serum-free minimum essential medium Eagle (MEM) and infected with HSV-1 at an MOI of 1. The whole-cell lysate samples were collected at the time point of HSV-1 infection as indicated. The samples were subjected to a Western blot assay to examine PML, ICP0, HCMV IE1 (or MCMV IE1), and tubulin. As shown in Fig. 3A, if the cells were not infected with HCMV before HSV-1 infection (left), SUMOylated PML was degraded (indicated by an asterisk). The SUMOylated PML from the MRC-5 cells treated by HCMV appears resistant to HSV-1 (Fig. 3A, middle). Previously, we showed that MCMV infection does not disperse ND10 of human cells (37), so we assume that MCMV infection does not protect human cell PML from HSV-1 degradation. As expected and shown on the right, the SUMOylated PML was degraded by HSV-1 infection. Therefore, HCMV infection can protect human PML (hPML) from degradation by a subsequent HSV-1 infection, but MCMV has no protective effects on hPML from degradation by HSV-1 in human cells.
FIG 3.
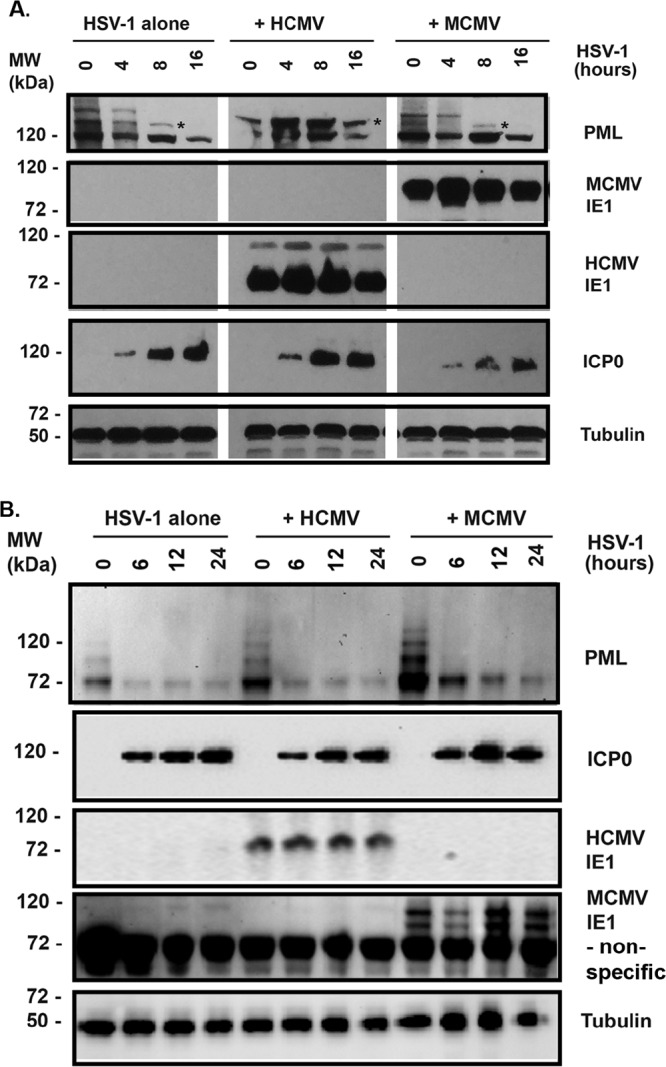
Effects of CMV infection on ICP0’s degradation of PML. (A) Human lung fibroblast cells (MRC-5) were mock infected (left) or infected with HCMV (middle) or MCMV at an MOI of 1 for 24 h. The cells then were superinfected with WT HSV-1 (strain 17) for different times, as indicated, at an MOI of 1. The whole-cell lysates were collected for Western blotting to detect the proteins PML, IE1, ICP0, and tubulin. (B) The same experiments were performed in SC1 mouse fibroblast cells.
We next set out to examine whether HCMV or MCMV infection could protect murine PML (mPML) from degradation by ICP0. The mouse fibroblast cells were mock infected or infected with HCMV or MCMV for 24 h. The cells were gently washed with MEM and infected with HSV-1 at an MOI of 1. The whole-cell lysate samples were collected at the time point indicated in Fig. 3B for Western blot assay to examine the mPML. Figure 3B showed that HCMV or MCMV infection did not deSUMOylate mPML, because the pattern of mPML in mock infection (first lane) was not different from that of MCMV or HCMV infection. It also showed that HCMV or MCMV failed to protect mPML from degradation by HSV-1 infection in mouse fibroblast cells. Interestingly, the non-SUMOylated mPML was also degraded by HSV-1 infection. Therefore, IE1’s protective effect on PML from ICP0 degradation was not observed in mouse cells.
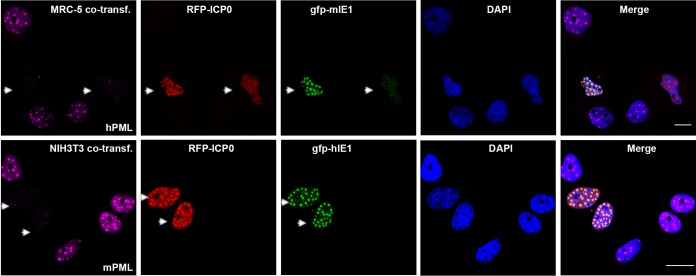
To further demonstrate our conclusion that HCMV IE1 cannot protect murine PML or MCMV IE1 cannot protect hPML from degradation by HSV-1 ICP0, we cotransfected green fluorescent protein (GFP)-tagged MCMV IE1 (gfp-mIE1) and red fluorescent protein (RFP)-tagged ICP0 to MRC-5 cells (Fig. 4, upper) or GFP-tagged HCMV IE1 (gfp-hIE1) and RFP-tagged ICP0 to NIH 3T3 cells (Fig. 4, lower). As indicated by arrows in Fig. 4, the ICP0-positve cells are also IE1 positive but PML was degraded. Therefore, MCMV IE1 did not protect human PML from ICP0-directed degradation, and HCMV IE1 did not protect mouse PML from degradation by ICP0. Interestingly, HCMV IE1 or MCMV IE1 colocalized with ICP0. Whether or not they interact will be investigated in the future.
FIG 4.
IFA to show that HCMV IE1 cannot protect mPML, or that MCMV IE1 cannot protect hPML, from degradation by ICP0. MRC-5 cells were cotransfected with plasmids expressing YFP-ICP0 and GFP-mIE1 (MCMV IE1) (upper) or NIH 3T3 cells were cotransfected with plasmids expressing YFP-ICP0 and GFP-hPML (lower) for 24 h. IFA was performed to visualize PML in purple, ICP0 in red, GFP in green, and DAPI in blue. Bar, 10 μm.
In vitro studies to demonstrate IE1’s function in protecting PML from degradation by ICP0.
We were initially interested in determining whether, as suggested from the work of Lee et al. (38), the PML degradation could be protected by IE1 in vitro. For that purpose, we utilized WT IE1 and its mutants: pYX118, which mutated the SUMOylation site of IE1, pYX145, which mutated L174 and failed to deSUMOylate PML, and pJHA423, which has a deletion of the C-terminal 68 aa. After transfection into HEK293T cells, we isolated and purified IE1 and its mutants (1-420, K450R, and L174P) by an antibody-mediated immunoprecipitation method. ICP0 was isolated and purified using the same method. The purified IE proteins were then incubated with HeLa cell nuclear extract (NE) for 30 min at 37°C. ICP0 was added or not added and the samples incubated for another 30 min. The samples were then lysed in Laemmli buffer for Western blotting for PML, ICP0, and IE1. As can be seen in Fig. 5, the mono-SUMOylated PML were not degraded by ICP0 in the groups of pJHA303, pJHA423, and pYX118. However, it was degraded by ICP0 in the presence of pYX145, which produces IE1 with an L174P mutation. Therefore, our in vitro experiments demonstrated that HCMV IE1 is able to protect hPML from degradation by ICP0 and that the deSUMOylation activity is needed for IE1’s protective effect.
FIG 5.
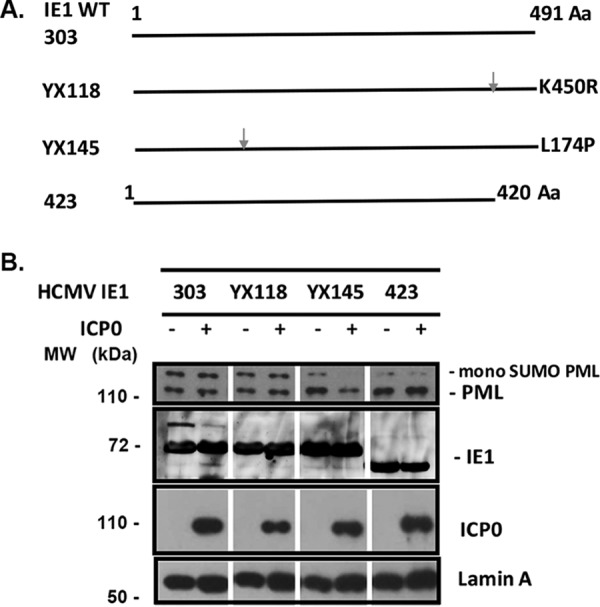
In vitro study to determine whether HCMV IE1 protected hPML from degradation by ICP0. (A) Diagram of the mutants of HCMV IE1. (B) Purified HCMV IE1 or its mutants were incubated with HeLa cell nuclear extracts for 30 min at 37°C. Purified ICP0 was then added or not added to the reaction mix, which was incubated for another 30 min. Western blot assay was then performed to detect PML, ICP0, IE1, and lamin A.
WT HSV-1 replication was not significantly reduced in IE1-expressing cells, but its ICP0 mutant (FXE) grew significantly better in HCMV IE1-expressing cell lines.
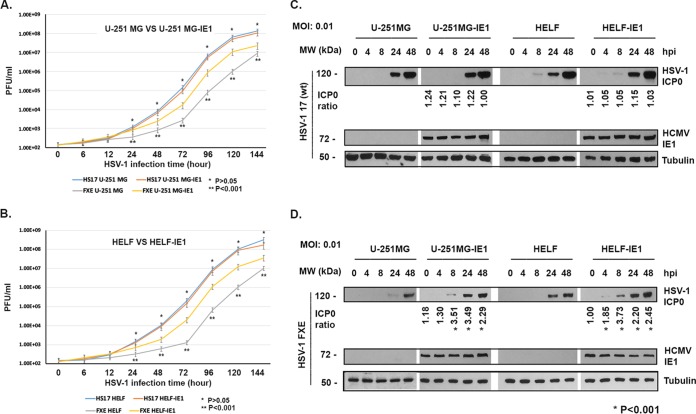
PML was shown to be a suppressive factor on viral gene expression and viral replication by several groups (14, 20, 37, 39–41). We asked whether the IE1-protected PML could play suppressive effects on HSV-1 replication. We infected 2 pairs of cell lines with WT HSV-1 (strain 17) at an MOI of 0.01: U-251MG versus U-251MG-IE1 and HELF versus HELF-IE1. The viral particle numbers were detected by PFU assay at the time points indicated in Fig. 6A and B. We found that WT HSV-1 (strain 17) replicated at the same level in IE1-expressing cells as in IE1-negative cells. Therefore, although IE1 protected the degradation of PML from HSV-1, it has no suppressive effects on WT HSV-1 replication. However, we cannot exclude the possibility that HCMV IE1 has enhancing effects on HSV-1 replication, which reduced the suppressive effects of protected deSUMOylated PML. We also infected 2 pairs of cells with the mutant HSV-1 (FXE), in which the ICP0 RING domain was mutated. HSV-1 FXE lost function of degrading PML and has a lower replicating ability than its wild type (42). As shown in Fig. 6A and B, HSV-1 FXE replicated significantly better in IE1-expressing cells than in IE1-negative cells. We also performed Western blot assay (Fig. 6C and D) to examine immediate-early protein (ICP0) production at a low MOI (0.01). The levels of ICP0 were compared, and the ratios are shown under each group. Statistical analysis was carried out using pairwise two-tailed t test to compare the two groups (IE1-expressing cells versus non-IE1 cells), and the differences were significant (*, P < 0.001). The results suggested that the deSUMOylated PML has no suppressive effects on wild-type HSV-1 gene expression in which ICP0 may have a counterdefensive effect on PML. This presumption is consistent with the results that IE1-expressing cells supported ICP0 RING finger-mutated HSV-1 (FXE) replication in which ICP0 lost the ability to degrade PML.
FIG 6.
HSV-1 replication in IE1-expressing or IE1-negative cell lines. (A and B) Growth curve assays. U-251 MG (or HELF) or U-251 MG-IE1 (or HELF-IE1) cells were infected with WT HSV-1 (strain 17) (A) or FXE strain (ICP0 RING finger helix mutation) (B) at an MOI of 0.01. The cells and supernatant samples were collected at the time points indicated. PFU assay was performed to count the viral particle numbers. Viral growth curves were determined. A Student's t test was applied to perform the comparison between two groups. Significance was set at a P value of <0.001 (*). (C and D) Western blot assays. The cells were infected with HSV-1 at an MOI of 0.01 for different times as shown. The whole-cell lysates were used for Western blot assays to examine the proteins as indicated. The ICP0 band was first compared to tubulin for normalization, and then the normalized ICP0 levels were compared between the non-IE1-expressing cells and the IE1-expressing cells. The ratios are shown under each group. Significance was set at a P value of <0.001 (*). Statistical analysis was carried out using pairwise two-tailed t test to compare the two groups (IE1-expressing cells versus non-IE1 cells) and the differences were significant, as shown by an asterisk (P < 0.001).
DISCUSSION
The initiation of the present study was based on previously published works from different groups. ICP0 of HSV-1 was the first identified viral protein to interact with ND10 and disrupt the ND10 structure in the nucleus (15, 16). Later, it was found that ICP0 serves as an E3 ligase to promote the protease-mediated degradation of PML (18) and that the RING finger helix of ICP0 is required for ICP0 to exert its degrading activity, because the ICP0 RING finger-mutated HSV-1 (FXE) failed to degrade PML (42). More interestingly, SUMOylation of PML is important in order for PML to be degraded by ICP0 (21). Another herpesvirus, CMV, was also found to disrupt ND10 immediately after infection, and IE1 was identified to be responsible for the biological function (24–26, 32, 43). HCMV IE1 interacts with hPML, deSUMOylates hPML, and disrupts ND10 by deSUMOylating hPML (30, 38, 44). Therefore, we hypothesized that HCMV IE1 could protect hPML from degradation by ICP0 because IE1 deSUMOylates hPML.
The salient findings of our study include that HCMV IE1 protected hPML from degradation by ICP0 (Fig. 1 and 2), that IE1’s activity of deSUMOylating PML is important for IE1 to protect hPML from being degraded by ICP0 because IE1 L174P failed to protect hPML from the ICP0-mediated degradation (Fig. 5), and that HCMV failed to protect mPML or MCMV failed to protect hPML from being degraded by ICP0 (Fig. 3 and 4). The protective effects of IE1 on PML were examined not only by Western blot assay but also by IFA. The interactions of PML with HSV-1 ICP0 or HCMV IE1 are summarized in Fig. 7. Poly-SUMO-PML is susceptible to degradation by ICP0. HCMV IE1 deSUMOylates PML from poly-SUMO-PML to mono-SUMO-PML, which becomes resistant to degradation by ICP0.
FIG 7.
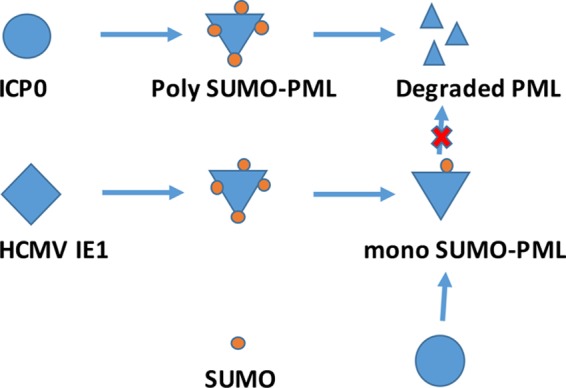
Model of interaction between HCMV IE1, hPML, and ICP0. HSV infection produces an immediate-early protein, ICP0, that degrades poly-SUMO-PML but not mono-SUMO-PML. HCMV immediate-early protein, hIE1, deSUMOylates hPML to generate mono-SUMO-PML, which cannot be degraded by ICP0. Therefore, hIE1 protects hPML from degradation by ICP0.
Although the finding that IE1 caused deSUMOylation of PML was not new, it is interesting that the deSUMOylation of PML by IE1 was incomplete, that mono-SUMO-PML still exists, and that mono-SUMO-PML is susceptible to ICP0-mediated degradation. Therefore, the interaction between HCMV IE1 and PML might be more complicated than we currently know. It is unknown how HCMV IE1 deSUMOylates hPML to generate mono-SUMO-PML.
PML has been widely investigated due to its relationship to cancer, especially APL (5, 6), and it has been accepted as a repressor of gene expression (9, 45). However, the protected PML in IE1-expressing cells failed to repress HSV-1 replication (Fig. 6), which was demonstrated in two different IE1-expressing cell lines. Our explanation for these results has two layers. First, IE1 is a gene expression enhancer and neutralizes the repressive effects of the PML on HSV-1 gene expression and viral replication. Second, IE1 holds PML and disrupts ND10 so that PML cannot cause its repressive effects on viral gene expression or viral replication. An IE1-expressing cell line slightly but significantly improved the viral replication of FXE HSV-1 (at an MOI of 0.01) in which the RING finger helix was deleted from ICP0. Interestingly, IE1-expressing cell lines dramatically supported FXE, not WT, HSV-1 replication, unlike non-IE1 cell lines. Therefore, PML’s repressive effects on HSV-1 can be diminished by HCMV IE1. This could also explain how IE1’s enhancive activities on viral gene expression occur through the IE1-mediated repression of PML in IE1-expressing cells.
It is interesting that IE1 deSUMOylated hPML from poly-SUMO-PML to mono-SUMO-PML but not to PML and that mono-SUMOylated PML is not degraded by ICP0 in the presence of IE1. Our explanation is that IE1 remains able to interact with mono-SUMOylated PML, hindering targeting by ICP0. This is supported by the finding that the L174P mutant IE1 does not bind PML and failed to protect PML from degradation. Importantly, we found that MCMV disperses ND10 of mouse cells but cannot cause deSUMOylation of mPML, and it cannot protect PML from degradation by ICP0. We plan to study the mechanisms through which mouse PML resists deSUMOylation by MCMV infection. This might result in the revelation of how MCMV disrupts ND10.
Due to the fact that both HCMV and HSV infect large populations, coinfection of the two viruses is possible. Even though the chance that HCMV and HSV infect the same cell or tissue is low, HCMV IE1 remains an activator of latent HSV. However, it is more important to know whether HSV-1 infection activates HCMV latency, because HAV-1 has broader cell permissiveness. This will be investigated in the future.
MATERIALS AND METHODS
Tissue culture, viruses and transfection.
The human neuronal glioblastoma (astrocytoma) cell line U-251MG was purchased from Sigma (number 09063001). U-251 MG-IE1 is a stable cell line producing HCMV IE1 (36). The human embryonic diploid lung fibroblast (HELF) cell line MRC-5 (ATCC CCL171) was purchased from ATCC. HELF-IE1 can stable express HCMV IE1 (46). The mouse fibroblast cell lines SC-1 (ATCC CRL-1404) and HEK 293T (ATCC CRL-11268) were purchased from the ATCC. The cells were maintained in minimum essential medium Eagle (MEM; M4655; Sigma) supplemented with 10% fetal calf serum (FCS) and penicillin (100 IU/ml)–streptomycin (100 μl/ml) and amphotericin B (2.5 μl/ml) (47). HCMV strain AD169 (ATCC VR538) and MCMV Smith strain (ATCC VR1399) were obtained from the ATCC. Wild-type HSV-1 17 and its mutant, FXE (RING finger deletion of ICP0), were obtained from R. D. Everett and were used previously (48).
For immunohistochemical staining, cells were grown on round coverslips (Corning Glass, Inc., Corning, NY) in 24-well plates (Falcon; Becton, Dickinson Labware, Lincoln Park, NJ). Plasmid DNA was transfected into cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Antibodies.
The antibodies used for Western blotting (WB) and immunofluorescence assay (IFA) are listed. Monoclonal antibody against tubulin (T-9026) and HCMV IE1 (MAB8131) was purchased from Sigma-Aldrich (1:1-000 for WB; St. Louis, MO). Polyclonal antibody against PML (sc-5621), mouse SP100 (M-75 and sc-25569), mouse Daxx (M-112 and sc-7152), ATRX (sc-15048; Santa Cruz, CA), and monoclonal anti-GFP (sc-9996) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA; dilutions were used according to the manufacturer’s manual). Monoclonal antibodies against MCMV IE1 and E1 were provided by Stipan Jonjic (Croatia; 1:50 for IF and 1:200 for WB) (49). Monoclonal antibodies against HSV-1 ICP0 were generously provided by R. D. Everett.
Molecular cloning and plasmids.
HCMV IE1-expressing plasmids (pJHA303 [WT; aa 1 to 491], pJHA423 [aa 1 to 420], pYX118 [K450R], and pYX145 [L174P]) were previously reported (38). The plasmid expressing an RFP- or YFP-tagged ICP0 was constructed by inserting the YFP-fused ICP0 to pcDNA3 vector. Plasmid expressing PML tagged with GFP was constructed by cloning the full-length PML isoform I that is fused with GFP at its N terminus into pcDNA3. Plasmids expressing GFP-tagged MCMV IE1 or HCMV IE1 were previously reported (50, 51).
Immunohistochemistry.
The localization of cellular or viral proteins by immunohistochemistry has been described (50). Briefly, cells were seeded on coverslips and washed twice with phosphate-buffered saline (PBS), fixed in 1% paraformaldehyde for 10 min at room temperature, washed again (twice) with PBS, and permeabilized with 0.2% Triton X-100 on ice for 20 min. Primary antibody was added and incubated for 30 min at room temperature. Cells were then washed twice with PBS. Secondary antibody (either anti-rabbit or anti-mouse IgG) labeled with Texas Red or fluorescein isothiocyanate (green) was added and incubated for an additional 30 min at room temperature. After a final wash with PBS, cells were stained with Hoechst 33258.
Western blot analysis.
Proteins were separated by 7.5% SDS-PAGE (10 to 20 µg loaded in each lane), transferred to nitrocellulose membranes (Amersham Inc., Piscataway, NJ), and blocked with 5% nonfat milk for 60 min at room temperature. Membranes were incubated overnight at 4°C with primary antibody, followed by incubation with horseradish peroxidase-coupled secondary antibody and then detection with enhanced chemiluminescence (Pierce, Rockford, IL) according to standard methods (for regular WB, we used secondary antibody from Amersham; for the detection of protein in the immunoprecipitation, we used secondary antibodies from TrueBlot ULTRA [18-8817 for mouse or 18-8816 for rabbit] from eBioscience). To detect additional proteins, membranes were stripped with stripping buffer (100 mM beta-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.8), washed with PBS–0.1% Tween 20, and reprobed as described above.
In vitro experiments to test IE1 protection of PML degradation by ICP0.
We first purified ICP0 or IE1 and its mutants from transfected HEK293T cells. Briefly, HEK293T cells were transfected with expression plasmid of ICP0 or IE1 (pJHA303, pJHA423, pYX118, or pYX145). Whole-cell lysate was made at 48 h posttransfection with Pierce immunoprecipitation lysis buffer (number 87788). The lysates sat on ice for 10 min and were then centrifuged at 3,000 × g for 5 min, and the supernatants were transferred to new tubes. The supernatants were incubated with antibodies overnight in a cold room with rolling. The incubations were coupled to protein G-Sepharose beads (Amersham Pharmacia Biotech AB, Sweden) according to the manufacturer's instructions for 3 h. The beads were washed three times in PBS–0.1% bovine serum albumin. The bound protein (ICP0 or IE1s) was eluted from the beads using Pierce IgG elution buffer. The elute was then neutralized with neutralization buffer (number) to bring the pH to physiological state. The purified proteins were stored at −80°C until use. HeLa cell nuclear extracts (12-309; Sigma-Aldrich) were used as a PML resource and reaction system to examine the protective effects of IE1 on PML degradation by ICP0.
PFU assay.
To detect viral growth, MRC-5 cells were infected with virus at a multiplicity of infection of 0.1 or 0.01 PFU/cell. Medium and cells from infected cultures were collected on different days postinfection, and virus was obtained after the collected culture underwent 3 freeze-thaw cycles. Virus titers were determined on MRC-5 cells after analyzing PFU. Student's t test was used to statistically analyze the difference between 2 groups; a P value lower than 0.01 was used as the threshold for significant difference.
Confocal microscopy.
Cells were examined at ×100 magnification with a Leica TCS SPII confocal laser scanning system equipped with a water-cooled argon-krypton laser. Two wavelength channels (495 and 590 nm) were recorded simultaneously or sequentially. Power and integration were adjusted to minimize bleed-through between the green and far-red channels prior to data acquisition. Digital images obtained were cropped and adjusted for contrast with Photoshop.
ACKNOWLEDGMENTS
We acknowledge the instrumental support of the PSM Molecular Biology Core Laboratory. This study was supported by NIH/NIAID grant SC1AI112785 (Q.T.), National Natural Science Foundation of China (no. 81701999) (W.H.), and National Institute on Minority Health and Health Disparities of the National Institutes of Health under award number G12MD007597. We thank Philip Roane for editing the English and critical reading of the manuscript.
REFERENCES
- 1.Fagioli M, Alcalay M, Pandolfi PP, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci PG. 1992. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7:1083–1091. [PubMed] [Google Scholar]
- 2.Borrow J, Goddard AD, Gibbons B, Katz F, Swirsky D, Fioretos T, Dube I, Winfield DA, Kingston J, Hagemeijer A. 1992. Diagnosis of acute promyelocytic leukaemia by RT-PCR: detection of PML-RARA and RARA-PML fusion transcripts. Br J Haematol 82:529–540. doi: 10.1111/j.1365-2141.1992.tb06463.x. [DOI] [PubMed] [Google Scholar]
- 3.Goddard AD, Borrow J, Solomon E. 1992. A previously uncharacterized gene, PML, is fused to the retinoic acid receptor alpha gene in acute promyelocytic leukaemia. Leukemia 6:117S–119S. [PubMed] [Google Scholar]
- 4.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. 1991. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66:675–684. doi: 10.1016/0092-8674(91)90113-D. [DOI] [PubMed] [Google Scholar]
- 5.Kakizuka A, Miller WH Jr, Umesono K, Warrell RP Jr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. 1991. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66:663–674. doi: 10.1016/0092-8674(91)90112-C. [DOI] [PubMed] [Google Scholar]
- 6.Alcalay M, Zangrilli D, Fagioli M, Pandolfi PP, Mencarelli A, Lo Coco F, Biondi A, Grignani F, Pelicci PG. 1992. Expression pattern of the RAR alpha-PML fusion gene in acute promyelocytic leukemia. Proc Natl Acad Sci U S A 89:4840–4844. doi: 10.1073/pnas.89.11.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandolfi PP, Alcalay M, Fagioli M, Zangrilli D, Mencarelli A, Diverio D, Biondi A, Lo Coco F, Rambaldi A, Grignani F. 1992. Genomic variability and alternative splicing generate multiple PML/RAR alpha transcripts that encode aberrant PML proteins and PML/RAR alpha isoforms in acute promyelocytic leukaemia. EMBO J 11:1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. 2013. Differential roles of PML isoforms. Front Oncol 3:125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera-Molina YA, Martínez FP, Tang Q. 2013. Nuclear domain 10 of the viral aspect. World J Virol 2:110–122. doi: 10.5501/wjv.v2.i3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Physiol 147:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honoré N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de Thé H. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med 193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishov AM, Vladimirova OV, Maul GG. 2004. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci 117:3807–3820. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- 13.Chen GQ, Zhu J, Shi XG, Ni JH, Zhong HJ, Si GY, Jin XL, Tang W, Li XS, Xong SM, Shen ZX, Sun GL, Ma J, Zhang P, Zhang TD, Gazin C, Naoe T, Chen SJ, Wang ZY, Chen Z. 1996. In vitro studies on cellular and molecular mechanisms of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia: As2O3 induces NB4 cell apoptosis with downregulation of Bcl-2 expression and modulation of PML-RAR alpha/PML proteins. Blood 88:1052–1061. [PubMed] [Google Scholar]
- 14.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. 2000. The function of PML in p53-dependent apoptosis. Nat Cell Biol 2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 15.Maul GG, Guldner HH, Spivack JG. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J Gen Virol 74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 16.Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol 75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 17.Everett RD, Maul GG, Orr A, Elliott M. 1995. The cellular RING finger protein PML is not a functional counterpart of the herpes simplex virus type 1 RING finger protein Vmw110. J Gen Virol 76:791–798. doi: 10.1099/0022-1317-76-4-791. [DOI] [PubMed] [Google Scholar]
- 18.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol 72:6581–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maul GG, Negorev D, Bell P, Ishov AM. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol 129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 20.Gu H, Roizman B. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci U S A 100:8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutell C, Orr A, Everett RD. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J Virol 77:8686–8694. doi: 10.1128/JVI.77.16.8686-8694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelhardt OG, Boutell C, Orr A, Ullrich E, Haller O, Everett RD. 2003. The homeodomain-interacting kinase PKM (HIPK-2) modifies ND10 through both its kinase domain and a SUMO-1 interaction motif and alters the posttranslational modification of PML. Exp Cell Res 283:36–50. doi: 10.1016/S0014-4827(02)00025-3. [DOI] [PubMed] [Google Scholar]
- 23.Cuchet-Lourenco D, Vanni E, Glass M, Orr A, Everett RD. 2012. Herpes simplex virus 1 ubiquitin ligase ICP0 interacts with PML isoform I and induces its SUMO-independent degradation. J Virol 86:11209–11222. doi: 10.1128/JVI.01145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res 229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 25.Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol 71:4599–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Q, Maul GG. 2003. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J Virol 77:1357–1367. doi: 10.1128/JVI.77.2.1357-1367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn JH, Hayward GS. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39–55. doi: 10.1006/viro.2000.0448. [DOI] [PubMed] [Google Scholar]
- 29.Ahn JH, Brignole EJ III, Hayward GS. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol 18:4899–4913. doi: 10.1128/MCB.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Ahn JH, Cheng M, apRhys CM, Chiou CJ, Zong J, Matunis MJ, Hayward GS. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J Virol 75:10683–10695. doi: 10.1128/JVI.75.22.10683-10695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang H, Kim ET, Lee HR, Park JJ, Go YY, Choi CY, Ahn JH. 2006. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J Gen Virol 87:2181–2190. doi: 10.1099/vir.0.81787-0. [DOI] [PubMed] [Google Scholar]
- 32.Kelly C, Van Driel R, Wilkinson GW. 1995. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol 76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 33.Schilling EM, Scherer M, Reuter N, Schweininger J, Muller YA, Stamminger T. 2017. The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J Virol 91:e02049-16. doi: 10.1128/JVI.02049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherer M, Reuter N, Wagenknecht N, Otto V, Sticht H, Stamminger T. 2013. Small ubiquitin-related modifier (SUMO) pathway-mediated enhancement of human cytomegalovirus replication correlates with a recruitment of SUMO-1/3 proteins to viral replication compartments. J Gen Virol 94:1373–1384. doi: 10.1099/vir.0.051078-0. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol 74:10006–10017. doi: 10.1128/JVI.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Q, Maul GG. 2006. Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J Virol 80:7510–7521. doi: 10.1128/JVI.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosme RC, Martinez FP, Tang Q. 2011. Functional interaction of nuclear domain 10 and its components with cytomegalovirus after infections: cross-species host cells versus native cells. PLoS One 6:e19187. doi: 10.1371/journal.pone.0019187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol 78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu XJ, Yang B, Huang SN, Wu CC, Li XJ, Cheng S, Jiang X, Hu F, Ming YZ, Nevels M, Britt WJ, Rayner S, Tang Q, Zeng WB, Zhao F, Luo MH. 2017. Human cytomegalovirus IE1 downregulates Hes1 in neural progenitor cells as a potential E3 ubiquitin ligase. PLoS Pathog 13:e1006542. doi: 10.1371/journal.ppat.1006542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagenknecht N, Reuter N, Scherer M, Reichel A, Muller R, Stamminger T. 2015. Contribution of the major ND10 proteins PML, hDaxx and Sp100 to the regulation of human cytomegalovirus latency and lytic replication in the monocytic cell line THP-1. Viruses 7:2884–2907. doi: 10.3390/v7062751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everett R, O'Hare P, O'Rourke D, Barlow P, Orr A. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol 69:7339–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishov AM, Maul GG. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Physiol 134:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherer M, Klingl S, Sevvana M, Otto V, Schilling EM, Stump JD, Muller R, Reuter N, Sticht H, Muller YA, Stamminger T. 2014. Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLoS Pathog 10:e1004512. doi: 10.1371/journal.ppat.1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maul GG. 2008. Initiation of cytomegalovirus infection at ND10. Curr Top Microbiol Immunol 325:117–132. [DOI] [PubMed] [Google Scholar]
- 46.Hofmann H, Sindre H, Stamminger T. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol 76:5769–5783. doi: 10.1128/JVI.76.11.5769-5783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Bruyn Kops A, Knipe DM. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 48.Tang Q, Li L, Ishov AM, Revol V, Epstein AL, Maul GG. 2003. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J Virol 77:5821–5828. doi: 10.1128/JVI.77.10.5821-5828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Q, Li L, Maul GG. 2005. Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter. J Virol 79:257–263. doi: 10.1128/JVI.79.1.257-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez FP, Cosme RS, Tang Q. 2010. Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J Gen Virol 91:2664–2676. doi: 10.1099/vir.0.022301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez FP, Cruz R, Lu F, Plasschaert R, Deng Z, Rivera-Molina YA, Bartolomei MS, Lieberman PM, Tang Q. 2014. CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J Virol 88:7389–7401. doi: 10.1128/JVI.00845-14. [DOI] [PMC free article] [PubMed] [Google Scholar]