Nonlytic release of hepatitis A virus (HAV) as exosome-like quasi-enveloped virions is a unique but incompletely understood aspect of the hepatovirus life cycle. Several lines of evidence indicate that the host protein ALIX is essential for this process. Tandem YPX3L “late domains” in the VP2 capsid protein could be sites of interaction with ALIX, but they are not accessible on the surface of an X-ray model of the extracellular capsid, raising doubts about this putative late domain function. Here, we describe YPX3L domain mutants that assemble capsids normally but fail to bind ALIX and be secreted as quasi-enveloped eHAV. Our data support late domain function for the VP2 YPX3L motifs and raise questions about the structure of the HAV capsid prior to and following quasi-envelopment.
KEYWORDS: ESCRT, exosome, picornavirus, quasi-envelope
ABSTRACT
The quasi-envelopment of hepatitis A virus (HAV) capsids in exosome-like virions (eHAV) is an important but incompletely understood aspect of the hepatovirus life cycle. This process is driven by recruitment of newly assembled capsids to endosomal vesicles into which they bud to form multivesicular bodies with intraluminal vesicles that are later released at the plasma membrane as eHAV. The endosomal sorting complexes required for transport (ESCRT) are key to this process, as is the ESCRT-III-associated protein, ALIX, which also contributes to membrane budding of conventional enveloped viruses. YPX1or3L late domains in the structural proteins of these viruses mediate interactions with ALIX, and two such domains exist in the HAV VP2 capsid protein. Mutational studies of these domains are confounded by the fact that the Tyr residues (important for interactions of YPX1or3L peptides with ALIX) are required for efficient capsid assembly. However, single Leu-to-Ala substitutions within either VP2 YPX3L motif (L1-A and L2-A mutants) were well tolerated, albeit associated with significantly reduced eHAV release. In contrast, simultaneous substitutions in both motifs (L1,2-A) eliminated virus release but did not inhibit assembly of infectious intracellular particles. Immunoprecipitation experiments suggested that the loss of eHAV release was associated with a loss of ALIX recruitment. Collectively, these data indicate that HAV YPX3L motifs function as redundant late domains during quasi-envelopment and viral release. Since these motifs present little solvent-accessible area in the crystal structure of the naked extracellular capsid, the capsid structure may be substantially different during quasi-envelopment.
IMPORTANCE Nonlytic release of hepatitis A virus (HAV) as exosome-like quasi-enveloped virions is a unique but incompletely understood aspect of the hepatovirus life cycle. Several lines of evidence indicate that the host protein ALIX is essential for this process. Tandem YPX3L “late domains” in the VP2 capsid protein could be sites of interaction with ALIX, but they are not accessible on the surface of an X-ray model of the extracellular capsid, raising doubts about this putative late domain function. Here, we describe YPX3L domain mutants that assemble capsids normally but fail to bind ALIX and be secreted as quasi-enveloped eHAV. Our data support late domain function for the VP2 YPX3L motifs and raise questions about the structure of the HAV capsid prior to and following quasi-envelopment.
INTRODUCTION
Hepatitis A virus (HAV) is an unusual hepatotropic picornavirus. Classified within the genus Hepatovirus, it is an ancient human pathogen that remains a common cause of enterically transmitted hepatitis globally (1). It infects susceptible individuals in a stealth-like manner, replicating efficiently in the liver and releasing newly replicated virus through the biliary track to the intestines from which it is shed in feces (2, 3). Virus-specific antibodies generally first appear after 3 to 4 weeks of clinically silent infection and typically herald both the onset of acute liver injury and resolution of the infection (4). Sudden increases in serum alanine aminotransferase (ALT) activity are associated with necrosis and apoptosis of hepatocytes and intrahepatic portal inflammatory cell infiltrates comprised of lymphocytes, macrophages, and plasma cells (5, 6). Although the disease can be fulminant, leading to death in a small fraction of those infected, the acute inflammatory phase of the illness is typically short-lived and self-limited. Fecal shedding and intrahepatic replication of the virus diminish rapidly after the appearance of antibody, and ALT levels return to normal over several weeks. Persistent infections occur rarely, if ever, and neutralizing anti-HAV antibodies provide durable protection against second episodes of disease (6).
Despite the acute necroinflammatory changes within the liver that accompany HAV infection, wild-type (wt) virus is noncytopathic; the acute liver injury that it causes results from innate and possibly also adaptive immune responses to the infection (7). Many mammalian cell lines are permissive for HAV replication (e.g., Huh-7, HepG2, MRC-5, and BSC-1), but wild-type and low-passage-number isolates induce no cytopathology (8). Virus is released from such cells without lysis in small extracellular vesicles (EVs) that protect the HAV capsid from neutralizing antibodies (9). These “quasi-enveloped” virions (eHAV) are the only form of virus found in sera from infected humans. They have a specific infectivity similar to that of naked HAV virions, are ∼50 to 110 nm in diameter, and possess a buoyant density of ∼1.100 g/cm3 in iodixanol (9). In many ways, these eHAV vesicles resemble exosomes, small EVs that function in intercellular communications (10). Recent quantitative proteomics studies have shown that intracellular HAV capsids are recruited for export in these vesicles via a highly specific sorting process and that the eHAV membrane is associated with a variety of endolysosomal proteins typically found in exosomes (11). Also present in eHAV are multiple proteins associated with endosomal sorting complexes required for transport (ESCRT), including ALIX, CHMP1A, and CHMP4B (11). In aggregate, these findings support an exosome-like mechanism of eHAV biogenesis, in which assembled intracellular capsids are recruited to the cytosolic surface of endosomes into which they bud in an ESCRT-mediated process to form multivesicular bodies (MVB) that ultimately fuse with the plasma membrane, releasing intraluminal eHAV to the extracellular environment (12).
How HAV capsids are recruited to ESCRT is incompletely understood, but a tantalizing clue is the presence of tandem YPX3L “late domain” motifs, separated by 28 amino acid (aa) residues, in the VP2 capsid protein (9) (Fig. 1A). The structural proteins of conventional enveloped viruses typically contain late domains [such as YPX1or3L, PPXY, P(S/T)AP, or GPPX3Y] that mediate interactions with ESCRT machinery (13, 14). The two YPX3L motifs in VP2 are conserved in all 94 human HAV sequences in GenBank as well as in novel hepatovirus species recently identified in bats and other small mammals (15). YPX3L motifs mediate interactions with ALIX, which in turn binds CHMP4 (ESCRT-III complex) in a step that is critical for membrane scission (16). Both CHMP4 and ALIX are associated with eHAV, and the release of eHAV from infected cell cultures is dependent upon ALIX (9, 11). Although previous studies of HAV mutants in which the Tyr residues in either YPX3L motif were replaced with Ala suggested that anti-ALIX antibodies bind the capsid in a YPX3L motif-dependent fashion (9, 11), the interpretation of these studies was confounded by disruption of capsid assembly by either Tyr-to-Ala substitution. A Tyr177Ala mutant (Y2-A, Fig. 2A) produced minimal amounts of capsid reactive with the anticapsid monoclonal antibody (MAb) K24F2, whereas a Tyr144Ala mutant (Y1-A) produced none (9).
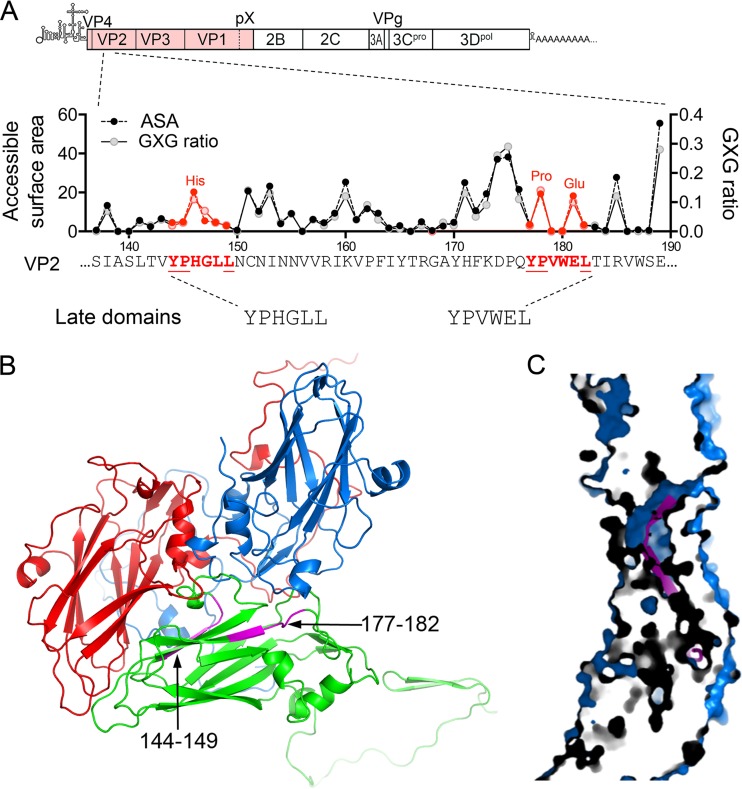
FIG 1.
HAV VP2 YPX3L late domain motifs. (A) Schematic representation of the two tandem late domain motifs in the HAV VP2 capsid protein. The organization of the HAV genome is shown at the top with the polyprotein coding region displayed as an extended box. Below is shown a segment of the VP2 protein with the two conserved YPX3L motifs in red font and with the accessible surface area (ASA) of each residue plotted above along with the ratio to calculated GXG value (surface area of the residue relative to that in a peptide in which it is flanked on each side by Gly) based on the X-ray model of the naked HAV capsid (17). (B) Protomer subunit of the HAV capsid showing the adjacent, antiparallel orientation of the two late domain motifs (L1, residues 144 to 149, and L2, residues 177 to 182, highlighted in magenta) within the VP2 (green) β-barrel (17). VP1 is shown in blue, and VP3 is shown in red. (C) Section through the HAV capsid wall, showing the buried position of residues within the late domain motifs (magenta).
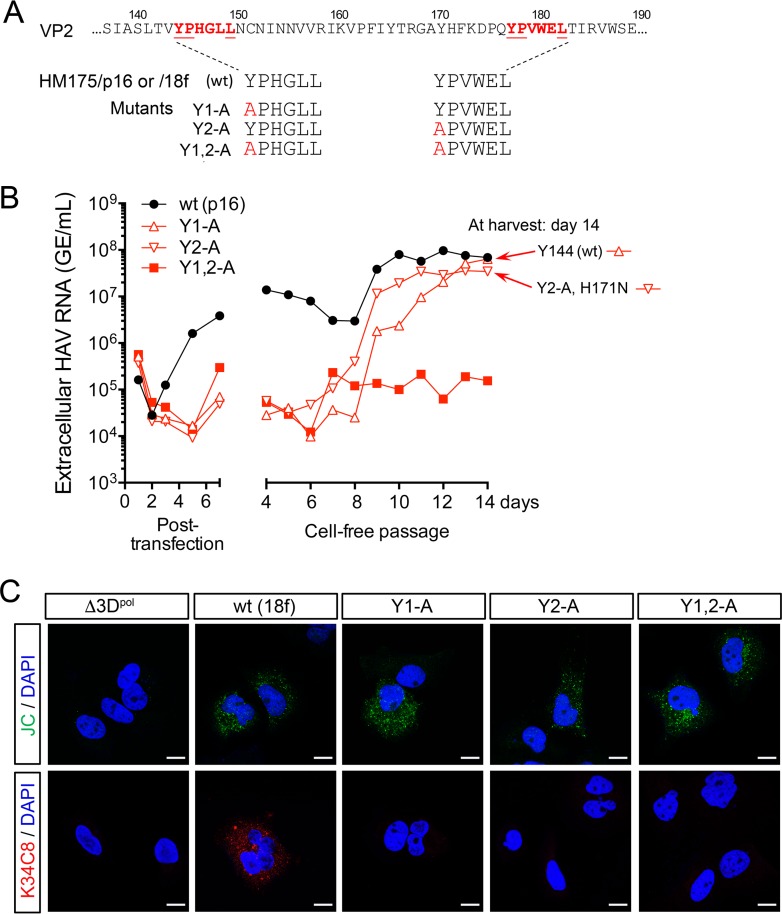
FIG 2.
Tyr-to-Ala mutations in the HAV VP2 YPX3L motifs ablate capsid assembly. (A) YPX3L late domain Tyr-to-Ala mutants constructed in the background of p16 and 18f virus (Fig. 1A). (B) Extracellular virus release following electroporation of Huh-7.5 cells with wt (HM175/p16) or related Y1-A, Y2-A, and Y1,2-A late domain mutants. Medium was replaced daily, and virus was quantified by reverse transcription-quantitative PCR (RT-qPCR). Cell-free virus was passaged from lysates harvested on day 7 postelectroporation, with supernatant fluid virus titers followed for an additional 14 days. Sequencing of viruses at day 14 postpassage revealed reversion of the Y1-A mutation to wt (Tyr144) and the presence of a second-site substitution in Y2-A (VP2 H171N). (C) Laser scanning confocal fluorescence microscopy of Huh-7.5 cells electroporated with the indicated wt (HM175/18f) or mutant RNA and fixed and stained 48 h later with either JC polyclonal human (top row) or K34C8 murine monoclonal anticapsid (bottom row) antibodies. The absence of K34C8 fluorescence despite abundant JC fluorescence in cells transfected with the mutant RNAs is consistent with the absence of capsid assembly. Nuclear counterstaining was with DAPI (blue). Δ3Dpol is a genome-length HAV RNA in which Gly-Ala-Ala has replaced the Gly-Asp-Asp motif in the 3Dpol RNA-dependent RNA polymerase. Bars, 10 µm.
Further confusion about the role of these VP2 YPX3L motifs in the HAV life cycle stems from the crystallographic structure of the naked, nonenveloped extracellular HAV particle, solved shortly after the discovery of quasi-enveloped eHAV (17) (Fig. 1B). Both VP2 motifs are largely buried beneath the surface of the capsid in the structure, with little solvent-accessible surface available for interaction with ALIX (Fig. 1A and C). This makes it difficult to understand how these putative late domains could function in ESCRT-mediated release of quasi-enveloped virus. The protein composition of the eHAV capsid differs from that of the naked capsid that has been studied crystallographically, as eHAV contains an unprocessed, 345-aa-long VP1pX capsid protein rather than the 274-aa VP1 present in naked extracellular capsids (9), but whether this results in significant conformational differences between the capsids of quasi-enveloped and naked capsids is unknown. There is no direct evidence for this, and the positioning of the YPX3L motifs within the naked capsid thus calls into question the proposed role of these motifs in recruitment of ALIX.
With these issues in mind, we carried out additional mutational studies of the YPX3L motifs of HAV using a recently developed, low-cell-culture-passage-number, noncytopathic infectious molecular HAV clone. We identified amino acid substitutions within these motifs that do not block capsid assembly and assessed their impact on both nonlytic release of quasi-enveloped eHAV from cells and ALIX interactions with the HAV capsid. These new studies provide additional support for a role of these conserved VP2 motifs in the biogenesis of quasi-enveloped virus and suggest indirectly that there may be significant differences in the structures of quasi-enveloped and naked HAV capsids.
RESULTS
Tyrosine substitutions within the VP2 YPX3L motifs disrupt capsid assembly.
Previous reverse molecular genetics studies (9) demonstrated that Ala substitutions for the N-terminal Tyr residues of either of the two VP2 YPX3L late domains motifs have no effect on replication of the HAV genome but substantially reduce release of infectious virus from cells transfected with synthetic viral RNA. However, the interpretation of these experiments was complicated by limited evidence for capsid assembly, especially when the first late domain motif (VP2 residues 144 to 149) was mutated, as well as the use of a parental infectious molecular clone that is highly cell culture adapted and that replicates rapidly with a strong cytopathic effect (HM175/18f [here called 18f]) (18, 19). To better assess the impact of such mutations on the nonlytic release of virus over longer periods of time, we used a recently constructed infectious molecular clone of a low-passage-number virus (HM175/p16 [here called p16]) that is adapted to cell culture but replicates like wild-type virus without cytopathic effect (20). The 18f and p16 viruses are derived from a common HM175 virus isolate made from fecal material in African green monkey kidney cells (8) but have different passage histories and different levels of adaptation to growth in cell culture. The sequences of their complete polyproteins are 99% identical, differing at 16 of 2,227 residues but only 1 of 836 residues in the 4 capsid proteins. Although 18f induces apoptotic cell death in a large proportion of infected cells, accounting for its cytopathic effect (19, 21), most extracellular 18f virus released from infected cells, like p16, is quasi-enveloped (9). The release of both quasi-enveloped 18f and p16 viruses is dependent upon ALIX, indicating that these closely related viral clones share a mechanism of quasi-envelopment (9).
As we had found previously with mutants in the cytopathic 18f virus background (9), single Tyr-to-Ala substitutions in either putative late domain (Fig. 2A) eliminated the release of extracellular p16 virus, which was first evident 3 to 4 days after electroporation of the parental p16 RNA containing wild-type motif sequences (Fig. 2B). However, blind, cell-free passage of lysates prepared 7 days after transfection of the p16 mutants resulted in the emergence of robust virus release 8 to 9 days later (Fig. 2B). Sequencing of virus collected from cell culture supernatant fluids following blind passage revealed that the Y1-A mutant had reverted to the wild-type sequence, restoring the first late domain motif. The VP2 Tyr177Ala substitution persisted in the Y2-A mutant but was now accompanied by a second substitution (His171Asn) in VP2. Only low levels of viral RNA, possibly reflecting inapparent cell lysis, were found in supernatant fluids of cells following blind passage of the double mutant Y1,2-A (Fig. 2B).
To more fully assess the impact of the original Tyr-to-Ala substitutions on the capacity of the mutated genomes to express HAV proteins and direct the intracellular assembly of capsids, we used laser scanning confocal fluorescence microscopy to image cells 48 h after electroporation of the viral RNA. For these short-term imaging experiments, we used mutants derived from the 18f clone, as it provides substantially higher levels of viral protein expression than the p16 clone under these conditions. Importantly, the amino acid sequences of the capsid proteins (including the pX domain of VP1) are identical in these clones, with the exception of a VP1 Ser271Pro substitution in 18f, 4 residues upstream of the VP1/pX cleavage site. We stained cells either with polyclonal, post-convalescent-phase human antibodies (JC plasma) or with a capsid-specific MAb (K34C8) that has been shown previously to recognize a conformational epitope present on 70S capsids but not 14S pentamer subunits (22). Under these conditions, the polyclonal JC antibody recognized only structural proteins, as it did not label cells transfected with a replication-competent subgenomic RNA replicon (18f-FLuc) that does not encode capsid proteins but replicates robustly, expressing abundant double-stranded RNA (dsRNA) identified with a dsRNA-specific MAb, J2 (23) (Fig. 3). Labeling with the polyclonal human antibodies revealed readily detectable antigen in cells transfected with each of the Tyr-to-Ala mutants, confirming capsid protein expression, whereas K34C8 stained only cells transfected with the parental virus containing wild-type (wt) late domain motif sequences (Fig. 2C).
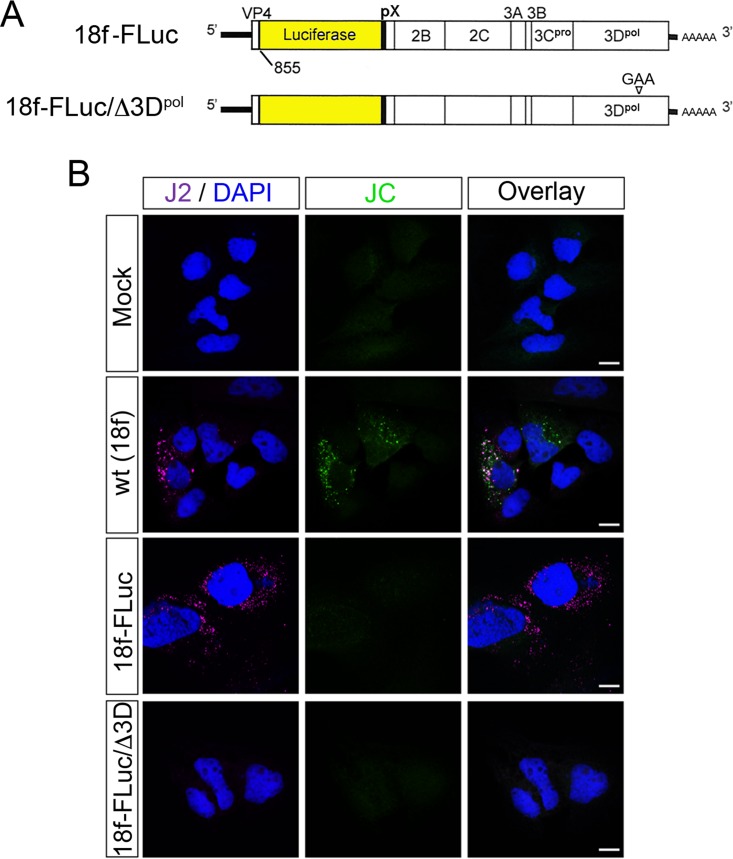
FIG 3.
JC polyclonal human convalescent antibody labels only HAV structural proteins in viral RNA-transfected cells examined by confocal fluorescence microscopy. (A) Schematic representation of the subgenomic 18f-FLuc RNA derived from the HM175/18f virus used in this study, in which the firefly luciferase sequence has replaced most of the VP2-VP1 coding sequence. Gly-Ala-Ala has replaced the 3Dpol Gly-Asp-Asp sequence in the replication-incompetent 18f-FLuc/Δ3Dpol mutant. (B) Cells were either mock electroporated or electroporated with wt (HM175/18f) virus RNA, the replicon 18f-FLuc, or 18f-FLuc/Δ3Dpol and then fixed and stained 72 h later with either J2, a murine monoclonal antibody that binds dsRNA in a sequence-independent fashion (23), or the JC polyclonal antibody. The absence of JC fluorescence in cells transfected with 18f-FLuc RNA, despite abundant J2 fluorescence indicative of RNA replication, indicates that JC fails to detect nonstructural proteins of the virus under the conditions used. Bars, 10 µm.
Collectively, these data confirm that Tyr-to-Ala substitutions in either late domain motif strongly impair capsid assembly. However, the ability of the Y2-A/H171N second-site revertant to replicate and efficiently egress from cells in the absence of cytopathic effect (Fig. 2B) indicates that the second late domain motif is not essential for eHAV release. This suggests that these late domains could function redundantly, if in fact they do function to recruit ALIX during quasi-envelopment of capsids.
Because the late domain Tyr residues are buried in the structure of the naked virus particle (Fig. 1A) (17), we considered that it might be important for assembly or stability of the capsid to replace them with residues that have side chains of approximately equal size and hydrophobicity. We thus created additional sets of mutants in which Tyr144 was replaced with Glu (Y1-E mutant) or Tyr177 was replaced with Trp (Y2-W), or both (Y1,2-E/W) (Fig. 4A). When constructed in the rapidly replicating 18f background, each of the mutants expressed capsid proteins that were readily detected 48 h after transfection by staining with the polyclonal JC antibody, but none expressed capsid antigen detectable with K34C8 (Fig. 4B). Moreover, in the more slowly replicating, noncytopathic p16 background, the release of extracellular virus was reduced more than 99% by either the single or double mutation as determined by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 4C). Intracellular viral RNA abundance was similarly reduced with each of the mutants 9 days after transfection (data not shown). Thus, these substitutions for the N-terminal Tyr residues also appear to prevent efficient capsid assembly.
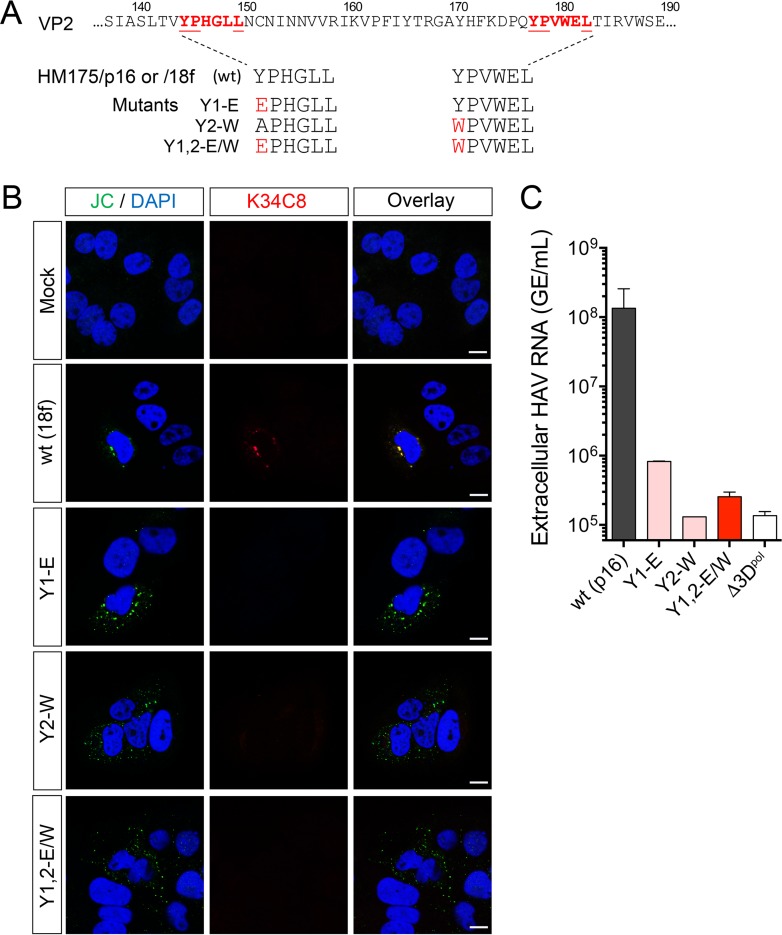
FIG 4.
Late domain mutants with Y1-E, Y2-W, and Y1,2-E/W substitutions are unable to efficiently assemble capsids and thus severely handicapped in replication. (A) YPX3L late domain mutants constructed in the background of p16 and 18f viruses (Fig. 1A). (B) Confocal fluorescence microscopic images of cells mock electroporated or electroporated 48 h previously with wt (HM175/18f) or related mutant RNAs as shown. Absence of K34C8 fluorescence despite readily detectable expression of structural proteins labeled with JC polyclonal antibody suggests a failure of capsid assembly. Bars, 10 µm. (C) Extracellular HAV RNA in supernatant of cells 9 days after electroporation with wt (HM175/p16) or related Y1-E, Y2-W, or Y1,2-E/W RNAs. Cells transfected with each of the mutants released less than 1% of the amount of virus released by cells transfected with the wt control. GE, genome equivalents.
Capsid assembly is not impaired by Leu-to-Ala substitutions in the late domain motifs.
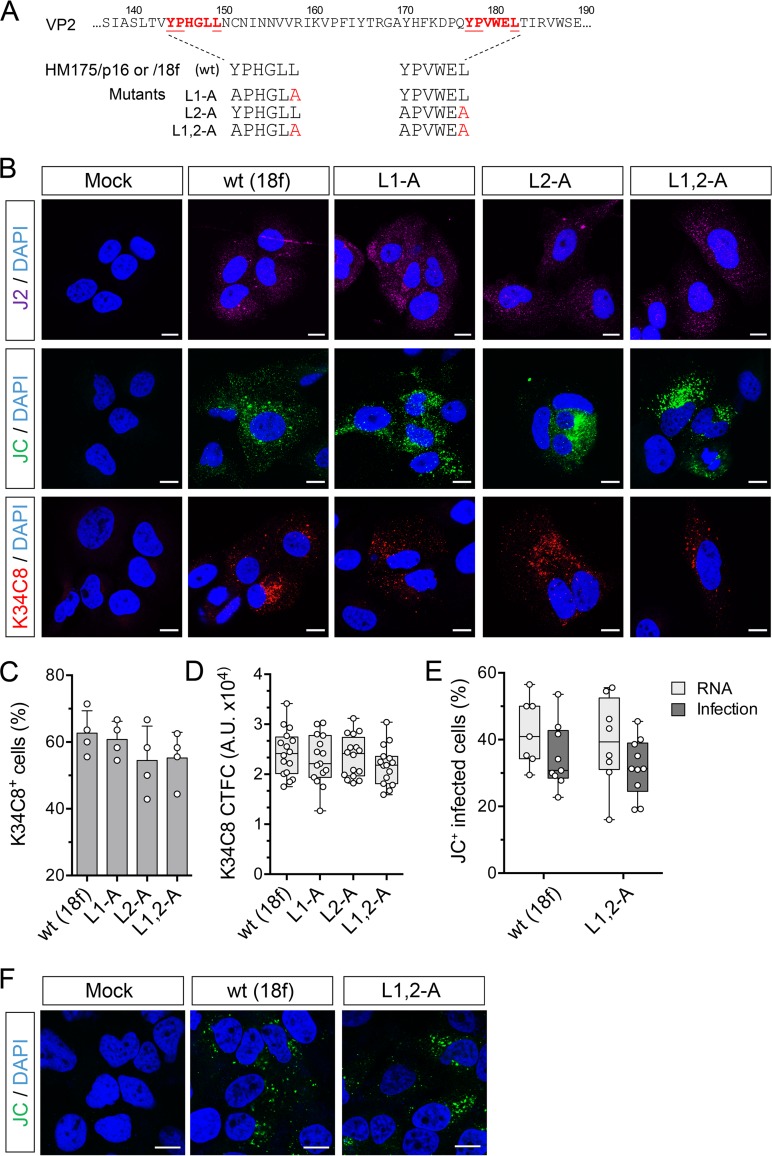
As an alternative to amino acid substitutions involving the N-terminal Tyr residues in the YPX3L motifs, we constructed mutants with single Leu-to-Ala motifs in the C-terminal Leu residues of the motifs (L1-A and L2-A) as well as a double mutant with substitutions in both (L1,2-A) (Fig. 5A). Each of these mutants replicated as well as the parental construct when transfected into cells as RNA, in both the p16 and 18f genetic backgrounds (data not shown). Consistent with that, all three 18f mutants generated strong staining for dsRNA with J2 antibody 48 h after transfection (Fig. 5B). These mutants generated strong fluorescence with both JC and K34C8 labeling also, the latter of which has been shown to uniquely recognize only fully assembled capsids and not 14S pentamer subunits which are recognized by the polyclonal JC antibody (22) (Fig. 5B). The strong fluorescent signal obtained with K34C8 is thus indicative of capsid assembly by each of the mutants. The proportion of transfected cells staining positively with K34C8 (Fig. 5C) and the corrected total K34C8 fluorescence intensity per cell (CTFC) (Fig. 5D) were similar for cells transfected with 18f RNA containing wt late domain motifs and each of the three mutants.
FIG 5.
Capsid assembly is not impaired by carboxy-terminal Leu-to-Ala substitutions within the VP2 YPX3L motifs of HAV. (A) YPX3L late domain Leu-to-Ala mutants constructed in the background of p16 and 18f viruses (Fig. 1A). (B) Confocal immunofluorescence microscopy of Huh-7.5 cells mock electroporated or electroporated with wt (HM175p16) or related L1-A, L2-A, or L1,2-A double mutant RNAs. Cells were fixed 48 h postelectroporation and stained with J2 (monoclonal anti-dsRNA), JC (polyclonal human anti-HAV), or K34C8 (monoclonal HAV anticapsid) antibodies. Strong K34C8 fluorescence in cells transfected with each of the mutants is indicative of efficient capsid assembly (22). Nuclear counterstaining was with DAPI (blue). Bars, 10 µm. (C) Percentage of cells stained with the K34C8 anticapsid monoclonal antibody following transfection with each of the mutants. Data shown are means ± standard deviations from 4 independent experiments. (D) Intensity of K34C8 fluorescence (CTFC, corrected total fluorescence intensity per cell) in cells transfected with wt or the indicated mutant viral RNA. All comparisons between cells transfected with mutant RNAs and those transfected with wt control RNAs were nonsignificant statistically (P > 0.29 by one-way ANOVA). A.U., arbitrary units. (E) Assay for infectious virus produced in cells transfected with wt (18f) or L1,2-A RNA. Results shown represent percent cells staining positively with JC antibody 48 h after RNA electroporation (RNA) or 48 h after inoculation of fresh cells with lysates of the electroporated cells (Infection). Data are from 7 to 10 low-power microscopy fields of cells under each condition and are representative of two independent experiments. (F) Confocal microscopic images of Huh-7.5 cells inoculated with cell-free lysates prepared from cells 48 h after electroporation with wt or L1,2-A RNA or no RNA (mock). Cells were stained with polyclonal JC antibody to HAV 48 h after inoculation.
Next, we confirmed that the L1,2-A double mutant is capable of assembly and production of intracellular infectious virus. We lysed cells 48 h after electroporation of the RNA and demonstrated the presence of infectious virus in the lysates by inoculating naive cells with the lysates and confirming the subsequent presence of replicating virus by confocal microscopy (Fig. 5E and F). Lysates from cells electroporated with wt or L1,2-A RNAs generated similar numbers of infected cells upon passage, suggesting that the double mutant is fully capable of capsid assembly. Collectively, these data show that Ala substitutions for the C-terminal Leu in either or both late domains do not impede assembly of capsids or encapsidation of the RNA genome.
Leu-to-Ala substitutions in both late domain motifs ablate viral egress.
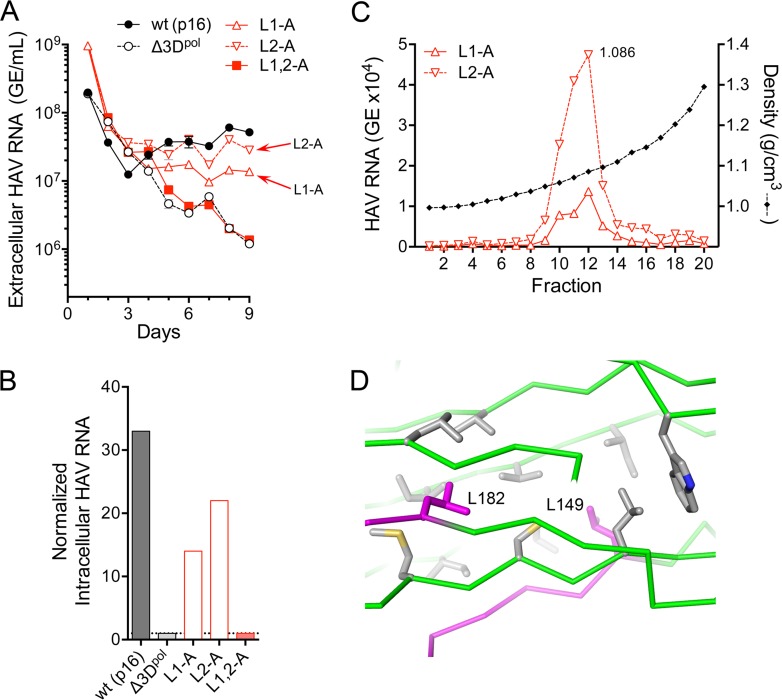
Single Leu-to-Ala substitutions in either late domain resulted in modest reductions in the nonlytic release of p16 virus from RNA-transfected cells (Fig. 6A). Eight to 9 days posttransfection, the amount of L1-A mutant released into supernatant culture fluids was 24% to 26% of the parental virus, and the amount of L2-A mutant was 55% to 64% of the parental virus, based on RT-qPCR quantitation of HAV RNA (L1-A versus p16, P = 0.0009, and L2-A versus p16, P = 0.0089, by two-way analysis of variance [ANOVA] with Sidak’s correction for multiple comparisons, from 5 to 9 days posttransfection). The intracellular abundance of these Leu-to-Ala mutant RNAs was also reduced compared to the parental p16 RNA-transfected cells (Fig. 6B), consistent with reductions in the spread of virus within the cultures. In contrast, there was no detectable release of virus from cells transfected with the L1,2-A double mutant (Fig. 6A), despite readily detectable K34C8 capsid antigen and recovery of virus from lysates of cells prepared 48 h after transfection (Fig. 5). The abundance of L1,2-A RNA in supernatant fluids at 8 to 9 days was only 3% of that of the parental p16 virus (P = 0.0004), which was indistinguishable from the amount of viral RNA present in supernatant fluids of cells transfected with a replication-incompetent 3Dpol mutant (Fig. 6A).
FIG 6.
Nonlytic viral egress is reduced by Ala substitutions of the carboxy-terminal Leu of the VP2 YPX3L late domain motifs. (A) Extracellular virus quantified by RT-qPCR following electroporation of wild-type (HM175/p16) or the related L1-A, L2-A, L1,2-A, or replication-incompetent Δ3Dpol mutant RNAs. See legend to Fig. 2B for details. (B) Intracellular viral RNA abundance 9 days after transfection of the indicated wt or mutant virus. Abundance was normalized to that present in cells transfected with the replication-incompetent Δ3Dpol RNA. (C) Isopycnic iodixanol gradients were loaded with virus present in supernatant fluids of cultures 9 days following electroporation of the single L1-A or L2-A motif mutants, and fractions were assayed after centrifugation for HAV RNA by RT-qPCR. Virus present in the peak fractions (fraction 12, 1.086 g/cm3) was sequenced, and the presence of each mutation was confirmed. (D) Orientation of the side chains of Leu149 (L149) and Leu182 (L182) (magenta) within the HAV capsid, showing the lack of a direct interaction between these residues.
Both the L1-A and L2-A virus populations present in the supernatant fluids of RNA-transfected cells were predominantly quasi-enveloped, with peak densities in isopycnic iodixanol gradients of ∼1.09 g/cm3 (Fig. 6C). Thus, the defect in release of these mutant viruses from cells was quantitative, not qualitative. The Leu residues that were altered in these mutants (Leu149 and Leu182) are relatively closely positioned within the structure of the naked capsid, but they have no direct interactions with each other (17) (Fig. 6D). These results are thus consistent with the mutations having independent, albeit redundant, effects on viral egress, with Leu-to-Ala substitutions in both late domains required for nonlytic egress of the virus to be disrupted.
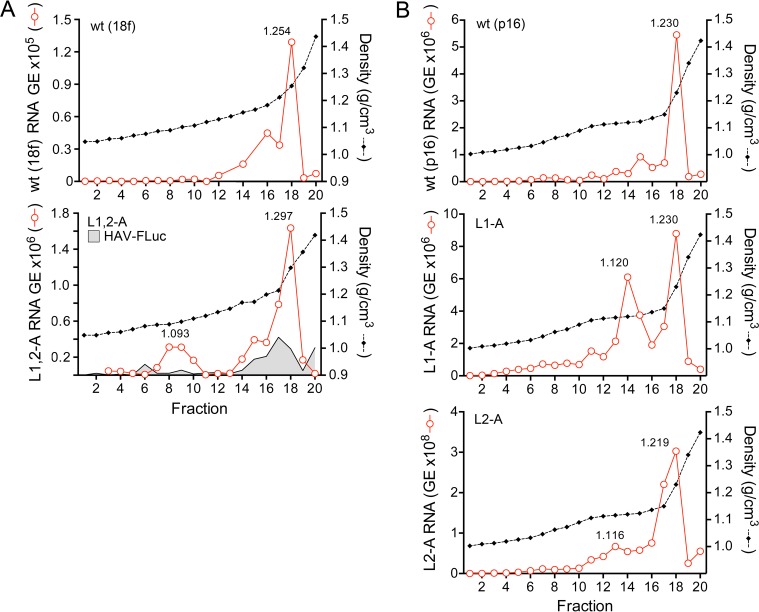
To better understand the handicap in release of the L1,2-A double mutant, we characterized the distribution of viral RNA in isopycnic iodixanol gradients loaded with lysates harvested 72 h after transfection of the RNA (18f virus background). RNA present in lysates of cells transfected with the parental 18f RNA or the 18f-FLuc subgenomic replicon banded at a predominant density of ∼1.25 g/cm3, with a somewhat lighter “shoulder” at ∼1.19 g/cm3 (Fig. 7A, top and bottom, respectively). Similar RNA species were present in lysates from cells transfected with the L1,2-A mutant, but also present was a unique minor species banding at 1.093 g/cm3 (Fig. 7A, bottom), which is similar to the density of extracellular quasi-enveloped virus (9). Similar light RNA peaks (at slightly higher densities) were evident in gradients loaded with lysates of cells supporting replication of the single L1-A and L2-A mutants (Fig. 7B, middle and bottom, respectively). The magnitude of this minor peak was greater with the L1-A than the L2-A mutant, which correlates with the greater handicap in release of L1-A than of L2-A virus (Fig. 6A and B). The viral RNA present in these peaks is likely associated with membranes and may represent a step in the egress pathway at which release is retarded (L1-A and L2-A mutants) or completely arrested (L1,2-A) by mutations in the late domains. Capsid proteins were not present in sufficient abundance in fractions to ascertain their positions in these gradients by immunoblotting or enzyme-linked immunosorbent assay (ELISA).
FIG 7.
Isopycnic iodixanol gradient profiles showing density distributions of intracellular wt and mutant viral RNAs. (A) Gradients loaded with lysates from cells electroporated 72 h previously with wt (HM175/18f) (top) or the related L1,2-A mutant (bottom). Also shown in the bottom panel is the density distribution of subgenomic replicon 18f-FLuc RNA (HAV-FLuc). A unique peak of viral RNA is present in the L1,2-A gradient (1.093 g/cm3). (B) Gradients loaded with lysates from cells electroporated 9 days previously with wt (HM175/p16) (top) or the related L1-A (middle) and L2-A (bottom) mutants. Unique, low-density RNA peaks are present in gradients loaded with lysates of cells transfected with either mutant.
Leu-to-Ala mutations in the late domain YPX3L motifs ablate capsid interactions with ALIX.
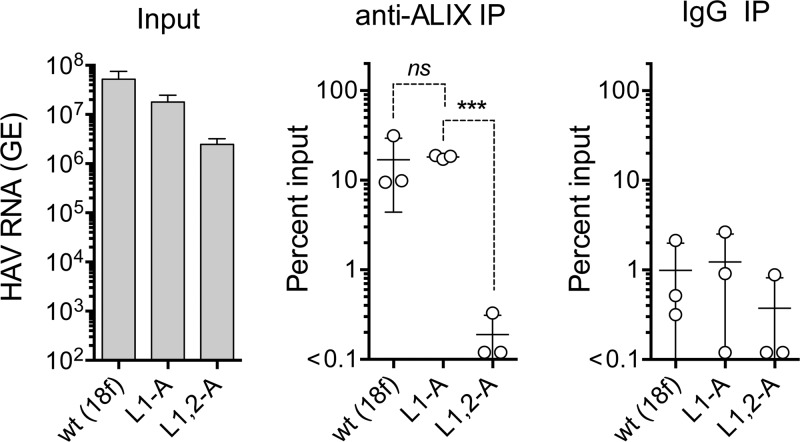
YPX1-3L late domain motifs in the structural proteins of enveloped viruses engage ALIX, an ESCRT-III-associated protein, during budding of virus from cellular membranes (24). Antibodies to ALIX can precipitate both intracellular and extracellular HAV capsids, and previous studies suggest that the VP2 Tyr177-Ala mutation (Y2-A, Fig. 2A) disrupts this association (9, 11). However, this amino acid substitution also disrupts capsid assembly (9) (Fig. 2C), as described above, clouding the interpretation of immunoprecipitation experiments. To determine whether Leu-to-Ala mutations in the late domains also interfere with ALIX binding to the HAV capsid, we used RT-PCR to quantify virus precipitated by anti-ALIX from cell lysates prepared 48 h after transfection of cells with parental (18f), L1-A (the single mutant with the greater defect in viral release), and double mutant (L1,2-A) viral RNAs. Antibody to ALIX robustly precipitated virus (>10-fold that precipitated by control IgG) from lysates of cells transfected with the wt and L1-A mutant RNAs (Fig. 8). However, anti-ALIX failed to precipitate more RNA-containing virus capsids than control IgG from the lysates of cells transfected with the L1,2-A double mutant (Fig. 8). These results are consistent either with a loss of capsid interactions with ALIX in the double mutant or with the L1,2-A double mutation impeding encapsidation of the RNA genome. This latter possibility is unlikely, however, as the results shown in Fig. 5E and F demonstrate that L1,2-A lysates contained as much infectious virus as lysates of cells transfected with the parental viral RNA. Whereas the difference between the amounts of the single mutant, L1-A, and double mutant, L1,2-A, precipitated by anti-ALIX was 2 orders of magnitude and highly significant (P = 0.0007 by two-sided paired t test), the variance in the immunoprecipitation experiments was too large to determine whether the single L1-A mutant capsid has a lesser defect in binding ALIX, consistent with the ∼4-fold reduction observed in release of eHAV (Fig. 6A).
FIG 8.
Immunoprecipitation of encapsidated viral RNA present in lysates of cells prepared 48 h following electroporation with wt (HM175/18f), L1-A, or L1,2-A mutant HAV RNAs. Input HAV RNA present in each lysate is shown on the left, quantified by RT-qPCR, followed (left to right) by the percentage of each RNA precipitated by anti-ALIX antibody and nonspecific immunoglobulin (IgG). Data shown represent means ± standard deviations from 3 independent experiments. Dashed lines indicate statistical comparisons by two-sided paired t test: ***, P = 0.0007; ns, nonsignificant.
DISCUSSION
The quasi-envelopment and extracellular release of nascent HAV capsids in exosome-like virions represent a key aspect of the hepatovirus life cycle (9, 12). They allow for nonlytic egress of the virus and, thus, noncytopathic replication with stealthy spread of virus within the liver, as well as extended shedding of infectious virus in feces via the biliary system prior to the onset of liver injury. The mechanism responsible for quasi-envelopment is incompletely understood, and in this report, we used a mutational approach to characterize two late domain motifs in the VP2 capsid protein of HAV that are likely to be major determinants of this process. The ESCRT complexes are critically involved in vesicular membrane budding events in cells (16). Previous studies indicate that the ESCRT-III-associated protein, ALIX (apoptosis-linked gene 2 [ALG2]-interacting protein X, otherwise known as PDCD6IP, programmed cell death 6 interacting protein), plays a central role in quasi-envelopment. RNA interference (RNAi)-mediated depletion of ALIX results in a loss of extracellular release of eHAV but has no impact on replication of viral RNA within infected cells (9). Immunoprecipitation and quantitative proteomics experiments have also demonstrated that ALIX is associated with encapsidated viral RNA in lysates of infected cells (9, 11). ALIX is present within quasi-enveloped eHAV virions, both those released from infected cell cultures and those circulating in the blood of infected humans (9, 11).
ALIX is also involved in the extracellular release of many conventionally enveloped viruses, some of which have YPX1or3L late domains in their structural proteins that represent important ALIX interaction domains (13, 14). The presence of two such YPX3L motifs, separated by only 28 aa in the VP2 capsid protein, is consistent with the important role played by ALIX in quasi-envelopment of the HAV capsid. These motifs are highly conserved among disparate hepatoviruses infecting a wide variety of small mammalian species, although Pro-to-Lys or -Arg substitutions are present in the second (downstream) motif in some bat viruses (15). As discussed above, however, the limited accessible surface area presented by these YPX3L residues within the crystallographic model of the naked extracellular capsid structure appears incompatible with a direct ALIX interaction (Fig. 1A) (17). Nonetheless, the mutational studies that we report here, summarized in Table 1 for late domain mutants with Tyr-to-Ala or Leu-to-Ala substitutions, provide strong genetic evidence that these motifs function in quasi-envelopment, as we predicted previously (9).
TABLE 1.
Impact of Tyr-to-Ala and Leu-to-Ala mutations in the VP2 late domains of HAV
Several studies have examined the structural basis of interactions between ALIX and peptides derived from the Gag proteins of retroviruses (24–26). ALIX is a large, 89-kDa multidomain protein that contributes to ESCRT functions in cytokinesis, endocytosis, exosome biogenesis, and budding of enveloped viruses (27, 28). Gag proteins interact through YPX1or3L motifs with a central V-shaped domain in ALIX (the “V” domain) that is formed by two multihelix bundles and flanked on its two sides by an N-terminal Bro1-like domain that binds ESCRT-III and a C-terminal Pro-rich protein interaction domain (25). Crystal structures of the V domain complexed with Gag peptides show that binding is stabilized by hydrophobic interactions involving a deep groove in the larger of the two helical bundles (24). The late domain Tyr residue is particularly important in this interaction, as its side chain extends deep into this hydrophobic pocket where it forms a hydrogen bond with a conserved Asp residue in ALIX. Additional hydrophobic interactions between the Gag late domains and ALIX involve conserved Leu residues immediately preceding the YPX1or3L motif (hence the retroviral late domain motif is LYPX1or3L) (24). Interestingly, the first VP2 late domain motif in VP2 is preceded in almost all hepatoviruses by Val143 (Ile or Met in a few nonprimate virus species) (Fig. 1A), which could contribute to similar stabilizing hydrophobic interactions with ALIX.
Mutational studies show that the late domain Tyr-Pro residues are critical for efficient HIV-1 release as well as the interaction of the HIV p6Gag YPX1or3L motif with ALIX (26). Similar studies aimed at determining whether this is also true for HAV have been confounded by a lack of capsid assembly when either late domain Tyr residue is replaced with an alternative amino acid (9) (Fig. 2C). Both of these Tyr residues are structurally important for capsid stability, but this is particularly true for Tyr144 as it stabilizes the protein core and a short stretch of 310 helix in an adjacent loop (17). It is not surprising that the Tyr144Ala substitution inhibits capsid assembly, as Ala cannot fulfill the same role. The importance of Tyr144 to capsid structure is consistent with the reversion to wild-type sequence that we observed on passage of the Y1-A mutant (Fig. 2B). Tyr177 in the second late domain stabilizes the conformation of the VP2 GH loop in the capsid structure by making bridging interactions across the loop (17). His171 is at the far end of the loop, and its replacement with Asn in the second-site revertant that we observed upon passage of the Y2-A mutant (Fig. 2B) would seem likely to increase the number of loop-stabilizing interactions at this position, achieving a similar effect in the absence of Tyr177. Importantly, the viability of the L2-A/H171N revertant and its efficient release from infected cells demonstrate that both late domain Tyr residues are not required for quasi-envelopment of HAV and that the VP2 late domains thus act redundantly. Similar redundancy has been noted among the late domains of enveloped viruses (14, 30).
The C-terminal Leu residues of the retroviral Gag YPX1or3L late domain motifs are also important for the interaction with ALIX, as they rest in hydrophobic contact with Phe and Ile residues in a shallow groove in the ALIX V domain (24). Ala substitution for the Leu44 residue in the HIV p6 Gag domain substantially impairs binding to ALIX (31). Consistent with this, single Ala substitutions at either Leu149 (L1-A mutant) or Leu182 (L2-A) resulted in modest reductions in the extracellular release of eHAV, while dual substitutions at both Leu residues (L1,2-A) resulted in a loss of detectable extracellular virus release (Fig. 6A and B). Importantly, these substitutions, either singly or together, had no apparent impact on the expression of K34C8-specific fluorescence or production of infectious intracellular virus in RNA-transfected cells (Fig. 5). These mutations thus do not inhibit capsid assembly but rather the quasi-envelopment and extracellular release of capsids. Gradient analyses demonstrated the presence of a novel RNA-containing particle banding at a density similar to extracellular eHAV in lysates of cells transfected with the dual motif L1,2-A mutant RNA (Fig. 7A), but such cells did not contain encapsidated RNA precipitable with anti-ALIX antibody (Fig. 8). Collectively, these data provide additional support for the VP2 YPX3L motifs functioning as late domains in the recruitment of ALIX to the HAV capsid and the subsequent budding of quasi-enveloped virus into MVB.
How can the lack of accessible surface area presented by the late domains in the crystal structure of the naked extracellular capsid (Fig. 1A) be reconciled with the notion that they interact with ALIX? The YPX1or3L Tyr residues in Gag proteins are critical for interactions with the V domain of ALIX (24), but in HAV, they play equally important roles in stabilizing the capsid structure, as discussed above, and are not available to be bound into the hydrophobic pocket of the V domain (17). Several possibilities could account for this apparent paradox. One is that the VP2 interaction with ALIX could occur prior to assembly of the capsid. This seems unlikely, however, as it fails to explain how all 4 capsid proteins would be selectively sorted for export, unless polyprotein processing and capsid assembly occurred within vesicles destined for release. A second possibility is that the assembled intracellular capsid might “breathe” in a fashion that renders one or more of the 60 copies of each late domain transiently accessible on the surface of the capsid. However, the likelihood of this is uncertain given the exceptionally high stability of the HAV capsid to heat and low pH (17, 32).
Yet a third scenario is that the structure of the intracellular capsids that are sorted for export might differ from that of the naked, extracellular capsid that has been studied crystallographically. Rather than containing the 30.2-kDa VP1 protein present in the naked capsid, quasi-enveloped eHAV virions contain unprocessed 38.5-kDa VP1pX (9), first described by Anderson and Ross in more slowly sedimenting intracellular particles that they named pre-provirions (33). The 8-kDa pX domain (sometimes inappropriately referred to as the hepatovirus “2A” protein) plays an essential role in pentamer assembly (34, 35), but much of it is surprisingly dispensable for replication (34, 36). It is rapidly lost from eHAV virions following exposure to detergents, although the identity of the cellular protease responsible for cleaving it from VP1 remains uncertain. Whether the structure of the naked capsid differs from that of the quasi-enveloped capsid, with the tandem VP2 YPX3L motifs accessible on the surface for interactions with ALIX, and whether the capsid undergoes a final major conformational rearrangement following loss of pX are important questions to be addressed.
MATERIALS AND METHODS
Cells.
Huh-7.5 human hepatoma cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin G, and 100 µg/ml streptomycin at 37°C in a 5% CO2 atmosphere.
Generation and transfection of HAV VP2 mutants.
Mutations altering the putative VP2 late domains (Fig. 2A, 4A, and 5A) were made in two closely related infectious molecular HAV clones: pHM175/18f.2 (high cell culture passage number, rapidly replicating, and cytopathic; GenBank sequence accession number KP879216.1) and pHM175/p16.2 (low cell culture passage number, noncytopathic, GenBank sequence accession number KP879217.1). The QuikChange II site-directed mutagenesis kit (Agilent) was used to generate both single and double mutants shown. Newly generated infectious clones were sequenced to confirm the mutations. For RNA transcription, plasmids were linearized at the MluI site at the 3ʹ end of the HAV sequence. RNA transcription was carried out using the T7 RiboMAX Express large-scale RNA production system (Promega) according to the manufacturer’s protocol. A total of 5 µg of synthetic RNA was electroporated into 2.5 × 106 Huh-7.5 cells using 4-mm cuvettes at 250 V, 950 μF, 50 Ω, in a Gene Pulser XCell total electroporation system (Bio-Rad). Following electroporation, the cells were maintained in DMEM supplemented with 3% FBS.
Viral RNA harvest and quantification.
Infected-cell supernatant fluids were cleared of dead cells and debris by centrifugation at 10,000 × g for 5 min, followed by RNA extraction as previously described (37). To quantify intracellular virus, total RNA was extracted using the RNeasy kit (Qiagen). Viral quantification was carried out using a two-step RT-qPCR as previously published (37). To confirm the production of intracellular infectious virus in cells transfected with L1,2-A RNA (see Results), approximately 1 × 106 electroporated cells were seeded into a 60-mm culture dish. Forty-eight hours later, cells were subjected to freeze-thaw lysis in 200 µl of 1× phosphate-buffered saline (PBS). Naive Huh-7.5 cells were inoculated with 50 µl of lysate, fixed, and stained with anti-HAV antibody for fluorescence microscopy (see below) 48 h later.
Immunofluorescence microscopy.
Eight-well chamber slides containing Huh-7.5 cells were fixed with 4% paraformaldehyde followed by blocking with 10% goat serum. The cells were then incubated with primary antibodies at the appropriate dilution in saponin solution (1 mg/ml in water; Sigma) for 2 h at room temperature. Antibodies used for immunofluorescence were anti-HAV capsid K34C8 (1:600; CSL Limited, Melbourne, Australia) (38), hepatitis A convalescent-phase human plasma JC (1:1,000) (9), and anti-dsRNA antibody J2 (1:300; English & Scientific Consulting kft, Hungary). Cells were washed and incubated with secondary antibodies—Alexa Fluor 488–goat anti-mouse, Alexa Fluor 594–goat anti-mouse, and Alexa Fluor 488–goat anti-human (Invitrogen)—for 1 h at room temperature, followed by additional washing and sealing under coverslips using Vectashield antifade mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Images were collected using an Olympus FV10000 laser scanning confocal microscope equipped with a supercorrected 60×/1.4-numerical-aperture (NA) oil-immersion objective (MPLAPLON 60×) and a dichroic mirror, DM405/488/543/635. The pinhole was maintained at 1 Airy unit, and laser power and image acquisition parameters were maintained at constant levels within independent experiments. The excitation/emission wavelengths were 405 nm/425 to 520 nm for DAPI, 488 nm/500 to 520 nm for Alexa Fluor 488, and 543 nm/555 to 647 nm for Alexa Fluor 594. Values of mean corrected total fluorescence per cell (CTFC) were obtained using ImageJ software and calculated as equal to integrated intensity – (cell area × mean background fluorescence). Colocalization indexes were obtained with the Just Another Co-localization Plugin (JACoP) module for ImageJ (39). Images were processed for presentation using Photoshop CS4.
Isopycnic gradient centrifugation of virus.
Isopycnic gradient fractionation of virus released into cell culture supernatant fluids was carried out as previously described (9). For gradient analysis of intracellular virus, infected cells were subjected to freeze-thaw lysis in 1× PBS. The lysate was cleared by centrifugation for 5 min at 10,000 × g and loaded onto an 8 to 40% iodixanol (Opti-Prep; Sigma) step gradient identical to that described previously (9).
Coimmunoprecipitation.
Cell extracts were prepared for immunoprecipitation (IP) using radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo). A total of 30 µl extract was used for each IP assay, adjusting the volume where necessary with 1× phosphate-buffered saline (PBS) supplemented with 1× protease inhibitor cocktail (Sigma). IP was carried out using anti-ALIX (1:50; sc-53540; Santa Cruz Biotechnologies) or control immunoglobulin, incubating the immunoglobulin with extracts overnight at 4°C in constant rotation. For antibody elution, 25 µl of Pierce protein A/G magnetic beads (Thermo) was added to extracts and incubated overnight at 4°C in constant rotation. Antibody-virus-bead complexes were collected with a magnet and washed three times with washing buffer (Tris-buffered saline containing 0.05% Tween 20). Antibody-virus complexes were then eluted from the magnetic beads by adding 100 µl low-pH elution buffer (0.1 M glycine, pH 2.0) for 10 min, followed by neutralization with 15 µl of 1 M Tris, pH 8. RNA was extracted using the QIAamp viral RNA minikit (Qiagen) and quantified using two-step RT-qPCR.
Statistical calculations.
Statistical calculations were made using Prism 6.0 for Mac OS X version 6 (GraphPad Software, Inc., La Jolla, CA).
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (R01-AI103083 and R01-AI131685 to S.M.L. and T32-AI007151 to E.E.R.-S.) and the UK Medical Research Council (MR/N00065X/1; D.I.S. and E.E.F.). The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Jacobsen KH. 2018. Globalization and the changing epidemiology of hepatitis A virus. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanford RE, Feng Z, Chavez D, Guerra B, Brasky KM, Zhou Y, Yamane D, Perelson AS, Walker CM, Lemon SM. 2011. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Natl Acad Sci U S A 108:11223–11228. doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin EC, Jeong SH. 2018. Natural history, clinical manifestations, and pathogenesis of hepatitis A. Cold Spring Harb Perspect Med 8:a031708. doi: 10.1101/cshperspect.a031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemon SM. 1985. Type A viral hepatitis: new developments in an old disease. N Engl J Med 313:1059–1067. doi: 10.1056/NEJM198510243131706. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Callendret B, Xu D, Brasky KM, Feng Z, Hensley LL, Guedj J, Perelson AS, Lemon SM, Lanford RE, Walker CM. 2012. Dominance of the CD4+ T helper cell response during acute resolving hepatitis A virus infection. J Exp Med 209:1481–1492. doi: 10.1084/jem.20111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker CM. 2018. Adaptive immune responses in hepatitis A virus and hepatitis E virus infections. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a033472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker CM, Feng Z, Lemon SM. 2015. Reassessing immune control of hepatitis A virus. Curr Opin Virol 11:7–13. doi: 10.1016/j.coviro.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binn LN, Lemon SM, Marchwicki RH, Redfield RR, Gates NL, Bancroft WH. 1984. Primary isolation and serial passage of hepatitis A virus strains in primate cell cultures. J Clin Microbiol 20:28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong S-H, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bobrie A, Colombo M, Raposo G, Thery C. 2011. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 11.McKnight KL, Xie L, González-López O, Chen X, Lemon SM. 2017. Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci U S A 114:6587–6592. doi: 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. 2014. Naked viruses that aren't always naked: quasi-enveloped agents of acute hepatitis. Annu Rev Virol 1:539–560. doi: 10.1146/annurev-virology-031413-085359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren X, Hurley JH. 2011. Proline-rich regions and motifs in trafficking: from ESCRT interaction to viral exploitation. Traffic 12:1282–1290. doi: 10.1111/j.1600-0854.2011.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drexler JF, Corman VM, Lukashev AN, van den Brand JMA, Gmyl AP, Brünink S, Rasche A, Seggewiβ N, Feng H, Leijten LM, Vallo P, Kuiken T, Dotzauer A, Ulrich RG, Lemon SM, Drosten C, Hepatovirus Ecology Consortium. 2015. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci U S A 112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurley JH. 2010. The ESCRT complexes. Crit Rev Biochem Mol Biol 45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2015. Hepatitis A virus and the origins of picornaviruses. Nature 517:85–88. doi: 10.1038/nature13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang HC, Chao SF, Ping LH, Grace K, Clarke B, Lemon SM. 1995. An infectious cDNA clone of a cytopathic hepatitis A virus: genomic regions associated with rapid replication and cytopathic effect. Virology 212:686–697. doi: 10.1006/viro.1995.1526. [DOI] [PubMed] [Google Scholar]
- 19.Lemon SM, Murphy PC, Shields PA, Ping LH, Feinstone SM, Cromeans T, Jansen RW. 1991. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol 65:2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen RW, Newbold JE, Lemon SM. 1988. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology 163:299–307. doi: 10.1016/0042-6822(88)90270-X. [DOI] [PubMed] [Google Scholar]
- 21.Brack K, Frings W, Dotzauer A, Vallbracht A. 1998. A cytopathogenic, apoptosis-inducing variant of hepatitis A virus. J Virol 72:3370–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapleton JT, Raina V, Winokur PL, Walters K, Klinzman D, Rosen E, McLinden JH. 1993. Antigenic and immunogenic properties of recombinant hepatitis A virus 14S and 70S subviral particles. J Virol 67:1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonin M, Oberstrass J, Lukacs N, Ewert K, Oesterschulze E, Kassing R, Nellen W. 2000. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA 6:563–570. doi: 10.1017/S1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. 2008. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol 15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 25.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. 2007. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. 2011. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol 85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bissig C, Gruenberg J. 2014. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Mercier V, Laporte MH, Destaing O, Blot B, Blouin CM, Pernet-Gallay K, Chatellard C, Saoudi Y, Albiges-Rizo C, Lamaze C, Fraboulet S, Petiot A, Sadoul R. 2016. ALG-2 interacting protein-X (Alix) is essential for clathrin-independent endocytosis and signaling. Sci Rep 6:26986. doi: 10.1038/srep26986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Fujii K, Hurley JH, Freed EO. 2007. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol 5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 31.Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem 282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 32.Siegl G, Frosner GG, Gauss-Muller V, Tratschin JD, Deinhardt F. 1981. The physicochemical properties of infectious hepatitis A virions. J Gen Virol 57:331–341. doi: 10.1099/0022-1317-57-2-331. [DOI] [PubMed] [Google Scholar]
- 33.Anderson DA, Ross BC. 1990. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J Virol 64:5284–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen L, Benichou D, Martin A. 2002. Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis. J Virol 76:7495–7505. doi: 10.1128/JVI.76.15.7495-7505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Probst C, Jecht M, Gauss-Müller V. 1999. Intrinsic signals for the assembly of hepatitis A virus particles. Role of structural proteins VP4 and 2A. J Biol Chem 274:4527–4531. doi: 10.1074/jbc.274.8.4527. [DOI] [PubMed] [Google Scholar]
- 36.Harmon SA, Emerson SU, Huang YK, Summers DF, Ehrenfeld E. 1995. Hepatitis A viruses with deletions in the 2A gene are infectious in cultured cells and marmosets. J Virol 69:5576–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das A, Hirai-Yuki A, González-López O, Rhein B, Moller-Tank S, Brouillette R, Hensley L, Misumi I, Lovell W, Cullen JM, Whitmire JK, Maury W, Lemon SM. 2017. TIM1 (HAVCR1) is not essential for cellular entry of either quasi-enveloped or naked hepatitis A virions. mBio 8:e00969-17. doi: 10.1128/mBio.00969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacGregor A, Kornitschuk M, Hurrell JGR, Lehmann NI, Coulepis AG, Locarnini SA, Gust ID. 1983. Monoclonal antibodies against hepatitis A virus. J Clin Microbiol 18:1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]