Human cytomegalovirus (HCMV) is the most common congenital infection worldwide, causing deafness, blindness, and other serious birth defects. CXCR4 is a human chemokine receptor that is crucial for both fetal development and immune responses. We found that the HCMV protein US27 stimulates increased expression of CXCR4 through activation of the transcription factor nuclear respiratory factor 1 (NRF-1). NRF-1 regulates stress response genes that contain the antioxidant response element (ARE), and HCMV infection is associated with increased expression of many stress response genes when US27 is present. Our results show that the US27 protein activates the NRF-1/ARE pathway, stimulating higher expression of CXCR4 and other stress response genes, which is likely to be beneficial for virus replication and/or immune evasion.
KEYWORDS: CXCL12, CXCR4, G protein-coupled receptors, HCMV, NRF, US27, chemokine signaling, chemokines, cytomegalovirus
ABSTRACT
Human cytomegalovirus (HCMV) is a widespread pathogen that modulates host chemokine signaling during persistent infection in the host. HCMV encodes four proteins with homology to the chemokine receptor family of G protein-coupled receptors (GPCRs): US27, US28, UL33, and UL78. Each of the four receptors modulates host CXCR4 signaling. US28, UL33, and UL78 impair CXCR4 signaling outcomes, while US27 enhances signaling, as evidenced by increased calcium mobilization and cell migration to CXCL12. To investigate the effects of US27 on CXCR4 during virus infection, fibroblasts were infected with bacterial artificial chromosome-derived clinical strain HCMV TB40/E-mCherry (wild type [WT]), mutants lacking US27 (TB40/E-mCherry-US27Δ [US27Δ]) or all four GPCRs (TB40 E-mCherry-allΔ), or mutants expressing only US27 but not US28, UL33, or UL78 (TB40/E-mCherry-US27wt [US27wt]). CXCR4 gene expression was significantly higher in WT- and US27wt-infected fibroblasts. This effect was evident at 3 h postinfection, suggesting that US27 derived from the parental virion enhanced CXCR4 expression. Reporter gene assays demonstrated that US27 increased transcriptional activity regulated by the antioxidant response element (ARE), and small interfering RNA treatment indicated that this effect was mediated by NRF-1, the primary transcription factor for CXCR4. Increased translocation of NRF-1 into the nucleus of WT-infected cells compared to mock- or US27Δ-infected cells was confirmed by immunofluorescence microscopy. Chemical inhibitors targeting Gβγ and phosphoinositide 3-kinase (PI3K) ablated the increase in ARE-driven transcription, implicating these proteins as mediators of US27-stimulated gene transcription. This work identifies the first signaling pathway activated by HCMV US27 and may reveal a novel regulatory function for this orphan viral receptor in stimulating stress response genes during infection.
IMPORTANCE Human cytomegalovirus (HCMV) is the most common congenital infection worldwide, causing deafness, blindness, and other serious birth defects. CXCR4 is a human chemokine receptor that is crucial for both fetal development and immune responses. We found that the HCMV protein US27 stimulates increased expression of CXCR4 through activation of the transcription factor nuclear respiratory factor 1 (NRF-1). NRF-1 regulates stress response genes that contain the antioxidant response element (ARE), and HCMV infection is associated with increased expression of many stress response genes when US27 is present. Our results show that the US27 protein activates the NRF-1/ARE pathway, stimulating higher expression of CXCR4 and other stress response genes, which is likely to be beneficial for virus replication and/or immune evasion.
INTRODUCTION
Human cytomegalovirus (HCMV) is a widespread pathogen that infects the majority of the adult population worldwide (1). HCMV is able to spread systemically throughout the host and can replicate in a wide variety of cell types, including epithelial cells of gland and mucosal tissue, smooth muscle cells, fibroblasts, macrophages, dendritic cells, hepatocytes, and vascular endothelial cells (2). Infection is generally asymptomatic in otherwise healthy individuals, but despite a robust immune response, HCMV is never cleared (3). Like other herpesviruses, HCMV establishes persistent infection by maintaining a reservoir of latent virus with occasional periods of active virus replication. Lifelong persistence in the host may be attributed in part to modulation of the host immune system via virally encoded cytokine, chemokine, and chemokine receptor orthologs (4). However, this delicate balance between the host and the virus is skewed in individuals with weakened immune systems. The immunocompromised, including organ transplant recipients and HIV-positive individuals, have a greater risk of experiencing debilitating disease upon virus reactivation (5). The virus can also cross the placenta when the mother experiences either primary infection during pregnancy, reactivation of latent virus, or superinfection with a new viral strain (6, 7). Congenital HCMV infection is a major cause of mental retardation and other neurodegenerative disorders, such as hearing loss and blindness, to children born in the United States (8, 9).
Comprising over 236 kb of DNA and 192 open reading frames (ORFs), the HCMV genome is one of the largest among human viruses (10). Only a subset of HCMV genes is essential for virus replication (11), however, while others function to modify host cellular activities in ways that promote virus dissemination, persistence, and evasion of immune clearance. In particular, HCMV encodes four proteins with homology to the chemokine receptor family of G protein-coupled receptors (GPCRs): US27, US28, UL33, and UL78 (12, 13).
GPCRs constitute the single largest family of membrane-bound proteins involved in transducing intracellular signaling from extracellular stimuli, including neurotransmitters, hormones, and chemokines (14). The US27 gene product exhibits many features of chemokine receptors, including seven α-helices that span the plasma membrane, conserved cysteine residues in the extracellular loop, and extensive glycosylation on extracellular asparagine residues (15). US27 has a DRY (aspartic acid, arginine, tyrosine) motif in the second intracellular loop, which is conserved in many GPCRs and facilitates interactions with intracellular G proteins upon receptor activation (16). Additionally, the carboxy-terminal domain of US27 contains a dileucine motif that mediates receptor endocytosis (17). Despite having many features of chemokine receptors, US27 is considered an orphan, and no human chemokines have been found to bind or signal through this receptor to date (18). An HCMV mutant lacking US27 (TB40/E-mCherry-US27Δ [US27Δ]) replicates to lower titers than the wild type (WT), and this mutant limits the virus to direct cell-to-cell spread, suggesting that US27 plays a role in extracellular dissemination (19).
Unlike US27, US28 is regarded as a true chemokine receptor and binds to multiple host chemokines, such as CX3CL1/fractalkine (20), CCL2/monocyte chemotactic protein 1 (MCP-1) (21), CCL5/RANTES (21, 22), and CCL7/MCP-3 (22). US28 also has immune evasion properties and is labeled as a chemokine sink, able to sequester chemokines from the extracellular environment, thereby decreasing exposure of the immune system to these chemokine signals (4, 23). In addition to ligand-induced signaling, US28 signals through multiple G proteins to constitutively activate a range of transcription factors, including NF-κB, CREB, nuclear factor of activated T cells, and STAT3 (24–27). While CCL5/RANTES and CCL2/MCP-1 have no effect on this constitutive activation, CX3CL1/fractalkine acts as an inverse agonist, partially inhibiting both NF-κB and phospholipase C activity. US28 has been studied in a wide range of cell types, and outcomes may vary depending on the specific intracellular signaling proteins available.
The UL33 and UL78 gene products are also considered orphan receptors with no known chemokine ligands (18, 28). However, UL33 promotes constitutive ligand-independent accumulation of inositol phosphates and activation of cAMP response element-mediated transcription in transfected cells (29). UL33 and UL78 homologs exist in the genomes of rodent CMVs and play roles in virus dissemination (27, 30–32) and latency (33, 34). In contrast, US27 and US28 are found only in primate CMVs and have been studied mainly in vitro. In the temporal cascade of viral replication, US28 and UL78 are expressed with early-late-phase kinetics, while US27 and UL33 are expressed with late kinetics (15, 28, 35, 36). Furthermore, incorporation of US27 (15, 37, 38), US28 (39), UL33 (37), and UL78 (40) into virus particles has been confirmed. Thus, upon virus fusion with the cell membrane, these receptors could potentially affect host cell signaling immediately.
We previously observed that when expressed in HEK293 cells, US27 augments the mRNA and protein levels of CXCR4, leading to greater calcium flux and migration to CXCL12/SDF-1α (18). CXCR4 is a human chemokine receptor that functions as a coreceptor for HIV entry and plays a role in promoting metastasis in several cancers (41–43). CXCR4 is essential for vascular development, and mice lacking CXCR4 die in utero (44). Nuclear respiratory factor 1 (NRF-1) is the primary transcription factor regulating CXCR4 expression (45, 46), and it belongs to the “cap-n-collar” subfamily of basic leucine zipper (b-Zip) transcription factors, along with NRF-2, NRF-3, and nuclear factor erythroid derived 2 (NFE2) (47–49). These transcription factors bind to the antioxidant response element (ARE) in the promoter region of key metabolic genes with essential functions involved with respiration (50, 51), heme biosynthesis (52, 53), mitochondrial gene transcription (54, 55), and DNA replication and cell cycle progression (56, 57). Since expression of US27 in transfected cells increased CXCR4 mRNA levels (18), we wondered whether the underlying mechanism might involve stimulating NRF-1 binding to the ARE in the CXCR4 promoter. Here, we report that US27 is a constitutively active receptor that drives transcription of ARE-regulated genes through NRF-1, leading to increased CXCR4 expression during HCMV infection.
RESULTS
Generation of viral recombinants.
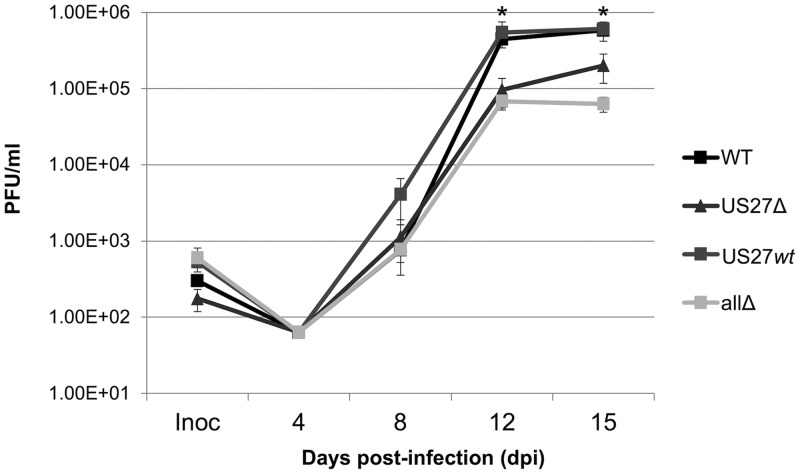
In order to investigate how US27 influences CXCR4 expression during infection, we generated a panel of viral mutants utilizing the galK bacterial recombineering system (58). We constructed each recombinant in the wild-type (WT) background of the bacterial artificial chromosome (BAC)-derived clinical isolate TB40/E-mCherry (40). We have previously described the mutants in which the entire US27 ORF was deleted (US27Δ) (19) or all four viral GPCRs were deleted (TB40/E-mCherry-allΔ [allΔ]) (26). For this study, we created an additional recombinant, TB40/E-mCherry-US27wt (US27wt). In this recombinant virus, we deleted the ORFs for UL33, UL78, and US28, resulting in a virus that expresses only one of the four viral GPCRs, US27, from its native location in the genome. At low multiplicities of infection (MOI) (MOI = 0.01 50% tissue culture infectious doses [TCID50]/cell), US27Δ and allΔ infection of fibroblasts resulted in titers lower than those of the WT at this MOI (Fig. 1), consistent with our earlier findings (19, 26). Infection with US27wt resulted in titers comparable to those of the WT virus over the course of lytic infection in fibroblasts. This is in agreement with previous findings showing that US28 (26, 59), UL33 (60), and UL78 (36, 40) are dispensable for lytic replication in fibroblasts.
FIG 1.
Growth kinetics of wild-type and mutant viruses. Fibroblasts were infected at a multiplicity of 0.01 TCID50/cell with the indicated viruses, samples of medium were collected at the indicated time points, and viral progeny were assayed by infecting fibroblasts and quantifying IE1-positive cells 24 h later by immunofluorescence. Each sample was measured in triplicate, and error bars represent the standard error. *, P < 0.05 by paired Student's t test versus US27Δ- and allΔ-infected cells.
US27 promotes increased CXCR4 expression during HCMV infection.
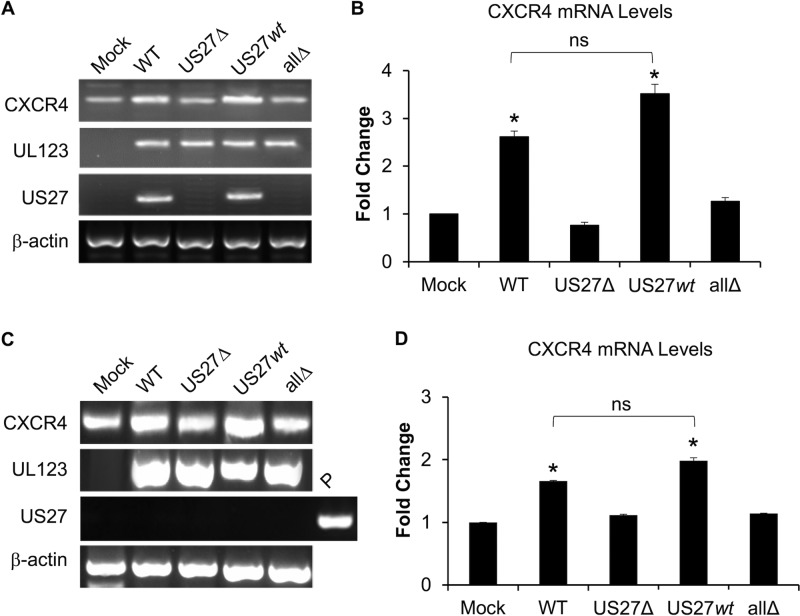
We and others have observed that transfection of US27 results in increased CXCR4 mRNA and protein levels in multiple cell types (18, 61, 62). Here, we examined the impact of US27 on CXCR4 expression in the context of HCMV infection. Fibroblasts were mock infected or infected with WT, US27Δ, allΔ, or US27wt virus (MOI = 0.2). At 120 h postinfection (hpi), expression of UL123 (encoding immediate early 1 [IE1]) in cells infected with all four recombinant viruses was confirmed by standard PCR (Fig. 2A), demonstrating that infection had occurred. In contrast to UL123, US27 was expressed only in WT- and US27wt-infected cells, as expected. CXCR4 was expressed in both mock- and virus-infected cultures, but we observed increased expression in WT- and US27wt-infected cultures compared to that in mock-, US27Δ-, and allΔ-infected cultures (Fig. 2A). Importantly, quantitative real-time PCR (qPCR) for CXCR4 showed a significantly higher level of expression in WT- and US27wt-infected cultures than either mock-, US27Δ-, or allΔ-infected cultures (Fig. 2B), suggesting that the increase in CXCR4 levels during HCMV infection requires the presence of US27.
FIG 2.
CXCR4 expression is upregulated by US27 during infection of fibroblasts. NuFF cells were infected with HCMV BAC-derived strain TB40/E-mCherry (WT) or the indicated recombinants, and CXCR4 expression was evaluated. (A) Cells were infected at an MOI of 0.2, and at 120 hpi RNA was harvested and expression was assessed by RT-PCR with gene-specific primers. PCR products were visualized via agarose gel electrophoresis. (B) RT-qPCR was performed on the same cDNA with normalization to the β-actin level, and the results are expressed as the fold change relative to the levels of expression in mock-infected cells. (C) Cells were infected at an MOI of 3, RNA was purified at 3 hpi, and RT-PCR was performed, as described above. P represents plasmid DNA (p3XFLAG-US27) used as a positive control. (D) RT-qPCR was performed with normalization to the β-actin level, and the results are expressed as the fold change relative to the level of expression in mock-infected cells. Error bars represent the standard error among 3 replicates. *, P < 0.01 by paired Student's t test versus mock-, US27Δ-, and allΔ-infected cells; ns, not significant.
Since US27 is present in the virus particle (15, 37), we next asked whether US27 could influence CXCR4 levels upon virus fusion and entry. To address this, fibroblasts were mock infected or infected with WT, US27Δ, US27wt, or allΔ virus (MOI = 3), and RNA was harvested at 3 hpi. Again, we observed an increase in CXCR4 expression for WT- and US27wt-infected cells compared to either mock-, US27Δ-, or allΔ-infected cells (Fig. 2C and D). While immediate early (IE) viral gene expression (UL123) was detected during infection with all the viruses, there was no evidence of US27 gene expression at this time, which is consistent with reports that US27 is expressed as a late gene (15, 28). Plasmid DNA (P) containing the US27 gene served as a positive control for PCR amplification of US27 in this experiment (Fig. 2C). Even within 3 h of infection, qPCR analysis indicated that CXCR4 levels had nearly doubled in cells infected with either WT or US27wt virus (Fig. 2D). Taken together, these data indicate that US27 upregulates CXCR4 gene expression during infection of fibroblasts.
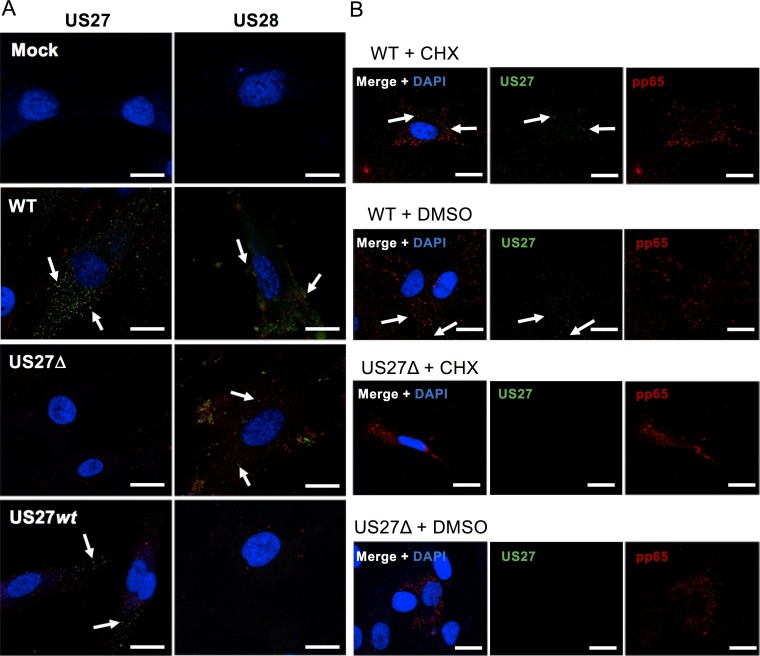
To confirm the presence of US27 early following infection, we performed immunofluorescence microscopy in infected fibroblasts at 3 hpi. Newborn human foreskin fibroblast-1 (NuFF) cells were cultured on glass coverslips, infected with the indicated virus for 3 h (MOI = 3), and then fixed and stained with anti-US27 or anti-US28 antibody followed by Alexa Fluor 514-conjugated secondary antibody. Multiple green fluorescent punctae representing the US27 protein were present in WT- and US27wt-infected cells (Fig. 3, white arrows), while US28 was detected in WT- and US27Δ-infected cells, consistent with the incorporation of these viral GPCRs in the mature, incoming virion. To verify that US27 was derived from the parental virion and not a result of de novo synthesis, we treated cells with cycloheximide overnight to block protein synthesis prior to virus infection. Both US27 and pp65, an abundant tegument protein, were detected in infected cells treated with cycloheximide (Fig. 3B). Coupled with our findings detailed in Fig. 2, these results demonstrate that US27 from the virus particle is sufficient to promote an increase in CXCR4 mRNA levels during HCMV infection.
FIG 3.
US27 and US28 are present in the cell immediately postinfection. (A) NuFF cells were seeded onto glass coverslips and then infected with the indicated viruses (red) for 3 h at an MOI of 3. The cells were then fixed and stained with antibodies specific for US27 or US28, followed by the Alexa Fluor 514-conjugated secondary antibody (green). White arrows highlight green punctae. (B) NuFF cells were treated with 25 μg/ml cycloheximide (CHX) or dimethyl sulfoxide (DMSO) for 24 h prior to infection with the indicated virus (MOI = 3, 3 h) and then stained for US27 (green) and pp65 (red). Nuclei were stained with DAPI (blue). Bars, 20 μm. Images are representative of those from one of three independent experiments.
US27 causes an increase in CXCR4 cell surface levels during infection.
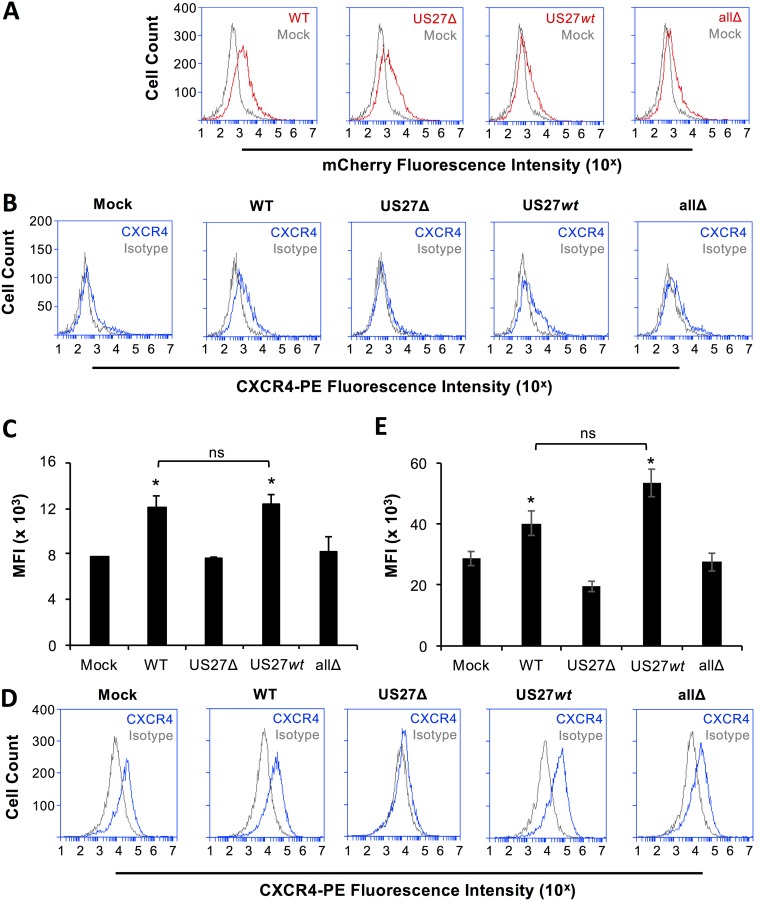
While an increase in CXCR4 gene expression was evident within 3 hpi, we predicted that a corresponding increase in surface protein levels would take additional time for protein translation and trafficking. In order to determine whether the observed increase in CXCR4 mRNA levels resulted in increased protein expression at the cell surface, fibroblasts were mock infected or infected with WT or mutant viruses (MOI = 3) for 18 h and then stained with anti-CXCR4 antibody and examined via flow cytometry. Red fluorescence from the mCherry tag was evident in cells infected with each of the viruses (Fig. 4A). CXCR4 surface levels were higher in cells infected with WT or US27wt virus than in mock-, US27Δ-, or allΔ-infected cells (Fig. 4B). Evaluation of the mean fluorescence intensity for CXCR4 staining revealed significant differences between the surface levels found on cells infected with viruses that do express US27 (WT, US27wt) and mock-infected cells or cells infected with viruses lacking US27 (US27Δ, allΔ) (Fig. 4C). We also observed increased CXCR4 surface levels on cells infected for longer times (120 hpi) with viruses containing US27 (Fig. 4D and E). These results demonstrate that US27 influences the surface protein levels of CXCR4 during HCMV infection of fibroblasts.
FIG 4.
US27 promotes increased cell surface expression of CXCR4. (A) NuFF cells were infected with the indicated HCMV strains for 18 h (MOI = 3). Cells were harvested, and mCherry fluorescence (red line) was measured via flow cytometry and compared to that in mock-infected cells (gray line). (B) Cells were stained with anti-CXCR4-PE antibody (blue line) or the IgG2b-PE isotype control (gray line) and evaluated via flow cytometry. These results are representative of those from three independent experiments. (C) The mean fluorescence intensity (MFI) for CXCR4-PE staining for each cell type from 3 independent experiments was normalized and averaged. (D) NuFF cells were infected with the indicated HCMV strains for 120 h (MOI = 0.2) and stained for CXCR4 as described above. (E) The mean fluorescence intensity for CXCR4-PE staining for each cell type from 3 independent experiments was normalized and averaged. Error bars represent the standard error of the mean. *, P < 0.01 by paired Student's t test versus mock-, US27Δ-, or allΔ-infected cells; ns, not significant.
US27 increases transcription of ARE-regulated genes.
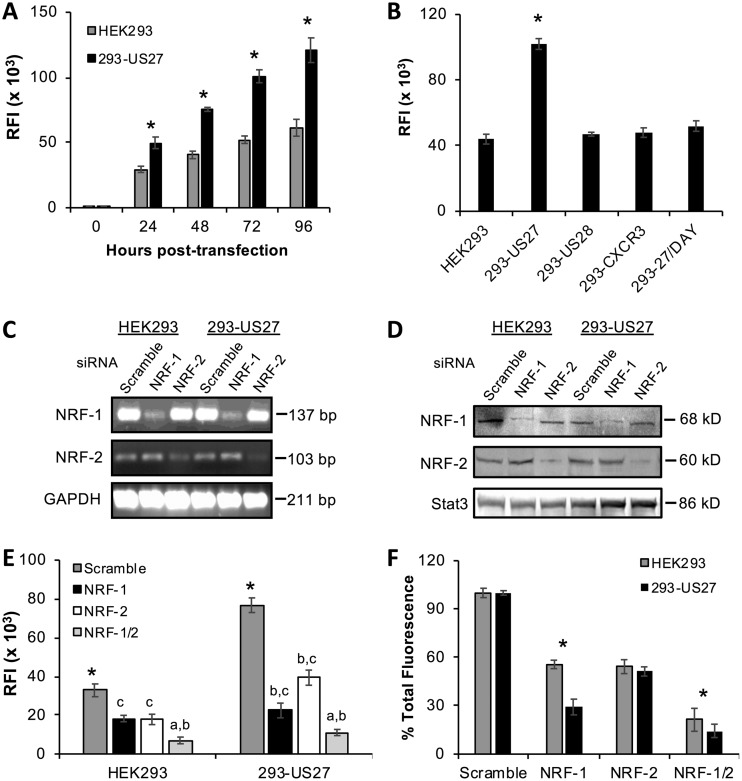
Since CXCR4 expression is regulated primarily by transcription factor NRF-1 binding to the antioxidant response element (ARE) found in the CXCR4 promoter (46, 63), we used a green fluorescent protein (GFP) reporter gene assay to investigate whether US27 had any impact on ARE-mediated transcription. HEK293 cells, either parent cells or a cell line stably expressing US27 (17, 18, 64, 65), were transfected with reporter plasmid pCignal-ARE-GFP, and GFP levels were evaluated using flow cytometry (Fig. 5A). GFP expression was evident in both cell types by 24 h posttransfection, but the level was significantly higher in cells expressing US27 (293-US27). This difference was notable after 24 h and became more prominent over longer time periods. To confirm that the effect was specific to US27, HEK293 cell lines stably expressing other GPCRs, such as HCMV US28 or human CXCR3, were also transfected with pCignal-ARE-GFP. After 48 h, flow cytometry revealed that cells expressing US27 had significantly higher GFP levels than cells expressing either US28 or CXCR3 (Fig. 5B). Additionally, we used a cell line expressing a mutant form of US27 with an alanine substitution in the DRY box domain (R128A) (18, 64, 65), which is known to play a role in signaling for many GPCRs (16). Upon transfection with pCignal-ARE-GFP, the 293-27/DAY cell line exhibited GFP expression that was comparable to that of parent HEK293 cells, as well as 293-US28 and 293-CXCR3 cells, indicating that basal ARE-mediated gene expression was intact (Fig. 5B). However, the elevated level of GFP expression observed in 293-US27 cells was not evident in the 293-27/DAY cells. These results demonstrate that US27 specifically stimulates ARE-driven gene expression and that an intact DRY motif is essential for this effect.
FIG 5.
US27 promotes ARE-driven transcription. (A) HEK293 or 293-US27 stable cell lines were transfected with the pCignal-ARE-GFP plasmid, and the relative fluorescence intensity (RFI) was measured by flow cytometry. (B) HEK293 and stable cell lines were transfected as described above, and the relative fluorescence intensity was measured after 48 h. *, P < 0.01 by paired Student's t test. (C to F) HEK293 and 293-US27 cells were transfected with the pCignal-ARE-GFP plasmid and after 24 h were transfected with the indicated siRNAs for 48 h. (C) RNA was extracted, RT-PCR was performed with the indicated primers, and the products were visualized via agarose gel electrophoresis. (D) Immunoblotting of cell lysates with the indicated siRNA treatments. (E) The relative fluorescence intensity was measured by flow cytometry. *, P < 0.01 by paired Student's t test versus all antibodies; a, significance versus NRF-1; b, significance versus NRF-2; c, significance versus NRF-1/2. (F) Data from panel E are expressed as percent total fluorescence. *, P < 0.01 by paired Student's t test. Error bars represent the standard error for three replicates.
To determine if the effects of US27 on ARE-driven gene expression were mediated by NRF-1, we utilized small interfering RNA (siRNA) to reduce NRF-1 expression. Since the related transcription factor NRF-2 can similarly bind to and regulate transcription from the ARE (47), we also transfected the cells with siRNA directed against NRF-2. HEK293 and 293-US27 cells were transfected with the pCignal-ARE-GFP plasmid for 24 h, followed by siRNA directed against NRF-1, NRF-2, or a scrambled control. The cells were harvested 48 h after siRNA transfection, we confirmed NRF-1/2 knockdown using PCR to evaluate mRNA levels (Fig. 5C) and Western blotting to visualize protein levels (Fig. 5D), and then we evaluated ARE-driven GFP expression using flow cytometry (Fig. 5E). As expected, the scrambled control siRNA had no impact on fluorescence intensity for either cell type, and 293-US27 cells still exhibited greater fluorescence than the parent HEK293 cells (Fig. 5E; see Fig. 5A for comparison). NRF-1 siRNA treatment reduced total GFP expression in HEK293 cells by 45% compared to that in the scrambled control-treated cells (Fig. 5E and F). For 293-US27 cells, NRF-1 siRNA treatment reduced the total fluorescence by 70% (Fig. 5E and F). In HEK293 cells, GFP levels were reduced by 45% with each siRNA treatment, suggesting that NRF-1 and NRF-2 likely contribute equally to ARE-mediated gene expression. Treatment with a combination of the NRF-1 and NRF-2 siRNAs resulted in dramatically decreased fluorescence in both cell types. In 293-US27 cells, NRF-2 siRNA reduced GFP expression, but not to the same extent as NRF-1 siRNA (Fig. 5E and F). While there remains the possibility that not all cells transfected with the pCignal-ARE-GFP plasmid were also transfected with the siRNA, these results indicate that US27 stimulates ARE-mediated gene transcription due mainly to the activity of NRF-1, with some contribution from NRF-2 as well.
US27 increases the nuclear translocation of NRF-1 in transfected cells.
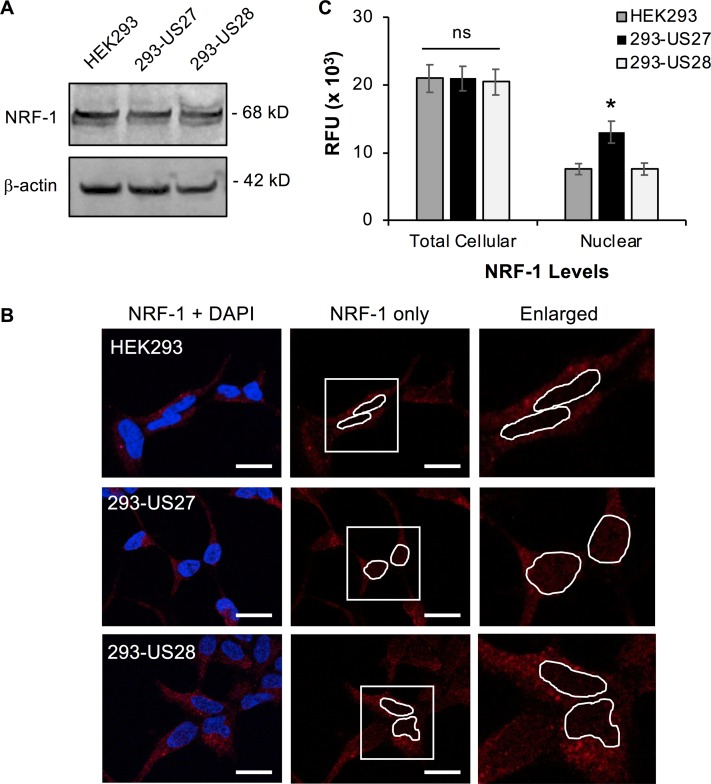
US27 could mediate increased NRF-1 transcriptional activity via several possible mechanisms, such as increasing total NRF-1 protein levels or stimulating protein trafficking into the nucleus. To evaluate total NRF-1 protein levels, Western blotting was performed on lysates from HEK293, 293-US27, or 293-US28 cells (Fig. 6A). A band of 68 kDa, the expected size for NRF-1, was evident and appeared to be comparable for each cell line, suggesting that US27 does not trigger an increase in total NRF-1 protein levels.
FIG 6.
US27 promotes the nuclear translocation of NRF-1. NRF-1 protein levels were evaluated in HEK293, 293-US27, and 293-US28 cells. (A) Whole-cell lysates were immunoblotted with antibodies directed against NRF-1 or β-actin as a total protein loading control. (B) Cells were cultivated on glass coverslips and then fixed and stained with anti-NRF-1 antibody followed by Alexa Fluor 594-conjugated secondary antibody (red). Nuclei are stained with DAPI (blue). Bars, 20 μm. Enlarged images (right) represent the areas in the white boxes in the center panels. These images are representative of those from one of three independent experiments. (C) Average total cellular and nuclear fluorescence due to NRF-1 was quantified for individual cells (n = 10). RFU, relative fluorescent units. Error bars represent the standard error. *, P < 0.0001 by paired Student's t test versus HEK293 or 293-US28 cells; ns, not significant.
Since US27 did not appear to affect the amount of NRF-1 protein in the cell, we next investigated the cellular distribution of NRF-1. In order to visualize intranuclear NRF-1, cells were cultured on glass coverslips and then fixed, permeabilized, and stained with anti-NRF-1 antibody followed by Alexa Fluor 594-conjugated secondary antibody and mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) to demarcate the nucleus (Fig. 6B). We observed an increase in red fluorescent punctae in the nuclear region of 293-US27 cells, indicating higher levels of NRF-1 inside the nucleus. Quantification of red fluorescence from the nuclear region of individual cells in maximum-intensity projection images revealed a 2-fold increase in intranuclear fluorescence in 293-US27 cells compared to the HEK293 or 293-US28 cells (Fig. 6C). We observed comparable levels of total cellular NRF-1 expression by fluorescence across the three cell lines, confirming that there is no significant difference between total NRF-1 protein levels (Fig. 6C). These results suggest that US27 promotes expression of ARE-regulated genes by stimulating increased nuclear translocation of the transcription factor NRF-1.
US27 promotes NRF-1 nuclear translocation during infection.
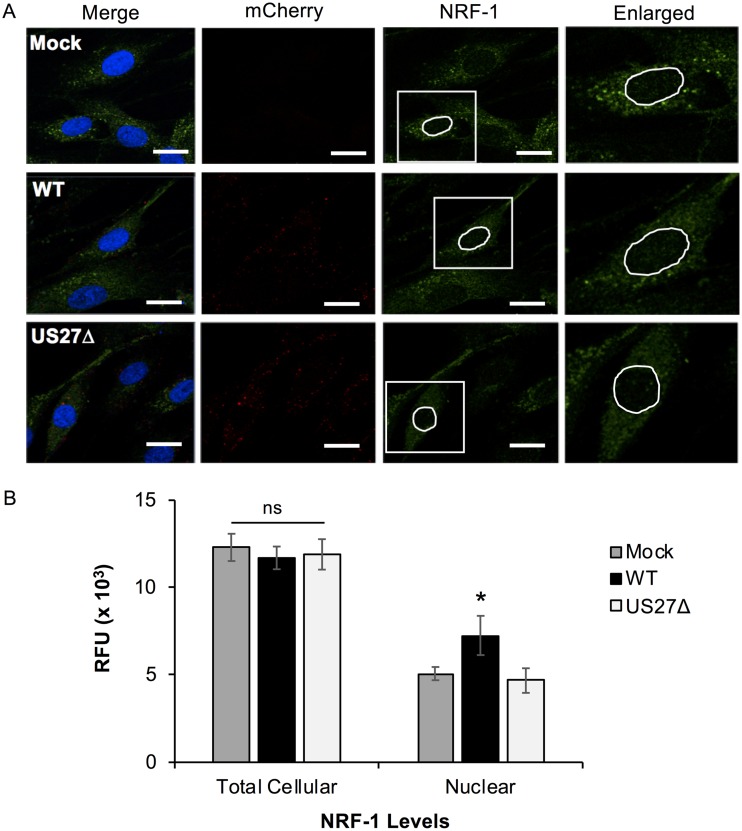
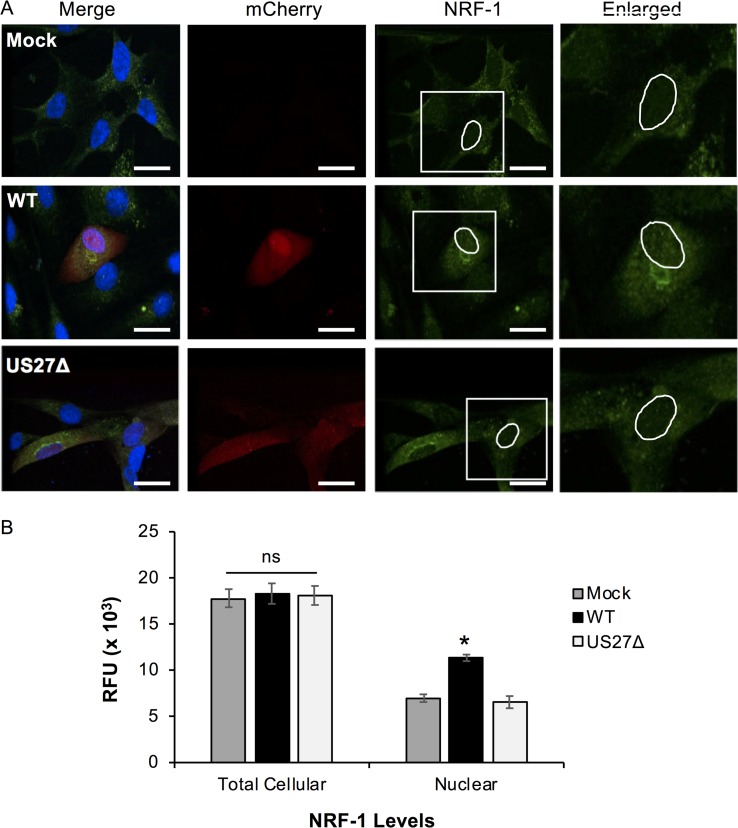
In order to confirm that US27 also increased the nuclear translocation of NRF-1 during virus infection, fibroblasts were cultured on glass coverslips, infected with WT or US27Δ virus for 3 h (MOI = 3), and stained with anti-NRF-1 antibody followed by Alexa Fluor 514-conjugated secondary antibody and DAPI, as described above (Fig. 7A). The red punctae indicate viral infection, as the virus contains a simian virus 40-driven mCherry cassette. Quantification of NRF-1 by green fluorescence revealed an increase in the intranuclear signal intensity in WT-infected cells compared to mock- or US27Δ-infected cells (Fig. 7B). Total fluorescence throughout the cells for each sample was comparable. These data suggest that US27 is constitutively active immediately postinfection and can induce NRF-1 translocation into the nucleus as early as 3 h after virus entry. To investigate the impact of US27 on the NRF-1 cellular distribution at later time points, fibroblasts were infected with WT or US27Δ virus for 72 h (MOI = 0.2) and stained with anti-NRF-1 antibody and DAPI, as described above (Fig. 8A). Quantification of nuclear NRF-1 levels at 72 hpi indicated a 2.5-fold increase in intranuclear green fluorescence in WT-infected cells compared to mock- or US27Δ-infected cells (Fig. 8B). These results demonstrate that during infection, US27 stimulates the movement of transcription factor NRF-1 into the nucleus.
FIG 7.
US27 promotes NRF-1 nuclear translocation immediately postinfection. (A) NuFF cells grown on glass coverslips were infected with TB40-mCherry WT or US27Δ (MOI = 3) and then fixed at 3 hpi and stained with anti-NRF-1 antibody and Alexa Fluor 514-cojugated secondary antibody (green) as well as DAPI (blue). mCherry (red) is a marker of infection. Bars, 20 μm. Enlarged images (right) represent the areas in white boxes in the NRF-1 panels. These images are representative of those from one of three independent experiments. (B) The average total cellular and nuclear fluorescence was quantified for individual cells (n = 10). RFU, relative fluorescent units. Error bars represent the standard error. *, P < 0.0001 by paired Student's t test versus mock- or US27Δ-infected cells; ns, not significant.
FIG 8.
US27 increases NRF-1 translocation at later times during infection. (A) NuFF cells grown on glass coverslips were infected with TB40-mCherry WT or US27Δ (MOI = 0.2) and then fixed at 72 hpi and stained with anti-NRF-1 antibody and Alexa Fluor 514-conjugated secondary antibody (green) as well as DAPI (blue). mCherry (red) is a marker of infection. Bars, 20 μm. Enlarged images (right) represent the areas in the white boxes in the NRF-1 panels. These images are representative of those from one of three independent experiments. (B) Average total cellular and nuclear fluorescence was quantified for individual cells (n = 10). RFU, relative fluorescent units. Error bars represent the standard error. *, P < 0.0001 by paired Student's t test versus mock- or US27Δ-infected cells; ns, not significant.
US27 requires Gβγ and PI3K to increase transcriptional activation via the ARE in transfected cells.
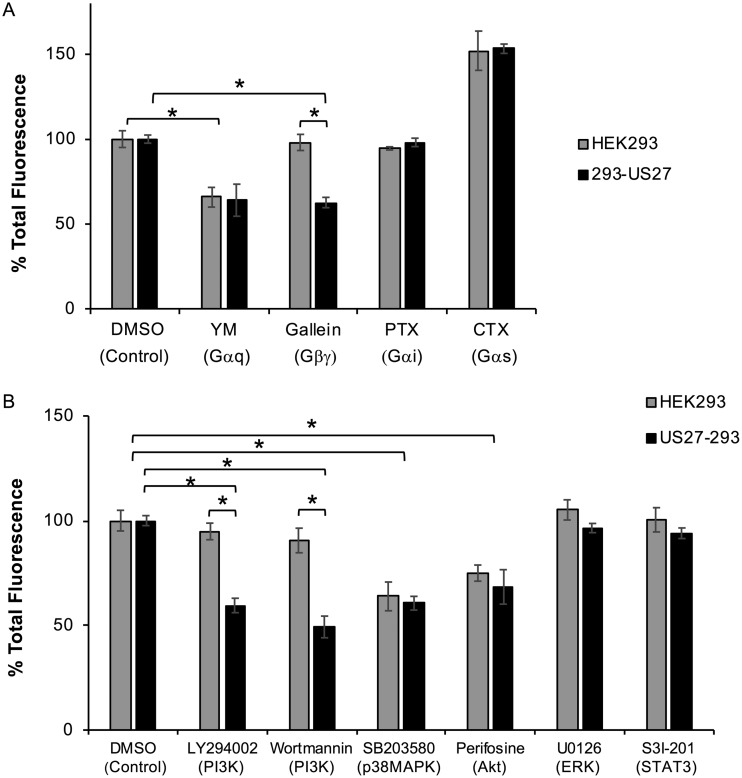
To investigate components of the signaling pathway that enable US27 to constitutively activate ARE-mediated transcription, we used a series of well-characterized inhibitors (Table 1). We transfected HEK293 or 293-US27 cells with pCignal-ARE-GFP for 24 h, treated the cells overnight with standard doses of the indicated compounds, and then assessed GFP levels by flow cytometry. Treatment with pertussis toxin (PTX) had no effect on GFP levels, suggesting that Gαi is not required for ARE-mediated transcription (Fig. 9A). In contrast, YM254890 (YM) treatment resulted in a decrease in total GFP levels in both HEK293 and 293-US27 cells, indicating that Gαq/11 is involved in the natural pathway for ARE-driven gene expression, irrespective of US27 (Fig. 9A, YM). Treatment with cholera toxin, a potent activator of Gαs, increased GFP fluorescence in both cell lines, suggesting that Gαs may also contribute to ARE-mediated transcription. Specific inhibition of GFP levels occurred only when 293-US27 cells were treated with gallein, an inhibitor of Gβγ protein signaling. These results indicate that US27 promotes NRF-1 translocation and transcription of ARE-regulated genes through the activity of Gβγ proteins.
TABLE 1.
Cell signaling inhibitors
| Inhibitor | Target | Concn (reference) | Source |
|---|---|---|---|
| Cholera toxin | Gαs | 200 ng/ml (92) | Sigma-Aldrich (St. Louis, MO) |
| Gallein | Gβγ | 10 μM (93) | Santa Cruz Biotechnology (Dallas, TX) |
| LY294002 | Phosphoinositide 3-kinases (PI3K) | 10 μM (94, 95) | Cell Signaling Technology (Danvers, MA) |
| Perifosine | Akt | 30 μM (96) | Cell Signaling Technology |
| Pertussis toxin | Gαi/o | 200 ng/ml (97) | Sigma-Aldrich |
| S3I-201 | STAT3 | 50 μM (98) | Santa Cruz Biotechnology |
| SB203580 | p38 MAPK | 100 μM (99) | Sigma-Aldrich |
| U0126 | MEK1/2 | 10 μM (100) | Cell Signaling Technology |
| Wortmannin | Phosphoinositide 3-kinases (PI3K) | 50 μM (101, 102) | Enzo Life Sciences (Farmingdale, NY) |
| YM-254890 | Gαq/11 | 20 ng/ml (103) | Wako Chemicals (Richmond, VA) |
FIG 9.
US27 activation of NRF-1 requires Gβγ and phosphatidylinositol 3-kinase (PI3K). HEK293 or 293-US27 cells were transfected with the pCignal-ARE-GFP plasmid for 24 h and then cultured overnight in the presence of the indicated chemical inhibitors, and total GFP levels were evaluated by flow cytometry. The results for a panel of inhibitors targeting G proteins (A) and other intracellular signaling proteins (B) are shown. Results are represented as the percentage of total fluorescence relative to the total fluorescence for control cells. Error bars represent the standard error. *, P < 0.001 by paired Student's t test. CTX, cholera toxin.
To further elucidate downstream proteins involved in US27 signaling, we employed inhibitors targeting additional intracellular signaling proteins (Table 1). Treatment with U0126 and S31-201 had no impact on GFP levels, suggesting that neither extracellular signal-regulated kinase 1/2 nor STAT3 plays a role in US27 signaling in these cells (Fig. 9B). Cultivation with perifosine and SB203580 (SB) resulted in a decrease in fluorescence in both cell lines, indicating that Akt and p38 mitogen-activated protein kinase (p38MAPK) are involved in the natural activation of ARE-governed transcription (Fig. 9B). The only compound that we tested that showed specific inhibition of US27-mediated GFP expression was LY294002, suggesting that phosphoinositide 3-kinase (PI3K) influences US27 signaling. To confirm this, we treated cells with wortmannin, an irreversible inhibitor of PI3K, and again, the GFP levels were decreased in the 293-US27 cells only (Fig. 9B). Taken together, these results demonstrate that US27 is a constitutively active GPCR that stimulates ARE-mediated transcription through activation of Gβγ and PI3K.
US27 increases transcription of other ARE-regulated genes.
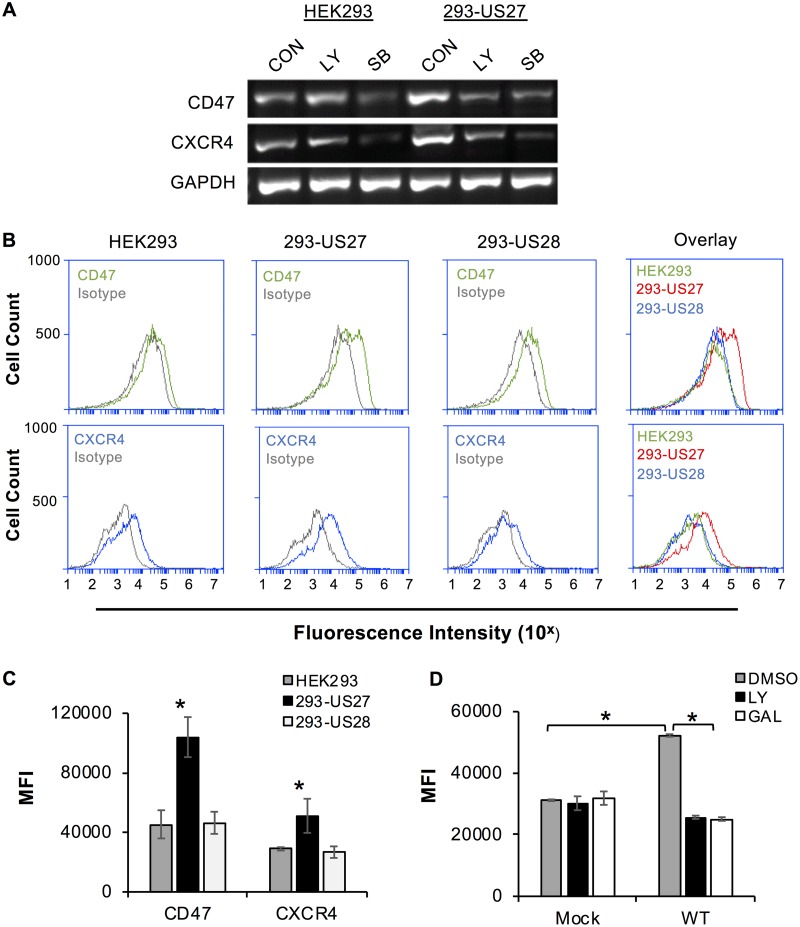
Although our initial observation was that US27 promoted an increase in CXCR4 levels, ARE sequences are found in the promoters of many cellular genes. To determine if US27 affects transcription of other ARE-regulated genes, we examined expression of CD47 (66, 67). CD47 is a widely expressed integrin-associated member of the immunoglobulin superfamily that has the ability to block phagocytosis through binding of signal regulatory protein alpha (SRPα) on myeloid cells, sending the message that the cell should not be engulfed (68, 69). To evaluate gene expression, we performed reverse transcription (RT)-PCR on total RNA harvested from HEK293 and 293-US27 cells. As expected, CXCR4 expression was higher in 293-US27 cells than in HEK293 cells (Fig. 10A). In addition, the band intensity for CD47 appeared brighter in the 293-US27 cells than in HEK293 cells, suggesting that expression of this gene was also upregulated by US27 (Fig. 10A). Treatment with the PI3K inhibitor LY294002 (LY) reduced the CXCR4 and CD47 band intensity in 293-US27 cells but had no effect on expression levels in HEK293 cells. In contrast, p38MAPK inhibitor SB203580 (SB) drastically reduced the band intensity for CD47 and CXCR4 in both cell types, demonstrating that p38MAPK plays a role in natural activation of this transcriptional pathway in the presence or absence of US27, as noted earlier (Fig. 9B). These data demonstrate that US27 expression increases the transcription of not only CXCR4 but also other ARE-regulated genes, such as CD47, in a PI3K-dependent manner in transfected cells.
FIG 10.
US27 increases expression of ARE-regulated gene CD47. (A) HEK293 or 293-US27 cells were treated with dimethyl sulfoxide (control [CON]), 20 μM LY294002 (LY), or 100 μM SB203580 (SB) for 5 h. RNA was harvested, and RT-PCR was performed with gene-specific primers. PCR products were visualized on a 2% agarose gel. (B) Cells were stained with CD47-FITC (green), CXCR4-PE (blue), or the appropriate isotype control (gray), and the fluorescence intensity was measured via flow cytometry. The right panels show overlays of receptor expression for all three cell types. (C) The mean fluorescence intensity (MFI) for CD47 or CXCR4 staining for each cell type from 3 independent experiments was normalized and averaged. *, P < 0.001 by paired Student's t test versus HEK293 and 293-US28 cells. (D) NuFF cells were treated with 10 μM LY or gallein (GAL) for 1 h prior to infection (MOI = 3, 18 h), and CXCR4 surface levels were measured by flow cytometry. Error bars represent the standard error of the mean. *, P < 0.05, paired Student's t test versus mock, LY, and gallein treatment.
Since CD47 is a cell surface receptor, we asked whether the increase in CD47 mRNA expression levels in 293-US27 cells correlated with an increase in membrane protein levels. To address this question, we stained cells with fluorochrome-conjugated antibodies directed against either CD47 or CXCR4 and examined them via flow cytometry. HEK293 cells endogenously expressed both CD47 and CXCR4, as indicated by the right-shifted fluorescence histograms relative to those for isotype control antibodies (Fig. 10B). As determined previously, 293-US27 cells exhibited a greater shift in fluorescence for CXCR4 than HEK293 or 293-US28 cells, indicating higher CXCR4 surface levels (18). For CD47, we observed significantly higher fluorescence intensity in 293-US27 cells than in HEK293 or 293-US28 cells (Fig. 10B and C). For CXCR4, we further examined surface levels during virus infection of fibroblasts in the presence of LY or gallein (Fig. 10D). CXCR4 levels were significantly higher 18 h after infection, but treatment with either LY or gallein blocked this increase. Taken together, these results indicate that US27 specifically increases the mRNA and surface protein levels of ARE-regulated genes (CXCR4 and CD47) through the action of Gβγ and PI3K.
US27 increases CD47 expression during infection.
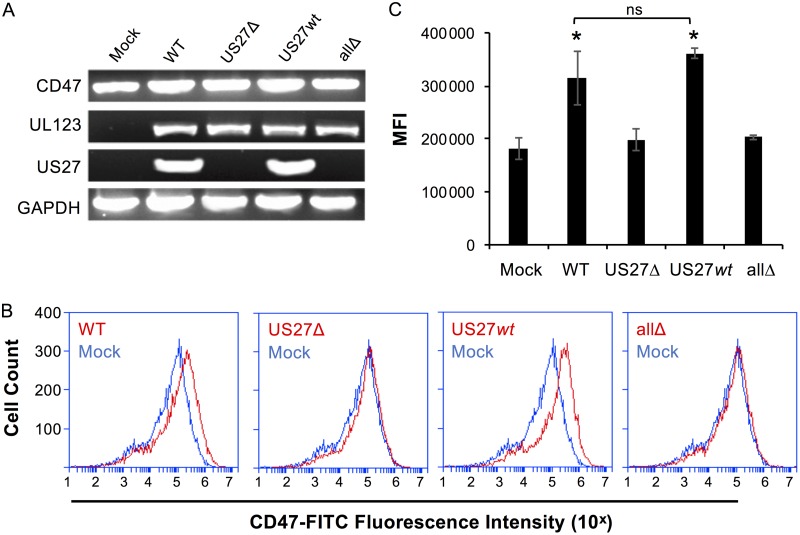
We had already observed increased CXCR4 levels upon infection of fibroblasts (Fig. 2, 4, and 10D), so we next evaluated infected fibroblasts (MOI = 0.2) for CD47 expression. At 120 hpi, expression of UL123 was evident in cells infected with each virus strain, while US27 was observed only in cells infected with the WT and US27wt (Fig. 11A). CD47 was expressed in both mock-infected and infected cells, but the levels were higher when US27 was present (WT- and US27wt viruses). Cell surface expression of CD47 was analyzed via flow cytometry, and the levels were significantly increased upon infection with WT and US27wt viruses (Fig. 11B and C). These findings demonstrate that CD47 expression is increased by US27 during infection of fibroblasts, which is in agreement with previous reports of CD47 upregulation during HCMV infection of monocytes (70).
FIG 11.
US27 increases expression of CD47 during infection. (A) NuFF cells were infected with the indicated viruses (MOI = 0.2). At 120 hpi, RNA was harvested and RT-PCR was performed with gene-specific primers. PCR products were visualized on a 2% agarose gel. (B) Mock-infected cells (blue) or infected cells (red) were stained with CD47-FITC, and the fluorescence intensity was measured via flow cytometry. (C) The mean fluorescence intensity (MFI) for CD47 staining for each cell type from 3 independent experiments was normalized and averaged. Error bars represent the standard error of the mean. *, P < 0.001 by paired Student's t test versus mock-, US27Δ-, and allΔ-infected cells; ns, not significant.
US27 impacts CXCR4 and CD47 expression during infection of endothelial cells.
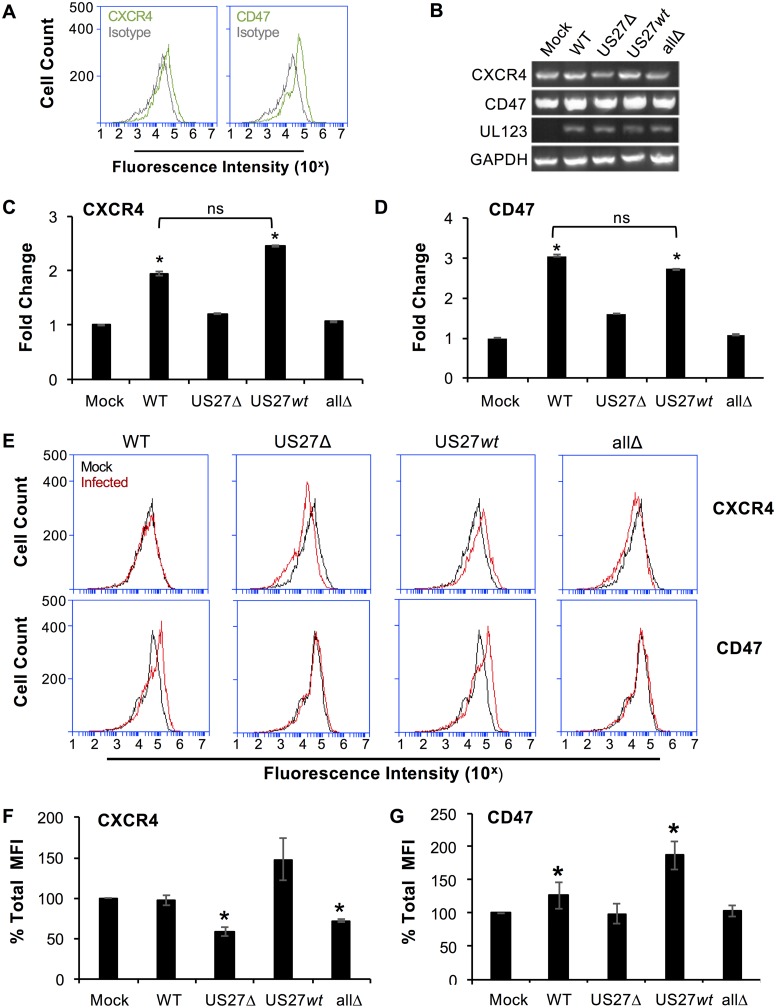
While fibroblasts are a useful model for infection in the laboratory, we wondered whether US27 would exhibit the same effects on ARE gene expression in other cell types that are also physiologically relevant for infection in vivo. We evaluated human umbilical vein endothelial cells (HUVECs) using flow cytometry, and the results confirmed that CXCR4 and CD47 were present on the cell surface (Fig. 12A). To investigate the effect of US27 on gene expression, HUVECs were infected (MOI = 2), RNA was harvested at 24 hpi, and standard PCR (Fig. 12B) and qPCR (Fig. 12C and D) were performed. The results show that both CXCR4 and CD47 were expressed at higher levels in WT- and US27wt-infected cells than in mock-, US27Δ-, or allΔ-infected cells (Fig. 12B to D). We next examined cell surface receptor levels in the infected HUVECs via flow cytometry. As expected, CD47 surface levels were higher in WT- and US27wt-infected cells than in mock-, US27Δ-, and allΔ-infected cells (Fig. 12E, bottom, red histograms). However, we were surprised to find that CXCR4 cell surface levels were not any higher in WT-infected cells than in mock-infected cells (Fig. 12E, top, red histograms). Thus, the observed increase in CXCR4 gene expression did not correlate with cell surface levels in WT-infected HUVECs, and this was confirmed by evaluation of the fluorescence intensity from three independent experiments (Fig. 12F). In contrast, infection with US27Δ and allΔ viruses resulted in significantly reduced CXCR4 surface levels, suggesting that US27 does have some influence on the level of CXCR4 surface expression. Moreover, US27wt-infected cells showed a trend toward higher CXCR4 surface levels than mock-infected cells, suggesting that the difference in WT infection of HUVECs may be attributable to one of the other HCMV GPCRs. CD47 surface levels were increased in response to both WT and US27wt infection (Fig. 12G). The infections lacking US27 displayed a significant decrease in CXCR4 cell surface levels (Fig. 12F), yet CD47 surface levels were unaffected by the absence of US27 (Fig. 12G). Taken together, these findings suggest that there may be multiple levels of regulation of cell surface expression with intracellular trafficking and interaction among the viral GPCRs involved in these processes.
FIG 12.
US27 increases gene expression of CXCR4 and CD47 during infection of HUVECs. (A) Cell surface staining with CXCR4-FITC, CD47-FITC (green), or the isotype control (gray), analyzed by flow cytometry. (B to D) HUVECs were infected with the indicated HCMV strains (MOI = 2), and RNA was harvested at 24 hpi. (B) RT-PCR was performed with gene-specific primers. PCR products were visualized on a 2% agarose gel. (C) RT-qPCR was performed using CXCR4 primers on the same cDNA with normalization to the amount of GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and the results are expressed as the fold change in expression relative to that in mock-infected cells. (D) RT-qPCR using CD47 primers as described above. (E) Mock-infected (black) or infected (red) HUVECs were stained with CXCR4-FITC (top) or CD47-FITC (bottom) at 96 hpi, and the fluorescence intensity was measured via flow cytometry. (F, G) The mean fluorescence intensity (MFI) for CXCR4 staining (F) or CD47 staining (G) for each infection from 3 independent experiments was normalized to that for mock-infected cells and averaged. Error bars represent the standard error of the mean. *, P < 0.05 by paired Student's t test versus mock-infected cells.
DISCUSSION
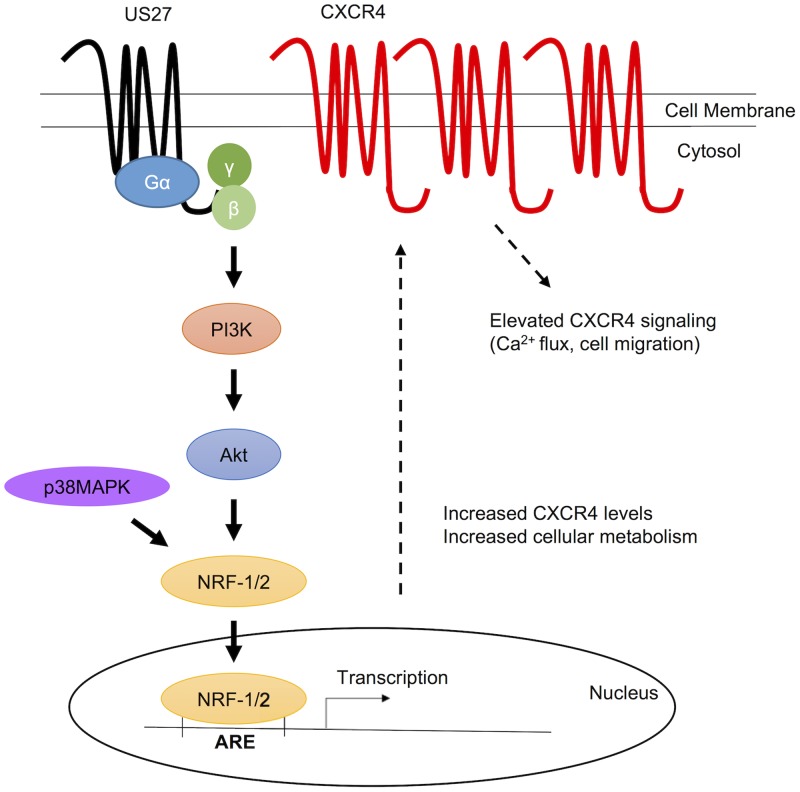
Virus infection induces a multitude of changes in the host cell, many of which are critical for virus replication and dissemination. Enhancement of CXCR4 signaling by the HCMV US27 gene product was first observed during a ligand screen for chemokines that bind to US27. While no chemokine ligands were identified, it was noted that US27 expression not only increased CXCR4 signaling but also led to elevated CXCR4 expression levels (18). The results presented here define the mechanistic basis for this observation and demonstrate for the first time that US27 is a constitutively active receptor that signals through Gβγ, PI3K, and NRF-1 to activate transcription of ARE-regulated genes during HCMV infection, as depicted in Fig. 13. Although elevated CXCR4 expression has been our primary readout for US27 signaling, we also found increased CD47 expression during infection, and it is likely that we will identify additional target genes that contribute to virus persistence in our future studies.
FIG 13.
Model depicting the proposed US27 pathway for activation of ARE gene transcription. US27 signals constitutively through Gβγ and PI3K to activate NRF-1 and promote its translocation to the nucleus, where expression of ARE-regulated genes, such as CXCR4, is enhanced.
While most cellular chemokine receptors require ligand binding to activate downstream signaling, it is common for viral GPCRs to exhibit ligand-independent constitutive signaling activity (71). The crystal structure of US28 was recently solved to 2.9-Å resolution and revealed molecular features that contribute to constitutive signaling activity (72). In most chemokine receptors, the aspartic acid in the DRY motif interacts with another arginine residue on the second intracellular loop, which stabilizes the inactive state (73). In US28, the interaction between D128 of the DRY motif and R139 appears to be prevented by E124, which acts as an ionic hook to pull R139 away from D128 (72). This destabilizes the inactive state of the receptor and favors the active conformation. Indeed, we found that the DRY motif of US27 was necessary for constitutive activation of ARE-mediated gene transcription (Fig. 5B). In US27, a polar asparagine residue (N123) occupies the position equivalent to E124 in US28 and may function similarly to maintain the US27 DRY motif in the active receptor conformation. These ionic hooks are not present in cellular chemokine receptors, suggesting that viral GPCRs may have modified functional domains to promote constitutive activity that is beneficial to the virus.
Previous studies that examined US27 signaling did not show constitutive activation of NF-κB or CREB pathways (27). This is not surprising given that US28 is already highly adept at activating these pathways. Phylogenetic analyses indicate that US27 and US28 are products of gene duplication and divergence following the capture of a single host gene that shares a common ancestor with human CX3CR1 (74). US28 has a retained higher degree of similarity to CX3CR1, as well as the ability to bind chemokine ligand CX3CL1/fractalkine (20), whereas US27 has diverged considerably. This divergence may represent viral evolution allowing HCMV to gain the ability to manipulate additional cell signaling pathways. For example, HCMV infection induces oxidative stress in cell culture and systemic inflammatory responses in patients experiencing primary infection (75–77). Oxidative stress can be damaging to the host cell and is likely to negatively impact virus replication. HCMV may have developed a mechanism to activate NRF-1 and NRF-2 via US27 to increase cellular resistance to oxidative stress. This is not unprecedented, as other viruses, notably, hepatitis B virus (HBV), avoid oxidative stress through activation of NRF-2/ARE-regulated genes (78).
In this study, infection of fibroblasts with either wild-type or mutant strains of HCMV TB40/E-mCherry resulted in the upregulation of CXCR4 mRNA and surface protein levels at both early and extended time points postinfection when US27 was present. We have also observed increased CXCR4 protein levels and heightened chemotaxis to CXCL12 in HCMV-infected monocytes (79). Since CXCL12 is highly expressed in stromal tissues and bone marrow, HCMV may upregulate CXCR4 to direct infected cells to favorable microenvironments to enhance virus dissemination, promote virus persistence, or avoid immune clearance. Intriguingly, upregulation of CXCR4 activity by US27 is in direct contrast to the effects of the other HCMV GPCRs. Monocytes transfected with UL33 or UL78 exhibited a decrease in CXCR4 surface levels and chemotaxis toward CXCL12 and CCL5, although it is important to note that these studies were performed in the absence of the other viral GPCRs (80). In the context of virus infection, CXCR4 surface levels and signaling were decreased in HUVECs infected with HCMV TB40/E but not TB40/E-ΔUS28, demonstrating that US28 also impacts CXCR4 expression and activity (62). In agreement with this study, we observed increased CXCR4 gene expression in HUVECs that did not correlate with increased cell surface protein levels when the other viral GPCRs were present (Fig. 12). While US27 clearly has the ability to stimulate CXCR4 gene expression, additional work is necessary to investigate factors that influence surface protein levels, which could be reduced due to protein degradation, sequestration in intracellular vesicles, or other factors.
Interestingly, there are conflicting reports describing the effect of HCMV infection on CXCR4 expression or overall cell migration in different cell types, many of which did not specifically investigate roles for viral GPCRs. Smith et al. (81) found that infection of primary human monocytes promoted transendothelial migration and increased cell motility. In contrast, Frascaroli et al. (82) reported that monocyte-derived macrophages (MDMs) were paralyzed following infection and unable to migrate toward inflammatory chemokines. However, CXCL12 was not among the chemokines tested, and no change in the level of CXCR4 expression was detected. Lecointe et al. (83) observed that HCMV infection led to downregulation of CXCR4 in microglial cells but not in astrocytes or MDMs; however, signaling outcomes were not examined. Warner et al. (84) reported that HCMV infection inhibited migration of cytotrophoblasts toward CXCL12. This result is particularly intriguing because the authors also noted an increase in mRNA and total cell surface expression levels of both CXCR4 and CXCR7, and higher CXCR4 cell surface levels are generally associated with a promigratory phenotype (85, 86). However, CXCR7 has the ability to heterodimerize with CXCR4 and impair its ability to activate G proteins upon CXCL12 binding (87). The reduction of chemotaxis observed in cytotrophoblasts may be a result of CXCR7 modulating CXCR4's response to CXCL12 binding. This suggests that different phenotypes observed across cell types (2) during HCMV infection could be dependent on the cellular machinery and transcriptome specific to each cell type; thus, it is possible that the potentiating effect of US27 on CXCR4 may be more pronounced in some cell types and less evident in others. This is not unprecedented, as the HCMV GPCR US28 also displays differential signaling phenotypes across cell types (26).
Activation of PI3K/Akt signaling by US27 suggests that multiple signal transduction cascades are activated in addition to NRF-1/2. We previously determined that US27 conveys proliferative and survival properties (64, 65). The cell cycle regulator p21 (cyclin-dependent kinase inhibitor 1 [CDK1NA]) (88) was decreased at the mRNA and protein levels in 293-US27 cells. Akt phosphorylates p21 at Thr145 (89), which reduces complex formation of p21Cip1 with Cdk2 and Cdk4 to promote cell proliferation. The proliferative properties that US27 conveys may be due to increased activation of Akt above basal levels. Higher Akt activity leads to more p21 phosphorylation, such that p21 is inhibited from blocking cell cycle progression.
The functions of US27 and how this viral GPCR affects the host cell have only recently been identified. To the best of our knowledge, the results of this study reveal for the first time a signaling pathway modulated by US27: constitutive activation of ARE-regulated genes. As the ARE is present in the promoter of a battery of genes, US27 activation during HCMV infection could heavily affect host cell machinery and metabolism, including potential roles in immune evasion, virus dissemination, and oncomodulation. Further studies will undoubtedly reveal more details of the contribution of US27 to HCMV infection.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Human embryonic kidney (HEK293) cells were grown in Eagle's minimal essential media (EMEM) with 10% fetal bovine serum (FBS) (Cellgro, Herndon, VA) in a humidified incubator at 37°C in a 5% CO2 atmosphere. HEK293 cell lines expressing 3×-FLAG-tagged human CXCR3 or HCMV wild-type US27, the US27-DAY mutant, or US28 were generated as previously described (17) and maintained as described above. Newborn human foreskin fibroblast-1 (NuFF) cells (MTI-Global Stem, Gaithersburg, MD) were cultivated in Dulbecco's modified essential medium (DMEM) supplemented with 10% FBS, 1% nonessential amino acids (NEAA), and 10 mM HEPES buffer. Human umbilical vein endothelial cells (HUVECs; ATCC, Manassas, VA) were cultivated in endothelial cell growth base media (R&D Systems, Minneapolis, MN) supplemented with 5% FBS and endothelial growth supplement (R&D Systems) and maintained as described above.
The BAC-derived clinical strain HCMV TB40/E-mCherry (19) (which was generated from TB40/E-BAC clone 4 [90]), TB40/E-mCherry-US27Δ (US27Δ) (19), and TB40/E-mCherry-allΔ (allΔ) (26) are described elsewhere. To generate TB40/E-mCherry-US27wt (US27wt), we used galK recombineering techniques, as previously described (58, 91). We used TB40/E-mCherry to generate two independently derived clones in which the ORFs for UL33, UL78, and US28 were excised in a single background, similar to the methods described by Humby and O'Connor (39). The resulting virus, US27wt, was generated using the primers in Table 2. Each US27wt clone was verified by sequence analysis. All viral stocks were generated on NuFFs and concentrated by ultracentrifugation through a 20% sorbitol cushion once a 100% cytopathic effect was observed, as described previously (91). The resulting viral pellets were resuspended in X-VIVO15 medium (Lonza, Walkersville, MD) containing 1.5% bovine serum albumin (BSA) and were flash frozen in liquid nitrogen before storage at −80°C. The titers of the viral stocks were subsequently determined by 50% tissue culture infectious dose (TCID50) assays on NuFFs. To determine the growth properties of the recombinant viruses compared to each other and the parental strain, NuFFs were infected at a low multiplicity of infection (MOI) of 0.01 and infectious supernatants were harvested over time. The titers viral in the supernatants were subsequently determined by IE staining, as described in detail previously (19).
TABLE 2.
Primers used for generating TB40/E-mCherry-US27wt (US27wt)
| Recombinant virus (TB40/E-mCherry) | Primer direction or characteristica | Primer sequence (5′ → 3′)b |
|---|---|---|
| US28Δ-galK insertion | FOR | GGTGCGTGGACCAGACGGCGTCCATGCACCGAGGGCAGAACTGGTGCTATCcctgttgacaattaatcatcggca |
| REV | AGAGGGGCGGACACGGGGTTTGTATGAAAAGGCCGAGGTAGCGCTTTTTTAtcagcactgtcctgctcctt | |
| US28Δ | ds oligo | CAGACGGCGTCCATGCACCGAGGGCAGAACTGGTGCTATCTAAAAAAGCGCTACCTCGGCCTTTTCATACAAACCCCGTG |
| UL33Δ-galK insertion | FOR | TTCCGCCCAGACCCGCAACAACACTCCTCCGCACATCAATGACACTTGCAACcctgttgacaattaatcatcggca |
| REV | GGGGAAATGGCGACGGGTTCTGGTGCTTTCTGAATAAAGTAACAGGAAAGCtcagcactgtcctgctcctt | |
| UL33Δ | ds oligo | CCGCAACAACACTCCTCCGCACATCAATGACACTTGCAACGCTTTCCTGTTACTTTATTCAGAAAGCACCAGAACCCGTC |
| UL78Δ-galK insertion | FOR | GTCCCCGGAGAGGGTATATTCGTTCGGCGAGAGCGGGCGGCGGTGGTGGGTcctgttgacaattaatcatcggca |
| REV | TAACGTGATTTATCTGCCACTTTTCTCCCCGCTGCCGTACAGCGCCGCCGCtcagcactgtcctgctcctt | |
| UL78Δ | ds oligo | GGGTATATTCGTTCGGCGAGAGCGGGCGGCGGTGGTGGGTGCGGCGGCGCTGTACGGCAGCGGGGAGAAAAGTGGCAGAT |
FOR, forward primer; REV, reverse primer; ds oligo, double-stranded oligonucleotide.
Lowercase sequences are complementary to the galK template plasmid.
PCR.
RNA was purified from cells using an RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions, and then cDNA was synthesized from the RNA template using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). For standard PCR, each reaction mixture contained cDNA template, primers, a deoxynucleoside trisphosphate mix, Ex Taq buffer, and Ex Taq polymerase (Clontech, Mountain View, CA) in a final volume of 25 μl. The gene-specific primers for standard PCR are listed in Table 2. The reaction underwent the following protocol on a MyCycler thermal cycler (Bio-Rad): 95°C for 1 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, followed by 1 cycle of 72°C for 10 min and a final hold at 4°C. The PCR products were visualized on a 2% agarose gel. For quantitative real-time PCR (qPCR), each reaction mixture contained cDNA template, RT2 SYBR green master mix (SABiosciences, Frederick, MD), and PrimeTime qPCR primers (Integrated DNA Technologies, San Diego, CA), and PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad). The gene-specific primers for CXCR4 and β-actin were the same as those used in the standard PCR (Table 3). The following protocol was used: 95°C for 1 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Data were analyzed using the ΔΔCT threshold cycle (CT) method according to the SABiosciences web portal. The same threshold value was used across all plates to ensure accurate reading of quality controls.
TABLE 3.
Primers for evaluating gene expression via standard PCR and qPCRa
| Target gene | Primer direction | Primer sequence (5′ → 3′) |
|---|---|---|
| CD47 | FOR | TCCACTGTCCCCACTGACTT |
| REV | TGGCAATGACGAAGGAGGTT | |
| CXCR4 | FOR | CCGTGGCAAACTGGTACTTT |
| REV | CCCTTGGAGTGTGACAGCTT | |
| GAPDH | FOR | ACCACAGTCCATGCCATCAC |
| REV | TCCACCACCCTGTTGCTGTA | |
| IE1 | FOR | GGTCACTAGTGACGCTTGTATGATGACCAGTACCGA |
| REV | GATAGTCGCGGGTACAGGGGACTCT | |
| NRF-1 | FOR | AGCAAAAGCAGAGGGTTTCA |
| REV | CTGTGTTTGCGTTTGCTGAT | |
| NRF-2 | FOR | GAGAGCCCAGTCTTCATTGC |
| REV | TGCTCAATGTCCTGTTGCAT | |
| US27 | FOR | GACGGACAAGCTTCGATGACCACCT |
| REV | CGACGAGCTGCAGTTACAACAGAAA | |
| β-Actin | FOR | ATTAAGGAGAAGCTGTGCTACG |
| REV | TGTTGGCGTACAGGTCTTTG |
FOR, forward primer; REV, reverse primer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Flow cytometry.
Cells were washed with phosphate-buffered saline (PBS), harvested with cell stripper (Corning, Corning, NY), resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS with 1% BSA and 1% sodium azide), fixed with 4% paraformaldehyde for 20 min on ice, and blocked in blocking buffer (FACS buffer with 10% FBS) for 1 h on ice. The cells were washed with FACS buffer and stained with fluorochrome-conjugated primary antibody or the isotype control for 1 h at 4°C with protection from light. The antibodies used were anti-CXCR4-phycoerythrin (PE) (R&D Systems, Minneapolis, MN), the IgG2b-PE isotype, anti-CD47-fluorescein isothiocyanate (FITC) (Santa Cruz Biotechnology, Dallas, TX), and the IgG1-FITC isotype. Cells were then washed three times and analyzed using a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
Reporter gene assay.
Cells were seeded in 24-well plates at a density of 5 × 104 cells per well and incubated for 48 h, followed by transfection, in triplicate, of the Cignal antioxidant response GFP reporter plasmid (pCignal-ARE-GFP; Qiagen) using the Fugene 6 transfection reagent (Roche Biosciences, Basel, Switzerland) at a ratio of 3:1 (the number of microliters of the Fugene reagent to the number of micrograms of plasmid DNA, per the manufacturer's instruction). For chemical inhibitor assays, wells were treated with the indicated chemical compound and concentration (Table 1) overnight prior to harvesting. For siRNA treatment, cells were transfected with the pCignal plasmid for 24 h, followed by treatment with NRF-1, NRF-2, or control siRNA according to the manufacturer's protocol for 48 h (Santa Cruz). Cells were washed with PBS, harvested with a cell stripper, resuspended in FACS buffer, washed twice, and analyzed using the BD Accuri C6 flow cytometer.
Immunofluorescence microscopy.
Cells were seeded in 6-well plates containing FBS-coated glass coverslips at a density of 2 × 105 cells per well and then incubated for 72 h at 37°C in 5% CO2. Cell monolayers were washed with PBS, fixed with 4% paraformaldehyde, and then permeabilized with 0.2% (wt/vol) Triton X-100. Cells were then blocked with PBS supplemented with 10% FBS for 1 h at 37°C and stained with anti-NRF-1 rabbit polyclonal primary antibody (Santa Cruz) at a 1:100 dilution for 1 h at 37°C. Following three PBS washes, the coverslips were incubated with the appropriate fluorochrome-conjugated secondary antibody at a 1:100 dilution for 30 min. The wells were washed again and then mounted on a glass slide using Prolong Gold antifade reagent with DAPI (Invitrogen, Carlsbad, CA). z-stack images were taken with a Zeiss LSM700 laser scanning confocal fluorescence microscope equipped with an AxioCAM imaging system and Zen software (Carl Zeiss, Inc., Oberkochen, Germany) using 63× objective magnification with immersion oil. The fluorescence intensity of each maximum-intensity projection image was quantified using ImageJ software. Individual whole cells were outlined to obtain a fluorescence signal value for total NRF-1 per cell, and then the DAPI-stained nucleus was outlined and the fluorescence signal value for just the nuclear region was determined. This was done for 10 cells per condition in each experiment, and the values were averaged to arrive at total cellular NRF-1 levels compared to nuclear levels.
NuFF cells were prepared and imaged as described above, except that they were seeded at a density of 3 × 105 cells per well and then infected after 48 h with the indicated virus, at the indicated MOI, and for the indicated length of time. The primary antibodies used were anti-NRF-1, anti-US27, or anti-US28 (Santa Cruz), each of which was used at a 1:100 dilution for 1 h at 37°C, as described previously (79). Where indicated, cells were treated with 25 μg/ml cycloheximide (Sigma) overnight prior to infection and stained with either anti-US27 or anti-pp65 (Sigma).
Western blotting.
HEK293 and stably transfected cells were seeded in a 10-cm dish at a density of 3 × 106 cells/dish and incubated for 48 h. The cells were harvested by incubating with 1 ml 0.25% trypsin for 3 min, followed by centrifugation. Cells were lysed via the addition of 200 μl lysis buffer (150 mM NaCl, protease cocktail inhibitor, 20 mM HEPES, 1 mM NaOV4, 1 mM EDTA, 0.1% NaN3, 4 M urea) for 15 min on ice. The lysate was centrifuged at 20,800 × g for 15 min at 4°C and incubated for 10 min at 70°C, followed by addition of loading dye. Samples were loaded onto a 4 to 12% bis-Tris gel and separated using SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane, which was blocked in Tris-buffered saline with 0.05% Tween 20 (TBS-T) and 5% milk for 1 h. The membranes were incubated with anti-NRF-1, NRF-2, β-actin, or STAT3 (Cell Signaling Technology, Danvers, MA) antiserum at a 1:1,000 dilution overnight at 4°C, followed by incubation with alkaline phosphatase-conjugated secondary antibody (Santa Cruz). Bands were visualized via the addition of Western blue substrate reagent (Promega, Madison, WI).
ACKNOWLEDGMENTS
We thank Kate Allaire and Jeff Oda for excellent technical assistance and Rhonda Cardin for helpful discussions.
This work was supported by NIH grants AI111232 (to J.V.S.) and AI119415 (to C.M.O.), American Heart Association grant 15SDG23000029 (to C.M.O.), and USF Faculty Development Funds (to J.V.S.).
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Cannon MJ, Schmid DS, Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20:202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 2.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325:63–83. [DOI] [PubMed] [Google Scholar]
- 3.Goodrum F, Caviness K, Zagallo P. 2012. Human cytomegalovirus persistence. Cell Microbiol 14:644–655. doi: 10.1111/j.1462-5822.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McSharry BP, Avdic S, Slobedman B. 2012. Human cytomegalovirus encoded homologs of cytokines, chemokines and their receptors: roles in immunomodulation. Viruses 4:2448–2470. doi: 10.3390/v4112448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wreghitt TG, Teare EL, Sule O, Devi R, Rice P. 2003. Cytomegalovirus infection in immunocompetent patients. Clin Infect Dis 37:1603–1606. doi: 10.1086/379711. [DOI] [PubMed] [Google Scholar]
- 6.Kenneson A, Cannon MJ. 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 7.Lanzieri TM, Dollard SC, Bialek SR, Grosse SD. 2014. Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. Int J Infect Dis 22:44–48. doi: 10.1016/j.ijid.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev 26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanshaw JB. 1971. Congenital cytomegalovirus infection: a fifteen year perspective. J Infect Dis 123:555–561. doi: 10.1093/infdis/123.5.555. [DOI] [PubMed] [Google Scholar]
- 10.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. 2003. Reevaluation of human cytomegalovirus coding potential. Proc Natl Acad Sci U S A 100:13585–13590. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA III, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol 154:125–169. [DOI] [PubMed] [Google Scholar]
- 13.Chee MS, Satchwell SC, Preddie E, Weston KM, Barrell BG. 1990. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature 344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 14.Pierce KL, Premont RT, Lefkowitz RJ. 2002. Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 15.Margulies BJ, Gibson W. 2007. The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles. Virus Res 123:57–71. doi: 10.1016/j.virusres.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan CA. 2005. A GPCR that is not “DRY.” Mol Pharmacol 68:1–3. [DOI] [PubMed] [Google Scholar]
- 17.Stapleton LK, Arnolds KL, Lares AP, Devito TM, Spencer JV. 2012. Receptor chimeras demonstrate that the C-terminal domain of the human cytomegalovirus US27 gene product is necessary and sufficient for intracellular receptor localization. Virol J 9:42. doi: 10.1186/1743-422X-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnolds KL, Lares AP, Spencer JV. 2013. The US27 gene product of human cytomegalovirus enhances signaling of host chemokine receptor CXCR4. Virology 439:122–131. doi: 10.1016/j.virol.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connor CM, Shenk T. 2011. Human cytomegalovirus pUS27 G protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J Virol 85:3700–3707. doi: 10.1128/JVI.02442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kledal TN, Rosenkilde MM, Schwartz TW. 1998. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett 441:209–214. doi: 10.1016/S0014-5793(98)01551-8. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn DE, Beall CJ, Kolattukudy PE. 1995. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem Biophys Res Commun 211:325–330. doi: 10.1006/bbrc.1995.1814. [DOI] [PubMed] [Google Scholar]
- 22.Billstrom MA, Johnson GL, Avdi NJ, Worthen GS. 1998. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J Virol 72:5535–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beisser PS, Goh CS, Cohen FE, Michelson S. 2002. Viral chemokine receptors and chemokines in human cytomegalovirus trafficking and interaction with the immune system. CMV chemokine receptors. Curr Top Microbiol Immunol 269:203–234. [DOI] [PubMed] [Google Scholar]
- 24.Casarosa P, Bakker RA, Verzijl D, Navis M, Timmerman H, Leurs R, Smit MJ. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J Biol Chem 276:1133–1137. doi: 10.1074/jbc.M008965200. [DOI] [PubMed] [Google Scholar]
- 25.Miller WE, Houtz DA, Nelson CD, Kolattukudy PE, Lefkowitz RJ. 2003. G-protein-coupled receptor (GPCR) kinase phosphorylation and beta-arrestin recruitment regulate the constitutive signaling activity of the human cytomegalovirus US28 GPCR. J Biol Chem 278:21663–21671. doi: 10.1074/jbc.M303219200. [DOI] [PubMed] [Google Scholar]
- 26.Miller WE, Zagorski WA, Brenneman JD, Avery D, Miller JL, O'Connor CM. 2012. US28 is a potent activator of phospholipase C during HCMV infection of clinically relevant target cells. PLoS One 7:e50524. doi: 10.1371/journal.pone.0050524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldhoer M, Kledal TN, Farrell H, Schwartz TW. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J Virol 76:8161–8168. doi: 10.1128/JVI.76.16.8161-8168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodaghi B, Jones TR, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier JL, Michelson S. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J Exp Med 188:855–866. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casarosa P, Gruijthuijsen YK, Michel D, Beisser PS, Holl J, Fitzsimons CP, Verzijl D, Bruggeman CA, Mertens T, Leurs R, Vink C, Smit MJ. 2003. Constitutive signaling of the human cytomegalovirus-encoded receptor UL33 differs from that of its rat cytomegalovirus homolog R33 by promiscuous activation of G proteins of the Gq, Gi, and Gs classes. J Biol Chem 278:50010–50023. doi: 10.1074/jbc.M306530200. [DOI] [PubMed] [Google Scholar]
- 30.Gruijthuijsen YK, Casarosa P, Kaptein SJ, Broers JL, Leurs R, Bruggeman CA, Smit MJ, Vink C. 2002. The rat cytomegalovirus R33-encoded G protein-coupled receptor signals in a constitutive fashion. J Virol 76:1328–1338. doi: 10.1128/JVI.76.3.1328-1338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beisser PS, Grauls G, Bruggeman CA, Vink C. 1999. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J Virol 73:7218–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira SA, Shenk TE. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc Natl Acad Sci U S A 98:3237–3242. doi: 10.1073/pnas.051629898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardin RD, Schaefer GC, Allen JR, Davis-Poynter NJ, Farrell HE. 2009. The M33 chemokine receptor homolog of murine cytomegalovirus exhibits a differential tissue-specific role during in vivo replication and latency. J Virol 83:7590–7601. doi: 10.1128/JVI.00386-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farrell HE, Abraham AM, Cardin RD, Sparre-Ulrich AH, Rosenkilde MM, Spiess K, Jensen TH, Kledal TN, Davis-Poynter N. 2011. Partial functional complementation between human and mouse cytomegalovirus chemokine receptor homologues. J Virol 85:6091–6095. doi: 10.1128/JVI.02113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zipeto D, Bodaghi B, Laurent L, Virelizier JL, Michelson S. 1999. Kinetics of transcription of human cytomegalovirus chemokine receptor US28 in different cell types. J Gen Virol 80(Pt 3):543–547. [DOI] [PubMed] [Google Scholar]
- 36.Michel D, Milotic I, Wagner M, Vaida B, Holl J, Ansorge R, Mertens T. 2005. The human cytomegalovirus UL78 gene is highly conserved among clinical isolates, but is dispensable for replication in fibroblasts and a renal artery organ-culture system. J Gen Virol 86:297–306. doi: 10.1099/vir.0.80436-0. [DOI] [PubMed] [Google Scholar]
- 37.Fraile-Ramos A, Pelchen-Matthews A, Kledal TN, Browne H, Schwartz TW, Marsh M. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218–232. doi: 10.1034/j.1600-0854.2002.030307.x. [DOI] [PubMed] [Google Scholar]
- 38.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG II, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humby MS, O'Connor CM. 2015. Human cytomegalovirus US28 is important for latent infection of hematopoietic progenitor cells. J Virol 90:2959–2970. doi: 10.1128/JVI.02507-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor CM, Shenk T. 2012. Human cytomegalovirus pUL78 G protein-coupled receptor homologue is required for timely cell entry in epithelial cells but not fibroblasts. J Virol 86:11425–11433. doi: 10.1128/JVI.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A. 2013. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther 6:1347–1361. doi: 10.2147/OTT.S36109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teicher BA, Fricker SP.. 2010. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res 16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 43.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 44.Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, Kishimoto T, Nagasawa T. 1998. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 45.Moriuchi M, Moriuchi H, Fauci AS. 1999. HTLV type I Tax activation of the CXCR4 promoter by association with nuclear respiratory factor 1. AIDS Res Hum Retroviruses 15:821–827. doi: 10.1089/088922299310728. [DOI] [PubMed] [Google Scholar]
- 46.Moriuchi M, Moriuchi H, Turner W, Fauci AS. 1997. Cloning and analysis of the promoter region of CXCR4, a coreceptor for HIV-1 entry. J Immunol 159:4322–4329. [PubMed] [Google Scholar]
- 47.Moi P, Chan K, Asunis I, Cao A, Kan YW. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A 91:9926–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1–12. doi: 10.1016/S0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 49.Blank V. 2008. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol 376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 50.Chau CM, Evans MJ, Scarpulla RC. 1992. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2 alpha, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem 267:6999–7006. [PubMed] [Google Scholar]
- 51.Evans MJ, Scarpulla RC. 1990. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev 4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 52.Aizencang GI, Bishop DF, Forrest D, Astrin KH, Desnick RJ. 2000. Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. J Biol Chem 275:2295–2304. [DOI] [PubMed] [Google Scholar]
- 53.Braidotti G, Borthwick IA, May BK. 1993. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J Biol Chem 268:1109–1117. [PubMed] [Google Scholar]
- 54.Virbasius JV, Scarpulla RC. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A 91:1309–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gleyzer N, Vercauteren K, Scarpulla RC. 2005. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol 25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huo L, Scarpulla RC. 2001. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol 21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scarpulla RC. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 58.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira J, Schall TJ, Corey L, Geballe AP. 1998. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J Virol 72:8158–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margulies BJ, Browne H, Gibson W. 1996. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boeck JM, Spencer JV. 2017. Effect of human cytomegalovirus (HCMV) US27 on CXCR4 receptor internalization measured by fluorogen-activating protein (FAP) biosensors. PLoS One 12:e0172042. doi: 10.1371/journal.pone.0172042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank T, Reichel A, Larsen O, Stilp AC, Rosenkilde MM, Stamminger T, Ozawa T, Tschammer N. 2016. Attenuation of chemokine receptor function and surface expression as an immunomodulatory strategy employed by human cytomegalovirus is linked to vGPCR US28. Cell Commun Signal 14:31. doi: 10.1186/s12964-016-0154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wegner SA, Ehrenberg PK, Chang G, Dayhoff DE, Sleeker AL, Michael NL. 1998. Genomic organization and functional characterization of the chemokine receptor CXCR4, a major entry co-receptor for human immunodeficiency virus type 1. J Biol Chem 273:4754–4760. doi: 10.1074/jbc.273.8.4754. [DOI] [PubMed] [Google Scholar]
- 64.Lares AP, Tu CC, Spencer JV. 2013. The human cytomegalovirus US27 gene product enhances cell proliferation and alters cellular gene expression. Virus Res 176:312–320. doi: 10.1016/j.virusres.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu CC, Spencer JV. 2014. The DRY box and C-terminal domain of the human cytomegalovirus US27 gene product play a role in promoting cell growth and survival. PLoS One 9:e113427. doi: 10.1371/journal.pone.0113427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang WT, Huang AM. 2004. Alpha-Pal/NRF-1 regulates the promoter of the human integrin-associated protein/CD47 gene. J Biol Chem 279:14542–14550. doi: 10.1074/jbc.M309825200. [DOI] [PubMed] [Google Scholar]
- 67.Jacob WF, Silverman TA, Cohen RB, Safer B. 1989. Identification and characterization of a novel transcription factor participating in the expression of eukaryotic initiation factor 2 alpha. J Biol Chem 264:20372–20384. [PubMed] [Google Scholar]
- 68.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. 2009. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai RK, Rodriguez PL, Discher DE. 2010. Self inhibition of phagocytosis: the affinity of ‘marker of self’ CD47 for SIRPalpha dictates potency of inhibition but only at low expression levels. Blood Cells Mol Dis 45:67–74. doi: 10.1016/j.bcmd.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan G, Bivins-Smith ER, Smith MS, Smith PM, Yurochko AD. 2008. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J Immunol 181:698–711. doi: 10.4049/jimmunol.181.1.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vischer HF, Siderius M, Leurs R, Smit MJ. 2014. Herpesvirus-encoded GPCRs: neglected players in inflammatory and proliferative diseases? Nat Rev Drug Discov 13:123–139. doi: 10.1038/nrd4189. [DOI] [PubMed] [Google Scholar]
- 72.Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, Feinberg EN, Angelini A, Waghray D, Dror RO, Ploegh HL, Garcia KC. 2015. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. 2013. Molecular signatures of G-protein-coupled receptors. Nature 494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 74.Scarborough JA, Paul JR, Spencer JV. 2017. Evolution of the ability to modulate host chemokine networks via gene duplication in human cytomegalovirus (HCMV). Infect Genet Evol 51:46–53. doi: 10.1016/j.meegid.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Speir E, Shibutani T, Yu ZX, Ferrans V, Epstein SE. 1996. Role of reactive oxygen intermediates in cytomegalovirus gene expression and in the response of human smooth muscle cells to viral infection. Circ Res 79:1143–1152. doi: 10.1161/01.RES.79.6.1143. [DOI] [PubMed] [Google Scholar]
- 76.van de Berg PJ, Heutinck KM, Raabe R, Minnee RC, Young SL, van Donselaar-van der Pant KA, Bemelman FJ, van Lier RA, ten Berge IJ. 2010. Human cytomegalovirus induces systemic immune activation characterized by a type 1 cytokine signature. J Infect Dis 202:690–699. doi: 10.1086/655472. [DOI] [PubMed] [Google Scholar]
- 77.Lee J, Koh K, Kim YE, Ahn JH, Kim S. 2013. Upregulation of Nrf2 expression by human cytomegalovirus infection protects host cells from oxidative stress. J Gen Virol 94:1658–1668. doi: 10.1099/vir.0.052142-0. [DOI] [PubMed] [Google Scholar]
- 78.Schaedler S, Krause J, Himmelsbach K, Carvajal-Yepes M, Lieder F, Klingel K, Nassal M, Weiss TS, Werner S, Hildt E. 2010. Hepatitis B virus induces expression of antioxidant response element-regulated genes by activation of Nrf2. J Biol Chem 285:41074–41086. doi: 10.1074/jbc.M110.145862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tu CC, Arnolds KL, O'Connor CM, Spencer JV. 2018. Human cytomegalovirus UL111A and US27 gene products enhance the CXCL12/CXCR4 signaling axis via distinct mechanisms. J Virol 92:e01981-17. doi: 10.1128/JVI.01981-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tadagaki K, Tudor D, Gbahou F, Tschische P, Waldhoer M, Bomsel M, Jockers R, Kamal M. 2012. Human cytomegalovirus-encoded UL33 and UL78 heteromerize with host CCR5 and CXCR4 impairing their HIV coreceptor activity. Blood 119:4908–4918. doi: 10.1182/blood-2011-08-372516. [DOI] [PubMed] [Google Scholar]
- 81.Smith MS, Bentz GL, Alexander JS, Yurochko AD. 2004. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J Virol 78:4444–4453. doi: 10.1128/JVI.78.9.4444-4453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frascaroli G, Varani S, Blankenhorn N, Pretsch R, Bacher M, Leng L, Bucala R, Landini MP, Mertens T. 2009. Human cytomegalovirus paralyzes macrophage motility through down-regulation of chemokine receptors, reorganization of the cytoskeleton, and release of macrophage migration inhibitory factor. J Immunol 182:477–488. doi: 10.4049/jimmunol.182.1.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lecointe D, Dugas N, Leclerc P, Hery C, Delfraissy JF, Tardieu M. 2002. Human cytomegalovirus infection reduces surface CCR5 expression in human microglial cells, astrocytes and monocyte-derived macrophages. Microbes Infect 4:1401–1408. doi: 10.1016/S1286-4579(02)00022-9. [DOI] [PubMed] [Google Scholar]
- 84.Warner JA, Zwezdaryk KJ, Day B, Sullivan DE, Pridjian G, Morris CA. 2012. Human cytomegalovirus infection inhibits CXCL12-mediated migration and invasion of human extravillous cytotrophoblasts. Virol J 9:255. doi: 10.1186/1743-422X-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mego M, Cholujova D, Minarik G, Sedlackova T, Gronesova P, Karaba M, Benca J, Cingelova S, Cierna Z, Manasova D, Pindak D, Sufliarsky J, Cristofanilli M, Reuben JM, Mardiak J. 2016. CXCR4-SDF-1 interaction potentially mediates trafficking of circulating tumor cells in primary breast cancer. BMC Cancer 16:127. doi: 10.1186/s12885-016-2143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mukherjee D, Zhao J. 2013. The role of chemokine receptor CXCR4 in breast cancer metastasis. Am J Cancer Res 3:46–57. [PMC free article] [PubMed] [Google Scholar]
- 87.Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. 2009. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood 113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- 88.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675–684. doi: 10.1016/0092-8674(95)90039-X. [DOI] [PubMed] [Google Scholar]
- 89.Rössig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. 2001. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol 21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 91.O'Connor CM, Miller WE. 2014. Methods for studying the function of cytomegalovirus GPCRs. Methods Mol Biol 1119:133–164. doi: 10.1007/978-1-62703-788-4_10. [DOI] [PubMed] [Google Scholar]
- 92.Zhang RG, Scott DL, Westbrook ML, Nance S, Spangler BD, Shipley GG, Westbrook EM. 1995. The three-dimensional crystal structure of cholera toxin. J Mol Biol 251:563–573. doi: 10.1006/jmbi.1995.0456. [DOI] [PubMed] [Google Scholar]
- 93.Lehmann DM, Seneviratne AM, Smrcka AV. 2008. Small molecule disruption of G protein beta gamma subunit signaling inhibits neutrophil chemotaxis and inflammation. Mol Pharmacol 73:410–418. doi: 10.1124/mol.107.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang H, Fan D, Zhou G, Li X, Deng H. 2010. Phosphatidylinositol 3-kinase inhibitor (LY294002) induces apoptosis of human nasopharyngeal carcinoma in vitro and in vivo. J Exp Clin Cancer Res 29:34. doi: 10.1186/1756-9966-29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gharbi SI, Zvelebil MJ, Shuttleworth SJ, Hancox T, Saghir N, Timms JF, Waterfield MD. 2007. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem J 404:15–21. doi: 10.1042/BJ20061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Momota H, Nerio E, Holland EC. 2005. Perifosine inhibits multiple signaling pathways in glial progenitors and cooperates with temozolomide to arrest cell proliferation in gliomas in vivo. Cancer Res 65:7429–7435. doi: 10.1158/0008-5472.CAN-05-1042. [DOI] [PubMed] [Google Scholar]
- 97.Burns DL. 1988. Subunit structure and enzymic activity of pertussis toxin. Microbiol Sci 5:285–287. [PubMed] [Google Scholar]
- 98.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, Sebti SM, Turkson J. 2007. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A 104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beyaert R, Cuenda A, Vanden Berghe W, Plaisance S, Lee JC, Haegeman G, Cohen P, Fiers W. 1996. The p38/RK mitogen-activated protein kinase pathway regulates interleukin-6 synthesis response to tumor necrosis factor. EMBO J 15:1914–1923. doi: 10.1002/j.1460-2075.1996.tb00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem 273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 101.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. 1996. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol 16:1722–1733. doi: 10.1128/MCB.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Powis G, Bonjouklian R, Berggren MM, Gallegos A, Abraham R, Ashendel C, Zalkow L, Matter WF, Dodge J, Grindey G, Vlahos C. 1994. Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res 54:2419–2423. [PubMed] [Google Scholar]
- 103.Takasaki J, Saito T, Taniguchi M, Kawasaki T, Moritani Y, Hayashi K, Kobori M. 2004. A novel Galphaq/11-selective inhibitor. J Biol Chem 279:47438–47445. doi: 10.1074/jbc.M408846200. [DOI] [PubMed] [Google Scholar]