EBV encodes 44 mature microRNAs, which have been proven to promote EBV-associated diseases by targeting host genes and self-viral genes. In EBV-associated carcinomas, the expression of viral protein is limited but the expression of BART microRNAs is extremely high, suggesting that they could be major factors in the contribution of EBV-associated tumorigenesis. p53 is a critical tumor suppressor. Unlike in most human solid tumors, TP53 mutations are rare in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues, suggesting a possibility that some EBV-encoded products suppress the functions of p53. This study provides the first evidence that a BART microRNA can suppress p53 expression by directly targeting its 3′-UTR. This study implies that EBV can use its BART microRNAs to modulate the expression of p53, thus maintaining its latency and contributing to tumorigenesis.
KEYWORDS: Epstein-Barr virus, p53, miR-BART5-3p, gastric cancer, nasopharyngeal carcinoma
ABSTRACT
Epstein-Barr virus (EBV) is the first human virus found to encode many microRNAs. It is etiologically linked to nasopharyngeal carcinoma and EBV-associated gastric carcinoma. During the latent infection period, there are only a few EBV proteins expressed, whereas EBV microRNAs, such as the BamHI-A region rightward transcript (BART) microRNAs, are highly expressed. However, how these BART miRNAs precisely regulate the tumor growth in nasopharyngeal carcinoma and gastric carcinoma remains obscure. Here, we report that upregulation of EBV-miR-BART5-3p promotes the growth of nasopharyngeal carcinoma and gastric carcinoma cells. BART5-3p directly targets the tumor suppressor gene TP53 on its 3′-untranslated region (3′-UTR) and consequently downregulates CDKN1A, BAX, and FAS expression, leading to acceleration of the cell cycle progress and inhibition of cell apoptosis. BART5-3p contributes to the resistance to chemotherapeutic drugs and ionizing irradiation-induced p53 increase. Moreover, BART5-3p also facilitates degradation of p53 proteins. BART5-3p is the first EBV-microRNA to be identified as inhibiting p53 expression and function, which suggests a novel mechanism underlying the strategies employed by EBV to maintain latent infection and promote the development of EBV-associated carcinomas.
IMPORTANCE EBV encodes 44 mature microRNAs, which have been proven to promote EBV-associated diseases by targeting host genes and self-viral genes. In EBV-associated carcinomas, the expression of viral protein is limited but the expression of BART microRNAs is extremely high, suggesting that they could be major factors in the contribution of EBV-associated tumorigenesis. p53 is a critical tumor suppressor. Unlike in most human solid tumors, TP53 mutations are rare in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues, suggesting a possibility that some EBV-encoded products suppress the functions of p53. This study provides the first evidence that a BART microRNA can suppress p53 expression by directly targeting its 3′-UTR. This study implies that EBV can use its BART microRNAs to modulate the expression of p53, thus maintaining its latency and contributing to tumorigenesis.
INTRODUCTION
Epstein-Barr virus (EBV) is the first virus shown to be associated with human cancers (1). It belongs to the γ-subfamily of the herpesvirus and infects approximately more than 90% of the worldwide adult population. Persistent latent infection of EBV is etiologically related to a series of human lymphocytic malignancies, including Burkitt's lymphoma, Hodgkin's lymphoma, extranodal nasal natural killer/T cell lymphoma (2–6), and several epithelial carcinomas, such as nasopharyngeal carcinoma and gastric carcinoma (7–15). EBV can be found in most of the tumor cells of nasopharyngeal carcinoma patients, which is a rare type of head and neck cancer in most parts of the world, whereas it is common in Southern China and Southeast Asia. In 1990, Burke et al. (16) reported that EBV was associated with gastric cancer for the first time. Then, Tokunaga et al. (17) confirmed the presence of EBV-encoded small RNAs (EBER) in gastric cancer tissues by in situ hybridization assay and defined the EBER-positive gastric cancer as EBV-associated gastric carcinoma (EBVaGC). EBV is found within tumor cells in 9% of gastric cancer cases (18). However, the role of EBV in the development of nasopharyngeal carcinoma and EBVaGC is still a matter of debate.
The tumor suppressor p53 is a critical cellular protein in response to various stresses and dictates various responses in cells, including apoptosis, DNA repair, cell cycle arrest, and cell differentiation (19–22). Many human tumors harbor TP53 mutations or suffer p53 loss, supporting p53's pivotal role as a tumor suppressor. Unlike in most human solid tumors, TP53 mutations are rare in nasopharyngeal carcinoma tissues, even in recurrent radioresistant cases (23–25). Moreover, evidence from comprehensive analysis of gastric carcinoma also rarely shows TP53 mutation in EBVaGC, which is quite different from other subtypes of gastric carcinoma displaying more-common p53 mutations (18, 26). The evidence suggests a possibility that there may be some EBV-encoded products that suppress the functions of p53 as a tumor suppressor in certain types of EBV-associated malignancies, contributing to virus latency and tumorigenesis. Until now, several EBV-encoded products have been found to be involved in inactivating the p53-mediated pathway. For instance, Jha et al. found that EBV-encoded EBNA3C can (i) upregulate Aurora kinase-B transcription (and Aurora kinase-B can lead to p53 degradation) (27) and (ii) attenuate H2AX expression (and H2AX knockdown leads to downregulation of p53) (28). EBV EBNA1 decreases p53 activation and results in impaired responses to DNA damage and promotes cell survival (7).
MicroRNAs (miRNAs) are a class of small noncoding endogenous RNAs containing 18 to 23 nucleotides that repress targeted gene expression by forming imperfect complementary duplexes with their target mRNAs in the 3′-untranslated region (3′-UTR), leading to mRNA degradation and/or translational inhibition (29). Accumulating evidence has suggested that miRNAs play crucial roles in cancer initiation and progression. EBV is the first virus shown to encode miRNAs (30, 31). To date, a total of 25 EBV miRNA precursors with 44 mature EBV miRNAs have been identified within two regions of the EBV genome, the BamHI fragment H rightward open-reading frame (BHRF) region and the BamHI-A region rightward transcript (BART) region (30, 32–35). Four EBV miRNAs are encoded from the BHRF region, and the remainders are from the BART region. Interestingly, BART miRNAs expressions were shown to be at a relative low level in lymphocytic malignancies but extremely high in nasopharyngeal carcinoma and EBVaGC (36–43), suggesting a pathogenic role in the development of such epithelial malignancies.
Recent studies demonstrated that BART miRNAs can regulate both the EBV genes and host genes expression, playing important roles in viral latency, host cell survival, and tumorigenesis (4, 6, 9–13, 44–49). However, their functions in tumorigenesis and tumor progression of nasopharyngeal carcinoma and EBVaGC remain elusive. In this study, we chose EBV-miR-BART5-3p (BART5-3p), which is significantly increased in clinical specimens of nasopharyngeal carcinoma and EBVaGC (42, 43), to explore its pathophysiology function. We found that EBV-miR-BART5-3p significantly downregulates the protein level of p53. Alterations of the expression levels of BART5-3p demonstrated that it induces the tumorigenesis of NPC and GC cells both in vitro and in vivo. The underlying molecular mechanisms by which BART5-3p promotes tumor growth are revealed to directly target two sites of TP53 mRNA on the 3′-UTR. Our findings provide new insights into the carcinogenesis of nasopharyngeal carcinoma and EBVaGC regulated by EBV miRNAs.
RESULTS
BART5-3p suppresses the expression and transcriptional activity of p53 in gastric cancer and nasopharyngeal carcinoma cells.
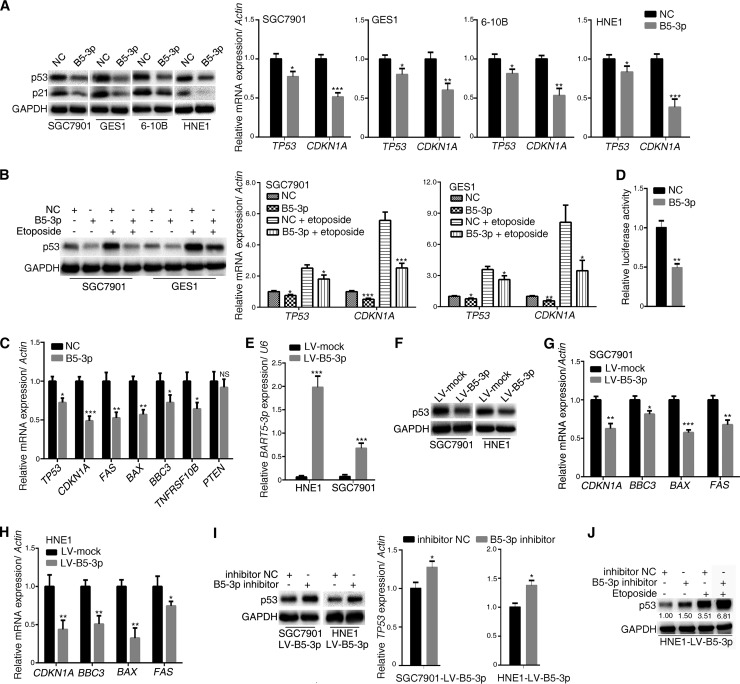
To investigate the effect of BART5-3p on p53, we transfected BART5-3p mimics into different epithelial cell lines, SGC7901 (a gastric cancer cell line), GES1 (a gastric epithelial cell line), and HNE1 and 6-10B (two nasopharyngeal carcinoma cell lines), and then analyzed the p53 expression changes of the cells. Figure 1A showed that BART5-3p mimics significantly reduced the endogenous p53 protein and mRNA levels and its target p21 gene (CDKN1A). As a chemotherapy medication, etoposide causes DNA strands to break and thus induces p53 expression. When p53 expression was induced by etoposide treatment in SGC7901 and GES1 cells, BART5-3p mimics significantly reduced the expression levels of p53 and p21 (Fig. 1B). BART5-3p mimics also significantly decreased the mRNA levels of TP53 and its target genes, including CDKN1A, FAS, BAX, BBC3 (also known as PUMA), and TNFRSF10B (Fig. 1C). Further study showed that BART5-3p reduced the p53 transcriptional activity (Fig. 1D). In order to further investigate the roles of BART5-3p on p53 modulation, we constructed two stable BART5-3p expression cell lines (SGC7901-LV-B5-3p, HNE1-LV-B5-3p) by lentivirus-mediated transduction (Fig. 1E). The protein levels of p53 and the mRNA levels of p53 target genes (CDKN1A, BBC3, BAX, and FAS) were significantly reduced in both SGC7901-LV-B5-3p and HNE1-LV-B5-3p cell lines compared to mock-transducted cells (Fig. 1F, G, and H). In contrast, downregulation of BART5-3p by means of BART5-3p inhibitor increased the protein and mRNA levels of p53 in these two cell lines (Fig. 1I and J). These data provide solid evidence that BART5-3p suppresses p53 transcription and expression in some gastric cancer and nasopharyngeal carcinoma cells.
FIG 1.
BART5-3p inhibits the expression and transcriptional activity of p53 in multiple cell lines. (A) SGC7901, GES1, 6-10B, and HNE1 cells were transfected with either negative-control mimics (NC) or BART5-3p mimics (B5-3p) for 36 h. The protein levels of p53 and p21 were determined by Western blotting, and the mRNA levels of TP53 and CDKN1A were analyzed by qPCR. (B) SGC7901 and GES1 cells were transfected with either NC mimics or BART5-3p mimics for 12 h initially and then treated (or not treated) with 10 μM etoposide for another 24 h; the protein levels of p53 were determined by Western blotting, and the mRNA levels of TP53 and CDKN1A were analyzed by qPCR. (C) SGC7901 cells were transfected with NC or BART5-3p mimics for 24 h. The mRNA expression levels of the indicated genes were analyzed by qPCR. (D) SGC7901 cells were cotransfected with mimics and firefly luciferase pp53-TA-luc reporter plasmid along with Renilla luciferase reporter plasmid for 24 h before the luciferase reporter assay was performed. The data are shown as the relative firefly luciferase activity normalized to the value of Renilla luciferase. (E to H) Two stable BART5-3p-expressing cell lines were constructed by lentivirus-mediated transduction in HNE1 and SGC7901 cells. The transcriptional levels of BART5-3p were assayed by qPCR (E), and the p53 protein levels were determined by Western blotting (F); mRNA expression levels of the indicated genes were analyzed by qPCR (G, H). (I) HNE1-LV-B5-3p and SGC7901-LV-B5-3p cells were transfected with NC inhibitors or BART5-3p inhibitors for 36 h. The p53 protein levels were determined by Western blotting, and the mRNA levels of TP53 were analyzed by qPCR. (J) HNE1-LV-B5-3p cells were transfected with either NC inhibitor or BART5-3p inhibitor for 12 h initially and then treated (or not treated) with 10 μM etoposide for another 24 h. The protein expression levels of p53 were determined by Western blotting. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as protein loading control. Actin was used for normalizing the expression of mRNA. RPU6B (U6) was used for normalizing the expression of BART5-3p. Both the mean mRNA expression and mean luciferase activity were calculated from 3 independent experiments, and data are shown as the means ± standard errors of the means (SEM). NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with the control group (NC or mock).
BART5-3p shows promotion of cell growth activity.
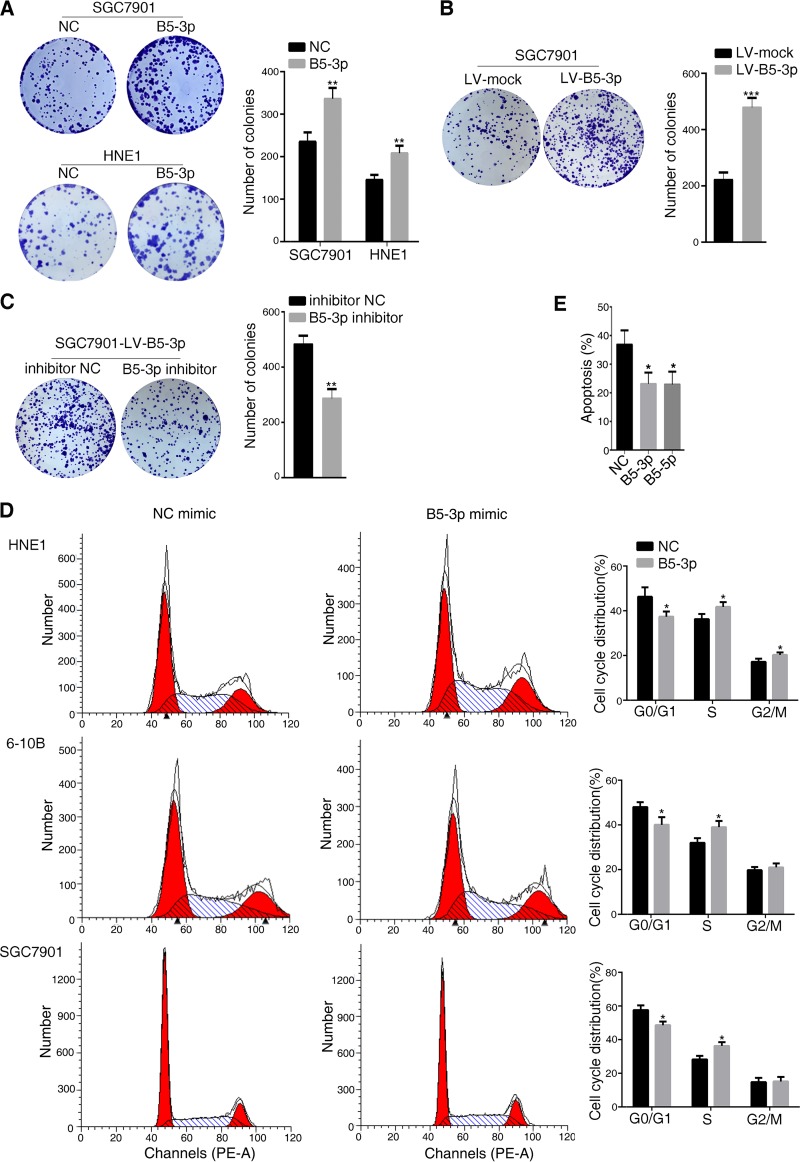
Since p53 is a master molecule that regulates cell cycle, apoptosis, senescence, metabolism, and DNA damage repair, here, we investigated the potential biological effects of BART5-3p on gastric cancer and nasopharyngeal carcinoma cells. Colony formation assays confirmed that upregulation of BART5-3p strongly promotes colony formation ability of SGC7901 and HNE1 cells, showing more colonies than the negative-control cells (Fig. 2A). The same phenomenon was observed in SGC7901-LV-B5-3p cells, a BART5-3p stable-expressing cell line, compared to the negative-control cells (Fig. 2B). In contrast, downregulation of BART5-3p by BART5-3p inhibitor reduced the colony formation ability of the SGC7901-LV-B5-3p cells (Fig. 2C). To examine whether BART5-3p-induced cell growth was associated with the regulation of cell cycle and apoptosis, the cell cycle distribution and cell apoptosis ratio in the presence of BART5-3p were analyzed by flow cytometry. Compared to the negative-control cells, BART5-3p mimic-transfected cells exhibited a shorter G0/G1 phase with a concomitantly longer S phase or G2/M phase in the cycle distribution (Fig. 2D). Etoposide-treated SGC7901 cells showed a high apoptosis percentage, which was reduced by BART5-3p mimics, as well as by BART5-5p, a known cell apoptosis-inhibitory EBV-encoded miRNA (Fig. 2E) (44). These data suggest that BART5-3p can promote cell growth partially by accelerating cell cycle progression and suppressing cell apoptosis.
FIG 2.
BART5-3p propels cell growth in vitro. Colony formation assays were performed as described in Materials and Methods, and representative images (A, B, and C, left panels) and number of colonies (A, B, and C, right panels) are shown. (A) SGC7901 and HEN1cells were transfected with either NC or BART5-3p mimics for 6 h, and then cells were reseeded. (B) Colony formation assays were performed in SGC7901-LV-mock and SGC7901-LV-B5-3p cells. (C) SGC7901-LV-B5-3p cells were transfected with either NC or BART5-3p inhibitors for 6 h, and then cells were reseeded. (D) HEN1, 6-10B, and SGC7901 cells were transfected with either NC or BART5-3p mimics for 36 h, and the cell cycle distribution was detected by a flow cytometer. (E) SGC7901 cells were transfected with the indicated mimics for 12 h and then treated with 20 μM etoposide for another 48 h. The number of apoptosis cells was determined by a flow cytometer. Three independent experiments were performed, and data are shown as the means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with the control group (NC or mock).
BART5-3p-expressing cells exhibit more resistance and a lower sensitivity to chemotherapeutic drug and ionizing irradiation-induced p53 increase.
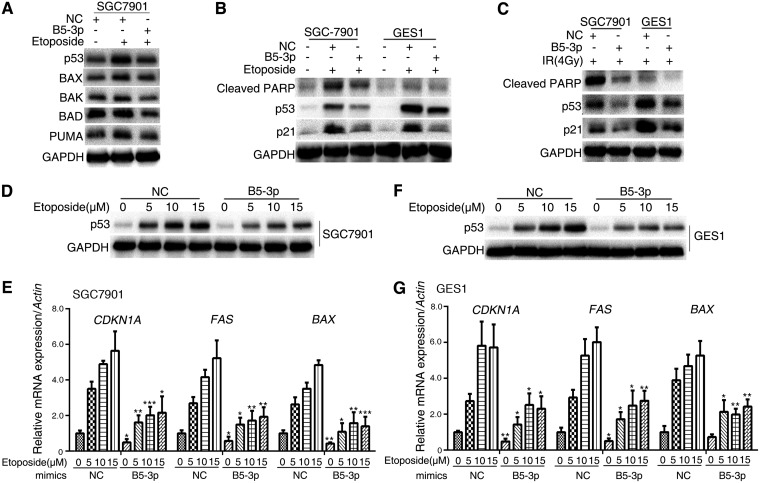
p53 transcription regulates multiple apoptosis-related genes such as BAX, BAK, BAD, and PUMA. Etoposide is a chemotherapy medication used for treatment of a number of types of cancer. It forms a ternary complex with DNA and the topoisomerase II enzyme, causes errors in DNA synthesis, and promotes apoptosis of the cancer cells. Treatment of SGC7901 cells with etoposide induced p53 expression as well as its downstream targets (BAX, BAK, BAD, and PUMA), but BART5-3p exhibited a strong ability to resist these increases (Fig. 3A). Cleaved poly(ADP-ribose) polymerase (PARP) is considered an indicator of apoptosis. It was observed that the cleaved PARP was increased upon etoposide treatment, whereas BART5-3p can reverse this effect (Fig. 3B). Radiotherapy significantly induces p53 expression and cell apoptosis. BART5-3p also can repress 4Gy ionizing irradiation-induced expression of cleaved PARP, p53, and p21 (Fig. 3C). Then, the effect of BART5-3p on cell sensitivity to etoposide was examined. SGC7901 and GES1 cells were each treated with 5, 10 and 15 μM etoposide, and the protein levels of p53 accordingly increased in a dose-dependent manner, whereas BART5-3p can reduce the upregulation of p53 induced by etoposide (Fig. 3D and F). At the same time, the mRNA levels of related genes (CDKN1A, FAS, and BAX) showed a dose-dependent increase in the control groups, whereas this effect was compromised in BART5-3p-overexpressing cells (Fig. 3E and G). These data suggest that BART5-3p-overexpressing cells show resistance to chemotherapeutic drugs and ionizing irradiation-induced p53 increase.
FIG 3.
BART5-3p protects cells from apoptosis. (A) SGC7901 cells were transfected with NC or BART5-3p mimics for 12 h and then treated with 10 μM etoposide for 24 h. The protein levels of p53, BAX, BAK, BAD, and PUMA were determined by Western blotting. (B) SGC7901 and GES1 cells were transfected with NC or BART5-3p mimics for 12 h and then treated with 20 μM etoposide for 48 h. The protein levels of p53, p21, and cleaved PARP were determined by Western blotting. (C) SGC7901 and GES1 cells were transfected with NC or BART5-3p mimics for 12 h and then treated with 4Gy IR. The cells were harvested 24 h after irradiation. The protein levels of p53, p21, and cleaved PARP were determined by Western blotting. (D to G) SGC7901 and GES1 cells were transfected with NC or BART5-3p mimics for 12 h and then treated with the indicated concentrations of etoposide for 24 h. The protein levels of p53 were determined by Western blotting. The mRNA levels of CDKN1A, FAS, and BAX were detected by qPCR. GAPDH was used as the protein loading control. Actin was used for normalizing the expression of mRNA. Three independent experiments were performed, and data are shown as the means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with the control group (NC).
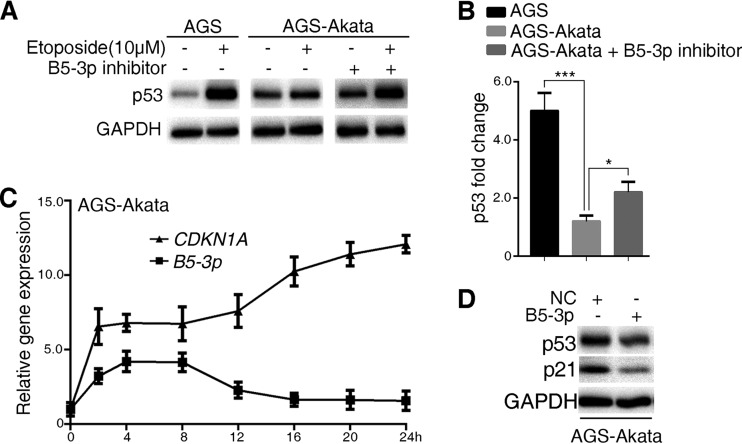
We further examined the sensitivity of AGS (an EBV-negative gastric cancer cell line) and AGS-Akata (an EBV-positive gastric cancer cell line) cells to etoposide. AGS-Akata cells showed a strong resistance to etoposide-induced p53 increase compared to AGS cells, whereas the BART5-3p inhibitor can reverse this resistance (Fig. 4A and B). Interestingly, it was noted that the levels of BART5-3p in AGS-Akata cells maintain a relatively high level within 8 h of etoposide treatment and then drastically decline to a little higher than the basic level. Meanwhile, the levels of CDKN1A was rapidly increased, showing a relatively high and stable level from 4 to 8 h, and then further rose up to 10 more times than the basic level at 24 h (Fig. 4C). It seems that the relatively high level of BART5-3p contributed to impeding the further rise of CDKN1A within 8 h of etoposide treatment, and when BART5-3p declined, CDKN1A correspondingly increased. To support this explanation, Western blotting was performed to reveal the inhibitory effect of BART5-3p on p53 and its target p21 in AGS-Akata cells (Fig. 4D). These results indicate that in EBV-positive gastric cancer cells, BART5-3p contributed to cell resistance to the chemotherapeutic drug-induced p53 increase.
FIG 4.
EBV confers gastric cancer cells resistance to apoptosis, and BART5-3p is involved. (A, B) Cells were treated (or not treated) with 10 μM etoposide for 24 h. AGS-Akata cells were transfected (or not) with BART5-3p inhibitor for 12 h before being treated with etoposide. The protein levels of p53 were detected by Western blotting (A); the relative p53 protein level was normalized to GAPDH, and the fold changes (after/before etoposide treatment) are shown. *, P < 0.05; ***, P < 0.001 (B). (C) AGS-AKATA cells were treated with 10 μM etoposide for the indicated times. The RNA levels of CDKN1A and BART5-3p were detected by qPCR. Actin was used for normalizing the expression of CDKN1A. RPU6B (U6) was used for normalizing the expression of BART5-3p. (D) AGS-AKATA cells were transfected with NC or BART5-3p mimics for 36 h. The protein levels of p53 and p21 were detected by Western blotting.
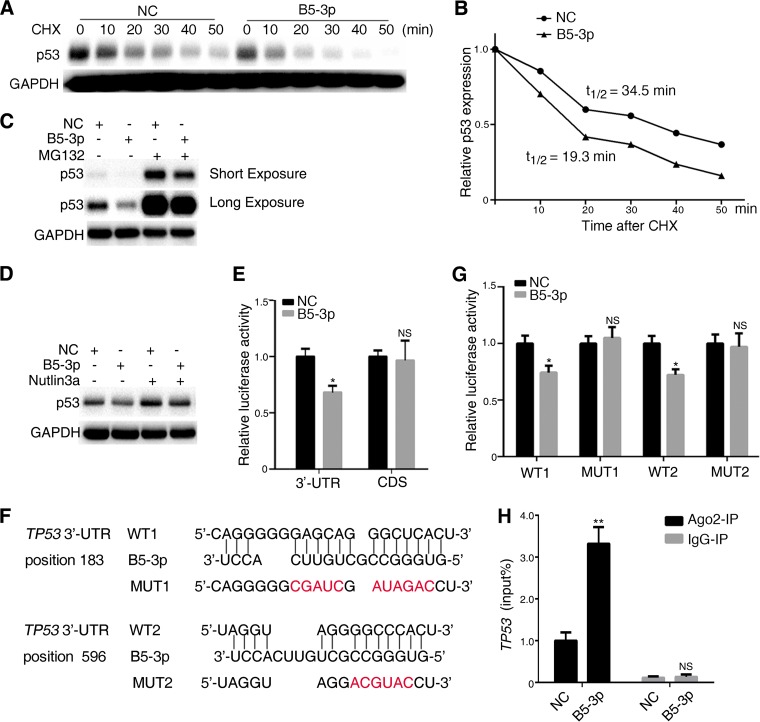
EBV-miR-BART5-3p accelerates p53 protein degradation and directly targets TP53.
Proteasome-mediated ubiquitination is an important posttranslational modification of p53, resulting in p53 degradation. We investigated whether BART5-3p modulates p53 expression is dependent on proteasome-mediated protein degradation. SGC7901 cells were transfected with a BART5-3p mimic or a negative-control mimic and then treated with cycloheximide (CHX), a potent inhibitor of protein translation. Western blot analysis was performed at indicated time periods after CHX treatment to measure p53 protein levels (Fig. 5A). Quantification of Western blot results showed that BART5-3p mimics did cause a decrease in the degradation time of the p53 protein, with the half-life shortened from 34.5 to 19.3 min (Fig. 5B). Furthermore, treatment of SGC7901 cells with the proteasome inhibitor MG132 did not influence the ability of BART5-3p to decrease p53 protein levels (Fig. 5C). MDM2 is the most important E3 ubiquitin ligase of p53, leading to p53 degradation by the proteasome. However, nutlin3a, an inhibitor of MDM2-p53 binding, did not influence the ability of BART5-3p to decrease p53 protein levels (Fig. 5D).
FIG 5.
BART5-3p directly targets TP53 and promotes the degradation of p53 protein. (A) SGC7901 cells were transfected with NC or BART5-3p mimics for 36 h, were subsequently treated with cycloheximide (20 μg/ml), and were collected after the indicated periods of time. The protein levels of p53 were determined by Western blotting. (B) p53 protein half-life was calculated by quantification of Western blots shown in panel A using Image Lab software; the half-life of p53 is indicated. (C) SGC7901 cells were transfected with NC or BART5-3p mimics for 36 h and were subsequently treated with MG132 (25 μM) for 4 h. The protein levels of p53 were determined by Western blotting. (D) GES1 cells were transfected with NC or BART5-3p mimics for 12 h and were subsequently treated with Nutlin3a (10 μM) for 24 h. The protein levels of p53 were determined by Western blotting. (E) HEK293 cells were cotransfected with NC (or BART5-3p) mimics and TP53 3′-UTR or TP53 CDS dual-luciferase reporter plasmid for 48 h before the luciferase reporter assay was performed. The data are shown as the relative firefly luciferase activity normalized to the value of Renilla luciferase. (F) Bioinformatics predictions of two binding sites by BART5-3p in the TP53 3′-UTR region. Wild-type (WT1 and WT2) and mutant (MUT1 and MUT2) sequences are indicated. (G) HEK293 cells were cotransfected with NC (or BART5-3p) mimics and TP53 3′-UTR WT (or MUT) dual-luciferase reporter plasmid for 48 h before the luciferase reporter assay was performed. The data are shown as the relative firefly luciferase activity normalized to the value of Renilla luciferase. (H) SGC7901 cells were transfected with NC or BART5-3p mimics for 36 h, and cell lysates were immunoprecipitated using either a normal rabbit IgG or anti-AGO2 antibody, and the mRNA level of TP53 binding to AGO2 was determined by qPCR. Immunoprecipitation (IP) with anti-IgG antibody was used as a negative control. Both mean luciferase activity and the mean TP53 expression were calculated from 3 independent experiments, and data are shown as the means ± SEM. NS, not significant; *, P < 0.05; **, P < 0.01 compared with the control group (NC).
To clarify whether TP53 is a direct cellular target gene for BART5-3p, luciferase reporter assays were performed by cotransfection of a BART5-3p mimic with a full-length of TP53 3′-UTR or coding DNA sequence (CDS)-containing luciferase reporter vector into HEK293 cells, respectively. The luciferase activity of the 3′-UTR but not CDS was significantly reduced by the BART5-3p mimic compared with the control mimic (NC) (Fig. 5E), suggesting that the 3′-UTR of TP53 mRNA may contain target sites directly targeted by BART5-3p. RNAhybrid, an online tool for microRNA target prediction, revealed that two possible BART5-3p binding sites existed in the 3′-UTR of TP53 mRNA, demonstrating perfect base pairing between the seed sequence of mature BART5-3p and the 3′-UTR of TP53 mRNA from nucleotide positions 183 and 596, respectively (Fig. 5F). Two mutant vectors (mut 1 or mut 2) were constructed to disrupt the binding of BART5-3p to the TP53 3′-UTR (Fig. 5F), and the result showed that the luciferase activity of the wild-type (WT) TP53 3′-UTR but not the mutant was significantly reduced by the BART5-3p mimic but not by the negative-control (NC) mimic (Fig. 5G). Argonaute 2 (Ago2) is a major component of the RNA-induced silencing complex and binds miRNAs along with their mRNA targets in ribonucleotide complexes. SGC7901 cells were transfected with BART5-3p mimics, and then immunoprecipitation assays were performed by using an anti-Ago2 antibody. BART5-3p mimics significantly increase the level of TP53 mRNA that binds to Ago2 compared to the level seen with NC mimics (Fig. 5H). Taken together, these results suggest that TP53 is a potential target gene of BART5-3p.
BART5-3p impels the growth of SGC7901 cells in vivo.
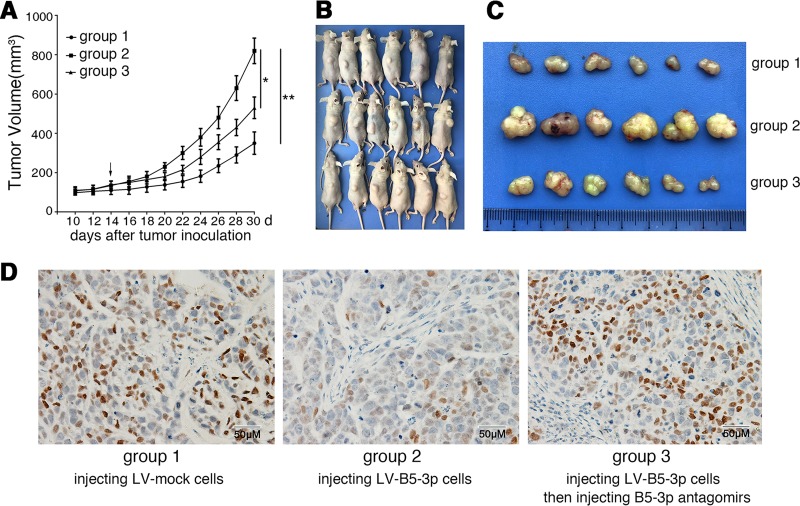
The in vitro study showed a solid effect of BART5-3p on p53 expression; we then examined the in vivo effect of this discovery by xenografting SGC7901-LV-BART5-3p cells or SGC7901-LV-mock cells into nude mice. As shown in Fig. 6A, B, and C, BART5-3p overexpression significantly increased the tumor growth of LV-BART5-3p cells compared with the LV-mock cells. Further, the tumor growth of LV-B5-3p cells was significantly inhibited in the presence of BART5-3p antagomir, a cholesterol-conjugated BART5-3p inhibitor. The results indicate that BART5-3p impels the growth SGC7901 cells in vivo. Immunohistochemical staining on tumor sections of xenografts demonstrated that BART5-3p significantly reduced the expression of p21 in LV-B5-3p tumors compared with LV-mock tumors. In contrast, downregulation of BART5-3p by BART5-3p antagomir increased the protein levels of p21 (Fig. 6D).
FIG 6.
BART5-3p promotes tumor growth in the nude mice xenograft model. (A) Tumor growth curves of SGC7901 xenografts in nude mice (n = 6 each). Group 1, 2, and 3 mice were injected with LV-mock, LV-BART5-3p, and LV-BART5-3p cells, respectively (see Materials and Methods for details). The arrow indicates the first administration of BART5-3p antagomir in group 3 mice. (B, C) Xenograft tumors collected on day 30 following subcutaneous implantation. (D) The representatives of immunohistochemistry staining for p21 in tumors from xenograft nude mice. Original magnification, ×200; bar, 50 μm.
BART5-3p is expressed in EBVaGC specimens.
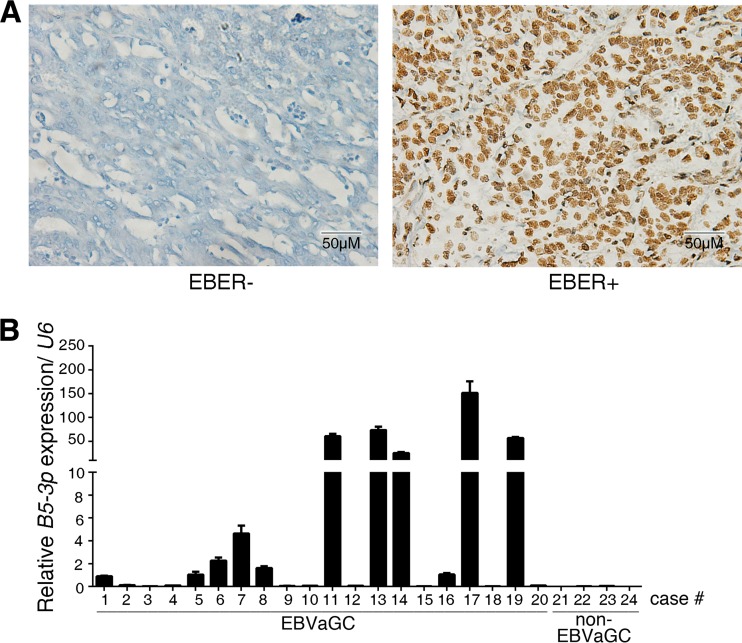
We collected tissue samples from 20 cases of EBVaGC and 4 cases of non-EBVaGC for analyzing the expression of BART5-3p within the tumor tissues. The clinicopathological parameters of gastric carcinoma specimens are listed in Table 1. EBVaGC was defined as positive EBER-staining by means of in situ hybridization assay (Fig. 7A). It was found that, taking the background level into account, about one-half of the EBVaGC specimens had different levels of expression of BART5-3p (Fig. 7B), whereas the non-EBVaGC tumors had no BART5-3p expression. This clinical finding suggests a potential role for BART5-3p in EVBaGC tumorigenesis and therapies.
TABLE 1.
Clinicopathological parameters of gastric carcinoma patients and specimens
| Variable | No. of cases |
|---|---|
| Age (yr) | |
| ≥55 | 13 |
| <55 | 11 |
| Gender | |
| Male | 21 |
| Female | 3 |
| Degree of differentiation | |
| Well differentiated | 1 |
| Moderately differentiated | 8 |
| Poorly differentiated | 15 |
| TNMa stages | |
| I and II | 7 |
| III and IV | 17 |
| EBER status | |
| Positive (EBER+) | 20 |
| Negative (EBER−) | 4 |
The TNM (tumor, nodes, metastasis) staging system describes the progression of the cancer.
FIG 7.
BART5-3p expression in EBVaGC clinical specimens. (A) Representative images of EBER staining by means of in situ hybridization. Shown are EBER-negative (left) and EBER-positive (right) gastric cancer specimens. (B) The transcriptional levels of BART5-3p in specimens from 20 cases of EBVaGC (numbered 1 to 20) and 4 cases of non-EBVaGC (numbered 21 to 24) were determined by stem-loop RT-qPCR. The expression values of case number 1 are set as 1 for BART5-3p. U6 was used for normalizing the expression of BART5-3p.
DISCUSSION
EBV infection is tightly linked to the development of various human lymphocytic and epithelial malignancies. Twenty-five EBV miRNA precursors and 44 mature EBV miRNAs have been identified, which could be divided into two main clusters, the BHRF1 and BART clusters. EBV miRNAs display a cell type-specific expression pattern. BHRF1 cluster miRNAs are highly expressed in EBV latency Ⅲ and lytic replication cells, such as B lymphoma cells, but are almost undetectable in latency I and II cells, such as EBVaGC and nasopharyngeal carcinoma cells. In contrast, BART miRNAs account for a large proportion of the total EBV miRNAs in EBVaGC and nasopharyngeal carcinoma cells. Considering the highly restricted patterns of viral protein expression in most EBV-associated tumors, the abundance of BART miRNAs in carcinomas suggests that they could be major factors in the contribution of EBV to tumorigenesis.
Several studies have demonstrated that BART miRNAs played crucial roles in tumor progression of EBV-associated carcinomas. Choy et al. (44) reported that BART5-5p conferred resistance to apoptosis by directly targeting the proapoptotic gene PUMA. Cai et al. (11, 12) demonstrated that BART1-3p, BART1-5p, and BART7-3p directly targeted the cellular tumor suppressor PTEN and consequently promoted NPC cells metastasis. Shinozaki-Ushiku et al. (41) found that BART4-5p reduced Bid expression in gastric cancer cell lines, leading to reduced apoptosis. BART22 was proved to target MAP3K5 and promote nasopharyngeal carcinoma cells' proliferative and invasive abilities (50). Lung et al. (13) revealed that BART5-5p, BART7-3p, BART9-3p, and BART14-3p could negatively regulate the expression of ATM by binding to multiple sites on its 3′-UTR in response to DNA damage, contributing to the tumorigenesis of NPC cells. BART20-5p was found to reduce apoptosis, enhance cell growth, and contribute to tumorigenesis of EBVaGC by directly targeting the BAD (47). Choi et al. (45) demonstrated that BART15-3p promotes cell apoptosis partially by targeting BRUCE in AGS-EBV cells. In the current study, we showed that an EBV miRNA, BART5-3p, suppresses the protein levels of p53 in several gastric cancer and nasopharyngeal carcinoma cell lines. Overexpression and knockdown experiments were performed to characterize the function of BART5-3p, highlighting the importance of BART5-3p in the control of p53 expression and p53-related phenotypes.
The modulation of p53 by BART5-3p was not surprising. The p53 protein is a sequence-specific DNA damage-inducible transcription factor that controls cell growth by regulating cell cycle and apoptosis, mainly by upregulating the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 and Bax, respectively (51). p53 is also involved in cellular responses to viral infection. The replicated genomes of DNA viruses, including adenoviruses, polyomavirus, and herpesviruses, are recognized by cellular DNA damage sensors, triggering activation of DNA damage responses (52–56). The TP53 mutation has been found to be the most frequent genetic alteration in human malignancy. However, unlike other types of cancer, TP53 accumulation with a rare mutation has been identified in nasopharyngeal carcinoma and EBVaGC (18, 23–26). Thus, EBV should develop countermeasures against p53 activation to maintain latent infection and promote the development of EBV-associated carcinomas. Limited evidence has been provided in this context. The EBV latent protein EBNA1 contributes to repression of p53-dependent DNA damage signaling by competition for the USP7 binding site with p53 (7, 57). EBNA3C can directly bind to p53 at the DNA binding and tetramerization domain, forming a complex to repress the p53 transactivation activity (58). EBNA3C also directly interacts with ING4, ING5, and Gemin3, negatively regulating p53-mediated functions (59, 60). Moreover, EBNA3C could enhance the intrinsic ubiquitin ligase activity of Mdm2 toward p53, which in turn facilitated p53 ubiquitination and degradation (61). Several EBV-encoded miRNAs have been proved to be involved in negatively regulating p53 activity. Huang and Lin (4) found that BART20-5p directly inhibited gamma interferon (IFN-γ) and BART8 directly inhibited STAT1, which resulted in secondary suppression of TP53. However, our study reports that TP53 was directly targeted by an EBV-encoded miRNA for the first time. As shown in this study, BART5-3p can inhibit the mRNA and protein levels of p53, downregulate p53 transcriptional activity, and promote cell growth of nasopharyngeal carcinoma and gastric cancer cells (Fig. 1 and 2). We noticed that the inhibition effect is more profound in protein levels than mRNA levels (Fig. 1A and B). We further showed that BART5-3p promoted p53 protein degradation, decreasing p53 protein's half-life from 34.5 min to 19.3 min (Fig. 5A and B). p53 protein degradation is regulated mainly by the ubiquitin proteasome pathway, and Mdm2 was identified as the principal endogenous E3 ligase with high specificity for p53 (62–64). Surprisingly, even under the influence of MG132 (a proteasome inhibitor) and nutlin3a (a selective Mdm2 antagonist), BART5-3p still can inhibit the protein levels of p53. So, the specific mechanism of BART5-3p facilitating p53 protein degradation remains to be revealed. We further identified that BART5-3p directly targets the 3′-UTR of the TP53, leading to reduced p53 expression.
p53 is activated upon exposure to genotoxic stress, which then upregulates the expression of p21, resulting in a halt in cell cycle progression to allow repair of damaged DNA. However, if the damage is too severe, p53 then induces apoptosis. BART5-3p significantly reduced the number of cells in G0/G1 phase and increased the number of cells in S phase. To test the effect of BART5-3p on p53 activation by genotoxic stress, cells were treated with etoposide and ionizing irradiation, respectively. Etoposide significantly increased the protein levels of p53 and promoted its transcriptional targets' expression, such as CDKN1A, BAX, and FAS, which were the crucial genes involved in cell cycle arrest and apoptosis. Importantly, BART5-3p significantly repressed the expression of these genes induced by genotoxic stress and reduced the cell apoptosis percentage. Therefore, we concluded that BART5-3p targets TP53 and consequently downregulates the expression of CDKN1A, BAX, and FAS, contributing to cell growth and tumorigenesis. Notably, several studies indicated that the expression level of BART5-3p is relatively low in multiple EBV-positive cancer cell lines but is normal (compared to other BART microRNAs) in clinical specimens of nasopharyngeal carcinoma and gastric carcinoma (41–43, 65, 66). Our study found that the expression of BART5-3p can be induced by etoposide (Fig. 4C) and the expression levels of BART5-3p are reasonable and common in EBVaGC specimens (Fig. 7B), indicating that the expression of BART5-3p is regulated by a complicated and tight regulation network, though the mechanism remains obscure. To date, there are few studies demonstrating the mechanism that regulates BART expression. Notably, the NF-κB signaling was proved to regulate the expression of BARTs (67). The dysregulation of BART5-3p suggests the possible biological meaning of BART5-3p in EBVaGC and nasopharyngeal carcinoma tumorigenesis.
The evidence provided in this study suggests a hypothesis that EBV carefully chooses multiple countermeasures against p53 activation to maintain latent infection and promote the development of EBV-associated carcinomas. Our work has derived one new mechanism for EBV modulation of cell growth. Based on this mechanism, new strategies can be devised to restrict the initiation and progression of nasopharyngeal carcinoma and EBVaGC. BART5-3p is a potential therapeutic target in nasopharyngeal carcinoma and EBVaGC. Considering the inhibitory effect of BART5-5p on PUMA and apoptosis, RNA interference with pre-miR-BART5 in nasopharyngeal carcinoma and EBVaGC cells to control cellular growth may be a feasible strategy in EBV-associated carcinomas.
MATERIALS AND METHODS
Cell cultures and reagents.
SGC7901 (EBV-negative gastric cancer cell line), GES1 (nonmalignant EBV-negative gastric epithelial cell line), and HNE1 and 6-10B (EBV-negative nasopharyngeal carcinoma cell lines) cells were maintained in RPMI 1640 (HyClone) supplemented with 10% fetal bovine serum (HyClone); AGS (EBV-negative gastric cancer cell line) cells were maintained in Ham's F-12 medium (HyClone) supplemented with 10% fetal bovine serum; AGS-Akata (Strain Akata, EBV-positive gastric cancer cell line) cells also received 400 μg/ml of G418 to maintain the EBV genome. HEK293 cells were maintained in Dulbecco's modified Eagle medium (DMEM; HyClone) supplemented with 10% fetal bovine serum. Etoposide and MG132 were obtained from Sigma-Aldrich. miRNA mimics and inhibitors were purchased from GenePharma Inc. (Shanghai, China). Sequences were as follows: for BART5-3p mimic, 5′-GUGGGCCGCUGUUCACCU-3′(sense) and 5′-GUGAACAGCGGCCCACUU-3′ (antisense); for BART5-5p mimic, 5′-CAAGGUGAAUAUAGCUGCCCAUCG-3′(sense) and 5′-AUGGGCAGCUAUAUUCACCUUGUU-3′(antisense); for negative-control (NC) mimic, 5′-UUCUCCGAACGUGUCACGUTT-3′(sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense); for BART5-3p inhibitor, 5′-AGGUGAACAGCGGCCCAC-3′; and for inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′. BART5-3p antagomir was purchased from Ribobio (Guangzhou, China). The cells were transfected with RNAs and/or plasmids using Lipofectamine 3000 (Invitrogen).
Patient samples.
Formalin-fixed paraffin-embedded (FFPE) surgical gastric cancer specimens (see Table 1 for patients' information) were obtained from the Xiangya Hospital (Changsha, China). Written informed consent was obtained from all study participants. Collections and using of tissue samples were approved by the ethical review committees of the appropriate institutions. In the present study, EBVaGC was defined as positive EBER staining by means of in situ hybridization assay, as described later in the text.
Lentivirus transduction.
The lentivirus packaging system was made by GenePharma Inc. (Shanghai, China). A lentiviral vector plasmid, LV3 (H1/GFP&Puro), was used in this study to construct the stable cell lines. The randomized flanking sequence control (mock) and EBV-miR-BART5-3p were purchased from GenePharma and transduced into cells according to the manufacturer's instructions.
Western blotting.
Protein extracts were resolved by 8% to 15% SDS-polyacrylamide gel, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with antibodies against p53 (DO-1) (1:500; Santa Cruz), p21 (1:500; Cell Signaling Technology), BAX (1:1,000; Cell Signaling Technology), BAK (1:1,000; Cell Signaling Technology), BAD (1:500; Cell Signaling Technology), cleaved-PARP (1:1,000; Cell Signaling Technology), GAPDH (1:5,000; Sangon Biotech, China). Horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz) was used as the secondary antibody. The antigen-antibody reaction was visualized by enhanced chemiluminescence assay. Western blot quantification was performed with Image lab software, analyzing the intensity of the grayscale images.
Flow cytometry.
For cell apoptosis detection, cells were plated into 6-well plates, transfected with miRNA mimics for 12 h, and after an additional 48 h treated with 20 μM etoposide. The number of apoptotic cells was determined using a fluorescein isothiocyanate (FITC)-annexin V apoptosis detection kit (BD Pharmingen) and a flow cytometer as per the manufacturer's instructions. For cell cycle detection, cells were plated into 6-well plates and transfected with miRNA mimics for 36 h. Cells were harvested and fixed in 70% ethanol overnight at 4°C. Then, cells were counterstained in the dark with 50 μg/ml phosphatidylinositol and 0.1% RNase A in 400 μl of phosphate-buffered saline (PBS) at 25°C for 30 min. Stained cells were assayed and quantified using a flow cytometer.
Plasmids and dual-luciferase reporter assays.
The full lengths of TP53 3′-UTR or CDS were amplified and subcloned into the pmirGLO dual-luciferase miRNA target expression vector. Two wild-type and mutant sites in TP53 3′-UTR were produced by GeneCreate Biotech (Wuhan, China). The pp53-TA-luc luciferase reporter plasmid was purchased from Beyotime (China) to monitor the transcriptional activity of p53. For miRNA target gene verification, HEK293 cells were seeded in 48-well plates and cotransfected with 200 ng of the luciferase reporter plasmid (TP53 3′-UTR, CDS, wild-type or mutant plasmid) along with 20 pmol of BART5-3p or negative-control (NC) mimics. For p53 transcriptional activity detection, SGC7901 cells were cotransfected with 200 ng of pp53-TA-luc luciferase reporter plasmid and 40 ng of Renilla plasmid along with 20 pmol of BART5-3p or NC mimics. Firefly and Renilla luciferase activities were measured via a Dual-GLO luciferase assay system (Promega). The data were expressed as the relative firefly luciferase activity normalized to the value of Renilla luciferase.
RT and real-time PCR.
Total RNAs were extracted using TRIzol reagent (Invitrogen). For mRNA reverse transcription (RT), 2 μg of RNA was used to synthesize cDNA using a RevertAid first-strand cDNA synthesis kit (Thermo) according to the manufacturer's protocol. For miRNA reverse transcription, 1 μg of RNA was used to synthesize cDNA using a Mir-X miRNA First-Strand synthesis kit (TaKaRa) according to the manufacturer's protocol. The levels of gene transcripts were determined by quantitative real-time PCR (qPCR) using specific primers and a SYBR premix Ex Taq II kit (TaKaRa). The expression levels of miRNA and mRNA were quantified by measuring cycle threshold (CT) values and normalized to U6 and Actin, respectively. The data were further normalized to the negative control, unless otherwise indicated. The primers used for qPCR are as follows. TP53, forward, 5′-CAGCACATGACGGAGGTTGT-3′, and reverse, 5′-TCATCCAAATACTCCACACGC-3′; BAX, forward, 5′-CCCGAGAGGTCTTTTTCCGAG-3′, and reverse, 5′-CCAGCCCATGATGGTTCTGAT-3′; BBC3, forward, 5′-GACCTCAACGCACAGTACGAG-3′, and reverse, 5′-AGGAGTCCCATGATGAGATTGT-3′; FAS, forward, 5′-TCTGGTTCTTACGTCTGTTGC-3′, and reverse, 5′-CTGTGCAGTCCCTAGCTTTCC-3′; CDKN1A, forward, 5′-TGTCCGTCAGAACCCATGC-3′, and reverse, 5′-AAAGTCGAAGTTCCATCGCTC-3′; TNFRSF10B, forward, 5′-GCCCCACAACAAAAGAGGTC-3′, and reverse, 5′-AGGTCATTCCAGTGAGTGCTA-3′; Actin, forward, 5′-GAGCTACGAGCTGCCTGACG-3′, and reverse, 5′-GTAGTTTCGTGGATGCCACAG; EBV-miR-BART5-3p, forward, 5′-GTGGGCCGCTGTTCACCT, and reverse, Universal Reverse Primer (TaKaRa); U6 primers were provided in the Mir-X miRNA First-Strand synthesis kit (TaKaRa).
TaqMan microRNA assay.
Total RNAs from formalin-fixed paraffin-embedded (FFPE) surgical specimens were extracted using the RecoverAll total nucleic acid isolation kit for FFPE (Invitrogen) according to the manufacturer's protocol. The miRNA (mature BART5-3p or U6)-specific RT primers and miRNA-specific TaqMan probes were purchased from Thermo, and the miRNAs' expression was detected by TaqMan microRNA assays according to the manufacturer's protocol. The results were normalized to U6.
RNA immunoprecipitation assays.
SGC7901 cells were harvested and washed twice in ice-cold PBS. The cell pellet was resuspended in gentle lysis buffer and incubated on ice for 20 min. After a further 15 min of centrifugation at 12,000 rpm at 4°C, the supernatants were incubated with anti-Argonaute-2 (AGO2) antibody (Abcam) or control anti-IgG antibody conjugated to protein A agarose beads (Santa Cruz) and rotated at 4°C for 4 h. After incubation, the supernatant was removed by brief centrifugation, and the beads were washed extensively by wash buffer, followed by adding 0.5 ml of TRIzol. Then, RNAs were extracted and analyzed by reverse-transcription quantitative PCR (RT-qPCR).
Colony formation assay.
Cells with different treatments were planted into 6-well cell culture plates (1,000 per well) and incubated for 12 days. Plates were washed with PBS and stained with crystal violet. Colonies with more than 50 cells were counted.
Immunohistochemistry staining.
Paraffin sections prepared from experiments with nude mice were applied to immunohistochemistry staining for the determination of protein levels of p21. In brief, tumor tissues embedded in paraffin were dewaxed in xylene and hydrated in ethanol. By placing the slides in boiling sodium citrate buffer (10 mmol/liter, pH 6.0) for 5 min, the antigen was retrieved. The endogenous peroxidase activity in tumor tissues was blocked with 3% H2O2. After incubation with goat serum to block the unspecific binding sites, the slides were incubated with anti-p21 antibodies at 4°C overnight. Biotinylated goat anti-rabbit IgG antibodies were added at a 1:100 dilution and incubated. Finally, the slides were incubated with HRP-conjugated streptavidin and then developed with 3,3′-diaminobenzidine (DAB) solution. After counterstaining with hematoxylin, the sections were dehydrated and mounted.
Tumor xenografts in nude mice.
Nude mice (4- to 5-week-old male nude mice) were used for examining tumorigenicity. To evaluate the role of BART5-3p in tumor formation in vivo, a group of 6 mice (group 1) and a group of 12 mice were subcutaneously injected with SGC7901-LV mock control cells and LV-B5-3p cells, respectively, into the left or right side on the back (1.5 × 106 cells per mouse). The tumor volumes were measured regularly and calculated using the formula (A × B2)/2, where A is the largest diameter and B is the perpendicular diameter. Two weeks after inoculation, the mice injected with SGC7901-LV-B5-3p cells were randomly divided into two groups (groups 2 and 3, each with six mice): group 3 mice were locally injected with BART5-3p antagomir (10 nmol/mouse) into the tumor mass twice a week for 2 weeks; group 2 mice were injected with the equal volumes of saline buffer. All mice were killed on day 30 after inoculation. Tumor tissues were fixed in 4% paraformaldehyde for 24 h and transferred to gradient ethanol. Tumors were embedded in paraffin, sectioned with LeiCa RM2245, and processed for histological examinations. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). The protocol was approved by the Animal Ethics Committee of Central South University. All surgeries were performed under sodium pentobarbital anesthesia, and all efforts were made to minimize animal suffering.
In situ hybridization.
EBER in situ hybridization was performed on the FFPE tissue sample slides with the EBER1 probe in situ hybridization kit (Zsbio, China). Only those samples with a universal and unequivocal nuclear staining within almost all tumor cells were interpreted as showing EBER positivity.
Statistical analysis.
Statistical analysis was determined by independent t test or analysis of variance (ANOVA) using SPSS17.0 and GraphPad Prism. Significance parameters were set at P values of <0.05.
ACKNOWLEDGMENTS
We thank Ya Cao and Lun-Quan Sun for providing reagents.
This work was supported by National Natural Science Foundation of China (81472694, 81672889, 81672993), China 111 Project (111-2-12), and Hunan Province Science and Technology Project (2016JC2035).
We declare that we have no conflict of interest.
Author contributions: Xiang Zheng, Jia Wang, and Jian Ma designed and conceived the experiments. Guiyuan Li supervised research. Xiang Zheng, Jia Wang, Lingyu Wei, Qiu Peng, Yingxue Gao, Yuxin Fu, Yuanjun Lu, Zailong Qin, Xuemei Zhang, Chunlin Ou, Zhengshuo Li, Xiaoyue Zhang, and Peishan Liu performed the experiments and analyzed the data. Jianhong Lu, Wei Xiong, Guiyuan Li, and Qun Yan analyzed data. Xiang Zheng and Jian Ma wrote the paper.
REFERENCES
- 1.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 2.Gruhne B, Kamranvar SA, Masucci MG, Sompallae R. 2009. EBV and genomic instability—a new look at the role of the virus in the pathogenesis of Burkitt's lymphoma. Semin Cancer Biol 19:394–400. doi: 10.1016/j.semcancer.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Portis T, Dyck P, Longnecker R. 2003. Epstein-Barr virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood 102:4166–4178. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- 4.Huang WT, Lin CW. 2014. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol 184:1185–1197. doi: 10.1016/j.ajpath.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Tsai SC, Lin SJ, Chen PW, Luo WY, Yeh TH, Wang HW, Chen CJ, Tsai CH. 2009. EBV Zta protein induces the expression of interleukin-13, promoting the proliferation of EBV-infected B cells and lymphoblastoid cell lines. Blood 114:109–118. doi: 10.1182/blood-2008-12-193375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin TC, Liu TY, Hsu SM, Lin CW. 2013. Epstein-Barr virus-encoded miR-BART20-5p inhibits T-bet translation with secondary suppression of p53 in invasive nasal NK/T-cell lymphoma. Am J Pathol 182:1865–1875. doi: 10.1016/j.ajpath.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Sivachandran N, Dawson CW, Young LS, Liu FF, Middeldorp J, Frappier L. 2012. Contributions of the Epstein-Barr virus EBNA1 protein to gastric carcinoma. J Virol 86:60–68. doi: 10.1128/JVI.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwakiri D, Eizuru Y, Tokunaga M, Takada K. 2003. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res 63:7062–7067. [PubMed] [Google Scholar]
- 9.He B, Li W, Wu Y, Wei F, Gong Z, Bo H, Wang Y, Li X, Xiang B, Guo C, Liao Q, Chen P, Zu X, Zhou M, Ma J, Li X, Li Y, Li G, Xiong W, Zeng Z. 2016. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis 7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B, Song Y, Li Q, Zeng Y, Liao Q, Chen P, Shi L, Fan S, Xiang B, Ma J, Zhou M, Li X, Yang J, Xiong W, Li G. 2015. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget 6:41766–41782. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, Wang S, Liu T, Cai H, Yao K, Li JL, Li X. 2015. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun 6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF, Yuan CC, Peng QX, Wu DH, Liu TF, Wang E, Marincola FM, Yao KT, Fang WY, Cai HB, Li X. 2015. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 34:2156–2166. doi: 10.1038/onc.2014.341. [DOI] [PubMed] [Google Scholar]
- 13.Lung RW, Hau PM, Yu KH, Yip KY, Tong JH, Chak WP, Chan AW, Lam KH, Lo AK, Tin EK, Chau SL, Pang JC, Kwan JS, Busson P, Young LS, Yap LF, Tsao SW, To KF, Lo KW. 2018. EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma. J Pathol 244:394–407. doi: 10.1002/path.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J, Tang M, Li H, Xu Z, Weng X, Li J, Yu X, Zhao L, Liu H, Hu Y, Tan Z, Yang L, Zhong M, Zhou J, Fan J, Bode AM, Yi W, Gao J, Sun L, Cao Y. 2016. EBV-LMP1 suppresses the DNA damage response through DNA-PK/AMPK signaling to promote radioresistance in nasopharyngeal carcinoma. Cancer Lett 380:191–200. doi: 10.1016/j.canlet.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 15.Endo K, Kondo S, Shackleford J, Horikawa T, Kitagawa N, Yoshizaki T, Furukawa M, Zen Y, Pagano JS. 2009. Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene 28:1725–1735. doi: 10.1038/onc.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke AP, Yen TS, Shekitka KM, Sobin LH. 1990. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol 3:377–380. [PubMed] [Google Scholar]
- 17.Tokunaga M, Land CE, Uemura Y, Tokudome T, Tanaka S, Sato E. 1993. Epstein-Barr virus in gastric carcinoma. Am J Pathol 143:1250–1254. [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherr CJ. 2004. Principles of tumor suppression. Cell 116:235–246. doi: 10.1016/S0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 20.Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M. 2017. Census and evaluation of p53 target genes. Oncogene 36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris SL, Levine AJ. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 23.Hoe SL, Lee ES, Khoo AS, Peh SC. 2009. p53 and nasopharyngeal carcinoma: a Malaysian study. Pathology 41:561–565. doi: 10.1080/00313020903071504. [DOI] [PubMed] [Google Scholar]
- 24.Effert P, McCoy R, Abdel-Hamid M, Flynn K, Zhang Q, Busson P, Tursz T, Liu E, Raab-Traub N. 1992. Alterations of the p53 gene in nasopharyngeal carcinoma. J Virol 66:3768–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang KP, Hao SP, Lin SY, Tsao KC, Kuo TT, Tsai MH, Tseng CK, Tsang NM. 2002. A lack of association between p53 mutations and recurrent nasopharyngeal carcinomas refractory to radiotherapy. Laryngoscope 112:2015–2019. doi: 10.1097/00005537-200211000-00019. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro J, Malta M, Galaghar A, Silva F, Afonso LP, Medeiros R, Sousa H. 2017. P53 deregulation in Epstein-Barr virus-associated gastric cancer. Cancer Lett 404:37–43. doi: 10.1016/j.canlet.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Jha HC, Yang K, El-Naccache DW, Sun Z, Robertson ES. 2015. EBNA3C regulates p53 through induction of Aurora kinase B. Oncotarget 6:5788–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jha HC, Aj MP, Saha A, Banerjee S, Lu J, Robertson ES. 2014. Epstein-Barr virus essential antigen EBNA3C attenuates H2AX expression. J Virol 88:3776–3788. doi: 10.1128/JVI.03568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 31.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 32.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol 83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundhoff A, Sullivan CS, Ganem D. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. 2010. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 5:e12745. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, Tinguely M, Faggioni A, Trivedi P, Meister G, Renner C, Grasser FA. 2011. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res 39:1880–1893. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. 2009. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol 83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alles J, Menegatti J, Motsch N, Hart M, Eichner N, Reinhardt R, Meister G, Grasser FA. 2016. miRNA expression profiling of Epstein-Barr virus-associated NKTL cell lines by Illumina deep sequencing. FEBS Open Bio 6:251–263. doi: 10.1002/2211-5463.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, Gel B, Abrisqueta P, Lopez-Guillermo A, Artells R, Montserrat E, Monzo M. 2008. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood 111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 41.Shinozaki-Ushiku A, Kunita A, Isogai M, Hibiya T, Ushiku T, Takada K, Fukayama M. 2015. Profiling of virus-encoded microRNAs in Epstein-Barr virus-associated gastric carcinoma and their roles in gastric carcinogenesis. J Virol 89:5581–5591. doi: 10.1128/JVI.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai CY, Liu YY, Liu KH, Hsu JT, Chen TC, Chiu CT, Yeh TS. 2017. Comprehensive profiling of virus microRNAs of Epstein-Barr virus-associated gastric carcinoma: highlighting the interactions of ebv-Bart9 and host tumor cells. J Gastroenterol Hepatol 32:82–91. doi: 10.1111/jgh.13432. [DOI] [PubMed] [Google Scholar]
- 43.Zeng Z, Huang H, Huang L, Sun M, Yan Q, Song Y, Wei F, Bo H, Gong Z, Zeng Y, Li Q, Zhang W, Li X, Xiang B, Li X, Li Y, Xiong W, Li G. 2014. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci 57:315–326. doi: 10.1007/s11427-013-4577-y. [DOI] [PubMed] [Google Scholar]
- 44.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. 2008. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi H, Lee H, Kim SR, Gho YS, Lee SK. 2013. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol 87:8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung YJ, Choi H, Kim H, Lee SK. 2014. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J Virol 88:9027–9037. doi: 10.1128/JVI.00721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H, Choi H, Lee SK. 2015. Epstein-Barr virus miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett 356:733–742. doi: 10.1016/j.canlet.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Qiu J, Thorley-Lawson DA. 2014. EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci U S A 111:11157–11162. doi: 10.1073/pnas.1406136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X, Wei L, Peng Q, Zheng Y, Ou C, Ye Q, Xiong W, Li G, Fu Y, Yan Q, Ma J. 2017. Epstein-Barr virus miR-BART6-3p inhibits the RIG-I pathway. J Innate Immun 9:574–586. doi: 10.1159/000479749. [DOI] [PubMed] [Google Scholar]
- 50.Chen R, Zhang M, Li Q, Xiong H, Liu S, Fang W, Zhang Q, Liu Z, Xu X, Jiang Q. 2017. The Epstein-Barr virus-encoded miR-BART22 targets MAP3K5 to promote host cell proliferative and invasive abilities in nasopharyngeal carcinoma. J Cancer 8:305–313. doi: 10.7150/jca.15753. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 52.Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 53.Dahl J, You J, Benjamin TL. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol 79:13007–13017. doi: 10.1128/JVI.79.20.13007-13017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirata N, Kudoh A, Daikoku T, Tatsumi Y, Fujita M, Kiyono T, Sugaya Y, Isomura H, Ishizaki K, Tsurumi T. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J Biol Chem 280:30336–30341. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 55.Gaspar M, Shenk T. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc Natl Acad Sci U S A 103:2821–2826. doi: 10.1073/pnas.0511148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem 280:8156–8163. doi: 10.1074/jbc.M411405200. [DOI] [PubMed] [Google Scholar]
- 57.Sivachandran N, Sarkari F, Frappier L. 2008. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog 4:e1000170. doi: 10.1371/journal.ppat.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi F, Saha A, Murakami M, Kumar P, Knight JS, Cai Q, Choudhuri T, Robertson ES. 2009. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 388:236–247. doi: 10.1016/j.virol.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai Q, Guo Y, Xiao B, Banerjee S, Saha A, Lu J, Glisovic T, Robertson ES. 2011. Epstein-Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis. PLoS Pathog 7:e1002418. doi: 10.1371/journal.ppat.1002418. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Saha A, Bamidele A, Murakami M, Robertson ES. 2011. EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J Virol 85:2079–2088. doi: 10.1128/JVI.02279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saha A, Murakami M, Kumar P, Bajaj B, Sims K, Robertson ES. 2009. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J Virol 83:4652–4669. doi: 10.1128/JVI.02408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kruse JP, Gu W. 2009. Modes of p53 regulation. Cell 137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 64.Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 65.Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. 2014. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J Virol 88:1389–1393. doi: 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hooykaas MJ, Kruse E, Wiertz EJ, Lebbink RJ. 2016. Comprehensive profiling of functional Epstein-Barr virus miRNA expression in human cell lines. BMC Genomics 17:644. doi: 10.1186/s12864-016-2978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verhoeven RJ, Tong S, Zhang G, Zong J, Chen Y, Jin DY, Chen MR, Pan J, Chen H. 2016. NF-kappaB signaling regulates expression of Epstein-Barr virus BART microRNAs and long noncoding RNAs in nasopharyngeal carcinoma. J Virol 90:6475–6488. doi: 10.1128/JVI.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]