ERVs in the host genome are highly informative about the long-term interactions of retroviruses and hosts. They are also interesting because they have influenced the evolution of mammalian genomes in various ways. In this study, we derive a calibrated timeline describing the process through which ERV diversity has been generated in the equine germ line. We determined the distribution and diversity of perissodactyl ERV lineages and inferred their retrotranspositional activity during evolution, thereby gaining insight into the long-term coevolutionary history of retroviruses and mammals. Our study provides a platform for future investigations to identify equine ERV loci involved in physiological processes and/or pathological conditions.
KEYWORDS: endogenous, equid, evolution, horse, perissodactyl, retrovirus, rhinoceros
ABSTRACT
The evolution of mammalian genomes has been shaped by interactions with endogenous retroviruses (ERVs). In this study, we investigated the distribution and diversity of ERVs in the mammalian order Perissodactyla, with a view to understanding their impact on the evolution of modern equids (family Equidae). We characterize the major ERV lineages in the horse genome in terms of their genomic distribution, ancestral genome organization, and time of activity. Our results show that subsequent to their ancestral divergence from rhinoceroses and tapirs, equids acquired four novel ERV lineages. We show that two of these ERV lineages proliferated extensively in the lineage leading to modern horses, and one contains loci that are actively transcribed in specific tissues. In addition, we show that the white rhinoceros has resisted germ line colonization by retroviruses for more than 54 million years—longer than any other extant mammalian species. The map of equine ERVs that we provide here will be of great utility to future studies aiming to investigate the potential functional roles of equine ERVs and their impact on equine evolution.
IMPORTANCE ERVs in the host genome are highly informative about the long-term interactions of retroviruses and hosts. They are also interesting because they have influenced the evolution of mammalian genomes in various ways. In this study, we derive a calibrated timeline describing the process through which ERV diversity has been generated in the equine germ line. We determined the distribution and diversity of perissodactyl ERV lineages and inferred their retrotranspositional activity during evolution, thereby gaining insight into the long-term coevolutionary history of retroviruses and mammals. Our study provides a platform for future investigations to identify equine ERV loci involved in physiological processes and/or pathological conditions.
INTRODUCTION
The genomes of mammalian species contain thousands of sequences derived from retroviruses (1, 2). Retroviruses are characterized by a replication strategy in which the viral genome is stably integrated into the genome of the host cell (a form referred to as provirus) (3). Thus, when retroviral infection occurs in cells of the host germ line (i.e., sperm, eggs, or early embryo), integrated proviruses can be vertically inherited as host alleles. These endogenous retrovirus (ERV) loci may subsequently increase their copy number within host species genome—either through reinfection of germ line cells or retrotransposition within them—leading to the generation of multicopy ERV lineages (4–6). A subset of ERV copies have been fixed in host genomes, and these sequences constitute a genomic “fossil record” from which the long-term evolutionary history of retroviruses can be inferred (6).
ERV insertions that are only slightly deleterious or selectively neutral may be fixed through chance or genetic hitchhiking (6). However, some appear to have been fixed because they have been domesticated and/or neofunctionalized by host genomes to perform important physiological functions (7–9). Furthermore, even ERV sequences that do not encode proteins can play important physiological roles. For example, ERV loci can exert important impacts on the regulation of gene expression through their impact on epigenetic machinery (10, 11) or by expression of long noncoding RNAs (lncRNAs) (12).
Comparative studies indicate that the myriad of ERV lineages found in the genomes of modern mammals arose from multiple independent genome invasion events. As many of these events occurred after the divergence of mammalian orders, each mammalian order typically has its own distinct ERV composition and history. In fact, some ERVs are unique to individual genera or species. For example, ERVs derived from retroviruses in the genus Gammaretrovirus are present in chimpanzees (Pan trogolodytes) and gorillas (Gorilla gorilla), but closely related ERVs are absent from the human genome (13). Each distinct mammalian lineage has its own characteristic history of ERV activity (e.g., infection, fixation, and expansion). Consequently, characterization of ERVs and investigation of their potential physiological roles have to be performed separately in distinct mammalian groups.
The domestic horse (Equus caballus) is an economically and scientifically important mammal that contributed significantly to the development of modern societies. Horses belong to the family Equidae, which comprises extant species of strict herbivores adapted for running and dietary specialization. The family Equidae, in turn, belongs to the order Perissodactyla (odd-toed ungulates) (14, 15). Living perissodactyls represent a small remnant of a diverse group of mammals that apparently arose in North America ∼54 million years ago (Mya), and subsequently became widespread on all continents apart from Australia and Antarctica (16–18). They are divided into two suborders: Hippomorpha containing the Equidae (horses, donkeys, and zebras) and Ceratomorpha containing the Tapiridae (tapirs) and Rhinocerotidae (rhinoceroses).
Several previous studies have examined ERV diversity in the horse genome (19–21). In this study, we use a range of bioinformatic approaches to characterize ERVs across a broad range of perissodactyl genomes, including several equid species and the white rhinoceros (Ceratotherium simum). We identify the major ERV lineages in the perissodactyl germ line, recover representative genomes for each, and examine the dynamics of their expansion in the branch leading to modern horses. We also investigate the transcriptional profiles among ERV loci in equine-derived cells and tissues.
RESULTS
Identification and phylogenetic classification of equine ERV lineages.
We used a phylogenetic screening (3) approach to characterize perissodactyl ERV lineages in silico. Because reverse transcriptase (RT) is relatively refractory to mutation, similarity searches using the RT protein sequence will typically recover all ERV loci that contain an RT gene (22). Moreover, because the RT protein can be used to reconstruct evolutionary relationships across the entire Retroviridae (23), phylogenetic approaches can be used to classify RT loci identified by screening (3).
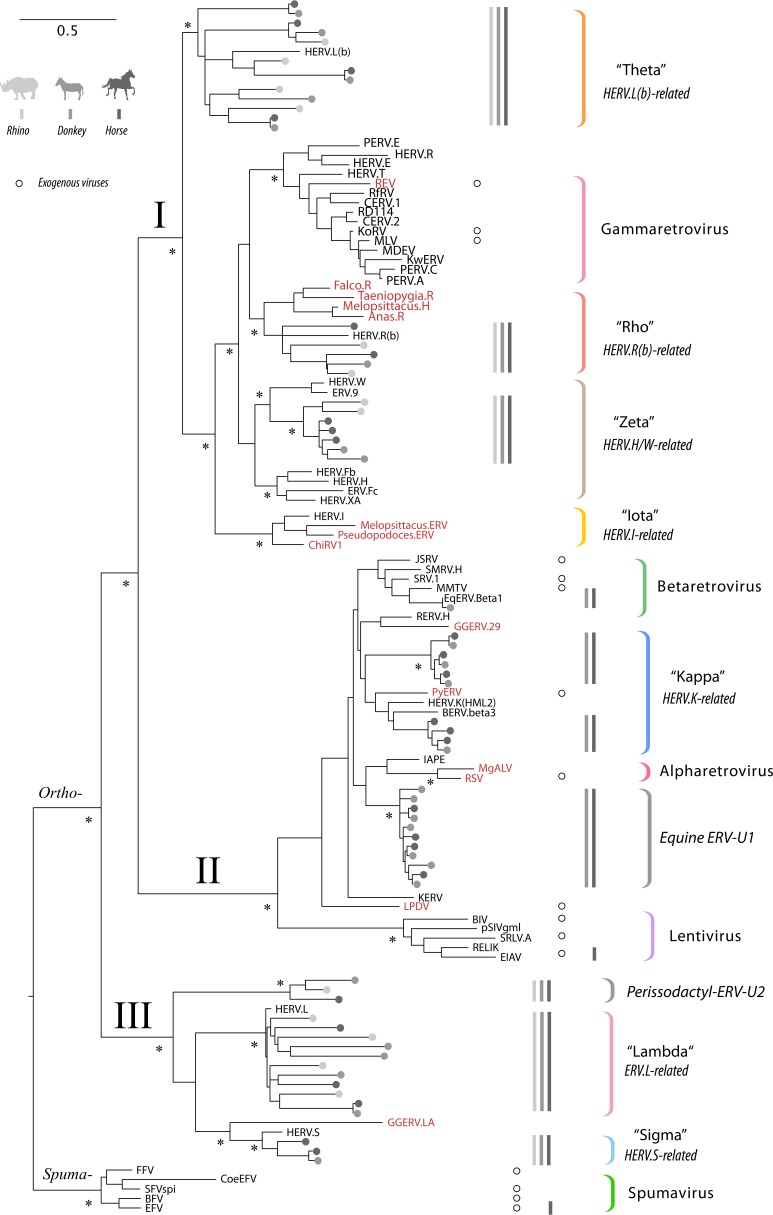
We used this approach to identify and phylogenetically classify ERV RT sequences in 17 published perissodactyl genome sequences, representing 10 distinct species, and 7 distinct breeds of the domestic horse (24, 25) (see Table S1 in the supplemental material). We constructed phylogenies of the RT sequences identified in these screens and identified all clades comprised exclusively of perissodactyl ERVs. Where these lineages were robustly separated from one another by RT sequences derived from ERVs or exogenous retroviruses found in nonmammalian hosts, we assumed that they had arisen in independent germ line invasion events (3). On this basis, we estimate that there are at least nine distinct ERV lineages present in the perissodactyl germ line. All nine lineages are present in equids, whereas only five are found in the rhinoceros. We did not identify any ERV lineages that were unique to the rhinoceros or any that were specific to particular equid species or breeds. The RT phylogeny in Fig. 1 provides an overview of our findings.
FIG 1.
Evolutionary relationships between perissodactyl endogenous, previously characterized ERVs, and exogenous retroviruses. The figure shows a maximum likelihood phylogeny reconstructed from an alignment of retroviral reverse transcriptase (RT) peptide sequences. Sequences extracted from horse, donkey, and rhino genomes are indicated by gray circles, following the color key shown in the top left corner of the figure. For previously characterized ERVs and exogenous retroviruses, taxon labels show the abbreviated names (see Table S8 in the supplemental material for complete details). Sequences derived from exogenous virus references are marked by open circles aligned with taxon labels. Sequences identified in nonmammalian hosts are indicated by red font. Retrovirus subfamilies and orthoretroviral clades (I, II, and III) are indicated on basal branches. Established retroviral genera and ERV lineages defined in this study are indicated by colored brackets. For each of these groups, the presence of sequences in the rhinoceros, donkey, and horse in each genus is indicated by gray bars, following the color key (top left corner of figure). Nodes with bootstrap support of ≥70% are indicated by an asterisk. The bar shows evolutionary distance in substitutions per site.
Notably, we observed a complete absence in perissodactyl genomes of ERVs that group robustly within the Gammaretrovirus clade (as defined sensu stricto by exogenous gammaretroviruses). The perissodactyl germ line also appears to lack any ERVs carrying RT genes that group with human endogenous virus type I (HERV-I), despite such ERVs being present in most other mammal groups and broadly distributed throughout vertebrates as a whole (26). While we did not identify any true members of the Gammaretrovirus genus in perissodactyls, we did identify several distinct lineages of clade I ERVs (Gammaretrovirus-related lineages). These ERV lineages appear to be more closely related to human endogenous retroviruses (HERVs) than to any known exogenous retroviruses. Here, we refer to these three lineages as Rho [HERV.R(b)-related], Zeta (HERV.H/HERV.W-related), and Theta [HERV.L(b)-related]; see Table 1 for further details of these ERV lineages and the HERV references they are based upon.
TABLE 1.
Profile of nine perissodactyl ERV lineages in the domestic horse genome
| Lineage or genus | Clade | Prototype | Prototype reference(s) |
ERV lineagea | PBSb | RepBase LTR subgroup(s)b | Copy no. |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| RTc | Provirusd | env+ proviruse | Solo LTRf | |||||||
| Rho | I | HERV.R(b) | 3 | Rho.1 | Arg(CCG) | 1-2, 1-3, 15, 45, 72A, 72B, 8B, 8E, 8F | 151 | 20 | 6 | 4,057 |
| Zeta | I | HERV.W | 70 | Zeta.1 | Leu(TAA) | 1, 14, 1420 | 37 | 13 | 5 | 3,862 |
| Theta | I | HERV.L(b) | 71–73 | Theta.1 | ND | 1-4, 27_FC | 251 | 11 | 2 | 351 |
| I | Theta.2 | ND | 1-4B, 1-6, 13A, 19, 23B, 6, 6B, MER34A_CF, MER34A1 |
67 | 9 | 6 | 8,675 | |||
| Betaretrovirus | II | MMTV | Beta.1 | Lys(TTT) | [4] | 10 | 3* | 3* | 350 | |
| Kappa | II | HERV.K(HML2) | 33 | Kappa.1 | Lys(CTT) | 2-2 | 5 | 4 | 4 | 79 |
| II | Kappa.2 | Lys(CTT) | This study | 3 | 1 | 1 | 35 | |||
| U1 | II | NA | U1 | Trp(CCA) | 2-1 | 45 | 32 | 32 | 705 | |
| U2 | III | NA | U2 | ND | ND | 54 | NA | NA | NA | |
| Lambda | III | HERV.L | 28 | Lambda | ND | None identified | 691 | NA | 0 | NA |
| Sigma | III | HERV.S | 3 | Sigma | Ser(AGA) Ser(CGA) | 3-1C, 74 | 67 | 1 | 0 | 296 |
| Total | 1,381 | 92 | 57 | 18,410 | ||||||
ERV lineages demarcated in Fig. 1.
PBS, primer binding site. ND, not detected.
Number of RT loci.
Only loci that contained RT plus at least two retroviral coding domains represented in PFAM (64) and that were flanked by paired LTRs of >100 bp in length were counted as proviruses. NA, not available.
Number of proviruses for which we detected the presence of env genes (intact or fragmentary).
Number of solo LTRs.
Strikingly, clade II ERVs were completely absent from the rhinoceros genome. In equids, by contrast, four clade II (Betaretrovirus-related) lineages are present, one of which (EqERV.b1) represents a bona fide Betaretrovirus, and has previously been described in detail (21). We identified two additional clade II lineages that grouped together with representatives of the HERV-K supergroup, which we refer to here as Kappa (27). Accordingly, we named these two lineages EqERV.Kappa.1 (Eq stands for equine) and EqERV.Kappa.2. The fourth and final lineage of clade II ERVs we identified was found to be distinct from all previously characterized retroviruses and ERVs and was named unclassified equine ERV 1 (EqERV.U1).
The ERV.L lineage (referred to here as Lambda) is an ancient group of clade III ERVs that is widespread throughout mammalian genomes and entered the mammalian germ line >105 million years ago (Mya) (28, 29). We identified numerous RT sequences belonging to this lineage in perissodactyls (Table 1). In addition, we identified a second lineage of clade III RT sequences that were related to the primate HERV.S lineage (referred to here as Sigma) (Fig. 1) (3). A potential third lineage of clade III ERVs was also identified, grouping immediately basal to the Lambda lineage. However, all sequences within this low-copy-number group were highly degraded, and we could not determine with confidence whether they should be regarded as genuinely distinct from the Lambda lineage, and thus, were not analyzed further.
The ERV lineages identified here are represented by approximately similar numbers of RT sequences in almost all distinct equid species (Table S2). A few equid genomes had lower overall numbers of insertions, but the relative proportion of loci in each lineage was broadly equivalent to those in other equid species, suggesting that differences were related to low coverage. The Rho and Theta lineages were found to have slightly higher copy numbers in the white rhino genome than in equids. Notably, a total of 908 Lambda RT sequences were identified in the rhino genome versus between 400 and 713 identified in equids.
Distribution and diversity of ERVs in perissodactyl genomes.
Having identified distinct, monophyletic lineages of perissodactyl ERVs using the RT gene, we next sought to characterize the genome structure and evolutionary history of those lineages in greater depth. Retroviral proviruses typically encode three principal coding domains (gag, pol, and env), flanked at either side by long terminal repeat (LTR) sequences, which are identical at the time of integration. However, many ERV loci are comprised of “solo LTRs,” generated when recombination between the 5’ and 3’ LTRs deletes internal coding sequences (30). To associate ERV RT sequences with full-length proviruses (and thereby map associations between RT lineages and LTRs), we performed a second round of screening using the ERV annotation pipeline (ERVAP) (Materials and Methods). Using this approach, we estimated the total number of proviruses (internal regions bounded by paired LTRs) and solo LTRs associated with each lineage of perissodactyl ERVs, as delineated by RT phylogeny (Fig. 1). Table 1 summarizes our findings.
All clade I lineages (Rho, Zeta, and Theta) and the single clade III lineage for which we could identify LTRs (Sigma) were associated with multiple, distinct LTR types. Furthermore, these lineages were clearly present in the ancestral perissodactyl germ line (i.e., prior to the Hippomorpha-Ceratomorpha divergence), since we identified loci that were orthologous between rhinos and equids (Fig. 2a). Notably, we found far fewer proviruses than RT sequences in the Rho and Theta lineages and no proviruses at all for the Lambda lineage. This likely reflects that the expansion of these ERV sequences in perissodactyls has been driven primarily by non-LTR mechanisms. These mechanisms commonly entail reverse transcription and integration of ERV transcripts by non-LTR retrotransposons such as LINE-1 (4, 5), in which case ERV insertions are generated with truncated LTR sequences (e.g., see reference 31). Such truncated sequences would in many cases not meet the criteria for classification as LTRs in our analysis pipeline (see Materials and Methods).
FIG 2.
Examples of orthologous and polymorphic ERV loci in perissodactyls. The DNA sequences of the extreme 5′ and 3′ ends of orthologous ERV internal genes are shown enclosed by red boxes (with the majority of intervening ERV sequence being omitted). Target site duplication (TSD) sequences flanking ERV insertions are shown enclosed by blue boxes. Regions (20 to 30 bp) of upstream and downstream flanking genomic DNA sequence are shown for each locus. (a) Examples of insertions belonging to the Theta, Rho, and Sigma lineages that occur at orthologous loci in the horse, donkey, and rhinoceros genomes. (b) Examples of the ERVs in EqERV.b1 and EqERV.U1 lineages that are polymorphic within horses.
We obtained further evidence that non-LTR mechanisms have been involved in amplification of ancestral ERV RTs when attempting to infer representative/consensus internal sequences for each of the five ancestral lineages: we found that a high proportion of RT sequences belonging to the Lambda lineage are located within 1,000 bp of open reading frames (ORFs) encoding L1 domains of >40 amino acids (aa) in length (Table S5 and Table S6), indicating that they have been amplified as components of LINE-1 (L1) transcripts. Multiple L1 lineages are believed to have been simultaneously active during the evolution of perissodactyls (32). Interestingly, we identified some L1 sequences encoding a chimeric protein containing ERV Lambda RT sequence fused to an L1 gene product (data not shown).
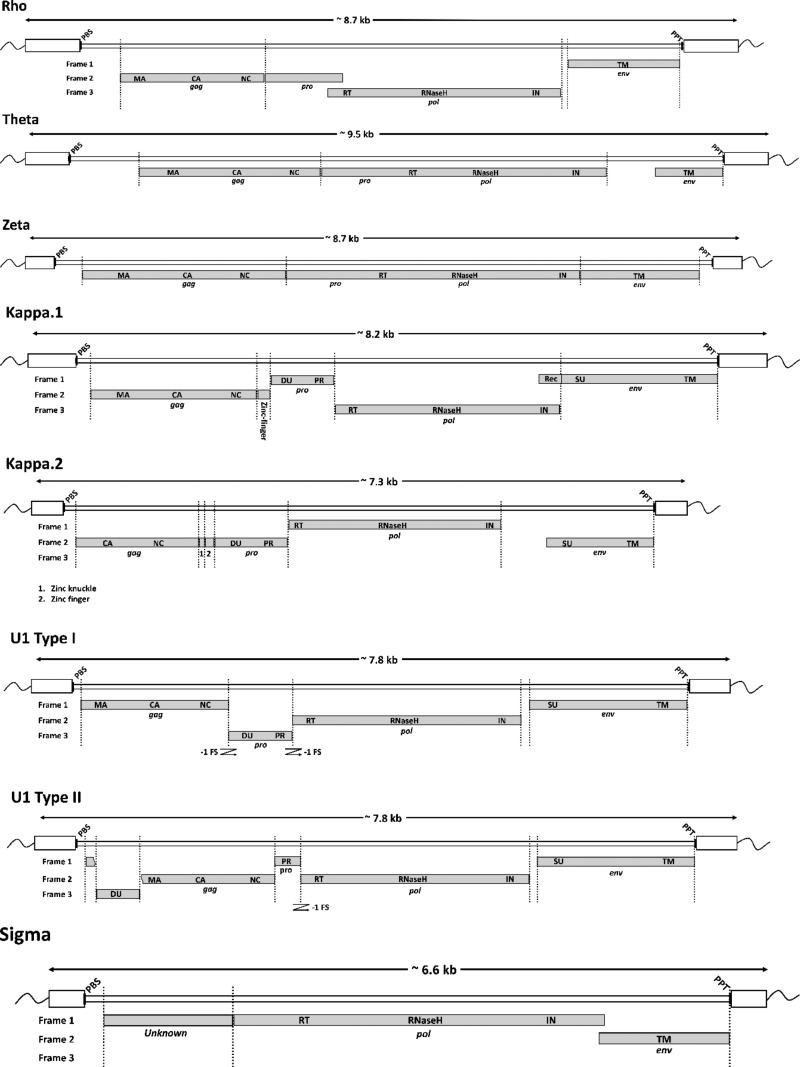
Since we could not confidently link RT sequences in the Lambda lineage with any LTRs, we could not count LTRs in this lineage. Furthermore, we were able to generate only a poor-quality, truncated consensus sequence (data not shown). For the remaining eight remaining lineages, however, we recovered full-length consensus sequences encoding putative env genes and established the links between RT lineages and LTR groups defined in RepBase (Table 1 and Fig. 3; see also Data Set S1 in the supplemental material).
FIG 3.
Schematic representation of proviruses. The putative locations of gag-, pro-, pol-, and env-coding domains within consensus proviral genomes are indicated by gray boxes. Long terminal repeat (LTR) sequences are shown as white boxes. The estimated positions of primer binding sequence (PBS) and polypurine tract (PPT) sequences are indicated by black bars. A bar indicating the length in kilobases is shown above each genome diagram. Abbreviations: PBS, primer binding site; MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease; DU, dUTPase; RT, reverse transcriptase; IN, integrase; SU, surface glycoprotein; TM, transmembrane domain; PPT, polypurine tract.
We found no evidence for the presence of any modern ERV lineages in the rhinoceros genome. Indeed, we could not identify any ERVs in the rhinoceros that were not derived from one of the five ancient lineages present in both rhinos and horses, suggesting that the rhinoceros has resisted ERV germ line invasions for more than 54 million years (16–18). To the best of our knowledge, this is the longest time that any mammalian lineage has existed without newly acquired ERVs becoming fixed in the germ line. The only other exception is humans, which have not acquired fixed insertions from any novel ERV lineages since diverging from other great ape species.
We used data recovered via ERVAP to search equid genomes for ERV loci that were specific to particular breeds or species. We performed this analysis in the awareness that, in general, such loci cannot be comprehensively or rigorously mapped solely comparing whole-genome sequences generated using short-read sequencing. First, assemblies constructed using a reference genome can include false-positive “pseudologs” (ERV insertions that are present in the reference but actually missing in assembled genome [due to multiply mapped reads]). Similarly, ERV insertions that are present only in individual horse breeds may not be detected, as reads from these loci may be incorrectly mapped to other loci in the reference genome. However, it is possible to identify a proportion of the loci that are absent from genomes assembled de novo (Table S1), but present in the reference genome. We identified a total of 10 such ERV loci (Table S4), all of which were derived from modern ERV lineages. These loci included an EqERV.b1 insertion that is absent from the genome of the Mongolian horse (an ancient breed of domestic horse) and an EqERV.U1 insertion that is absent from the genome of Przewalski’s horse (Fig. 2b).
Evolution of ERV lineages in the horse germ line.
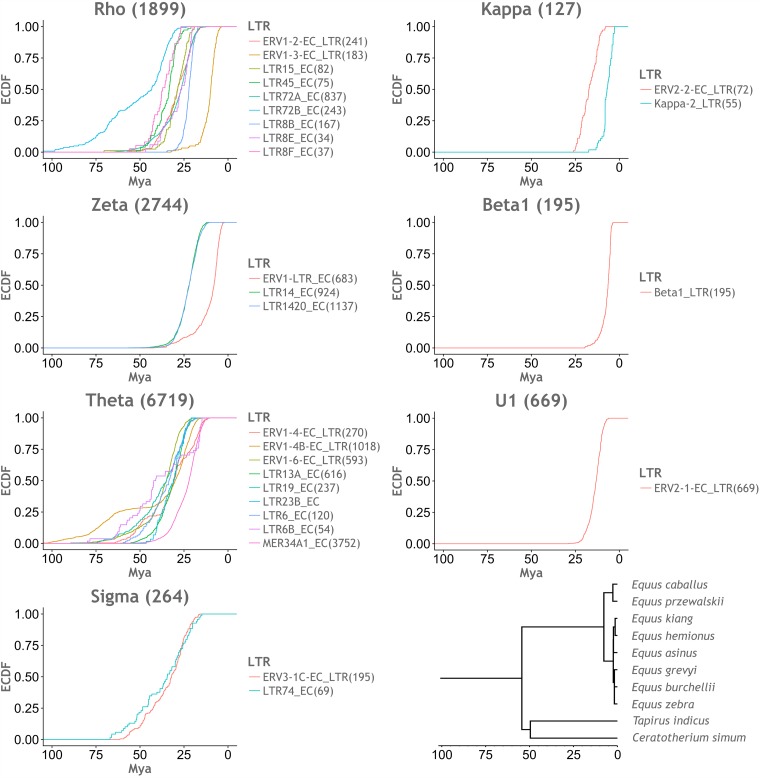
We used a molecular clock-based approach, first described by Subramanian et al. (33), to investigate the historical activity of ERV lineages. For each LTR group listed in Table 1, we created alignments including all LTR sequences identified by our screen (subject to sequence quality). We used these alignments to construct consensus LTR sequences for each LTR group. We then calculated pairwise distances between individual LTR loci and their corresponding LTR group consensus sequence. We converted pairwise distances into age estimates by assuming a neutral molecular clock and generated plots of estimated lineage activity over time (Fig. 4).
FIG 4.
Inferred timeline of ERV lineage expansions. Empirical cumulative distribution function (ECDF) plots, representing the accumulation of observed LTRs over time. The ages of LTRs were inferred by estimating divergence from an LTR consensus sequence and applying a molecular clock-based calibration. The x axes show time in millions of years before the present (millions years ago [Mya]), and the y axes show the proportion of LTR sequences accumulated. Distinct LTR groups found to occur within the same ERV lineage are shown within the same plot, using distinct colors as indicated by the color key for the plot. All x axes were adjusted to the same scale. (Bottom right) Time-scaled perissodactyl phylogeny obtained from the TimeTree website (69).
We categorized perissodactyl ERV lineages that entered the germ line prior to the rhino-equid divergence as “ancient” and those that entered after as “modern.” LTR dating (Fig. 4) indicated that germ line expansions of ancestral perissodactyl ERV lineages largely occurred in the Paleogene period (66 to 23 Mya) and continued for many millions of years after the divergence of the Hippomorpha and Ceratomorpha. In fact, some LTR groups associated with ancestral ERV lineages have undergone more recent expansions. In particular, the Rho and Zeta lineages include LTR groups (LTR1.3 and LTR1, respectively) that appear to have expanded much more recently (from ∼25 to 5 Mya) (Fig. 4).
Studies of mammalian ERVs indicate that intragenomic proliferation can occur through LTR-driven, intracellular retrotransposition (22). This process is characterized by proviral loci with paired LTRs and intact gag and pol genes but truncated or missing env genes. Several ancestral lineages (Rho, Theta, and Zeta) and at least one modern lineage (Kappa.1) contained loci with such genome structures (Table S3). However, we also identified proviruses carrying envelope genes in all four ancestral ERV lineages (Table 1), and furthermore, each of these lineages contains at least one locus that encodes a near intact envelope protein (Table S3). Notably, expression of env RNA derived from the Zeta lineage has been reported previously in reproductive tissue (34). We found 252 ancestral ERV loci and 14 modern ERV lineage loci that overlapped with lncRNA loci (same strand with >1 bp overlapping) annotated by Scott et al. (35), including representatives of the Kappa2, Beta1, and U1 lineages.
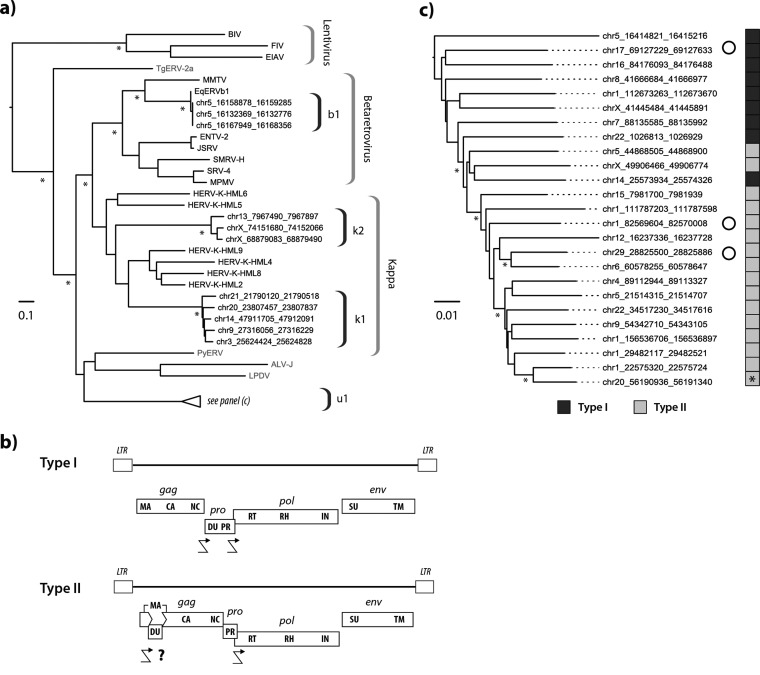
The four modern ERV lineages identified in equid genomes grouped robustly within clade II. By narrowing the focus of our evolutionary investigations to this group, we could reconstruct phylogenetic relationships using longer alignments (Fig. 5a). Among the four lineages, one is a bona fide Betaretrovirus called EqERV.b1, and has been described previously (21). The EqERV.b1 lineage is relatively closely related to mouse mammary tumor virus (MMTV) and shares some of its characteristic features (e.g., LTRs of >1,000 bp in length). We established that orthologous EqERV.b1 insertions are shared in the horse and donkey genomes, demonstrating that the lineage was present in the equid germ line prior to the divergence of horses and donkeys ∼6 to 10 Mya (36, 37). This establishes a minimum age for the EqERV.b1 lineage that is considerably more ancient than the 0.5 million years (Myr) suggested previously (21). Furthermore, by extension, the identification of this ortholog demonstrates a minimum age of 9 Myr for the entire lineage of MMTV-related retroviruses. The EqERV.b1 family contains a relatively large number of solo LTRs (Table 1), and when these sequences are used to estimate lineage activity, they indicate that EqERV.b1 expansion occurred in the late Neogene period, from ∼12 to 5 Mya (Fig. 4).
FIG 5.
Characteristics of modern equine ERVs. (a) Maximum likelihood phylogeny representing the estimated evolutionary relationships between Pol sequences derived from clade II ERVs in perissodactyl genomes and those of previously characterized ERVs and exogenous retroviruses. Taxon labels for RT sequences detected in this study indicate the species in which they were identified. Other taxon labels show the abbreviated name of the virus or ERV. Sequences identified in nonmammalian hosts are indicated in gray. Brackets on the right indicate ERV lineages and retroviral genera. Nodes with bootstrap support above 70% are indicated by asterisks. The bar shows evolutionary distance in substitutions per site. Details of taxa are provided in Table S8. chr5, chromosome 5. (b) Consensus genome structures of EqERV.U1 proviruses. Viral coding domains are shown as dark gray bars. Long terminal repeats (LTRs) are shown as boxes. Crooked arrows indicate where we have inferred translational frameshifting. For type II proviruses, we show a putative frameshift site (indicated with a question mark) that would allow expression of a matrix-dUTPase fusion protein. Abbreviations: LTR, long terminal repeat; MA, matrix; CA, capsid; NC, nucleocapsid; DU, dUTPase; PR, protease; RT, reverse transcriptase; IN, integrase; SU, surface; TM, transmembrane. (c) Maximum likelihood phylogeny of EqERV.U1 loci based on the aligned nucleotide sequences of 25 full-length proviruses. The sidebar boxes to the right of the taxa indicate the type of genome found in the element (see panel b) as indicated in the key below the tree. An asterisk on the sidebar shows the youngest provirus based on the paired LTR dating. Open circles indicate loci that show evidence of transcription based on analysis of transcriptomic data sets. Asterisks indicate nodes with bootstrap support above 70%. The bar shows evolutionary distance in substitutions per site.
The remaining three Betaretrovirus-like lineages group outside the clade defined by exogenous betaretroviruses (Fig. 5a) and together with members of the HERV.K supergroup, which comprises 10 distinct groups of ERVs identified in primate genomes and labeled HML1 to HML10. These groups, which were originally defined using DNA hybridization, have since been shown to comprise at least two phylogenetically distinct lineages: one containing the HML5 and HML6 lineages and one containing all the other HML lineages (Fig. 5a). Here we refer to the clade that contains both these lineages and the related equine ERV lineages as Kappa. Phylogenies based on pol show that both equine Kappa lineages (k1 and k2) are clearly distinct from related lineages in the human genome. Notably, we found that the EqERV.k1 genome contains a potential homolog of the HERV-K(HML2) rec gene with predicted splice sites in the expected locations (data not shown).
The EqERV.U1 lineage is not closely related to any previously characterized retrovirus or ERV, and in phylogenetic trees based on pol (Fig. 5a), it groups as a robustly supported sister clade to ERVs and exogenous betaretroviruses found in birds and reptiles (38, 39). The EqERV.U1 lineage contains the largest number of proviruses (n = 45) and solo LTRs (n = 705) of any modern perissodactyl ERV lineage in the horse genome, and intriguingly, also shows indications of relatively recent activity. We therefore investigated the evolution of the EqERV.U1 lineage in greater depth.
Genomic and phylogenetic characterization of the EqERV.U1 lineage.
The alignment of full-length proviruses was used to infer a consensus genome structure for the EqERV.U1 lineage. This revealed that there were, in fact, two, distinct types of genomic organizations among EqERV.U1 insertions (Fig. 5b). In the first of these (type I), the pro ORF encodes a dUTPase domain at its 5′ end, as is found in betaretroviruses (40). However, the majority of EqERV.U1 insertions had a more unusual genome structure (type II) in which the dUTPase was encoded by an ORF inserted into the 5′ end of gag. This second type of genome structure has not previously been reported in any retrovirus.
We used a combination of approaches to calibrate the time scale of EqERV.U1 activity. Where paired LTRs were present, we estimated the age of loci by calculating the divergence between these sequences (which are derived from identical copies) and applying a neutral rate for the host genome. In addition, we examined published genome assemblies of other Perissodactyl species and subspecies for the presence of orthologous EqERV loci. We annotated information about locus ages and genome structure onto a phylogeny constructed from an alignment of EqERV.U1 proviruses (with dUTPase-encoding regions removed). We then annotated information about genome structure (type I versus type II) and insertion age onto this phylogeny (Fig. 5b). Notably, the midpoint-rooted phylogeny showed the oldest insertions clustering toward the root of the tree. Furthermore, insertions with the more typical type I genome organization were found almost exclusively toward the root, whereas all proviruses that exhibited a type II genome structure clustered together in a single derived clade with robust bootstrap support. We identified two proviral loci that were unique to the horse, both of which exhibited a type II genome structure (Fig. 5c). All other EqERV-U1 loci in the horse genome had orthologs in the donkey genome.
Together, these data indicate that the germ line invasion event that originally generated the EqERV.U1 lineage occurred somewhere between 25 and 30 Mya (Fig. 4). The initial expansion of this lineage involved ERVs with type I genome structures. Approximately 15 Mya (Fig. 4), one EqERV copy underwent the genome rearrangements that generated the type II genome structure, and this element gave rise to a lineage that has been expanding until relatively recently (∼1 Mya based on integration dates estimated by LTR comparisons).
Analysis of publicly available transcriptome data revealed that 21 EqERV.U1 loci showed evidence of expression and that for 9 of these loci, the entire provirus appeared to be transcribed (based on read coverage). However, we did not have sufficient resolution in this data set to determine whether all expressed genes were from the same locus. The transcriptome data sets analyzed here encompassed 17 derived from specific equine tissues and one derived from an equine-derived cell line (E-derm). We found that brain stem, spinal cord, and oviduct have only type I provirus expression, whereas E-derms and skin expressed only type II proviruses. Trophectoderm has both type I and type II provirus transcripts. In E-derms, only one complete EqERV.U1 locus on chromosome 29 is transcribed.
DISCUSSION
In this study, we examined ERV diversity in the order Perissodactyla, with the aim of understanding how interactions with retroviruses have shaped equid evolution. We used a phylogenetic screening approach to characterize ERV lineages; with this method, evolutionary relationships between RT-encoding proviral sequences were used as the primary basis for classifying loci. This established that there have been at least nine distinct genome invasion events in the perissodactyl lineage (Fig. 1). We provide a minimum estimate because it is difficult to be certain that the nine lineages described here are comprised entirely of ERV insertions that arose from the same ancestral founder. This is particularly challenging when ERV lineages have undergone numerous separate expansions; for example, many of the ancestral lineages identified here contain multiple LTR subgroups (Table 1). These subgroups might reflect multiple distinct genome invasions by related viruses utilizing distinct LTRs, or recombination events wherein preexisting ERV lineages acquire novel LTRs, enabling further waves of intragenomic expansion.
Our efforts to recover representative proviral loci were instructive with regard to determining which equine ERV lineages were more ancient. Proviruses in the Lambda, Rho, Zeta, Theta, and Sigma lineages all exhibited multiple frameshifts, in-frame stop codons, and indels. Moreover, for four of these lineages, we identified examples of loci that were orthologous between the Hippomorpha and Ceratomorpha (Fig. 2a), establishing that they entered the mammalian germ line >54 Mya. Given that no intact or nearly intact proviruses were identified for any ancestral ERV lineage, it is likely that amplification in trans (probably via non-LTR mechanisms) accounts for the differences in RT copy number observed for these lineages and the relatively low number of proviruses versus RT sequences.
Overall, the ERV landscape of perissodactyl genomes broadly resembles that found in other large-bodied placental mammal groups (e.g., hominids, cetaceans, and artiodactyls). These species generally have lower numbers of ERV sequences in their genomes compared with many smaller-bodied mammal species (e.g., rodents and bats) (41). Furthermore, all the lineages we have defined as ancestral within perissodactyls (i.e., Lambda, Sigma, Rho, Theta, and Zeta) have relatively closely related counterparts in humans, carnivores, and artiodactyls. Importantly, when examined in the context of the entire retrovirus family, retroviral lineages that are in fact only distantly related can appear superficially similar, even though they in fact diverged a long time ago. For example, due to the time-dependent phenomenon observed for rates of evolutionary change in virus sequences (42), it is entirely possible that the retroviruses that gave rise to the avian and mammalian Rho lineages (Fig. 1) are as distantly related to one another as the host species they infect.
Although the ERV composition of the horse genome shares broad similarities with other large-bodied mammals, it also exhibits some intriguing differences. Perhaps the most conspicuous of these is the total absence of ERVs grouping within the Gammaretrovirus genus (as defined by exogenous isolates) in any of the genomes we screened. In addition, the rhinoceros genome exhibits a total absence of clade II (Betaretrovirus-related) ERVs, despite these being present in the genomes of most other mammalian species, including equids. The absence of these groups is surprising when considered in the light of previous studies, which have shown that they are extremely widespread in mammalian genomes (43–46). Given the diversity of species that appear to have harbored gamma- and betaretroviruses in the past, it seems likely that perissodactyl ancestors would have been exposed to these viruses. Potentially, the absence of these viruses from all or some perissodactyl lineages might reflect the existence of perissodactyl-specific antiviral factors that potently restrict these particular retrovirus groups, and experimental studies challenging equine cells with gammaretroviruses might allow these factors to be identified. However, it is also important to interpret the distribution of ERVs cautiously. Because it is highly statistically unlikely that any ERV locus will reach fixation, it is entirely possible that perissodactyl genomes have been invaded by ERV lineages that are not represented in the genomes of extant perissodactyl species. This may also have occurred in the case of the rhino, which has acquired no fixed ERV loci from retroviruses that entered the germ line after the Hippomorpha-Ceratomorpha divergence (∼54 Mya).
The horse and human genomes are similar in that the only ERV lineages that appear likely to have been active recently are related to betaretroviruses. In humans and apes, the HERV.K(HML2) lineage contains some intact proviral loci that are capable of producing infectious particles and are present only at a low frequency in the human population (47). In horses, two clade II (Betaretrovirus-related) lineages (EqERV.U1 and EqERV.b1) have generated high numbers of fixed loci in the past 20 million years. We identify insertions belonging to these lineages that are polymorphic among horse subspecies and breeds (see Table S3 in the supplemental material), indicating that the EqERV.b1 and EqERV.U1 lineages have remained active up until relatively recently. The annotations generated in our study (Table S4) can inform future efforts to map the distribution of polymorphic equine ERV (EqERV) loci more precisely (e.g., by using PCR to amplify insertion sites from a range of breeds and subspecies).
Over recent years, it has become increasingly clear that ERVs have played an important role in shaping mammalian genome evolution. One way that ERVs can impact their hosts is by providing genes that are coopted by host genomes to perform physiological functions in their host species (7–9). For example, syncytins are proteins derived from retroviral envelope (env) genes that have been domesticated by mammals to carry out an essential function in placental development (48, 49). We identified intact or nearly intact env genes in several ancient ERVs, and some of these might represent genes or pseudogenes that have (or had) syncytin-like properties. Indeed, one of the env genes identified in our study (belonging to the Zeta lineage) is highly expressed in the placenta, and on this basis has previously been identified as a candidate syncytin-like gene (34). Alternatively, some (or all) of these env genes might encode proteins that restrict related retroviruses from infecting the cell via a receptor interference mechanism, as has been described for exogenous retroviruses (50), as well as endogenous env genes in other species (51–53). Intriguingly, one modern lineage (EqERV.U1) contains actively transcribed loci, consistent with a potential physiological role. In this lineage, expansion has been associated with the transposition of the dUTPase gene into the 5′ end of the gag gene (Fig. 4), and we found evidence that some of these rearranged forms might express a Gag-dUTPase fusion protein via ribosomal frameshifting (Fig. 5c). The significance of the patterns of genomic rearrangement and transcription in the EqERV.U1 lineage remains unclear. However, to the extent that these patterns have been shaped by selection pressures related to the dUTPase gene, they might provide insight into the functions of this poorly understood retroviral enzyme (54).
Genomic changes mediated by ERV activity are also thought to have facilitated mammalian evolution by providing a platform for the emergence of new layers of epigenetic gene regulation during development (10). Notably, we found that many of the ERVs identified in our study overlapped lncRNAs (Table S7), indicating a potential role for equine ERVs in lncRNA-mediated gene regulation (55). We do not yet know to what extent ERV activity has mediated adaptive changes during equid evolution. Nonetheless, insofar as it has, our study offers some insight into which groups of ERVs are likely to have been involved. Equid evolution during the Miocene (15 to 20 Mya) was associated with physiological adaptations that arose as equine ancestors shifted from being small forest-dwelling animals feeding on leafy vegetation into larger-bodied herbivores adapted for life in open grassland (56). Our investigation indicates that during this period, loci belonging to specific ERV lineages and sublineages were being fixed in the equid germ line at an elevated rate. As shown in Fig. 4, these include several modern ERV lineages (EqERV.U1, EqERV.b1, and EqERV.K1) as well as certain LTR subgroups of the ancestral Rho, Zeta, and Theta lineages (in particular, the ERV1-2, ERV1, and MER34A1 subgroups of these lineages, respectively). Whereas in the case of the modern ERV lineages, expansion appears to have been driven by a mixture of reinfection and intracellular retrotransposition, the expansion of ancestral ERV lineages is more clearly associated with non-LTR mechanisms, particularly within the most ancient ERV groups found in the perissodactyl germ line, Lambda, Rho, and Theta.
MATERIALS AND METHODS
Genome assembly.
The reference genome of the domestic horse (Equus caballus) (equCab2, GCF_000002305.2) and the white rhinoceros (Ceratotherium simum) (cerSim1, GCF_000283155.1) were downloaded from the NCBI Genome database (1). The donkey genome sequences (assembly willy) were downloaded from the Centre for GeoGenetics website (25). Whole-genome sequencing short reads of the Somali wild ass (Equus asinus somalicus), Onager (Equus hemionus), Kiang (Equus kiang), plains zebra (Equus burchellii boehmi), Burchell’s zebra (Equus burchellii quagga), Grevy’s zebra (Equus grevyi), and Hartmann’s mountain zebra (Equus zebra hartmannae) were obtained from the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra; accession no. PRJEB7446) (24, 25). Read trimming was performed by Trim Galore (57), and reads were mapped to the horse or donkey reference genomes using Bowtie2 with a very-sensitive–local option (equal to –D 20 –R 3 –N 0 –L 20 –i S,1,0.50) (58). Consensus genomes were generated using a combination of SAMtools and BCFtools (59).
Genome screening in silico.
As a first step toward more definitively characterizing the evolutionary history of equine ERVs, we implemented a phylogenetic screening strategy based on analysis of the reverse transcriptase (RT) peptide sequence. We collated a representative set of RT sequences derived from ERVs and exogenous retroviruses. These sequences were conceptually translated to peptide sequences. RT peptide sequences representing established retrovirus groups and ERV lineages were used as probes for in silico screening of perissodactyl genomes.
The phylogenetic screening was performed using the database-integrated genome screening (DIGS) tool (60). Genomic sequences that disclosed statistically significant similarity to RT probes were extracted and classified by BLAST comparison to the RT reference library. A subset of these RT sequences was extracted and entered into a multiple-sequence alignment (MSA) with RT sequences from our reference set. This MSA was then used as input for a maximum likelihood (ML) phylogenetic analysis. We used the phylogeny to identify well-supported clades that were comprised entirely of perissodactyl ERVs. We then created RT reference sequences based on recovered equine RT sequences to represent these clades and repeated the DIGS process.
This enabled us to identify a complete set of RT-encoding ERV genes in each of the species examined. For these loci, we then attempted to recover a more complete provirus, using the ERV annotation pipeline (ERVAP) pipeline (Fig. 5). In this pipeline, RT sequences were extracted along with 10 kb of flanking sequence on each side. The LTRharvest (61) program is used to search for potential LTR sequences flanking RT matches. To be counted as LTRs, sequences were required to be >100 bp long and <20% divergent from one another. Where putative LTRs were identified, these were classified by BLAST comparison to a library of repetitive sequences obtained from RepBase (62). For proviral sequences with paired LTRs, the LTRdigest program (63) is used to annotate internal regions (i.e., by demarcating putative coding domains). For sequences that exhibited similarity to retroviral RTs but were not flanked by identifiable LTRs, the HMMR program was used to search for these domains. Annotations generated by LTRdigest and HMMR were based on retrovirus protein libraries obtained from PFAM (64) and a tRNA library obtained from GtRNAdb (65).
We used BLAST to search for ERV loci that were unique to individual species or breeds. We generated probe sequences that comprised 100 bp of insertion site sequences and 30 bp of ERV sequence. Potential empty insertion sites were identified as genomic sequences that matched the probe in the flanking sequence region but not in the ERV region.
Phylogenetic analysis.
Maximum likelihood phylogenies were generated using RAxML (66), and model parameters were selected using IQ-TREE model selection function (67). Support for phylogenies was assessed via 1,000 nonparametric bootstrap replicates. A phylogeny based on RT was used to infer the relationships of equine ERVs to one another and to previously characterized retroviral RT sequences. This phylogeny was based on an alignment spanning 135 amino acid residues in RT and was reconstructed using the rtREV amino acid substitution matrix as selected by IQ-TREE (68). To investigate the evolutionary relationship of EqERV.U1 to other, closely related retroviruses, we constructed a second data set by aligning complete Pol polyprotein sequences. Phylogenies were reconstructed using a codon-based alignment spanning RT, RNase H, and integrase domains.
Dating.
For LTR comparisons we excluded pairs that did not group together in LTR phylogenies since these pairs could reflect proviruses that have undergone nonhomologous recombination in the internal region or artefacts generated during genome assembly. To date solo LTRs, we applied a molecular clock approach described by Subramanian et al. (33), in which each LTR is dated by measuring divergence from a subgroup consensus and applying a neutral rate calibration.
Transcriptomics.
Equine transcriptome data were obtained from the European Nucleotide Archive (ENA) (see Table S10 in the supplemental material). Adapter sequences were removed using the Trim Galore! script. Trimmed reads were aligned to the E. caballus reference using TopHat and an annotation file generated in-house from ENSEMBL 84 gene annotations combined with ERV annotations obtained via genome screening. Expression levels were inferred using Cuffquant, and values obtained from distinct experiments were normalized using Cuffnorm. Approximately 4,551 million reads were obtained, which were then mapped to the equine reference genome (EquCab2). Mapping to Ensembl and ERV annotation resulted in 80.91% of reads (∼3,683 million) being assigned to host genes or ERV loci.
Supplementary Material
ACKNOWLEDGMENTS
Robert J. Gifford and Pablo R. Murcia were supported by grants from the UK Medical Research Council (grants MC_UU_12014/10 and MC_UU_12014/9, respectively).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00927-18.
REFERENCES
- 1.Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blocker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MC, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guerin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Roed KH, Ryder OA, Searle S, Skow L, Swinburne JE, et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Tristem M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol 74:3715–3730. doi: 10.1128/JVI.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M. 2005. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol 22:814–817. doi: 10.1093/molbev/msi088. [DOI] [PubMed] [Google Scholar]
- 5.de Parseval N, Heidmann T. 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res 110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 6.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315. doi: 10.1023/A:1024455415443. [DOI] [PubMed] [Google Scholar]
- 7.Varela M, Spencer TE, Palmarini M, Arnaud F. 2009. Friendly viruses: the special relationship between endogenous retroviruses and their host. Ann N Y Acad Sci 1178:157–172. doi: 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci 368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redelsperger F, Raddi N, Bacquin A, Vernochet C, Mariot V, Gache V, Blanchard-Gutton N, Charrin S, Tiret L, Dumonceaux J, Dupressoir A, Heidmann T. 2016. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet 12:e1006289. doi: 10.1371/journal.pgen.1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imbeault M, Helleboid PY, Trono D. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543:550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- 11.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 12.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yohn CT, Jiang Z, McGrath SD, Hayden KE, Khaitovich P, Johnson ME, Eichler MY, McPherson JD, Zhao S, Paabo S, Eichler EE. 2005. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol 3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radinsky LB. 1966. The adaptive radiation of the phenacodontid condylarths and the origin of the Perissodactyla. Evolution 20:408–417. doi: 10.1111/j.1558-5646.1966.tb03375.x. [DOI] [PubMed] [Google Scholar]
- 15.Wilson DE, Reeder DM. 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd ed Johns Hopkins University Press, Baltimore, MD. doi: 10.1108/09504120610673024. [DOI] [Google Scholar]
- 16.Bowen GJ, Clyde WC, Koch PL, Ting S, Alroy J, Tsubamoto T, Wang Y, Wang Y. 2002. Mammalian dispersal at the Paleocene/Eocene boundary. Science 295:2062–2065. doi: 10.1126/science.1068700. [DOI] [PubMed] [Google Scholar]
- 17.Rose KD, Holbrook LT, Rana RS, Kumar K, Jones KE, Ahrens HE, Missiaen P, Sahni A, Smith T. 2014. Early Eocene fossils suggest that the mammalian order Perissodactyla originated in India. Nat Commun 5:5570. doi: 10.1038/ncomms6570. [DOI] [PubMed] [Google Scholar]
- 18.Steiner CC, Ryder OA. 2011. Molecular phylogeny and evolution of the Perissodactyla. Zoolog J Linnean Soc 163:1289–1303. doi: 10.1111/j.1096-3642.2011.00752.x. [DOI] [Google Scholar]
- 19.Brown K, Moreton J, Malla S, Aboobaker AA, Emes RD, Tarlinton RE. 2012. Characterisation of retroviruses in the horse genome and their transcriptional activity via transcriptome sequencing. Virology 433:55–63. doi: 10.1016/j.virol.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Etxebarria K, Jugo BM. 2012. Detection and characterization of endogenous retroviruses in the horse genome by in silico analysis. Virology 434:59–67. doi: 10.1016/j.virol.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 21.van der Kuyl AC. 2011. Characterization of a full-length endogenous beta-retrovirus, EqERV-beta1, in the genome of the horse (Equus caballus). Viruses 3:620–628. doi: 10.3390/v3060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magiorkinis G, Gifford RJ, Katzourakis A, De Ranter J, Belshaw R. 2012. Env-less endogenous retroviruses are genomic superspreaders. Proc Natl Acad Sci U S A 109:7385–7390. doi: 10.1073/pnas.1200913109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y, Eickbush TH. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonsson H, Schubert M, Seguin-Orlando A, Ginolhac A, Petersen L, Fumagalli M, Albrechtsen A, Petersen B, Korneliussen TS, Vilstrup JT, Lear T, Myka JL, Lundquist J, Miller DC, Alfarhan AH, Alquraishi SA, Al-Rasheid KA, Stagegaard J, Strauss G, Bertelsen MF, Sicheritz-Ponten T, Antczak DF, Bailey E, Nielsen R, Willerslev E, Orlando L. 2014. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc Natl Acad Sci U S A 111:18655–18660. doi: 10.1073/pnas.1412627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas A-S, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AMV, Cahill J, Rasmussen M, Wang X, Min J, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Røed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KAS, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MTP, Kjær K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 26.Martin J, Herniou E, Cook J, Waugh O'Neill R, Tristem M. 1997. Human endogenous retrovirus type I-related viruses have an apparently widespread distribution within vertebrates. J Virol 71:437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lower R, Lower J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A 93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benit L, Lallemand JB, Casella JF, Philippe H, Heidmann T. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J Virol 73:3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee A, Nolan A, Watson J, Tristem M. 2013. Identification of an ancient endogenous retrovirus, predating the divergence of the placental mammals. Philos Trans R Soc Lond B Biol Sci 368:20120503. doi: 10.1098/rstb.2012.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. 2007. Rate of recombinational deletion among human endogenous retroviruses. J Virol 81:9437–9442. doi: 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grandi N, Cadeddu M, Blomberg J, Mayer J, Tramontano E. 2018. HERV-W group evolutionary history in non-human primates: characterization of ERV-W orthologs in Catarrhini and related ERV groups in Platyrrhini. BMC Evol Biol 18:6. doi: 10.1186/s12862-018-1125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sookdeo A, Hepp CM, Boissinot S. 2018. Contrasted patterns of evolution of the LINE-1 retrotransposon in perissodactyls: the history of a LINE-1 extinction. Mob DNA 9:12. doi: 10.1186/s13100-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefanetti V, Marenzoni ML, Passamonti F, Cappelli K, Garcia-Etxebarria K, Coletti M, Capomaccio S. 2016. High expression of endogenous retroviral envelope gene in the equine fetal part of the placenta. PLoS One 11:e0155603. doi: 10.1371/journal.pone.0155603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott EY, Mansour T, Bellone RR, Brown CT, Mienaltowski MJ, Penedo MC, Ross PJ, Valberg SJ, Murray JD, Finno CJ. 2017. Identification of long non-coding RNA in the horse transcriptome. BMC Genomics 18:511. doi: 10.1186/s12864-017-3884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PC, Yang Z. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc Biol Sci 279:3491–3500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilstrup JT, Seguin-Orlando A, Stiller M, Ginolhac A, Raghavan M, Nielsen SC, Weinstock J, Froese D, Vasiliev SK, Ovodov ND, Clary J, Helgen KM, Fleischer RC, Cooper A, Shapiro B, Orlando L. 2013. Mitochondrial phylogenomics of modern and ancient equids. PLoS One 8:e55950. doi: 10.1371/journal.pone.0055950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henzy JE, Gifford RJ, Johnson WE, Coffin JM. 2014. A novel recombinant retrovirus in the genomes of modern birds combines features of avian and mammalian retroviruses. J Virol 88:2398–2405. doi: 10.1128/JVI.02863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huder JB, Boni J, Hatt JM, Soldati G, Lutz H, Schupbach J. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J Virol 76:7607–7615. doi: 10.1128/JVI.76.15.7607-7615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petropoulos C. 1997. Retroviral taxonomy, protein structures, sequences, and genetic maps, 757–805 In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 41.Katzourakis A, Magiorkinis G, Lim AG, Gupta S, Belshaw R, Gifford R. 2014. Larger mammalian body size leads to lower retroviral activity. PLoS Pathog 10:e1004214. doi: 10.1371/journal.ppat.1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aiewsakun P, Katzourakis A. 2016. Time-dependent rate phenomenon in viruses. J Virol 90:7184–7195. doi: 10.1128/JVI.00593-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. 1998. Retroviral diversity and distribution in vertebrates. J Virol 72:5955–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gifford R, Kabat P, Martin J, Lynch C, Tristem M. 2005. Evolution and distribution of class II-related endogenous retroviruses. J Virol 79:6478–6486. doi: 10.1128/JVI.79.10.6478-6486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayward A, Cornwallis CK, Jern P. 2015. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci U S A 112:464–469. doi: 10.1073/pnas.1414980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuo X, Feschotte C. 2015. Cross-species transmission and differential fate of an endogenous retrovirus in three mammal lineages. PLoS Pathog 11:e1005279. doi: 10.1371/journal.ppat.1005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. 2016. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci U S A 113:E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A 106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 50.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson HL, Lamoreux WF. 1976. Expression of endogenous ALV antigens and susceptibility to subgroup E ALV in three strains of chickens (endogenous avian C-type virus). Virology 69:50–62. doi: 10.1016/0042-6822(76)90193-8. [DOI] [PubMed] [Google Scholar]
- 52.Spencer TE, Mura M, Gray CA, Griebel PJ, Palmarini M. 2003. Receptor usage and fetal expression of ovine endogenous betaretroviruses: implications for coevolution of endogenous and exogenous retroviruses. J Virol 77:749–753. doi: 10.1128/JVI.77.1.749-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco-Melo D, Gifford RJ, Bieniasz PD. 2017. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife 6:e22519. doi: 10.7554/eLife.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hizi A, Herzig E. 2015. dUTPase: the frequently overlooked enzyme encoded by many retroviruses. Retrovirology 12:70. doi: 10.1186/s12977-015-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon JH, Abdelmohsen K, Gorospe M. 2013. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacFadden BJ. 1994. Fossil horses: systematics, paleobiology, and evolution of the family Equidae. Cambridge University Press, Cambridge, United Kingdom. doi: 10.1002/gea.3340090507. [DOI] [Google Scholar]
- 57.Krueger F. 2017. Trim Galore!, v0.4.5. Bioinformatics Group, Babraham Institute, Cambridge, United Kingdom: www.bioinformatics.babraham.ac.uk/projects/trim_galore/. [Google Scholar]
- 58.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu H, Dennis T, Hughes J, Gifford RJ. 2018. Database-integrated genome screening (DIGS): exploring genomes heuristically using sequence similarity search tools and a relational database. BioRxiv. doi: 10.1101/246835. [DOI]
- 61.Ellinghaus D, Kurtz S, Willhoeft U. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinbiss S, Willhoeft U, Gremme G, Kurtz S. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res 37:7002–7013. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dimmic MW, Rest JS, Mindell DP, Goldstein RA. 2002. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol 55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 70.Blond JL, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. 1999. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol 73:1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katzourakis A, Tristem M. 2005. Phylogeny of human endogenous and exogenous retroviruses, p 186–203. In Sverdlov E. (ed), Retroviruses and primate genome evolution. Landes Bioscience, Austin, TX. [Google Scholar]
- 72.Brown K, Emes RD, Tarlinton RE. 2014. Multiple groups of endogenous epsilon-like retroviruses conserved across primates. J Virol 88:12464–12471. doi: 10.1128/JVI.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vargiu L, Rodriguez-Tome P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, Tramontano E, Blomberg J. 2016. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.