African swine fever virus causes a highly contagious and lethal disease of swine that currently affects many countries of sub-Saharan Africa, the Caucasus, the Russian Federation, and Eastern Europe and has very recently spread to China. Despite extensive research, effective vaccines or antiviral strategies are still lacking, and many basic questions on the molecular mechanisms underlying the infective cycle remain. One such gap regards the composition and structure of the infectious virus particle. In the study described in this report, we identified the set of viral and host proteins that compose the virion and determined or inferred the localization of many of them. This information significantly increases our understanding of the biological and structural features of an infectious African swine fever virus particle and will help direct future research efforts.
KEYWORDS: African swine fever virus, giant virus, mass spectrometry, NCLDV, proteome, proteomic analysis, virus composition, virus structure, immunoelectron microscopy
ABSTRACT
African swine fever virus (ASFV) is a large and complex DNA virus that causes a highly lethal swine disease for which there is no vaccine available. The ASFV particle, with an icosahedral multilayered structure, contains multiple polypeptides whose identity is largely unknown. Here, we analyzed by mass spectroscopy the protein composition of highly purified extracellular ASFV particles and performed immunoelectron microscopy to localize several of the detected proteins. The proteomic analysis identified 68 viral proteins, which account for 39% of the genome coding capacity. The ASFV proteome includes essentially all the previously described virion proteins and, interestingly, 44 newly identified virus-packaged polypeptides, half of which have an unknown function. A great proportion of the virion proteins are committed to the virus architecture, including two newly identified structural proteins, p5 and p8, which are derived from the core polyproteins pp220 and pp62, respectively. In addition, the virion contains a full complement of enzymes and factors involved in viral transcription, various enzymes implicated in DNA repair and protein modification, and some proteins concerned with virus entry and host defense evasion. Finally, 21 host proteins, many of them localized at the cell surface and related to the cortical actin cytoskeleton, were reproducibly detected in the ASFV particle. Immunoelectron microscopy strongly supports the suggestion that these host membrane-associated proteins are recruited during virus budding at actin-dependent membrane protrusions. Altogether, the results of this study provide a comprehensive model of the ASFV architecture that integrates both compositional and structural information.
IMPORTANCE African swine fever virus causes a highly contagious and lethal disease of swine that currently affects many countries of sub-Saharan Africa, the Caucasus, the Russian Federation, and Eastern Europe and has very recently spread to China. Despite extensive research, effective vaccines or antiviral strategies are still lacking, and many basic questions on the molecular mechanisms underlying the infective cycle remain. One such gap regards the composition and structure of the infectious virus particle. In the study described in this report, we identified the set of viral and host proteins that compose the virion and determined or inferred the localization of many of them. This information significantly increases our understanding of the biological and structural features of an infectious African swine fever virus particle and will help direct future research efforts.
INTRODUCTION
African swine fever virus (ASFV) is the causative agent of a highly lethal hemorrhagic disease of domestic pigs and wild boars for which there is no vaccine or antiviral strategy available. The disease, which has been endemic for many years in sub-Saharan Africa, has spread over the past decade through many countries of the Caucasus, the Russian Federation, and Eastern Europe, posing a serious risk of further expansion (1). Indeed, very recent outbreaks have been detected in China (2).
ASFV is the only member of the family Asfarviridae and the only known DNA arbovirus. Thus, the virus can be transmitted through direct contact with infected swine or their products or by soft ticks of the genus Ornithodoros. The family Asfarviridae belongs to the group of nucleocytoplasmic large DNA viruses (NCLDV), a monophyletic clade of genetically and structurally complex eukaryotic viruses (3, 4). The ASFV genome is a double-stranded DNA molecule of 170 to 190 kbp that contains between 151 and 167 open reading frames (ORFs), depending of the virus strain (5, 6). Like other NCLDVs, ASFV encodes many proteins dedicated not only to virus assembly but also to DNA replication and repair as well as gene expression. Also, the ASFV genome encodes a number of proteins involved in the evasion of host defenses, including type I interferon and cell death pathways (7). About half of ASFV genes lack any known or predictable function.
The ASFV particle possesses a multilayered structure with an overall icosahedral morphology and a diameter of about 200 nm. It consists of a genome-containing nucleoid, which is successively wrapped by a thick protein core shell, an inner lipid membrane, an icosahedral protein capsid, and an outer lipid membrane (8).
ASFV infects mainly swine monocytes and macrophages. The infective cycle commences with virus entry into the host cell by either clathrin-mediated endocytosis or macropinocytosis (9). Once internalized, the endocytosed particles undergo a stepwise, low-pH-driven disassembly process leading to inner envelope fusion at late endosomes and core delivery in the cytoplasm (10). The transcription of viral genes takes place in a temporally and spatially controlled sequence that is coordinated with viral DNA replication. Thus, the immediate early and early genes are expressed by the virion-packaged transcriptional machinery before the onset of DNA replication, whereas the intermediate and late genes are expressed afterwards (11). DNA replication and virus morphogenesis take place in specialized cytoplasmic areas close to the nucleus referred to as viral factories. In these areas, the assembling particle acquires its inner lipid envelope from membrane fragments derived from the endoplasmic reticulum (12, 13). Viral membranes become icosahedral particles by the progressive building of the outer capsid, while, concomitantly, the core material is enclosed (14). The intracellular mature particles move to the cell surface by microtubule-mediated transport (15) and exit by budding at the plasma membrane, where they acquire the outer envelope (16).
The protein composition of the ASFV virion is largely unknown. Early studies using both one- and two-dimensional gel electrophoresis of highly purified extracellular particles detected between 34 and 54 polypeptides ranging in size from 10 to 150 kDa (17, 18). Currently, about 25 virion proteins have been identified, and some of them have been localized in the virus structure by immunoelectron microscopy (8). Many of the known virion proteins were identified by N-terminal sequencing of the major protein bands detected after gel electrophoresis of purified virus. Other virus-packaged polypeptides, particularly some membrane proteins and viral enzymes, were detected in the virion by immunological approaches. No systematic proteomic approach has been employed so far to determine the protein content of the infectious ASFV particle.
In the study described in this report, we employed a mass spectrometry (MS)-based analysis to determine the proteins that compose the infectious ASFV particle. This proteomic analysis detected 68 viral polypeptides, including essentially all those previously described, as well as 21 cellular proteins. More importantly, 44 novel viral polypeptides, many of them with unknown function, were identified. By using immunoelectron microscopy, we determined the subviral localization of a significant proportion of the detected virion components, including host proteins. Altogether, our results provide a comprehensive model of the ASFV particle that combines both compositional and structural information and may guide future research on the biology of this complex virus.
RESULTS AND DISCUSSION
Purification and quality control of ASFV particles.
Extracellular ASFV particles were obtained using the Percoll gradient sedimentation method (17), which represents the gold standard for ASFV purification. In brief, supernatants of infected Vero cells were collected at 48 h postinfection, clarified to remove cell debris, and centrifuged through two successive Percoll density gradients. Then, the visible virus band was subjected to a gel filtration chromatography step.
The purified virions were examined by negative-staining electron microscopy to check both purity and virus integrity. As shown in Fig. 1A, the virus preparations were essentially free of cellular debris, and the virus particles presented their normal multilayered icosahedral structure. When examined by SDS-PAGE and Coomassie blue staining, they exhibited the characteristic protein profile of highly purified particles, with molecular masses ranging from approximately 150 to 10 kDa. As shown in Fig. 1B, prominent bands corresponding to the major capsid protein p72, the inner envelope protein p17, and the major DNA-binding protein p10, as well as several core shell proteins derived from the polyprotein precursors pp220 (p150, p37, and p34) and pp62 (p35), were visualized. In relation to this, the absence of the polyprotein precursors is considered a reliable indicator of virus purity. To test this, the virus stocks were analyzed in parallel with extracts of mock- and ASFV-infected cells by Western immunoblotting with antibodies that recognize the proteins pp220/p150 or pp62/p35. As shown in Fig. 1C, unique bands corresponding to the mature products were detected in the virus samples, while the polyprotein precursors were detected only in infected cell extracts.
FIG 1.
Quality control of purified ASFV particles. (A) Percoll-purified ASFV particles were analyzed by electron microscopy after negative staining. Note that ASFV virions look intact and essentially free of contaminant structures. Bar, 100 nm. (B) The virion proteins were analyzed by SDS-PAGE and stained with colloidal Coomassie blue. Some of the most prominent protein bands are indicated. (C) Western immunoblotting of extracts of mock-infected cells (lanes M) and ASFV-infected cells (lanes I) as well as purified ASFV (lanes V) with antibodies against the structural protein p150, derived from polyprotein pp220, and p35, derived from polyprotein pp62. Note the absence of the polyprotein precursors in the virus fraction, which are present in the infected cell extract.
Proteomic analysis of the ASFV particle.
Three independent stocks of Percoll-purified virus were analyzed by mass spectrometry. To this end, virion proteins were first electrophoresed by reducing SDS-PAGE followed by in-gel digestion with trypsin. The resulting tryptic peptide mixtures were subjected to reversed-phase high-performance liquid chromatography (HPLC) coupled via a nanospray source to a quadrupole time of flight (QTOF) spectrometer for tandem mass spectrometry (MS/MS), as described in Materials and Methods. Individual MS and MS/MS peptide spectra were analyzed using a modified ASFV protein database containing 157 putative viral proteins encoded by the BA71V strain. This includes all potential ORFs with 60 or more amino acids previously described (5), with the exception that ORFs CP2475L and CP530R, encoding polyproteins pp220 and pp62, respectively, were replaced by the amino acid sequences of their predicted mature products.
Mass spectrometry analysis identified 59 viral proteins present in all three virus samples (Table 1; marked in bold) and 9 additional viral proteins found in 2 of 3 virus stocks (Table 1; shown in plain text). We decided to include these 68 proteins in the list of virion components, as some of the additional proteins mentioned had been previously identified as such. Table 1 provides for each identified protein its corresponding ORF; a brief description, including available functional and structural information; the number of unique peptides matching the target protein in each virus sample; the maximal percentage of sequence coverage in the triplicate analysis; the estimated abundance; and the relevant reference(s). ASFV ORFs are designated according to the EcoRI fragment containing the 5′ end of the ORF, followed by the number of amino acids encoded and a letter, either L or R, indicating the direction of transcription (leftwards or rightwards, respectively). The letter P included in certain ORF names is used as a substitute for the prime symbol (i.e., CP530R is located in the EcoRI C′ fragment).
TABLE 1.
ASFV proteins identified by LC-MS/MSa
| ORF | Description/localization/function | Mass | No. of unique peptides |
Sequence coverage (%) | Abundance (%) | Previously detectede | Reference(s) | ||
|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | |||||||
| CP2475L | Protein p150, polyprotein pp220-derived/core shell/morphogenesis | 181.2 | 95 | 70 | 68 | 65.2 | 19.66 | Y | 14, 24, 32, 34 |
| CP2475L | Protein p34, polyprotein pp220-derived/core shell/morphogenesis | 36.6 | 24 | 22 | 21 | 68.2 | 10.43 | Y | 14, 24, 32, 34 |
| B646L | Major capsid protein p72/capsid/morphogenesis | 73.6 | 29 | 27 | 27 | 48.0 | 9.55 | Y | 27, 75 |
| CP2475L | Protein p37, polyprotein pp220-derived/core shell/morphogenesis | 41.5 | 21 | 19 | 21 | 62.5 | 7.15 | Y | 14, 24, 32, 34 |
| CP2475L | Protein p14, polyprotein pp220-derived/core shell/morphogenesis | 17.9 | 13 | 13 | 12 | 62.3 | 5.46 | Y | 14, 24, 32, 34 |
| CP530R | Protein p35, polyprotein pp62-derived/core shell/morphogenesis | 35.2 | 18 | 18 | 18 | 73.8 | 5.04 | Y | 24, 33, 35 |
| A104R | Histone-like DNA-binding protein/nucleoid/morphogenesisd | 11.6 | 5 | 7 | 6 | 69.2 | 4.64 | Y | 34, 39 |
| B962L | VACV I8-like RNA helicase/nucleoidd/transcriptiond | 110.2 | 12 | 11 | 12 | 14.6 | 2.48 | N | 76 |
| D1133L | VACV D6-like RNA helicase/nucleoidd/transcriptiond | 129.8 | 34 | 17 | 27 | 32.1 | 2.33 | N | 76 |
| NP1450L | RNA polymerase subunit 1/nucleoidd/transcriptiond | 164.6 | 31 | 20 | 28 | 22.3 | 2.16 | N | 77 |
| D117L | Major transmembrane protein p17/inner envelope/morphogenesis | 13.2 | 6 | 3 | 3 | 65.8 | 2.12 | Y | 31, 42 |
| M1249L | Uncharacterized protein | 145.3 | 33 | 20 | 20 | 32.0 | 2.07 | N | |
| CP123L | Uncharacterized protein/transmembrane domain | 14.1 | 11 | 7 | 5 | 50.4 | 1.96 | N | |
| CP530R | Protein p15, polyprotein pp62-derived/core shell/morphogenesis | 17.9 | 8 | 8 | 6 | 45.6 | 1.67 | Y | 24, 33, 35 |
| EP1242L | RNA polymerase subunit 2/nucleoidd/transcriptiond | 140.8 | 24 | 15 | 21 | 19.5 | 1.57 | N | 77 |
| G1340L | VACV A7 early transcription factor large subunit-like/nucleoidd/transcriptiond | 155.8 | 23 | 12 | 21 | 19.8 | 1.39 | N | 4 |
| C129R | Uncharacterized protein | 15.0 | 7 | 7 | 7 | 65.1 | 1.25 | N | |
| C717R | Uncharacterized protein | 84.1 | 14 | 13 | 13 | 18.1 | 1.12 | N | |
| A224L | Inhibitor of apoptosis protein (IAP)/nucleoid/host defense | 27.1 | 7 | 10 | 7 | 34.8 | 1.03 | Y | 60–62 |
| I177L | Uncharacterized protein; transmembrane domain | 20.5 | 6 | 6 | 9 | 63.3 | 1.01 | N | |
| K145R | Uncharacterized protein | 17.2 | 1 | 3 | 10 | 63.4 | 0.95 | N | |
| B438L | Protein p49/capsid/morphogenesis | 49.6 | 10 | 10 | 10 | 23.3 | 0.93 | Y | 29 |
| E248R | Transmembrane myristoylated protein pE248R/inner envelope/entry | 27.7 | 7 | 7 | 7 | 46.0 | 0.91 | Y | 10, 44 |
| Q706L | VACV D11-like helicase/nucleoidd/transcriptiond | 80.9 | 16 | 9 | 8 | 22.4 | 0.85 | N | 78 |
| K78R | DNA-binding protein p10/nucleoid/morphogenesisd | 8.4 | 3 | 2 | 3 | 39.7 | 0.85 | Y | 38 |
| NP868R | mRNA-capping enzyme/nucleoid/transcriptiond | 100.4 | 19 | 7 | 9 | 21.3 | 0.84 | Y | 53 |
| KP177R | Membrane protein p22/inner envelope | 20.7 | 7 | 5 | 7 | 46.9 | 0.79 | Y | 49 |
| C475L | Poly(A) polymerase/nucleoidd/transcriptiond | 55.1 | 11 | 7 | 6 | 21.7 | 0.63 | N | 4 |
| K421R | Uncharacterized protein | 49.7 | 6 | 7 | 8 | 17.8 | 0.58 | N | |
| E146L | Uncharacterized protein; transmembrane domain | 16.3 | 4 | 4 | 6 | 47.3 | 0.54 | N | |
| H359L | RNA polymerase subunit 3-11/nucleoidd/transcriptiond | 41.6 | 9 | 4 | 6 | 28.4 | 0.50 | N | 5 |
| F317L | Uncharacterized protein | 36.8 | 5 | 6 | 6 | 22.7 | 0.48 | N | |
| R298L | Serine/threonine-protein kinase 1 | 35.2 | 5 | 4 | 7 | 25.2 | 0.44 | Y | 66 |
| CP530R | Protein p8, polyprotein pp62-derived/core shelld/morphogenesisd | 7.8 | 3 | 3 | 3 | 56.7 | 0.41 | N | 33 |
| A137R | Protein p11.5 | 16.2 | 1 | 4 | 5 | 31.4 | 0.39 | Y | 63 |
| H240R | Uncharacterized protein | 27.7 | 6 | 4 | 4 | 27.9 | 0.38 | N | |
| CP312R | Uncharacterized protein | 35.2 | 2 | 2 | 8 | 33.3 | 0.35 | N | |
| E423R | Uncharacterized protein | 48.2 | 7 | 3 | 4 | 21.5 | 0.35 | N | |
| D205R | RNA polymerase subunit 5/nucleoidd/transcriptiond | 23.7 | 4 | 4 | 4 | 26.3 | 0.34 | N | 5 |
| O174L | DNA polymerase X/nucleoidd/DNA integrity | 20.4 | 3 | 3 | 3 | 15.5 | 0.24 | Y | 57, 58 |
| E199L | Cysteine-rich transmembrane protein pE199L/inner enveloped/entryd | 22.7 | 3 | 3 | 3 | 13.1 | 0.23 | Y | 41 |
| NP419L | DNA ligase/nucleoidd/DNA integrityd | 48.5 | 3 | 2 | 5 | 8.8 | 0.23 | N | 56 |
| E184L | Uncharacterized protein | 22.0 | 2 | 2 | 4 | 19.6 | 0.23 | N | |
| O61R | Virus attachment membrane protein p12/inner envelope/entry | 6.9 | 1 | 1 | 2 | 27.9 | 0.21 | Y | 40, 43 |
| S273R | Polyprotein processing protease/core shell/morphogenesis | 32.0 | 2 | 3 | 3 | 12.1 | 0.20 | Y | 36, 37 |
| D339L | RNA polymerase subunit 7/nucleoidd/transcriptiond | 39.1 | 2 | 3 | 3 | 11.2 | 0.20 | N | 5 |
| C257L | Uncharacterized protein; transmembrane domain | 30.2 | 2 | 2 | 3 | 12.1 | 0.19 | N | |
| H171R | Uncharacterized protein | 19.9 | 2 | 2 | 3 | 19.3 | 0.18 | N | |
| EP152R | Protein EP152R; transmembrane domain | 18.0 | 1 | 1 | 3 | 19.1 | 0.17 | N | 79 |
| B117L | Uncharacterized protein; transmembrane domain | 13.4 | 2 | 2 | 2 | 12.0 | 0.16 | N | |
| EP424R | Putative RNA methyltransferase/nucleoidd/transcriptiond | 49.6 | 2 | 1 | 4 | 10.8 | 0.15 | N | 4 |
| B169L | Uncharacterized protein; two transmembrane domains | 18.8 | 2 | 2 | 2 | 16.6 | 0.15 | N | |
| H339R | Alpha-NAC binding protein pH339R | 40.0 | 1 | 1 | 4 | 10.6 | 0.14 | N | 65 |
| CP204L | Early phosphoprotein p32 | 23.6 | 2 | 1 | 3 | 11.8 | 0.13 | Y | 64, 80 |
| E296R | AP endonuclease/nucleoid/DNA integrity | 33.9 | 2 | 2 | 2 | 6.4 | 0.13 | N | 55 |
| E183L | Transmembrane protein pE183L/inner envelope/morphogenesis | 19.9 | 1 | 1 | 1 | 6.6 | 0.12 | Y | 30, 81 |
| EP84R | Uncharacterized protein; two transmembrane domains | 9.0 | 1 | 1 | 1 | 23.8 | 0.12 | N | |
| C147L | RNA polymerase subunit 6/nucleoidd/transcriptiond | 16.8 | 2 | 1 | 2 | 15.0 | 0.11 | N | 82 |
| QP383R | Uncharacterized protein | 42.9 | 2 | 2 | 1 | 7.0 | 0.11 | N | |
| E165R | dUTPase/nucleoidd/DNA integrity | 18.6 | 1 | 3 | 22.4 | 0.16 | N | 59 | |
| I73R | Uncharacterized protein | 8.5 | 1 | 2 | 26.0 | 0.14 | N | ||
| B119L | FAD-linked sulfhydryl oxidase | 14.5 | 2 | 2 | 16.0 | 0.14 | N | 67, 68 | |
| C122R | Uncharacterized protein | 12.4 | 1 | 2 | 11.4 | 0.11 | N | ||
| CP2475L | Protein p5, polyprotein pp220 derived/core shell/morphogenesis | 4.7 | 1 | 1b | 43.2 | 0.11 | N | 32 | |
| E120R | Protein pE120R/capsid/morphogenesis | 13.6 | 1 | 1 | 9.2 | 0.08 | Y | 28 | |
| H124R | Uncharacterized protein | 15.0 | 1 | 1 | 4.8 | 0.08 | N | ||
| EP402R | Membrane glycoprotein similar to host CD2/outer envelope/host defense | 46.5 | 1 | 1 | 2.5 | 0.06 | Y | 45–47 | |
| M448R | Uncharacterized protein | 52.4 | 1 | 1 | 2.0 | 0.05 | N | ||
| H108Rc | Transmembrane protein pH108R/inner envelope | 12.5 | 1 | 13.9 | 0.05 | Y | 26 | ||
The data presented in Table 1 were compiled from three different ASFV samples (S1, S2, S3). ORFs present in all three samples (S1, S2, and S3) are in bold.
Protein p5 was detected in the global proteome of one virus stock and in the low-molecular-weight analysis of a second one.
Protein pH108R was detected in only one of the analyzed stocks but is included for reference as it was previously described as a structural component.
Proteins with a predicted function or with a subviral localization inferred from their known or predicted role.
Y, yes; N, no.
The 68 identified proteins represent 39% of the genome coding capacity of ASFV strain BA71V. A somewhat larger number of proteins has been described for the intracellular form of vaccinia virus (VACV), which contains 75 to 80 out of more than 200 putative encoded proteins (19, 20). The proteome of other related NCLDVs with a larger coding capacity, such as Cafeteria roebergensis virus or faustovirus, contains 141 (out of 544 predicted proteins) or up to 164 polypeptides (out of 451 predicted proteins), respectively (21, 22).
More than 90% of the detected ASFV proteins were identified by at least two unique peptides covering a protein sequence that ranges from 6 to 74%. The identified proteins are sorted in Table 1 in order of decreasing estimated abundance (percentage of the virion protein mass), according to the exponentially modified protein abundance index (emPAI) score, which is based on the correlation of the peptides identified by mass spectrometry with the predicted peptides obtained by in silico digestion (23). Using these estimates of protein abundance and the corresponding molecular weights, the major capsid protein p72 (B646L) together with the core shell proteins p150, p37, p34, and p14, derived from polyprotein pp220 (CP2475L), and p35 and p15, derived from polyprotein pp62 (CP530R), accounts for about two-thirds of the virion protein mass. This value is in general agreement with the values presented in previous reports of studies based on gel densitometry of ASFV proteins labeled with [14C]amino acids or stained with Coomassie blue (24). In addition, the quantitative analysis revealed that most of the virion proteins were present at a low abundance. Indeed, only 20 proteins were present with an estimated abundance higher than 1%.
Interestingly, this ASFV proteome essentially contains all the virus proteins previously reported to be present in the virus particle (8), supporting the confidence in this proteomic approach. It includes the major capsid protein p72 (ORF B646L); the mature proteins derived from polyprotein pp220 (CP2474L) and polyprotein pp62 (CP530R); the major DNA-binding proteins p10 (K78R) and pA104R; the membrane proteins p12 (O61R), p17 (D117L), p22 (KP177R), pE183L, pE248R, pE199L, and pEP402R; the phosphoproteins p32 (CP204L) and pE120R; the apoptosis inhibitor IAP (A224L); the proteins p49 (B438L) and p11.5 (A137R); and the enzymes encoded by ORFs S273R (cysteine proteinase), R298L (serine/threonine-protein kinase 1), and O174L (DNA polymerase X). To our knowledge, only two proteins previously described as virion components, the ubiquitin-conjugating enzyme pI215L (25) and the membrane protein pH108R (26), are not present in this ASFV proteome. Thus, protein pH108R was detected in just 1 out of 3 analyzed virus samples (Table 1), and protein pI215L was not detected in any virus stock. Importantly, the virus proteome contains 44 new virus-packaged polypeptides, which represents a very significant increase in relation to our previous knowledge on the ASFV protein content. Among these, more than 20 uncharacterized proteins have no significant similarity with proteins of known function.
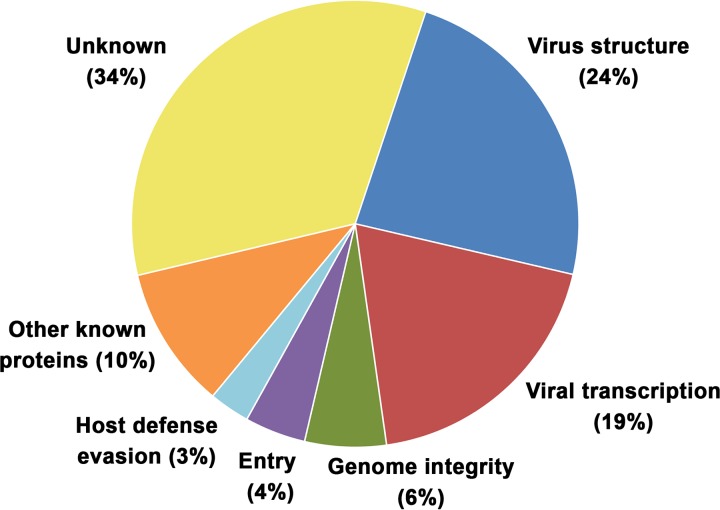
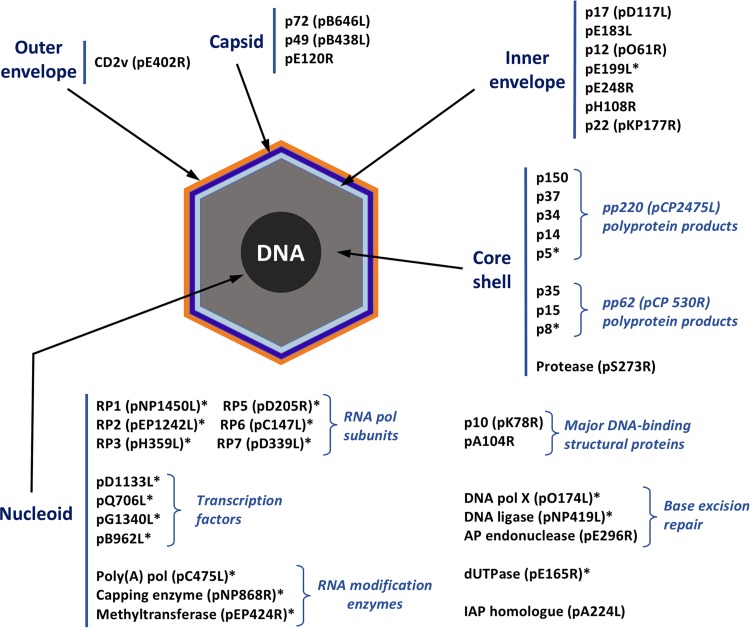
Taking into account the available information on the identified virus-packaged proteins, these can be grouped into several functional categories (Fig. 2). Most of the virion proteins are involved in virus morphogenesis (24%) and viral gene transcription (19%). Other categories with fewer representatives include maintenance of genome integrity (6%), virus entry (4%), evasion of host defenses (3%), and other known proteins (10%). The group of proteins of unknown function remained, however, the most abundant category (34%).
FIG 2.
Functional classification of ASFV virion proteins. The viral proteins identified in the ASFV proteome are assigned to the following functional categories: virus structure and morphogenesis (p72, p150, p37, p34, p14, p5, p35, p15, p8, pS273R, p17, pE183L, p49, pE120R, p10, and pA104R), viral transcription and RNA modification (pB962L, pD1133L, pNP1450L, pEP1242L, pG1340L, pQ706L, pNP868R, pC475L, pH359L, pD205R, pD339L, pEP424R, and pC147L), maintenance of genome integrity (pO174L, pNP419L, pE296R, and pE165R), virus entry (p12, pE248R, and pE199L), evasion from host defense (pEP402R and pA224R), other known proteins (pR298L, pB119L, p22, p32, p11.5, pEP152R, and pH339R), and unknown proteins (pM1249L, pCP123L, pC129R, pC717R, pI177L, pK145R, pK421R, pE146L, pF317L, pH240R, pCP312R, pE423R, pE184L, pC257L, pH171R, pB117L, pB169L, pEP84R, pI73R, pC122R, pQP383R, pM448R, and pH124R).
Virion proteins involved in morphogenesis.
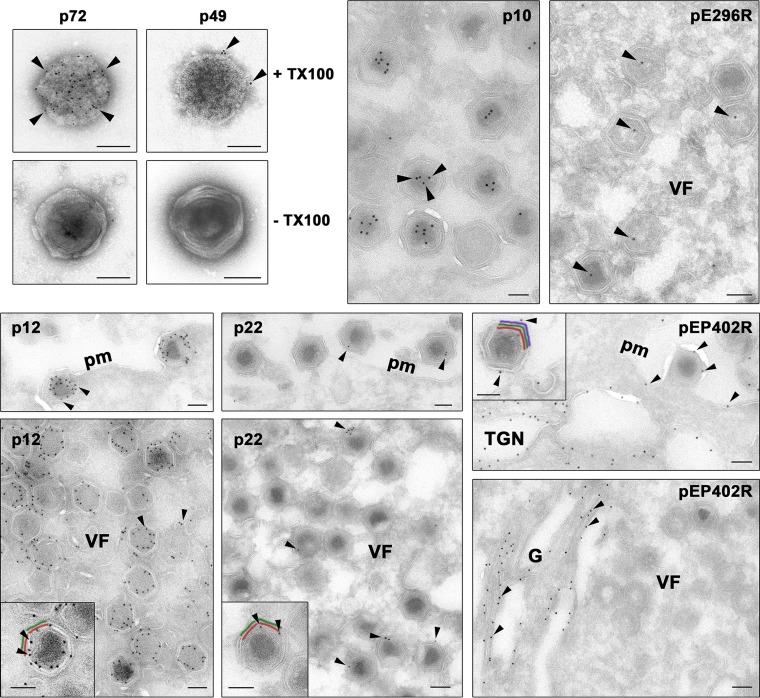
At least 16 of the 68 detected ASFV proteins are thought to be involved in the assembly of the virus particle (8). Consistent with their structural role, most of them have higher estimated abundance values and sequence coverages than other ASFV components (Table 1). The function of many of these proteins has been analyzed by using IPTG (isopropyl-β-d-thiogalactopyranoside)-dependent ASFV recombinants which inducibly express the protein of interest (8). Thus, it has been shown that the assembly of the icosahedral capsid on the inner envelope requires the major capsid protein p72 (ORF B646L) (27) but not the capsid protein pE120R, which is actually involved in the microtubule-mediated transport of the assembled particles from the virus factories to the cell surface (28). Another putative capsid component is protein p49 (B438L), which has been shown to be required for the formation of the capsid vertices but has never been localized (29). To determine the distribution of protein p49 in the virus particle, we performed immunoelectron microscopy on intact virions or on virus particles treated with 0.1% Triton X-100 to remove the lipid membranes and expose the capsid layer (13). As a control, the major capsid protein p72 was also analyzed. As shown in Fig. 3, the detergent treatment considerably increased the signal observed with both p72 (from 1.5 ± 2.0 to 36.8 ± 12.1 gold particles/virion, n = 100) and p49 (from 0.1 ± 0.5 to 6.3 ± 2.7 gold particles/virion, n = 100) antibodies. Interestingly, whereas protein p72 appeared homogeneously distributed on the viral capsid, protein p49 was located in close proximity to the capsid vertices, which suggests that it may be a component of those structures.
FIG 3.
Immunoelectron microscopy of viral proteins in the virus particle. To localize protein p49, ASFV particles were treated or not with 0.1% Triton X-100 (TX100) and incubated with a control antibody against the major capsid protein p72 or an antibody against p49, followed by protein A-gold (5 nm). Note that both anti-p72 and anti-p49 antibodies stained the capsid on detergent-treated particles. To localize proteins p10, pE296R, p12, p22, and pEP402R, cryosections of ASFV-infected cells were incubated with specific antibodies followed by protein A-gold (10 nm). Note that both anti-p10 and anti-pE296R labeled the nucleoid region, whereas the anti-p12 and anti-p22 antibodies labeled the inner viral envelope of virus particles at the virus factories (VF) and the plasma membrane (pm). The anti-pEP402R serum mainly stained the Golgi stack (G) as well as presumed trans-Golgi network membranes. A minor fraction of pEP402R was also found on the plasma membrane and on the outer viral envelope of budding ASFV particles. (Insets) Details of labeling of intracellular (p12 and p22) and extracellular (pEP402R) particles. The arrowheads indicate, for each case, examples of the described distribution patterns. To facilitate the interpretation, the inner membrane (red), capsid (green), and outer membrane (purple) are depicted in color. Bars, 100 nm.
With regard to the viral membrane polypeptides, protein pE183R has been found to be essential for the formation of the inner envelope (30), while another inner envelope component, protein p17 (ORF D117L), is required for the assembly of the capsid layer on this membrane (31).
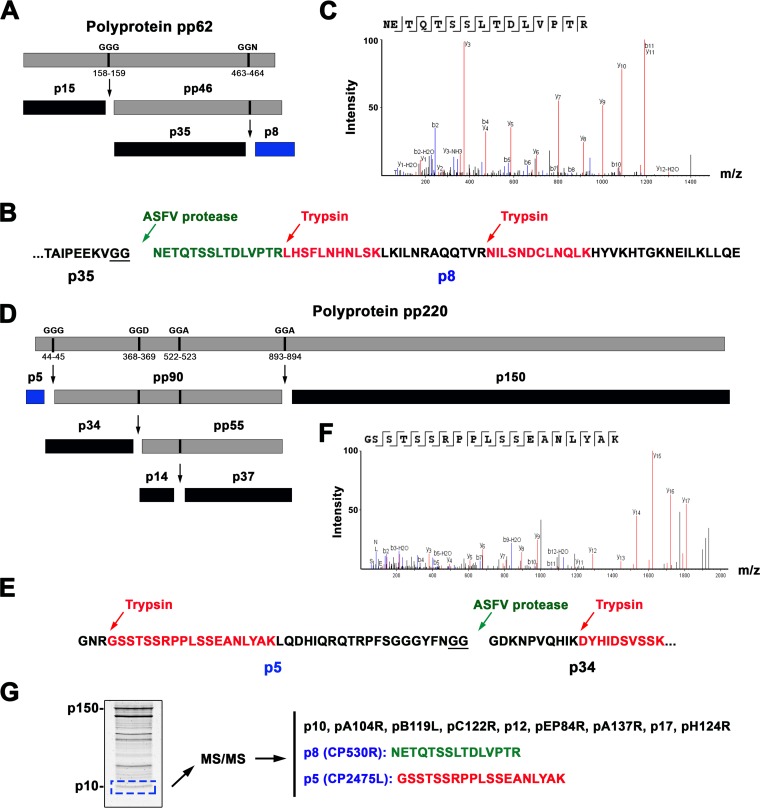
As mentioned before, ASFV encodes two polyprotein precursors, pp220 and pp62 (32, 33), whose mature products coassemble into the core shell that surrounds the DNA-containing nucleoid (24). Indeed, inducible ASFV recombinants for either of the polyproteins lead to the assembly of nearly empty icosahedral particles (34, 35). Importantly, the proteomic analysis identified all the predicted mature products derived from both polyprotein precursors, including two small proteins, p8 and p5, which had never been previously detected. Protein p8 is a 67-amino-acid mature product derived from the proteolytic processing of polyprotein pp62 (ORF CP530R). It is known that this processing takes place through an ordered cascade of cleavages by the viral proteinase pS273R at the consensus sequence GGX (32, 33). As shown in Fig. 4A, a first cleavage at the GGG site at positions 158 and 159 gives rise to the mature protein p15 and an intermediate precursor (pp46), which, after a subsequent cleavage at the GGR site at positions 463 and 464, generates the mature proteins p35 and p8. Interestingly, while p15 and p35 are easily detected with specific antibodies in both infected cells and purified virus, p8 has never been detected by either immunoprecipitation or immunoblotting using antibodies raised against the whole pp46 region (33). The proteomic analysis identified protein p8 with high abundance and significance scores (Table 1) through three matching peptides (Fig. 4B). Two of them correspond to tryptic fragments, whereas the other one (NETQTSSLTDLVPTR), which matches exactly the N terminus of p8 (Fig. 4B and C), would derive from a cleavage by the viral protease at the GGR site at positions 463 and 464. This finding rules out the possibility that this peptide derives from contaminant traces of the polyprotein precursor rather than from the mature product.
FIG 4.
Two novel structural ASFV proteins, p8 and p5, are derived from the core polyproteins pp62 and pp220, respectively. (A) Schematic representation of polyprotein pp62 processing at GGX cleavage sites. The polyprotein pp62, the intermediate precursor pp46, and the mature products p15, p35, and p8 (in blue) are indicated. (B) Detection of p8 in the ASFV proteome. Protein p8 was identified by two tryptic peptides (highlighted in red) and one peptide (green) matching the N-terminal p8 sequence, which is generated by cleavage by the ASFV proteinase at positions 463 and 464 of the GGR site. (C) MS/MS spectrum of the N-terminal nontryptic p8 peptide NETQTSSLTDLVPTR. (D) Schematic representation of polyprotein pp220 processing at GGX cleavage sites. The polyprotein pp220, the intermediate precursors pp90 and pp55, and the mature products p5 (in blue), p34, p14, p150 are indicated. (E) Detection of protein p5 through one tryptic peptide (in red) covering 43% of its amino acid sequence. (F) MS/MS spectrum of the tryptic peptide GSSTSSRPPLSSEANLYAK. (G) Proteomic analysis of the low-molecular-weight virion proteins. ASFV proteins were separated by SDS-PAGE, and the low-molecular-weight region was excised and in-gel digested with trypsin. MS/MS analysis identified all the small viral proteins, including p8 and p5.
The other newly detected polyprotein product, p5, is a predicted protein of 43 amino acids derived from the precursor pp220 after cleavage at the GGG site at positions 44 and 45 (32) (Fig. 4D). Protein p5 had not been previously detected, and, as for the p8 protein, this was interpreted to be a consequence of its small size, its low immunogenicity, its rapid degradation, or a combination of these factors (32). Protein p5 was identified by a unique tryptic peptide covering 43% of its amino acid sequence (Fig. 4E and F). To discard the possibility that this peptide derives from the precursor pp220 and not from a genuine p5 protein, we performed an additional mass spectrometry analysis specifically targeting the region of low molecular weight (ranging from 12 to 5 kDa) of the electrophoretic ASFV profile. As shown in Fig. 4G, the MS/MS analysis confirmed the presence of proteins p5 and p8, together with the remaining small polypeptides previously detected in the whole ASFV proteome (p10 [pK78R], pA104R, pB119L, pC122R, p12 [pO61R], pEP84R, p11.5 [pA137R], p17 [pD117L], and pH124R). Altogether, these results indicate that the polyprotein-derived products p5 and p8 are structural components of the virus particle. In this context, it is also interesting to note the presence in the virion of the proteinase pS273R, responsible for polyprotein processing. Indeed, it localizes at the core shell of the virus particle (36), maybe as a reminiscence of its key late role in core assembly or because of a putative early role at the first stages of ASFV infection (37).
Two detected major DNA-binding proteins, pA104R and p10 (pK78R), might play a role in the assembly of the DNA-containing nucleoid (38). We analyzed the localization of protein p10 in the virion by immunogold labeling on cryosections of ASFV-infected cells fixed at 18 h postinfection (hpi). As shown in Fig. 3, antibodies against p10 strongly labeled the electron-dense DNA-containing nucleoid of mature ASFV particles, which is consistent with a hypothetical structural role in this viral domain that deserves further research. The second structural DNA-binding protein, pA104R, is a histone-like protein, also located at the virus nucleoid (34), that, on the basis of knockdown experiments with small interfering RNAs, has been involved in viral transcription, DNA replication, and genome packaging (39). Thus, proteins p10 and pA104R are ideal targets to study unexplored ASFV processes relating to nucleoid formation and genome encapsidation.
Membrane proteins.
Of the 26 virus-encoded proteins with predicted transmembrane domains (5), 15 were detected in the virus proteome, which represents 22% (15 of 68) of the total virus-packaged proteins. Whereas some have been studied with some detail (p17 [pD117L], pE183L, p12 [pO61R], p22 [pKP177L], pE248R, pE199L, pEP152R, pEP402R) (30, 31, 40–47), about half of them remain uncharacterized. This includes proteins pCP123L, pI177L, pE146L, pC257L, and pB117L, which contain a single putative transmembrane segment, and proteins pB169L and pEP84R with two membrane domains.
It is interesting to note that the majority of the previously studied virus-packaged membrane proteins localize at the inner viral envelope. Indeed, immunoelectron microscopy studies have shown that proteins p17, pE183L, p12, and pE248R, as well as pH108R, localize at precursor viral membranes and at intracellular icosahedral particles within the viral factories but not at the cell surface (8, 26, 31, 44). As commented before, proteins p17 and pE183L are required for the assembly process, while the proteins p12, pE248R, and pE199L have been related to virus entry. Thus, an inducible ASFV recombinant defective in the pE248R protein is not able to fuse its inner envelope with the endosomal membrane to deliver cytoplasmic cores (10). Interestingly, protein pE248R and also protein pE199L share sequence similarity with members of the poxvirus entry/fusion complex (10, 48), which suggest that they may be components of the ASFV fusion machinery. In relation to protein p12 (pO61R), it was initially described to be involved in the virus attachment process since the purified protein binds to host cells, preventing virus binding (43). However, immunoelectron microscopy studies have shown that p12 is actually located at the inner viral envelope (8). Further research is hence needed to clarify its role as an attachment protein.
The viral membrane protein p22 (pKP177L) has been suggested to be externally located in the virion because it is solubilized from virus particles treated with a nonionic detergent (49). However, such treatment is known to disrupt both the inner and outer membranes, solubilizing even some core proteins, such as protein p35 (13, 40). One known viral membrane protein whose role is compatible with an external location is the protein pEP402R, a homologue of the T-lymphocyte surface antigen CD2 (46). Protein pEP402R has been shown to be responsible for the adhesion of erythrocytes to infected cells and for the binding of ASFV particles to erythrocytes. Intriguingly, it has been localized to the Golgi network around the virus factories but not at the cell surface (47, 50). To ascertain the subviral localization of membrane proteins p22 and pEP402R, we performed immunogold labeling on cryosections of ASFV-infected cells. As a control, the subcellular distribution of the inner membrane protein p12 was also analyzed. As shown in Fig. 3, anti-p12 antibodies strongly labeled curved membrane precursors of the inner envelope as well as intracellular particles at the virus factories but not the cell surface. On the other hand, protein p22 was weakly detected throughout the cytoplasm, including the virus factories, where the signal was mainly associated with the periphery of assembling and mature icosahedral particles. Interestingly, the p22 labeling at the budding particles on the cell surface was similar to that found on the virus particles at the virus factories. Altogether, these results are consistent with a localization of protein p22 at the inner envelope. Finally, anti-pEP402R antibodies intensely labeled the membrane stacks of the Golgi complex as well as intracellular membrane vesicles compatible with trans-Golgi network cisternae but not the virus factories. Remarkably, a minor fraction of the pEP402R protein was also detected on the cell surface and on the outer viral membrane of the budding particles. At present, the viral CD2 homologue remains the sole characterized viral protein that is externally located.
ASFV transcriptional machinery.
One interesting finding of this study is the detection in the ASFV particle of a set of viral enzymes and transcription factors involved in viral transcription and mRNA modification. These results are in full agreement with previous evidences showing that purified ASFV particles are able to synthesize viral mRNAs with methylated cap structures at their 5′ ends and poly(A) tails at their 3′ ends (11, 51, 52). Thus, all the predicted RNA polymerase subunits (RPBs) (3, 4, 11), namely, pNP1450L (RPB1), pEP1242L (RPB2), pH359L (RPB3-11), pD205R (RPB5), pC147L (RPB6), and pD339L (RPB7) but not pCP80R (RPB10), were detected in the virus proteome. Additionally, the proposed capping enzyme (pNP868R), methyltransferase (pEP424R), and polyadenylation enzyme (pC475L) (4, 53) were also identified.
Besides this transcriptional machinery, the proteomic analysis identified several proteins potentially involved in early viral transcription on the basis of their similarity with VACV enzymes (3, 4, 11). These included protein pG1340L and the putative helicase pD1133L, which are similar to the large (A7L) and small (D6R) subunits, respectively, of the VACV early gene transcription factor. Other detected helicase domain-containing proteins (ORFs B962L and Q706L) display similarity to VACV I8 and D11 proteins (11), which have been involved in the initiation and termination, respectively, of early poxviral transcription.
The presence of a complex transcriptional machinery with several common elements packaged in the viral particle is a hallmark of the NCLDVs and serves to ensure its independence from the host machinery for early viral transcription.
Maintenance of genome integrity.
All three components of a proposed base excision repair (BER) system (54) were detected in the ASFV particle. It includes a class II apurinic/apyrimidinic (AP) endonuclease (pE296R) (55), an ATP-dependent DNA ligase (pNP419L) (56), and a type X DNA polymerase (pO174L) that can efficiently fill single nucleotide gaps and is the smallest DNA polymerase described to date (57, 58). It is though the ASFV BER system corrects the DNA lesions induced in the highly oxidative environment of the host macrophage (54). Consequently, its presence in the virion could ensure the protection of the incoming viral genome from genotoxic agents. It is worth noting that the virion of faustovirus, the closest evolutionary relative of ASFV among the NCLDVs described to date, contains orthologues of these three ASFV enzymes (22).
To further support the possible presence of the BER system in the virus particle, we performed immunogold staining with an antibody against the viral AP endonuclease (pE296R) on cryosections of ASFV-infected cells. As shown in Fig. 3, a weak but significant signal was detected in the core region of immature icosahedral particles and in the nucleoid of mature virions.
Another enzyme that can contribute to ensure the fidelity of genome replication is the viral dUTPase (pE165R), which seems to play a role in lowering the dUTP concentration to minimize misincorporation of deoxyuridine into the viral DNA (59). The viral dUTPase activity has been shown to be dispensable for virus replication in dividing Vero cells but not in nondividing swine macrophages. Interestingly, no other viral enzymes involved in DNA replication, such as DNA polymerase type B (ORF G1207R), DNA primase (C962R), or DNA topoisomerase type II (P1192R), were detected in the ASFV proteome. In general, DNA replication enzymes are synthesized from early mRNAs produced by the viral transcriptional machinery upon core penetration in the cytoplasm. Their absence from the proteomic analysis therefore supports the low level of contaminant proteins in the analyzed virus samples.
Proteins involved in modulating host defenses.
The ASFV proteome contains at least two viral proteins, pA224L and pEP402R, which might interfere with host defense mechanisms. Protein pA224L is a nucleoid-packaged, nonessential homologue of the cellular inhibitor of the apoptosis protein (IAP) (60, 61) which has been shown to be able to protect cells from apoptosis by inhibition of caspase 3 (62). Interestingly, IAPs are found in other insect-infecting viruses, like baculoviruses and entomopoxviruses, supporting the proposed notion that it may be of importance during ASFV infection of its tick host.
As mentioned before, the presence of pEP402R at the outer viral envelope mediates the attachment of extracellular virions to red blood cells, which can facilitate virus spread. Interestingly, this viral CD2 homologue also possesses immunosuppressive activity in vitro by inhibiting lymphocyte proliferation (45).
Other viral proteins.
Other described virus-packaged polypeptides are the late viral protein p11.5 (pA137R), which localizes at the virus factories (63); the phosphoprotein p32 (pCP204L), an abundant and highly immunogenic cytoplasmic protein (64); and the late protein pH339R, which binds to the alpha chain of the nascent polypeptide-associated complex (alpha-NAC) (65). The list of detected ASFV enzymes also includes a serine/threonine-protein kinase 1 (pR298L) (66) and a sulfhydryl oxidase (pB119L) that is involved in disulfide bond formation (67) and whose deletion in the virus genome causes attenuation (68). It is interesting to note that the virion proteins pE296R and pE248R contain cytoplasmic intramolecular disulfide bonds and could be substrates for the viral sulfhydryl oxidase (67).
Host proteins.
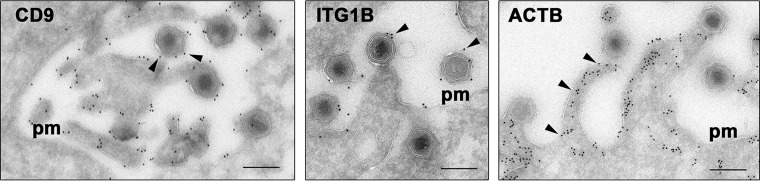
In addition to the ASFV-encoded proteins, 21 cellular proteins were identified in the three viral stocks analyzed (Table 2). The list of host proteins includes several metabolic enzymes (enolase 1, glyceraldehyde-3-phosphate dehydrogenase, triosephosphate isomerase, and gamma-glutamyltransferase 1), two major cytoskeletal proteins (beta-tubulin and beta-actin), various small GTPases (RAB5C, CDC42, and RHOA), and a large number of membrane proteins (tetraspanin CD9; glucose transporter SLC2A1; annexins A1, A2, and A4; and integrins beta 1, alpha 3, and V). Given the high sensitivity of the proteomic analysis, the detected host proteins might represent copurified contaminants. However, it is also possible that the some of these cellular proteins are incorporated in the virion during its assembly, transport, or egress. In this respect, it is known that the intracellular mature particles exit by budding through virus-induced actin-mediated filopodia (69). Interestingly, many of the identified host proteins transiently or stably localize at the cell surface (RAB5C, CDC42, RHOA, EEF1A1, CD9, SLC2A1, BSG, ANXA1, ANXA2, ANXA4, ITGB1, ITGA3, and ITGAV) and interact with the cortical actin cytoskeleton (CDC42, RHOA, EEF1A1, CD9, ANXA1, ANXA2, ITGAB1, ITGA3, and ITGAV). We therefore performed immunoelectron microscopy to test the recruitment of some of the detected host proteins (tetraspanin CD9, integrin beta 1, and beta-actin) to the outgoing ASFV particles. As shown in Fig. 5, antibodies against CD9 and integrin beta 1 abundantly labeled the cell surface as well as the outer envelope of budding virus particles. Interestingly, a large proportion of these budding particles were detected at the tip of membrane protrusions strongly decorated with antiactin antibodies (Fig. 5). These observations warrant the study of the potential roles of virus-packaged host proteins in the ASFV replication cycle.
TABLE 2.
Host proteins identified in ASFV particles by LC-MS/MSa
| UniProt accession no. | Description | Symbol | No. of unique peptides | Sequence coverage (%) |
|---|---|---|---|---|
| A0A0D9RI07 | Annexin A2 | ANXA2 | 17 | 47.5 |
| A0A0D9S9M0 | Beta-actin | ACTB | 16 | 52.5 |
| A0A0D9RG78 | Integrin beta | ITGB1 | 12 | 15.4 |
| A0A0D9QYT4 | Integrin subunit alpha 3 | ITGA3 | 9 | 9.4 |
| A0A0D9RED0 | Tetraspanin CD9 | CD9 | 4 | 9.6 |
| A0A0D9RNJ1 | Elongation factor 1-alpha | EEF1A1 | 10 | 23.2 |
| A0A0D9S441 | Heat shock protein family A (Hsp70) member 8 | HSPA8 | 13 | 22.3 |
| A0A0D9R7E7 | Tubulin beta chain | TUBB | 11 | 30.6 |
| A0A0D9RDR4 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 6 | 19.7 |
| A0A0D9REB3 | Gamma-glutamyltransferase 1 | GGT1 | 6 | 9.6 |
| A0A0D9S8A9 | Cell division cycle 42 | CDC42 | 4 | 25.7 |
| A0A0D9RRC2 | Annexin A4 | ANXA4 | 7 | 23.1 |
| A0A0D9R740 | Annexin A1 | ANXA1 | 5 | 15.6 |
| A0A0D9RJD3 | Integrin subunit alpha V | ITGAV | 5 | 5.0 |
| A0A0D9S8Q7 | Enolase 1 | ENO1 | 4 | 7.6 |
| A0A0D9S247 | RAB5C, member RAS oncogene family | RAB5C | 4 | 19.2 |
| A0A0D9R6C1 | Histone H2B | HIST1H2BD | 2 | 15.9 |
| A0A0D9S7K0 | Solute carrier family 2 member 1 | SLC2A1 | 2 | 3.4 |
| A0A0D9RKP4 | Basigin (Ok blood group) | BSG | 3 | 7.5 |
| A0A0D9RRD7 | Ras homolog family member A | RHOA | 2 | 13.0 |
| A0A0D9RCV3 | Triosephosphate isomerase | TPI1 | 1 | 4.5 |
The number of unique peptides and the percentages of sequence coverage correspond to the maximal values among the 3 virus proteomes.
FIG 5.
Immunoelectron microscopy of host proteins recruited to the virus particle. Thawed cryosections of ASFV-infected cells were stained with mouse MAbs against the tetraspanin CD9, integrin beta 1 (ITGB1), and beta-actin (ACTB), followed by rabbit anti-mouse immunoglobulins and protein A-gold (10 nm). Note that anti-CD9 and integrin beta 1 antibodies decorated the plasma membrane (pm) and the outer envelope of the virus particles released by budding. Note also that antiactin antibodies stained the membrane protrusions induced by the budding ASFV particles. The arrowheads indicate, for each case, examples of the described distribution patterns. Bars, 200 nm.
Distribution of ASFV proteins in the virion structure.
On the basis of the published evidence as well as the results presented here, we propose an ASFV model that combines both compositional and structural information. This virion atlas (Fig. 6) provides the distribution of 40 viral proteins in the five virus structural domains. In this model, most of the detected proteins are localized at the DNA-containing nucleoid, including the major DNA-binding proteins pA104R (34, 39) and p10 (pK78R), whose localization has been determined by immunoelectron microscopy (Fig. 3), along with a set of putative transcription factors and viral enzymes involved in mRNA transcription and modification. It is known that ASFV entry leads to the delivery of naked cores into the cytoplasm (10). Therefore, it seems reasonable to hypothesize that this transcriptional machinery is located at the viral nucleoid in order to ensure that early virus genes are properly transcribed and modified for subsequent translation. In this connection, VACV enzymes involved in early transcription are known to be associated with the viral core (70). Similarly, the viral dUTPase (pE165R) along with the set of three enzymes involved in DNA repair can also be tentatively located in this viral domain on the basis of its putative function. The localization of the viral A/P endonuclease (pE296R) at the nucleoid (Fig. 3) supports this assumption. Finally, the viral IAP homologue (pA224L) has also been located at the virus nucleoid (60).
FIG 6.
ASFV atlas. The subviral localization of 40 viral proteins among the five structural domains of the ASFV particle is shown. The distribution of proteins marked with an asterisk was inferred from the predicted or known role, while that of the remaining proteins was determined by immunoelectron microscopy. pol, polymerase.
The core shell is essentially composed of the mature products derived from the proteolytic processing of polyproteins pp220 (ORF CP2475L) (14) and pp62 (CP530R) (24), which represent about one-third of the total virion mass (24), and the viral protease (36). The protein icosahedral capsid consists of the major capsid component p72 (27) and the minor capsid protein p49 (Fig. 3), both of which are involved in the assembly of this domain (27, 29), as well as the protein pE120R, which mediates the intracellular virus transport (28).
At least seven known membrane viral proteins (p17 [pD117L], pE183L, p12 [pO61R], p22 [pKP177L], pH108R, pE199L, and pE248R) localize at the inner envelope, while only one viral protein, the CD2 homologue pEP402R, has been unambiguously located at the outermost membrane (Fig. 3). A possible reason for this uneven distribution is the pivotal role of the inner envelope in both the assembly and the entry of the ASFV particle. Thus, the inner envelope should contain the proteins needed to transform and stabilize an endoplasmic reticulum-derived membrane into a viral membrane (8). Also, the inner membrane acts as a scaffold to support the multiple interactions required to assemble the outer capsid and the internal core shell (8). Finally, this membrane contains the fusion machinery required for the cytoplasmic core delivery that follows virus entry (9), which in the case of the related VACV includes up to 12 different polypeptides (71). At variance with many other enveloped viruses, the budding process of ASFV is not concomitant with the assembly of a mature infectious particle. Indeed, the intracellular mature particles produced at the virus factories are infectious (28). This could explain, at least partly, the minor presence of viral proteins in the outer envelope. Despite this, it will be highly relevant to identify in future, specifically targeted studies the components of the outer viral membrane. These will most probably be involved in virus exit and attachment of the extracellular virions to the host cell, hence playing crucial roles in virus dissemination and pathogenesis.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Vero cells (Chlorocebus sabaeus kidney fibroblasts; ATCC number CCL-81) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), which was reduced to 2% during viral infection. The Vero cell-adapted ASFV strain BA71V has already been described (72). The following antibodies against ASFV proteins have been described previously: rabbit anti-p49 (29), rabbit anti-p10 (34), rabbit anti-pE296R (55), rabbit anti-CD2v (73), rat anti-p22 (49), mouse monoclonal antibody (MAb) 24BB7 against the structural protein p12 (40), and mouse MAb 19BB2 against capsid protein p72 (27). The mouse MAb against beta-actin (clone AC-15) was purchased from Sigma, and the mouse MAbs against integrin beta 1 (clone TS2/16) and CD9 (clone VJ1/20) were obtained from Thermo Fisher Scientific and Abnova, respectively.
ASFV purification.
Extracellular ASFV particles were purified from supernatants of infected Vero cells by using the Percoll gradient sedimentation method (17). In brief, ASFV particles from clarified infection supernatants were collected at 48 h postinfection (hpi) and concentrated by high-speed centrifugation. Then, the virus sample was subjected to two successive Percoll (GE Healthcare) density gradient centrifugations, and the final virus band was further purified by Sephacryl S-1000 (GE Healthcare) gel filtration chromatography. The resulting virus-enriched fractions were collected and concentrated by centrifugation. An average virus yield of 200 μg of protein with a specific infectivity of 1 × 106 PFU/μg was obtained from 109 infected cells. Purified virus particles were resuspended in phosphate-buffered saline and stored as small aliquots at −80°C until they were analyzed.
Western blotting.
Cell and virus samples were dissociated in 2× Laemmli buffer (2% SDS, 100 mM dithiothreitol [DTT], 125 mM Tris-HCl, pH 6.8), heated at 90°C for 5 min, and electrophoresed on 7% or 12% polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride membranes (pore size, 0.2 μm; Bio-Rad). The membranes were incubated overnight at 4°C with the primary antibodies and for 1 h at room temperature with anti-rabbit immunoglobulin secondary antibodies conjugated to horseradish peroxidase (GE Healthcare). Bands were developed with the enhanced chemiluminescence Prime Western blotting detection reagent (GE Healthcare).
Electron microscopy.
To check ASFV purification, virus particles were adsorbed to ionized collodion-carbon-coated grids, washed with water, and negatively stained with 2% uranyl acetate or 2% phosphotungstic acid before visualization. For immunoelectron microscopy of purified virions, grid-adsorbed particles were treated or not with 0.1% Triton X-100 for 5 min at room temperature (RT). Then, the particles were incubated with mouse MAb anti-p72 (the 18BA2 clone) or rabbit anti-p49 antibodies, followed by protein A conjugated to 5-nm gold particles (EM Laboratory, Utrecht University, Utrecht, The Netherlands). Finally, the specimens were negatively stained with 2% phosphotungstic acid before visualization.
For immunogold labeling of cell cryosections, ASFV-infected cells were fixed at 18 hpi with 4% paraformaldehyde in PHEM buffer (60 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 25 mM HEPES, 10 mM EGTA, 2 mM MgSO4), pH 6.9, for 4 h at RT. Subsequently, the cells were embedded in 10% (wt/vol) gelatin and cryoprotected overnight in a mixture of 2.3 M sucrose and 20% polyvinylpyrrolidone. Specimens were rapidly frozen in liquid nitrogen and cryosectioned with a Leica EM FCS cryoultramicrotome at −120°C. For immunogold labeling, thawed 90-nm-thick cryosections were incubated with 20 mM glycine for 5 min to quench the free aldehyde groups and with 10% FBS for 5 min to block nonspecific binding. Then, primary rabbit antibodies were incubated for 30 min at RT, followed by addition of protein A conjugated to 10-nm gold particles (EM Laboratory, Utrecht University, Utrecht, The Netherlands). For rat antibodies, a goat anti-rat IgG conjugated to 10-nm gold particles (British Biocell International, UK) was used. For mouse monoclonal antibodies, the signal was amplified in some cases with rabbit anti-mouse IgG (Dako) followed by protein A conjugated to 10-nm gold particles.
Sections were stained with a mix of 1.8% methylcellulose and 0.4% uranyl acetate before their visualization. Specimens were examined at 80 kV with a JEOL JEM-1010 or a JEOL JEM-1400Flash electron microscope equipped with TVIPS F416 or Gatan One View CMOS 4K cameras, respectively.
Proteomic analysis and database searching.
Approximately 2 μg of purified virus samples from three different preparations was individually analyzed by electrophoresis on 12% SDS-PAGE gels. Each sample was divided into 3 to 4 bands, and these were digested with trypsin, using an automatic robot (Proteineer; Bruker). In all cases, digestion was performed according to the protocol described previously (74). In summary, gel plugs were washed with 50 mM ammonium bicarbonate, and samples were reduced with 10 mM DTT. Alkylation was carried out with 55 mM iodoacetamide at room temperature before adding recombinant sequencing-grade trypsin (0.1 μg; Promega). Digestion took place at 37°C for 18 h. Then, the peptides were extracted, pooled, dried by speed vacuum centrifugation, and stored at −20°C until needed. About 1 μg of each digested virus sample was analyzed by mass spectrometry.
Nano-liquid chromatography (LC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) analysis was performed using an Eksigent 1D nano-HPLC coupled to a 5600TripleTOF quadrupole time of flight (QTOF) mass spectrometer (Sciex). The analytical column used was a silica-based reversed-phase column (Waters nanoACQUITY ultraperformance liquid chromatograph; 75 μm by 15 cm; particle size, 1.7 μm). The trap column was an Acclaim PepMap 100 (particle diameter, 5 μm; pore size, 100 Å) switched on-line with the analytical column. The loading pump delivered a solution of 0.1% formic acid in 98% water–2% acetonitrile (Scharlab, Barcelona, Spain) at 3 μl/min. The nanopump provided a flow rate of 250 nl/min and was operated under gradient elution conditions, using 0.1% formic acid (Fluka) in water as mobile phase A and 0.1% formic acid in 100% acetonitrile as mobile phase B. Gradient elution was performed according to the following scheme: isocratic conditions of 96% mobile phase A and 4% mobile phase B for 5 min, a linear increase to 40% mobile phase B in 25 min, a linear increase to 95% mobile phase B in 2 min, isocratic conditions of 95% mobile phase B for 5 min, and a return to the initial conditions in 10 min. The injection volume was 5 μl. The LC system was coupled via a nanospray source to the mass spectrometer. Automatic data-dependent acquisition using dynamic exclusion allowed the collection of both full-scan (m/z 350 to 1,250) MS spectra followed by tandem MS collision-induced dissociation spectra of the 15 most abundant ions. The acquisition time was 250 ms and 100 ms for the MS and MS/MS spectra, respectively. The MS and MS/MS data were used to search against a Chlorocebus sabaeus and African swine fever virus (strain BA71V) combined database containing 19,453 sequences downloaded from UniProtKB (https://www.uniprot.org/). The ASFV protein list, which contains 157 putative viral proteins, includes all potential ORFs with 60 or more amino acids previously described (5), with the exception that ORFs CP2475L and CP530R, encoding polyproteins pp220 and pp62, respectively, were replaced by the amino acid sequences of their predicted mature products (from the N to the C terminus, p5, p34, p14, p37, and p150 for precursor pp220 and p15, p35, and p8 for precursor pp62) (32, 33). The database also includes an exclusion list of about 50 common contaminant proteins (i.e., human keratins, serum albumin, trypsin, casein, etc.) that was used to remove contaminant peptides.
Database searches were done using a licensed version of the Mascot (v.2.6) program, and search parameters were set as follows: carbamidomethyl cysteine as a fixed modification and acetyl (protein N terminus), asparagine and glutamine deamidation, glutamic acid to pyro-glutamic, and oxidized methionine as a variable one. Peptide mass tolerance was set at 25 ppm and 0.1 Da for MS and MS/MS spectra, respectively, and 2 missed cleavages were allowed. Peptides with individual Mascot ion scores indicating identity or extensive homology (P < 0.05) were used for protein identification. Only those proteins with at least one unique peptide were considered.
The estimated abundance of each protein was determined from the exponentially modified protein abundance index (emPAI) score, which is based on the correlation of the peptides identified by MS/MS with the predicted peptides obtained by in silico digestion (23). The abundance of each protein is presented as the weight percentages of the total virion protein mass.
Data availability.
The raw data derived from the proteomic analyses used in this report are available from the corresponding author upon request.
ACKNOWLEDGMENTS
The expert technical advice of Alberto Paradela at the Proteomics Facility of the National Centre for Biotechnology (CNB; Madrid, Spain) is gratefully acknowledged. We thank Carlos Cabañas (CBMSO) for providing useful reagents and helpful comments. We especially acknowledge José Antonio Tercero (CBMSO) for his continued support.
Germán Andrés is supported by the Amarouto Program for senior scientists from the Comunidad de Madrid.
REFERENCES
- 1.Sanchez-Cordon PJ, Montoya M, Reis AL, Dixon LK. 2018. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet J 233:41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou X, Li N, Luo Y, Liu Y, Miao F, Chen T, Zhang S, Cao P, Li X, Tian K, Qiu H, Hu R. 13 August 2018. Emergence of African swine fever in China, 2018. Transbound Emerg Dis. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Yanez RJ, Rodriguez JM, Nogal ML, Yuste L, Enriquez C, Rodriguez JF, Vinuela E. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 6.Dixon LK, Chapman DA, Netherton CL, Upton C. 2013. African swine fever virus replication and genomics. Virus Res 173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Reis AL, Netherton C, Dixon LK. 2017. Unraveling the armor of a killer: evasion of host defenses by African swine fever virus. J Virol 91:e02338-16. doi: 10.1128/JVI.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salas ML, Andres G. 2013. African swine fever virus morphogenesis. Virus Res 173:29–41. doi: 10.1016/j.virusres.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Andres G. 2017. African swine fever virus gets undressed: new insights on the entry pathway. J Virol 91:e01906-16. doi: 10.1128/JVI.01906-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernaez B, Guerra M, Salas ML, Andres G. 2016. African swine fever virus undergoes outer envelope disruption, capsid disassembly and inner envelope fusion before core release from multivesicular endosomes. PLoS Pathog 12:e1005595. doi: 10.1371/journal.ppat.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez JM, Salas ML. 2013. African swine fever virus transcription. Virus Res 173:15–28. doi: 10.1016/j.virusres.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Suarez C, Andres G, Kolovou A, Hoppe S, Salas ML, Walther P, Krijnse Locker J. 2015. African swine fever virus assembles a single membrane derived from rupture of the endoplasmic reticulum. Cell Microbiol 17:1683–1698. doi: 10.1111/cmi.12468. [DOI] [PubMed] [Google Scholar]
- 13.Andres G, Garcia-Escudero R, Simon-Mateo C, Vinuela E. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J Virol 72:8988–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andres G, Simon-Mateo C, Vinuela E. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J Virol 71:2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouvenet N, Monaghan P, Way M, Wileman T. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J Virol 78:7990–8001. doi: 10.1128/JVI.78.15.7990-8001.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breese SS Jr, Pan IC. 1978. Electron microscopic observation of African swine fever virus development in Vero cells. J Gen Virol 40:499–502. doi: 10.1099/0022-1317-40-2-499. [DOI] [PubMed] [Google Scholar]
- 17.Carrascosa AL, del Val M, Santaren JF, Vinuela E. 1985. Purification and properties of African swine fever virus. J Virol 54:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteves A, Marques MI, Costa JV. 1986. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology 152:192–206. doi: 10.1016/0042-6822(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 19.Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J Virol 80:2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. 2007. Protein composition of the vaccinia virus mature virion. Virology 358:233–247. doi: 10.1016/j.virol.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 21.Fischer MG, Kelly I, Foster LJ, Suttle CA. 2014. The virion of Cafeteria roenbergensis virus (CroV) contains a complex suite of proteins for transcription and DNA repair. Virology 466-467: 82–94. doi: 10.1016/j.virol.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 22.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol 89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Andres G, Alejo A, Salas J, Salas ML. 2002. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J Virol 76:12473–12482. doi: 10.1128/JVI.76.24.12473-12482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hingamp PM, Leyland ML, Webb J, Twigger S, Mayer RJ, Dixon LK. 1995. Characterization of a ubiquitinated protein which is externally located in African swine fever virions. J Virol 69:1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brookes SM, Sun H, Dixon LK, Parkhouse RM. 1998. Characterization of African swine fever virion proteins j5R and j13L: immuno-localization in virus particles and assembly sites. J Gen Virol 79(Pt 5):1179–1188. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Escudero R, Andres G, Almazan F, Vinuela E. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J Virol 72:3185–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andres G, Garcia-Escudero R, Vinuela E, Salas ML, Rodriguez JM. 2001. African swine fever virus structural protein pE120R is essential for virus transport from assembly sites to plasma membrane but not for infectivity. J Virol 75:6758–6768. doi: 10.1128/JVI.75.15.6758-6768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epifano C, Krijnse-Locker J, Salas ML, Salas J, Rodriguez JM. 2006. Generation of filamentous instead of icosahedral particles by repression of African swine fever virus structural protein pB438L. J Virol 80:11456–11466. doi: 10.1128/JVI.01468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez JM, Garcia-Escudero R, Salas ML, Andres G. 2004. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J Virol 78:4299–4313. doi: 10.1128/JVI.78.8.4299-4313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suarez C, Gutierrez-Berzal J, Andres G, Salas ML, Rodriguez JM. 2010. African swine fever virus protein p17 is essential for the progression of viral membrane precursors toward icosahedral intermediates. J Virol 84:7484–7499. doi: 10.1128/JVI.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon-Mateo C, Andres G, Vinuela E. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J 12:2977–2987. doi: 10.1002/j.1460-2075.1993.tb05960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon-Mateo C, Andres G, Almazan F, Vinuela E. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J Virol 71:5799–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andres G, Garcia-Escudero R, Salas ML, Rodriguez JM. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J Virol 76:2654–2666. doi: 10.1128/JVI.76.6.2654-2666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suarez C, Salas ML, Rodriguez JM. 2010. African swine fever virus polyprotein pp62 is essential for viral core development. J Virol 84:176–187. doi: 10.1128/JVI.01858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andres G, Alejo A, Simon-Mateo C, Salas ML. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J Biol Chem 276:780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- 37.Alejo A, Andres G, Salas ML. 2003. African swine fever virus proteinase is essential for core maturation and infectivity. J Virol 77:5571–5577. doi: 10.1128/JVI.77.10.5571-5577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz M, Freije JM, Salas ML, Vinuela E, Lopez-Otin C. 1993. Structure and expression in E. coli of the gene coding for protein p10 of African swine fever virus. Arch Virol 130:93–107. doi: 10.1007/BF01318999. [DOI] [PubMed] [Google Scholar]
- 39.Frouco G, Freitas FB, Coelho J, Leitao A, Martins C, Ferreira F. 2017. DNA-binding properties of African swine fever virus pA104R, a histone-like protein involved in viral replication and transcription. J Virol 91:e02498-16. doi: 10.1128/JVI.02498-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrascosa AL, Sastre I, Vinuela E. 1991. African swine fever virus attachment protein. J Virol 65:2283–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun H, Jenson J, Dixon LK, Parkhouse ME. 1996. Characterization of the African swine fever virion protein j18L. J Gen Virol 77(Pt 5):941–946. [DOI] [PubMed] [Google Scholar]
- 42.Simon-Mateo C, Freije JM, Andres G, Lopez-Otin C, Vinuela E. 1995. Mapping and sequence of the gene encoding protein p17, a major African swine fever virus structural protein. Virology 206:1140–1144. doi: 10.1006/viro.1995.1039. [DOI] [PubMed] [Google Scholar]
- 43.Angulo A, Vinuela E, Alcami A. 1993. Inhibition of African swine fever virus binding and infectivity by purified recombinant virus attachment protein p12. J Virol 67:5463–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez I, Nogal ML, Redrejo-Rodriguez M, Bustos MJ, Salas ML. 2009. The African swine fever virus virion membrane protein pE248R is required for virus infectivity and an early postentry event. J Virol 83:12290–12300. doi: 10.1128/JVI.01333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borca MV, Carrillo C, Zsak L, Laegreid WW, Kutish GF, Neilan JG, Burrage TG, Rock DL. 1998. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J Virol 72:2881–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez JM, Yanez RJ, Almazan F, Vinuela E, Rodriguez JF. 1993. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J Virol 67:5312–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Nunez D, Garcia-Urdiales E, Martinez-Bonet M, Nogal ML, Barroso S, Revilla Y, Madrid R. 2015. CD2v interacts with adaptor protein AP-1 during African swine fever infection. PLoS One 10:e0123714. doi: 10.1371/journal.pone.0123714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senkevich TG, Ojeda S, Townsley A, Nelson GE, Moss B. 2005. Poxvirus multiprotein entry-fusion complex. Proc Natl Acad Sci U S A 102:18572–18577. doi: 10.1073/pnas.0509239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camacho A, Vinuela E. 1991. Protein p22 of African swine fever virus: an early structural protein that is incorporated into the membrane of infected cells. Virology 181:251–257. doi: 10.1016/0042-6822(91)90490-3. [DOI] [PubMed] [Google Scholar]
- 50.Goatley LC, Dixon LK. 2011. Processing and localization of the African swine fever virus CD2v transmembrane protein. J Virol 85:3294–3305. doi: 10.1128/JVI.01994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuznar J, Salas ML, Vinuela E. 1980. DNA-dependent RNA polymerase in African swine fever virus. Virology 101:169–175. doi: 10.1016/0042-6822(80)90493-6. [DOI] [PubMed] [Google Scholar]
- 52.Salas ML, Kuznar J, Vinuela E. 1981. Polyadenylation, methylation, and capping of the RNA synthesized in vitro by African swine fever virus. Virology 113:484–491. doi: 10.1016/0042-6822(81)90176-8. [DOI] [PubMed] [Google Scholar]
- 53.Pena L, Yanez RJ, Revilla Y, Vinuela E, Salas ML. 1993. African swine fever virus guanylyltransferase. Virology 193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- 54.Redrejo-Rodriguez M, Salas ML. 2014. Repair of base damage and genome maintenance in the nucleo-cytoplasmic large DNA viruses. Virus Res 179:12–25. doi: 10.1016/j.virusres.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Redrejo-Rodriguez M, Garcia-Escudero R, Yanez-Munoz RJ, Salas ML, Salas J. 2006. African swine fever virus protein pE296R is a DNA repair apurinic/apyrimidinic endonuclease required for virus growth in swine macrophages. J Virol 80:4847–4857. doi: 10.1128/JVI.80.10.4847-4857.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamarche BJ, Showalter AK, Tsai MD. 2005. An error-prone viral DNA ligase. Biochemistry 44:8408–8417. doi: 10.1021/bi047706g. [DOI] [PubMed] [Google Scholar]
- 57.Oliveros M, Yanez RJ, Salas ML, Salas J, Vinuela E, Blanco L. 1997. Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J Biol Chem 272:30899–30910. doi: 10.1074/jbc.272.49.30899. [DOI] [PubMed] [Google Scholar]
- 58.Redrejo-Rodriguez M, Rodriguez JM, Suarez C, Salas J, Salas ML. 2013. Involvement of the reparative DNA polymerase Pol X of African swine fever virus in the maintenance of viral genome stability in vivo. J Virol 87:9780–9787. doi: 10.1128/JVI.01173-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveros M, Garcia-Escudero R, Alejo A, Vinuela E, Salas ML, Salas J. 1999. African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. J Virol 73:8934–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chacon MR, Almazan F, Nogal ML, Vinuela E, Rodriguez JF. 1995. The African swine fever virus IAP homolog is a late structural polypeptide. Virology 214:670–674. doi: 10.1006/viro.1995.0083. [DOI] [PubMed] [Google Scholar]
- 61.Neilan JG, Lu Z, Kutish GF, Zsak L, Burrage TG, Borca MV, Carrillo C, Rock DL. 1997. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growth in vitro and viral virulence. Virology 230:252–264. doi: 10.1006/viro.1997.8481. [DOI] [PubMed] [Google Scholar]
- 62.Nogal ML, Gonzalez de Buitrago G, Rodriguez C, Cubelos B, Carrascosa AL, Salas ML, Revilla Y. 2001. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J Virol 75:2535–2543. doi: 10.1128/JVI.75.6.2535-2543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alcami A, Angulo A, Vinuela E. 1993. Mapping and sequence of the gene encoding the African swine fever virion protein of M(r) 11500. J Gen Virol 74(Pt 11):2317–2324. [DOI] [PubMed] [Google Scholar]
- 64.Prados FJ, Vinuela E, Alcami A. 1993. Sequence and characterization of the major early phosphoprotein p32 of African swine fever virus. J Virol 67:2475–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goatley LC, Twigg SR, Miskin JE, Monaghan P, St-Arnaud R, Smith GL, Dixon LK. 2002. The African swine fever virus protein j4R binds to the alpha chain of nascent polypeptide-associated complex. J Virol 76:9991–9999. doi: 10.1128/JVI.76.19.9991-9999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baylis SA, Banham AH, Vydelingum S, Dixon LK, Smith GL. 1993. African swine fever virus encodes a serine protein kinase which is packaged into virions. J Virol 67:4549–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez I, Redrejo-Rodriguez M, Rodriguez JM, Alejo A, Salas J, Salas ML. 2006. African swine fever virus pB119L protein is a flavin adenine dinucleotide-linked sulfhydryl oxidase. J Virol 80:3157–3166. doi: 10.1128/JVI.80.7.3157-3166.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Donnell V, Holinka LG, Krug PW, Gladue DP, Carlson J, Sanford B, Alfano M, Kramer E, Lu Z, Arzt J, Reese B, Carrillo C, Risatti GR, Borca MV. 2015. African swine fever virus Georgia 2007 with a deletion of virulence-associated gene 9GL (B119L), when administered at low doses, leads to virus attenuation in swine and induces an effective protection against homologous challenge. J Virol 89:8556–8566. doi: 10.1128/JVI.00969-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jouvenet N, Windsor M, Rietdorf J, Hawes P, Monaghan P, Way M, Wileman T. 2006. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol 8:1803–1811. doi: 10.1111/j.1462-5822.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- 70.McFadden BD, Moussatche N, Kelley K, Kang BH, Condit RC. 2012. Vaccinia virions deficient in transcription enzymes lack a nucleocapsid. Virology 434:50–58. doi: 10.1016/j.virol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moss B. 2012. Poxvirus cell entry: how many proteins does it take? Viruses 4:688–707. doi: 10.3390/v4050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Enjuanes L, Carrascosa AL, Moreno MA, Vinuela E. 1976. Titration of African swine fever (ASF) virus. J Gen Virol 32:471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- 73.Galindo I, Almazan F, Bustos MJ, Vinuela E, Carrascosa AL. 2000. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology 266:340–351. doi: 10.1006/viro.1999.0080. [DOI] [PubMed] [Google Scholar]
- 74.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 75.Lopez-Otin C, Freije JM, Parra F, Mendez E, Vinuela E. 1990. Mapping and sequence of the gene coding for protein p72, the major capsid protein of African swine fever virus. Virology 175:477–484. doi: 10.1016/0042-6822(90)90432-Q. [DOI] [PubMed] [Google Scholar]
- 76.Yanez RJ, Rodriguez JM, Boursnell M, Rodriguez JF, Vinuela E. 1993. Two putative African swine fever virus helicases similar to yeast ‘DEAH’ pre-mRNA processing proteins and vaccinia virus ATPases D11L and D6R. Gene 134:161–174. doi: 10.1016/0378-1119(93)90090-P. [DOI] [PubMed] [Google Scholar]
- 77.Yanez RJ, Boursnell M, Nogal ML, Yuste L, Vinuela E. 1993. African swine fever virus encodes two genes which share significant homology with the two largest subunits of DNA-dependent RNA polymerases. Nucleic Acids Res 21:2423–2427. doi: 10.1093/nar/21.10.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baylis SA, Twigg SR, Vydelingum S, Dixon LK, Smith GL. 1993. Three African swine fever virus genes encoding proteins with homology to putative helicases of vaccinia virus. J Gen Virol 74(Pt 9):1969–1974. [DOI] [PubMed] [Google Scholar]
- 79.Borca MV, O'Donnell V, Holinka LG, Rai DK, Sanford B, Alfano M, Carlson J, Azzinaro PA, Alonso C, Gladue DP. 2016. The Ep152R ORF of African swine fever virus strain Georgia encodes for an essential gene that interacts with host protein BAG6. Virus Res 223:181–189. doi: 10.1016/j.virusres.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 80.Afonso CL, Alcaraz C, Brun A, Sussman MD, Onisk DV, Escribano JM, Rock DL. 1992. Characterization of p30, a highly antigenic membrane and secreted protein of African swine fever virus. Virology 189:368–373. doi: 10.1016/0042-6822(92)90718-5. [DOI] [PubMed] [Google Scholar]
- 81.Rodriguez F, Alcaraz C, Eiras A, Yanez RJ, Rodriguez JM, Alonso C, Rodriguez JF, Escribano JM. 1994. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J Virol 68:7244–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu Z, Kutish GF, Sussman MD, Rock DL. 1993. An African swine fever virus gene with a similarity to eukaryotic RNA polymerase subunit 6. Nucleic Acids Res 21:2940. doi: 10.1093/nar/21.12.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data derived from the proteomic analyses used in this report are available from the corresponding author upon request.