Abstract
The 5′-terminus of eukaryotic mRNAs comprises a 7-methylguanosine cap linked to the first transcribed nucleotide via a 5′-5′ triphosphate bond. This cap structure facilitates numerous interactions with molecules participating in mRNA processing, turnover and RNA translation. Here, we report the synthesis and biochemical properties of a set of biotin-labelled cap analogues modified within the triphosphate bridge and increasing mRNA stability while retaining biological activity. Successful co-transcriptional incorporation of the cap analogues allowed for the quantification of cap-dependent translation efficiency, capping efficiency and the susceptibility to decapping by Dcp2. The utility of such cap-biotinylated RNAs as molecular tool was demonstrated by ultraviolet-cross-linking and affinity capture of protein–RNA complexes. In conclusion, RNAs labelled with biotin via the 5′ cap structure can be applied to a variety of biological experiments based on biotin–avidin interaction or by means of biotin-specific antibodies, including protein affinity purification, pull-down assays, in vivo visualization, cellular delivery and many others.
This article is part of the theme issue ‘5′ and 3′ modifications controlling RNA degradation’.
Keywords: mRNA cap, biotin, RNA degradation
1. Introduction
The 7-methylguanosine (m7G) residue linked to the first nucleoside of the mRNA transcript, often referred to as the cap, contributes greatly to eukaryotic mRNA stability and recognition by translation initiation factors [1,2]. Protein complexes interacting with this structure govern important events in the life cycle of mRNAs, including splicing, subcellular localization, initiation of RNA translation and turnover [3,4]. Most proteins known to interact with the cap structure share a common pattern and rely on the positively charged 7-methylguanosine for specificity of recognition [2,5], whereas the oligophosphate bridge often serves as a site for enzymatic cleavage [6]. Of particular interest are cap-specific mRNA-decapping pyrophosphatase, Dcp1–Dcp2, initiating mRNA degradation in the cell, and the eukaryotic translation initiation factor 4E (eIF4E), which directly binds the cap during initiation of translation [7]. Chemically modified cap derivatives aid in the studies on mRNA-related processes, serving as structure, activity and capture probes. Their potential as therapeutic agents is also under investigation [8], especially as factors stabilizing mRNA and improving translation efficiency in gene therapy, repressors of RNA translation in cancer cells, inhibitors of decapping scavenger enzyme, an enzyme implicated in spinal muscular atrophy, and in antiviral immunotherapy against cancer [9].
To gain a comprehensive understanding of many fundamental gene expression processes, knowledge about specific mRNA–protein interactions is indispensable. Exploration of these interactions can be facilitated by the attachment of a biotin tag to a biomolecule specifically binding to target molecules of interest, thus allowing efficient protein and nucleic acid isolation, detection and visualization by using biotin-(strept)avidin technology [10–13]. Biotin-(strept)avidin technology and biotin-specific antibodies can be used to identify RNA-binding partners under a wide range of experimental conditions, including cell lysates and living cells. While designing biotinylated probes, it is important to minimize the influence of the biotin tag on crucial interactions of the modified molecule (so that it retains to a reasonable degree its intrinsic biological activity) by proper selection of the biotin attachment site. Numerous biotinylating methods have been made available, allowing for convenient biotinylation of proteins, nucleic acids, carbohydrates and other biomolecules, [12] most of them relying on conjugation via formation of stable amide bonds. We have previously reported a biotinylated mRNA cap analogue that was suitable as a reagent for enzymatic production of 5′-biotinylated and 5′-capped mRNA that remained translationally active (as demonstrated in a rabbit reticulocyte lysate, RRL, system), albeit less efficient than unmodified mRNA [14]. Such biotinylated mRNAs are applicable for electrophoretic mobility shift assays and protein capture experiments. However, the presence of the unmodified triphosphate bridge made such mRNAs susceptible for decapping by Dcp1–Dcp2 and therefore, arguably, of limited utility under conditions where mRNA decapping takes place such as in living cells. It has been previously shown that the introduction of certain modifications within the triphosphate bridge of cap derivatives can overcome some of these problems as mRNAs capped with such modifications are less susceptible to decapping by Dcp1–Dcp2 [15]. We envisaged that combining an affinity label (biotin) with decapping-resistance conferring modifications (triphosphate bridge substitutions) should improve the molecular tools applicable to a variety of studies such as gene expression and protein–mRNA interactions.
Here, we report the synthesis and biochemical properties of four novel biotin-labelled dinucleotide cap analogues with modified triphosphate bridge, designed as decapping-resistant reagents for simultaneous mRNA capping and biotinylation. Three types of moieties were incorporated into the triphosphate: imidodiphosphate, methylene-bisphosphonate and thiophosphate (figure 1). The biotin-labelled cap analogues, along with the previously reported phosphate-unmodified cap, were subjected to extensive biochemical testing in order to select for compounds with superior enzymatic resistance, translational activity and applicability to approaches with streptavidin. To this end, we determined the capping efficiencies of in vitro synthesized RNAs that can be achieved with the new analogues, mRNA translation efficiencies in vitro and in living cells, and susceptibility of RNAs to decapping by the catalytic subunit of the human mRNA-decapping enzyme Dcp1/Dcp2 complex. Finally, using the cap structure with most beneficial biochemical properties, we demonstrated the utility of our biotinylated-capped mRNAs as baits in cross-linking-assisted protein capture experiments.
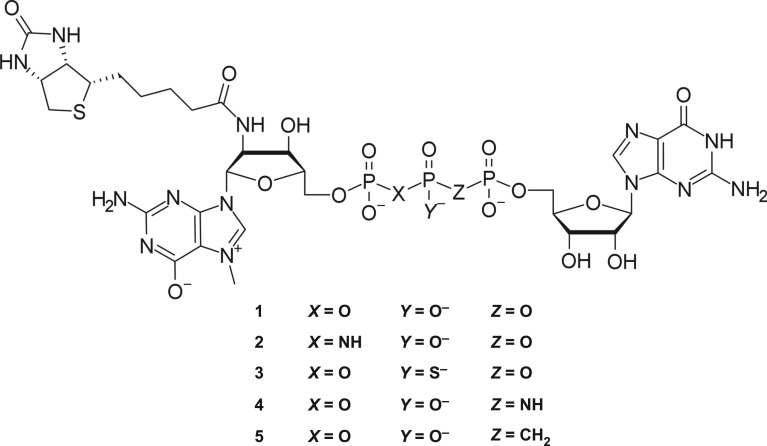
Figure 1.
Structure of biotinylated cap analogues with modified triphosphate bridge.
2. Results and discussion
(a). Chemical synthesis
The affinity tag was attached at the 2′ position of m7G via an amide bond, which was possible due to the presence of the primary aliphatic amino group in the starting material—2′-amino-2′-deoxyguanosine—and in situ biotin activation with O-(N-succinimidyl)-1,1,3,3-tetramethyluronium tetrafluoroborate (TSTU). The site for biotin moiety attachment was chosen in a way that ensures the least possible interference with eIF4E interaction, because the hydroxyl groups of 7-methylguanosine in the cap-eIF4E complex are free and solvent exposed [16] (electronic supplementary material, figure S1). The ability of the cap analogues conjugated with biotin to serve as substrates for RNA polymerase in an in vitro transcription reaction was of equally high importance. In this context, the additional advantage of introducing the tag at the 2′ position is that, similar to 2′-O-methylated dinucleotides [17], the resulting compound can only be incorporated into RNA in the correct orientation (and, as such, belongs to a class of the so-called Anti-Reverse Cap Analogs or ARCAs [17]).
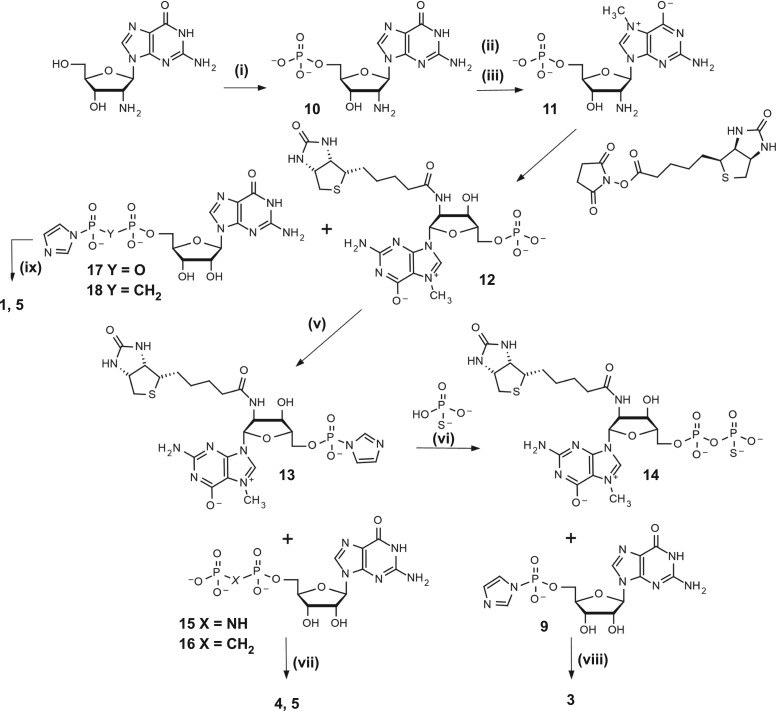
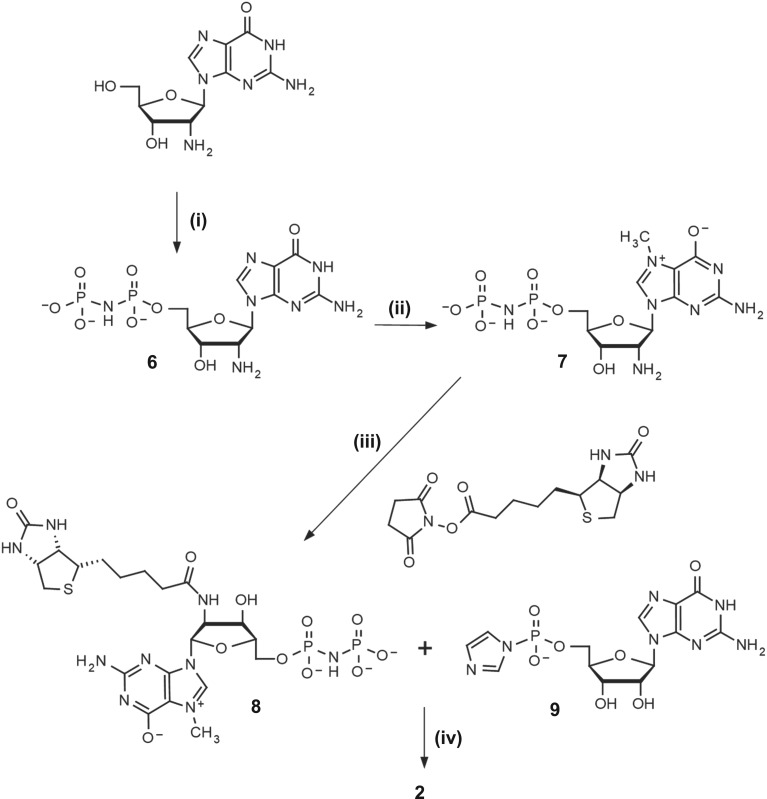
The general synthesis strategy we applied differs slightly from the one reported earlier [14]—the order of steps was changed to overcome problems arising from the incorporation of different phosphate-modified moieties, resulting in a highly variable yield. Attaching the biotin tag at an earlier step in the synthesis pathway results in faster and more reliable coupling reactions. The synthesis was based on a commercially available 2′-amino-2′-deoxyguanosine, which was 5′-phosphorylated with POCl3 and N7-methylated as described previously [14] (figure 2). The product was subsequently biotinylated in a two step–one pot reaction, employing in situ biotin activation with N-succinimidyl ester formation and dropwise addition to a buffered solution of 11 (pH 8.0). A similar synthetic route was designed to obtain 8 (figure 3), wherein the initial compound was subjected to a modified phosphorylation procedure with tri-chloro[(dichlorophosphoryl)imido]phosphorane in place of POCl3, leading to the target nucleoside imidodiphosphate (6) [18]. Biotinylated derivatives of 7-methylguanosine (8 and 12) were then coupled [19] with P-imidazolides of guanosine monophosphate, diphosphate or methylenebisphosphonate [20], yielding final compounds 2, 1 and 5, respectively. Activation of 12 using the dithiodipyridine-triphenylphosphine system was used to obtain P-imidazolide, 13, which was further coupled with thiophosphate [21] or O-to-NH/O-to-CH2 substituted guanosine diphosphate [22,23] producing 14, 4 and 5. The β-S modified cap analogue 3 was obtained from 14 in reaction with imidazole-activated GMP, in the form of a mixture of two P-diastereoisomers. We could not find HPLC conditions to resolve the two diastereoisomers of 3, likely due to high lipophilicity of biotin, and thus the compound was used in further studies as a diastereomeric mixture. Overall, five different biotin-labelled cap derivatives were synthesized, bearing thiophosphate substitution at the β position, methylenebisphosphonate substitution at the α,β position, imidodiphosphate substitution at either α,β or β,γ position, or no bridge modifications (figure 1). 1H NMR analysis of final compounds confirmed their structure and homogeneity (details on synthesis and purification in electronic supplementary material).
Figure 2.
Synthesis of cap analogues 1 and 3–5. (i) 1. POCl3 (6 equiv.), (MeO)3PO, 0°C, 8 h; 2. H2O, NaHCO3; (ii) CH3I (8 equiv.), DMSO, 6 h; (iii) (CH3O)2SO2 (15 equiv.), H2O pH 4, 8 h; (iv) 0.5 M borate buffer pH 8.5, 1 h; (v) 1. Imidazole (10 equiv.), 2,2′-DTDP (3 equiv.), Ph3P (3 equiv.), TEA, N,N-dimethylformamide (DMF), 4 h; 2. NaClO4, acetone; (vi) PSO33−/TEA (2 equiv.), ZnCl2 (8 equiv.), DMF, 40 min; (vii) ZnCl2 (8 equiv.), DMF, 9 h; (viii) ZnCl2 (16 equiv.), DMF, 24 h; (ix) ZnCl2 (8 equiv.), DMF, 9 h.
Figure 3.
Synthesis of cap analogue 2 (i) 1. Cl3PNP(O)Cl2, (MeO)3PO, −8°C, 6 h; 2. H2O, NaHCO3; (ii) (CH3O)2SO2 (5 equiv.), H2O pH 4, 6 h; (iii) 0.5 M borate buffer pH 8.5, 1 h; (iv) ZnCl2 (8 equiv.), DMF, 8 h.
(b). Affinity for eukaryotic translation initiation factor 4E
To evaluate the new cap analogues in the context of interaction with eIF4E, a series of fluorescence quenching titration (FQT) experiments was carried out. A standard procedure was used taking into consideration: protein stability, fluorescence intensity decline due to dilution, the inner filter effect, and overlapping bands of absorption and emission of the ligand and protein under investigation [24]. Table 1 summarizes the results from replicate FQT experiments, featuring both the determined KAS values and the calculated free energies of binding. Overall, the affinity of biotinylated cap derivatives to eIF4E is slightly diminished as compared to the reference cap–m7GpppG, yet the specificity of binding is clearly retained, with free energy of binding values differing by less than 1 kcal mole−1 in the edge case. As expected, introduction of the thiophosphate modification had a stabilizing effect on the binding, and it increased the KAS value compared to other tested compounds.
Table 1.
Binding affinity constants (KAS) of biotin-labelled cap analogues for murine eIF4E.
| cap analogue | KAS (µM−1) | ΔGc (kcal mol−1) |
|---|---|---|
| m7GpppG | 9.4 ± 0.4b | −9.35 ± 0.03 |
| m72′-N-BiotGpppG (1) | 3.8 ± 0.1b | −8.82 ± 0.02 |
| m72′-N-BiotGpNHppG (2) | 3.3 ± 0.1a | −8.74 ± 0.02 |
| m72′-N-BiotGppSpG (3) | 5.7 ± 0.2a | −9.06 ± 0.02 |
| m72′-N-BiotGppNHpG (4) | 3.0 ± 0.2a | −8.68 ± 0.02 |
| m72′-N-BiotGppCH2pG (5) | 3.6 ± 0.2a | −8.79 ± 0.02 |
aConditions for FQT experiments: 50 mM HEPES/KOH, 100 mM KCl, 0.5 mM EDTA, pH 7.2.
bData from Jemielity et al. [14].
The assessment of binding constants of biotin-labelled analogues to eIF4E confirmed the expected complex formation with stability levels similar to those registered for a natural dinucleotide analogue [17]. The biotin tag does not impede cap–eIF4E interaction to a significant degree, and the specificity of binding is conserved.
(c). In vitro transcription
The incorporation of biotin-labelled cap analogues into RNAs of varying length was probed by using in vitro transcription. Full-length mRNAs encoding firefly luciferase, as well as short (approximately 35 nt) RNAs, were obtained using the SP6 RNA polymerase. All analogues were incorporated with efficiency ranging from 40 to 70% (table 2), which is lower than that usually achieved for unmodified cap analogues (80% or more), but still sufficient for experimental purposes. Moreover, if necessary, the fraction of uncapped RNA can be selectively removed from the in vitro transcription reaction by consecutive treatment with 5′-polyphosphatase and the RNAse Xrn1 [25].
Table 2.
In vitro biochemical properties of cap analogues assessed in this study.
| cap analogue | capping efficiency (%) | translation efficiency (RRL) | hDcp2 decapping susceptibility |
|---|---|---|---|
| m7GpppG | 76 | 1 | fast degradation |
| m27,2′-OGpppG (ARCA) | 69 | 1.56 ± 0.48 | fast degradation |
| ApppG | 43 | 0.03 ± 0.01 | resistant |
| m72′-N-BiotGpppG (1) | 61 | 0.68 ± 0.16 | slow degradation |
| m72′-N-BiotGpNHppG (2) | 64 | 0.37 ± 0.12 | slow degradation |
| m72′-N-BiotGppSpG (3) | 61 | 0.93 ± 0.14 | partially resistant |
| m72′-N-BiotGppNHpG (4) | 39 | 0.11 ± 0.04 | resistant |
| m72′-N-BiotGppCH2pG (5) | 47 | 0.28 ± 0.08 | resistant |
(d). In vitro translation
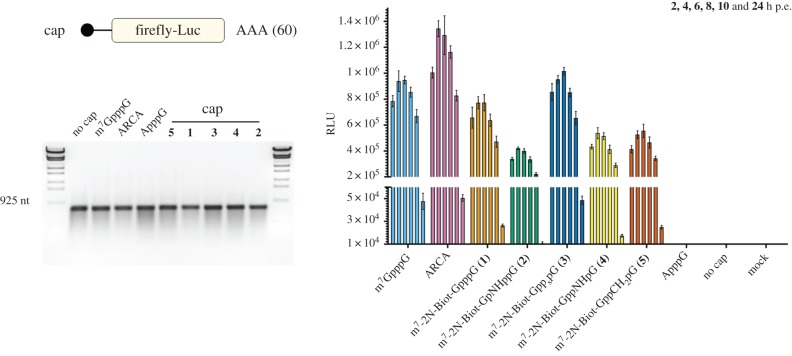
Transcripts encoding firefly luciferase and 5′-capped with the novel cap analogues were prepared to determine translation efficiency by using the RRL system optimized for cap-dependent translation [14]. mRNAs capped with the standard residue (m7GpppG) or with ARCA [17] were used as references along with the non-functional negative control Biot-NHEX-ApppG-capped RNA (electronic supplementary material, figure S2). Translation efficiency was determined based on luminescence of the sample after 1 h of incubation at 37°C, and immediately after the addition of luciferin substrate. Final values were calculated as a mean of three independent replicates and normalized to the values obtained with the m7GpppG-capped mRNA (figure 4). Cap-independent translation, assessed with the negative control, was at the background level. Efficiency of translation obtained with the biotin-labelled cap analogues ranged from 11 to 93% (table 2), with the highest level noted for the β-S analogue. mRNAs with lowest levels of translation were those capped with α,β modified analogues (11–28%), for which the capping efficiency was also less efficient (39–47%).
Figure 4.
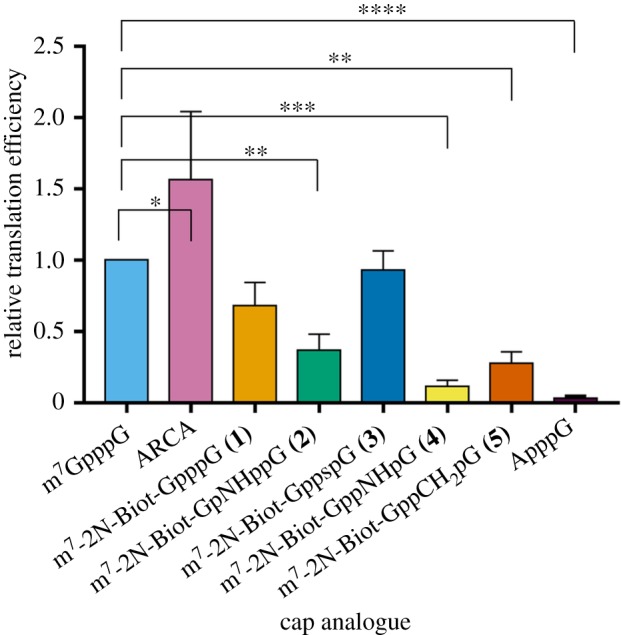
Translation efficiencies in RRL of luciferase-encoding mRNAs capped with biotin-labelled analogues 1–5 or reference caps. mRNAs with biotinylated 5′ termini specified on the bottom were generated by in vitro transcription and their translation efficiency was measured after 60 min of incubation in RRL. Translation efficiencies were normalized to the values obtained with the m7GpppG-capped mRNA and calculated as mean values from three independent experiments ±s.d. (Online version in colour.)
While the RRL system contains all the components necessary for the biosynthesis of proteins, it fails to mirror important aspects of a live cell such as the balance between mRNA translation and degradation. Therefore, we studied the overall protein expression from firefly luciferase-encoding mRNAs containing polyA tail and capped with biotinylated cap analogues 1–5 in human hepatoma cells. Polyadenylated mRNAs with non-functional cap (Biotin-ApppG; electronic supplementary material, figure S2) or lacking the cap structure at the 5′ end were not translated in transfected cells (figure 5). By contrast, the mRNAs containing either unmodified or modified biotinylated caps were efficiently translated. We measured luciferase activity at different time points after transfection to study the influence of the cap structures on overall protein expression. Translation of input mRNA (2 h after transfection) increased from 2 to 4 h, being maximal with ARCA-capped mRNA, and slightly lower for m7GpppG-capped mRNA. Notably, protein expression from the mRNA capped with biotinylated β-S analogue (3) was superior to that of mRNA carrying phosphate-unmodified biotinylated analogue (1) and comparable to that of the non-biotinylated m7GpppG-capped mRNA. The mRNAs containing cap analogues modified with –CH2 (5) and –NH (2, 4) groups in the triphosphate bridge were twofold less efficiently translated compared with the unmodified m7GpppG RNA. These results indicate that biotinylation of the cap structure is well tolerated and does not interfere with translation of mRNA in human hepatoma cells. Moreover, introduction of a β-S group in the triphosphate bridge increases the overall protein expression from the biotinylated mRNA.
Figure 5.
Translation efficiency of polyadenylated RNAs encoding firefly luciferase and capped with 1–5 analogues, or reference caps, in mammalian cells. Electroporation was performed with 2.5 µg of each RNA in Huh-7 cells and luciferase expression was measured at time points specified on the top. Prior to transfection, RNA quality was validated by agarose electrophoresis (left panel). Data from duplicate experiments (two measurements per sample) are depicted ±s.d. (Online version in colour.)
(e). Decapping susceptibility
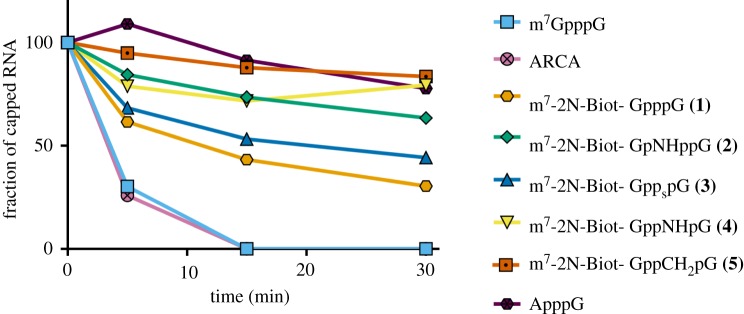
Short RNAs were subjected to enzymatic degradation by hDcp2 in order to test the susceptibility of biotin-labelled cap analogues to cleavage by this RNA-dependent decapping pyrophosphatase. The reactions were stopped at four time intervals and the reaction products were resolved on a denaturing polyacrylamide gel (electronic supplementary material, figure S3), revealing the fraction of decapped RNA at a given time point (figure 6). Reference cap analogues (m7GpppG and ARCA) underwent complete digestion in the first 15 min of reaction, contrary to 2′-biotin-labelled analogues, for which the conversion was slowed down to a degree depending on the modification present in the triphosphate bridge. The highest resistance to degradation was observed for the analogue bearing methylenebisphosphonate modification at the α,β-position (analogue 5). The O-to-NH substitution at the α,β- and β,γ positions of the triphosphate bridge (analogue 4 and 2) improved durability of the RNA 5′ end to a significant degree as well. Surprisingly, our experiment showed little difference in the susceptibility to hDcp2 of β-S-modified cap 3 and unmodified biotin-labelled cap analogue.
Figure 6.
Susceptibility of cap analogues to degradation by hDcp2. The percentage of undigested short capped RNA is plotted as a function of time. (Online version in colour.)
Prior to the design and synthesis of compounds 1–5 no experimentally determined complexes of cap-derived ligand with Dcp1–Dcp2 were available, therefore no assumptions about interactions of biotin-labelled cap analogues with this enzyme could have been made. However, several crystal structures of Dcp1–Dcp2 in complex with cap analogues have become available for analysis recently [26–28]. The fact that biotin-conjugated 5′ mRNA shows improved in vitro stability as a group can be partially explained by the mode of binding, where the 2′-OH group of 7-methylguanosine is enclosed in a binding cavity of Dcp2, with little space left for a bulky substituent [26]. Based on this mechanism of cap recognition, the biotin tag attached at the 2′ position of m7G might hinder the 5′ mRNA cleavage reaction catalysed by Dcp2, thereby increasing the stability of the biotinylated transcripts, without affecting the translation process.
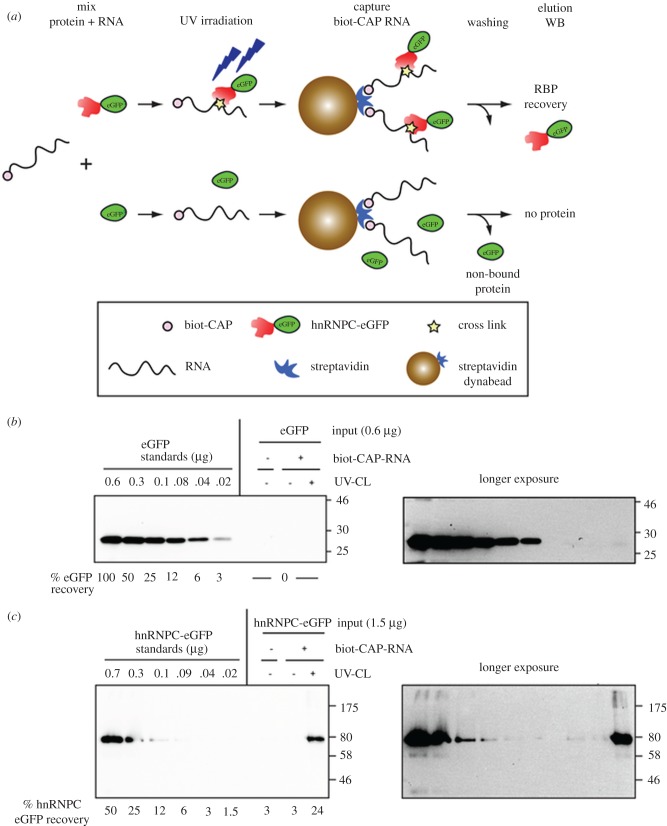
(f). UV-cross-linking and affinity capture of protein–RNA complexes using single-biotin-labelled RNA
The efficient and specific incorporation of a biotinylated cap structure at the 5′ end of RNA was exploited here as a simple method to selectively label RNA and study its interaction with RNA-binding proteins in vitro. Incorporation of biotinylated nucleosides during in vitro transcription by recombinant polymerases is a technique widely used for labelling short RNAs of interest [29–31]. The RNA synthesized contains multiple biotin moieties distributed randomly in its sequence [32]. By contrast, RNA labelling by addition of a biotinylated cap analogue ensures the incorporation of a single biotin molecule at a specific site in the RNA. With this method, the structure and composition of the RNA remains intact and free of additional biotin. Alternative site-specific approaches to capturing RNA–protein complexes include introduction of photo-caging groups at the 5′ termini via enzymatic transfer [33], use of unnatural base pairs [34], labelling based on inverse electron-demand Diels–Alder reactions [35] and other procedures based on biorthogonal chemistry [36,37].
Biotinylated RNAs are useful probes for the identification of RNA-binding proteins from different cells and tissues. The isolation of protein–RNA complexes is achieved by the ability of biotin to bind streptavidin with high affinity. Streptavidin coupled to a solid matrix of beads allows the specific capture of biotinylated RNA easily. To demonstrate the suitability of biotinylated cap analogues for the study of protein–RNA interactions we established a combined protocol including ultraviolet (UV) cross-linking, and affinity purification of biotinylated-capped RNA. For this purpose, we studied the interaction of heterogeneous nuclear ribonucleoprotein C (hnRNPC) with the 3′-non-translated region (NTR) of the hepatitis C virus (HCV) genome. hnRNPC has been described to interact with poly U tracts present in their RNA targets. The HCV 3′-NTR is highly structured but contains a long stretch of polyU/UC. A previous study suggested that the interaction of hnRNPC with the HCV RNA genome is mediated by the binding of its RNA recognition motif to the poly U sequence [38].
Initially, we synthesized a single-biotin-labelled 3′-NTR of HCV by in vitro transcription in the presence of the biotinylated cap analogues 1–5. The percentage of biotinylated-capped RNA in the reaction was greater than or equal to 80% as determined by electrophoretic mobility shift assay (EMSA; electronic supplementary material, figure S4). Next, to optimize the affinity capture of biotinylated RNA, we carried out a titration experiment and determined the maximum binding capacity of streptavidin magnetic beads to the biotinylated RNA. Increasing amounts of biotinylated input RNA (capped with analogue 3) were incubated with a constant amount of magnetic streptavidin dynabeads for 45 min. After extensive washing, the immobilized biotinylated RNA was eluted (for details see Material and methods: UV-cross-linking and affinity capture of protein–RNA complexes) and analysed using a denaturing agarose gel (electronic supplementary material, figure S5). Non-capped RNA corresponding to the 3′-NTR of HCV was used to prepare reference standards of known concentration and also served as negative control in the RNA capture assay. The biotinylated RNA recovered after elution was quantified by densitometry of the RNA bands. We determined that the maximum capacity of streptavidin beads for binding the single-biotin-labelled 3′-NTR of HCV is 40–80 ng µl−1.
To evaluate in vitro the interaction of hnRNPC with the 3′-NTR of HCV, we first immunoisolated hnRNPC-eGFP from human hepatoma cells that stably express this fusion protein. eGFP, a protein that lacks RNA-binding capacity, was used as a negative control (figure 7a). The biotinylated HCV 3′-NTR (capped with analogue 3) was incubated either with hnRNPC-eGFP or eGFP for 30 min on ice. Controls containing only the proteins were used to evaluate the specificity of the capture procedure. The reaction mixtures were exposed or not to UV irradiation to create covalent bonds between the RNA-binding protein and the RNA at protein–RNA contact sites. Protein–RNA complexes were isolated by using our optimized single-biotin-capped RNA affinity capture protocol (electronic supplementary material, figure S5). After extensive and stringent washing, RNA–protein complexes were eluted by RNase treatment. The eluates were subjected to SDS-PAGE and the amounts of captured eGFP and hnRNPC-eGFP were determined by western blot using anti-GFP antibodies.
Figure 7.
Study of protein–RNA interactions by UV-cross-linking and afinity capture of single-biotin-capped RNA. Schematic of RNA-binding protein recovery using biotinylated 3′NTR of HCV (a) capped with analogue 3. The interaction of eGFP (b) and hnRNPC-eGFP (c) with the biotinylated RNA was evaluated by measuring the protein recovery upon RNA affinity capture using western blot and eGFP-specific antibodies. Standards of known amount (on the left of the panels) were used to calculate the percentage of protein recovery (numbers below). Panels on the right are longer exposures of the panels on the left. Protein markers (kDa) are indicated on the right side of each panel. For further details see text. (Online version in colour.)
As expected, eGFP, which is not an RNA-binding protein, was not cross-linked to RNA after UV-light irradiation, nor was it recovered upon affinity capture of the biotinylated RNA (figure 7b). By contrast, we clearly detected the interaction between hnRNPC-eGFP and the biotinylated 3′-NTR of HCV after UV-cross-linking (figure 7c). In the absence of UV-cross-link, this interaction was disrupted by washing with high salt- and detergent-containing buffers used for the RNA capture procedure. These results show that the combination of UV-cross-linking with affinity capture of single-biotin-labelled RNA in vitro is an easy and efficient method to determine the RNA-binding capacity of a protein to its target RNA.
3. Conclusion
In summary, we developed the synthesis of biotin-labelled cap analogues that are efficiently incorporated into RNA during in vitro transcription and that confer resistance against degradation by hDcp2. Those cap analogues, bearing additional modifications in the triphosphate bridge, were shown to retain biological activity with little effect on interaction with eIF4E. All analogues were found to promote RNA translation in vitro. Compound 3, modified with a thiophosphate moiety, was shown to confer better translational properties to capped mRNA than the previously reported biotinylated cap analogue without phosphate modifications (analogue 1). Analogues 4 and 5, modified at the α,β position, as well as compound 2 with β,γ modification, are characterized by strongly decreased susceptibility to hDcp2 degradation in comparison to the natural cap. We demonstrate the utility of biotin-labelled mRNAs obtained with our method for affinity capture experiments, in which specific interaction of heterogeneous nuclear ribonucleoprotein C (hnRNP C) with the polyU tract in an RNA sequence was demonstrated. Biotin-tagged mRNAs have a variety of potential biological applications, including protein affinity purification, pull-down assays, in vivo visualization, cellular delivery and many others. Our single-biotin labelling approach of the cap structure minimizes the risk of the tag (biotin) interference with the recognition of specific RNA motifs (or sequences) by RNA-binding proteins. Moreover, in contrast to other random biotin incorporation protocols, in which the position and amount of biotinylation is more difficult to control, biotinylation of the cap structure overcomes those problems and makes the labelling protocol highly reproducible.
4. Material and methods
(a). Preparation of compounds: supplementary
(i). Fluorescence assays (eIF4E)
Fluorescence quenching experiments were carried out in 50 mM HEPES/KOH (7.2 pH), 100 mM KCl, 0.5 mM EDTA, 1 mM DTT at 20°C in a quartz cuvette with optical lengths of 4 mm for absorption and 10 mm for emission. To a solution of 0.1 µM of eIF4E gradually increasing concentrations of cap analogue were added in 1 µl portions. With each addition, the solution was allowed to stir for 30 s, then the fluorescence intensity was recorded with 30 s time integration. The excitation wavelength was set to 280 nm (slit width 5 nm), with observation at 340 nm and cut-off filter at 290 nm (slit width 10 nm). The experiment was concluded once full saturation was achieved. The samples were thermostatted throughout the experiment. Collected data were corrected for dilution and the inner filter effect, and fitted to a previously described equation [24,39], relating total ligand concentration to fluorescence intensity. Consequently, the equilibrium association constant (KAS) and the concentration of active protein in solution were determined. The final value of KAS represents a weighted average of three independent titrations.
(ii). Enzymatic assays (hDcp2)
RNAs initiated with various cap analogues were synthesized by in vitro transcription with SP6 polymerase (New England BioLabs) as described previously [40]. Short RNAs were generated on templates of annealed oligonucleotides (ATACGATTTAGGTGACACTATAGAAGAAGC-GGGCATGCGGCCAGCCATAGCCGATCA and TGATCGGCTATGGCTGGCCGCA-TGCCCGCTTCTTCTATAGTGTCACCTAAATCGTAT), which contain SP6 promoter sequence (ATTTAGGTGAC-ACTATAGA) and encode modified 35-nt long sequence (GAAGAAGCGGGCA-TGCGGCCAGCCATAGCCGATCA) [41]. Typical in vitro transcription reaction (20 µl) was incubated in 40°C for 2 h and contained: 1 U µl−1 SP6 polymerase, 1 U µl−1 RiboLock RNase Inhibitor (ThermoFisher Scientific), 0.5 mM ATP/CTP/UTP, 0.125 mM GTP, 1.25 mM cap analogue and 0.1 µM annealed oligonucleotides as a template. After 2 h of incubation, 1 U µl−1 DNase I (Ambion) was added to the solution and incubation was resumed for another 30 m at 37°C. EDTA was then added to 25 µM final concentration. Short RNAs were purified using RNA Clean & Concentrator-25 (Zymo Research). Quality of transcripts was checked on 15% polyacrylamide–7 M urea gels; concentration was determined spectrophotometrically. In total, 35-nt long transcripts (1 µM) were incubated with 1 µM DNAzyme 10–23 (TGATCGGCTAGG-CTAGCTACAACGAGGCTGGCCGC) in 50 mM MgCl2 and 50 mM Tris–HCl pH 8.0 for 1 h at 37°C [41], in order to obtain 3′ homogeneous 25-nt RNAs. Prepared transcripts were purified with RNA Clean & Concentrator-25 (Zymo Research) and DNase on-column. Their quality and concentration were checked as before.
Susceptibility of 25 nt RNAs capped with biotin-labelled analogues to enzymatic degradation by human Dcp2 complex was measured with an in vitro assay. A total of 20 ng of RNA were subjected to digestion with 120 nM hDcp2 in decapping buffer (50 mM Tris–HCl pH 8.0, 50 mM NH4Cl, 0.01% NP-40, 1 mM DTT, 5 mM MgCl2 and 2 mM MnCl2) for 1 h at 37°C. Reactions were terminated by addition of equal volume of formamide stop dye (44% formamide, 5 M urea, 20 mM EDTA, 0.03% bromophenol blue, 0.03% xylene cyanol) and the RNA products were analysed by electrophoresis on a 15% denaturing polyacrylamide–7 M urea gel with SYBR Gold (Invitrogen) staining. Visualization was performed using the Storm 860 PhosphorImager system (GE Healthcare).
(iii). In vitro translation
Full-length mRNAs encoding firefly luciferase and capped with biotin-labelled analogues were synthesized in vitro with SP6 polymerase (New England BioLabs). PCR product from pGEM-luc plasmid (Promega) was used as template. Primers (ATTTAG GTGACA CTATAG AAGTAC TGTTGG TAAAGC CACCAT GGAAGA CGCCAA AAACAT and TTACAA TTTGGA CTTTCC GCCCT) allowed for the introduction of SP6 specific promoter sequence. Reactions, containing 1 U µl−1 SP6 polymerase, 1 U µl−1 RiboLock RNase inhibitor (ThermoFisher Scientific), 0.5 mM ATP/CTP/UTP, 0.125 mM GTP, 1.25 mM cap analogue and 25 µg µl−1 template, were incubated at 40°C for 2 h. The DNA template was then digested with 1 U µl−1 DNase I (Ambion) for 30 min at 37°C, followed by addition of EDTA to 25 µM final concentration. Products were purified with NucleoSpin RNA Clean-up XS (Macherey-Nagel). Obtained RNA transcripts were used to programme the RRL in vitro system optimized for cap-dependent translation as described previously [42]. Each translation reaction was performed in 10 µl volume mixture of 40% RRL (Promega), 0.01 mM mixture of amino acids, 1.2 mM MgCl2, 170 mM potassium acetate and one of four different concentrations of 5′-capped mRNA (0.25, 0.125, 0.075 or 0.0375 ng µl−1). Reactions were incubated at 37°C for 1 h. Translation efficiency was measured based on activity of luciferase expressed in the system with Luciferase Reporter Assay (Promega) in a microplate reader Synergy H1 (Biotech). Three independent sets of experiments were carried out for all cap analogues.
(iv). Luciferase assay in mammalian cells
Firefly luciferase in vitro transcripts were generated by using 10 µg pLuc-A60, previously linearized by digestion with HpaI for 3 h at 37°C, and purified. In vitro transcription reaction mixtures (total volume 50 µl; ratio biotinylated cap:GTP = 4 : 1) were prepared as indicated for 3′-NTR of HCV (see section 4a,viii). Luciferase assays were performed with Huh-7 (human hepatoma) cells. Single-cell suspensions were prepared by trypsinization, washed with phosphate-buffered saline (PBS) and resuspended at a concentration of 107 cells per ml in Cytomix supplemented with 2 mM adenosine triphosphate and 5 mM glutathione. An aliquot containing 2.5 µg of luciferase in vitro transcript was mixed with 200 µl cell suspension and transfected by electroporation using a GenePulser system (Bio-Rad, Hercules, CA) and a cuvette with a gap width of 0.2 cm (Bio-Rad). One pulse at 975 µF and 166 V was applied. Cells were immediately diluted in 20 ml complete DMEM and 1.5 ml of the suspension was seeded in each well of a 12-well plate. Translation of the luciferase RNA input was analysed at 4, 6, 8 and 10 h post electroporation by luciferase activity measurement. Cells were washed once with PBS and lysed by adding 350 µl of lysis buffer (0.1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA and 1 mM dithiothreitol, pH 7.8). Cells were frozen immediately at −70°C and, after thawing, the lysates were resuspended by pipetting. A volume of 100 µl lysate was mixed with 360 µl assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol, 2 mM adenosine triphosphate and 15 mM K2PO4, pH 7.8) and, after addition of 200 µl of a luciferin solution (200 mM luciferin, 25 mM glycylglycine [pH 8.0]), measured for 20 s in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany). Lysates in each well were measured in duplicate. Luciferase values from cells electroporated without RNA (background of the assay) were subtracted from the rest of values. Non-capped RNA and RNA capped with non-functional cap (ApppG) were used as negative controls of cap-dependent translation. RNAs capped with ARCA or m7GpppG were used as positive controls of cap-dependent translation.
(v). Recombinant protein expression and purification
Human Dpc2 was expressed and purified according to a previously published protocol by Liu et al. [43].
(vi). Generation of stable cell lines
To generate the lentiviral plasmid pWPI eGFP, an XbaI/BamHI (New England Biolabs) fragment containing eGFP sequence was amplified by PCR, digested and inserted into pcDNA 3.1. The pcDNA 3.1 eGFP construct was digested with PmeI/BamHI and the insert was cloned into pWPI plasmid. To obtain the plasmid pWPI hnRNPC-eGFP, an XbaI/XhoI fragment containing hnRNPC sequence was amplified by PCR from pcDNA5 FRT/TO hnRNPC-eGFP [44] digested and inserted into pcDNA 3.1 eGFP plasmid. The resulting pcDNA 3.1 hnRNPC-eGFP was digested with PmeI/BamHI and the insert was cloned into pWPI plasmid.
To generate cell lines that stably express hnRNPC-eGFP or eGFP, we used a lentiviral vector system. 293T cells (4 × 106 cells) were seeded in a 10 cm culture plate in complete DMEM. Next day, 5 µg of the lentiviral plasmid were transfected together with 5 µg of the packaging vector (pCMV) and 1.7 µg of the VSV envelope vector (pMD.G) into 293T cells by using polyethylenimine according to the instructions of the manufacturer. After 6 h, the medium was replaced by a fresh one, and 48 h later it was collected and filtered through a 0.45 µM pore size polyvinylidene difluoride syringe filter (Carl Roth GmbH, Karlsruhe Germany) and used for transduction. Huh7 cells (1.5 × 106 cells in 3 ml DMEM) were mixed with 4 ml of the filtered lentiviral supernatant, seeded in a 10 cm culture plate and incubated at 37°C. The next day the medium was replaced by fresh medium containing 10 µg ml−1 blasticidin to select infected cells. Cells were maintained in DMEM with blasticidin until they reached 100% confluency. Expression of eGFP and hnRNPC-eGFP was confirmed by visualization of eGFP fluorescence using an epifluorescence microscope, and analysed by western blot.
(vii). Immunoisolation of hnRNPC-eGFP and eGFP from mammalian cells
We used GFP-Trap A beads (Chromotek) to immunoisolate eGFP and hnRNPC-eGFP from Huh7 cell lysates. Initially, Huh7 cells (a 15 cm culture dish with 107 cells) were washed with PBS, collected with a cell scrapper in 1 ml ice-cold PBS and centrifuged. The cell pellet was washed twice with cold PBS and resuspended in 200 µl RIPA buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton X-100 and 1% deoxycholate) containing 9 U DNase, 2 µg RNase A, 5 U RNase T1 (RNase A/T1 mix, ThermoFisher Scientific), 2.5 mM MgCl2, 1X complete protease inhibitor cocktail (Roche) and 1 mM PMSF. Cells were lysed on ice for 30 min (with extensive pipetting every 10 min) and centrifuged at 20 000g for 10 min at 4°C. The supernatant was transfer to a pre-cooled tube, diluted with washing buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1X complete protease inhibitor cocktail and 1 mM PMSF) up to 500 µl and incubated with 25 µl GFP-Trap A beads (previously equilibrated in the same buffer) for 1 h at 4°C with rotation. Next, the beads were washed twice with washing buffer, and once in between with washing buffer containing 500 mM NaCl. Finally, eGFP and hnRNPC-eGFP were eluted by adding 0.2 M glycine (pH 2.5) and constant shaking for 30 s. The pH of the protein solution was immediately neutralized to pH 7.0 with 1 M Tris base (pH 10.4). Protein concentration was determined by the measurement of absorbance at 280 nm using a Nanodrop device (ThermoFisher Scientific). Bovine serum albumin standards of known concentration were used to represent a standard curve and extrapolate the protein concentration in the final eluates.
(viii). UV-cross-linking and affinity capture of protein–RNA complexes
To incorporate the biotinylated cap analogue modified with β-S group in the 3′-NTR of the HCV genome, in vitro transcription reactions (100 µl) were performed including the following components: 0.6 U µl−1 T7 RNA polymerase (Promega), 1 U µl−1 RNAsin (Promega), 1 mM ATP/CTP/UTP/GTP, 5 mM cap analogue, 1X transcription buffer (80 mM HEPES, pH 7.5, 12 mM MgCl2, 2 mM spermidine and 40 mM dithiothreitol), and 0.8 µg DNA template containing T7 RNA polymerase promoter preceding a sequence of 419 nt encoding the 3′-NTR of the HCV genome (isolate JFH1, GenBank accession no. AB047639). Initially, the cap analogue was preincubated for 5 min at 37°C with T7 RNA polymerase, RNAsin and transcription buffer. Next, the ribonucleotides and the DNA template were added and the reaction mixtures incubated at 37°C. After 2-3 h, 0.6 U µl−1 T7 RNA polymerase were added and the reaction continued for 2 h. Transcription was terminated by addition of 1.2 U of RNase-free DNase (Promega) and incubation for 1 h at 37°C. Obtained RNA was purified by extraction with acidic phenol and chloroform, precipitated with isopropanol and ethanol, and dissolved in RNase-free water. To get rid of possible free-biotin and free-ribonucleotides, the RNA was subsequently purified with Illustra MicroSpin G-25 columns (GE healthcare Life Sciences). The concentration of RNA was measured in a Nanodrop device and its integrity was analysed in a non-denaturing agarose gel. The percentage of HCV-derived RNA that incorporated the cap analogue was determined by RNA-EMSA using tetrameric streptavidin (Merck). Further details on the RNA-EMSA experiment are provided in the electronic supplementary material and methods.
We developed a method that combines single-biotin RNA labelling, UV-cross-linking and affinity capture of RNA to analyse protein–RNA interactions in vitro. The binding of hnRNPC, a well-known RNA-binding protein, to its RNA target was analysed to demonstrate the validity of our approach. A short RNA corresponding to the 3′-NTR of HCV was used as target, as it contains a long poly U stretch where hnRNPC preferentially binds [38]. First, the biotinylated cap analogue containing a β-S group (analogue 3) was incorporated at the 5′ end of the RNA during in vitro transcription. The single-biotin-labelled RNA was purified and its concentration and integrity were determined. Aliquots of biotin-labelled RNA were prepared and stored at −80°C until used. Affinity binding of single-biotin-capped RNA to magnetic streptavidin dynabeads MyOne C1 (Life Technologies) was initially optimized by titration experiments to maximize the capture of biotinylated RNA (see Results). The dynabeads were washed three times in pre-chilled binding and high salt washing buffer (B&hsW: 10 mM Tris–HCl, pH 7.5, 1 M NaCl, 1 mM EDTA, 1 mM DTT, 0.02% NP-40 and 40 U RNAsin) and blocked for 15 min at 4°C in the same buffer containing 200 µg ml−1 yeast tRNA (Sigma). To immobilize the biotinylated 3′-NTR of HCV, constant amounts of beads in B&hsW buffer were incubated with increasing amounts of RNA for 45 min at 4°C using gentle rotation. Next, the dynabeads were extensively washed, under stringent conditions that disrupt protein–protein interactions without affecting streptavidin-biotin binding, as follows: two times with B&hsW, two times with low salt buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.02% NP-40, 0.05% SDS and 40 U ml−1 RNAsin), two times with low salt buffer without SDS and two times with low salt buffer in the absence of detergents. Finally, the single-biotin-labelled RNA was eluted by incubation with 10 mM EDTA (pH 8.2) and 95% formamide for 5 min at 65°C. The RNA capture efficiency was assessed in 1% non-denaturing agarose gel. To determine the proportion of biotinylated RNA recovered upon affinity binding to streptavidin dynabeads, we compared RNA concentration before and after elution from dynabeads. For this, we used RNA standards of known concentrations. The intensity of bands from biotinylated RNA and reference RNA was quantified by densitometry using Quantity One software (Bio-Rad) and the amount of biotinylated RNA (% to the initial RNA input) was determined by extrapolation to the reference RNAs using Graph Prism software (GraphPad Software, Inc.).
To analyse the interaction between hnRNPC and the 3′-NTR of HCV, the single-biotin-labelled RNA (18 pmol) (previously denatured at 95°C for 3 min and refolded for 10 min at 20°C) was incubated with hnRNPC-eGFP (200 nM) or eGFP (200 mM) in reaction buffer (20 mM Tris–HCl, pH 7.5, 12.5 mM MgCl2, 25 mM KCl, 1 mM EDTA, 1 mM DTT and 40 U ml−1 RNasin) for 30 min on ice. The samples were transferred to a pre-chilled small plastic plate and irradiated with 0.15 J cm−2 at 254-nm UV light in a Stratalinker 1800 (Stratagene). Non-irradiated samples were used as controls to confirm that hnRNPC–RNA interaction is not preserved under the stringent conditions of the RNA capture protocol. Immediately after UV-cross-linking, the samples were transferred to fresh pre-chilled tubes and diluted with B&hsW buffer. At this point, we added 55 µl pre-washed streptavidin dynabeads to the samples and followed the affinity capture procedure we established previously with minor modifications. In this case, it is essential to perform the two last washing steps in the absence of RNAsin, as the elution of RNA-binding proteins is achieved by addition of 1 µl RNase A/T1 cocktail (2 µg µl−1 RNase A; 5 U µl−1 RNase T1) in 50 mM triethylamonium bicarbonate (pH 8.5) and 5 mM EDTA. After 30 min incubation at 37°C with shaking, we added 0.1% SDS to the samples and boiled them for 5 min. The supernatants were collected, mixed with 6X protein loading buffer and proteins were separated by SDS-polyacrylamide gel electrophoresis and electrotransferred onto nitrocellulose membranes (GE Healthcare). Membranes were blocked with PBS containing 5% dried milk for 1 h before overnight incubation (4°C) with specific rat anti-GFP 3H9 antibodies (Chromotek) diluted 1 : 1000 in PBS containing 1% dry milk. Membranes were washed three times with PBS supplemented with 0.2% Tween 20 and incubated for 1 h with horseradish-peroxidase-conjugated rat secondary antibodies (Sigma-Aldrich, St Louis, MO) diluted 1 : 10 000. Bound antibodies were detected by using the Western Lightning PlusECL reagent (Perkin-Elmer, Waltham, MA). Signals were visualized on BioMax Light films (Kodak, Rochester, MN). The presence of the protein in the eluates denotes its RNA-binding capacity.
Supplementary Material
Acknowledgements
We thank Christopher Lima for plasmid pet28-Dcp2 encoding hDcp2(1-350) protein. We also thank Dorota Kubacka for expressing and purifying the eIF4E protein.
Data accessibility
This article has no additional data.
Authors' contributions
J.J., R.B. and J.K. were involved in planning of the study and supervised the work. S.B. performed chemical syntheses. S.B. and P.S. planned, performed and analysed the experiments shown in figures 4, 6 and electronic supplementary material, figure S2. V.M. planned, performed and analysed the experiments shown in figures 5, 7 and electronic supplementary material, figures S3 and S4. J.J., S.B., V.M. and J.K. wrote the paper with input from all authors. All authors discussed the results and commented on the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Science Centre, Poland (UMO-2016/21/B/ST5/02556). R.B. was supported by the Deutsche Forschungsgemeinschaft, TRR179, TP9.
References
- 1.Curry S, Kotik-Kogan O, Conte MR, Brick P. 2009. Getting to the end of RNA : structural analysis of protein recognition of 5′ and 3′ termini. Biochim. Biophys. Acta Gene Regul. Mech. 1789, 653–666. ( 10.1016/j.bbagrm.2009.07.003) [DOI] [PubMed] [Google Scholar]
- 2.Quiocho FA, Hu G, Gershon PD. 2000. Structural basis of mRNA cap recognition by proteins. Curr. Opin. Struct. Biol. 10, 78–86. ( 10.1016/S0959-440X(99)00053-6) [DOI] [PubMed] [Google Scholar]
- 3.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. 2011. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298. ( 10.1002/wrna.52) [DOI] [PubMed] [Google Scholar]
- 4.Gu M, Lima CD. 2005. Processing the message: structural insights into capping and decapping mRNA. Curr. Opin. Struct. Biol. 15, 99–106. ( 10.1016/j.sbi.2005.01.009) [DOI] [PubMed] [Google Scholar]
- 5.von der Haar T, Gross JD, Wagner G, McCarthy JEG. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11, 503–511. ( 10.1038/nsmb779) [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Rodgers ND, Jiao X, Kiledjian M. 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21, 4699–4708. ( 10.1093/emboj/cdf448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graff J, Konicek B. 2007. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 117, 2638–2648. ( 10.1172/JCI32044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziemniak M, Strenkowska M, Kowalska J, Jemielity J. 2013. Potential therapeutic applications of RNA cap analogs. Future Med. Chem. 5, 1141–1172. ( 10.4155/fmc.13.96) [DOI] [PubMed] [Google Scholar]
- 9.Sahin U, Karikó K, Türeci Ö. 2014. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780. ( 10.1038/nrd4278) [DOI] [PubMed] [Google Scholar]
- 10.Wilchek M, Bayer EA, Livnah O. 2006. Essentials of biorecognition: the (strept)avidin-biotin system as a model for protein-protein and protein-ligand interaction. Immunology Lett. 103, 27–32. ( 10.1016/j.imlet.2005.10.022) [DOI] [PubMed] [Google Scholar]
- 11.Diamandis EP, Christopoulos TK. 1991. The biotin-(strept)avidin system: principles and applications in biotechnology. Clin. Chem. 37, 625–636. [PubMed] [Google Scholar]
- 12.Trippier PC. 2013. Synthetic strategies for the biotinylation of bioactive small molecules. ChemMedChem 8, 190–203. ( 10.1002/cmdc.201200498) [DOI] [PubMed] [Google Scholar]
- 13.Rodgers JT, Patel P, Hennes JL, Bolognia SL, Mascotti DP. 2000. Use of biotin-labeled nucleic acids for protein purification and agarose-based chemiluminescent electromobility shift assays. Anal. Biochem. 277, 254–259. ( 10.1006/abio.1999.4394) [DOI] [PubMed] [Google Scholar]
- 14.Jemielity J, Lukaszewicz M, Kowalska J, Czarnecki J, Zuberek J, Darzynkiewicz E. 2012. Synthesis of biotin labelled cap analogue—incorporable into mRNA transcripts and promoting cap-dependent translation. Org. Biomol. Chem. 10, 8570–8574. ( 10.1039/c2ob26060c) [DOI] [PubMed] [Google Scholar]
- 15.Jemielity J, Kowalska J, Rydzik AM, Darzynkiewicz E. 2010. Synthetic mRNA cap analogs with a modified triphosphate bridge—synthesis, applications and prospects. New J. Chem. 34, 829 ( 10.1039/c0nj00041h) [DOI] [Google Scholar]
- 16.Marcotrigiano J, Gingras C, Sonenberg N, Burley SK. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89, 951–961. ( 10.1016/S0092-8674(00)80280-9) [DOI] [PubMed] [Google Scholar]
- 17.Jemielity J, Fowler T, Zuberek J, Stepinski J, Lewdorowicz M, Niedzwiecka A, Stolarski R, Darzynkiewicz E, Rhoads RE. 2003. Novel ‘anti-reverse’ cap analogs with superior translational properties. RNA 9, 1108–1122. ( 10.1261/rna.5430403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomasz J, Vaghefi M. 1988. Nucleoside imidodiphospbates synthesis and biological activities. Nucleic Acids Res. 16, 8645–8664. ( 10.1093/nar/16.17.8645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michinori K, Wada T, Urashima C, Sekine M. 1997. Efficient synthesis of gamma-methyl-capped guanosine 5′-triphosphate as a 5′-terminal unique structure of U6 RNA via a new triphosphate bond formation involving activation of methyl phsphoroimidazolidate using ZnCl2 as a catalyst in DMF under anhydrous conditions. Tetrahedron Lett. 38, 8359–8362. ( 10.1016/S0040-4039(97)10263-5) [DOI] [Google Scholar]
- 20.Kalek M, Jemielity J, Darzynkiewicz ZM, Bojarska E, Stepinski J, Stolarski R, Davis RE, Darzynkiewicz E. 2006. Enzymatically stable 5′ mRNA cap analogs: synthesis and binding studies with human DcpS decapping enzyme. Bioorg. Med. Chem. 14, 3223–3230. ( 10.1016/j.bmc.2005.12.045) [DOI] [PubMed] [Google Scholar]
- 21.Kowalska J, et al. 2008. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA 14, 1119–1131. ( 10.1261/rna.990208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalek M, Jemielity J, Stepinski J, Stolarski R, Darzynkiewicz E. 2005. A direct method for the synthesis of nucleoside 5′-methylenebis(phosphonate)s from nucleosides. Tetrahedron Lett. 46, 2417–2421. ( 10.1016/j.tetlet.2005.02.069) [DOI] [Google Scholar]
- 23.Rydzik AM, Kulis M, Lukaszewicz M, Kowalska J, Zuberek J, Darzynkiewicz ZM, Darzynkiewicz E, Jemielity J. 2012. Synthesis and properties of mRNA cap analogs containing imidodiphosphate moiety--fairly mimicking natural cap structure, yet resistant to enzymatic hydrolysis. Bioorg. Med. Chem. 20, 1699–1710. ( 10.1016/j.bmc.2012.01.013) [DOI] [PubMed] [Google Scholar]
- 24.Niedzwiecka A, et al. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319, 615–635. ( 10.1016/S0022-2836(02)00328-5) [DOI] [PubMed] [Google Scholar]
- 25.Burgess HM, Mohr I. 2015. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17, 332–344. ( 10.1016/j.chom.2015.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mugridge JS, Ziemniak M, Jemielity J, Gross JD. 2016. Structural basis of mRNA-cap recognition by Dcp1Dcp2. Nat. Struct. Mol. Biol, 1–9. ( 10.1038/nsmb.3301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charenton C, Taverniti V, Gaudon-Plesse C, Back R, Séraphin B, Graille M.. 2016. Structure of the active form of Dcp1Dcp2 decapping enzyme bound to m7GDR and its Edc3 activator Nat. Struct. Mol. Biol. 23, 982–986. ( 10.1038/nsmb.3300) [DOI] [PubMed] [Google Scholar]
- 28.Mugridge JS, Tibble RW, Ziemniak M, Jemielity J, Gross JD. 2018. Structure of the activated Edc1-Dcp1-Dcp2-Edc3 mRNA decapping complex with substrate analog poised for catalysis. Nat. Commun. 9, 1152 ( 10.1038/s41467-018-03536-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KY, Kee MK, Chong SA, Nam MJ. 2007. Galanin is up-regulated in colon adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 16, 2373–2379. ( 10.1158/1055-9965.EPI-06-0740) [DOI] [PubMed] [Google Scholar]
- 30.Kirby R, Cho EJ, Gehrke B, Bayer T, Sok Y, Neikirk DP, Mcdevitt JT, Ellington AD, Park YS. 2004. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal. Chem. 76, 4066–4075. ( 10.1021/ac049858n) [DOI] [PubMed] [Google Scholar]
- 31.Li RW, et al. 2006. Identification of estrogen-responsive genes in the parenchyma and fat pad of the bovine mammary gland by microarray analysis. Physiol. Genomics 27, 42–53. ( 10.1152/physiolgenomics.00032.2006) [DOI] [PubMed] [Google Scholar]
- 32.Langer PR, Waldrop AA, Ward DC. 1981. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc. Natl Acad. Sci. USA 78, 6633–6637. ( 10.1073/pnas.78.11.6633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muttach F, Mäsing F, Studer A, Rentmeister A. 2017. New AdoMet analogues as tools for enzymatic transfer of photo-cross-linkers and capturing RNA–protein interactions. Chem. Eur. J. 23, 5988–5993. ( 10.1002/chem.201605663) [DOI] [PubMed] [Google Scholar]
- 34.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. 2005. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 33, 1–8. ( 10.1093/nar/gni128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoch J, Ameta S, Jäschke A. 2011. Inverse electron-demand Diels–Alder reactions for the selective and efficient labeling of RNA. Chem. Commun. 47, 12 536–12 537. ( 10.1039/c1cc15476a) [DOI] [PubMed] [Google Scholar]
- 36.Büttner L, Javadi-Zarnaghi F, Höbartner C. 2014. Site-specific labeling of RNA at internal ribose hydroxyl groups: terbium-assisted deoxyribozymes at work. J. Am. Chem. Soc. 136, 8131–8137. ( 10.1021/ja503864v) [DOI] [PubMed] [Google Scholar]
- 37.Holstein JM, Schulz D, Rentmeister A. 2014. Bioorthogonal site-specific labeling of the 5′-cap structure in eukaryotic mRNAs. Chem. Commun. 50, 4478–4481. ( 10.1039/C4CC01549E) [DOI] [PubMed] [Google Scholar]
- 38.Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott G, Del Vecchio AM. 1999. hnRNPC and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region with the 3′ NTR of the HCV genome. Nucleic Acids Res. 27, 1457–1463. ( 10.1093/nar/27.6.1457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eftink MR. 1997. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 278, 221–257. ( 10.1016/S0076-6879(97)78013-3) [DOI] [PubMed] [Google Scholar]
- 40.Warminski M, et al. 2017. Amino-functionalized 5′ cap analogs as tools for site-specific sequence-independent labeling of messenger RNA. Bioconjug. Chem. 28, 1978–1992. ( 10.1021/acs.bioconjchem.7b00291) [DOI] [PubMed] [Google Scholar]
- 41.Coleman TM, Wang G, Huang F. 2004. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 phi 2.5 promoter. Nucleic Acids Res. 32, e14 ( 10.1093/nar/gnh007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rydzik AM, Lukaszewicz M, Zuberek J, Kowalska J, Darzynkiewicz ZM, Darzynkiewicz E, Jemielity J. 2009. Synthetic dinucleotide mRNA cap analogs with tetraphosphate 5′,5′ bridge containing methylenebis(phosphonate) modification. Org. Biomol. Chem. 7, 4763–4776. ( 10.1039/b911347a) [DOI] [PubMed] [Google Scholar]
- 43.Liu S, Jiao X, Liu H, Gu M, Lima CD, Kiledjian M. 2004. Functional analysis of mRNA scavenger decapping enzymes. RNA 10, 1412–1422. ( 10.1261/rna.7660804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strein C, Alleaume A, Rothbauer U, Hentze MW, Castello A. 2014. A versatile assay for RNA-binding proteins in living cells. RNA 20, 1–11. ( 10.1261/rna.043562.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.