Abstract
RNA uridylation consists of the untemplated addition of uridines at the 3′ extremity of an RNA molecule. RNA uridylation is catalysed by terminal uridylyltransferases (TUTases), which form a subgroup of the terminal nucleotidyltransferase family, to which poly(A) polymerases also belong. The key role of RNA uridylation is to regulate RNA degradation in a variety of eukaryotes, including fission yeast, plants and animals. In plants, RNA uridylation has been mostly studied in two model species, the green algae Chlamydomonas reinhardtii and the flowering plant Arabidopsis thaliana. Plant TUTases target a variety of RNA substrates, differing in size and function. These RNA substrates include microRNAs (miRNAs), small interfering silencing RNAs (siRNAs), ribosomal RNAs (rRNAs), messenger RNAs (mRNAs) and mRNA fragments generated during post-transcriptional gene silencing. Viral RNAs can also get uridylated during plant infection. We describe here the evolutionary history of plant TUTases and we summarize the diverse molecular functions of uridylation during RNA degradation processes in plants. We also outline key points of future research.
This article is part of the theme issue ‘5′ and 3′ modifications controlling RNA degradation’.
Keywords: RNA degradation, RNA decay, terminal nucleotidyltransferase, uridylation, mRNAs
1. RNA uridylation, a key post-transcriptional regulatory process
RNA uridylation is a post-transcriptional modification, which consists of the addition of uridines to the 3′ end of RNA. RNA uridylation plays a key role in the regulation of gene expression across eukaryotes, with the exception to date of Saccharomyces cerevisiae, which has lost the capacity to uridylate RNAs. U-tailing has been reported for a variety of RNA substrates: from mitochondrial transcripts in trypanosomes, to mRNAs and a plethora of non-coding RNAs in diverse organisms, including fission yeast, amphibians, insects, plants or mammals [1–9]. Uridylation targets transcripts produced by RNA polymerases I, II and III (see accompanying articles by Zigackova and Vanacova [10], Warkocki et al. [11] and [12–14]) and it emerges as a pervasive post-transcriptional process.
RNA uridylation plays diverse roles, which depend on the cellular compartment, the identity of the terminal uridylyltransferase (TUTase) or the RNA substrate itself [1,2,4–9]. The 3′ untemplated addition of uridines may facilitate processing of primary transcripts, stabilize RNA and possibly control translation of mRNAs. Yet, its chief role is to trigger degradation both by the 5′–3′ and 3′–5′ RNA degradation pathways [1,2,4–9].
Here, we summarize our current knowledge on RNA uridylation and decay in plants. We begin by describing the terminal nucleotidyltransferase (TNTase) family, to which TUTases belong. As an example, the organization of the small multigenic TNTase family is detailed for the model plant Arabidopsis thaliana. We then present a comprehensive evolutionary history of TUTases in Archaeplastida (i.e. all plants), which reveals that two TUTases have been maintained in the whole green lineage, suggesting specific and critical functions. We then review our current knowledge on how uridylation by these TUTases impacts the degradation of various classes of RNAs in plants.
2. Characteristic features of TUTases in plants
RNA uridylation is catalysed by terminal uridylyltransferases (TUTases). TUTases belong to the superfamily of DNA polymerase beta-like nucleotidyltransferases [15]. This superfamily regroups enzymes that conjugate nucleotides to proteins, antibiotics or RNAs [15]. The nucleotidyltransferases that add untemplated nucleotides (adenosines, uridines, guanosines and cytidines) to the 3′ end of RNAs are called ribonucleotidyl transferases (rNTases) or terminal nucleotidyltransferases (TNTases).
(a). Classification of terminal nucleotidyltransferases
TNTases are split into two classes based on structural differences in their catalytic fold and in the domain responsible for nucleotide selection [15]. Class I includes the ‘canonical’ poly(A) polymerases (cPAPs), which are responsible for the co-transcriptional addition of stabilizing poly(A) tails to transcripts synthesized by RNA polymerase II (Pol II), the tRNA CCA-adding enzymes in Archaea, 2′-5′-oligo(A) synthetases (OAS) and a group of TNTases involved in the polyadenylation, uridylation, cytidylation and guanylation of diverse RNA substrates [3,15]. Class II TNTases correspond to bacterial poly(A) polymerases and tRNA CCA-adding enzymes found in eukaryotes and in certain bacteria.
(b). Eukaryotic TNTases are encoded by small multigenic families
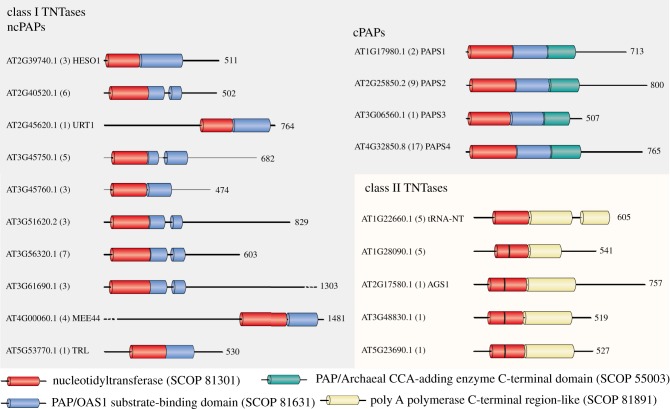
TNTases are encoded by small multigenic families, whose complexity varies across eukaryotes. For instance, three canonical and 11 non-canonical PAPs (ncPAPs) are expressed in humans [16] (see also accompanying paper by Warkocki et al. [11]). TNTases are also encoded by small multigenic families in plants, as reported for the green algae Chlamydomonas reinhardtii, or for two land plants: Zea mays (maize) and Arabidopsis thaliana [17–24]. In Arabidopsis, 19 TNTase genes have been identified on the basis of sequence homology: 14 class I TNTases and 5 class II TNTases (figure 1). The class II of Arabidopsis TNTases contains a single tRNA CCA-adding enzyme also called tRNA-nucleotidyltransferase (tRNA-NT), which processes tRNAs encoded by the nuclear, plastidial and mitochondrial genomes [26], and four bacterial PAP-like nucleotidyltransferases, which are predicted to localize in mitochondria and plastids [18] (figure 1). The class I of Arabidopsis TUTases is composed of 10 non-canonical PAPs and four PAPS that contain the characteristic domains of canonical PAPs (PAPS1 to 4) [17–20]. Yet, PAPS3 is localized in the cytosol, is mostly expressed in pollen and does not contain the C-terminal extension found in PAPS1, S2 and S4 (figure 1) [19]. The role of PAPS3 remains to be characterized. By contrast, PAPS1, S2 and S4 correspond to the canonical PAPs involved in the co-transcriptional polyadenylation of RNA Pol II transcripts. Interestingly, PAPS1, S2 and S4 are functionally specialized and preferentially target subpopulations of transcripts [27–30].
Figure 1.
Domain organization of class I and class II TNTases of A. thaliana. The class I TNTase family is composed of 10 non-canonical poly(A) polymerases (ncPAPs) and four canonical poly(A) polymerases (cPAPs). The class II TNTase family contains the tRNA nucleotidyltransferase (tRNA-NT), also called the tRNA CCA-adding enzyme, and four bacterial PAP-like nucleotidyltransferases. Boxes represent conserved structural domains identified using the structural classification of proteins (SCOP) according to the superfamily database [25]. Non-conserved regions are drawn as lines. Each TNTase is identified by its AGI (Arabidopsis Genome Initiative) reference gene model. The numbers of all gene models are shown in parentheses. Finally, names are shown for the 10 TNTases that have been studied to date. The numbers on the right indicate the number of amino acids for each TNTase. The colour code for the Superfamily SCOP domains is indicated on the figure. The vertical black bar drawn in the nucleotidyltransferase domain of four of the five class II TNTases represents a motif discriminating bacterial PAP-like nucleotidyltransferases from bacterial tRNA-NT [15].
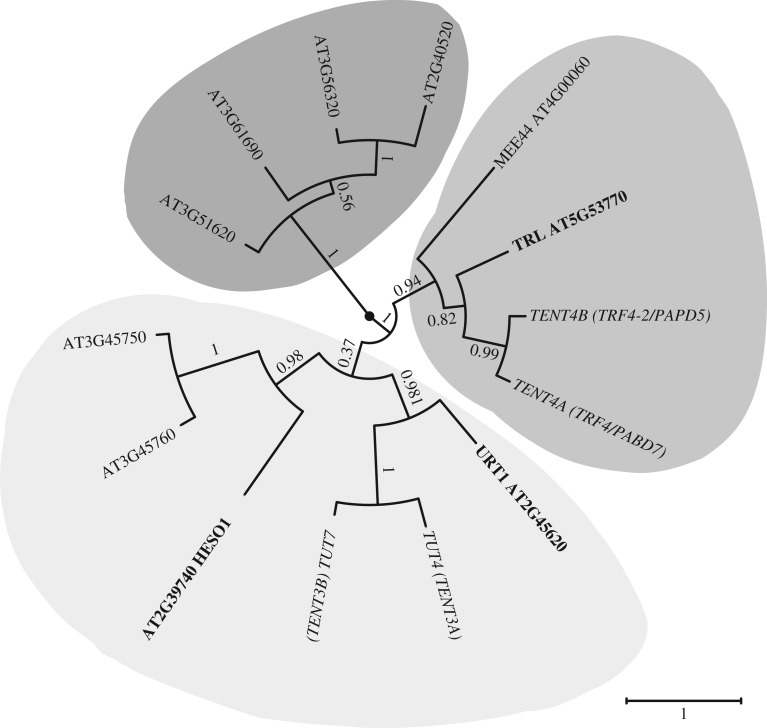
Much remains to be discovered about the function of the 10 Arabidopsis class I ncPAPs. To date, functional data have been reported for three of them: TRF4/5-LIKE (TRL), HEN1 SUPPRESSOR 1 (HESO1) and UTP:RNA URIDYLYLTRANSFERASE 1 (URT1) [20,22,31,32]. TRL is a nuclear ncPAP, which adenylates rRNA maturation by-products and precursors to facilitate their degradation or processing by the RNA exosome [22]. TRL is an orthologue of TRF4 in S. cerevisiae or TENT4B (hTRF4–2, PAPD5) in humans [33–36]. Another Arabidopsis class I ncPAP, MEE44, is evolutionarily close to TRL and TENT4A/B. Indeed, a phylogenetic analysis using the nucleotidyltransferase domains of the 10 Arabidopsis ncPAPs aligned with that of four human ncPAPs (TENT4A, TENT4B and the 2 TUTases TUT4 and TUT7) reveals that TRL and MEE44 cluster with TENT4A/4B (figure 2). This analysis also indicates that four uncharacterized Arabidopsis ncPAPs form a distinct clade and finally that four proteins including HESO1 and URT1 cluster with the human TUTases TUT4/7 (figure 2). The nucleotide specificity of AT3G45750 and AT3G45760 has not been reported yet, but HESO1 and URT1 are indeed bona fide TUTases. Altogether, HESO1 and URT1 uridylate small RNAs, mRNAs and the 5′ fragments resulting from cleavage by the RNA-induced silencing complex (RISC) [20,31,32,39,40]. Of note, HESO1 is the homologue of MUT68, which was discovered in C. reinhardtii, as a ncPAP involved in the degradation of RISC-cleaved transcripts and small RNAs [41,42].
Figure 2.
Phylogenetic relationship among A. thaliana class I ncPAPs and four human ncPAPs. The nucleotidyltransferase domains SCOP 81301 and the PAP/OAS1 substrate-binding domains SCOP 81631 of the 10 class I ncPAPs of Arabidopsis and four human ncPAPs were aligned with Muscle (v. 3.8.31) [37]. The maximum-likelihood tree was generated using PhyML (v. 3.1) on Phylogeny.fr [38] and edited using FigTree (v. 1.4.3, http://tree.bio.ed.ac.uk/software/figtree/). Arabidopsis and human ncPAPs are indicated in regular and italic characters, respectively. Support values (approximate likelihood-ratio statistical test, aLRT v 3.0) are shown on branches. The scale bar represents the number of substitutions per amino acid site. HESO1, URT1 and two other TNTases form a cluster with human TUT7 and TUT4. MEE44 and TRL form a cluster with human TENT4A and TENT4B. The remaining four TNTases form a separated cluster. HESO1, URT1 and TRL, the three class I ncPAPs that have been functionally characterized in Arabidopsis, are indicated in bold.
(c). Evolutionary analysis of TUTases in plants
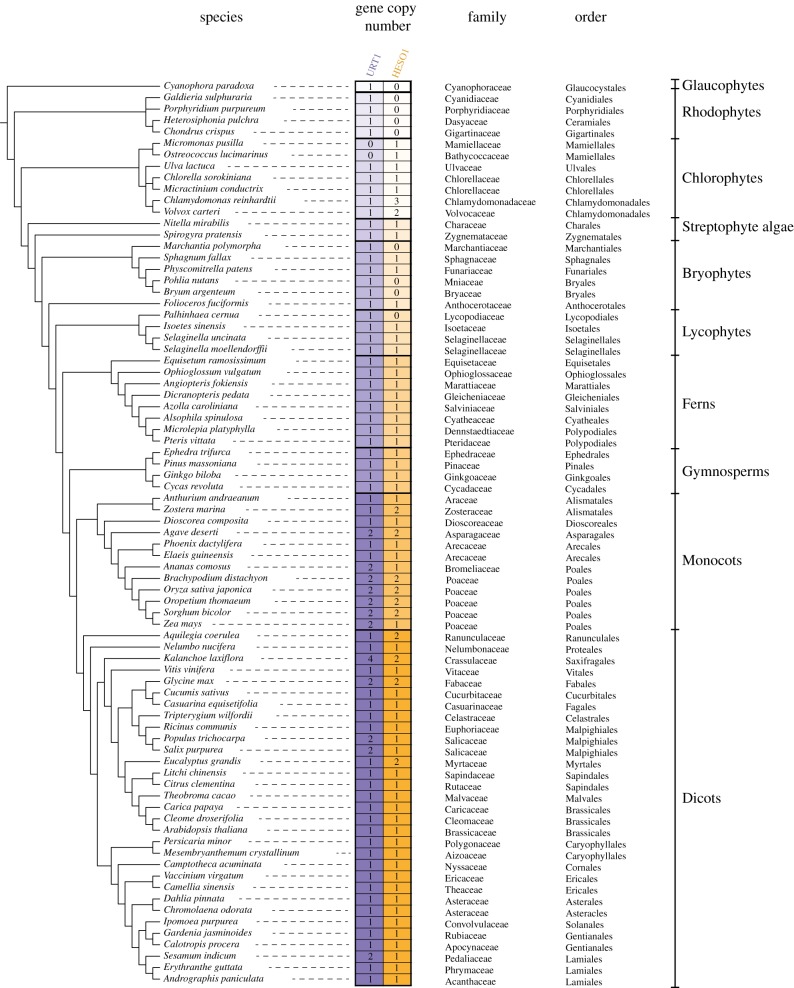
A recent phylogenetic analysis of HESO1 and URT1 homologues, mostly from bryophytes, lycophytes and ferns, revealed that each TUTase forms a monophyletic group [43]. To extend our knowledge on the evolutionary history of TUTases in Archaeplastida (i.e. all plants), a comprehensive phylogenetic analysis was performed using URT1 and HESO1 homologues from 79 species representing all major groups of Archaeplastida, including glaucophytes, rhodophytes (red algae), chlorophytes and streptophyte algae, bryophytes (liverworts, hornworts and mosses), lycophytes and pteridophytes (e.g. ferns), gymnosperms (e.g. conifers and Gingko), and angiosperms (flowering plants; figure 3). To retrieve URT1 and HESO1 homologue sequences, we screened phytozome, NCBI TSA and NCBI EST databases by using either BLASTP or TBLASTN algorithms with the amino acid sequence of Arabidopsis URT1 (AT2G45620) and HESO1 (AT2G39740). We also included in our analysis previously published URT1 and HESO1 sequences from bryophytes, lycophytes and ferns [43] (see electronic supplementary material, Dataset S1 and S2 for a compilation of HESO1 and URT1 sequences, respectively). URT1 and HESO1 homologous sequences for the representative species shown in figure 3 were separately aligned with MUSCLE [37]. Amino acids that did not align to the Catalytic Core Domain (CCD) (COG5260) identified in URT1 and HESO1 were trimmed. Finally, URT1 and HESO1 trimmed sequences were realigned altogether and the phylogenetic tree shown in figure 4 was generated using the maximum-likelihood method (v. 3.1/3.0 aLRT) and WAG substitution model implemented in PhyML [45,46] (see electronic supplementary material, Dataset S3 for the final alignment of all sequences). The main outcome of this analysis is that a monophyletic group is observed for each TUTase, indicating an early divergence of HESO1 and URT1 that have been maintained in the green lineage (figure 4). In most species, homologues of HESO1 and URT1 are present each as a single copy (figures 3 and 4), raising the possibility that they are orthologues, as previously proposed for bryophyte, lycophyte and fern species [43]. Yet, several species either have multiple URT1 and/or HESO1 homologues, or have only one TUTase: either HESO1 or URT1. Interestingly, species representing the phyla Glaucophyta and Rhodophyta lack a HESO1 homologue (figures 3 and 4). Because glaucophytes and rhodophytes are early diverging in the Archaeplastida lineage, the absence of HESO1 homologues suggests that either URT1 homologues predate the apparition of HESO1, or glaucophytes and rhodophytes have lost HESO1. In addition, HESO1 was not detected in representative species of Marchantiales, Bryales and Lycopodiales (figures 3 and 4). Conversely, two species of the order Mamiellales, Micromonas pusilla and Ostreococcus lucimarinus, have no URT1 homologues. We further checked that URT1 is not detected in Ostreococcus tauri, another species of the order Mamiellales. Those three Chlorophyta green algae of the order Mamiellales are unicellular organisms with a reduced nuclear genome that seem to have lost URT1 during speciation. Of note, Mamiellales is the only order of all Viridiplantae lacking a URT1 homologue. Thus, the absence of either HESO1 or URT1 homologues appears extremely rare in plant species, and in such species, a single TUTase could be responsible for the RNA uridylation catalysed by both HESO1 and URT1 in most plant species. Conversely, certain plant species have dual copies of URT1 and/or HESO1 because of either local or whole genome duplication (WGD) events, as for Glycine max (figures 3 and 4) [48]. Four out of five representative species of Poaceae chosen for our initial analysis shown in figures 3 and 4 also have at least two copies of URT1 and HESO1. To obtain a better view of the evolutionary history of TUTases in this family that regroups important crops such as maize (Zea mays), sorghum (Sorghum bicolor) or rice (Oryza sativa), a more focused phylogenetic analysis of URT1 and HESO1 homologues was performed from 11 species of Poales (10 Poaceae and the Bromeliaceae Ananas comosus) (figure 5; see electronic supplementary material, Datasets S4 and S5 for alignments of HESO1 and URT1 sequences, respectively). In fact, two copies of HESO1 were not systematically found (figure 5). For instance, maize and pineapple (A. comosus) are among the Poales species that lack the second HESO1 isoform, noted HESO1B. Usually, HESO1A, which is the isoform detected in all Poales, is constitutively expressed, and at a higher level than HESO1B (figure 6). Altogether, these data suggest that HESO1A could be the orthologue of the eudicot HESO1, while HESO1B is either dispensable in some Poales or may have acquired a specialized function or expression pattern in certain Poales species.
Figure 3.
Copy number of HESO1 and URT1 in 79 representative species of Archaeplastida. The phylogenic relationship between the 79 species analysed in this study was visualized using Phylostatic [44]. Full species names are grouped by taxonomic clades indicated on the right. The colour code for URT1 and HESO1 in the different clades is conserved in figures 4 and 5. The numbers of copies detected for HESO1 and URT1 are indicated for each species. Sequences in FASTA format are given in electronic supplementary material, Datasets S1 and S2 for HESO1 and URT1, respectively.
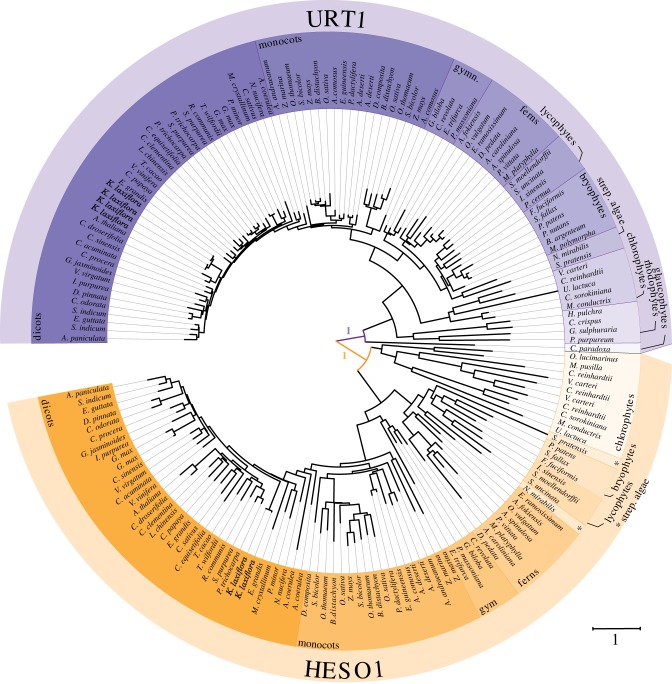
Figure 4.
Phylogenetic relationship between URT1 and HESO1 sequences among 79 representative species of Archaeplastida. The phylogenetic tree was generated using the maximum-likelihood method and WAG substitution model implemented in PhyML (v. 3.1) [45,46]; see electronic supplementary material, Dataset S3 for sequence alignment). The tree was edited using iTOL (v. 4.2.1) [47]. Colour code for taxonomic clades is defined in figure 3. Statistical values for the first branches (approximate likelihood-ratio test, aLRT v. 3.0) support that URT1 and HESO1 proteins form two distinct clades. The scale bar represents the number of substitutions per amino acid site.
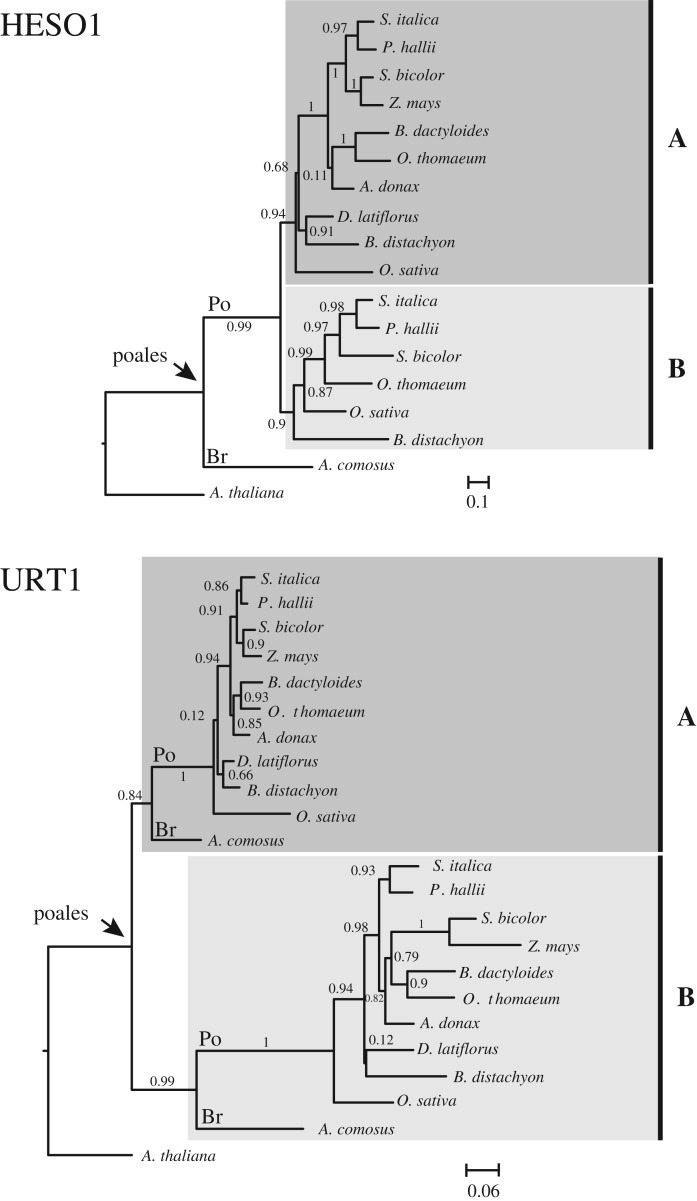
Figure 5.
Phylogenetic relationship of URT1 and HESO1 isoforms among 11 species of Poales. Sequences of HESO1 and URT1 isoforms from 10 Poaceae (Po) and 1 Bromeliaceae (Br) were aligned separately. Full names of species are given in figure 3. The analysis was performed as described for figure 4 except that the trees were edited with FigTree (v. 1.4.3, http://tree.bio.ed.ac.uk/software/figtree/). Support values (approximate likelihood-ratio statistical test, aLRT v. 3.0) are shown on branches. The sequence alignments for HESO1 and URT1 used to build the trees are given in electronic supplementary material, Datasets S4 and S5, respectively.
Figure 6.
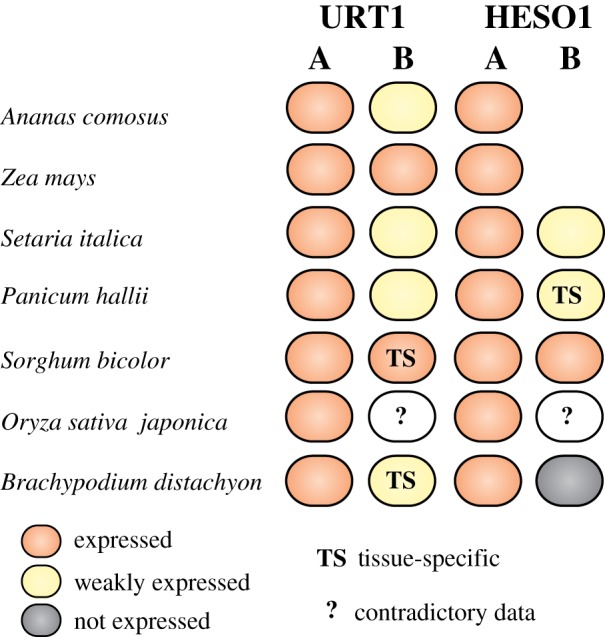
Expression of URT1 and HESO1 isoforms from selected species of the Poaceae family. The diagram was drawn based on the transcriptomic data deposited in Phytozome v. 12.1 (https://phytozome.jgi.doe.gov) [49], in Next-Gen Sequence DBs (https://mpss.danforthcenter.org/dbs) [50], in eFP browser (http://bar.utoronto.ca) [51] and from the RNA-seq data from [52]. Expression data were analysed for Poales species from the BOP clade (B. distachyon, O. sativa) and the PACMAD clade (P. hallii, S. italica, S. bicolor, Z. mays) of Poaceae and for a Bromeliaceae (A. comosus).
In contrast to HESO1 isoforms, two copies of URT1 are present in all 11 representative species chosen here and URT1 sequences form two well-defined clades, defining A and B isoforms (figure 5). Clearly, URT1A homologues are more closely related to eudicot URT1 than the B homologues (figure 5). In addition, transcriptomic data in Ananas comosus, Zea mays, Setaria italica, Panicum hallii, Sorghum bicolor, Oryza sativa japonica or Brachypodium distachyon indicate that URT1A isoforms are constitutively expressed in all tissues, whereas the expression of URT1B isoforms is generally lower and sometimes restricted to specific tissues (figure 6). Both the phylogenetic and transcriptomic studies support the hypothesis that URT1A isoforms are orthologues of URT1 from eudicots, whereas B isoforms may be neo-functionalized. We cannot exclude that certain URT1B isoforms are in the process of pseudogenization. For instance, we failed to amplify fully spliced URT1B mRNAs by RT-PCR analysis in Brachypodium seedlings, in contrast to URT1A mRNAs. Yet, it is possible that URT1B mRNAs are effectively spliced only in response to environmental stimuli or at particular developmental stages. Future experimental work is needed to determine whether URT1B homologues are indeed neo-functionalized.
Overall, URT1 homologues seem present in almost all species of Archaeplastida. Except for the early branching Glaucophyta and Rhodophyta species that contain only URT1 homologues, the genomes of the vast majority of green algae and land plants encode both URT1 and HESO1 homologues. URT1 and HESO1 homologues form a monophyletic group for each TUTase. Several species have multiple isoforms of URT1 and HESO1 that sometimes have tissue-specific patterns of expression. It remains to be experimentally investigated which of these isoforms of TUTases have evolved specialized functions, and which are in the process of pseudogenization following gene duplication.
3. Uridylation accelerates the decay of small RNAs
Small RNAs can be tailed by untemplated nucleotides, mostly uridines and adenosines. 3′ adenylation of miRNAs seemed to slow down degradation in vitro using Populus trichocarpa cell extracts [53], but the role of small RNA adenylation in regulating stability remains to be fully elucidated. By contrast, uridylation triggers the degradation of small RNAs. This process, which was discovered in Arabidopsis [54], constitutes the best-documented role of uridylation in plants. Plant small RNAs are mostly 20–24 nt in length and are divided in two main families: microRNAs (miRNAs) and short interfering RNAs (siRNAs). miRNAs are processed from primary transcripts that contain a hairpin with an imperfectly paired stem, while siRNAs are processed from near-perfect double-stranded RNAs (dsRNAs) or fully paired dsRNAs. The fully paired dsRNAs are produced by RNA-dependent RNA polymerase (RDR), which uses the sense strand as a template to generate the dsRNA precursor. miRNA and siRNA precursors are processed by DICER-like (DCLs) enzymes into small RNA duplexes. The 3′ end of each strand of the duplexes is 2′-O-methylated by the methyltransferase HUA ENHANCER1 (HEN1) [55,56]. Only one strand of the duplexes is finally retained in complex with an ARGONAUTE (AGO) protein, while the passenger strand is discarded (reviewed in [57]). Except for rare exceptions mentioned below, virtually all mature small RNAs are thus methylated on their 3′ terminal ribose in plants.
Mutations in HEN1 result in pleiotropic developmental abnormalities in Arabidopsis because miRNA levels are drastically reduced [55,56]. Methylation by HEN1 is indeed necessary to protect the 3′ end of small RNAs (both miRNAs and siRNAs) from uridylation, which triggers their decay [54,58]. Besides Arabidopsis, mutation of HEN1 orthologues also induces uridylation-mediated destabilization of small RNAs in maize and rice [59,60]. Of note, small RNAs can be uridylated and adenylated in a wild-type context as reported in many species including Arabidopsis, tomato, Medicago truncatula, rice, maize and in the moss Physcomitrella patens [61]. However, untemplated tailing is mostly detected for ‘off-size’ small RNAs [61]. In addition, ‘off-size’ 23 nt heterochromatic siRNAs (hc-siRNAs, also called het- or he-siRNAs), as compared to canonical 24 nt hc-siRNAs, are mostly adenylated rather than uridylated, and this tailing is not increased by hen1 mutation in Arabidopsis [61]. A possible explanation is that the untemplated nucleotides are added to hc-siRNAs precursors [61].
During the biogenesis of the vast majority of small RNAs, once small RNA duplexes have been generated by DCLs, the dsRNA binding domains of HEN1 position the 3′ ends of small RNA duplexes in the catalytic site to deposit the methyl group that prevents uridylation. It is worth noting that some miRNAs, like miR158 or miR319a in Arabidopsis and miR1510 in Phaseoleae species (e.g. soya bean), are substantially truncated and tailed even in a HEN1 wild-type context, suggesting that some small RNA duplexes could be poor substrates of HEN1 [32,60,62]. However, for the vast majority of small RNAs, the absence of HEN1 results in extensive nibbling and tailing, and the added nucleotides are mostly uridines [20,32,54–56,60,61]. Yet, the patterns of trimming and uridylation are different across miRNA families and specific patterns are conserved for the same miRNA family between maize, rice and Arabidopsis hen1 mutants [60]. Therefore, intrinsic features of miRNAs that are conserved between monocotyledons and dicotyledons could determine the extent of nibbling and tailing [60].
The TUTase HESO1 is the major TUTase uridylating both siRNAs and miRNAs to facilitate their decay in Arabidopsis [20,32]. Its orthologue in C. reinhardtii, MUT68, was previously shown to uridylate small RNAs to trigger their degradation, revealing the conservation of this process from algae to land plants [42]. HEN1 SUPPRESSOR 1 (HESO1) was identified in Arabidopsis by a forward genetic screen aiming at identifying suppressors of the hen1 phenotype, but also by systematically testing which of the T-DNA mutants for the 10 class I ncPAPs of Arabidopsis was able to partially rescue the hen1 phenotype [20,32]. In both studies, a heso1 mutation in a hen1 background partially restores miRNA levels and markedly reduces small RNA uridylation [20,32]. Altogether, these data show that HESO1 is the predominant TUTase uridylating small RNAs in plants. Yet, the residual uridylation of small RNAs in the double heso1 hen1 mutant indicates that TUTases other than HESO1 are able to target small RNAs. Both forward and reverse genetic strategies identified URT1 as a secondary TUTase able to uridylate miRNAs in the heso1 hen1 background [63,64]. URT1 was previously identified as the major TUTase uridylating deadenylated mRNAs in Arabidopsis (see §5) [31]. URT1 is localized in the cytosol, P-bodies and stress granules [31]. Its cytosolic localization likely explains why URT1 does not uridylate nuclear hc-siRNAs in a heso1 hen1 background, but is restricted to miRNAs [63,64]. Importantly, the residual uridylation of hc-siRNAs in a hen1 heso1 urt1 background reveals the existence of another TUTase yet to be characterized. In Arabidopsis, the two genes that cluster with HESO1 and URT1 in the phylogenetic analysis shown in figure 2 are possible candidates for encoding this additional TUTase activity.
HESO1 and URT1 uridylate miRNAs, but both TUTases act distinctively and cooperatively [63,64]. For instance, HESO1 uridylates full-length miR158 (which is poorly methylated by HEN1) while 1-nt truncated miR158 is mostly uridylated by URT1 [63]. Also, HESO1 has a broad role in uridylating miRNAs, while URT1 action seems restricted to fewer targets, likely explaining why the urt1 mutation does not rescue the hen1 phenotype, while the heso1 mutation does [32,63].
Also, because mono-uridylated miRNAs accumulate in hen1 heso1 background, it was proposed that URT1 could mono-uridylate unmethylated miRNAs to provide a U-terminating substrate, which is favoured by HESO1. HESO1 would then further tail the small RNA [20,32,57,63,64].
The role of HESO1 in uridylating small RNAs was identified in a hen1 mutant, and that of URT1 in a hen1 heso1 background, because both HESO1's and URT1's activities are inhibited by the methyl group deposited by HEN1 on the 3′ terminal ribose of small RNAs. In a wild-type context, tailing occurs mostly on nibbled small RNAs. In Arabidopsis, four 3′–5′ exoribonucleases, SMALL RNA DEGRADING NUCLEASES (SDN1 to 4) are responsible for nibbling small RNAs [65,66]. SDNs are only partially inhibited by 2′-O-methylation of small RNAs and they are able to remove the 3′ terminal methylated nucleotide of small RNAs, thereby generating truncated, unprotected substrates for HESO1 and URT1 [66]. In contrast to heso1 and urt1 single mutants, or a null double mutant (data not shown), which have no obvious phenotype, combining mutations in three out the four SDNs results in higher miRNA levels and pleiotropic developmental defects [65], indicating that nibbling by SDNs is a limiting step in controlling miRNA decay as compared to uridylation.
Both SDNs and TUTases interact with AGO proteins [39,63,66], explaining why nibbling and tailing of miRNAs are AGO1-dependent [60]. In addition, uridylation antagonizes nibbling of small RNAs, likely revealing a competition between TUTases and SDNs to access the 3′ end of small RNAs [64]. Importantly, SDNs are unlikely to degrade uridylated small RNAs [65] and therefore a yet unidentified activity is responsible for the degradation of uridylated small RNAs in Arabidopsis. In C. reinhardtii, RRP6, a cofactor of the RNA exosome, was proposed to degrade uridylated small RNAs [42]. Our current view of small RNA degradation based mostly on the work in Arabidopsis is summarized in the model presented in figure 7.
Figure 7.
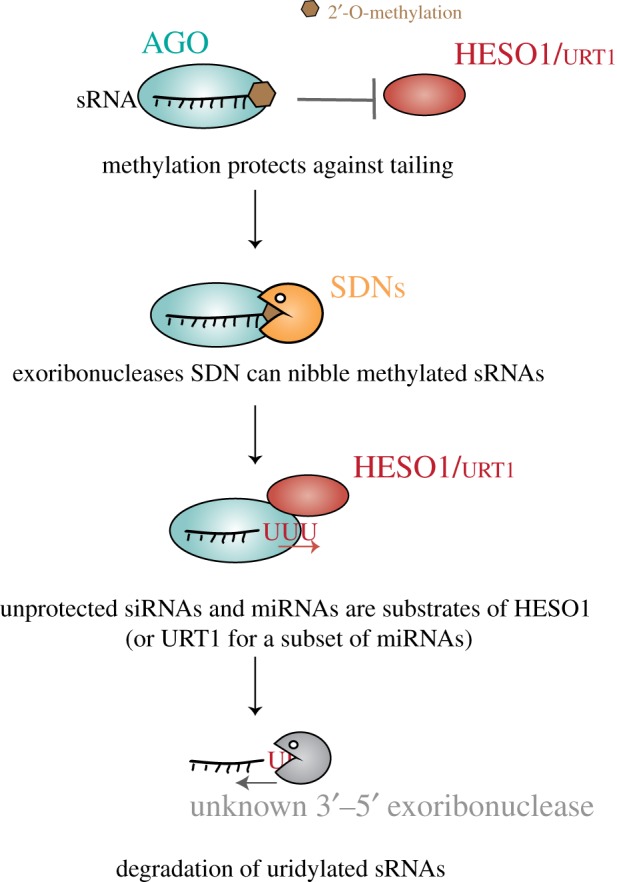
Small RNA uridylation and decay. 2′-O-methylation deposited by the methyltransferase HEN1 protects against uridylation by HESO1 or by URT1. The exoribonucleases' SDNs can nibble methylated sRNAs that are loaded in AGO, thereby generating nibbled, unprotected sRNAs. These unprotected siRNAs or miRNAs are targeted by HESO1 (or URT1 for a subset of miRNAs). The uridylated small RNAs are subsequently degraded by a 3′ to 5′ exoribonucleolytic activity(ies) that is unknown in Arabidopsis and was proposed to be RRP6 in C. reinhardtii [42].
Besides facilitating small RNA degradation, uridylation has additional roles in small RNA metabolism. The slicer activity of AGO1 is inhibited in vitro by the uridylation of miR165/6 in complex with AGO1 [63]. In addition, uridylation of miR158 by URT1 seems to impair its repression activity in a hen1 mutant [63]. These observations, made so far either in vitro or in a hen1 background, indicate that TUTases have the potential to control miRNA activity in plants. Such a regulatory role was reported in mice for miR-26, whose uridylation abrogates function without affecting stability [67]. A second alternative role of miRNA uridylation in plants is to control the biogenesis of secondary siRNAs, which are triggered by cleavage of a target by certain 22 nt miRNAs. Indeed, mono-uridylation of miR170/1 to 22 nt isoforms triggers the production of phased, secondary siRNAs (phasiRNAs) in a hen1 background [60]. Why other 22 nt miRNA isoforms also accumulating in hen1 do not trigger phasiRNA production is unknown. Interestingly, the control of phasiRNA biogenesis by uridylation was recently identified for miR1510 in Phaseoleae species, which include common bean and soya bean [62]. miR1510 regulates several nucleotide-binding and leucine-rich repeat protein (NB-LRR) genes by triggering the production of phasiRNAs. In soya bean, and most other Phaseoleae species, the miR1510 duplex has a terminal mispairing that inhibits HEN1 activity [62]. As a result, unmethylated miR1510 is mono-uridylated into a 22 nt species able to trigger phasiRNA production [62]. By recapitulating miR1510 biogenesis in Arabidopsis, HESO1 was identified as the TUTase that mono-uridylates miR1510 [62]. This example illustrates that uridylation of miRNAs might evolve functions distinct from merely promoting small RNA degradation.
4. Uridylation of 5′ fragments of mRNAs cleaved by RISC
The repression of gene expression by post-transcriptional gene silencing (PTGS) is achieved either by repressing translation or by inducing mRNA degradation (reviewed in [68]). In plant PTGS, mRNA degradation is initiated by AGO1-mediated cleavage that is guided by sequence complementarity between the small RNA loaded in RISC and its target mRNA (reviewed in [57]).
RISC generates a 5′-cleavage fragment (5′CF) and a 3′-cleavage fragment (3′CF). The 3′CF is degraded by XRN4, the cytosolic 5′–3′ exoribonuclease in plants [69]. The 5′CF is eliminated both by the 5′–3′ and the 3′–5′ RNA degradation pathways. Interestingly, uridylation participates in the clearance of this fragment by stimulating degradation from both its 5′ and its 3′ ends. The addition of uridines to the 3′ end of the 5′ fragment is an evolutionarily conserved mechanism detected in Arabidopsis, mice or humans [70,71].
In Arabidopsis, HESO1, which acts on siRNAs and miRNAs, was identified as the major TUTase uridylating 5′CF resulting from RISC cleavage [39]. The immunoprecipitation of AGO1 by recombinant HESO1 suggests that uridylation of 5′CF (and small RNAs) may occur in the AGO complex [39]. Importantly, 5′CF for MYB33 mRNAs accumulate in heso1 mutants, showing that uridylation by HESO1 promotes the degradation of this fragment [39]. Of note, URT1 is responsible for the residual uridylation of MYB33 5'CF although its activity is not required to stimulate the degradation of this 5'CF [40]
Several observations indicate that uridylated 5′CF are degraded by the 5′–3′ pathway. The simultaneous analysis of 5′ and 3′ ends of 5′CF in Arabidopsis identified the presence of oligo(U) stretches at the 3′ end and showed a diversity of 5′ end positions for the analysed targets [70]. The authors suggested that uridylated 5′CF can be degraded from their 5′ end, implying that uridylation enhances 5′–3′ decay of the 5′ mRNA fragment produced by RISC. Indeed, uridylated MYB33 5′CF are preferentially uncapped in Arabidopsis [39]. These uncapped RNAs could be produced either by endoribonucleolytic cleavages or by decapping. This latter possibility is coherent with in vitro decapping assays in mammalian cell extracts that revealed uridylation as promoting decapping [72]. Finally, the accumulation of 5′CF of MYB33 mRNAs in Arabidopsis xrn4 mutants shows that the 5′–3′ RNA degradation pathway indeed participates to the elimination of the 5′ fragments generated by RISC cleavage [39].
5′CF are also incontestably cleared by the 3′–5′ RNA degradation pathway in plants [73]. Indeed, several 5′CF accumulate in Arabidopsis ski2, ski3 and ski8 mutants, SKI2/3/8 forming the Ski complex, the activator of the RNA exosome in the cytosol [73]. This observation strongly suggests the involvement of the RNA exosome in degrading 5′CF in Arabidopsis, as shown in Drosophila [74]. However, this involvement remains to be formally demonstrated using mutants affected in the function of core subunits of the Arabidopsis RNA exosome. Interestingly, secondary siRNAs are produced in the absence of the SKI complex for a number of miRNA targets and it was proposed that SKI2 could promote the rapid dissociation of RISC from the target mRNA, thereby restricting the production of transitive siRNAs [73]. In addition to this study in Arabidopsis, the RNA exosome was also suggested to participate in the degradation of 5′CF in C. reinhardtii [40]. MUT68, the TUTase that uridylates small RNAs in C. reinhardtii, was reported to facilitate the degradation of a mRNA targeted by artificial siRNAs [40]. No uridylation was detected at the sites of cleavage by RISC but at that time, a low-depth analysis was performed. In addition, to our knowledge, no endogenous miRNA-mediated cleavage was investigated. It is therefore unknown at present whether MUT68 uridylates 5′CF. Interestingly, oligo(A)-tailing was detected upstream of the siRNA-induced RISC cleavage sites in the mut68 strain, suggesting that non-canonical polyadenylation tags 5′CF [40]. These oligo(A) tails were proposed to facilitate 3′–5′ degradation by the RNA exosome [40].
Of note, the degradation of RISC cleavage fragments likely promotes the dissociation of RISC from its target, which appears of prime importance for recycling RISC. This recycling involves recently identified 3′–5′ exoribonucleases that interact with AGO1 and AGO10: RICE1 and RICE2 [75]. The inactivation of catalytic residues of homohexameric RICE proteins leads to low levels of miRNAs and accumulation of 5′CF [75]. RICE1/2 likely initiates the degradation of 5′CF, thereby facilitating RISC dissociation and recycling [75].
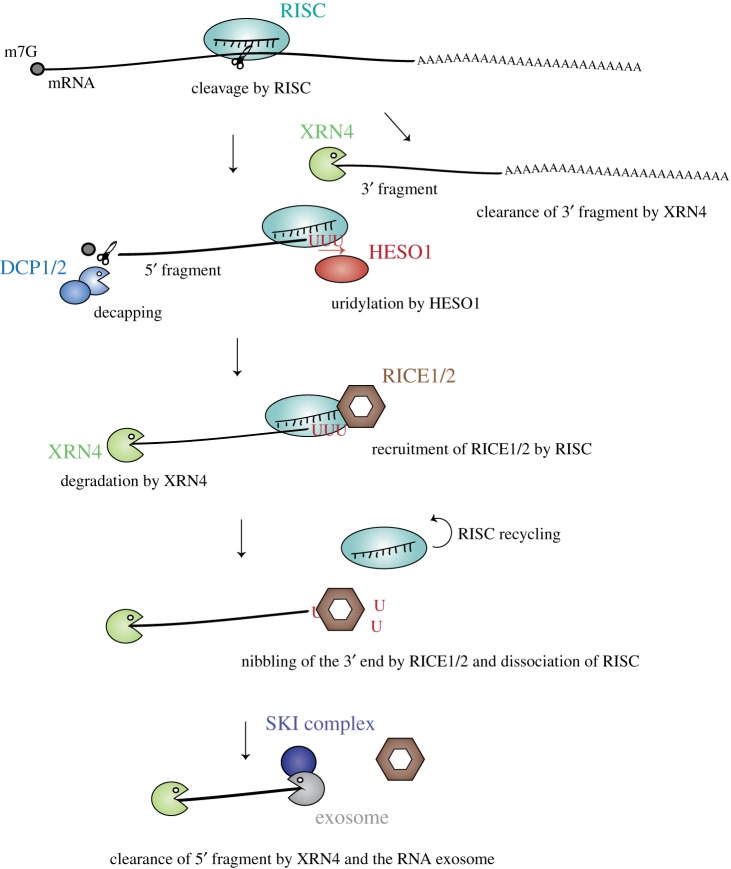
The current model for the degradation of 5′CF is shown in figure 8. HESO1 and RICEs are recruited to RISC through the interaction with AGO (figure 8). HESO1 catalyses the uridylation of 5′CF, possibly promoting decapping and subsequent degradation by XRN4. Concomitantly or alternatively, RICE1/2 ensures the initiation of 3′–5′ degradation of RISC-associated 5′ fragments by starting to nibble uridylated 3′ ends of 5′CF. RICEs'action promotes RISC dissociation and therefore its recycling, but RICEs unlikely fully degrade the 5′CF. This is ensured by the RNA exosome assisted by the SKI complex (figure 8).
Figure 8.
Uridylation by HESO1 promotes degradation of 5′ fragments of RISC-cleaved mRNAs. RISC cleavage of mRNAs generates a 3′ cleavage fragment that is degraded by XRN4, and a 5′ cleavage fragment. The 5′ cleavage fragment is uridylated by HESO1, which binds RISC, but can also be decapped by the DCP1/2 complex. The exoribonucleases RICE1/2, which are recruited by RISC, nibble the uridylated 5′ cleavage fragment. This nibbling helps RISC dissociation and RISC recycling. Finally, the 5′ cleavage fragment is degraded by XRN4 and the RNA exosome.
5. Intricate function for uridylation in the decay of plant mRNAs
Uridylation of deadenylated mRNAs is widely conserved among eukaryotes, including plants but excluding S. cerevisiae that has lost the capacity to uridylate all RNAs. Studies over the past years in S. pombe, X. laevis, Aspergillus nidulans, A. thaliana, M. musculus, Patiria pectinifera (starfish), D. melanogaster or H. sapiens have revealed that uridylation must be considered as an integral step in the degradation of mRNAs [3,13,31,76–83]. Tailing oligo(A) tails with a few uridines likely facilitates the binding of LSm1-7 complex, which binds preferentially to short oligo(A) tails and oligo(U) tails [72,84–86]. Binding of the LSm1-7 complex leads to the recruitment of the decapping complex and subsequent degradation by the cytosolic 5′ to 3′ exoribonuclease Xrn1. A similar process could occur in plants: the binding of the LSm1-7 complex could promote the recruitment of the decapping machinery triggering degradation of the uncapped RNA by XRN4. Moreover, U-tails can be directly recognized by Dis3L2 or the RNA exosome to promote 3′ to 5′ decay. Therefore, uridylation influences both the 5′–3′ and 3′–5′ degradation of mRNAs (see accompanying articles by Zigackova and Vanacova [10], Warkocki et al. [11] and recent reviews [2,4,5,8]).
In Arabidopsis, uridylation of oligo-adenylated mRNAs is mainly performed by URT1 [31,83]. Uridylation in Arabidopsis can be detected on uncapped mRNAs, as shown for CCR2 and LOM1 mRNAs [31,80] and originally described in S. pombe [82]. Yet, no experiment has demonstrated so far an influence of URT1 on global mRNA half-lives, possibly because its direct targets correspond to deadenylated mRNAs, which represent a very minor population among all mRNAs. Also, deadenylation is likely a rate-limiting step in the bulk decay of mRNAs, thereby masking the potential impact of URT1 on mRNA degradation, which is restricted to its targets, i.e. the minor sub-population of deadenylated mRNAs. Although an impact of uridylation in accelerating mRNA decay remains to be shown formally in plants, uridylation definitely has a role in the mRNA degradation process. URT1 prevents excessive deadenylation, as mRNAs with shorter oligo(A) tails accumulate in urt1 mutants, whereas the complementation and overexpression of URT1 in Arabidopsis increase oligo(A) tail sizes [31,83]. Importantly, a global analysis of mRNA uridylation by TAIL-seq revealed that URT1 repairs oligo(A) tails to an average extension length of 16 nucleotides (nt) [83]. A similar sub-population of mRNAs with a oligo(A) tail size distribution centered at 16 nt exists for non-uridylated mRNAs in wild-type plants [83]. Both these uridylated and non-uridylated 16 nt extensions are recognized and bound by a Poly(A) Binding Protein (PABP) in vitro and in vivo [83]. It is at present unknown in plants whether translation can be initiated on mRNAs with uridylated oligo(A) tails bound by PABP, or whether uridylation would inhibit translation as proposed for reporter mRNAs co-expressed with TUTases in Xenopus oocytes [87] or for Nonsense-Mediated Decay (NMD) targets in A. nidulans [80]. Besides a link between uridylation and translation, which remains to be explored in Arabidopsis, the recognition of uridylated oligo(A) tails by PABPs could modulate deadenylation [83]. Moreover, terminal uridines per se are likely to impede deadenylase activities, thereby contributing to slowing down deadenylation. By impeding deadenylation at the 3′ end and possibly stimulating decapping at the 5′ end as in other eukaryotes, URT1 could favour the 5′–3′ directionality of mRNA degradation. Such 5′–3′ directionality is crucial during co-translational decay and in line with this, uridylated mRNAs were detected on polysomes [31].
mRNA uridylation drops by 70–80% in null urt1 mutants [31,83]. This shows that URT1 is the main TUTase uridylating mRNAs, but the residual uridylation observed in null urt1 mutants also reveals the involvement of at least another TUTase [31,83]. HESO1 is a likely candidate as the secondary TUTase able to uridylate mRNAs. This possibility remains to be experimentally demonstrated. The TAIL-seq analysis of urt1 xrn4 double mutant suggested that this URT1-independent uridylation has a different function in mRNA metabolism. URT1-independent uridylation targets mostly very short oligo(A) tails and, unlike URT1, does not seem able to restore a nucleotide extension of sufficient length allowing PABP binding. Interestingly, in xrn4 mutant, only these short uridylated oligo(A) tails accumulate compared to WT, suggesting that this population could be targeted by XRN4 and rapidly degraded [83]. Hence, a complex role of uridylation in the metabolism of mRNAs is emerging in Arabidopsis. Uridylation by distinct TUTases or of different oligo(A) sizes could favour decapping, impede deadenylation or restore a PABP binding site. Whether mRNA uridylation impacts translation or mRNA storage has to be explored in plants.
6. Conclusion and future key points in plant RNA uridylation
The primary function of RNA uridylation in controlling RNA stability is conserved across eukaryotes, including plants. HESO1 and URT1 homologues are present in most plant species, and their roles in the metabolism of small RNAs and mRNAs could be conserved. Yet, we are just beginning to apprehend the diversity of roles played by RNA uridylation in plants. In addition, our current knowledge of RNA uridylation in plants has been gathered using mostly two model species, the flowering plant Arabidopsis thaliana and the green algae Chlamydomonas reinhardtii. Therefore, the diversity of specialized biological functions involving RNA uridylation remains to be explored in various plant species. The discovery that mono-uridylation of a miRNA triggers the biogenesis of phased secondary siRNAs in Phaseoleae species to regulate disease resistance genes illustrates the potential of exploring uridylation in diverse plant species [62].
The just-emerging picture drawn from our knowledge in A. thaliana, and to a lesser extent in C. reinhardtii, is that uridylation targets short and long non-coding RNAs, as well as mRNAs. It is certain that the RNA substrates identified to date represent just a fraction of what is left to discover. For instance, the uridylation of ribosomal or viral RNAs has been reported in plants, with no clues about the impact of U-tailing on these RNAs [22,88]. The systematic identification of RNA substrates of uridylation is a mandatory step towards discovering all functions of RNA uridylation in plants. In addition to the substrates, the identification of all the factors ‘reading’ the uridylation status of RNA and translating this information into biological outputs is required to decipher the molecular basis for the multiple roles played by uridylation in plant RNA metabolism.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
D.G. and C.d.A. wrote the paper; H.S.,. C.d.A. and A.G. performed the evolutionary analyses; V.F., F.M. and H.Z. analysed TUTase expression patterns; C.d.A., H.S. and H.Z. prepared illustrations; D.G. acquired funding.
Competing interests
We declare we have no competing interests.
Funding
Work in D.G.'s laboratory is currently supported by the Centre National de la Recherche Scientifique (CNRS, France) and the Agence Nationale de Recherche (ANR, France) as part of the program d'Investissements d'Avenir in the frame of the LabEx NetRNA (ANR-2010-LABX-36) and ANR-15-CE12-0008-01 to D.G.
References
- 1.Aphasizhev R, Suematsu T, Zhang L, Aphasizheva I. 2016. Constructive edge of uridylation-induced RNA degradation. RNA Biol. 13, 1078–1083. ( 10.1080/15476286.2016.1229736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Almeida C, Scheer H, Zuber H, Gagliardi D. 2018. RNA uridylation: a key post-transcriptional modification shaping the coding and non-coding transcriptome. WIREs RNA 9, e1440 ( 10.1002/wrna.1440) [DOI] [PubMed] [Google Scholar]
- 3.Kwak JE, Wickens M. 2007. A family of poly(U) polymerases. RNA 13, 860–867. ( 10.1261/rna.514007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Łabno A, Tomecki R, Dziembowski A. 2016. Cytoplasmic RNA decay pathways - enzymes and mechanisms. Biochim. Biophys. Acta 1863, 3125–3147. ( 10.1016/j.bbamcr.2016.09.023) [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Tello P, Rajappa L, Coquille S, Thore S. 2015. Polyuridylation in eukaryotes: a 3′-end modification regulating RNA life. BioMed Res. Int. 2015, 968127 ( 10.1155/2015/968127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norbury CJ. 2010. 3′ Uridylation and the regulation of RNA function in the cytoplasm. Biochem. Soc. Trans. 38, 1150–1153. ( 10.1042/BST0381150) [DOI] [PubMed] [Google Scholar]
- 7.Norbury CJ. 2013. Cytoplasmic RNA: a case of the tail wagging the dog. Nat. Rev. Mol. Cell Biol. 14, 643–653. ( 10.1038/nrm3645) [DOI] [PubMed] [Google Scholar]
- 8.Scheer H, Zuber H, De Almeida C, Gagliardi D. 2016. Uridylation earmarks mRNAs for degradation… and more. Trends Genet. 32, 607–619. ( 10.1016/j.tig.2016.08.003) [DOI] [PubMed] [Google Scholar]
- 9.Scott DD, Norbury CJ. 2013. RNA decay via 3′ uridylation. Biochim. Biophys. Acta 1829, 654–665. ( 10.1016/j.bbagrm.2013.01.009) [DOI] [PubMed] [Google Scholar]
- 10.Zigáčková D, Vaňáčová S. 2018. The role of 3' end uridylation in RNA metabolism and cellular physiology. Phil. Trans. R. Soc. B 373, 20180171 ( 10.1098/rstb.2018.0171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warkocki Z, Liudkovska V, Gewartowska O, Mroczek S, Dziembowski A. 2018. Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism. Phil. Trans. R. Soc. B 373, 20180162 ( 10.1098/rstb.2018.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Łabno A, Warkocki Z, Kuliński T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A. 2016. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res. 44, 10 437–10 453. ( 10.1093/nar/gkw649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reimão-Pinto MM, Manzenreither RA, Burkard TR, Sledz P, Jinek M, Mechtler K, Ameres SL. 2016. Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J. 35, 2417–2434. ( 10.15252/embj.201695164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, Zavolan M, Vanacova S. 2016. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J. 35, 2179–2191. ( 10.15252/embj.201694857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin G, Keller W. 2007. RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849. ( 10.1261/rna.652807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mroczek S, et al. 2017. The non-canonical poly(A) polymerase FAM46C acts as an onco-suppressor in multiple myeloma. Nat. Commun. 8, 619 ( 10.1038/s41467-017-00578-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt AG, et al. 2008. Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein–protein interactions and gene expression profiling. BMC Genomics 9, 220 ( 10.1186/1471-2164-9-220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange H, Sement FM, Canaday J, Gagliardi D. 2009. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 14, 497–504. ( 10.1016/j.tplants.2009.06.007) [DOI] [PubMed] [Google Scholar]
- 19.Meeks LR, Addepalli B, Hunt AG. 2009. Characterization of genes encoding Poly(A) polymerases in plants: evidence for duplication and functional specialization. PLoS ONE 4, e8082 ( 10.1371/journal.pone.0008082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G, Chen X, Yu B. 2012. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. 22, 695–700. ( 10.1016/j.cub.2012.02.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salinas-Giegé T, Cavaiuolo M, Cognat V, Ubrig E, Remacle C, Duchêne A-M, Vallon O, Maréchal-Drouard L. 2017. Polycytidylation of mitochondrial mRNAs in Chlamydomonas reinhardtii. Nucleic Acids Res. 45, 12 963–12 973. ( 10.1093/nar/gkx903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sikorski PJ, Zuber H, Philippe L, Sement FM, Canaday J, Kufel J, Gagliardi D, Lange H. 2015. Distinct 18S rRNA precursors are targets of the exosome complex, the exoribonuclease RRP6L2 and the terminal nucleotidyltransferase TRL in Arabidopsis thaliana. Plant J. 83, 991–1004. ( 10.1111/tpj.12943) [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Song J, Yue L, Mo X, Song J, Mo B. 2017. Identification and expression profiling of Oryza sativa nucleotidyl transferase protein (NTP) genes under various stress conditions. Gene 628, 93–102. ( 10.1016/j.gene.2017.06.038) [DOI] [PubMed] [Google Scholar]
- 24.Zimmer SL, Schein A, Zipor G, Stern DB, Schuster G. 2009. Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase. Plant J. 59, 88–99. ( 10.1111/j.1365-313X.2009.03853.x) [DOI] [PubMed] [Google Scholar]
- 25.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. 2009. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37, D380–D386. ( 10.1093/nar/gkn762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt von Braun S, Sabetti A, Hanic-Joyce PJ, Gu J, Schleiff E, Joyce PBM. 2007. Dual targeting of the tRNA nucleotidyltransferase in plants: not just the signal. J. Exp. Bot. 58, 4083–4093. ( 10.1093/jxb/erm267) [DOI] [PubMed] [Google Scholar]
- 27.Czesnick H, Lenhard M. 2016. Antagonistic control of flowering time by functionally specialized poly(A) polymerases in Arabidopsis thaliana. Plant J. 88, 570–583. ( 10.1111/tpj.13280) [DOI] [PubMed] [Google Scholar]
- 28.Kappel C, et al. 2015. Genome-wide analysis of PAPS1-dependent polyadenylation identifies novel roles for functionally specialized poly(A) polymerases in Arabidopsis thaliana. PLoS Genet. 11, e1005474 ( 10.1371/journal.pgen.1005474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trost G, Vi SL, Czesnick H, Lange P, Holton N, Giavalisco P, Zipfel C, Kappel C, Lenhard M. 2014. Arabidopsis poly(A) polymerase PAPS1 limits founder-cell recruitment to organ primordia and suppresses the salicylic acid-independent immune response downstream of EDS1/PAD4. Plant J. 77, 688–699. ( 10.1111/tpj.12421) [DOI] [PubMed] [Google Scholar]
- 30.Vi SL, et al. et al. 2013. Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl Acad. Sci. USA 110, 13 994–13 999. ( 10.1073/pnas.1303967110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon J-M, Bousquet-Antonelli C, Lange H, Gagliardi D. 2013. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 41, 7115–7127. ( 10.1093/nar/gkt465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. 2012. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694. ( 10.1016/j.cub.2012.02.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 121, 713–724. ( 10.1016/j.cell.2005.04.029) [DOI] [PubMed] [Google Scholar]
- 34.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 43, 624–637. ( 10.1016/j.molcel.2011.06.028) [DOI] [PubMed] [Google Scholar]
- 35.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3, e189 ( 10.1371/journal.pbio.0030189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyers F, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737. ( 10.1016/j.cell.2005.04.030) [DOI] [PubMed] [Google Scholar]
- 37.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. ( 10.1093/nar/gkn180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B. 2014. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl Acad. Sci. USA 111, 6365–6370. ( 10.1073/pnas.1405083111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuber H, Scheer H, Joly A-C, Gagliardi D. 2018. Respective contributions of URT1 and HESO1 to the uridylation of 5′ fragments produced from RISC-cleaved mRNAs. Front Plant Sci. ( 10.3389/fpls.2018.01438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibrahim F, Rohr J, Jeong W-J, Hesson J, Cerutti H. 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314, 1893 ( 10.1126/science.1135268) [DOI] [PubMed] [Google Scholar]
- 42.Ibrahim F, Rymarquis LA, Kim E-J, Becker J, Balassa E, Green PJ, Cerutti H. 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl Acad. Sci. USA 107, 3906–3911. ( 10.1073/pnas.0912632107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You C, Cui J, Wang H, Qi X, Kuo L-Y, Ma H, Gao L, Mo B, Chen X. 2017. Conservation and divergence of small RNA pathways and microRNAs in land plants. Genome Biol. 18, 158 ( 10.1186/s13059-017-1291-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoltzfus A, et al. 2013. Phylotastic! Making tree-of-life knowledge accessible, reusable and convenient. BMC Bioinf. 14, 158 ( 10.1186/1471-2105-14-158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. ( 10.1080/10635150600755453) [DOI] [PubMed] [Google Scholar]
- 46.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. ( 10.1093/nar/gkw290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panchy N, Lehti-Shiu M, Shiu S-H. 2016. Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. ( 10.1104/pp.16.00523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. ( 10.1093/nar/gkr944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. 2006. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 34, D731–D735. ( 10.1093/nar/gkj077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718 ( 10.1371/journal.pone.0000718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu S-H, Jiang N, Buell CR. 2012. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 71, 492–502. ( 10.1111/j.1365-313X.2012.05005.x) [DOI] [PubMed] [Google Scholar]
- 53.Lu S, Sun Y-H, Chiang VL. 2009. Adenylation of plant miRNAs. Nucleic Acids Res. 37, 1878–1885. ( 10.1093/nar/gkp031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Yang Z, Yu B, Liu J, Chen X. 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507. ( 10.1016/j.cub.2005.07.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z, Ebright YW, Yu B, Chen X. 2006. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675. ( 10.1093/nar/gkj474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. ( 10.1126/science.1107130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu Y, Jia T, Chen X. 2017. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216, 1002–1017. ( 10.1111/nph.14834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu B, et al. 2010. siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 38, 5844–5850. ( 10.1093/nar/gkq348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe M, Yoshikawa T, Nosaka M, Sakakibara H, Sato Y, Nagato Y, Itoh J. 2010. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 154, 1335–1346. ( 10.1104/pp.110.160234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhai J, et al. et al. 2013. Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25, 2417–2428. ( 10.1105/tpc.113.114603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang F, Johnson NR, Coruh C, Axtell MJ. 2016. Genome-wide analysis of single non-templated nucleotides in plant endogenous siRNAs and miRNAs. Nucleic Acids Res. 44, 7395–7405. ( 10.1093/nar/gkw457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fei Q, Yu Y, Liu L, Zhang Y, Baldrich P, Dai Q, Chen X, Meyers B. 2018. Biogenesis of a 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation. Proc. Natl Acad. Sci. USA 115, 8037–8042. ( 10.1073/pnas.1807403115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tu B, et al. et al. 2015. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 11, e1005119 ( 10.1371/journal.pgen.1005119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Zhang S, Dou Y, Zhang C, Chen X, Yu B, Ren G. 2015. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet. 11, e1005091 ( 10.1371/journal.pgen.1005091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramachandran V, Chen X. 2008. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 321, 1490–1492. ( 10.1126/science.1163728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu Y, et al. 2017. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 15, e2001272 ( 10.1371/journal.pbio.2001272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 11, 1157–1163. ( 10.1038/ncb1931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwakawa H, Tomari Y. 2015. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 25, 651–665. ( 10.1016/j.tcb.2015.07.011) [DOI] [PubMed] [Google Scholar]
- 69.Souret FF, Kastenmayer JP, Green PJ. 2004. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell. 15, 173–183. ( 10.1016/j.molcel.2004.06.006) [DOI] [PubMed] [Google Scholar]
- 70.Shen B, Goodman HM. 2004. Uridine addition after microRNA-directed cleavage. Science. 306, 997 ( 10.1126/science.1103521) [DOI] [PubMed] [Google Scholar]
- 71.Xu K, Lin J, Zandi R, Roth JA, Ji L. 2016. MicroRNA-mediated target mRNA cleavage and 3′-uridylation in human cells. Sci. Rep. 6, 30242 ( 10.1038/srep30242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song M-G, Kiledjian M. 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 13, 2356–2365. ( 10.1261/rna.765807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O, Brodersen P. 2015. SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 43, 10 975–10 988. ( 10.1093/nar/gkv1014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orban TI, Izaurralde E. 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 11, 459–469. ( 10.1261/rna.7231505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Z, et al. 2017. RISC-interacting clearing 3′–5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife. 6, e24466 ( 10.7554/eLife.24466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53, 1044–1052. ( 10.1016/j.molcel.2014.02.007) [DOI] [PubMed] [Google Scholar]
- 77.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 159, 1365–1376. ( 10.1016/j.cell.2014.10.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 32, 1842–1854. ( 10.1038/emboj.2013.63) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. 2010. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol. Cell. Biol. 30, 460–469. ( 10.1128/MCB.00997-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morozov IY, Jones MG, Gould PD, Crome V, Wilson JB, Hall AJW, Rigden DJ, Caddick MX. 2012. mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol. Cell. Biol. 32, 2585–2595. ( 10.1128/MCB.00316-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ochi H, Chiba K. 2016. Hormonal stimulation of starfish oocytes induces partial degradation of the 3′ termini of cyclin B mRNAs with oligo(U) tails, followed by poly(A) elongation. RNA. 22, 822–829. ( 10.1261/rna.054882.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rissland OS, Norbury CJ. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16, 616–623. ( 10.1038/nsmb.1601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuber H, Scheer H, Ferrier E, Sement FM, Mercier P, Stupfler B, Gagliardi D. 2016. Uridylation and PABP cooperate to repair mRNA deadenylated ends in Arabidopsis. Cell Rep. 14, 2707–2717. ( 10.1016/j.celrep.2016.02.060) [DOI] [PubMed] [Google Scholar]
- 84.Chowdhury A, Mukhopadhyay J, Tharun S. 2007. The decapping activator Lsm1p-7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13, 998–1016. ( 10.1261/rna.502507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou L, Zhou Y, Hang J, Wan R, Lu G, Yan C, Shi Y. 2014. Crystal structure and biochemical analysis of the heptameric Lsm1–7 complex. Cell Res. 24, 497–500. ( 10.1038/cr.2014.18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharif H, Conti E. 2013. Architecture of the Lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep. 5, 283–291. ( 10.1016/j.celrep.2013.10.004) [DOI] [PubMed] [Google Scholar]
- 87.Lapointe CP, Wickens M. 2013. The nucleic acid-binding domain and translational repression activity of a Xenopus terminal uridylyl transferase. J. Biol. Chem. 288, 20 723–20 733. ( 10.1074/jbc.M113.455451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huo Y, Shen J, Wu H, Zhang C, Guo L, Yang J, Li W. 2016. Widespread 3′-end uridylation in eukaryotic RNA viruses. Sci. Rep. 6, e25454 ( 10.1038/srep25454) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.