Abstract
Most eukaryotic RNAs are posttranscriptionally modified. The majority of modifications promote RNA maturation, others may regulate function and stability. The 3′ terminal non-templated oligouridylation is a widespread modification affecting many cellular RNAs at some stage of their life cycle. It has diverse roles in RNA metabolism. The most prevalent is the regulation of stability and quality control. On the cellular and organismal level, it plays a critical role in a number of pathways, such as cell cycle regulation, cell death, development or viral infection. Defects in uridylation have been linked to several diseases. This review summarizes the current knowledge about the role of the 3′ terminal oligo(U)-tailing in biology of various RNAs in eukaryotes and describes key factors involved in these pathways.
This article is part of the theme issue ‘5′ and 3′ modifications controlling RNA degradation’.
Keywords: RNA uridylation, tutase, RNA modification, RNA surveillance, RNA processing, RNA degradation
1. Recognition of RNA uridylation as an essential posttranscriptional modification
Most of the RNA molecules are synthetized in a precursor form, which needs to be further processed to form a functional molecule. Processing of RNAs involves a series of posttranscriptional modifications, of which the best characterized are 5′ capping, 3′ polyadenylation and splicing of pre-mRNAs. However, coding and non-coding RNAs (ncRNAs) contain a much broader repertoire of internal and terminal modifications. The roles of some of them have been characterized to a great detail, whereas the function of many other modifications still remains largely unknown. On a cellular level, they affect a broad range of processes from transcription, RNA processing, nuclear export, translation and stability. In this review, we focus on the modification occurring at the 3′ RNA termini. RNAs with 3′ terminal tails consisting of each of the four RNA nucleotides (A, G, C, U) or their combination (e.g. CCA tail of tRNAs) have been observed in different organisms [1]. The homomeric poly(A)-tails are the most widespread and best understood [2–7]. The second most prevalent appears to be the homomeric oligo(U)-tails. RNA 3′ terminal addition of uridines, so-called uridylation, is catalysed by terminal uridylyltransferases (TUTases). They belong to a group of terminal nucleotidyltrasferases (TENT), sometimes also referred to as non-canonical poly(A)-polymerases (ncPAPs). For a detailed review of the biology, biochemistry and terminology of TUTases and ncPAPs, see articles by de Almeida et al. [8] and Warkocki et al. [9]. The 3′ terminal non-templated oligo(U) extensions have been detected in diverse eukaryotes in a wide range of transcripts produced by all three nuclear RNA polymerases, respectively (summarized in table 1). It plays a role in RNA maturation of some small RNAs (sRNAs) [23,24,37–41], but its major role appears to be in the regulation of gene expression via (m)RNA stability [4,27,48].
Table 1.
Involvement of uridylated RNAs in cellular processes.
| organism | process | uridylated RNA | references |
|---|---|---|---|
| Ceanorhabditis elegans, Danio rerio, mammals | development | let-7 microRNA | [10–12] |
| mammals | apoptosis | mRNA | [13] |
| mammals | cell cycle | histone mRNA | [14–20] |
| eukaryotes | host–virus interaction | ncRNAs, viral RNA | [21,22] |
| trypanosomes | RNA editing | guide RNA | [23,24] |
| trypanosomes | translation activation | mRNA | [25] |
| Xenopus, mammals | translation repression | mRNA | [4,26] |
| Schizosaccharomyces pombe, plants, mammals | RNA degradation | various types of RNAs | [4,7,27–36] |
| trypanosomes, mammals | RNA biogenesis | U6 snRNA, guide RNA, let-7 miRNA | [23,24,37–41] |
| plants, mammals | RNA stabilization | histone mRNA, mRNAs | [7,18,42] |
| mammals | immunity | miRNAs | [43,44] |
| mammals | sorting of RNA into extracellular exosomes | miRNA, Y RNA | [45] |
| mammals | oogenesis | mRNA | [30] |
| Drosophila melanogaster | mirtron elimination | mirtrons | [46,47] |
In multicellular organisms, RNA uridylation is crucial for germ cell maturation, differentiation and development, response to infections (table 1) and mutations in the factors involved have been linked to several cancers. Here, we review the current knowledge of the mechanisms, factors involved and the role of uridylation in RNA quality control and decay.
2. The factors involved in uridylation pathways
(a). Terminal uridyltransferases
Enzymes that catalyse oligo(U)-tailing belong to the protein superfamily of polymerase β-like nucleotidyl transferases [49]. They display terminal nucleotidyltransferase activity, therefore they were recently termed TENTs. TENTs typically possess the conserved nucleotidyltransferase core domain conserved with canonical PAP, however, they mostly lack a typical RNA recognition motif (RRM) and display distinct substrate specificity [50,51]. Based on the nucleotide specificity (ATP or UTP), they can be classified as either PAPs or TUTases. For more details, see article by Warkocki et al. [9]. Most of the ncPAPs display a distributive terminal transferase activity, which can be significantly enhanced by their association with either RNA-binding proteins or other small molecule cofactors [52–55]. Moreover, the ncPAP activity can significantly differ in vivo and in vitro or when associated with cofactors [56,57]. They are usually selective for specific nucleotides in vivo, but flexible to process also other nucleotides in vitro [57]. For instance, S. pombe Cid1 uridylates mRNAs in vivo [57], however, purified Cid1 also possesses polyadenylation activity in vitro [57,58].
Mammalian genomes contain at least 11 members of TENTs and their function and structure are described in detail by Warkocki et al. [9].
(b). The oligo(U)-binding factors
Finding the ‘readers’ of non-canonical tailing is crucial for understanding the fate of modified RNA and its biological relevance. To date, there are only few factors known to specifically recognize oligo(U)-tails. They include the two oligo(U)-specific exoribonucleases DIS3L2 and 3′hExo (ERI1) and the exosome complex [14,15,28,59].
Exoribonuclease 3′hExo is a 3′ exonuclease that is required for replication-dependent histone mRNA degradation [14]. It binds to the 3′ terminal stem-loop of histone mRNA to promote its uridylation-dependent degradation [14]. The heteroheptameric LSM1–7 complex forms a donut-shaped LSM1-2-3-6-5-7-4 ring that binds the 3′ end of the histone mRNA and interacts with 3′hExo [14,60]. The LSM complex most probably provides the specificity of 3′hExo for oligouridylated histone mRNA, because no internal affinity of 3′hExo toward U-tails has been uncovered [14]. After the stem-loop removal, the mRNA is accessible for 3′–5′ degradation by the exosome and owing to the decapping, 5′–3′ degradation also can take place. The 3′–5′ degradation is supposed to be dominant, however, the relative processivity of the two directions is unclear [14].
The cytoplasmic exoribonuclease, DIS3L2, belongs to the RNaseII/R 3′–5′ exonuclease superfamily and was first identified as a ‘reader’ of uridylation decay mark for pre-let-7 microRNA (miRNA) [28,59]. Furthermore, it was described to convey 3′–5′ degradation of hundreds of mRNAs independently of the exosome, unlike its homologues DIS3 and DIS3L that belong to the exosome complex [28,61]. The list of substrates of DIS3L2 was lately extended via transcriptome-wide methods to various kinds of preferentially misprocessed and highly structured uridylated RNAs [27,29,48,62]. To understand the substrate preference, the crystal structure of mouse and yeast DIS3L2 binding oligo(U) RNA was determined [63,64]. It revealed a unique shape of RNA-binding funnel enabling U-tail-specific interactions and processing of structured RNA molecules. Thus, TUTase-DIS3L2 was established as a novel cytoplasmic RNA surveillance pathway. Furthermore, mutation of DIS3L2 is associated with a severe congenital overgrowth Perlman syndrome [65]. Perlman syndrome is characteristic among other symptoms of organomegaly, renal anomalies, delayed neurodevelopment, frequent neonatal mortality and a strong predisposition to Wilms tumour and bilateral tumours [65–67]. Moreover, the loss of DIS3L2 causes severe mitotic errors in human cell culture [65]. The cause of Perlman syndrome and its precise link to DIS3L2 remain to be uncovered. One can speculate a connection to let-7 regulation or a harmful accumulation of aberrant RNAs. Perlman syndrome, however, provides an important insight into uridylation and DIS3L2 function and their impact on phenotype.
DIS3L2 was initially identified in an unbiased screen for oligo(U) RNA-binding factors [28]. This search identified several other oligo(U) candidates, e.g. the nuclear exosome targeting complex (NEXT). NEXT targets ncRNAs, such as snRNAs and aberrant by-products of RNA polymerase II activity, such as promoter upstream transcripts (PROMPTs), and recruits the exosome for their trimming or degradation. The structural analysis of an RRM of RBM7 subunits of the NEXT revealed a binding preference for U-rich regions [68]. However, it is yet to be further investigated whether NEXT can mediate exosomal degradation of terminally uridylated RNAs in the nucleus.
3. Non-templated uridylation of mRNAs and mRNA cleavage products
The advent of high-throughput methods designed for 3′ RNA termini has exposed the global occurrence of uridylation in mRNAs and has helped to begin to explain the role of uridylation in specific biological tasks [7,30,42]. Oligo(U)-tails of various lengths were identified on diverse forms of mRNAs, including the full-length polyadenylated transcripts, cleavage products and other short forms in a wide range of eukaryotes (table 1) [4,69,70]. In general, uridylation of mRNA appears connected to its polyadenylation status. Polyadenylation is the most prevalent 3′ RNA tailing since the presence of a poly(A)-tail is mandatory for crucial aspects of RNA life, such as their stability, processing, localization and translation.
In eukaryotes, mRNA polyadenylation mostly serves stability and the shortening of poly(A)-tail, so-called deadenylation, is an integral step in mRNA turnover. Usually, RNA degradation is mediated by a deprotection of RNA and theoretically can be triggered either from the 5′ end by decapping, 3′ end by deadenylation or internally by endonucleases (e.g. induced by RNAi). Several studies have indicated that uridylation plays an important part in mRNA decay. Non-templated uridine extensions were detected on both: mRNAs with shortened poly(A)-tails as well as on diverse upstream cleavage products or short forms of mRNAs (figure 1) [4,13,27,29,30,42,57,61,69,71–74]. Most of these studies connected mRNA uridylation to a decay.
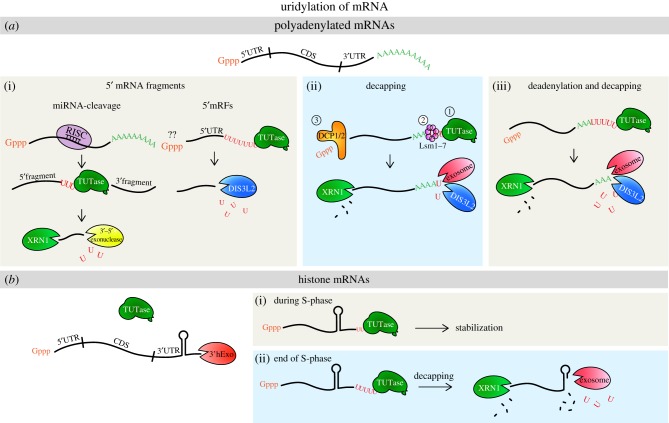
Figure 1.
The role of 3′ uridylation in mRNA metabolism. (a) Uridylation of polyadenylated mRNAs, (i) the upstream cleavage mRNA 5′ fragments resulting from miRNA-directed mRNA cleavage are uridylated, decapped and degraded from 5′ and 3′ termini by exonuclease XRN1 and DIS3L2, respectively. The 5′ terminal mRNA fragments (5′mRFs) are uridylated and chopped in the 3′–5′ direction by RICE1/2 in Arabidopsis, and DIS3L2 in mammals. (ii) In S. pombe, uridylation of mRNA with naturally short poly(A)-tails promotes binding of the Lsm1–7 complex, which triggers decapping by DCP1/2 and mRNA is then degraded in the 5′–3′ direction by XRN1 or in 3′–5′ direction by exosome and DIS3L2. (iii) In mammals, mRNAs with shortened poly(A)-tails are uridylated by TUT4/7, which triggers decapping and degradation in 5′–3′ direction by XRN1 and by exosome and DIS3L2 in 3′–5′ direction. CDS, coding sequence; UTR, untranslated region; Gppp, 5'guanosine-triphosphate cap. (b) Uridylation of histone mRNAs has opposing roles. (i) During S-phase of the cell cycle, when histone mRNAs are trimmed by 3′hExo, they are stabilized via addition of 1–2 Us. (ii) At the end of S-phase of the cell cycle histone mRNAs are partially trimmed by 3′hExo, uridylated by TUT7 and upon decapping degraded in 5′–3′ direction by XRN1 and the exosome in 3′–5′ direction.
(a). Uridylation-mediated mRNA decapping and decay
The first link between uridylation and mRNA degradation was reported by the Norbury lab. In genetic screens in S. pombe they discovered that ncPAP Cid1 suppresses the combined toxicity of hydroxyurea and caffeine on DNA-replication and S-M checkpoint, respectively [75], by regulating the stability of several cell cycle-dependent mRNAs [57,69]. In contrast to other eukaryotes (see below), Cid1 does not require prior deadenylation as oligo(U) stretches were detected on decapped mRNAs often with substantial poly(A)-tails [69]. This indicated that uridylation-mediated decay in S. pombe is independent of deadenylation [69,76].
Moreover, yeast mutants in decapping and deadenylation, respectively, display an increase in oligouridylated mRNAs indicating that uridylation acts upstream of both decapping and deadenylation in S. pombe [69]. The bypass of deadenylation in mRNA degradation in S. pombe might occur owing to its relatively short poly(A)-tails (the median is 28 nt) compared with other eukaryotes [42,77]. The median poly(A)-tail length for mammalian cells is 50–100 nt, 51 nt for Arabidopsis leaves and 50 nt for Drosophila S2 cells [7,77]. Nevertheless, the coinciding presence of oligo(U)-tails on deadenylated mRNAs suggested that deadenylation-dependent and deadenylation-independent decapping coexists in S. pombe [69]. Uridylated mRNAs are then directly targeted by the yeast homologue of the oligo(U)-specific exoribonuclease Dis3l2 and degraded in 3′–5′ direction (figure 1a(ii)) [61]. However, this work did not address the capping status of uridylated mRNAs and the extent of the contribution of the 5′–3′ decay [61]. In vitro, the addition of only two Us to poly(A)-tract was enough to recruit Dis3l2 for destabilization, which was even more prominent with longer U-tails [61].
In contrast to S. pombe, in mammals and plants uridylation was mostly observed on partially or fully deadenylated 3′ ends [7,42]. Shortening of poly(A)-tails is an integral step in the decay of poly(A)+ mRNAs. The mRNA turnover typically starts with deadenylation, catalysed mostly by Ccr4-Not and Pan2–Pan3 complexes [78]. Next, the LSM1–7/Pat1 complex binds the shortened poly(A)-tail and promotes decapping, which then allows the 5′–3′ degradation by exonuclease XRN1 [79,80]. In parallel, since the shortened 3′ end is not protected by poly(A)-binding proteins (PABPs), it is exposed to the 3′–5′ degradation via a robust multisubunit RNA degradation complex, the exosome [81]. The LSM1–7 complex possesses a strong affinity towards uridine-rich regions [16,60,82,83]. It was proposed that up to two uridine residues downstream of oligo(A)-tail are sufficient to promote binding of the LSM1–7 complex. The LSM1–7 complex then recruits Dcp1–Dcp2 complex that triggers decapping and uncapped mRNAs are subsequently degraded by XRN1 in the 5′–3′ direction [69,84]. This model is supported by in vitro reconstitution assays with human cell extracts, which showed decapping activity on oligouridylated synthetic mRNA [16]. Altogether, these studies implied uridylation as a trigger of decapping and subsequent mRNA decay in as diverse species as S. pombe and humans.
In Aspergillus nidulans, TUTases CutA and CutB add a CUCU tag to mRNAs with diverse length of poly(A)-tails. This then also triggers decapping and degradation [5,85]. This tailing was linked to the nonsense-mediated decay (NMD) because CUCU-tailing is facilitated by the presence of premature stop codon and is dependent on NMD factors, Upf1 and NmdA [85]. Hence, CUCU 3′ modification presumably accelerates the NMD-based degradation.
(b). Uridylation of deadenylated mRNAs
The development of the TAIL-seq method for sequencing analysis of poly(A)-tails demonstrated on a transcriptome-wide scale that uridylation occurs prevalently on mRNAs with shortened poly(A)-tails (figure 1a(iii)) [7]. The study revealed that mRNA uridylation is much more prevalent than was previously anticipated. Eighty per cent of mammalian mRNAs were uridylated at a frequency above 2% and certain mRNAs were uridylated with frequency of up to 41%. Furthermore, this high-throughput analysis recapitulated the previous findings that poly(A)-tail length and its uridylation are the key factors of mRNA turnover [4,7,69]. Considering the human transcriptome, about 20% of poly(A)-tails shorter than 25 nt are uridylated, which targets the mRNA for a decay [4,7]. The seminal work by Lim et al. revealed that most if not all of the uridylation of mRNA 3′ ends is mediated by the TUTases, TUT4 and TUT7 [4]. The failure to generate the double-knockout Hela cells for TUT4 and TUT7 indicates the lethality of the double-knockout and the essential function of TUT4/7. However, this study did not further address whether the uridylation activity was the key feature, as they did not attempt to rescue lethality with catalytically inactive forms.
In agreement with the TAIL-seq analyses, TUT4 and TUT7 display higher activity on mRNAs with short or no poly(A)-tails in vitro [4]. In vivo, uridylation of long poly(A)-tails is probably obstructed by PABP, as even low concentrations of recombinant PABP are able to abolish uridylation of long poly(A)-tail mRNAs in vitro [4]. Furthermore, depletion of TUT4/7 led to an overall increased stability of mRNA and uridylation frequency negatively correlated with the stability, demonstrating the global involvement of uridylation in mRNA decay [4]. In turn, uridylation can be also considered as a suppressor of protein translation via mRNA destabilization, as was confirmed by luciferase reporter assay [4]. Next, depletion of both 5′–3′ and 3′–5′ degradation factors, including XRN1, DCP1 and LSM1 for 5′–3′ direction and DIS3L2 and exosomal RRP41 for 3′–5′ direction, causes the enrichment of uridylated mRNAs, indicating the involvement of these factors in degradation [4]. Taking all together, uridylation was found to be much more prevalent than previously thought and to be a step in a turnover of deadenylated transcripts.
In Arabidopsis, uridylation of mRNAs serves complex functions dependent on the poly(A)-tail length and the TUTase. Interesting features and consequences of uridylation in plants are reviewed in detail by De Almeida et al. [8].
(c). Uridylation-mediated regulation of histone expression
Replication-dependent histone mRNAs occupy a special place among mRNAs because they are the only known eukaryotic mRNAs that are not polyadenylated and instead end with a conserved encoded 3′ hairpin structure [86,87]. Interestingly, only metazoan replication-dependent histone mRNAs are not polyadenylated, but plants and most of the unicellular eukaryotes have polyadenylated histone mRNAs [15]. The unique 3′ end processing is crucial for the tight regulation of histone expression to produce high levels only during S-phase of the cell cycle [87]. At the end of S-phase, histone expression needs to be rapidly and efficiently abolished [86,88]. One of the key steps is the fast decay of histone mRNAs and uridylation is an important player in this process [15]. At the end of S-phase or upon the DNA-replication arrest, 5–7 3′ terminal nucleotides are partly trimmed by the 3′–5′ exoribonuclease 3′ hExo (ERI1) and the intermediate cleavage products are oligouridylated by TUT7, which further facilitates 3′–5′ degradation by the exosome and 3′ hExo (figure 1b(i)) [17]. The decapping and XRN1-mediated cleavage also takes place but is not as important as for the decay of polyadenylated mRNAs [15].
Interestingly, uridylation may also have a correcting function on histone mRNAs. The high-throughput sequencing analysis of histone mRNA degradation intermediates (EnD-seq) uncovered the presence of 1–2 non-templated U additions at histone mRNA terminal stem-loops. These extra non-templated Us apparently restore the functional length of histone mRNAs after 3′ hExo trimming (figure 1b(ii)) [18]. In addition, DIS3L2 exoribonuclease was shown to bind uridylated histone mRNAs in vivo [27]. However, it does not seem responsible for their degradation [19]. It is possible that DIS3L2 trims initially longer oligo(U)-tails to 1–2 Us to also form a functional and stable molecule. For details about histone mRNA uridylation, see the article by Meaux et al. [89].
(d). Uridylation-mediated removal of different short forms of mRNAs
In addition to uridylation at the oligo(A)-tails, several studies have identified that different forms of cleaved or truncated mRNAs contain oligo(U)-tails. Several cellular processes, such as NMD, RNAi, apoptosis, ribosome stalling, etc., lead to endonucleolytic cleavage of mRNAs and subsequent degradation of upstream and downstream cleavage products. Uridylation appears to facilitate the removal of the upstream cleavage fragments [27,70,72,73,90–92]. mRNA fragments resulting from miRNA-cleavage are uridylated, which targets them for degradation in diverse organisms [70,72,90–92]. Uridylation presumably facilitates the NMD-based degradation, see §3a [5,85]. Recent studies have implied uridylation in surveillance of 5′ UTR mRNA fragments and in a clearance of mRNAs during apoptosis, oogenesis and zygotic development [13,27,30,74].
(i). Uridylation-mediated decay of RISC-cleaved mRNAs
The miRNA-directed mRNA cleavage generates 5′ and 3′ mRNA fragments [93]. It was shown that uridylation facilitates the clearance of the 5′ cleavage products (figure 1a(i)). In total, 5–24 nt uridine extensions were initially detected on host 5′ mRNA cleavage fragments in Epstein-Barr (EB) virus-infected human cells [94]. Subsequently, similar observations were made in other systems, such as plants and mouse [70,90]. In Arabidopsis, the 5′ fragments are uridylated by the TUTase HESO1, which triggers their degradation [72,92]. The degradation is conducted by XRN4 in the 5′–3′ direction and in the 3′–5′ direction it is initiated by RISC-interacting clearing 3′–5′ exoribonucleases 1 and 2 (RICE1/2) and further degradation is mediated by the SKI complex and the RNA exosome [73,95]. This also serves to relieve and recycle the RISC complex [73]. For further details on RNA uridylation in plants, see article by De Almeida et al. [8].
Similar mechanisms seem to operate in human cells [71]. In this study, knockdown of TENT2 (TUT2) and TENT4B (TUT3) revealed that these TUTases seem to be responsible for oligouridylation (up to 15 Us) of mRNA fragments derived from let-7 miRNA-mediated cleavage [71]. TENT2 was implied in uridylation of the primary 5′ cleavage fragment, whereas TENT4b and perhaps other TUTases modify secondary 3′–5′ intermediate degradation products [71]. Alike in Arabidopsis, the uridylated 5′ cleavage products appear to be degraded in the 5′–3′ direction by a yet unidentified exonuclease [71]. However, this study, except for another on truncated pre-miRNAs [96], remains the only one reporting the uridylation activity of TENT2 and TENT4b. A number of previous studies characterized TENT2 as a non-canonical poly(A) polymerase [50,51,53,97–100]. TENT4b also displays features rather more similar to ncPAP than TUTase. It displays the highest preference for ATP in vitro [101], although in vivo, it catalyses also the addition of guanosines to poly(A)-tails [3].
(ii). The role of mRNA uridylation in apoptosis
During apoptosis, a tightly controlled cell death, cellular components are degraded in a programmed way. The apoptotic stimulus triggers the mitochondrial outer membrane permeabilization (MOMP) and the caspase cascade [102]. One of the earliest effectors is translation shutdown and rapid mRNA decay. In human cells, mRNA degradation is activated by MOMP within one hour after the start of apoptosis, while ncRNAs remain largely unaffected [13]. TUT4/7 and 3′ to 5′ exoribonuclease DIS3L2 appear to play an important role in facilitating this process. mRNA decay intermediates are modified by oligo(U)-tails near the stop codon. Knockdown of both TUT4/7 and DIS3L2 negatively affects mRNA decay as well as apoptosis [13]. In addition, mRNA decay leads to an apoptotic translational arrest. Widespread mRNA decay is dependent on MOMP, which means it occurs only in the classical apoptosis. mRNA is not rapidly degraded in apoptosis when MOMP or caspase activation is inhibited or during oxidative stress. A new study of the fate of RNA in apoptosis revealed another step, in which exoribonuclease PNPT1, which is released from mitochondria upon MOMP, triggers an extensive decay of poly(A) RNA species from the 3′ end [103]. Knockdown of either exoribonuclease, PNPT1 and DIS3L2, caused similar reduction of mRNA decay and apoptosis. Depletion of both nucleases did not show an additive effect, suggesting they act sequentially and non-redundantly. It was proposed that PNPT1 initiates RNA decay from the 3′ mRNA end to a stop codon, where it might be blocked by ribosome, and TUTase-DIS3L2 facilitates further decay [103].
(iii). Uridylated fragment of 5′ termini of mRNAs
Another type of mRNA fragments is produced during faulty transcription, e.g. as a byproduct of RNA polymerase II stalling owing to different obstacles. Recently, uridylation was detected on a group of fragments of mammalian mRNAs originating from 5′ UTRs and parts of intronic and coding regions, so-called 5′mRFs (figure 1a(i)) [27,29]. 5′mRFs were detected by sRNA-seq and a high-throughput cross-linking and immunoprecipitation (CLIP) method using catalytically inactive DIS3L2 as the bait [27,29]. Uridylated 5′mRFs are enriched in the cytoplasm in cells overexpressing catalytically inactive form of 3′–5′ exoribonuclease DIS3L2, indicating they are aimed for degradation or processing by a uridylation-DIS3L2-dependent manner. 5′mRFs often contain regions of the first intron, which suggests that uridylation occurs also on unspliced pre-mRNAs [27]. The mechanism leading to the formation of 5′mRFs is currently unknown. However, the position of uridylation overlaps with the sites of RNA Pol II stalling [27,104,105]. RNA Pol II stalling is known to produce promoter proximal transcripts known as TSSas or tiny RNAs resembling the 5′mRFs [27,104,105]. Moreover, the same study uncovered oligo(U)-tails also on another type of transcripts produced by aberrant transcription from Pol II promoters, the promoter-associated transcripts, so-called PROMPTs [106]. It is possible that the cytoplasmic TUTase-DIS3L2 pathway clears RNA Pol II by-products that are exported to the cytoplasm. However, it is also possible that 5′mRFs are generated from improperly processed mRNAs in the cytoplasm, e.g. in an NMD-like process. Further detailed studies are needed to address these questions.
(e). The role of mRNA uridylation in development
Several recent studies demonstrated that uridylation is a key factor in germline development, early embryogenesis and differentiation [30,74]. The expression of TUT4/7 is elevated during embryogenesis, which indicates their importance in development. TUT4/7 play a major role in the establishment of the mammalian maternal transcriptome during oogenesis [30]. Each stage of oogenesis expresses a specific transcriptome. Oocytes need to have the ability to selectively degrade mRNAs that are specific to the previous stage. This selective removal is regulated by deadenylation and uridylation [30]. The uridylation activities of TUT4 and TUT7 are required to mediate the selective removal of mRNA pools with short poly(A)-tails in several stages during oogenesis.
Uridylation further regulates events after fertilization via targeting coding [74] as well as ncRNAs [107,108]. The early zygote is transcriptionally inactive and its gene expression is ruled by the maternal transcriptome pool of the oocyte, which is formed during folliculogenesis [109,110]. Early during embryogenesis, the zygote undergoes maternal-to-zygotic transition (MTZ) when zygotic transcription replaces the maternal pool. The critical step in MTZ is an efficient and specific removal of maternal mRNAs. A genome-wide study revealed that the clearance of maternal mRNA is also facilitated by TUT4/7 uridylation and that uridylation is necessary for early embryo development [74]. Altogether, mRNA uridylation is a key component in shaping the maternal transcriptome and in its subsequent removal, and in turn, in the regulation of proper embryogenesis.
(f). Uridylation of viral RNA as an antiviral defence mechanism
A recent genetic screen in C. elegans identified CDE-1, a homologue of mammalian TUT4/7, as a part of an antiviral innate immunity mechanism [111]. The genome RNA of the Orsay virus, which is a natural pathogen of C. elegans, was found to be monouridylated by CDE-1 and targeted to degradation by XRN proteins and the RNA exosome. Similarly, in mammalian cells infected by influenza A virus, viral mRNAs are highly uridylated by TUT4/7 [111]. The knockout of TUT4/7 results in the accumulation of viral nucleoprotein mRNAs and in an increased number of infected cells. Together these findings demonstrate CDE-1- and TUT4/7-mediated uridylation as one of the critical players of antiviral immune response in worms and mammals, respectively [111].
4. The role of 3′ terminal uridylation in the metabolism of ncRNAs
(a). Uridylation-mediated degradation of microRNA precursors
Uridylation has opposing roles in miRNA biology. Whereas monouridylation is an integral step in maturation of the group II let-7 family miRNAs (see §5b) [37,38], oligouridylation of the group I pre-let-7 blocks maturation and triggers degradation [28,37,54,55,107,108,112]. Let-7 miRNAs are an exceptional class of miRNAs, with a key role in the regulation of differentiation during early embryogenesis. Let-7 miRNAs target a number of pluripotency mRNAs and cell cycle factors (reviewed in [113]). They are expressed in differentiated cells, but their expression needs to be downregulated in pluripotent cells. Defective expression of let-7 was also linked to certain cancers (reviewed in [113]).
The regulation of let-7 expression occurs on both transcription and processing levels (reviewed in [114]). Oligouridylation by TUT4/7 of pre-let-7 miRNAs plays a critical role in silencing of let-7 expression in pluripotent cells by inhibiting further processing by Dicer and enhancing pre-let-7 destabilization [37,54,55,107,108,112]. For this activity, TUT4/7 require the let-7-specific binding cofactor LIN28 [37,54,55,107,108,112]. In undifferentiated cells, LIN28 can block let-7 maturation at both primary and pre-let-7 stages. The mechanism has been studied to a great molecular detail. LIN28 possesses specific affinity to pre-let-7 miRNA and recruits TUT4/7 for oligouridylation, which in turn suppresses the processing by Dicer. Uridylated pre-let-7 is subsequently targeted for degradation by DIS3L2 [28,59,63]. The co-crystal structure of LIN28/pre-let-7 revealed an interaction between LIN28 and two sequences in the pre-let-7 terminal loop, GNGAY and GGAG, by its N-terminal cold-shock domain (CSD) and C-terminal CCHC zinc knuckle domain, respectively [115,116]. LIN28 CSD binds an additional motif in a subclass of pre-let-7, (U)GAU [117]. Binding between LIN28 zink knuckle and pre-let-7 motif GGAG is necessary to create a stable formation with TUT4/7 to induce TUTase processivity [118,119]. The processivity of the complex is further enhanced by the second cofactor, E3 ligase TRIM25, which binds the conserved terminal loop of pre-let-7 and reinforces the processivity of LIN28A/TUT4 heterodimer [120]. The oligo(U)-tail then hinders the interaction with Dicer, because Dicer is incapable of binding long 3′ overhangs [54,55,107]. Several CLIP-seq-based analyses showed that LIN28 also binds other miRNAs and mRNAs, but the interaction does not appear to promote their uridylation [121–125]. In the absence of LIN28, TUTase interacts with pre-let-7 transiently, resulting in the addition of a single uridine.
In mammalian cells, oligouridylation also promotes degradation of aberrantly processed, truncated pre-miRNAs [96]. Such aberrant forms of pre-miRNAs may originate from cleavage by ribonucleases, such as Ago2, Regnase-1, IRE1alpha or complex Translin/Trax [126–129]. Oligouridylation of these truncated pre-miRNAs is carried out by TUT4/7 and presumably also TENT2, but without the help of LIN28 [96]. In this case, the oligouridylation is achieved in a distributive manner by frequent reassociation of TUTase with its target RNA [96].
(b). Uridylation of mirtrons in Drosophila
Besides the canonical miRNA biogenesis, there are alternative pathways for the production of miRNAs [130]. In flies, worms and mammals, some miRNAs are produced in a Drosha-independent manner. For instance, mirtrons are miRNAs encoded by and processed from intronic regions of pre-mRNAs [131]. In Drosophila, mirtrons are generally poorly conserved, and rather unstable. High-throughput sequencing analysis of sRNAs revealed that Drosophila's mirtrons, in contrast to other miRNAs, are predominantly uridylated [46,47]. Uridylation is catalysed by the TUTase Tailor [46,47], which forms a stable complex with the exoribonuclease DIS3L2, called the TRUMP [62]. This association enhances the mirtron elimination. Tailor specificity for mirtrons is given by the very 3′ end nucleotide. Substrates ending with G or U are preferentially tailed rather than those ending with A or C. As mirtrons are generated by splicing with 3′–AG ends, they are optimal Tailor targets. Canonical miRNAs are depleted in 3′ Gs, and thus escape the TRUMP targeting [47]. This mechanism is a way to prevent de novo biogenesis of miRNAs in Drosophila [46,47].
(c). Uridylation of mature sRNAs
Oligouridylation mediates decay of mature forms of miRNAs (figure 2b(i)). Here we summarize several such examples acting in particular physiological pathways, studied mostly in mammalian cells. High-throughput sequencing analyses uncovered that 3′ tailing by uridylation and adenylation is a common feature of siRNAs and miRNAs in a number of eukaryotes, such as algae, higher plants, worms and mammals [10,11,31,32,43,44,132–135]. Uridylation of mature sRNAs has different implications. For instance, it serves as a destabilization tag in the absence of 3′ terminal 2′–O-methylation (discussed in detail below) [31,135].
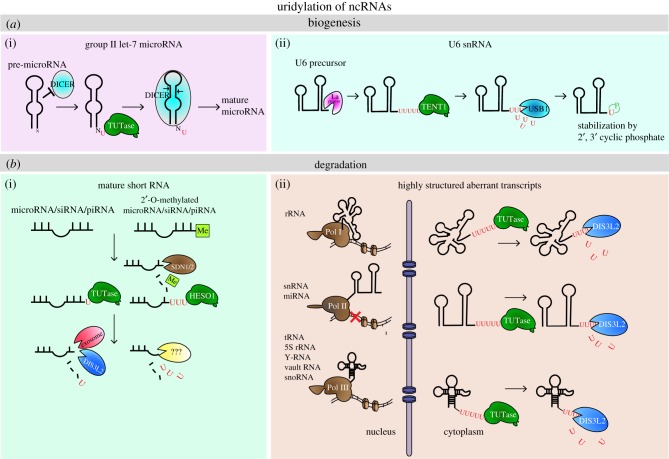
Figure 2.
The role of 3′ uridylation in metabolism of ncRNAs. (a) Uridylation in biogenesis of let-7 miRNA (i) Group II let-7 miRNAs are monouridylated by TUT4/7, which promotes processing by Dicer, and of U6 snRNA (ii) TENT1 uridylates precursors of U6 snRNA, exonuclease USB1 trims oligo(U)-tails down to 4 Us and leaves the terminal U with 2′, 3′cyclic phosphate. (b) Uridylation in snRNA decay. (i) mature short RNAs can be monouridylated by TUTases and degraded in 3′–5′ direction by exosome and/or DIS3L2. In Arabidopsis, SDN1/2 first removes 2′–O-methylation, then TUTase HESO1 adds oligo(U)-tail and small RNAs (sRNAs) are degraded by yet unknown 3′–5′ exonuclease. (ii) TDS mediates surveillance of various highly structured aberrant ncRNAs in the cytoplasm.
Uridylation has been implicated in the regulation of inflammatory response via uridylation of miR-26, a regulator of cytokine expression in lymphocytes. Uridylation of mature miR-26 plays a role in macrophage activation [43]. Upon activation, TUT4 adds oligo(U)-tails to miR-26a and miR-26b, which inhibits their activity on a number of cytokine mRNAs, such as interleukin-6 (IL-6) [43,136]. Accordingly, TUT4 depletion results in downregulation of cytokines [43].
In T-cells, TUT4/7 mediate clearance of a specific subset of miRNAs upon antigen-mediated activation [44]. Uridylated miRNAs are specifically degraded after T-cell activation by yet unidentified exonuclease.
Furthermore, TUT4-deficient mice suffer from impaired growth and early lethality [10]. Apart from the role in differentiation, Jones et al. [10] proposed that TUT4 targets miR-126-5p and miR-379 that downregulate an important growth factor, IGF-1. The TUT4 depletion leads to the stabilization of miR-126-5p and miR-379, and in turn to downregulation of IGF-1 [10]. The downregulation of IGF-1 may at least partially explain the phenotype of the TUT4-deficient mouse.
In addition to let-7, mammalian TUT4/7 monouridylate other miRNAs, such as those involved in cell differentiation and Homeobox regulation [11]. This mechanism was further investigated in zebrafish, where TUT4/7-deficient animals displayed developmental defects and dysregulated Homeobox expression [11]. Interestingly, TUT4/7 knockdown was accompanied by increased non-templated adenylation of given miRNAs. Adenylation might presumably be catalysed by TENT2 as a compensatory mechanism to maintain a steady-state level of modified versus unmodified miRNAs and to regulate their stability [11]. In summary, non-templated tailing of rather stable miRNAs leads to their destabilization. In future, it will be interesting to uncover whether this is a general mechanism for the turnover of most miRNAs and whether extensions with different nucleotides have distinct consequences for miRNA stability and function. Another interesting and important question that remains is whether specific cofactors recognize these extensions and recruit still largely unknown and uncharacterized nucleases.
(d). Target-directed miRNA degradation
Another mechanism of miRNA degradation involves target-based oligouridylation, so-called target RNA-directed miRNA degradation (TDMD). In this process, mature miRNAs are eliminated by 3′ tailing and trimming, when base-paired with a highly complementary target in vivo [33–35]. The screen for factors involved in TDMD identified TENT1 and exoribonuclease DIS3L2, although the involvement of other TUTases was not excluded [33]. TDMD is for instance exploited by some viruses to inhibit antiviral RNAi response [137–139]. For instance, mouse cytomegalovirus expresses long ncRNA m169, which serves as a sponge for host cell miR-27 [138]. When base-paired with m169, miR-27 is modified by TUTase and chopped by DIS3L2 [33].
Regarding the high number of reported uridylated mature miRNAs, it is likely that a portion of them undergoes TDMD by using yet unknown target RNAs [10,11,27,43,44].
(e). Uridylation targets unmethylated sRNAs
2′–O-methylation of the ribose of the last nucleotide in sRNAs, such as siRNAs, miRNAs and piRNAs, is an integral step in sRNA biogenesis in a number of organisms. This modification was initially observed in Arabidopsis and later was found e.g. in Chlamydomonas, Trypanosoma brucei, C. elegans, Drosophila and zebrafish [132,135,140–144]. It is catalysed by HEN1 methyltransferase [134,140,141,145]. In principle, 2′–O-methylation protects sRNAs from 3′ end uridylation that would otherwise lead to 3′–5′ decay (figure 2b(i)) [140]. In plants, unmethylated sRNAs are uridylated by HESO1, which further promotes their degradation [32,36].
A similar pathway operates in highly diverse single-cell eukaryotes, such as algae or trypanosomes [31,134]. In Chlamydomonas, TUTase, MUT68, preferentially uridylates sRNAs, miRNAs and siRNAs and stimulates their decay by the exosome [31]. Chlamydomonas siRNAs and miRNAs are constitutively 2′–O-methylated at their 3′ ends by the HEN1 orthologue and thus protected from uridylation [31]. In addition, MUT68 appears to oligoadenylate and promote decay of the 5′ miRNA-based cleavage mRNA fragments [146]. However, this activity is yet to be validated by enzymatic assays.
In C. elegans, HEN1 activity is crucial for development. It 2′–O-methylates piRNAs and siRNAs and its loss leads to impaired fertility [147–149]. Surprisingly, HEN1 does not appear to target miRNAs in C. elegans. Similarly, the HEN1 orthologue in Drosophila, called DmHen1 or Pimet (piRNA methyltransferase), causes 2′–O-methylation of piRNAs and siRNAs [143,150]. Without Pimet, piRNAs are downregulated and possess heterogeneous 3′ ends.
Finally, in zebrafish, HEN1 mediated 2′–O-methylation is found mainly in germ cells on piRNAs [135,151]. HEN1 mutations lead to increased piRNA 3′ uridylation and subsequent 3′–5′ degradation [135]. Danio hen1 mutants display delayed ovary formation and sex determination, pointing to the importance of counteraction of 3′ terminal 2′–O-methylation and uridylation.
(f). TUT-DIS3L2 surveillance of structured aberrant transcripts, TDS
Most ncRNAs form complex secondary and tertiary structures, that are important for their function, RNP formation, but also to protect them from nucleolysis. DIS3L2 exoribonuclease is a potent enzyme processively degrading highly structured RNAs, such as tRNAs [28,152]. DIS3L2 possess specificity to oligo(U) RNA [28]. Some of the first evidence for TUT-DIS3L2 surveillance (TDS) was observed in cells infected with Moloney leukaemia virus [21]. Interestingly, uridylated aberrant ncRNAs, such as tRNAs and snRNAs are packed into viral particles. This work found Exportin5 responsible for the nuclear export of these uridylated RNAs [21]. Recent findings from several studies uncovered that evolutionarily distant eukaryotes such as flies and mammals use the TDS to remove a wide spectrum of aberrant highly structured ncRNAs in the cytoplasm (figure 2b(ii)) [27,29,48,62]. The targets of this pathway include transcripts from all three nuclear RNA polymerases, such as vault RNAs (vtRNAs), Y RNAs, tRNAs, 7SK, 7SL, Rmrp (RNaseMRP) and Rpph1 (RNaseP), snRNAs, rRNAs, lncRNAS, etc. In many cases, the TDS targets misprocessed, extended or shorter forms, with the oligo(U)-tail in a proximity of a stable secondary structure. These transcripts obtain oligo(U)-tails in an average of 8–9 nt, which is a length favoured by DIS3L2 [27,63,64]. In summary, the TDS is a conserved RNA quality control mechanism responsible for the removal of aberrant structured RNAs in the cytoplasm. TUT4/7 are the likely responsible TUTases, however, it is unknown whether LIN28 or similar cofactors are needed to promote the processivity of oligo(U)-tailing.
5. The role of uridylation in RNA biogenesis
Most of the studies in the past 10 years linked non-templated 3′ uridylation to RNA decay. However, the first reports of RNA uridylation were on uridylation-mediated RNA maturation. Maturation typically involves other TUTases than those acting in the decay, and in some organisms these mechanisms reside in specific organelles, such as mitochondria. In the next section, we summarize examples where the activity of TUTases is crucial for the formation of functional mature RNA molecules.
(a). U6 snRNA maturation
U6 snRNA is a small RNA essential for pre-mRNA splicing. It is transcribed by RNA polymerase III. The transcription of U6 snRNA terminates on four templated Us [153,154]. However, the 3′ end needs to be extended by up to 16 additional Us. This oligouridylation is catalysed by the TUTase TENT1 (also known as TUT6/U6 TUTase/TUT1/PAPD2/RBM21) [39–41]. To stabilize the transcript, U-tails are then trimmed down to only one U by the oligo(U)-specific distributive exoribonuclease, USB1 (also called Mpn1) [155,156]. USB1 leaves only one uridine residue with 2′, 3′ cyclic phosphate at the ribose (figure 2a(ii)) [155,156]. Mature U6 snRNAs then comprise four templated Us and one non-templated U at the 3′ terminus. The 3′ terminal 2′, 3′ cyclic phosphate is specifically bound by the LSM2-8 complex, which is crucial for U6 snRNA stabilization [157]. U6 snRNA transcripts that are not protected by the terminal cyclic phosphate are polyadenylated and degraded by the nuclear exosome [155]. Interestingly, oligouridylation appears to be linked to other modifications, such as m6A. The methyltransferase METTL16 binds U6 snRNAs with extended oligo(U)-tails [158]. In summary, the U6 snRNA oligouridylation is a key process to promote 3′ stabilization.
(b). Uridylation promotes maturation of the group II family of let-7 miRNAs
MiRNAs are potent regulators of gene expression through RNA-interference. Therefore their own expression needs to be tightly controlled, as dysregulation of multiple miRNAs is connected to developmental defects and diseases, such as cancer. Uridylation is one of the regulators of miRNA expression, not only providing stability but also helping to promote processing. Monouridylation of some pre-miRNAs facilitates Dicer processing (figure 2a(i)), whereas oligouridylation prevents Dicer processing and marks miRNAs for degradation (see §4a).
The best-studied example is the role in the let-7 family of miRNAs, which is a key factor in embryonic stem cell differentiation, pluripotency and tumour-suppression (reviewed in [114]). miRNAs are classified into two groups, based on the structure of the 3′ overhang of their precursor. Group I represents pre-miRNAs with a 2 nt 3′ terminal overhang resulting from the Drosha cleavage in the nucleus. This type is further processed by Dicer in the cytoplasm [38]. Group II pre-miRNAs contain only 1 nt 3′ terminal overhang, which causes poor Dicer activity. Such termini typically originate from an unusual mismatched nucleotide at the Drosha cleavage site [38]. The let-7 family has 12 members in humans: three of them generate canonical precursors of the group I and nine belong to group II. TUTases TUT4 and TUT7 can restore the 3′ overhang of the group II pre-let-7 miRNAs. They add a single uridine to the 3′ end, forming the 2 nt overhang optimal for further Dicer processing [37,38]. Alternatively, the cytoplasmic ncPAP TENT2 can also facilitate the group II pre-let-7 processing by monoadenylation [97]. Importantly, the maturation through monouridylation occurs only in the absence of LIN28 [38]. In cells expressing LIN28, pre-let-7 miRNAs are oligouridylated and degraded (see §4a).
Deep sequencing analyses revealed other miRNAs, such as miR-105 and miR-449b-3p, that are also frequently monouridylated [38]. Based on the secondary structure prediction, they might be produced by Drosha-cleavage only with 1 nt 3′overhang, which would classify them as group II miRNAs.
TUTases and ncPAPs are mostly ubiquitously expressed. It is likely that the repertoire of group II miRNAs is much larger and tissue- or development-specific. Further, the growing evidence of defects and cancers linked to aberrant TUTase expression reflects possible defects in yet uncharacterized group II miRNAs.
(c). TUTases in the mRNA metabolism of mitochondrial RNAs in Trypanosomes
Historically the first role of uridylation was identified in the mitochondrion of the parasitic protist T. brucei. Trypanosomes display many unique features of RNA metabolism. The most obscure is the processing of mRNAs via an editing process in the kinetoplast, a single large mitochondrion. mRNA editing comprises massive addition and removal of Us from the precursor transcripts. The specificity is dictated by 50–60 nt long guide RNAs (gRNAs), which themselves possess up to 20 nt oligo(U)-tails [159,160]. gRNAs are transcribed from DNA minicircles and maxicircles as 800–1200 nt precursors [23,161,162]. Their transcription is bidirectional and creates 50 nt long 5′ complementary regions forming stable duplexes. The gRNA uridylation proceeds in two steps. At first, RET1 TUTase uridylates pre-gRNA, which triggers trimming by the 3′–5′ exoribonuclease DSS1 [23,24]. RET1 and DSS1 group together with three additional proteins to form MPSS1-3, a mitochondrial 3' processome (MPsome). DSS1 stops the trimming a few nucleotides downstream of the duplex. The second round of uridylation by RET1 then promotes disassembling of MPsome from the duplex and the antisense strand is degraded [24]. The uridylated sense transcript forms the mature gRNA. The mRNA editing process then involves additional TUTase KRET2/RET2, as part of a large editosome complex, which catalyses the insertion of one or more Us within pre-mRNA based on the complementarity with gRNAs (reviewed in [163]).
6. Uridylation regulates translation and RNA localization and targeting
On the cellular and organism level, RNA uridylation has an impact on events as diverse as translation, viral infection and intracellular and extracellular RNA targeting (such as vesicular exosomes), etc. [22,45].
(a). Uridylation regulates translation efficiency
The role of uridylation in translation differs depending on the organism and subcellular localization. In trypanosome mitochondria, it is required for proper translation of mitochondrial mRNAs [25]. Trypanosomal mitochondrial mRNAs comprise 200–300 nt long A/U-tails, added by poly(A)-polymerases KPAFs and TUTase RET1 [25]. The small ribosomal subunit interacts with the long A/U-tail and initiates the translation.
In vertebrates, uridylation appears to serve mainly for translation downregulation. For instance, TUT7-mediated mRNA uridylation in Xenopus negatively affects reporter mRNAs' translation without changes in mRNA stability [26]. It was proposed that the oligo(U)-tail forms an intramolecular duplex with the upstream poly(A), which prevents poly(A) recognition by poly(A)-dependent factors, such as PABP, which in turn inhibits translation stability [26]. On the other hand, human TUT4/7 were demonstrated to repress translation via mRNA destabilization [4]. In addition, uridylation can inhibit translation by also facilitating turnover of rRNAs and tRNAs. Oligo(U)-tails were detected also on decay intermediates of rRNAs and tRNAs [27], however, its role in translation regulation has not yet been addressed.
(b). Uridylation in RNA sorting to viral and other extracellular particles
Several independent studies linked RNA uridylation to sorting to different extracellular particles, such as viral or secreted exosomes. Modification by oligo(U)-tailing appears to be rather widespread among RNA viruses [22]. Different types of 3′ termini were detected on all positive-strand, negative-strand and double-stranded RNA viruses with hosts ranging from fungi to plants and animals [22]. The impact of 3′ oligo(U)-tailing of viral RNAs remains to be elucidated.
Interestingly, not only oligouridylated viral genomes were found in viral particles. The retrovirus Moloney leukaemia virus, which assembles in the cytoplasm, packages unprocessed forms of snRNAs, snoRNAs and tRNAs to virions [21]. This process requires the nuclear export receptor, Exportin-5. Interestingly, tailed (A and U) forms of these aberrant RNAs accumulate in virions upon downregulation of the exoribonucleases DIS3L2 and RNA exosome. The reason for packaging these RNAs is as yet unknown, although it is hypothesized to contribute, e.g. to Gag oligomerization, the interaction with host cell sensors or to retrovirus replication [21].
In addition to viruses, uridylated RNAs were also detected in the extracellular vesicles called exosomes [45]. Detailed analysis of the composition of exosomes originating from human B-cells revealed the enrichment of uridylated species of miRNAs and Y RNAs, whereas, oligoadenylated miRNAs were preferentially retained in the cells. It was hypothesized that specific 3′ end tailing might regulate the distribution of released versus retained sRNAs [45]. However, this study did not address any particular factors, e.g. TUTases, RNA-binding factors or exonucleases. Further mechanistic studies are needed to validate the role of tailing in RNA sorting to exosomes.
7. Outlook
There are still many open questions that need to be addressed. For instance, it is not yet known what causes the cellular phenotypes of DIS3L2 KO, such as the gene expression changes or disease phenotype on the molecular level. Are there any cell type-specific or tissue-specific cofactors that modulate TUTase specificity and activity on particular substrates? Can the TUT-exonuclease pathway be a drug target for the treatment of associated cancers and developmental defects? Lin & Gregory [164] have identified potential inhibitors of TUTase activity, but we are still far from knowing whether they have therapeutic potential.
Acknowledgements
We thank Viacheslav Zemlianski for initial help with figure preparation.
Data accessibility
This article has no additional data.
Authors' contributions
D.Z. and Š.V. wrote the paper; D.Z. prepared illustrations; Š.V. acquired funding.
Competing interests
We have no competing interests.
Funding
This work was supported by the Czech Science Foundation 16-21341S to Š.V., and D.Z. was supported from 305/11/1095, and by the Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (LQ1601).
References
- 1.Labno A, Tomecki R, Dziembowski A. 2016. Cytoplasmic RNA decay pathways - enzymes and mechanisms. Biochim. Biophys. Acta 1863, 3125–3147. ( 10.1016/j.bbamcr.2016.09.023) [DOI] [PubMed] [Google Scholar]
- 2.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724. ( 10.1016/j.cell.2005.04.029) [DOI] [PubMed] [Google Scholar]
- 3.Lim J, et al. 2018. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science 361, 701–704. ( 10.1126/science.aam5794) [DOI] [PubMed] [Google Scholar]
- 4.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159, 1365–1376. ( 10.1016/j.cell.2014.10.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. 2010. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol. Cell. Biol. 30, 460–469. ( 10.1128/MCB.00997-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaňáčova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3, e189 ( 10.1371/journal.pbio.0030189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol. Cell 53, 1044–1052. ( 10.1016/j.molcel.2014.02.007) [DOI] [PubMed] [Google Scholar]
- 8.de Almeida C, Scheer H, Gobert A, Fileccia V, Martinelli F, Zuber H, Gagliardi D. 2018. RNA uridylation and decay in plants. Phil. Trans. R. Soc. B 373, 20180163 ( 10.1098/rstb.2018.0163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warkocki Z, Liudkovska V, Gewartowska O, Mroczek S, Dziembowski A. 2018. Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism. Phil. Trans. R. Soc. B 373, 20180162 ( 10.1098/rstb.2018.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones MR, et al. 2012. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS Genet. 8, e1003105 ( 10.1371/journal.pgen.1003105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thornton JE, Du P, Jing L, Sjekloca L, Lin S, Grossi E, Sliz P, Zon LI, Gregory RI. 2014. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 42, 11 777–11 791. ( 10.1093/nar/gku805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, 403–414. ( 10.1016/j.devcel.2005.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas MP, Liu X, Whangbo J, McCrossan G, Sanborn KB, Basar E, Walch M, Lieberman J. 2015. Apoptosis triggers specific, rapid, and global mRNA decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Rep. 11, 1079–1089. ( 10.1016/j.celrep.2015.04.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. 2013. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 20, 73–81. ( 10.1038/nsmb.2450) [DOI] [PubMed] [Google Scholar]
- 15.Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22, 50–65. ( 10.1101/gad.1622708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song MG, Kiledjian M. 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13, 2356–2365. ( 10.1261/rna.765807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lackey PE, Welch JD, Marzluff WF. 2016. TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA 22, 1673–1688. ( 10.1261/rna.058107.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch JD, Slevin MK, Tatomer DC, Duronio RJ, Prins JF, Marzluff WF. 2015. EnD-Seq and AppEnD: sequencing 3′ ends to identify nontemplated tails and degradation intermediates. RNA 21, 1375–1389. ( 10.1261/rna.048785.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval PL, Su W, Rhoads RE, Prins JF, Marzluff WF. 2014. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol. Cell 53, 1020–1030. ( 10.1016/j.molcel.2014.02.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XC, Purdy M, Marzluff WF, Dominski Z. 2006. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J. Biol. Chem. 281, 30 447–30 454. ( 10.1074/jbc.M602947200) [DOI] [PubMed] [Google Scholar]
- 21.Eckwahl MJ, Sim S, Smith D, Telesnitsky A, Wolin SL. 2015. A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev. 29, 646–657. ( 10.1101/gad.258731.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huo Y, Shen J, Wu H, Zhang C, Guo L, Yang J, Li W. 2016. Widespread 3′–end uridylation in eukaryotic RNA viruses. Sci. Rep. 6, 25454 ( 10.1038/srep25454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aphasizheva I, Aphasizhev R. 2010. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol. Cell. Biol. 30, 1555–1567. ( 10.1128/MCB.01281-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suematsu T, Zhang L, Aphasizheva I, Monti S, Huang L, Wang Q, Costello CE, Aphasizhev R. 2016. Antisense transcripts delimit exonucleolytic activity of the mitochondrial 3′ processome to generate guide RNAs. Mol. Cell 61, 364–378. ( 10.1016/j.molcel.2016.01.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. 2011. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol. Cell 42, 106–117. ( 10.1016/j.molcel.2011.02.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapointe CP, Wickens M. 2013. The nucleic acid-binding domain and translational repression activity of a Xenopus terminal uridylyl transferase. J. Biol. Chem. 288, 20 723–20 733. ( 10.1074/jbc.M113.455451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigáčková D, Fortova A, Zavolan M, Vaňáčova S. 2016. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J. 35, 2179–2191. ( 10.15252/embj.201694857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ustianenko D, et al. 2013. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 19, 1632–1638. ( 10.1261/rna.040055.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labno A, Warkocki Z, Kulinski T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A. 2016. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res. 44, 10 437–10 453. ( 10.1093/nar/gkw649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan M, et al. 2017. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548, 347–351. ( 10.1038/nature23318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl Acad. Sci. USA 107, 3906–3911. ( 10.1073/pnas.0912632107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. 2012. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694. ( 10.1016/j.cub.2012.02.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J, Kuhn L, Hammann P, Pfeffer S. 2016. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 44, 2873–2887. ( 10.1093/nar/gkw040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539. ( 10.1126/science.1187058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, Grosshans H. 2015. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 16, 500–511. ( 10.15252/embr.201540078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren G, Chen X, Yu B. 2012. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. 22, 695–700. ( 10.1016/j.cub.2012.02.052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C. 2011. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep. 12, 690–696. ( 10.1038/embor.2011.100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. 2012. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151, 521–532. ( 10.1016/j.cell.2012.09.022) [DOI] [PubMed] [Google Scholar]
- 39.Trippe R, Sandrock B, Benecke BJ. 1998. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 26, 3119–3126. ( 10.1093/nar/26.13.3119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trippe R, Richly H, Benecke BJ. 2003. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur. J. Biochem. 270, 971–980. ( 10.1046/j.1432-1033.2003.03466.x) [DOI] [PubMed] [Google Scholar]
- 41.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12, 1494–1504. ( 10.1261/rna.87706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuber H, Scheer H, Ferrier E, Sement FM, Mercier P, Stupfler B, Gagliardi D. 2016. Uridylation and PABP cooperate to repair mRNA deadenylated ends in Arabidopsis. Cell Rep. 14, 2707–2717. ( 10.1016/j.celrep.2016.02.060) [DOI] [PubMed] [Google Scholar]
- 43.Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP. 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 11, 1157–1163. ( 10.1038/ncb1931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutierrez-Vazquez C, et al. 2017. 3′ Uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA 23, 882–891. ( 10.1261/rna.060095.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koppers-Lalic D, et al. 2014. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 8, 1649–1658. ( 10.1016/j.celrep.2014.08.027) [DOI] [PubMed] [Google Scholar]
- 46.Reimao-Pinto MM, et al. 2015. Uridylation of RNA hairpins by Tailor confines the emergence of microRNAs in Drosophila. Mol. Cell 59, 203–216. ( 10.1016/j.molcel.2015.05.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bortolamiol-Becet D, Hu F, Jee D, Wen J, Okamura K, Lin CJ, Ameres SL, Lai EC. 2015. Selective suppression of the splicing-mediated microRNA pathway by the terminal uridyltransferase Tailor. Mol. Cell 59, 217–228. ( 10.1016/j.molcel.2015.05.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pirouz M, Du P, Munafo M, Gregory RI. 2016. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep. 16, 1861–1873. ( 10.1016/j.celrep.2016.07.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin G, Keller W. 2007. RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849. ( 10.1261/rna.652807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl Acad. Sci. USA 101, 4407–4412. ( 10.1073/pnas.0400779101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwak JE, Wickens M. 2007. A family of poly(U) polymerases. RNA 13, 860–867. ( 10.1261/rna.514007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451, 1013–1017. ( 10.1038/nature06666) [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312–316. ( 10.1038/nature01039) [DOI] [PubMed] [Google Scholar]
- 54.Thornton JE, Chang HM, Piskounova E, Gregory RI. 2012. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18, 1875–1885. ( 10.1261/rna.034538.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708. ( 10.1016/j.cell.2009.08.002) [DOI] [PubMed] [Google Scholar]
- 56.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626. ( 10.1038/sj.emboj.7600070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rissland OS, Mikulasova A, Norbury CJ. 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 27, 3612–3624. ( 10.1128/Mcb.02209-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Read RL, Martinho RG, Wang SW, Carr AM, Norbury CJ. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl Acad. Sci. USA 99, 12 079–12 084. ( 10.1073/pnas.192467799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang HM, Triboulet R, Thornton JE, Gregory RI. 2013. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28–let-7 pathway. Nature 497, 244–248. ( 10.1038/nature12119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharif H, Conti E. 2013. Architecture of the Lsm1–7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep 5, 283–291. ( 10.1016/j.celrep.2013.10.004) [DOI] [PubMed] [Google Scholar]
- 61.Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 32, 1842–1854. ( 10.1038/emboj.2013.63) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reimao-Pinto MM, Manzenreither RA, Burkard TR, Sledz P, Jinek M, Mechtler K, Ameres SL. 2016. Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J. 35, 2417–2434. ( 10.15252/embj.201695164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faehnle CR, Walleshauser J, Joshua-Tor L. 2014. Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway. Nature 514, 252–256. ( 10.1038/nature13553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv H, Zhu Y, Qiu Y, Niu L, Teng M, Li X. 2015. Structural analysis of Dis3l2, an exosome-independent exonuclease from Schizosaccharomyces pombe. Acta Crystallogr. D Biol. Crystallogr. 71, 1284–1294. ( 10.1107/S1399004715005805) [DOI] [PubMed] [Google Scholar]
- 65.Astuti D, et al. 2012. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat. Genet. 44, 277–284. ( 10.1038/ng.1071) [DOI] [PubMed] [Google Scholar]
- 66.Morris MR, Astuti D, Maher ER. 2013. Perlman syndrome: overgrowth, Wilms tumor predisposition and DIS3L2. Am. J. Med. Genet. C Semin. Med. Genet. 163C, 106–113. ( 10.1002/ajmg.c.31358) [DOI] [PubMed] [Google Scholar]
- 67.Wegert J, et al. 2015. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 27, 298–311. ( 10.1016/j.ccell.2015.01.002) [DOI] [PubMed] [Google Scholar]
- 68.Hrossova D, Sikorsky T, Potesil D, Bartosovic M, Pasulka J, Zdrahal Z, Stefl R, Vaňáčova S. 2015. RBM7 subunit of the NEXT complex binds U-rich sequences and targets 3′–end extended forms of snRNAs. Nucleic Acids Res. 43, 4236–4248. ( 10.1093/nar/gkv240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rissland OS, Norbury CJ. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16, 616–623. ( 10.1038/nsmb.1601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen B, Goodman HM. 2004. Uridine addition after microRNA-directed cleavage. Science 306, 997 ( 10.1126/science.1103521) [DOI] [PubMed] [Google Scholar]
- 71.Xu K, Lin J, Zandi R, Roth JA, Ji L. 2016. MicroRNA-mediated target mRNA cleavage and 3′–uridylation in human cells. Sci. Rep. 6, 30242 ( 10.1038/srep30242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren G, Wang X, Yu B. 2017. Analysis of the uridylation of both ARGONAUTE-bound MiRNAs and 5′ cleavage products of their target RNAs in plants. Methods Mol. Biol. 1640, 23–37. ( 10.1007/978-1-4939-7165-7_2) [DOI] [PubMed] [Google Scholar]
- 73.Zhang Z, et al. 2017. RISC-interacting clearing 3′– 5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife 6, e24466 ( 10.7554/eLife.24466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang H, et al. 2018. Terminal uridylyltransferases execute programmed clearance of maternal transcriptome in vertebrate embryos. Mol. Cell 70, 72–82. ( 10.1016/j.molcel.2018.03.004) [DOI] [PubMed] [Google Scholar]
- 75.Wang SW, Toda T, MacCallum R, Harris AL, Norbury C. 2000. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase δ or ɛ is inactivated. Mol. Cell. Biol. 20, 3234–3244. ( 10.1128/Mcb.20.9.3234-3244.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheer H, Zuber H, De Almeida C, Gagliardi D. 2016. Uridylation earmarks mRNAs for degradation and more. Trends Genet. 32, 607–619. ( 10.1016/j.tig.2016.08.003) [DOI] [PubMed] [Google Scholar]
- 77.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71. ( 10.1038/nature13007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. ( 10.1038/nrg3965) [DOI] [PubMed] [Google Scholar]
- 79.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19, 1661–1671. ( 10.1093/emboj/19.7.1661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tharun S, Parker R. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell 8, 1075–1083. ( 10.1016/S1097-2765(01)00395-1) [DOI] [PubMed] [Google Scholar]
- 81.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91, 457–466. ( 10.1016/S0092-8674(00)80432-8) [DOI] [PubMed] [Google Scholar]
- 82.Chowdhury A, Mukhopadhyay J, Tharun S. 2007. The decapping activator Lsm1p–7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13, 998–1016. ( 10.1261/rna.502507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou L, Zhou Y, Hang J, Wan R, Lu G, Yan C, Shi Y. 2014. Crystal structure and biochemical analysis of the heptameric Lsm1–7 complex. Cell Res. 24, 497–500. ( 10.1038/cr.2014.18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Norbury CJ. 2013. Cytoplasmic RNA: a case of the tail wagging the dog. Nat. Rev. Mol. Cell Biol. 14, 643–653. ( 10.1038/nrm3645) [DOI] [PubMed] [Google Scholar]
- 85.Morozov IY, Jones MG, Gould PD, Crome V, Wilson JB, Hall AJ, Rigden DJ, Caddick MX. 2012. mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol. Cell. Biol. 32, 2585–2595. ( 10.1128/MCB.00316-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pandey NB, Marzluff WF. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7, 4557–4559. ( 10.1128/MCB.7.12.4557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marzluff WF, Koreski KP. 2017. Birth and death of histone mRNAs. Trends Genet. 33, 745–759. ( 10.1016/j.tig.2017.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graves RA, Pandey NB, Chodchoy N, Marzluff WF. 1987. Translation is required for regulation of histone mRNA degradation. Cell 48, 615–626. ( 10.1016/0092-8674(87)90240-6) [DOI] [PubMed] [Google Scholar]
- 89.Meaux SA, Holmquist CE, Marzluff WF. 2018. Role of oligouridylation in normal metabolism and regulated degradation of mammalian histone mRNAs. Phil. Trans. R. Soc. B 373, 20180170 ( 10.1098/rstb.2018.0170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pfeffer S, et al. 2004. Identification of virus-encoded microRNAs. Science 304, 734–736. ( 10.1126/science.1096781) [DOI] [PubMed] [Google Scholar]
- 91.Lin J, Xu K, Roth JA, Ji L. 2016. Detection of siRNA-mediated target mRNA cleavage activities in human cells by a novel stem-loop array RT-PCR analysis. Biochem. Biophys. Rep. 6, 16–23. ( 10.1016/j.bbrep.2016.02.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B. 2014. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl Acad. Sci. USA 111, 6365–6370. ( 10.1073/pnas.1405083111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349. ( 10.1038/nature02873) [DOI] [PubMed] [Google Scholar]
- 94.Furnari FB, Adams MD, Pagano JS. 1993. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc. Natl Acad. Sci. USA 90, 378–382. ( 10.1073/pnas.90.2.378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O, Brodersen P. 2015. SKI2 mediates degradation of RISC 5′–cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 43, 10 975–10 988. ( 10.1093/nar/gkv1014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim B, et al. 2015. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 34, 1801–1815. ( 10.15252/embj.201590931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung CZ, Jo DH, Heinemann IU. 2016. Nucleotide specificity of the human terminal nucleotidyltransferase Gld2 (TUT2). RNA 22, 1239–1249. ( 10.1261/rna.056077.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee M, et al. 2014. Adenylation of maternally inherited microRNAs by Wispy. Mol. Cell 56, 696–707. ( 10.1016/j.molcel.2014.10.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. 2006. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 289, 115–126. ( 10.1016/j.ydbio.2005.10.017) [DOI] [PubMed] [Google Scholar]
- 100.Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. 2005. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11, 1117–1130. ( 10.1261/rna.2630205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rammelt C, Bilen B, Zavolan M, Keller W. 2011. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17, 1737–1746. ( 10.1261/rna.2787011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taylor RC, Cullen SP, Martin SJ. 2008. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241. ( 10.1038/nrm2312) [DOI] [PubMed] [Google Scholar]
- 103.Liu X, Fu R, Pan Y, Meza-Sosa KF, Zhang Z, Lieberman J. 2018. PNPT1 release from mitochondria during apoptosis triggers decay of poly(A) RNAs. Cell 174, 187–201. ( 10.1016/j.cell.2018.04.017) [DOI] [PubMed] [Google Scholar]
- 104.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322, 1849–1851. ( 10.1126/science.1162253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taft RJ, et al. 2009. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 41, 572–578. ( 10.1038/ng.312) [DOI] [PubMed] [Google Scholar]
- 106.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854. ( 10.1126/science.1164096) [DOI] [PubMed] [Google Scholar]
- 107.Hagan JP, Piskounova E, Gregory RI. 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025. ( 10.1038/nsmb.1676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276–284. ( 10.1016/j.molcel.2008.09.014) [DOI] [PubMed] [Google Scholar]
- 109.Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. ( 10.1242/dev.033183) [DOI] [PubMed] [Google Scholar]
- 110.Svoboda P, Franke V, Schultz RM. 2015. Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr. Top. Dev. Biol. 113, 305–349. ( 10.1016/bs.ctdb.2015.06.004) [DOI] [PubMed] [Google Scholar]
- 111.Le Pen J, et al. 2018. Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nat. Struct. Mol. Biol. 25, 778–786. ( 10.1038/s41594-018-0106-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. 2008. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 283, 21 310–21 314. ( 10.1074/jbc.C800108200) [DOI] [PubMed] [Google Scholar]
- 113.Balzeau J, Menezes MR, Cao S, Hagan JP. 2017. The LIN28/let-7 pathway in cancer. Front Genet 8, 31 ( 10.3389/fgene.2017.00031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee H, Han S, Kwon CS, Lee D. 2016. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 7, 100–113. ( 10.1007/s13238-015-0212-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. 2006. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384, 51–61. ( 10.1016/j.gene.2006.07.011) [DOI] [PubMed] [Google Scholar]
- 116.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. 2011. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091. ( 10.1016/j.cell.2011.10.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ustianenko D, Chiu HS, Treiber T, Weyn-Vanhentenryck SM, Treiber N, Meister G, Sumazin P, Zhang C. 2018. LIN28 selectively modulates a subclass of Let-7 microRNAs. Mol. Cell 71, 271–283. ( 10.1016/j.molcel.2018.06.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Faehnle CR, Walleshauser J, Joshua-Tor L. 2017. Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis. Nat. Struct. Mol. Biol. 24, 658–665. ( 10.1038/nsmb.3428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang L, Nam Y, Lee AK, Yu C, Roth K, Chen C, Ransey EM, Sliz P. 2017. LIN28 zinc knuckle domain is required and sufficient to induce let-7 oligouridylation. Cell Rep. 18, 2664–2675. ( 10.1016/j.celrep.2017.02.044) [DOI] [PubMed] [Google Scholar]
- 120.Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G. 2014. Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation. Cell Rep. 9, 1265–1272. ( 10.1016/j.celrep.2014.10.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wilbert ML, et al. 2012. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell 48, 195–206. ( 10.1016/j.molcel.2012.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. 2012. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765–777. ( 10.1016/j.cell.2012.10.019) [DOI] [PubMed] [Google Scholar]
- 123.Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. 2013. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 19, 613–626. ( 10.1261/rna.036491.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Towbin H, Wenter P, Guennewig B, Imig J, Zagalak JA, Gerber AP, Hall J. 2013. Systematic screens of proteins binding to synthetic microRNA precursors. Nucleic Acids Res. 41, e47 ( 10.1093/nar/gks1197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Graf R, Munschauer M, Mastrobuoni G, Mayr F, Heinemann U, Kempa S, Rajewsky N, Landthaler M. 2013. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol. 10, 1146–1159. ( 10.4161/rna.25194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diederichs S, Haber DA. 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131, 1097–1108. ( 10.1016/j.cell.2007.10.032) [DOI] [PubMed] [Google Scholar]
- 127.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. 2011. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell 44, 424–436. ( 10.1016/j.molcel.2011.09.012) [DOI] [PubMed] [Google Scholar]
- 128.Upton JP, et al. 2012. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 338, 818–822. ( 10.1126/science.1226191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Asada K, et al. 2014. Rescuing dicer defects via inhibition of an anti-dicing nuclease. Cell Rep. 9, 1471–1481. ( 10.1016/j.celrep.2014.10.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang JS, Lai EC. 2011. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 43, 892–903. ( 10.1016/j.molcel.2011.07.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Westholm JO, Lai EC. 2011. Mirtrons: microRNA biogenesis via splicing. Biochimie 93, 1897–1904. ( 10.1016/j.biochi.2011.06.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207. ( 10.1016/j.cell.2006.10.040) [DOI] [PubMed] [Google Scholar]
- 133.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 21, 1450–1461. ( 10.1101/gr.118059.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi H, Barnes RL, Carriero N, Atayde VD, Tschudi C, Ullu E. 2014. Role of the Trypanosoma brucei HEN1 family methyltransferase in small interfering RNA modification. Eukaryot. Cell 13, 77–86. ( 10.1128/EC.00233-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 29, 3688–3700. ( 10.1038/emboj.2010.233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Minoda Y, Saeki K, Aki D, Takaki H, Sanada T, Koga K, Kobayashi T, Takaesu G, Yoshimura A. 2006. A novel zinc finger protein, ZCCHC11, interacts with TIFA and modulates TLR signaling. Biochem. Biophys. Res. Commun. 344, 1023–1030. ( 10.1016/j.bbrc.2006.04.006) [DOI] [PubMed] [Google Scholar]
- 137.Marcinowski L, et al. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8, e1002510 ( 10.1371/journal.ppat.1002510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. 2012. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl Acad. Sci. USA 109, 279–284. ( 10.1073/pnas.1114204109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cazalla D, Yario T, Steitz JA. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328, 1563–1566. ( 10.1126/science.1187197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li J, Yang Z, Yu B, Liu J, Chen X. 2005. Methylation protects miRNAs and siRNAs from a 3′–end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507. ( 10.1016/j.cub.2005.07.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. ( 10.1126/science.1107130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang Z, Ebright YW, Yu B, Chen X. 2006. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675. ( 10.1093/nar/gkj474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′–O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608. ( 10.1101/gad.1563607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. 2007. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447, 1126–1129. ( 10.1038/nature05903) [DOI] [PubMed] [Google Scholar]
- 145.Boutet S, et al. 2003. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13, 843–848. ( 10.1016/S0960-9822(03)00293-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ibrahim F, Rohr J, Jeong W.-J., Hesson J, Cerutti H. 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314, 1893 ( 10.1126/science.1135268) [DOI] [PubMed] [Google Scholar]
- 147.Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. 2012. Differential impact of the HEN1 homolog HENN-1 on 21 U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 8, e1002702 ( 10.1371/journal.pgen.1002702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G. 2012. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616 ( 10.1371/journal.pgen.1002616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. 2012. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 8, e1002617 ( 10.1371/journal.pgen.1002617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272. ( 10.1016/j.cub.2007.06.030) [DOI] [PubMed] [Google Scholar]
- 151.Houwing S, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82. ( 10.1016/j.cell.2007.03.026) [DOI] [PubMed] [Google Scholar]
- 152.Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. 2013. Exonuclease hDIS3L2 specifies an exosome-independent 3′–5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 32, 1855–1868. ( 10.1038/emboj.2013.135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kunkel GR, Maser RL, Calvet JP, Pederson T. 1986. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc. Natl Acad. Sci. USA 83, 8575–8579. ( 10.1073/pnas.83.22.8575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Reddy R, Henning D, Das G, Harless M, Wright D. 1987. The capped U6 small nuclear-RNA is transcribed by RNA polymerase-III. J. Biol. Chem. 262, 75–81. [PubMed] [Google Scholar]
- 155.Hilcenko C, et al. 2013. Aberrant 3′ oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood 121, 1028–1038. ( 10.1182/blood-2012-10-461491) [DOI] [PubMed] [Google Scholar]
- 156.Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. 2012. Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep. 2, 855–865. ( 10.1016/j.celrep.2012.08.031) [DOI] [PubMed] [Google Scholar]
- 157.Licht K, Medenbach J, Luhrmann R, Kambach C, Bindereif A. 2008. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA 14, 1532–1538. ( 10.1261/rna.1129608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, Sloan KE, Bohnsack MT. 2017. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. ( 10.15252/embr.201744940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blum B, Bakalara N, Simpson L. 1990. A model for RNA editing in kinetoplastid mitochondria: ‘guide’ RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60, 189–198. ( 10.1016/0092-8674(90)90735-W) [DOI] [PubMed] [Google Scholar]
- 160.Seiwert SD, Stuart K. 1994. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science 266, 114–117. ( 10.1126/science.7524149) [DOI] [PubMed] [Google Scholar]
- 161.Clement SL, Mingler MK, Koslowsky DJ. 2004. An intragenic guide RNA location suggests a complex mechanism for mitochondrial gene expression in Trypanosoma brucei. Eukaryot. Cell 3, 862–869. ( 10.1128/EC.3.4.862-869.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grams J, McManus MT, Hajduk SL. 2000. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 19, 5525–5532. ( 10.1093/emboj/19.20.5525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Aphasizheva I, Aphasizhev R. 2016. U-insertion/deletion mRNA-editing holoenzyme: definition in sight. Trends Parasitol. 32, 144–156. ( 10.1016/j.pt.2015.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lin S, Gregory RI. 2015. Identification of small molecule inhibitors of Zcchc11 TUTase activity. RNA Biol. 12, 792–800. ( 10.1080/15476286.2015.1058478) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.