Abstract
A polyA (pA) tail is an essential modification added to the 3′ ends of a wide range of RNAs at different stages of their metabolism. Here, we describe the main sources of polyadenylation and outline their underlying biochemical interactions within the nuclei of budding yeast Saccharomyces cerevisiae, human cells and, when relevant, the fission yeast Schizosaccharomyces pombe. Polyadenylation mediated by the S. cerevisiae Trf4/5 enzymes, and their human homologues PAPD5/7, typically leads to the 3′-end trimming or complete decay of non-coding RNAs. By contrast, the primary function of canonical pA polymerases (PAPs) is to produce stable and nuclear export-competent mRNAs. However, this dichotomy is becoming increasingly blurred, at least in S. pombe and human cells, where polyadenylation mediated by canonical PAPs may also result in transcript decay.
This article is part of the theme issue ‘5′ and 3′ modifications controlling RNA degradation’.
Keywords: RNA polyadenylation, transcription termination, TRAMP complex, RNA export, RNA decay, polyA binding proteins
1. Introduction
The RNA 3′-end polyA (pA) tail is a post-transcriptional modification discovered in the 1970s to occur on both eukaryotic and prokaryotic transcripts [1–5]. Polyadenylation of eukaryotic mRNAs, mediated by canonical pA polymerases (PAPs), was found to result in the production of long polyadenosine chains and initially shown to contribute to transcript stability, processing, nuclear export and translation [6,7]. Prokaryotic RNA pA tails, on the other hand, were found to be shorter and only present in a small fraction of the RNA population, making their initial studies more cumbersome [6,8,9]. Moreover, prokaryotic pA tails were shown to mainly be involved in the degradation and quality control of RNA and, in addition to their production by bacterial PAP, to also be produced by polynucleotide phosphorylase (PNPase) in a sequential process of degradation and readenylation. This marked divide between eukaryotic and prokaryotic pA-tail biology was later narrowed considerably when it was discovered that the Saccharomyces cerevisiae Trf4/5-Air1/2-Mtr4 Polyadenylation (TRAMP) complex can destabilize a range of transcripts by the addition of short, and ‘bacterial-like’, oligoA tails, targeting RNAs to the 3'-5′ exonucleolytic nuclear exosome complex [10–12]. Additionally, it is now clear that pA binding proteins (PABs) can participate in the decay of transcripts with longer pA-tails both in yeast and in mammals [13–19]. In this review, we focus on the nuclear roles of the pA tail in yeast and human cells and how it directs RNAs to different fates, depending on its kinetics of synthesis, its length and its context of associated factors.
2. The pA tail stimulates mRNP formation and nuclear export
(a). Finding the cleavage site
Cleavage and polyadenylation of the mRNA 3' end is an intrinsic part of the transcription termination process for eukaryotic protein-coding loci and is operated by a conserved complex called cleavage and polyadenylation factor (CPF) in S. cerevisiae and cleavage and polyadenylation specificity factor (CPSF) in human. The multi-subunit nature of CPF/CPSF confers RNA binding, RNA endonuclease, protein phosphatase and PAP activities to the complex (figure 1a for details). In S. cerevisiae, CPF is assisted by the cleavage factor I (CFI) complex, while this activity in human cells is divided between the cleavage stimulation factor (CstF) and the cleavage factors I and II (CFIm/CFIIm). These CFI-like complexes dually interact with RNA polymerase II (RNAPII) and the nascent RNA, which contributes to the proper selection of the RNA cleavage site [20,24,25]. Saccharomyces cerevisiae encodes one PAP enzyme called Pap1, while this activity in human cells is divided between the α/γPAPs (PAPOLA and PAPOLG) and the recently identified Star-PAP (TUT1). Additionally, human cells encode for the testis-specific PAPOLB protein. All these enzymes bind to the Cft1/CPSF160 subunit of CPF/CPSF (figure 1a). Pap1 and its orthologues PAPOLA/PAPOLG also contact Fip1/FIP1, which is particularly important for the regulation of PAP processivity [26,27]. By contrast, Star-PAP interacts with the CPSF73 orthologue of the yeast Ysh1 endonuclease [28,29].
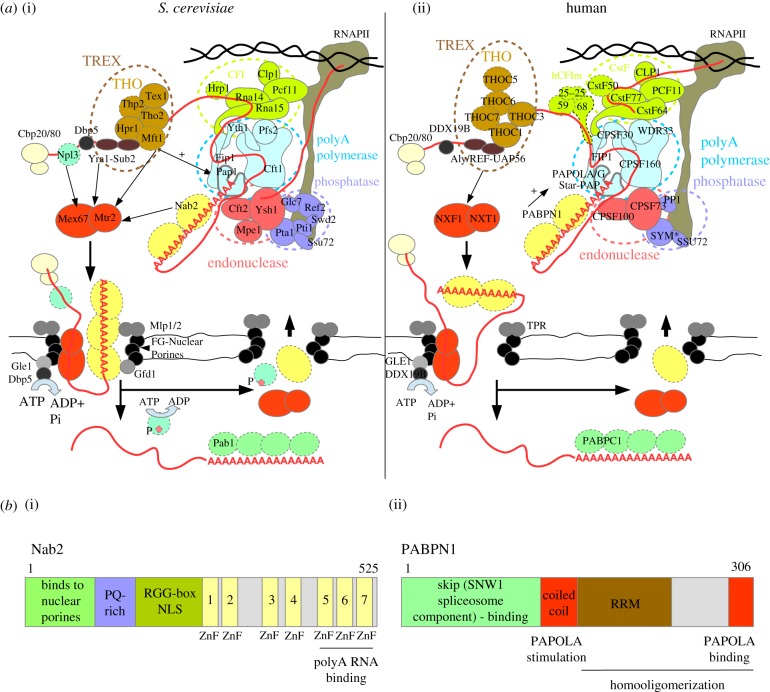
Figure 1.
Model of CPF/CPSF-mediated mRNA 3'-end formation and nuclear export. (a) Schematic overview of 3′-end cleavage, polyadenylation and export factors from S. cerevisiae (a(i)) and human (a(ii)) cells. Factors with no human or S. cerevsiae orthologues with a similar function in polyadenylation and export, respectively, are marked with dashed outlines. In some cases, the human orthologue has not yet been studied in the context of cleavage and polyadenylation. The CPF and CPSF complexes are composed of three basic modules (inspired from model in [20]): (i) the endonuclease module organized around the Ysh1/CPSF73 enzyme supported by Cft2/CPSF100 and Mpe1; (ii) the RNAPII phosphatase module, the catalytic activity of which is mediated by the Glc7/PP1 and Ssu72 subunits of the ‘associated with Pta1′ (human symplekin protein – SYM*) complex (APT: Pta1, Swd2, Ref2, Pti1, Syc1); and (iii) the pA polymerase module, containing the Pap1/PAPOLA/PAPOLG/Star-PAP enzymes, which are recruited to this module mainly by an interaction with Cft1/CPSF160 but also with Fip1/FIP1, Pfs2/WDR33 and Yth1/CPSF30. The yeast CPF complex is further assisted by Cleavage Factor I (CFI: Clp1, Hrp1, Pcf11, Rna14, Rna15), while the human CPSF complex is supported by the CFI-like sub-complexes: cleavage stimulation factor (CstF: CstF50/77/64), cleavage factor II (CFIIm: CLP1, PCF11) and the human-specific cleavage factor I (CFIm: CFIm25/59/68). The CPF and CPSF complexes are recruited dually by binding to specific sequences in the nascent RNA and to the RNAPII CTD. In S. cerevisiae cells, polyadenylation is shown to be stimulated by the THO complex (indicated by the arrow) and in human cells by PABPN1 (arrow). The formation of an export-competent mRNP starts co-transcriptionally with association of the cap-binding complex (CBC, CBP20/80). In yeast Npl3, TREX and Nab2 loading enhances recruitment of the export adaptor Mex67/Mtr2, while in human only the TREX subunit Aly/REF stimulates nuclear export. Successful synthesis of a long pA tail and mRNP assembly leads to release of the newly made RNA from the site of transcription and its translocation to the nuclear pore. Mex67/NXF1 interaction with core FG-nuclear pore factors and Nab2 interactions with the Gfd1 NPC subunit stimulates export. Nab2 binding to Mlp1/Mlp2 promotes retention of unspliced transcripts. An mRNP associated helicase Dbp5/DDX19B is activated by interaction with the Gle1/GLE1 NPC subunit located at the cytoplasmic face and contributes to the displacement of Nab2 and possibly other nuclear mRNP subunits from the transcript. Other mechanisms also contribute to recycling of export factors, such as phosphorylation in case of Npl3. This allows for the release of the transcript into the cytoplasm. Human PABPN1 shuttles between the nucleus and the cytoplasm, but the mechanism by which it is exchanged by cytoplasmic PABPC1 is still unclear. (b) Schematic overview of domain and motif organizations of the main CPF-associated non-orthologous PABs in S. cerevisiae (b(i) Nab2) and human (b(ii) PABPN1) cells highlights the diverse functions of these proteins in pA tail-length control and RNA nuclear export. PABPN1 binds RNA using an RNA Recognition Motif (RRM), while Nab2 recognizes RNA via its three distal zinc fingers (ZnFs) [21]. PABPN1 interacts directly with and stimulates PAPOLA activity, while Nab2 binds the NPC subunits Mlp1/2 and Gfd1 through its N-terminal domain. Moreover, PABPN1 interacts with the spliceosome subunit SKIP [22]. The region responsible for nuclear import of Nab2 is located within the RGG domain [23].
The cleavage/polyadenylation process initiates by the recruitment of CPF/CPSF and the associated complexes to the pA site within the nascent RNA and to the C-terminal domain (CTD) of RNAPII. The nature of an optimal pA site sequence varies between species but generally is composed of several short motifs, which for efficient recruitment of the 3′-end processing complex need to be sequential and strictly separated spatially. In human cells, the cleavage site is located between an AAUAAA hexamer consensus sequence and a GU/U-rich downstream sequence element (DSE). The strength of the pA site also depends on other upstream U-rich and downstream G-rich elements. In S. cerevisiae, this consensus motif is organized differently, with the RNA cleavage site located between two U-rich sequences and assisted by upstream A- and AU-rich elements [24]. Recruitment of CPF/CPSF to the pA site is further facilitated by the interaction of Pcf11/PCF11 with RNAPII (figure 1a). Taken together, these events lead to Ysh1/CPSF73-mediated endocleavage of the RNA. pA sites are often degenerate with several motifs occurring sequentially, so that a locus can produce RNA isoforms with different 3′ ends, often displaying distinct half-lives and localizations. In human cells such different isoforms can be preferentially polyadenylated by either PAPOLA/G or Star-PAP [28–31].
(b). Synthesizing the tail
After endocleavage, a pA tail is added to the 3′ end of the upstream cleavage fragment. Nascent pA tails have a tightly constrained species-specific length, which is required for efficient mRNA nuclear export. Any alterations to the required number of adenosines added may lead to nuclear retention of the transcript and its ensuing decay. Hence, mechanisms exist that regulate pA tail length, but although this process has been studied for over two decades, any in vivo mechanism is still speculative. This is in part due to deadenylation processes, which occur in the nucleus as well as in the cytoplasm, making it challenging to experimentally distinguish the impact of nuclear versus cytoplasmic factors.
In vitro and in vivo studies show that the efficiency of PAP-mediated polyadenylation depends on the strength and composition of the pA site and on the activity of regulatory factors. Pap1/PAPOLA activity in vitro is greatly enhanced in the context of the CPF/CPSF complex [27,32,33], most probably due to tethering of the PAP to the substrate. Indeed, the AAUAAA sequence element has been shown in vitro to be required for specific recruitment of the CPSF complex. Moreover, the upstream and downstream elements can additionally enhance CPSF binding to the substrate [27,34,35]. Fip1/FIP1 may also regulate PAP enzymatic activity, although the underlying mechanism seems to differ between organisms. In the case of S. cerevisiae, Fip1 can conditionally inhibit Pap1 activity in vitro on an A12 oligonucleotide, presumably serving to reduce any unspecific polyadenylation [36,37]. By contrast, human PAPOLA polyadenylation is strongly distributive and FIP1 appears to be required to stimulate PAP activity and does so most optimally on targets containing a strong U-rich upstream element [27]. Human PAP activity in vitro and in vivo is also stimulated by the nuclear PAB, PABPN1 [33,38]. The presence of both CPSF and PABPN1 synergistically stimulates polyadenylation by decreasing the off rate of the PAP enzyme from the target RNA. In doing so, this complex is capable of synthesizing an approximately 250 nt long pA tail without dissociating from the transcript substrate. It is, therefore, likely that PAPOLA, in the early stages of polyadenylation, only interacts with CPSF, leaving the polyadenylation reaction less processive until the first PABPN1 molecule binds the nascent tail. This may create a time-window for regulation of the process, possibly defining the fate of the RNA as discussed below. After synthesis of the polymer beyond these approximately 250 nt the pA addition reaction is strongly inhibited and changes from a highly processive to a distributive mode. The reason for this change in processivity is elusive. PABPN1 has been shown to form spherical structures when bound to long adenosine polymers, and it has been suggested that the formation of such structures during the polyadenylation reaction might interfere with PAP activity. Alternatively, long pA tails might impair the CPSF–PAP interaction, leading to a decrease in complex processivity [39].
PABPN1 has no obvious orthologue in S. cerevisiae (figure 1b), so it is possible that the CPF complex is sufficient for controlling Pap1 processivity. However, additional mechanisms of PAP-stimulation and tail-length restriction have also been proposed. Some of these efforts have employed whole cell extracts derived from selected mutant strains to study the chemistry of polyadenylation and have led to somewhat contradictory models, probably due to the mixing of nuclear and cytoplasmic factors. Saccharomyces cerevisiae harbours two major PABs, Pab1 and Nab2 [40,41]. Initial models postulated that Pap1/CPF would interact to processively polyadenylate the RNA 3' end, while an interaction of Pab1 with the growing oligoadenosine chain and with CFI would restrict the tail length to the 70–80 nt characteristic for S. cerevisiae cells [42]. It was later shown that Pab1 could inhibit A-tail extension in vitro on an A12 oligonucleotide but only in high concentrations [37], possibly reflecting competition of Pab1 with Pap1 in binding to the substrate. However, this seeming control of pA tail length was also suggested to result from Pab1-mediated recruitment of the Pan2/Pan3 deadenylase complex [43]; only Pab1 and Pan2/Pan3 are predominantly cytoplasmic and thus probably mediate tail length regulation in this compartment. Instead, the predominantly nuclear Nab2p was suggested as a possible tail-length restriction factor [21,44,45], and in a more recent study, Nab2 was shown to be required both in vivo and in vitro to protect newly made RNA from decay by the nuclear ribonucleolytic RNA exosome [17]. Pap1 and PAPOLA are both less processive when outside the context of the CPF complex and even though the yeast enzyme is more robust on its own [27,32] both PAPs rely on the CPF/CPSF for polyadenylation initiation. This feature might leave a window of opportunity for shifting the fate of the newly made RNA towards decay rather than full polyadenylation at the early steps of 3′-end processing [17]. Importantly, the models for Pab1 and Nab2 function in pA tail biogenesis are not mutually exclusive as both proteins shuttle between the nucleus and cytoplasm [46,47], potentially allowing Pab1-mediated pA tail restriction of at least some transcripts. Moreover, S. cerevisae pA tails can also be extended by Trf4-mediated polyadenylation [48]. This is, however, only apparent in nuclear exosome-impaired cells and it is, therefore, not clear whether nuclear 3′–5′ decay partakes in pA tail restriction in a wild-type context. In human cells, PABPN1 is functionally connected to the exosome (see below) and a nuclear exosome-dependent pA tail restriction process is, therefore, possible but has not been reported.
(c). pA tail-guided RNA export
It is generally assumed that proper synthesis of the pA tail stimulates the assembly of an export-competent mRNP. Studies in S. cerevisiae indicate that a minimum of 48 DNA-encoded adenosines enhances RNA export without any apparent contribution from the CPF complex [49]. Consistently, inhibition of PABPN1 by the influenza virus NS1A protein, or otherwise PABPN1 depletion, results in the nuclear accumulation of pre-mRNAs with short pA tails [50,51]. However, the pA tail itself is not the only player involved in RNA export. Several proteins are recruited to the nascent transcript by RNA binding and/or via interaction with RNAPII. These factors induce RNA remodelling steps to form mature export-competent mRNPs. Interestingly, some export factors are also required for proper pA tail synthesis. For example, in S. cerevisiae, the integrity of the conserved THO complex (harbouring the Tho2, Hpr1, Mft1, Thp2 and Tex1 proteins) [52] is important for maintaining normal levels of the polyadenylation factor Fip1 and deletion of THO components leads to inefficient polyadenylation and mRNA decay [53]. A generally accepted model explaining a contribution of the pA tail to mRNA export stipulates that recruitment of the main export adaptor, the Mex67/Mtr2 heterodimer in S. cerevisiae (NXF1/NXT1 or TAP-p15 in human), is enhanced by binding to Nab2 [54] (figure 1a). Other adaptors for Mex67/Mtr2 are the Yra1 protein (orthologue of human Aly/REF) and other subunits of the transcription-export (TREX) complex (composed also of the THO complex and the Sub2 helicase) [55,56], and Npl3, an abundant SR-like S. cerevisiae-specific RNA binding protein [57]. Moreover, several mRNP components, including Mex67 and Nab2, interact with constituents of the nuclear pore complex (NPC), contributing to export efficiency. Export directionality is achieved by the Dbp5/DDX19B helicase, which mediates the release of Nab2 and possibly other export factors from the RNA. This step is enhanced by other mechanisms such as Npl3 phosporylation [23,58–63]. Although the network of Nab2 interactions places the protein and the pA tail centrally in the RNA export process, Nab2 depletion on its own only slightly impairs export when, for example, compared with mutation of Mex67 [44,64]. In addition to the aforementioned activity of other Mex67/Mtr2 recruiters, this could also be due to redundant Nab2 and Pab1 export functions, as adding a nuclear localization signal (NLS) to Pab1 partially rescues viability of nab2Δ cells [44]. As Nab2 is not orthologous to human PABPN1 (figure 1b), it is not yet clear if this mechanism is fully conserved. PABPN1 was proposed to contribute to Aly/REF recruitment to the 3′ end of mRNAs [65]. PABPN1 also shuttles between the nucleus and the cytoplasm [50,66] and it has, therefore, been proposed that it accompanies the mRNA during export and is thereafter exchanged with cytoplasmic PABPC1 [67,68]. However, how this might occur is not fully understood and more investigation is required to elucidate the exact function of the pA tail and PABPN1 in mRNP export.
3. pA tails assist the 3′-end trimming and complete degradation of nuclear RNA
Polyadenylation can stimulate or mediate RNA decay in the nucleus by different means depending on its source. Transcripts that are terminated and polyadenylated by the CPF/CPSF complex might be directed towards degradation conditioned by their nuclear retention and such decay is often mediated by PABs. The mechanisms provoking nuclear retention have not been fully described, though some examples from both S. cerevisiae and human cells involve the sensing of pre-mRNA splicing defects. Other polyadenylated transcripts that fall prey to nuclear decay include lncRNAs as well as shorter transcripts like some ‘PROMoter uPstream Transcripts’ (PROMPTs) and products of premature cleavage and polyadenylation (PCPA). In contrast to CPF/CPSF-mediated events, non-canonical polyadenylation instigated by the TRAMP complexes can target RNAs produced by all three polymerases and is independent of PABs. Instead, this pathway is driven by the tight cooperation of the TRAMP complexes with the nuclear RNA exosome. RNAs that are targeted by TRAMP are mainly non-coding and range from stable RNAs such as rRNAs, sn-/snoRNAs and tRNAs to ‘cryptic unstable transcripts’ (CUTs).
(a). pA binding proteins target nuclear-retained transcripts
A clear example of the degradative function of pA tails mediated by PABs has been studied in Schizosaccharomyces pombe, which displays functional similarities to the human system and has yielded valuable insight. Pab2, an orthologue of human PABPN1, is seemingly not involved in the control of pA tail synthesis in S. pombe [38]. Instead, Pab2 and Pla1, the orthologue of human PAPOLA, are both part of an RNA exosome cofactor complex called ‘Mtl1-Red1 core’ (MTREC) or ‘nuclear RNA silencing’ (NURS) (figure 2a) [71–74]. MTREC/NURS is composed of several modules organized around Mtl1, a homologue of the Mtr4/MTR4 helicase, and the Red1 protein, and these factors mediate contact to the S. pombe exosome via the Rrp6 exonuclease The distinct MTREC/NURS modules are suggested to target different exosome substrates, including meiotic mRNAs during mitosis [75–77], CUTs and unspliced pre-mRNAs [74].
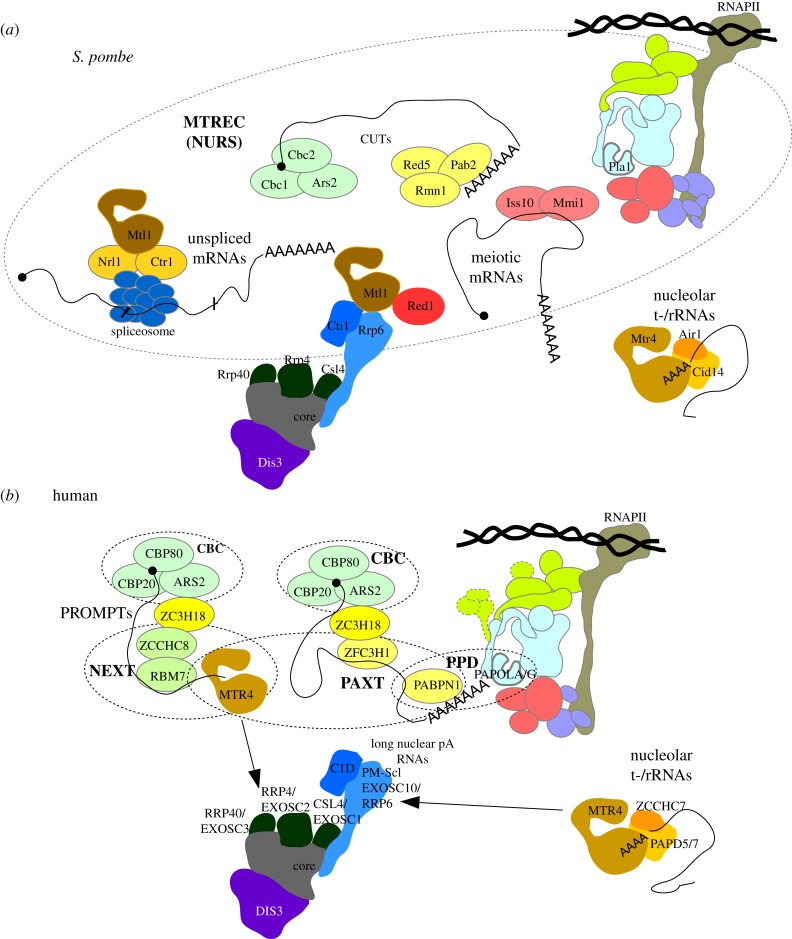
Figure 2.
Nuclear RNA decay pathways in S. pombe and human cells. (a) Schematic overview of the different modules of the S. pombe MTREC/NURS complex. Mtl1 and Red1 are central proteins that are suggested to facilitate the recruitment of the various modules to the RNA exosome. The Mmi1-Iss10 module, assisted by Pab2, plays a key role in the removal of meiotic mRNAs during vegetative growth. The modules binding the RNA 5′ cap (Cbc1-Cbc2-Ars2) and 3′-end pA tail (Pab2-Rmn1-Red5) mediate the removal of CUTs. Finally, the Mtl1–Ctr1–Nrl1 complex interacts with the spliceosome and is involved in the recognition and degradation of unspliced or mis-spliced RNA. Also depicted is the S. pombe TRAMP complex, composed of Cid14, Air1 and the Mtl1 paralog Mtr4. This complex targets nucleolar tRNAs and rRNAs for decay. (b) Schematic overview of nucleoplasmic human complexes involved in RNA targeting. The cap-binding complex (CBC), composed of CBC20 and CBC80, is bridged via the ARS2 and ZC3H18 proteins to the NEXT complex composed of the RNA-binding protein RBM7, the Zn-finger protein ZCCHC8 and MTR4, forming the CBC-NEXT (CBCN) complex [69,70]. The NEXT complex, which does not appear to have a S. pombe orthologous module, targets short cryptic RNAs, like PROMPTs and enhancer RNAs (eRNAs), for exosomal decay. PABPN1, on the other hand, targets longer ncRNAs and pre-mRNAs, which are polyadenylated by PAPOLA/PAPOLG, via the PPD and/or PAXT pathways. PAXT can also interact with the CBC via ARS2 and ZC3H18. The human TRAMP-like complexes, composed of the RNA-binding protein ZCCHC7, MTR4 and one of the pA-polymerases PAPD5 or PAPD7, mediate the decay and processing of nucleolar rRNAs.
A related interaction network has emerged from studies in human cells (figure 2b and table 1), where complexes functionally homologous to MTREC modules target distinct classes of RNAs. Interestingly, PABPN1 has been implicated, highlighting the likely relevance of pA tails. The protein was first described to target selected ncRNAs and pre-mRNAs for degradation by the exosome in a process depending on PAPOLA/PAPOLG and dubbed the ‘PABPN1 and PAPα/γ-mediated decay’ (PPD) pathway [14–16]. In subsequent work, PABPN1 was linked to the exosome via the Zn-finger protein ZFC3H1 and MTR4, forming a stable dimer, which associates with PABPN1 in a partially RNA-dependent manner in the so-coined ‘Poly(A) RNA eXosome Targeting’ (PAXT) connection [19,78]. As the PAXT and PPD pathways share substrates, they may act redundantly or even be overlapping. In addition to its functional interaction with the nuclear exosome, PABPN1 has been reported to partake in another process were polyadenylation is linked to a degradative activity, namely the mechanism that promotes maturation of the human telomerase RNA (hTR), where the pA-specific ribonuclease PARN, together with PABPN1, processes hTR into its mature form [79–81]. Of note, the PABPN1 dependent pathways are not solely responsible for degrading cryptic transcripts in human cells where a well-described parallel pathway is mediated by the ‘nuclear exosome targeting’ (NEXT) complex [69,70], which likely does not target polyadenylated RNA (figure 2b and see below). Nevertheless, in human, as in fission yeast, the MTR4/Mtl1 helicase provides a ‘hub’ that brings different decay complexes together, some of which contain PABs.
Table 1.
Orthologue proteins involved in nuclear RNA decay in S. cerevisiae, S. pombe and human cells.
|
S. cerevisiae |
S. pombe |
H. sapiens |
main activity | ||||
|---|---|---|---|---|---|---|---|
| protein | complex | protein | complex | protein | complex | ||
| Trf4 Trf5 |
TRAMP4 TRAMP5 |
Cid14 | TRAMP | PAPD5 (TRF4-2) PAPD7 (TRF4-1) |
TRAMP | non-canonical polyA-polymerase | |
| Air1/Air2 | Air1 | ZCCHC7 | RNA binding (zinc knuckle) | ||||
| Mtr4 | Mtr4 | MTR4 (SKIV2L2) | helicase | ||||
| Mtl1 | MTREC/NURS | ||||||
| — | Ars2 | ARS2 | CBCA | CBC binding | |||
| Sto1 (Cbp80) | Cbc1 | CBP80 | CBC | cap-binding complex large subunit | |||
| Cbp20 (Cbc2) | Cbc2 | CBP20 | cap-binding complex small subunit | ||||
| — | — | ZCCHC8 | NEXT (with MTR4) | zinc finger; linker between MTR4 and RBM7 | |||
| — | — | RBM7 | RNA binding (RNA recognition motif) | ||||
| — | — | ZC3H18 | — | RNA binding (zinc finger); linker between CBCA and NEXT or PAXT | |||
| — | Red1 | ZFC3H1 | PAXT (with MTR4) | RNA binding (zinc finger) | |||
| Sgn1 | Pab2 | PABPN1 | PPD | pA RNA binding (RNA recognition motif) | |||
| Pap1 | Pla1 | PAPOLA/G | canonical polyA polymerase | ||||
| — | Rmn1 | Rbm26/27 | pA RNA binding (RNA recognition motif) | ||||
| — | Red5 | ZC3H3 | RNA binding (zinc finger) | ||||
| — | Iss10 | — | associates with meiotic transcripts | ||||
| Pho94 | Mmi1 | YTHDF1/2/3 | YTH domain | ||||
| — | Ctr1 | CCDC174 | telomerase regulatory factor Ctr1 | ||||
| — | Nrl1 | NRDE2 | spliceosome-associated protein Nrl1 | ||||
| Hrp1 | Msi2 | MSI1/2 | mRNA cleavage factor complex subunit (predicted in human) | ||||
| Nrd1 | NNS | Seb1 | SCAF4 SCAF8 |
RNA binding (RNA recognition motif), Rpb1 binding: CTD-S5P in S. cerevisiae and CTD-S2P in S. pombe and human | |||
| Nab3 | Nab3 | RALY/RALYL HNRNPC/HNRNPCL1/2/3/4 |
RNA binding (RNA recognition motif) | ||||
| Sen1 | Sen1 | SETX | helicase | ||||
| — | Cid12 | RDRC | — | non-canonical polyA-polymerase | |||
| — | Hrr1 | ZNFX1 | Hrr1: helicase ZNFX1: RNA binding (zinc finger domain) |
||||
| — | Rdp1 | — | RNA-directed RNA polymerase | ||||
| Nab2 | Nab2 | ZC3H14 | RNA binding (zinc finger domain) interaction with nuclear porines |
||||
Finally, S. cerevisiae Nab2 has also been implicated in pre-mRNA decay via its interactions with the splicing machinery. Nab2 was suggested to exert quality control and prevent the ultimate export of pre-mRNA in a process requiring an interaction with the Mlp1-Mlp2 proteins of the NPC nuclear basket. The Nab2 human orthologue ZC3H14 was also shown to interact with the splicing machinery, though its participation in nuclear RNA decay has not yet been addressed (figure 1a and table 1) [18,48,62,82].
4. Oligoadenylation by TRAMP triggers exosome-mediated exonucleolysis
(a). The conserved TRAMP complex
The S. cerevisiae TRAMP complex is composed of a pA polymerase (PAP), Trf4 (alias Pap2) or Trf5 (DNA topoisomerase I related function), a zinc knuckle (ZnK) and RNA binding protein Air1 or Air2 (arginine methyltransferase interacting RING finger) and the Mtr4 helicase, belonging to the DExD/H family (figure 3). It can exist as two isoforms: TRAMP4 (Trf4/Air2/Mtr4) and TRAMP5 (Trf5/Air1/Mtr4), though some reports suggest that TRAMP4 might alternatively contain Air1. The Trf4/5 proteins are unable to bind and adenylate RNA without Air1 or Air2 [11], which contact a domain in Trf4/5 adjacent to their catalytic sites via the Air1/2 terminal ZnK4-5 [84]. Outside of their known domains, the Trf and Air proteins are both largely unstructured but contain short epitopes at their N-termini, that bind Mtr4 in a cooperative manner. This binding positions the exit of the helicase domain towards the PAP domain of Trf [85–87].
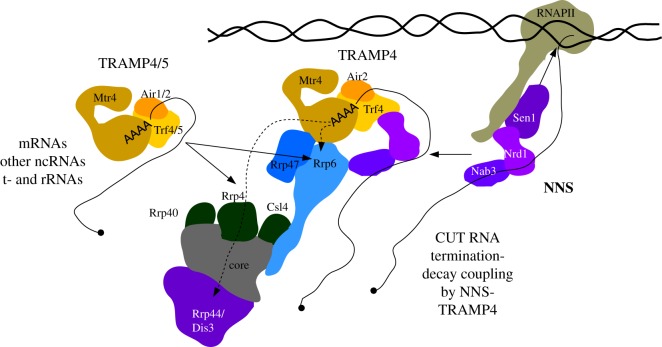
Figure 3.
Schematic overview of S. cerevisiae TRAMP4/5 complexes and their physical relationships with the NNS and exosome complexes. TRAMP4 and TRAMP5 can both be recruited to their targets via Air1/2-mediated RNA binding. In addition, TRAMP4 recruitment to some ncRNAs is enhanced by the direct binding of Trf4 to the Nrd1 subunit of the NNS complex. TRAMP4/5 complexes contact the exosome via Mtr4. TRAMP4 interaction with the exosome can be further enhanced via binding of Nab3 to Rrp6 [83].
In human cells, orthologues of TRAMP components (table 1) have been reported to interact with each other and with RRP6, which suggests that the overall structure of the TRAMP complex and its association with the nuclear exosome is conserved. The PAP activity is provided by the ‘PAP-associated domain-containing proteins 5 and 7′ (PAPD5 and PAPD7 aka TRF4-2 and hTRF4-1). PAPD5 and PAPD7 interact independently with ZCCHC7, orthologue of Air1/2 and with MTR4 and RRP6 [69,88]. PAPD5 contains a basic RNA binding motif at its C-terminus and is capable of adenylating RNA substrates in vitro without any cofactors [89]. However, it is likely that, for efficient activity and/or specificity, it requires ZCCHC7 in vivo. The TRAMP complex is also conserved in S. pombe and is composed of the pA polymerase Cid14, the RNA binding protein Air1 and the helicase Mtr4, a paralogue of Mtl1 [90]. Schizosaccharomyces pombe also contains another non-canonical PAP, Cid12, which is not part of a TRAMP-like complex (see below).
(b). TRAMP is an important cofactor of the RNA exosome
Transcripts adenylated by TRAMP are rapidly degraded by the RNA exosome (figure 3), a conserved complex that constitutes the main source of nuclease activity in eukaryotic nuclei and is also active in the cytoplasm. The exosome core consists of a ring of six catalytically inert proteins with polynucleotide phosphorylase (PNPase) homology and three RNA binding proteins attached to one side, coined the cap. The core and the cap form a central channel ending at the 3′–5′ hydrolytic exonuclease catalytic site of Rrp44 (alias Dis3) [91–93]. Rrp44 also contains an endonuclease site [94], that can access RNA independent of the core [95,96], but which has limited in vivo significance [97]. Rrp6, a nuclear-specific 3′–5′ exonuclease, and its cofactor Rrp47, are attached opposite to Rrp44 via interactions with the Csl4 cap subunit and both of the Rrp43 and Mtr3 core proteins [93]. In all studied species, the exosome requires cofactors for substrate handling and here the TRAMP complex is the best understood. In S. cerevisiae, Mtr4 physically connects Trf4/5-Air1/2 with the exosome via binding to Rrp6 and Rrp47 [98]. Curiously, in human cells, ZCCHC7 and PAPD5 interact with Rrp6 independently of MTR4 [88].
(c). Oligoadenylation enhances the unwinding activity of Mtr4
The S. cerevisiae and human TRAMP-associated PAPs adenylate RNA 3′ ends in a non-processive manner [10,89]. In the case of the S. cerevisiae complex, in vitro studies have shown that the TRAMP adenylation and helicase activities act in a cooperative manner to unwind structured RNAs. The Trf/Air dimer cannot adenylate blunt-ended dsRNA but requires an overhang of at least 1–3 nt [84,87,99]. On the other hand, Mtr4 requires a minimal 5 nt overhang, preferably of adenosines, to conduct its helicase activity. In turn, an A-stretch of 5 nt reduces the affinity of TRAMP towards ATP, limiting further adenylation [99]. Indeed, the average size of TRAMP-produced A tails in vivo appears to be 4–5 nt [100]; however, it is not clear whether this is only due to the chemistry of TRAMP-mediated adenylation or whether exosome-mediated tail shortening also plays a role [48]. Regardless, these data indicate that the TRAMP complex provides adenylation to stimulate the unwinding activity of Mtr4. The channel formed by the core and cap of the exosome can accommodate a 25–30 nt long RNA [93,101]. Thus, the primary function of TRAMP appears to be to produce an ssRNA long enough to be threaded through the exosome core.
(d). Non-canonical polyadenylation targets diverse RNAs
The S. cerevisiae Trf and Air proteins are found in both the nucleoplasm and nucleolus and impact the maturation and expression of transcripts produced by all three RNA polymerases. Individual deletion of TRF or AIR genes does not significantly affect yeast growth at permissive conditions, however, loss of both TRF4 and TRF5 is synthetically lethal and concomitant deletion of AIR1 and AIR2 results in severe growth impairment [102,103]. Thus, TRAMP4 and TRAMP5 appear to act redundantly in the processing of some essential RNAs, the identities of which remain unknown. Trf4 and Trf5 were both proposed to play a role in the decay of incorrectly processed rRNA and tRNA precursors [10,100,104–106], and to be required for the regulation of histone mRNA levels [107]. In addition, the Trf proteins have specialized sets of substrates with Trf4 targeting some mRNAs but mostly sn/snoRNAs, CUTs and Ty1 retrotransposons, while Trf5 is primarily involved in regulating mRNA abundance [10,12,108]. Interestingly, Trf4-mediated adenylation is dispensable for the maturation or decay of many substrates [108], which is probably because the Trf/Air dimer can stimulate Mtr4 unwinding activity in an adenylation-independent manner; at least in vitro [109]. Both of the Air proteins bind RNA rather unspecifically and may, therefore, rely on other factors for their substrate recruitment. Indeed, the specificity of TRAMP4 for ncRNAs is mediated by its association with the Nrd1-Nab3-Sen1 (NNS) complex, which recognizes short sequence motifs enriched in CUTs and snRNAs [100]. Simultaneously, Nrd1 binds to the RNAPII CTD and hereby mediates Sen1 helicase recruitment, which induces transcription termination. In a subsequent step, Nrd1 binds Trf4, which stimulates RNA decay, possibly by enhancing TRAMP4 adenylation activity or by stabilizing the TRAMP complex on the transcript, which allows efficient exosome recruitment (figure 3) [110–112].
The human Air homolog, ZCCHC7, is strictly localized to nucleoli [69]. By contrast, while also localized to nucleoli, human PAPD5 and MTR4 are robustly present in the cell nucleoplasm. This differential localization of human TRAMP subunits is reflected in the repertoire of their RNA targets. PAPD5 and ZCCHC7 interact with proteins involved in rRNA processing and have been shown to mediate turnover of pre-rRNA 5′-ETS fragments [69,88]. PAPD5 also binds to splicing factors [69,113], but the function of this connection still remains incompletely explored.
The S. pombe TRAMP subunit Cid14 is exclusively localized to nucleoli where it participates in the removal of aberrant tRNAs and rRNAs [114] (figure 2a). Another fission yeast polyA polymerase takes part in heterochromatin formation. In S. pombe, production of small interfereing RNAs (siRNAs) is mediated by the RNA-induced transcriptional silencing (RITS) complex, which interacts with the RNA-directed RNA polymerase complex (RDRC, table 1). RDRC is composed of the non-canonical polyA polymerase Cid12, the helicase Hrr1 and the RNA-directed RNA polymerase Rdp1. The Cid12 protein itself is required for heterochromatin formation, though it is unclear what is the role of its polyA-polymerase activity [115,116]. Curiously, impairment of nucleolar TRAMP function and its resulting accumulation of abundant aberrant tRNAs and rRNAs has been shown to direct the siRNA machinery towards nucleolar RNAs in an unspecific manner, thus perturbing heterochromatin formation at the usual loci [90,117].
In conclusion, all human and yeast TRAMP complexes participate in the decay of nucleolar targets [88,100,104–106,114,118]. However, it appears that S. cerevisiae Trf4/5 also have broad nucleoplasmic activities, which in S. pombe and human cells are mediated by other complexes organized around MTR4: MTREC, NEXT and PAXT, respectively. Additionally, in the case of S. pombe, non-canonical polyadenylation mediates heterochromatin formation.
5. How can pA tail status help distinguish unstable from stable RNA?
In this review, we have described two opposing outcomes deriving from the addition of a pA tail to the 3' end of an RNA. Some adenylation events are coupled to exonuclease-mediated 3′-end maturation or degradation, while others facilitate the stabilization of the RNA and its export to the cytoplasm. One critical question is, therefore, how RNAs that are destined for nuclear export, such as mRNAs, are distinguished from those that are normally degraded, such as PROMPTs, PCPA RNAs [119] and CUTs. S. cerevisiae has evolved an elegant and unique system in which cryptic transcripts are specifically terminated and marked for TRAMP/exosome degradation by the dedicated NNS complex. This, however, appears different in human and S. pombe cells where coding, and at least some cryptic, transcripts are 3′-end processed by the CPF/CPSF pathway. Thus, the difference between export-competent and nuclear-degraded RNAs must lie in the differential recruitment of proteins to these RNAs. However, though the question is still not fully explored, the answer does not seem to be fully provided by the differential recruitment of export versus degradation factors. Firstly, the PAXT subunit PABPN1 is supposedly recruited to all these RNAs as an intrinsic component of the CPSF complex, and secondly, the RBM7 subunit of the NEXT complex appears to bind all nascent RNAs alike [120]. Thus other features must contribute to selecting RNAs for decay. One to consider here is transcript length. Saccharomyces cerevisiae CUTs, human PROMPTs and PCPA transcripts are all short in comparison to their mRNA counterparts. For CUTs this feature has a key bearing in promoting their transcription termination as Nrd1 binds the RNAPII CTD phosphorylated at serine five residues, a hallmark of early transcription activity. A conceptually analogous mechanism might be operating in human cells through the CBC, which is physically close to the 3′ ends of the short PROMPT/PCPA RNAs and might elicit degradation by its efficient recruitment of the RNA exosome via the CBCN complex (figure 2b) [70].
Such probable propensity of short transcripts to be degraded does, however, not explain how nuclear decay of longer transcripts, which are terminated by the CPSF pathway and bound by PABPN1, can occur. A ‘nuclear-timer’ model has therefore been proposed, suggesting that PABPN1 binding to newly synthesized RNAs initially yields protection, while decay will only be promoted if the RNA has a prolonged residence time in the nucleus, providing sufficient time for RNA exosome recruitment [121,122]. Several mechanisms, such as export kinetics or splicing, could contribute to selective nuclear retention. With the exception of specific highly expressed genes, which upon activation might translocate towards the NPC [123], most mRNAs diffuse stochastically within the interchromatin matrix until reaching the nuclear exit [124]. Thus, some transcripts may reside in the nucleus for a sufficient amount of time to be targeted for decay. Studies in the human system have shown that various mRNAs exhibit different nuclear export rates and depending on the cell type and environmental condition some mRNAs might reside longer in the nucleus than in the cytoplasm, which was proposed to play a role in reducing the transcriptional noise resulting from gene-activation bursts [125]. Exact mechanistic aspects of selective transcript retention remain elusive but are probably also relevant for the degradation of improperly processed transcripts that are not exported with sufficient kinetics. An obvious feature contributing to nuclear retention time is splicing. In S. cerevisiae, the NPC is actively engaged in retaining unspliced mRNAs in the nucleus via the Mlp1 and Mlp2 proteins located on the nuclear face of the pore complex [82]. While this has not been reported to occur in the human system, it has been shown that intron excision stimulates RNA export [126]. Moreover, some poorly processed transcripts with nuclear localization are stabilized in cells overexpressing a dominant-negative PABPN1 mutant [127], which indicates that slow splicing could lead to nuclear retention and degradation mediated by PABPN1.
The length of the pA tail itself might also be a key factor that promotes nuclear retention as a critical length of the adenosine chain is required for promoting export [49]. In S. cerevisiae, CUTs, which are destined for nuclear decay, are on average short-tailed, which is a consequence of the low Trf4 processivity and the physical interaction of the TRAMP complex with the exosome. Human PAP is a distributive enzyme in the early phase of its polyadenylation activity and prior to PABPN1 recruitment. This feature might result in the formation of a fraction of RNAs harbouring only short pA tails, which would be more susceptible to decay. It is, therefore, tempting to speculate that polyadenylation of some unstable human ncRNAs might be negatively regulated to further their decay. This regulation could depend on sequence elements. The FIP1 protein is most efficient at stimulating PAPOLA activity when the RNA template is U-rich. In a similar manner, other CPSF associated proteins, like CFIm, CFIm68 and CFIm59, have the capacity to bind RNA in a sequence-specific manner and their recruitment regulates Fip1 function in alternative polyadenylation [35]. Polyadenylation efficiency can also be influenced by other activities such as splicing, which has been shown to stimulate 3'-end processing [35,128–130]. The CPSF complex is recruited to the nascent RNA by binding to RNA and to the RNAPII CTD phosphorylated at serine 2 [131]. It is, therefore, also possible that a polymerase that has not yet entered into a state of productive elongation, marked by robust serine 2 phosphorylation status, does not support the formation of a fully competent polyadenylation machinery. Further studies will show whether any of these aspects of the polyadenylation process explain the de-stabilization of transcripts destined for nuclear decay.
Polyadenylation is a 3′-end modification that can direct RNA fate towards stability and export or processing and decay, depending on the context in which it is produced. TRAMP-mediated adenylation is always coupled to exonucleolysis, whereas CPF/CPSF A-tailing may induce export or decay, depending on still ill-explained mechanisms. While some of the decisive potential for CPF/CPSF-targets appears to relate to PABs, an equally important, but less explored, role might be exercised by negative regulation of PAP activity, potentially impacting export efficiency and allowing more time for nuclear decay.
Acknowledgements
We thank Manfred Schmid and Toomas Silla for critical reading of the manuscript.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
Work in the authors' laboratory is supported by the ERC (grant 339953), the Lundbeck- and the Novo Nordisk-Foundations.
References
- 1.Darnell JE, Wall R, Tushinski RJ. 1971. An adenylic acid-rich sequence in messenger RNA of HeLa cells and its possible relationship to reiterated sites in DNA. Proc. Natl Acad. Sci. USA 68, 1321–1325. ( 10.1073/pnas.68.6.1321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmonds M, Vaughan MH, Nakazato H. 1971. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: possible evidence for a precursor relationship. Proc. Natl Acad. Sci. USA 68, 1336–1340. ( 10.1073/pnas.68.6.1336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SY, Mendecki J, Brawerman G. 1971. A polynucleotide segment rich in adenylic acid in the rapidly-labeled polyribosomal RNA component of mouse sarcoma 180 ascites cells. Proc. Natl Acad. Sci. USA 68, 1331–1335. ( 10.1073/pnas.68.6.1331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakazato H, Venkatesan S, Edmonds M. 1975. Polyadenylic acid sequences in E. coli messenger RNA. Nature 256, 144–146. ( 10.1038/256144a0) [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan PR, Ramanarayanan M, Rabbani E. 1975. Presence of polyriboadenylate sequences in pulse-labeled RNA of Escherichia coli. Proc. Natl Acad. Sci. USA 72, 2910–2914. ( 10.1073/pnas.72.8.2910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyfus M, Régnier P. 2002. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell 111, 611–613. ( 10.1016/S0092-8674(02)01137-6) [DOI] [PubMed] [Google Scholar]
- 7.Edmonds M. 2002. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 71, 285–389. ( 10.1016/S0079-6603(02)71046-5) [DOI] [PubMed] [Google Scholar]
- 8.Mohanty BK, Kushner SR. 2010. Bacterial/archaeal/organellar polyadenylation. Wiley Interdiscip. Rev. RNA 2, 256–276. ( 10.1002/wrna.51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66, 173–197. ( 10.1146/annurev.biochem.66.1.173) [DOI] [PubMed] [Google Scholar]
- 10.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724. ( 10.1016/j.cell.2005.04.029) [DOI] [PubMed] [Google Scholar]
- 11.Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3, e189 ( 10.1371/journal.pbio.0030189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyers F, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737. ( 10.1016/j.cell.2005.04.030) [DOI] [PubMed] [Google Scholar]
- 13.Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bähler J, Bachand F. 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol. Cell 44, 108–119. ( 10.1016/j.molcel.2011.06.035) [DOI] [PubMed] [Google Scholar]
- 14.Beaulieu YB, Kleinman CL, Landry-Voyer A-M, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 8, e1003078 ( 10.1371/journal.pgen.1003078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bresson SM, Conrad NK. 2013. The human nuclear poly(A)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 9, e1003893 ( 10.1371/journal.pgen.1003893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bresson SM, Hunter OV, Hunter AC, Conrad NK. 2015. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet. 11, e1005610-25 ( 10.1371/journal.pgen.1005610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid M, Olszewski P, Pelechano V, Gupta I, Steinmetz LM, Jensen TH. 2015. The nuclear polyA-binding protein Nab2p is essential for mRNA production. Cell Rep. 12, 128–139. ( 10.1016/j.celrep.2015.06.008) [DOI] [PubMed] [Google Scholar]
- 18.Soucek S, et al. 2016. The evolutionarily-conserved polyadenosine RNA binding protein, Nab2, cooperates with splicing machinery to regulate the fate of pre-mRNA. Mol. Cell. Biol. 36, 2697–2714. ( 10.1128/MCB.00402-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meola N, et al. 2016. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533. ( 10.1016/j.molcel.2016.09.025) [DOI] [PubMed] [Google Scholar]
- 20.Casañal A, et al. 2017. Architecture of eukaryotic mRNA 3′-end processing machinery. Science 358, 1056–1059. ( 10.1126/science.aao6535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly SM, Leung SW, Apponi LH, Bramley AM, Tran EJ, Chekanova JA, Wente SR, Corbett AH. 2010. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA-binding protein 2 (Nab2) is required for correct mRNA 3'-end formation. J. Biol. Chem. 285, 26 022–26 032. ( 10.1074/jbc.M110.141127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YJ, Noguchi S, Hayashi YK, Tsukahara T, Shimizu T, Arahata K. 2001. The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum. Mol. Genet. 10, 1129–1139. ( 10.1093/hmg/10.11.1129) [DOI] [PubMed] [Google Scholar]
- 23.Truant R, Fridell RA, Benson RE, Bogerd H, Cullen BR. 1998. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol. Cell. Biol. 18, 1449–1458. ( 10.1128/MCB.18.3.1449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millevoi S, Vagner S. 2009. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38, 2757–2774. ( 10.1093/nar/gkp1176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neve J, Patel R, Wang Z, Louey A, Furger AM. 2017. Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol. 14, 865–890. ( 10.1080/15476286.2017.1306171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murthy KG, Manley JL. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3'-end formation. Genes Dev. 9, 2672–2683. ( 10.1101/gad.9.21.2672) [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23, 616–626. ( 10.1038/sj.emboj.7600070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laishram RS, Anderson RA. 2010. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. EMBO J. 29, 4132–4145. ( 10.1038/emboj.2010.287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Laishram RS, Ji Z, Barlow CA, Tian B, Anderson RA. 2012. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIα and PKCδ signaling. Mol. Cell 45, 25–37. ( 10.1016/j.molcel.2011.11.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451, 1013–1017. ( 10.1038/nature06666) [DOI] [PubMed] [Google Scholar]
- 31.Kandala DT, Mohan N, Laishram RS. 2016. CstF-64 and 3'-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding PAPα. Nucleic Acids Res. 44, 811–823. ( 10.1093/nar/gkv1074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. 1997. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 16, 4727–4737. ( 10.1093/emboj/16.15.4727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, Wahle E. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22 803–22 814. ( 10.1074/jbc.M109.018226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheets MD, Ogg SC, Wickens MP. 1990. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18, 5799–5805. ( 10.1093/nar/18.19.5799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y, et al. 2018. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol. Cell 69, 62–74.e4. ( 10.1016/j.molcel.2017.11.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmling S, Zhelkovsky A, Moore CL. 2001. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 21, 2026–2037. ( 10.1128/MCB.21.6.2026-2037.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhelkovsky A, Helmling S, Moore C. 1998. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol. 18, 5942–5951. ( 10.1128/MCB.18.10.5942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kühn U, Buschmann J, Wahle E. 2017. The nuclear poly(A) binding protein of mammals, but not of fission yeast, participates in mRNA polyadenylation. RNA 23, 473–482. ( 10.1261/rna.057026.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kühn U, Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678, 67–84. ( 10.1016/j.bbaexp.2004.03.008) [DOI] [PubMed] [Google Scholar]
- 40.Sachs AB, Davis RW, Kornberg RD. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7, 3268–3276. ( 10.1128/MCB.7.9.3268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson JT, Wilson SM, Datar KV, Swanson MS. 1993. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13, 2730–2741. ( 10.1128/MCB.13.5.2730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17, 3694–3701. ( 10.1128/MCB.17.7.3694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mangus DA, Smith MM, McSweeney JM, Jacobson A. 2004. Identification of factors regulating poly(A) tail synthesis and maturation. Mol. Cell. Biol. 24, 4196–4206. ( 10.1128/MCB.24.10.4196-4206.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21, 1800–1810. ( 10.1093/emboj/21.7.1800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dheur S, Nykamp KR, Viphakone N, Swanson MS, Minvielle-Sebastia L. 2005. Yeast mRNA poly(A) tail length control can be reconstituted in vitroin the absence of pab1p-dependent poly(A) nuclease activity. J. Biol. Chem. 280, 24 532–24 538. ( 10.1074/jbc.M504720200) [DOI] [PubMed] [Google Scholar]
- 46.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277, 7752–7760. ( 10.1074/jbc.M110053200) [DOI] [PubMed] [Google Scholar]
- 47.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. 2005. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11, 517–531. ( 10.1261/rna.7291205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. 2012. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol. Cell 47, 267–280. ( 10.1016/j.molcel.2012.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dower K, Kuperwasser N, Merrikh H, Rosbash M. 2004. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA 10, 1888–1899. ( 10.1261/rna.7166704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3'-end processing machinery. EMBO J. 18, 2273–2283. ( 10.1093/emboj/18.8.2273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. 2010. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 19, 1058–1065. ( 10.1093/hmg/ddp569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peña A, et al. 2012. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 31, 1605–1616. ( 10.1038/emboj.2012.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saguez C, Schmid M, Olesen JR, Ghazy MAE-H, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. 2008. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell 31, 91–103. ( 10.1016/j.molcel.2008.04.030) [DOI] [PubMed] [Google Scholar]
- 54.Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Dach VE, Corbett AH, Dargemont C, Stutz F. 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24, 1927–1938. ( 10.1101/gad.583310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strässer K, Hurt E. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413, 648–652. ( 10.1038/35098113) [DOI] [PubMed] [Google Scholar]
- 56.Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. 2006. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl Acad. Sci. USA 103, 16 376–16 381. ( 10.1073/pnas.0607941103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilbert W, Guthrie C. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell 13, 201–212. ( 10.1016/S1097-2765(04)00030-9) [DOI] [PubMed] [Google Scholar]
- 58.Katahira J, Straesser K, Saiwaki T, Yoneda Y, Hurt E. 2002. Complex formation between Tap and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J. Biol. Chem. 277, 9242–9246. ( 10.1074/jbc.M110007200) [DOI] [PubMed] [Google Scholar]
- 59.Suntharalingam M, Alcázar-Román AR, Wente SR. 2004. Nuclear export of the yeast mRNA-binding protein Nab2 is linked to a direct interaction with Gfd1 and to Gle1 function. J. Biol. Chem. 279, 35 384–35 391. ( 10.1074/jbc.M402044200) [DOI] [PubMed] [Google Scholar]
- 60.Tran EJ, Zhou Y, Corbett AH, Wente SR. 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28, 850–859. ( 10.1016/j.molcel.2007.09.019) [DOI] [PubMed] [Google Scholar]
- 61.Grant RP, Marshall NJ, Yang J-C, Fasken MB, Kelly SM, Harreman MT, Neuhaus D, Corbett AH, Stewart M. 2008. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J. Mol. Biol. 376, 1048–1059. ( 10.1016/j.jmb.2007.11.087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fasken MB, Stewart M, Corbett AH. 2008. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J. Biol. Chem. 283, 27 130–27 143. ( 10.1074/jbc.M803649200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilbert W, Siebel CW, Guthrie C. 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7, 302–313. ( 10.1017/S1355838201002369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, Hurt E. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18, 6826–6838. ( 10.1128/MCB.18.11.6826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi M, Zhang H, Wu X, He Z, Wang L, Yin S, Tian B, Li G, Cheng H. 2017. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 45, 9640–9653. ( 10.1093/nar/gkx597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calado A, Kutay U, Kühn U, Wahle E, Carmo-Fonseca M. 2000. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 6, 245–256. ( 10.1017/S1355838200991908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemay J-F, Lemieux C, St-André O, Bachand F. 2010. Crossing the borders: poly(A)-binding proteins working on both sides of the fence. RNA Biol. 7, 291–295. ( 10.4161/rna.7.3.11649) [DOI] [PubMed] [Google Scholar]
- 68.Wigington CP, Williams KR, Meers MP, Bassell GJ, Corbett AH. 2014. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip. Rev. RNA 5, 601–622. ( 10.1002/wrna.1233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637. ( 10.1016/j.molcel.2011.06.028) [DOI] [PubMed] [Google Scholar]
- 70.Andersen PR, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 20, 1367–1376. ( 10.1038/nsmb.2703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sugiyama T, Sugioka-Sugiyama R. 2011. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 30, 1027–1039. ( 10.1038/emboj.2011.32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee NN, et al. 2013. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155, 1061–1074. ( 10.1016/j.cell.2013.10.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egan ED, Braun CR, Gygi SP, Moazed D. 2014. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA 20, 867–881. ( 10.1261/rna.044479.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Y, Zhu J, Schermann G, Ohle C, Bendrin K, Sugioka-Sugiyama R, Sugiyama T, Fischer T. 2015. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat. Commun. 6, 7050 ( 10.1038/ncomms8050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harigaya Y, et al. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50. ( 10.1038/nature04881) [DOI] [PubMed] [Google Scholar]
- 76.St-André O, Lemieux C, Perreault A, Lackner DH, Bähler J, Bachand F. 2010. Negative regulation of meiotic gene expression by the nuclear poly(A)-binding protein in fission yeast. J. Biol. Chem. 285, 27 859–27 868. ( 10.1074/jbc.M110.150748) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. 2010. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 29, 2173–2181. ( 10.1038/emboj.2010.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ogami K, Richard P, Chen Y, Hoque M, Li W, Moresco JJ, Yates JR, Tian B, Manley JL. 2017. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 31, 1257–1271. ( 10.1101/gad.302604.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, Tai AK, Agarwal S. 2015. Poly(A)-specific ribonuclease (PARN) mediates 3'-end maturation of the telomerase RNA component. Nat. Genet. 47, 1482–1488. ( 10.1038/ng.3423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen D, Grenier St-Sauveur V, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F. 2015. A polyadenylation-dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 13, 2244–2257. ( 10.1016/j.celrep.2015.11.003) [DOI] [PubMed] [Google Scholar]
- 81.Tseng C-K, Wang H-F, Burns AM, Schroeder MR, Gaspari M, Baumann P. 2015. Human telomerase RNA processing and quality control. Cell Rep. 13, 2232–2243. ( 10.1016/j.celrep.2015.10.075) [DOI] [PubMed] [Google Scholar]
- 82.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73. ( 10.1016/S0092-8674(03)01026-2) [DOI] [PubMed] [Google Scholar]
- 83.Fasken MB, Laribee RN, Corbett AH. 2015. Nab3 facilitates the function of the TRAMP complex in RNA processing via recruitment of Rrp6 independent of Nrd1. PLoS Genet. 11, e1005044 ( 10.1371/journal.pgen.1005044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamill S, Wolin SL, Reinisch KM. 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl Acad. Sci. USA 107, 15 045–15 050. ( 10.1073/pnas.1003505107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falk S, Weir JR, Hentschel J, Reichelt P, Bonneau F, Conti E. 2014. The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol. Cell 55, 856–867. ( 10.1016/j.molcel.2014.07.020) [DOI] [PubMed] [Google Scholar]
- 86.Losh JS, King AK, Bakelar J, Taylor L, Loomis J, Rosenzweig JA, Johnson SJ, van Hoof A. 2015. Interaction between the RNA-dependent ATPase and poly(A) polymerase subunits of the TRAMP complex is mediated by short peptides and important for snoRNA processing. Nucleic Acids Res. 43, 1848–1858. ( 10.1093/nar/gkv005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holub P, et al. 2012. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res. 40, 5679–5693. ( 10.1093/nar/gks223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sudo H, Nozaki A, Uno H, Ishida Y-I, Nagahama M. 2016. Interaction properties of human TRAMP-like proteins and their role in pre-rRNA 5'ETS turnover. FEBS Lett. 590, 2963–2972. ( 10.1002/1873-3468.12314) [DOI] [PubMed] [Google Scholar]
- 89.Rammelt C, Bilen B, Zavolan M, Keller W. 2011. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17, 1737–1746. ( 10.1261/rna.2787011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bühler M, Haas W, Gygi SP, Moazed D. 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129, 707–721. ( 10.1016/j.cell.2007.03.038) [DOI] [PubMed] [Google Scholar]
- 91.Liu Q, Greimann JC, Lima CD. 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237. ( 10.1016/j.cell.2006.10.037) [DOI] [PubMed] [Google Scholar]
- 92.Dziembowski A, Lorentzen E, Conti E, Séraphin B. 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Genes Dev. 14, 15–22. ( 10.1038/nsmb1184) [DOI] [PubMed] [Google Scholar]
- 93.Makino DL, Baumgärtner M, Conti E. 2013. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–75. ( 10.1038/nature11870) [DOI] [PubMed] [Google Scholar]
- 94.Lebreton A, Tomecki R, Dziembowski A, Séraphin B. 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996. ( 10.1038/nature07480) [DOI] [PubMed] [Google Scholar]
- 95.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. 2008. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell 29, 717–728. ( 10.1016/j.molcel.2008.02.018) [DOI] [PubMed] [Google Scholar]
- 96.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139, 547–559. ( 10.1016/j.cell.2009.08.042) [DOI] [PubMed] [Google Scholar]
- 97.Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell 48, 409–421. ( 10.1016/j.molcel.2012.08.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schuch B, Feigenbutz M, Makino DL, Falk S, Basquin C, Mitchell P, Conti E. 2014. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 33, 2829–2846. ( 10.15252/embj.201488757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jia H, Wang X, Liu F, Guenther U-P, Srinivasan S, Anderson JT, Jankowsky E. 2011. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell 145, 890–901. ( 10.1016/j.cell.2011.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wlotzka W, Kudla G, Granneman S, Tollervey D. 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 30, 1790–1803. ( 10.1038/emboj.2011.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wasmuth EV, Lima CD. 2012. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell 48, 133–144. ( 10.1016/j.molcel.2012.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Castaño IB, Heath-Pagliuso S, Sadoff BU, Fitzhugh DJ, Christman MF. 1996. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 24, 2404–2410. ( 10.1093/nar/24.12.2404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmidt K, Xu Z, Mathews DH, Butler JS. 2012. Air proteins control differential TRAMP substrate specificity for nuclear RNA surveillance. RNA 18, 1934–1945. ( 10.1261/rna.033431.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dez C, Houseley J, Tollervey D. 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 25, 1534–1546. ( 10.1038/sj.emboj.7601035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kadaba S, Wang X, Anderson JT. 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12, 508–521. ( 10.1261/rna.2305406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wery M, Ruidant S, Schillewaert S, Leporé N, Lafontaine DLJ. 2009. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 15, 406–419. ( 10.1261/rna.1402709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reis CC, Campbell JL. 2006. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics 175, 993–1010. ( 10.1534/genetics.106.065987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Paolo SS, Vanácová S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP. 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 5, e1000555-17 ( 10.1371/journal.pgen.1000555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jia H, Wang X, Anderson JT, Jankowsky E. 2012. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc. Natl Acad. Sci. USA 109, 7292–7297. ( 10.1073/pnas.1201085109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vasiljeva L, Buratowski S. 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21, 239–248. ( 10.1016/j.molcel.2005.11.028) [DOI] [PubMed] [Google Scholar]
- 111.Tudek A, et al. 2014. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol. Cell 55, 467–481. ( 10.1016/j.molcel.2014.05.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim K, Heo D-H, Kim I, Suh J-Y, Kim M. 2016. Exosome cofactors connect transcription termination to RNA processing by guiding terminated transcripts to the appropriate exonuclease within the nuclear exosome. J. Biol. Chem. 291, 13 229–13 242. ( 10.1074/jbc.M116.715771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nag A, Steitz JA. 2012. Tri-snRNP-associated proteins interact with subunits of the TRAMP and nuclear exosome complexes, linking RNA decay and pre-mRNA splicing. RNA Biol. 9, 334–342. ( 10.4161/rna.19431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang S-W. 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 26, 1710–1721. ( 10.1128/MCB.26.5.1710-1721.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802. ( 10.1016/j.cell.2004.11.034) [DOI] [PubMed] [Google Scholar]
- 116.Win TZ, Stevenson AL, Wang S-W. 2006. Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control. Mol. Cell. Biol. 26, 4435–4447. ( 10.1128/MCB.02205-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bühler M, Spies N, Bartel DP, Moazed D. 2008. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 15, 1015–1023. ( 10.1038/nsmb.1481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 11, 106–111. ( 10.1038/embor.2009.271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berg MG, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150, 53–64. ( 10.1016/j.cell.2012.05.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lubas M, Andersen PR, Schein A, Dziembowski A, Kudla G, Jensen TH. 2015. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 10, 178–192. ( 10.1016/j.celrep.2014.12.026) [DOI] [PubMed] [Google Scholar]
- 121.Libri D. 2010. Nuclear poly(A)-binding proteins and nuclear degradation: take the mRNA and run? Mol. Cell 37, 3–5. ( 10.1016/j.molcel.2009.12.029) [DOI] [PubMed] [Google Scholar]
- 122.Meola N, Jensen TH. 2017. Targeting the nuclear RNA exosome: poly(A) binding proteins enter the stage. RNA Biol. 14, 820–826. ( 10.1080/15476286.2017.1312227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Burns LT, Wente SR. 2014. From hypothesis to mechanism: uncovering nuclear pore complex links to gene expression. Mol. Cell. Biol. 34, 2114–2120. ( 10.1128/MCB.01730-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. 2005. Mechanism of mRNA transport in the nucleus. Proc. Natl Acad. Sci. USA 102, 17 008–17 013. ( 10.1073/pnas.0505580102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bahar Halpern K, Caspi I, Lemze D, Levy M, Landen S, Elinav E, Ulitsky I, Itzkovitz S. 2015. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13, 2653–2662. ( 10.1016/j.celrep.2015.11.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Valencia P, Dias AP, Reed R. 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. USA 105, 3386–3391. ( 10.1073/pnas.0800250105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mukherjee N, Calviello L, Hirsekorn A, de Pretis S, Pelizzola M, Ohler U. 2017. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 24, 86–96. ( 10.1038/nsmb.3325) [DOI] [PubMed] [Google Scholar]
- 128.Niwa M, Rose SD, Berget SM. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4, 1552–1559. ( 10.1101/gad.4.9.1552) [DOI] [PubMed] [Google Scholar]
- 129.Dye MJ, Proudfoot NJ. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3, 371–378. ( 10.1016/S1097-2765(00)80464-5) [DOI] [PubMed] [Google Scholar]
- 130.Rigo F, Martinson HG. 2008. Functional coupling of last-intron splicing and 3'-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol. Cell. Biol. 28, 849–862. ( 10.1128/MCB.01410-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meinhart A, Cramer P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature 430, 223–226. ( 10.1038/nature02679) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.