Abstract
We are developing the fluoropyrimidine polymer F10 to overcome limitations of 5-fluorouracil (5-FU) that result from inefficient metabolism to 5-fluoro-2′-deoxyuridine-5′-mono- and tri-phosphate, the deoxyribonucleotide metabolites that are responsible for 5-FU's anticancer activity. F10 is much more cytotoxic than 5-FU to colorectal cancer (CRC) cells; however, the mechanism of enhanced F10 cytotoxicity remains incompletely characterized. Using DNA fiber analysis, we establish that F10 decreases replication fork velocity and causes replication fork collapse, while 1000-fold excess of 5-FU is required to achieve similar endpoints. Treatment of HCT-116 cells with F10 results in Chk1 phosphorylation and activation of intra–S-phase checkpoint. Combining F10 with pharmacological inhibition of Chk1 with either PF-477736 or prexasertib in CRC cells enhanced DNA damage relative to single-agent treatment as assessed by γH2AX intensity and COMET assay. PF-477736 or prexasertib co-treatment also inhibited upregulation of Rad51 levels in response to F10, resulting in reduced homologous repair. siRNA knockdown of Chk1 also increased F10-induced DNA damage assessed and sensitized CRC cells to F10. However, Chk1 knockdown did not inhibit Rad51 upregulation by F10, indicating that the scaffolding activity of Chk1 imparts activity in DNA repair distinct from Chk1 enzymatic activity. Our results indicate that F10 is cytotoxic to CRC cells in part through DNA damage subsequent to replication fork collapse. F10 is ~1000-fold more potent than 5-FU at inducing replication-mediated DNA damage which correlates with the increased overall potency of F10 relative to 5-FU. F10 efficacy can be enhanced by pharmacological inhibition of Chk1.
Introduction
Fluoropyrimidine drugs (FPs) such as 5-fluorouracil (5-FU) form the backbone of multiagent chemotherapy regimen in the management of colorectal cancer (CRC), particularly since targeted therapies are not yet established to effectively treat the ~40% of CRC cases with KRAS mutations [1]. FP-based chemotherapy regimens [2], such as FOLFOX [3] and FOLFIRI [4], result in improved outcomes for patients with stage II [5], III [6], and IV CRC [7]. However, the prognosis for CRC patients with distant metastases remains dismal, and the 5-year survival rate for patients with stage IV CRC is <10% [8], underscoring the need to develop more potent FPs. The efficacy of 5-FU is limited, in part, by inefficient metabolism to 5-fluoro-2′-deoxynucleotide metabolites [9] such as 5-fluoro-2′-deoxyuridine-5′-monophosphate (FdUMP) and 5-fluoro-2′-deoxyuridine-5′-triphosphate (FdUTP) that are primarily responsible for antitumor activity.
Our laboratory has been involved in the development of FP polymers [10] to overcome some of the limitations of 5-FU that decrease its clinical efficacy. Specifically, 5-FU is rapidly degraded and excreted (~15-minute half-life; 85% degraded or excreted intact [11]), and it affects RNA function through misincorporation of the ribonucleotide metabolite FUTP into RNA, which causes gastrointestinal (GI) toxicities [12] that are often dose-limiting and may be life-threatening [13]. In contrast, our prototype FP polymer F10 is first converted to FdUMP [10], the nucleotide metabolite that specifically inhibits the folate-dependent enzyme thymidylate synthase (TS) [14], and then to the triphosphate metabolite FdUTP, which is subsequently incorporated into DNA and causes DNA topoisomerase 1 (Top1)–mediated DNA damage [15]. We have previously shown that F10 displays markedly improved anticancer activity relative to 5-FU in multiple preclinical models of acute leukemia [16], [17] through dual targeting of TS and Top1 [16]. F10 is also efficacious in an orthotopic model of glioblastoma multiforme [18] and caused minimal neurotoxicity. Importantly, in vivo studies showed that F10 causes minimal systemic toxicities [10], including minimal GI toxicity consistent with efficacy resulting from a nearly exclusive DNA-directed mechanism.
While the increased potency of F10 relative to 5-FU is evident based on the NCI60 cell line screen data [19], the mechanistic basis for this improved potency is not fully understood. F10 is a potent TS inhibitor [20], and reduced thymidine (Thy) levels may result in deoxy nucleotide triphosphate (dNTP) pool imbalances that decrease replication fork velocity leading to collapse of replication forks. We demonstrate that F10 (10 nM) decreases replication fork velocity and that 1000-fold higher concentrations of 5-FU are required to induce the same level of DNA-directed effects, a ratio similar to the 338-fold overall potency advantage for F10 in the NCI 60 cell line screen [19]. F10 causes DNA double-strand breaks (DSBs) but also induces Chk1 phosphorylation that activates DNA repair including homologous recombination repair (HR) via Rad51. Co-treatment of F10 with siChk1 or Chk1 inhibitors (PF-477736 or prexasertib) enhances F10-induced DNA damage sensitizing CRC cells to F10. The combination of F10 and Chk1 inhibition may provide a more effective strategy for treatment of advanced colon cancer.
Material and Methods
Cell Lines and Reagents
Human CRC cell lines HCT-116 and HT-29 were acquired from ATCC (Manassas, VA). Both cells were grown in Dulbecco's modified Eagle's medium (Corning, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Omega Scientifics, Tarzana, CA) and 1× penicillin/streptomycin (Corning, Manassas, VA). Cell lines were tested for mycoplasma contamination before each experiment. F10 compound was synthesized according to methods described previously [21]. 5-FU and Chk1 inhibitors PF-477736 and prexasertib were purchased from Sellechem (Houston, TX). Both the PF-477736 and prexasertib were dissolved in dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO), whereas F10 was dissolved in sterile H2O. 5-Chloro-2′-deoxyuridine (CldU) and 5-iodo-2′-deoxyuridine (IdU) were purchased from Sigma (St. Louis, MO) and dissolved in growth media. The following antibodies were used in this study: Santa Cruz Biotechnology (Dallas, TX): FANCD2 (catalog no. sc-20022), GAPDH (catalog no. sc-32233), Chk1 (catalog no. sc-8408), and Rad51 (catalog no. 8349); Cell Signaling (Beverly, MA): pChk1-317 (catalog no. 2344) and γH2AX (catalog no. 2577).
siRNA Transfection
siRNAs (siControl and siChk1) were purchased from Dharmacon (Lafayette, CO). Sequence for siControl is 5′-UAG CGA CUA AAC ACA UCA AUU-3′; sequence for siCHK1 is 5′-GCG UGC CGU AGA CUG UCC AUU -3′. siRNAs were transfected using Lipofectamine 3000 (Invitrogen, Eugene, OR) based on protocol supplied by the manufacturer.
DNA Fiber Assay
HCT-116 cells were treated with either DMSO or F10 or 5-FU for 30 hours and analyzed for replication dynamics using DNA fiber assay as described previously [22]. In brief, after drug treatment, cells were labeled with 25 μM CldU for 20 minutes and then followed by 20-minute labeling with 250 μM IdU. Cells were then trypsinized and resuspended in ice-cold phosphate-buffered saline (PBS). Cells were mixed well, and a 2-μl drop of cell suspension was placed on glass slide, followed by lysing the cells for 5 minutes with 8 μl of lysis buffer (0.5% SDS, 200 mM Tris–hydrochloric acid pH 7.4, 50 mM EDTA). DNA fibers were stretched by tilting slides to a 15° angle until the drop reaches the other end of the slide. Slides were then air dried and fixed in 3:1 methanol:acetic acid, denatured in 2.5 M hydrochloric acid, and blocked with 5% bovine serum albumin. Slides were then incubated with rat anti–5-bromo-2′-deoxyuridine (Abcam, USA, catalog no. 6326) and mouse anti–5-bromo-2′-deoxyuridine (BD, USA, catalog no. 347580) for 2 hours followed by fluorescent conjugated secondary antibody for an hour. Fork velocities were calculated by measuring the length of single-stained fibers, which was then multiplied by DNA extension factor (2.59 kbp/μm). This gives the DNA length in base pairs that was synthesized in 20-minute labeling of CldU followed by 20-minute labeling of IdU. Dividing the DNA length (kb) by 40 minutes will give the fork velocity achieved for a minute. For fork velocity, a total of 100 replication fibers were measured, and statistical significance were assessed using Mann–Whitney test, and Student’s t test was performed using Prism 5 (GraphPad Software, La Jolla, CA). To measure stalled/collapsed/terminal forks, a total of 100 replication structures (both red and green) from above experiment, red only tracks were counted and presented as stalled replication fork. The experiments were repeated three times, and the mean values with statistical significance were presented.
High-Throughput Alkaline Comet Assay
HCT-116 cells were treated with DMSO or F10 or PF-477736 for 24 or 48 hours. After treatment, cells were trypsinized and analyzed for % of DNA tail (DNA damage) using 96-well comet chip system as described by the manufacturer (Trevigen, Gaithersburg, MD).
Immunofluorescence
For immunofluorescence intensity studies, we used confocal microscopy as described previously [23]. Briefly, HCT-116 cells were seeded into 35-mm glass-bottom fluoro-dishes (World Precision Instruments, Sarasota, FL) and incubated overnight. After overnight incubation, cells were either treated with DMSO or F10 or PF-477736 and then fixed and permeabilized using 100% cold methanol on ice. Fixed cells were then blocked in 10% goat serum for 30 minutes followed by overnight incubation at 4°C with primary antibodies [γH2AX (Millipore, USA, catalog no. 05-636) and Rad51 (Cell Signaling, Beverly, MA, catalog no. 8875)]. Cells were then washed thrice with PBS and incubated with appropriate fluorophore-conjugated secondary antibody (Invitrogen, Eugene, OR) for 2 hours at room temperature followed by 4′,6-diamidino-2-phenylindole (Invitrogen, Eugene, OR) staining. γH2AX and Rad51 intensity was captured and analyzed for fluorescence intensity using Nikon Ti Eclipse confocal microscope (Nikon, USA).
Western Blotting
Proteins were isolated, and their differential expressions were analyzed using Western blot as described previously [24]. Briefly, cells were lysed in ice-cold cytoskeletal buffer [10 mM PIPES (pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF, and 0.1% Triton X-100] freshly supplemented with protease and phosphatase inhibitors (Roche, Mannheim, Germany). Concentration of the proteins was quantified using Bradford assay (Bio-Rad, USA), and samples were normalized for equal loading. Samples were then heated at 100°C for 10 minutes in the presence of 6× Laemmli buffer (Boston Bioproducts, Ashland, MA). Proteins were then resolved by SDS-PAGE, and their expression was analyzed by immunoblotting using specific antibodies.
Cell Cycle Analysis
HCT-116 cells were plated at 50% confluence and allowed to attach overnight. The following day, cells were treated with DMSO or F10 or PF-477736 for 24 hours. Cells were then trypsinized and fixed in ethanol overnight. After overnight incubation, cells were rehydrated with PBS wash and stained with propidium iodide solution (Invitrogen, Eugene, OR) and ribonuclease A (Invitrogen, Carlsbad, CA) at concentration of 40 μg/ml and 10 μg/ml, respectively, for 30 minutes at 37°C. Cells were then stored at 4°C until analyzed using BD FACSCanto II (BD Biosciences, San Jose, CA).
Clonogenic Survival Assay
For siRNA-mediated experiments, 48 hours after siRNA transfection (for siChk1), cells were trypsinized, and approximately 350 cells per well were seeded in 6-well cell culture plates and allowed to attach overnight. The following day, cells were treated with DMSO or indicated doses of F10 and allowed to form colonies up to 10 days. For Chk1i-mediated experiments, cells were trypsinized, and approximately 350 cells per well were seeded in 6-well cell culture plates and allowed to attach overnight. The following day, cells were treated with DMSO or indicated doses of F10 with and without or IC-50 concentration of PF-477736 or prexasertib and allowed to form colonies up to 10 days. The colonies were fixed with methanol, stained with 0.5% crystal violet (w/v) (Invitrogen, Eugene, OR) for 5 minutes, and counted as described previously [24].
Results
F10 Causes Replication Stress
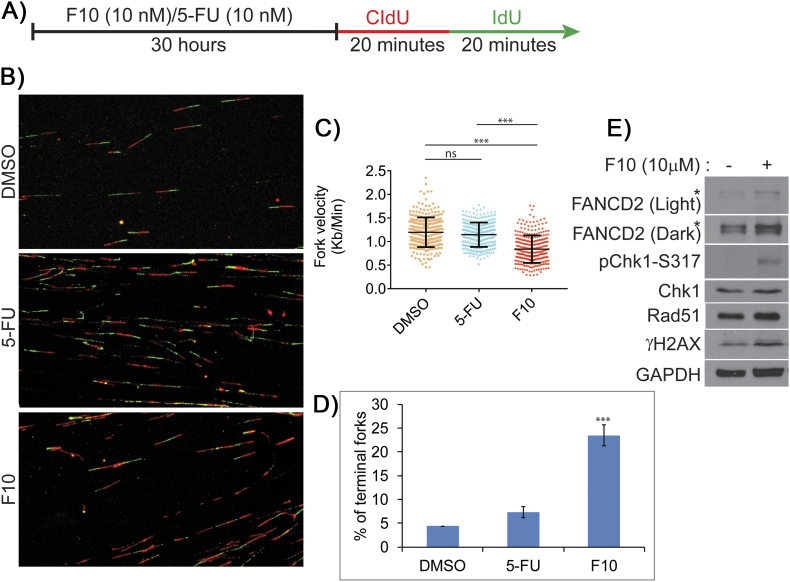
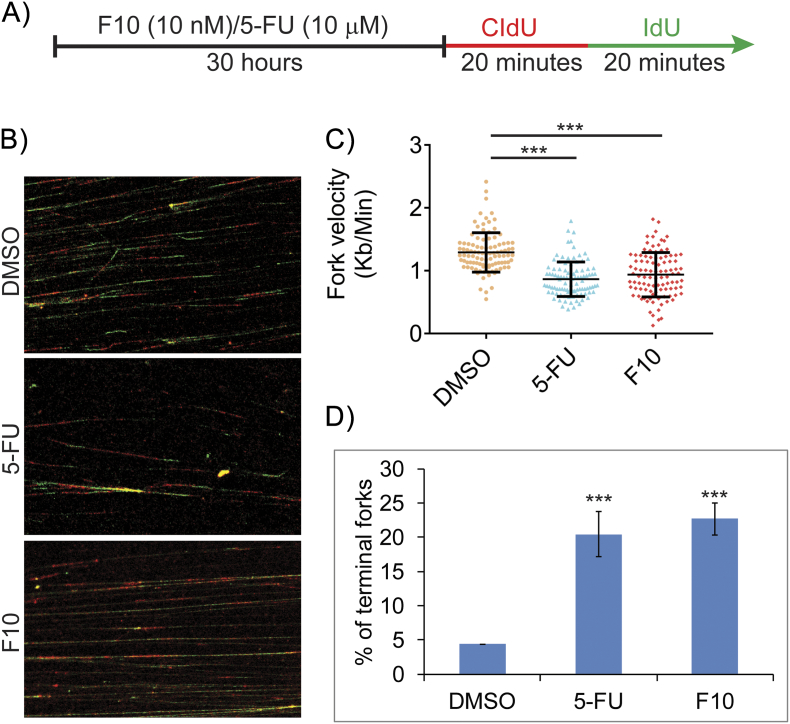
Previous studies with hydroxyurea demonstrated that depletion of deoxyribonucleotide pools slows replication fork velocity and causes replication fork collapse [25], [26]. In principle, decreased Thy levels resulting from TS inhibition by FPs may also decrease replication fork velocity by causing dNTP imbalances, and this may result in replication stress–mediated DNA damage resulting from stalled or collapsed forks. We performed DNA fiber analysis to determine if F10 and 5-FU are equally effective at decreasing fork velocity and causing replication fork collapse. To evaluate fork velocity, HCT-116 cells were treated with either F10 or 5-FU (both at 10 nM) for 30 hours and then pulse-labeled with CldU for 20 minutes followed immediately by pulse labeling with IdU for 20 minutes (Figure 1A). DNA spreads were then prepared from the pulse-labeled cells and analyzed by immunofluorescence (Figure 1B). F10 (10 nM) significantly decreased fork velocity relative to both DMSO-treated cells and cells treated with 5-FU (10 nM) (Figure 1C). In contrast, treatment with 5-FU (10 nM) did not significantly decrease fork velocity relative to control. A likely reason for decrease in fork velocity in F10 treated cells is that F10 more effectively reduces Thy levels, causing greater imbalance in dNTP pools compared to 5-FU. Consistent with the slow progression of replication, F10 treatment also caused significantly increased stalled/terminal forks when compared to 5-FU at equimolar concentrations (Figure 1D). To test if higher concentrations of 5-FU induce replication stress, we evaluated fork dynamics in HCT-116 cells treated with either 5-FU (10 μM) or F10 (10 nM) (Figure S1A). At 1000-fold higher concentration (10 μM), 5-FU decreased fork velocity to a similar extent as the 10 nM concentration of F10 (Figure S1, B and C). At this higher concentration, 5-FU also induced a similar increase in stalled/terminal forks as F10 (10 nM) (Figure S1D). However, it is not clear that such concentrations are therapeutically relevant in light of the GI-tract toxicity of 5-FU that is often dose-limiting.
Figure 1.
Equimolar concentration of F10 induces increased replication stress compared to 5-FU.
(A) HCT-116 cells were treated with similar concentration of F10 and 5-FU followed by CldU and IdU incorporation. (B) DNA fiber images were captured and analyzed. (C) F10 (10 nM) reduces fork velocity significantly compared to 5-FU (10 nM). (D) F10 (10 nM) increases terminal forks significantly compared to 5-FU (10 nM). Statistical significance represented is in comparison with the control. (E) HCT-116 cells were exposed to F10 (10 μm) for 2 hours, and evaluation of replication stress–associated proteins shows increased expression of Rad51, increased phosphorylation of Chk1 and γH2AX, and increased ubiquitination of FANCD2.
Figure S1.
IC-50 concentration of F10 induces increased replication stress compared to IC-50 5-FU.
(A) HCT-116 cells were treated with IC-50 concentrations of F10 and 5-FU followed by CldU and IdU incorporation. (B) DNA fiber images were captured and analyzed. (C) F10 (10 nM) reduces fork velocity significantly compared to 5-FU (10 μM). (D) F10 (10 nM) increases terminal forks significantly compared to 5-FU (10 μM). Statistical significance represented is in comparison with the control.
Replication fork collapse may result in DNA DSBs, genomic instability, and cell death [27], [28]. The adverse consequences of replication fork breakdown may be countered by ATR/Chk1 pathway–mediated activation of the intra–S-phase checkpoint which decreases initiation of new replication forks and stimulates HR-mediated repair of collapsed forks [29], [30]. F10 significantly stimulated induction of terminal forks and increased pChk1 Serine-317 (S317) levels consistent with ATR/Chk1 pathway activation (Figure 1E). Similarly, FANCD2 monoubiquitination also increased in F10-treated cells, consistent with its role in stabilizing stalled forks and facilitating repair of collapsed forks [31], [32]. Further, both γH2AX and Rad51 levels increased in F10-treated cells consistent with generation of DNA DSBs during the repair of collapsed forks, with repair ultimately occurring via HR. Our results are consistent with F10 cytotoxicity resulting from DNA damage associated with replication fork collapse with the extent of damage mitigated through activation of the ATR/Chk1 pathway and HR.
Chk1 Inhibition Increases F10-Induced DNA Lesions
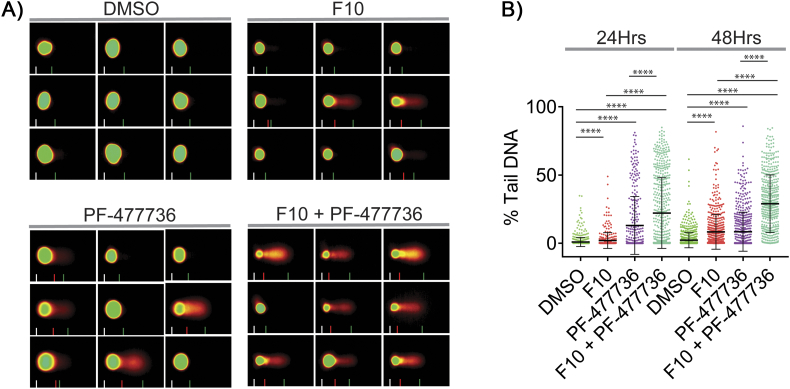
Since F10 cytotoxicity is mediated in part by DNA damage consequent to replication fork collapse and repair of this damage occurs downstream of ATR/Chk1 pathway activation, we investigated whether Chk1 inhibition would increase DNA damage in F10-treated cells. Using a high-throughput alkaline COMET assay, we demonstrated that both F10 and the Chk1 inhibitor PF-477736 significantly increased DNA damage relative to vehicle (Figure 2A). Further, the F10 + PF-477736 combination significantly increased DNA damage relative to either single agent. This is consistent with previous studies that showed Chk1 inhibition abrogates the S-phase checkpoint, which destabilizes stalled forks and increases fork collapse and generation of DSBs [33], [34]. The magnitude of DNA damage was also time-dependent for both F10 and for the F10 + PF-477736 combination, with greater damage at 48-hour relative to 24-hour treatment (Figure 2B).
Figure 2.
High throughput alkaline COMET assay shows increased DNA damage in F10 and Chk1i (PF-477736) combination.
(A) HCT-116 cells were treated with F10 (250 nM) and/or PF-477736 (1.65 μM) for 24 or 48 hours and analyzed for DNA damage by COMET assay. (B) Combination of F10 and PF-477736 shows increased % tail DNA (DNA damage) compared to the individual treatments.
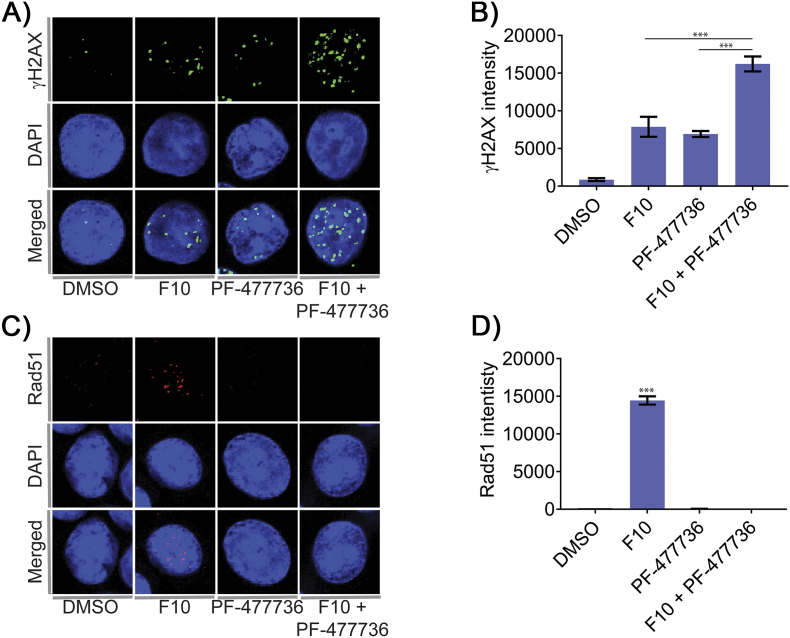
We further investigated the effects of Chk1 inhibition on DNA DSB formation in F10-treated cells by immunofluorescence using an antibody to detect γH2AX intensity (Figure 3A). F10 and PF-477736 both induced γH2AX intensity consistent with generation of DNA DSBs. The F10 + PF-477736 combination was, however, significantly more effective than either single agent at inducing DNA DSBs (Figure 3B). Since activated Chk1 is known to promote repair of replication-associated DSB by Rad51-mediated HR, we further evaluated Rad51 intensity formation in HCT-116 cells treated with either F10, PF-477736, or the combination. Consistent with our Western blots data that showed increased Rad51 levels in F10-treated cells (Figure 1E), the intensity of Rad51 also increased in F10-treated cells (Figure 3C). Interestingly, although PF-477736 treatment induced DNA damage as evidenced by comet assay and γH2AX intensity, these cells failed to show Rad51 intensity. Similarly, Chk1 inhibition also abrogated formation of F10-induced Rad51 intensity (Figure 3D). Similarly, PF-477736 treatment also suppresses Rad51 intensity in response to F10-induced DNA damage. The inhibition of Rad51-mediated HR by PF-477736 results in high levels of persistent DNA lesions in these cells at 48 hours, as indicated by increased comet tail (Figure 2B).
Figure 3.
Confocal analysis of HCT-116 cells exposed to F10 and/or CHK1i (PF-477736).
(A) HCT-116 cells were treated with F10 (250 nM) and/or PF-477736 (1.65 μM) for 24 hours and analyzed for γH2AX intensity by confocal microscopy. (B) Combination of F10 and PF-477736 shows increased DSB (γH2AX) compared to the individual treatments. (C) HCT-116 cells were treated with F10 (250 nM) and/or PF-477736 (1.65 μM) for 24 hours and analyzed for Rad51 intensity by confocal microscopy. (D) Cells exposed to F10 shows significant increase in the Rad51 intensity; however, cells exposed to PF-477736 attenuated the expression of Rad51 intensity. Statistical significance represented is in comparison with the control.
Chk1 Inhibition Abrogates F10-induced S-Phase Arrest
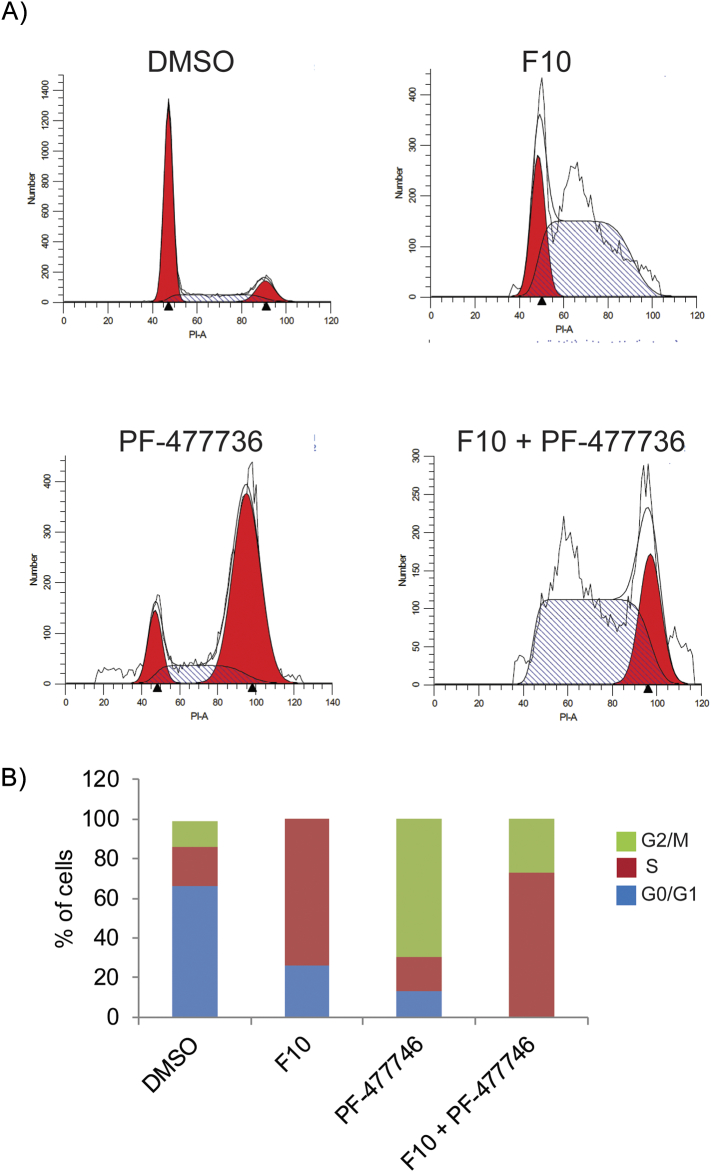
As our studies show that F10 induces replication stress (Figure 1C) and activates ATR/Chk1-mediated checkpoint response (Figure 1E), we analyzed the cell cycle profiles of these cells by flow cytometry. Consistent with replication stress–mediated DNA damage that activates the intra–S-phase checkpoint, F10 treatment for 24 hours increased accumulation of cells in S-phase from ~15% to >75% (Figure 4, A and B). Interestingly, Chk1 inhibition with PF-477736 resulted in accumulation of cells in G2/M (Figure 4, A and B). Accumulation in G2/M may be due to defect in intra–S-phase checkpoint in these cells, which prevents firing of late origins during endogenous replication stress, a common feature in tumor cells. However, Chk1 inhibition combined with F10 treatment resulted in most cells progressing through S-phase despite F10-induced DNA damage and accumulation in G2/M. Moreover, premature entry of tumor cells into G2/M phase with damaged DNA leads to cell death [35]. Consistently, most of the cells were dead and floating in the media by 48 hours.
Figure 4.
Cell cycle analysis of F10 and Chk1i (PF-477736) combination.
(A) HCT-116 cells were treated with F10 (250 nM) and/or PF-477736 (1.65 μM) for 24 hours and analyzed for cell cycle status. (B) Histogram shows more cells were entering G2/M phase in combination when compared to F10 treatment alone.
Chk1 Knockdown Sensitizes CRC Cells to F10
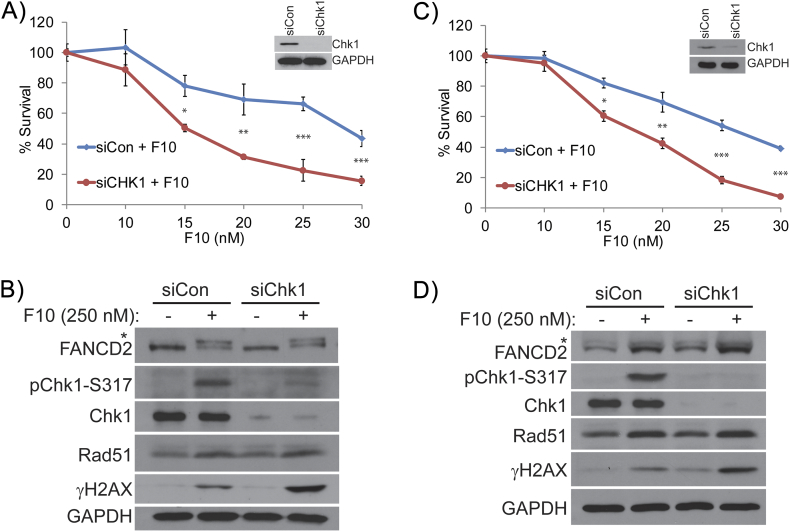
We next determined if Chk1 knockdown enhanced the cytotoxicity of F10 in both HCT-116 and HT-29 CRC cells (Figure 5). siRNA-mediated knockdown of Chk1 was validated by Western blot in both HCT-116 (Figure 5A) and HT-29 cells (Figure 5C). Consistently, phosphorylated Chk1 (pChk1 S317) levels were also considerably downregulated in F10-treated CRC cells (Figure 5, B and D). Interestingly, monoubiquitinated FANCD2 and Rad51 levels were increased in Chk1 downregulated HCT-116 and HT-29 cells following F10 treatment. This is consistent with the roles for FANCD2 and Rad51 proteins in repair of DSBs as a consequence of F10-induced collapsed replication forks. However, γH2AX levels were markedly higher in Chk1 knockdown cells treated with F10 consistent with repair being ineffective and/or lack of Chk1 enabling additional DNA damage. The higher levels of DNA damage in Chk1-knockdown cells following treatment with F10 are also associated with a significant increase in sensitivity to F10 (Figure 5, A and C; Table 1).
Figure 5.
Survival analysis of F10 and Chk1 inhibition by siRNA.
Colony survival assay performed in (A) HCT-116 and (C) HT-29 cells shows decreased survival of siChk1 colonies treated with F10 compared to siControl cells treated with F10. (B and D) Western blot analysis shows decreased pChk1-317 and increased γH2AX in the combination in both (B) HCT-116 and (D) HT-29 cell lines.
Table 1.
Comparison of IC-50 Values for F10 (nM) Alone Or in Combination with Chk1 siRNA/Inhibitors
| siCon | siChk1 | |
|---|---|---|
| HCT-116 | 29.05 | 14.17 |
| HT-29 | 26.12 | 17.59 |
| DMSO | PF-477736 | |
| HCT-116 | 27.9 | 12.31 |
| HT-29 | 23.47 | 13.79 |
| DMSO | Prexasertib | |
| HCT-116 | 27.51 | 14.13 |
Chk1 Inhibition Sensitizes CRC Cells to F10
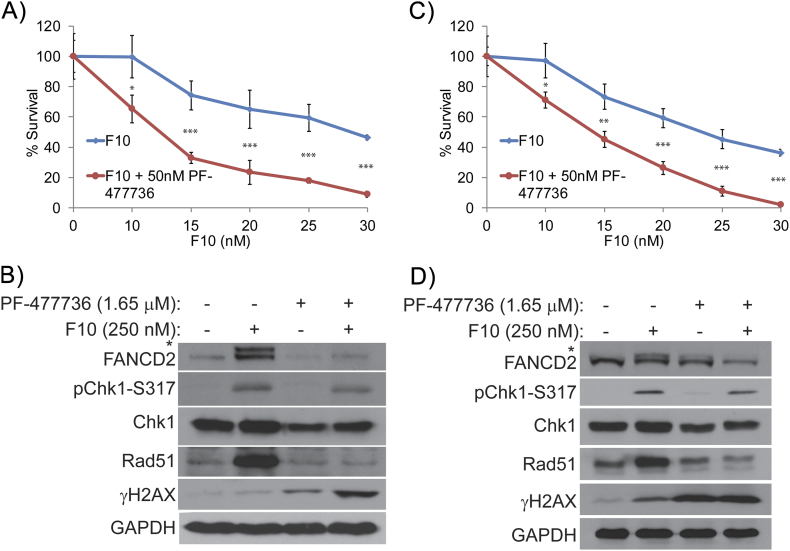
We further investigated whether pharmacological inhibition of Chk1 sensitized CRC cells to F10 through increased DNA DSB formation. We used two different types of Chk1 inhibitors, namely, PF-477736 and prexasertib. Both these inhibitors are currently in clinical trials for various cancers [36], [37]. The F10 + PF-477736 combination was significantly more potent in killing CRC cells compared to either F10 or PF-477736 alone (Figure 6, A and C), and this increased potency was associated with increased DNA DSBs as assessed by γH2AX (Figure 6, B and D). While pharmacological Chk1 inhibition and Chk1 siRNA knockdown both sensitized CRC cells to F10, a significant difference was observed in DNA damage responses. In contrast to Chk1 knockdown CRC cells (Figure 5, B and D), PF-477763 treatment decreased FANCD2 and Rad51 proteins (Figure 5, B and D). Thus, pharmacological Chk1 inhibition attenuates repair of F10-induced DNA DSBs resulting in increased γH2AX levels. Chk1 knockdown by siRNA has a similar effect as PF-477763 on F10 sensitivity and γH2AX levels; however, this occurs without attenuating repair via FANCD2 activation and increased Rad51 expression and thus may act by a distinct mechanism independent of Chk1 scaffolding.
Figure 6.
Survival analysis of F10 and Chk1 inhibition by PF-477736.
Colony survival assay performed in (A) HCT-116 and (C) HT-29 cells shows decreased survival of Chk1i treated colonies co-treated with F10 compared to DMSO-treated cells co-treated with F10. Western blot analysis shows decreased pChk1-317 and increased γH2AX in the combination in both (B) HCT-116 and (D) HT-29 cell lines. Additionally, decreased expression of Rad51 and ubiquitination of FANCD2 were observed in both the cell lines treated with PF-477736.
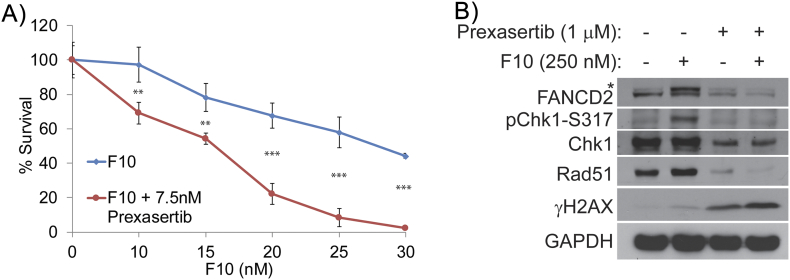
We went on to investigate Chk1 inhibition with prexasertib to determine if pharmacological Chk1 inhibition generally inhibits repair of F10-induced DNA DSBs generated from replication fork collapse. Results with this agent were similar to PF-477763 (Figure 7, A and B). Prexasertib also significantly increased sensitivity of HCT-116 cells to F10 (Table 1), and the F10 + prexasertib combination induced higher γH2AX levels. Prexasertib, like PF-477763, inhibited both FANCD2 ubiquitination and increased Rad51 levels, consistent with Chk1 inhibition generally increasing F10-induced DNA damage by inhibiting repair. Our results show that Chk1 inhibition is a promising strategy to enhance the efficacy of F10 for CRC treatment.
Figure 7.
Survival analysis of F10 and Chk1 inhibition by prexasertib.
Colony survival assay performed in (A) HCT-116 cells shows decreased survival of Chk1i-treated colonies co-treated with F10 compared to DMSO-treated cells co-treated with F10. (B) Western blot analysis shows decreased pChk1-317 and increased γH2AX in the combination in HCT-116 cells. Similar to PF-477736, decreased expression of Rad51 and ubiquitination of FANCD2 were observed in prexasertib-treated cells too.
Discussion
F10 displays promising anticancer activity in a number of preclinical models and is more potent than 5-FU to CRC cells [19] and is very well tolerated in vivo [16], [17], [18], [20]. In the present studies, we investigated the role of replication fork collapse [27] as a contributing factor for F10-induced DNA damage, and we assess the potential to improve the antitumor activity of F10 by combining it with Chk1 inhibitors [38] to attenuate cell cycle arrest. F10 caused significant levels of replication forks to collapse at concentrations near the IC-50, while these concentrations of 5-FU neither significantly affect replication fork velocity nor cause replication fork collapse. In fact, supraphysiological concentrations of 5-FU 1000-fold higher are required to induce similar effects as the 10-nM dose of F10. This 1000-fold difference in potency at inducing replication fork collapse parallels the improved cytotoxicity of F10 relative to 5-FU (338-fold over the NCI60 cell line screen), consistent with collapsed replication forks being contributory to F10-induced thymineless death.
The present studies suggest a role for replication fork collapse in F10-induced DNA DSBs; however, our previous studies have demonstrated that F10 induces Top1 cleavage complexes (Top1cc) [15], [16], which are an alternative source of DNA damage. It is likely that replication fork collapse and Top1cc are distinct processes that separately induce DNA DSBs in F10-treated cells, although these two processes are closely linked for Top1 poisons such as camptothecin [39]. Camptothecin causes replication fork collapse by inhibiting Top1-mediated relaxation of positive supercoils that are generated ahead of the advancing replication fork, causing fork stalling and collapse. Such a process is unlikely to occur for F10 because Top1 acts ahead of the advancing replication fork on DNA that has not yet undergone replication under thymineless conditions and lacks FdU sites at which Top1 may become trapped. The effects of F10 may more closely resemble hydroxyurea which induces fork stalling by depleting dNTP pools [25], [26]. While F10 depletes only Thy, the resulting imbalance in dNTP levels contributes to slowing of fork progression and causes fork collapse (Figure 1). Nonetheless, F10 is much more potent than hydroxyurea [19] consistent with multiple mechanisms contributing to F10-induced DNA damage. One possible scenario for how F10 can both induce replication fork progression and cause Top1cc is that after replication and FdU incorporation, DNA becomes a template for transcription later in S-phase. While 10 nM F10 induces significant levels of terminal forks, approximately three-fourths of all forks monitored by DNA fiber analysis were not terminated in our studies (Figure 1D), and this nascent DNA may be transcriptionally active later in S-phase and become a source of Top1-mediated DNA damage adding to DSBs resulting from F10-induced replication fork collapse.
Our studies show that F10 induces Chk1 S317 phosphorylation (Figure 1E) and activates the intra–S-phase checkpoint (Figure 4) under conditions that stimulate F10-induced replication fork collapse (Figure 1, A-D). Activation of the intra–S-phase checkpoint permits cells to repair DNA damage prior to entering G2/M and thus avoid mitotic catastrophe.
We have recently shown that the functional interactions between FANCD2, BRCA2, and Rad51 are important to repair DNA topoisomerase 1 poisons–induced lesions and to promote fork recovery [22]. Interestingly, CRC cells exposed to Chk1 inhibitors PF-477736 or prexasertib show downregulation of Rad51 and FANCD2. Similar results were observed for Rad51, where the ovarian cancer cells failed to form olaparib-induced Rad51 foci in the presence of prexasertib [40]. However, we for the first time observed that the Rad51 protein expression itself downregulated when the cells are exposed to PF-477736 or prexasertib. Additionally, we also observed that both the expression and ubiquitination of FANCD2 are attenuated in Chk1 inhibitors–treated cells.
Pharmacological inhibition of Chk1 enhances F10 cytotoxicity in part by inhibiting Rad51- and FANCD2-mediated HR of DNA DSBs (Figure 3). Combining F10 with either PF-477736 or prexasertib results in cells with higher levels of DNA DSBs and also permits ~25% of cells to enter G2/M with these high levels of DNA damage. siRNA knockdown of Chk1 has similar effects as pharmacological inhibition, although inhibition of Rad51 and FANCD2 upregulation is actually more effective with pharmacological inhibition than with siRNA knockdown.
Conclusion
Our studies demonstrate that Chk1 inhibition accentuates the cytotoxicity of F10 to both HCT-116 and HT-29 CRC cells and this occurs in part by inhibiting Rad51- and FANCD2-mediated HR to increase DNA DSBs. Thus, F10 has strong potential to be an effective new agent for the treatment of CRC, and combination with Chk1 inhibitors may be a particularly effective novel combination with clinical potential.
The following are the supplementary data related to this article.
Acknowledgement
We would like to thank Dr. Joel Andrews and Steve McClellan, Mitchell Cancer Institute, for their assistance in confocal and flow cytometry, respectively. We would like to acknowledge funding sources for K. P. (R01GM098956; R01CA219187) and W. G. (NIH-NCI CA R21; NIH-NCI P30 012197; NC Biotechnology Center; Wake Innovations).
Contributor Information
Komaraiah Palle, Email: komaraiah.palle@ttuhsc.edu.
William H. Gmeiner, Email: bgmeiner@wakehealth.edu.
References
- 1.Punt CJA, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 2.Gustavsson B, Carlsson G, Machover D, Petrelli N, Roth A, Schmoll H-J, Tveit K-M, Gibson F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin Colorectal Cancer. 2015;14:1–10. doi: 10.1016/j.clcc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Peixoto RD, Kennecke HF, Renouf DJ, Lim HJ, Gill S, Speers CH, Cheung WY. Effect of adjuvant FOLFOX chemotherapy duration on outcomes of patients with stage III colon cancer. Clin Colorectal Cancer. 2015;14:262-268.e1. doi: 10.1016/j.clcc.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Jeon EK, Hong SH, Kim TH, Jung SE, Park JC, Won H-S, Ko Y-H, Rho SY, Hong YS. Modified FOLFIRI as second-line chemotherapy after failure of modified FOLFOX-4 in advanced gastric cancer. Cancer Res Treat. 2011;43:148–153. doi: 10.4143/crt.2011.43.3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastos DA, Ribeiro SC, de Freitas D, Hoff PM. Combination therapy in high-risk stage II or stage III colon cancer: current practice and future prospects. Ther Adv Med Oncol. 2010;2:261–272. doi: 10.1177/1758834010367905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall JL, Haller DG, de Gramont A, Hochster HS, Lenz H-J, Ajani JA, Goldberg RM. Adjuvant therapy for stage II and III colon cancer: consensus report of the International Society of Gastrointestinal Oncology. Gastrointest Cancer Res. 2007;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- 7.Zacharakis M, Xynos ID, Lazaris A, Smaro T, Kosmas C, Dokou A, Felekouras E, Antoniou E, Polyzos A, Sarantonis J. Predictors of survival in stage IV metastatic colorectal cancer. Anticancer Res. 2010;30:653–660. [PubMed] [Google Scholar]
- 8.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 9.Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 10.Gmeiner WH, Debinski W, Milligan C, Caudell D, Pardee TS. The applications of the novel polymeric fluoropyrimidine F10 in cancer treatment: current evidence. Future Oncol. 2016;12:2009–2020. doi: 10.2217/fon-2016-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diasio RB, Harris BE. Clinical pharmacology of 5-fluorouracil. Clin Pharmacokinet. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 12.Pritchard DM, Watson AJ, Potten CS, Jackman AL, Hickman JA. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc Natl Acad Sci U S A. 1997;94:1795–1799. doi: 10.1073/pnas.94.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boussios S, Pentheroudakis G, Katsanos K, Pavlidis N. Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann Gastroenterol. 2012;25:106–118. [PMC free article] [PubMed] [Google Scholar]
- 14.Gmeiner WH. Novel chemical strategies for thymidylate synthase inhibition. Curr Med Chem. 2005;12:191–202. doi: 10.2174/0929867053363432. [DOI] [PubMed] [Google Scholar]
- 15.Liao Z-Y, Sordet O, Zhang H-L, Kohlhagen G, Antony S, Gmeiner WH, Pommier Y. A novel polypyrimidine antitumor agent FdUMP[10] induces thymineless death with topoisomerase I-DNA complexes. Cancer Res. 2005;65:4844–4851. doi: 10.1158/0008-5472.CAN-04-1302. [DOI] [PubMed] [Google Scholar]
- 16.Pardee TS, Gomes E, Jennings-Gee J, Caudell D, Gmeiner WH. Unique dual targeting of thymidylate synthase and topoisomerase1 by FdUMP[10] results in high efficacy against AML and low toxicity. Blood. 2012;119:3561–3570. doi: 10.1182/blood-2011-06-362442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardee TS, Stadelman K, Jennings-Gee J, Caudell DL, Gmeiner WH. The poison oligonucleotide F10 is highly effective against acute lymphoblastic leukemia while sparing normal hematopoietic cells. Oncotarget. 2014;5:4170–4179. doi: 10.18632/oncotarget.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gmeiner WH, Lema-Tome C, Gibo D, Jennings-Gee J, Milligan C, Debinski W. Selective anti-tumor activity of the novel fluoropyrimidine polymer F10 towards G48a orthotopic GBM tumors. J Neurooncol. 2014;116:447–454. doi: 10.1007/s11060-013-1321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gmeiner WH, Reinhold WC, Pommier Y. Genome-wide mRNA and microRNA profiling of the NCI 60 cell-line screen and comparison of FdUMP[10] with fluorouracil, floxuridine, and topoisomerase 1 poisons. Mol Cancer Ther. 2010;9:3105–3114. doi: 10.1158/1535-7163.MCT-10-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gmeiner WH, Willingham MC, Bourland JD, Hatcher HC, Smith TL, D'Agostino RB, Blackstock W. F10 inhibits growth of PC3 xenografts and enhances the effects of radiation therapy. J Clin Oncol Res. 2014;2 [PMC free article] [PubMed] [Google Scholar]
- 21.Gmeiner WH, Trump E, Wei C. Enhanced DNA-directed effects of FdUMP[10] compared to 5FU. Nucleosides Nucleotides Nucleic Acids. 2004;23:401–410. doi: 10.1081/ncn-120028336. [DOI] [PubMed] [Google Scholar]
- 22.Tripathi K, Mani C, Clark DW, Palle K. Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery. Oncotarget. 2016;7:12537–12553. doi: 10.18632/oncotarget.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somasagara RR, Spencer SM, Tripathi K, Clark DW, Mani C, Madeira da Silva L, Scalici J, Kothayer H, Westwell AD, Rocconi RP. RAD6 promotes DNA repair and stem cell signaling in ovarian cancer and is a promising therapeutic target to prevent and treat acquired chemoresistance. Oncogene. 2017;36:6680–6690. doi: 10.1038/onc.2017.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathi K, Mani C, Barnett R, Nalluri S, Bachaboina L, Rocconi RP, Athar M, Owen LB, Palle K. Gli1 protein regulates the S-phase checkpoint in tumor cells via Bid protein, and its inhibition sensitizes to DNA topoisomerase 1 inhibitors. J Biol Chem. 2014;289:31513–31525. doi: 10.1074/jbc.M114.606483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 26.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair. 2015;32:149–157. doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander JL, Orr-Weaver TL. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 2016;30:2241–2252. doi: 10.1101/gad.288142.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buisson R, Niraj J, Rodrigue A, Ho CK, Kreuzer J, Foo TK, Hardy EJ-L, Dellaire G, Haas W, Xia B. Coupling of homologous recombination and the checkpoint by ATR. Mol Cell. 2017;65:336–346. doi: 10.1016/j.molcel.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petermann E, Caldecott KW. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 2006;5:2203–2209. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Su F, Mukherjee S, Mori E, Hu B, Asaithamby A. FANCD2 influences replication fork processes and genome stability in response to clustered DSBs. Cell Cycle. 2015;14:1809–1822. doi: 10.1080/15384101.2015.1036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michl J, Zimmer J, Buffa FM, McDermott U, Tarsounas M. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat Struct Mol Biol. 2016;23:755–757. doi: 10.1038/nsmb.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DAF, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mc Gee MM. Targeting the mitotic catastrophe signaling pathway in cancer. Mediators Inflamm. 2015;2015:146282. doi: 10.1155/2015/146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doerr F, George J, Schmitt A, Beleggia F, Rehkämper T, Hermann S, Walter V, Weber J-P, Thomas RK, Wittersheim M. Targeting a non-oncogene addiction to the ATR/CHK1 axis for the treatment of small cell lung cancer. Sci Rep. 2017;7:15511. doi: 10.1038/s41598-017-15840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J-M, Nair J, Zimmer A, Lipkowitz S, Annunziata CM, Merino MJ, Swisher EM, Harrell MI, Trepel JB, Lee M-J. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: a first-in-class proof-of-concept phase 2 study. Lancet Oncol. 2018;19:207–215. doi: 10.1016/S1470-2045(18)30009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jennings-Gee J, Pardee TS, Gmeiner WH. Replication-dependent irreversible topoisomerase 1 poisoning is responsible for FdUMP[10] anti-leukemic activity. Exp Hematol. 2013;41:180-188.e4. doi: 10.1016/j.exphem.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brill E, Yokoyama T, Nair J, Yu M, Ahn Y-R, Lee J-M. Prexasertib, a cell cycle checkpoint kinases 1 and 2 inhibitor, increases in vitro toxicity of PARP inhibition by preventing Rad51 foci formation in BRCA wild type high-grade serous ovarian cancer. Oncotarget. 2017;8:111026–111040. doi: 10.18632/oncotarget.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]