Abstract
Bacteroides ovatus ELH-B2 is considered as a potential next-generation probiotic due to its preventive effects on lipopolysaccharides-associated inflammation and intestinal microbiota disorders in mice. To study safety issues associated with B. ovatus ELH-B2, we conducted comprehensive and systematic experiments, including in vitro genetic assessments of potential virulence and antimicrobial resistance genes, and an in vivo acute toxicity study of both immunocompetent and immunosuppressed mice via cyclophosphamide treatment. The results indicated that this novel strain is non-toxigenic, fragilysin is not expressed, and most of potential virulence genes are correlated with cellular structures such as capsular polysaccharide and polysaccharide utilizations. The antibiotic resistance features are unlikely be transferred to other intestinal microorganisms as no plasmids nor related genomic islands were identified. Side effects were not observed in mice. B. ovatus ELH-B2 also alleviated the damages caused by cyclophosphamide injection.
Keywords: Bacteroides ovatus, safety evaluation, antibiotic resistance, virulence genes, next-generation probiotics
Introduction
Probiotics, prebiotics, and antibiotics are the most relevant therapies for disorders induced by disturbed microbiota. Traditional probiotics mainly refer to Lactobacillus and Bifidobacterium, which are normally obtained from traditional fermented foods and are widely accepted as food ingredients or supplements for daily intake, with a prediction of global turnover value of US$46.55 billion by 2020 (O’Toole et al., 2017).
Beneficial strains other than the traditional probiotics have been discovered due to the developments in bacterial culture methodologies and sequencing techniques and have started to be authorized as ingredients in food, particularly from Bacteroides which is one of the most abundant genera in the human intestine. For example, Bacteroides xylanisolvens DSM23964, which promotes the maturation of natural antibodies against cancers in humans, has recently been permitted to be added to pasteurized milk products under Novel Food Regulation No. 258/97 by the European Commission (Brodmann et al., 2017); B. uniformis CECT7771 can improve overweight-induced disorders by reducing the levels of cholesterol and triglyceride (Cano et al., 2012), with no obvious damages identified in vivo (Fernandez-Murga and Sanz, 2016).
B. ovatus is another dominant species identified as next-generation probiotics either in its original form with sufficient tumor-specific Thomsen–Friedenreich antigen expressed for cancer prevention (Ulsemer et al., 2013), or genetically modified with genes encoding human keratinocyte growth factors (Hamady et al., 2010) or transforming growth factors (Hamady et al., 2011) to facilitate therapies for bowel diseases. However, the beneficial functions of Bacteroides are strain-dependent, such as the polysaccharides A (PSA)-producing B. fragilis is capable of relieving Helicobacter hepaticus associated inflammation and autism spectrum disorders (Mazmanian et al., 2008; Hsiao et al., 2013), but the fragilysin (bft)-carrying B. fragilis directly contributes to severe colitis (Yim et al., 2013), emphasizing the necessities for safety evaluations of each potentially beneficial strain.
Previously, we established an efficient method for purifying low-abundant Bacteroides species from the human intestine (Tan et al., 2018), and discovered that one of the isolates, B. ovatus ELH-B2 displaying promising potentials of modulating lipopolysaccharides (LPS)-induced disorders in cytokine secretions and intestinal microbiota through restoring the balance of regulatory T cells (Tregs) and T helper 17 (Th-17) cells (data unpublished). In this study, a pilot safety assessment of this novel strain was carried out, including explorations of its hemolytic and motile characteristics, antibiotic resistance, genetic virulence factors, and underlying side effects in both normal and immunosuppressed mice.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacteroides ovatus ELH-B2 was recovered from the in-house preservations at Culture Collections of Food Microbiology (CCFM), Jiangnan University (Wuxi, China). B. ovatus JCM5824 was purchased from RIKEN BioResource Center, Japan. Salmonella enterica CMCC50335 and Escherichia coli CMCC44102 were acquired from the National Center for Medical Culture Collections, China.
Salmonella enterica and E. coli were anaerobically cultured in brain heart infusion (BHI, Hopebio, China) at 37°C. The B. ovatus strains were cultured in BHI supplemented with hemin (Sangon Biotech, China) and vitamin K1 (BHIS) at 37°C in anaerobic chamber for further analysis of bacterial characterizations. The bacteria solutions for in vivo tests were prepared with cells at early stationary phase after centrifugation at 6000 rpm for 15 min and re-suspension in phosphate buffer saline supplemented with 20% glycerol, and maintained at -80°C. Cell viability after freezing and thawing was evaluated via colony-forming unit (cfu) enumeration on BHIS agar before use.
Bacterial Characterizations
Bacteroides ovatus type strain JCM5824 was used as control for the bacterial characterization assessments of ELH-B2. Hemolytic capabilities were examined by dropping 5 μl of overnight culture on Brucella agar (Hopebio, China) supplemented with hemin, vitamin K1 and 5% sheep blood (Nanjing SenBeiJia Biological Technology Co., Ltd., China) (Robertson et al., 2006). Motility was tested via standard motility agar assays using BHIS broth supplemented with 0.5% (w/v) agar (soft agar) (Cousin et al., 2015), inoculated with 5 μl of overnight culture and incubated anaerobically for 48 h. S. enterica CMCC50335 was adopted as the positive control and E. coli CMCC44102 as the negative control. All of the experiments were carried out in three biological replicates.
Genome Sequencing and Screening of Potential Virulence Factors
The genomic DNA was extracted from B. ovatus ELH-B2 culture at early stationary phase and sequenced with Illumina Hiseq system by Majorbio (China). Library of average insert size of 410 bp was generated with low-quality reads filtered. The genome was assembled using SOAPdenovo v2.041 (Li et al., 2008; Li et al., 2010) followed by gap closure and base correction using GapCloser v1.12. A K-mer value of 23 was determined according to the accuracy evaluation. Gene annotation was performed by blastp (BLAST 2.2.28+) against Nr, Swiss-prot, string and GO databases. In order to show the relationships between B. ovatus ELH-B2 and other B. ovatus isolates, whose genome sequences were available from the NCBI database2, a neighbor-joining phylogenetic tree (Bottacini et al., 2014) was established by phyML3 (Guindon et al., 2009) after alignment of homologous genes identified by graph theory-based Markov clustering algorithm using mafft4 (Katoh and Standley, 2013) (Figure 1).
FIGURE 1.
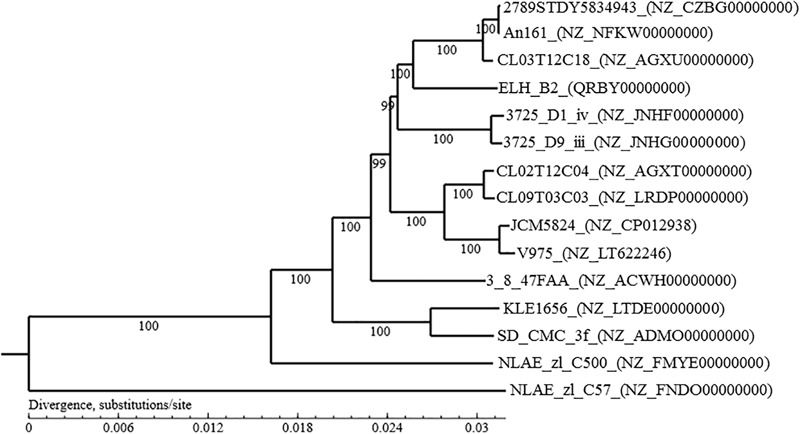
Phylogenetic tree based on the complete genome sequences of Bacteroides ovatus ELH-B2 and other B. ovatus strains.
Putative antibiotic resistance genes and virulence genes were identified in the genome of B. ovatus ELH-B2 by using a Protein-translated nucleotide Basic Local Search Tool (tblastn) according to the Comprehensive Antibiotic Resistance Database (CARD5 (McArthur et al., 2013) and the Virulence Factor Database (VFDB6) (Chen et al., 2012) respectively. Positive results were accepted with at least 30% identity and 70% coverage, and e-value less than 0.01 (Salvetti et al., 2016). Genetic islands were also predicted using IslandPath-DIMOB and Islander (Lu and Leong, 2016) for identifying putative virulence factors and possibilities of transportation of antibiotics resistance genes between bacteria. Moreover, Bacteroides-specific virulence factors including bft, ompW, upaY, upaZ, wcfR, wcfS, cfiA, cepA, and cfxA, of which the amino acid sequences were acquired from the NCBI database (Table 1B), were screened in B. ovatus ELH-B2 using tblastn based on BioEdit v7.2.5 with e-value less than 1e-5. The genome sequence of B. ovatus JCM5824 (GenBank accession number NZ_CP012938) was analyzed for comparison.
Table 1.
(A) Identification of potential virulence factors in the genome of Bacteroides ovatus ELH-B2 according to VFDB database; (B) prediction of putative genomic islands of over 30 kb in the genome of B. ovatus ELH-B2; (C) comparison of Bacteroides-specific virulence genes in B. ovatus ELH-B2 and JCM5824.
| (A) | ||||||
|---|---|---|---|---|---|---|
| VFDB_ID | Name | Description | Original source | Identity | Coverage | e-value |
| VFG000077 | clpP | ATP-dependent Clp protease proteolytic subunit | Listeria monocytogenes EGD-e | 54% | 76% | 3.00E-67 |
| VFG000165 | pchF | Pyochelin synthetase PchF | Pseudomonas aeruginosa PAO1 | 71% | 88% | 6.00E-04 |
| VFG000321 | rfbD | GDP-D-mannose dehydratase | Helicobacter pylori 26695 | 59% | 74% | 2.00E-136 |
| VFG000366 | ybtQ | Inner membrane ABC-transporter YbtQ | Yersinia pestis CO92 | 56% | 72% | 3.00E-04 |
| VFG000574 | mgtB | Mg2+ transport protein | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | 55% | 72% | 0.00E+00 |
| VFG000696 | bexA | ATP-dependent polysaccharide export protein BexA | Haemophilus influenzae str. 1007 | 49% | 78% | 1.00E-07 |
| VFG001266 | pchI | ABC transporter ATP-binding protein | Pseudomonas aeruginosa PAO1 | 53% | 76% | 1.00E-05 |
| VFG001269 | cyaB | Cyclolysin secretion ATP-binding protein | Bordetella pertussis Tohama I | 59% | 75% | 8.00E-04 |
| VFG001301 | cap8E | Capsular polysaccharide synthesis enzyme Cap8E | Staphylococcus aureus subsp. aureus MW2 | 63% | 78% | 1.00E-137 |
| VFG001303 | cap8G | Capsular polysaccharide synthesis enzyme Cap8G | Staphylococcus aureus subsp. aureus MW2 | 57% | 72% | 1.00E-140 |
| VFG001341 | cpsJ | Glycosyl transferase CpsJ(V) | Streptococcus agalactiae 2603V/R | 58% | 78% | 6.00E-10 |
| VFG001342 | cpsO | Glycosyl transferase CpsO(V) | Streptococcus agalactiae 2603V/R | 56% | 76% | 3.00E-14 |
| VFG001374 | cps4J | Capsular polysaccharide biosynthesis protein Cps4J | Streptococcus pneumoniae TIGR4 | 72% | 85% | 1.00E-160 |
| VFG001375 | cps4K | Capsular polysaccharide biosynthesis protein Cps4K | Streptococcus pneumoniae TIGR4 | 61% | 78% | 1.00E-169 |
| VFG001376 | cps4L | UDP-N-acetylglucosamine 2-epimerase | Streptococcus pneumoniae TIGR4 | 79% | 90% | 0.00E+00 |
| VFG001855 | htpB | Hsp60, 60K heat shock protein HtpB | Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | 59% | 77% | 2.00E-168 |
| VFG001937 | Cj1136 | Glucosyltransferase | Campylobacter jejuni NCTC11168 | 48% | 70% | 7.00E-14 |
| VFG001939 | Cj1138 | Glycosyltransferase | Campylobacter jejuni NCTC11168 | 47% | 74% | 2.00E-13 |
| VFG001940 | wlaN | Beta-1,3 galactosyltransferase | Campylobacter jejuni NCTC11168 | 50% | 76% | 1.00E-12 |
| VFG001968 | Cj1440c | Sugar transferase | Campylobacter jejuni NCTC11168 | 49% | 70% | 1.00E-11 |
| VFG002181 | cpsJ | ABC transporter, ATP-binding protein | Enterococcus faecalis V583 | 58% | 76% | 1.00E-03 |
| VFG002187 | cpsD | Glycosyl transferase, group 2 family protein | Enterococcus faecalis V583 | 36% | 71% | 3.00E-12 |
| VFG002225 | gmd | GDP-mannose 4,6-dehydratase | Brucella melitensis bv. 1 str. 16M | 69% | 81% | 2.00E-156 |
| VFG002365 | gmd | GDP-mannose 4,6-dehydratase | Yersinia enterocolitica 8081 | 64% | 78% | 2.00E-151 |
| VFG002368 | wbcG | Putative glycosyltransferase | Yersinia enterocolitica 8081 | 50% | 78% | 2.00E-09 |
| VFG002376 | ddhB | CDP-glucose 4,6-dehydratase | Yersinia enterocolitica 8081 | 49% | 71% | 5.00E-113 |
| VFG002377 | ddhA | Glucose-1-phosphate cytidylyltransferase | Yersinia enterocolitica 8081 | 58% | 74% | 2.00E-94 |
| VFG002440 | bprB | Two-component response regulator | Burkholderia pseudomallei K96243 | 50% | 72% | 6.00E-07 |
| VFG002480 | tssH-5/clpV | Clp-type ATPase chaperone protein | Burkholderia pseudomallei K96243 | 56% | 74% | 2.00E-83 |
| VFG002563 | wzt2 | ATP-binding ABC transporter capsular polysaccharide export protein | Burkholderia pseudomallei K96243 | 51% | 80% | 3.00E-06 |
| VFG005773 | cylZ | 3R-hydroxymyristoyl ACP dehydratase CylZ | Streptococcus agalactiae 2603V/R | 52% | 86% | 5.00E-04 |
| VFG011414 | kdsA | 2-dehydro-3-deoxyphosphooctonate aldolase | Brucella melitensis bv. 1 str. 16M | 53% | 70% | 1.00E-65 |
| VFG011430 | acpXL | Acyl carrier protein | Brucella melitensis bv. 1 str. 16M | 64% | 80% | 2.00E-24 |
| VFG012509 | iroC | ATP binding cassette transporter | Escherichia coli CFT073 | 45% | 71% | 4.00E-05 |
| VFG013354 | kfiC | Lipopolysaccharide biosynthesis protein | Haemophilus influenzae Rd KW20 | 47% | 83% | 2.00E-14 |
| VFG013368 | rffG | dTDP-glucose 46-dehydratase | Haemophilus influenzae Rd KW20 | 50% | 70% | 3.00E-99 |
| VFG013471 | lgtA | N-acetylglucosamine glycosyltransferase | Haemophilus influenzae Rd KW20 | 48% | 80% | 2.00E-10 |
| VFG037028 | katA | Catalase | Neisseria meningitidis MC58 | 71% | 82% | 0.00E+00 |
| VFG037386 | bauE | Ferric siderophore ABC transporter, ATP-binding protein BauE | Acinetobacter baumannii ACICU | 42% | 70% | 8.00E-05 |
| VFG038722 | AHA_1389 | CobQ/CobB/MinD/ParA family protein | Aeromonas hydrophila ATCC7966 | 51% | 71% | 5.00E-04 |
| VFG038918 | rtxE | RTX toxin transporter, ATPase protein | Aeromonas hydrophila ATCC7966 | 63% | 77% | 2.00E-05 |
| VFG043373 | cheA | Histidine kinase CheA | Helicobacter pylori 26695 | 39% | 70% | 5.00E-04 |
| VFG043394 | cheA | Chemotaxis histidine kinase | Campylobacter jejuni NCTC11168 | 34% | 70% | 7.00E-04 |
| VFG045340 | ricA | Rab2 interacting conserved protein A | Brucella melitensis bv. 1 str. 16M | 43% | 75% | 6.00E-07 |
| (B) | |||
|---|---|---|---|
| Island no. | Genes in island | Size (kb) | Potential functional genes within island |
| 1 | g0188–g0233 | 37.2 | Zeta-toxin (g0214); antitoxin (g0215) |
| 2 | g0842–g0866 | 35.3 | Arabinan endo-1,5-alpha-L-arabinosidase (g0844); SusC/RagA family TonB-linked outer membrane protein (g0846); glycosyl hydrolase (g0842, g0851, g0854) |
| 3 | g0978–g1164 | 170 | Thiamine biosynthesis protein ApbE (g0984); tonB-linked outer membrane, SusC/RagA family protein (g1005, g1062, g1092); CepA family class A extended-spectrum beta-lactamase (g1025); xylulokinase (g1107) |
| 4 | g1544–g1594 | 34.9 | Type IV secretory pathway TrbF components (g1575) |
| 5 | g1605–g1720 | 105.8 | Glycosyl transferase 2 family protein (g1616, g1619); two-component sensor histidine kinase (g1646);, TonB-dependent receptor (g1709) |
| 6 | g1837–g1864 | 32.2 | Membrane protein (g1855); iron ABC transporter ATP-binding protein (g1864) |
| 7 | g2583–g2631 | 33.3 | Redox-active disulfide protein (g2584); thioredoxin (g2585); conjugal transfer protein TraG (g2600, g2610), TraA (g2605), TraH (g2611), TraJ (g2613), TraK (g2614), TraM (g2616), TraN (g2617) |
| 8 | g2866–g2999 | 91.7 | Phage Mu F like family protein (g2947); phage tail tape measure protein, TP901 family, core region (g2955); toxin-antitoxin system, antitoxin component, MerR family (g2998) |
| 9 | g3114–g3160 | 41.2 | N-acetylmuramoyl-L-alanine amidase (g3133) |
| (C) | |||||
|---|---|---|---|---|---|
| Potential virulence factors | GenBank accession number | Size (aa) | % Similarity in B. ovatus ELH-B2 | % Similarity in B. ovatus JCM5824 | Reference |
| bft | AAF72837.1 | 63 | No hit | No hit | Gutacker et al., 2000 |
| ompW | AAL09385.1 | 947 | 95% (904/947) | 95% (901/947) | Wei et al., 2001 |
| upaY | AAK68912.1 | 172 | 29% (48/163) | 30% (48/160) | Coyne et al., 2001 |
| upaZ | AAK68913.1 | 157 | No hit | No hit | |
| wcfR | AAK68921.1 | 407 | 20% (88/407) | 25% (101/395) | |
| wcfS | AAK68922.1 | 198 | 48% (68/141) | 48% (68/141) | |
| cfiA | AAK71520 | 111 | No hit | No hit | Gutacker et al., 2002 |
| cfxA | CAP78899.1 | 306 | 40% (111/273) | 40% (110/273) | Garcia et al., 2008 |
| cepA | AAA21538.1 | 300 | 77% (233/299) | 77% (233/299) | Rogers et al., 1994 |
Minimum Inhibitory Concentration (MIC) of Different Antibiotics
Fourteen antibiotics, corresponding to ampicillin, cefoxitin, ceftriaxone, penicillin G, and vancomycin which suppress cell wall synthesis; chloromycetin, clindamycin, erythromycin, kanamycin, streptomycin, and tetracycline which restrain protein synthesis; ciprofloxacin and metronidazole which inhibit nucleic acid synthesis; and polymyxin B which suppresses cytoplasmic functions, were applied to determine the antibiotic resistance profiles of B. ovatus ELH-B2, with the type strain of JCM5824 as comparison. B. ovatus overnight culture (100 μl) at a concentration of 107 cfu/ml was treated with serially diluted antibiotics from 0.125 to 1024 μg/ml in sterile 96-well plates. The optical density at 600 nm was checked with a microplate reader (Multiskan GO, Thermo Scientific, United States) after anaerobic cultivation at 37°C for 48 h. The MIC of each antibiotic was determined by the lowest concentration that inhibited 90% of the growth of the tested B. ovatus strains (D’Aimmo et al., 2007). All of the experiments were carried out in three biological replicates.
Animals
Male C57 mice (7 weeks old, spf grade) were purchased from Shanghai Laboratory Animal Center (China) and raised within the IVC rodent caging system at Jiangnan University. The mice were maintained under a 12-h light/dark cycle with temperature and humidity strictly controlled. Treatment was initiated after acclimatization for at least 1 week. The entire experiment was approved by the Animal Ethics Committee of Jiangnan University (JN. No. 20180415c0450730[61]), and protocols for the care and use of experimental animals were based on the European Community guidelines (Directive 2010/63/EU).
Acute Toxicity to Immunocompetent and Immunosuppressed Mice
Both immunocompetent and immunosuppressed mice were involved in this acute toxicity assessment of B. ovatus ELH-B2. The immunocompetent mice, comprising control group (CTRL, 6 mice) and B. ovatus ELH-B2 group (BO, 6 mice), were given 150 μl of PBS/glycerol solution or 109 cfu B. ovatus ELH-B2 solution by gavage, respectively, every 24 h for 5 days. The other 12 mice were immunosuppressed by intraperitoneal injection with 250 mg/kg of cyclophosphamide (CTX, Sigma-Aldrich, United States), and were allocated to CTX group or CTX + BO group 3 days after followed by daily oral administration of 150 μl of PBS/glycerol solution or 109 cfu B. ovatus ELH-B2 solution, respectively, for 5 days (Hirsh et al., 2004; Salva et al., 2014).
The behavior and body weight of each mouse were monitored and recorded throughout the experiments. All of the mice were anesthetized with sodium pentobarbital and sacrificed by cervical dislocation. Liver, spleen and colon tissues and blood samples were collected immediately after sacrifice for further investigations.
Assays of Hematological and Liver Parameters
Hematological parameters were assessed using automatic hematology analyzer (BC-5000, Mindray, China) and associated buffers with fresh blood samples. The liver parameters were examined using automatic biochemical analyzer (BS-480) and corresponding kits (Mindray, China) with serum obtained by centrifuging the blood samples at 2500 rpm for 10 min. The serum standard (Shanghai Zhicheng Biological Technology Co. Ltd., China) was used for quality control.
Cytokine Concentrations in Serum
The secretions of tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), interleukin-8 (IL-8), and interleukin-10 (IL-10) were determined using mouse Elisa kits purchased from Nanjing SenBeiJia Biological Technology Co., Ltd. (China) with serum samples, according to the manufacturer’s instructions.
Histological Analysis
Liver, spleen and colon tissues were preserved in 4% paraformaldehyde solution and then embedded in paraffin. The histological analysis was conducted using Hematoxylin-Eosin (H&E) staining as published (Al-Hashmi et al., 2011). Images were recorded using Pannoramic digital slide scanner (Pannoramic MIDI II, 3DHISTECH Ltd., Hungary).
Statistical Analysis
Significant differences between groups were determined by unpaired Student’s t-test using Graphpad Prism v5.0 (Graph Pad Software Inc., United States), with p-values of less than 0.05. All of the data were presented as mean ± SD.
Results
Microbiological Properties
Bacteroides ovatus ELH-B2 grew well at 37°C on the Brucella agar supplemented with laked sheep blood under strict anaerobic conditions. The colonies were round, semi-opaque with smooth edges, and the bacteria were Gram-negative and rod shaped, which matches the description of B. ovatus in Bergey’s Manual (Krieg et al., 2001). Similar to the type strain B. ovatus JCM5824, ELH-B2 was confirmed to be non-motile, but slightly hemolytic.
Genetic Characteristics and Identification of Potential Virulence Factors
The size of the complete genome of B. ovatus ELH-B2 is 1 206 654 732 base pair, including 102 scaffolds and 5909 genes. The GC concent is 41.98%. The genomic information indicates the most similar strain to B. ovatus ELH-B2 is B. ovatus CL03T12C18 (Figure 1).
According to the blast against VFDB database, 44 virulence factor homologs were identified in B. ovatus ELH-B2 (Table 1A), most of which correlate with cellular structures like capsular polysaccharide and polysaccharide utilizations such as glycosyltransferase, and yet have been discovered as the pathogenesis of B. ovatus. And there are nine predicted genomic islands of over 30 kb (Table 1B), six of which are metabolism-related. Toxin-antitoxin system-associated genes were discovered two of the potential genomic islands, and one of the components in the Type IV secretion system (T4SS) were also identified.
As for the Bacteroides-specific virulence factors (Table 1C), Similar to B. ovatus JCM5824, B. ovatus ELH-B2 does not contain the diarrhea-associated B. fragilis enterotoxin bft. The coding gene of TonB-linked outer membrane protein (ompW), which has been implicated in inflammatory bowel disease (IBD) (Wei et al., 2001), were identified in both B. ovatus strains with high similarity. No hits or only low matches were identified in ELH-B2 for the highly conserved open reading frames, upaY and upaZ, and another two genes critical for synthesizing capsular PSA, wcfR and wcfS. And among the three β-lactamase-associated genes, only cepA was found in the two genomic sequences with high similarity.
Minimum Inhibitory Concentrations of Antibiotics
As shown in Table 2A, the potential antibiotic resistance genes indicated the possibilities of B. ovatus ELH-B2 to survive under the treatment of tetracycline, kanamycin, macrolide antibiotics like erythromycin, cationic antibiotics like polymyxin B, and glycopeptide like vancomycin. Accordingly, the MIC experiments verified that B. ovatus ELH-B2 was resistant to these antibiotics except tetracycline (Table 2B). It was also clinically susceptible to penicillin, cefoxitin, chloromycetin, and metronidazole with MICs of no more than 32 μg/ml (D’Aimmo et al., 2007). The lowest MIC of ELH-B2 was 4 μg/ml during treatment with metronidazole. Clindamycin and erythromycin were able to inhibit the growth of B. ovatus JCM5824 rather than ELH-B2.
Table 2.
(A) Predicted genes associated with antibiotic resistance in the genome of B. ovatus ELH-B2; (B) minimum inhibitory concentrations for antibiotics against B. ovatus ELH-B2 and B. ovatus JCM5824 (μg/ml).
| (A) | ||||||
|---|---|---|---|---|---|---|
| CARD_ID | Name | Description | Original source | Identity | Coverage | e-value |
| ARO:3000191 | tetQ | Tetracycline resistance | Bacteroides fragilis | 96% | 98% | 0.00E + 00 |
| ARO:3000197 | tet36 | Tetracycline resistance | Bacteroides coprosuis DSM 18011 | 60% | 79% | 0.00E + 00 |
| ARO:3000206 | emrK | Major facilitator superfamily (MFS) antibiotic efflux pump | Escherichia coli | 41% | 73% | 7.00E-03 |
| ARO:3000250 | ErmC | Erythromycin resistance | Staphylococcus epidermidis | 49% | 71% | 1.00E-72 |
| ARO:3000375 | ErmB | Erythromycin resistance | Enterococcus faecium | 98% | 99% | 4.00E-158 |
| ARO:3000499 | acrE | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Escherichia coli str. K-12 substr. MG1655 | 46% | 73% | 5.00E-05 |
| ARO:3000768 | abeS | Small multidrug resistance (SMR) antibiotic efflux pump | Acinetobacter baumannii AB307-0294 | 64% | 81% | 3.00E-26 |
| ARO:3000816 | mtrA | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Mycobacterium tuberculosis H37Rv | 40% | 75% | 5.00E-07 |
| ARO:3000828 | baeR | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Escherichia coli str. K-12 substr. W3110 | 50% | 71% | 1.00E-08 |
| ARO:3002522 | novA | Type III ABC transporter | Streptomyces niveus | 49% | 71% | 3.00E-03 |
| ARO:3002626 | ANT(6)-Ia | Aminoglycoside nucleotidyltransferase | Exiguobacterium sp. S3-2 | 62% | 78% | 2.00E-114 |
| ARO:3002627 | aadK | Aminoglycoside nucleotidyltransferase | Bacillus subtilis subsp. subtilis str. 168 | 58% | 76% | 1.00E-102 |
| ARO:3002628 | aad(6) | Aminoglycoside nucleotidyltransferase | Streptococcus oralis | 64% | 78% | 5.00E-107 |
| ARO:3002629 | ANT(6)-Ib | Aminoglycoside nucleotidyltransferase | Campylobacter fetus subsp. Fetus | 73% | 87% | 2.00E-133 |
| ARO:3002817 | carA | ABC transporter involved in macrolide resistance | Streptomyces thermotolerans | 46% | 74% | 5.00E-06 |
| ARO:3002825 | ErmY | Methyltransferase | Staphylococcus aureus | 52% | 72% | 2.00E-70 |
| ARO:3002827 | tlrC | Efflux pump | Streptomyces fradiae | 47% | 71% | 1.00E-02 |
| ARO:3002830 | vgaALC | Efflux protein | Staphylococcus haemolyticus | 56% | 75% | 3.00E-04 |
| ARO:3002845 | vatH | Acetyl transferase | Enterococcus faecium | 45% | 72% | 2.00E-04 |
| ARO:3002871 | tet37 | Tetracycline resistance | Uncultured bacterium | 65% | 79% | 1.00E-31 |
| ARO:3002925 | vanRF | Glycopeptide resistance | Paenibacillus popilliae ATCC14706 | 44% | 70% | 7.00E-15 |
| ARO:3003036 | oleB | ABC transporter | Streptomyces antibioticus | 46% | 72% | 7.00E-04 |
| ARO:3003048 | rosA | Efflux pump for resistance to cationic antimicrobial peptides such as polymyxin B | Yersinia enterocolitica (type O:8) | 53% | 72% | 1.00E-96 |
| ARO:3003109 | msrE | ABC-efflux pump for resistance to erythromycin | Escherichia coli | 65% | 81% | 2.00E-03 |
| ARO:3003548 | mdtN | Multidrug resistance efflux pump | Escherichia coli str. K-12 substr. W3110 | 43% | 70% | 5.00E-04 |
| ARO:3003559 | cepA | Beta-lactamase | Bacteroides fragilis | 78% | 89% | 3.00E-157 |
| ARO:3003577 | ugd | Resistance to cationic antimicrobial peptides | Escherichia coli str. K-12 substr. MG1655 | 58% | 72% | 7.00E-156 |
| ARO:3003728 | vanRI | Glycopeptide resistance gene | Desulfitobacterium hafniense | 42% | 70% | 3.00E-17 |
| ARO:3003744 | vatF | Streptogramin A acetyl transferase | Yersinia enterocolitica | 44% | 72% | 1.00E-08 |
| ARO:3003836 | qacH | Small multidrug resistance (SMR) antibiotic efflux pump | Vibrio cholerae | 53% | 71% | 1.00E-29 |
| ARO:3003841 | kdpE | Adaptive regulator involved in the virulence and intracellular survival of pathogenic bacteria | Escherichia coli str. K-12 substr. MG1655 | 43% | 74% | 6.00E-06 |
| ARO:3003922 | oqxA | RND efflux pump | Escherichia coli | 56% | 72% | 2.00E-05 |
| ARO:3003971 | erm(44) | Resistance to lincosamide and macrolide antibiotics | Staphylococcus saprophyticus | 49% | 73% | 1.00E-67 |
| ARO:3003986 | TaeA | Efflux pump | Paenibacillus sp. LC231 | 43% | 74% | 3.00E-05 |
| ARO:3003987 | VatI | Acetyltransferase for resistance to streptogramin A antibiotics | Paenibacillus sp. LC231 | 43% | 72% | 6.00E-04 |
| ARO:3004038 | emrE | Multidrug transporter for resistance to kanamycin | Pseudomonas aeruginosa PAO1 | 50% | 73% | 2.00E-10 |
| ARO:3004039 | emrE | Efflux pump for resistance to erythromycin | Escherichia coli | 53% | 71% | 4.00E-30 |
| ARO:3004042 | acrA | Resistance-nodulation-cell division (RND) antibiotic efflux pump | Enterobacter cloacae | 45% | 70% | 1.00E-03 |
| ARO:3004451 | Chloramphenicol acetyltransferase | Chloramphenicol acetyltransferase | Agrobacterium tumefaciens str. C58 | 53% | 71% | 2.00E-10 |
| (B) | ||
|---|---|---|
| Antibiotics | B. ovatus ELH-B2 | B. ovatus JCM5824 |
| Ampicillin | MIC ≤ 64 | MIC ≤ 128 |
| Penicillin G | MIC ≤ 32 | MIC ≤ 32 |
| Vancomycin | MIC ≤ 64 | MIC ≤ 64 |
| Cefoxitin | MIC ≤ 16 | MIC ≤ 32 |
| Ceftriaxone | MIC ≤ 256 | MIC ≤ 128 |
| Clindamycin | MIC ≤ 1024 | MIC ≤ 0.25 |
| Chloromycetin | MIC ≤ 16 | MIC ≤ 8 |
| Erythromycin | MIC ≤ 1024 | MIC ≤ 4 |
| Kanamycin | MIC > 1024 | MIC > 1024 |
| Streptomycin | MIC > 1024 | MIC > 1024 |
| Tetracycline | MIC ≤ 8 | MIC < 0.125 |
| Metronidazole | MIC ≤ 4 | MIC ≤ 4 |
| Ciprofloxacin | MIC ≤ 64 | MIC ≤ 16 |
| Polymyxin B | MIC ≤ 512 | MIC ≤ 128 |
In vivo Toxicity of B. ovatus ELH-B2
All of the animals were alive and healthy at the end of the experiment, and no abnormal behaviors were witnessed. Although the immunosuppressed mice displayed significantly less body weights, B. ovatus ELH-B2 treatment did not induce obvious alterations in the body mass of healthy or cyclophosphamide-injected mice (Figure 2A). Concerning the organ index, which refers to the ratio of organ weight to body weight, ELH-B2 had very little effect on liver indexes in immunocompetent mice, and did not enhance the enlarged spleen indexes of the drug-injected mice (Figure 2B). Colon length was also not notably influenced by the administration of ELH-B2.
FIGURE 2.
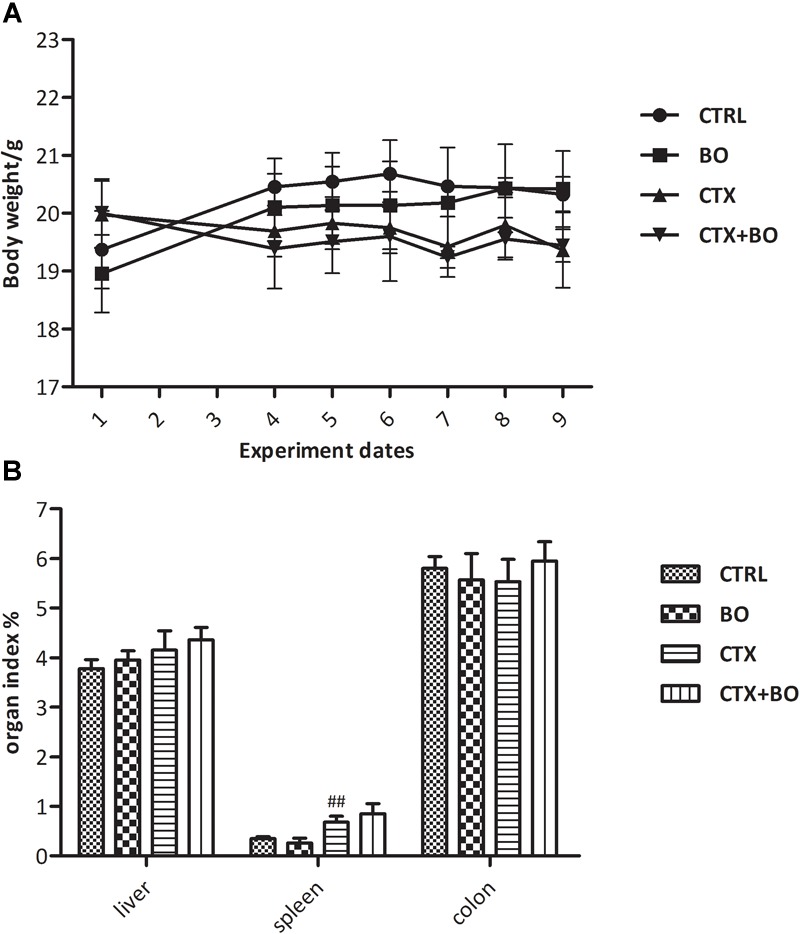
Impact of B. ovatus ELH-B2 on (A) the body weight and (B) the organ indexes of immunocompetent and immunosuppressed mice. The organ indexes of the liver and spleen are expressed as the ratio of the corresponding weight of the organ and body, while the colon index is expressed as the colon length of each animal. Data are displayed as mean ± SD, “##” indicates statistically significant differences between the CTX + BO group and the CTRL group (p < 0.01).
B. ovatus ELH-B2 intervention did not show significant alterations in the hematological (Table 3A) or liver parameters (Table 3B) of normal mice. After CTX injection, the percentages of lymphocytes (p < 0.01), hemoglobin concentration (p < 0.001), hematocrit value (p < 0.01) and platelets enumeration (p < 0.05) were markedly dropped, and the percentage of neutrophils were dramatically increased (p < 0.01). However, during the treatment with ELH-B2, the hematocrit value of the immunosuppressed mice was recovered (p < 0.01) and corpuscular volume was also significantly increased (p < 0.01). As for the liver parameters, CTX induced notable upregulation of alanine aminotransferase (p < 0.05), and concentration of alkaline phosphatase was dropped to normal level due to the administration of ELH-B2 in CTX-treated mice (p < 0.01).
Table 3.
Profiles of (A) hematological values and (B) liver parameters in immunocompetent and immunosuppressed mice after 5-day treatments with B. ovatus ELH-B2.
| (A) | ||||
|---|---|---|---|---|
| CTRL | BO | CTX | CTX + BO | |
| WBC (10ˆ9/L) | 2.47 ± 0.38 | 2.76 ± 0.64 | 2.19 ± 0.43 | 2.21 ± 0.92 |
| Neu (10ˆ9/L) | 0.73 ± 0.23 | 0.40 ± 0.20 | 0.92 ± 0.38# | 1.01 ± 0.65 |
| Lym (10ˆ9/L) | 2.56 ± 0.49 | 2.31 ± 0.48 | 1.15 ± 0.17## | 1.10 ± 0.31 |
| Mon (10ˆ9/L) | 0.05 ± 0.02 | 0.03 ± 0.02 | 0.08 ± 0.04 | 0.09 ± 0.05 |
| Eos (10ˆ9/L) | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.03 ± 0.01 | 0.04 ± 0.02 |
| Bas (10ˆ9/L) | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.02 |
| Neu (%) | 14.76 ± 5.61 | 13.90 ± 4.10 | 40.27 ± 10.93## | 39.44 ± 4.86 |
| Lym (%) | 83.16 ± 5.67 | 84.00 ± 4.58 | 54.17 ± 12.04## | 54.95 ± 4.41 |
| Mon (%) | 1.30 ± 0.60 | 1.10 ± 0.58 | 3.17 ± 1.23# | 2.93 ± 0.90 |
| Eos (%) | 0.36 ± 0.19 | 0.57 ± 0.24 | 1.33 ± 0.40## | 1.90 ± 0.37 |
| Bas (%) | 0.42 ± 0.13 | 0.43 ± 0.14 | 1.07 ± 0.66 | 1.35 ± 0.23 |
| RBC (10ˆ12/L) | 10.44 ± 0.23 | 10.07 ± 0.46 | 8.35 ± 0.20 | 8.88 ± 0.32 |
| HGB (g/L) | 171.40 ± 3.14 | 168.00 ± 3.92 | 141.33 ± 3.30### | 147.50 ± 6.60 |
| HCT (%) | 49.78 ± 0.93 | 48.65 ± 0.70 | 39.90 ± 0.73### | 44.98 ± 2.12** |
| MCV (fL) | 47.72 ± 0.28 | 48.08 ± 0.38 | 47.80 ± 0.29 | 50.17 ± 0.65** |
| MCH (pg) | 16.98 ± 0.17 | 17.27 ± 0.26 | 16.93 ± 0.25 | 16.43 ± 0.27 |
| MCHC (g/L) | 356.20 ± 4.71 | 358.67 ± 5.02 | 354.00 ± 4.55 | 356.50 ± 3.59 |
| RDW-CV (%) | 12.56 ± 0.24 | 12.83 ± 0.38 | 12.63 ± 0.09 | 13.07 ± 0.59 |
| RDW-SD (fL) | 26.88 ± 0.53 | 27.45 ± 0.73 | 29.07 ± 0.74 | 29.77 ± 1.06 |
| PLT (10ˆ9/L) | 1227.60 ± 171.16 | 1059.25 ± 49.44 | 907.33 ± 109.00# | 984.50 ± 160.82 |
| MPV (fL) | 5.50 ± 0.24 | 5.38 ± 0.07 | 6.00 ± 0.22 | 5.93 ± 0.22 |
| PDW (%) | 15.54 ± 0.12 | 15.43 ± 0.07 | 15.77 ± 0.25 | 15.98 ± 0.17 |
| PCT (%) | 0.68 ± 0.10 | 0.54 ± 0.05 | 0.55 ± 0.07 | 0.58 ± 0.09 |
WBC, white blood cells; Neu, neutrophils; Lym, lymphocytes; Mon, monocytes; Eos, eosinophils; Bas, basophils; RBC, red blood cells; HGB, hemoglobin concentration; HCT, hematocrit value; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red cell distribution width; CV, coefficient of variation; SD, standard deviation; PLT, platelet; MPV, mean platelet volume; PDW, platelet distribution width; PCT, thrombocytocrit. Data are displayed as mean ± SD, “∗∗” indicates statistically significant differences between the CTX + BO group and the CTX group (p < 0.01); “#”, “##,” and “###” indicate statistically significant differences between the CTX + BO group and the CTRL group (p < 0.05, p < 0.01 and p < 0.001, respectively).
| (B) | ||||
|---|---|---|---|---|
| CTRL | BO | CTX | CTX + BO | |
| Glu (mmol/L) | 6.40 ± 0.49 | 6.59 ± 0.56 | 7.07 ± 0.96 | 6.76 ± 0.40 |
| TC (mmol/L) | 1.87 ± 0.23 | 1.90 ± 0.28 | 1.78 ± 0.28 | 1.77 ± 0.17 |
| TG (mmol/L) | 1.51 ± 0.31 | 1.58 ± 0.11 | 1.56 ± 0.45 | 1.36 ± 0.46 |
| ALT (U/L) | 16.30 ± 1.41 | 17.85 ± 1.57 | 24.00 ± 3.82# | 22.08 ± 1.21 |
| AST (U/L) | 175.06 ± 1.42 | 178.44 ± 13.43 | 172.50 ± 36.94 | 180.48 ± 32.45 |
| TBIL (μmol /L) | 1.94 ± 0.64 | 1.96 ± 0.34 | 2.39 ± 0.43 | 2.27 ± 0.35 |
| ALP (U/L) | 5.25 ± 3.90 | 5.40 ± 2.24 | 8.25 ± 3.27 | 5.00 ± 1.41** |
| TP (g/L) | 40.80 ± 3.17 | 40.70 ± 4.99 | 44.10 ± 2.36 | 44.50 ± 1.47 |
| ALB (g/L) | 25.44 ± 2.62 | 25.05 ± 3.57 | 27.80 ± 1.81 | 27.60 ± 1.60 |
| CK (U/L) | 2072.58 ± 203.93 | 2262.52 ± 284.58 | 2431.00 ± 974.13 | 2256.23 ± 676.64 |
| LDH (U/L) | 423.83 ± 228.77 | 465.80 ± 69.24 | 511.00 ± 63.94 | 516.30 ± 62.95 |
Glu, glucose; TC, total cholesterol; TG, triglyceride; ALT, alanine aminotransferase; AST, aspartate aminotransferase; TBIL, total bilirubin; ALP, alkaline phosphatase; TP, total protein; ALB, albumin; CK, creatine kinase; LDH, lactic dehydrogenase.
No obvious modifications in cytokine productions were observed after the treatment of B. ovatus ELH-B2 in both healthy and immunosuppressed mice (Figure 3). Treatment with B. ovatus ELH-B2 did not lead to any histopathological damage in the liver, spleen or colon of the healthy mice (Figure 4). However, the CTX-treated mice suffered from hypertrophy of spleen, the histological structure of which was severely damaged with obvious fibrosis and hemorrhage. The red and white pulps could not be well-defined and splenocytes were irregularly aligned (Figure 4).
FIGURE 3.
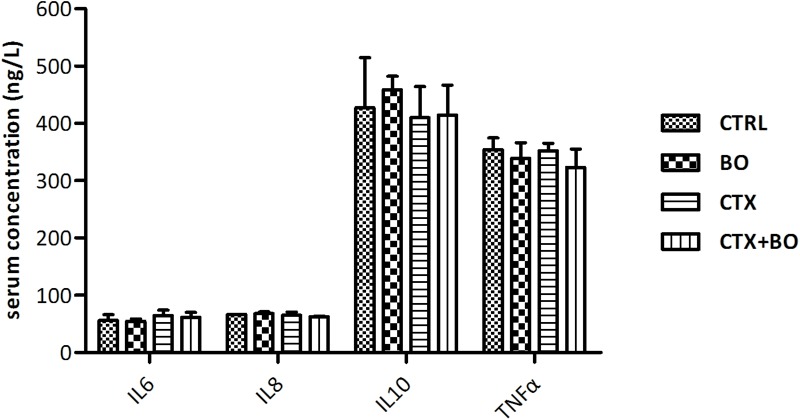
Impact of B. ovatus ELH-B2 on the cytokine productions of immunocompetent and immunosuppressed mice. Data are displayed as mean ± SD.
FIGURE 4.
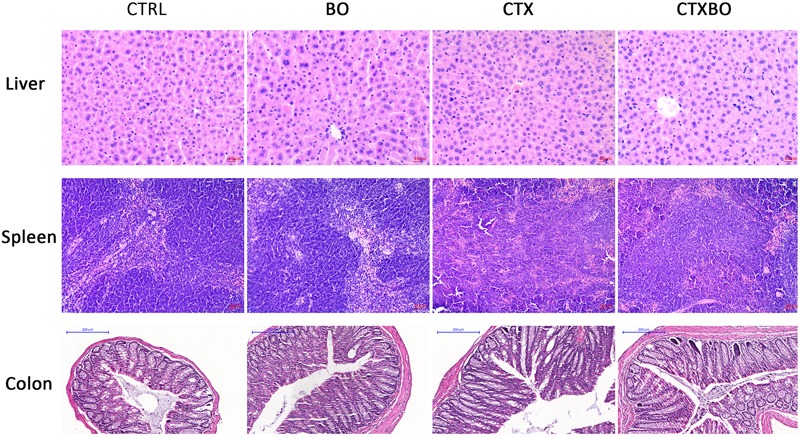
Impact of B. ovatus ELH-B2 on the tissue histology of immunocompetent and immunosuppressed mice.
Discussion
Evidences of indigenous and genetically modified intestinal commensals which obtain underlying efficacy in modulating immune and metabolic disorders, extend the range of probiotics, and are termed “next-generation probiotics” or “live biotherapeutic products.” The United States Food and Drug Administration (FDA) provide a definition for live biotherapeutic products as “a biological product that contains live organisms, such as bacteria, and is applicable to the prevention, treatment or cure of a disease or condition of human beings, but is not a vaccine,” which is also suitable for next-generation probiotics (O’Toole et al., 2017). In the meantime, the FDA drafted guidance that next-generation probiotics should be authorized as food ingredients when first entering the market. However, specific guidelines for applications of these promising microorganisms are yet to be developed. Therefore, a safety evaluation of B. ovatus ELH-B2 was carried out according to the regulations of the FAO/WHO for development of probiotics, in which explained the importance of complete bacterial characterizations such as original source, culture history, phenotype and genotype, antibiotic resistance, and manufacturing methods and three-step clinical trials, including safety assessment and functional characterization; double blind, randomized, placebo-controlled human studies; and efficiency comparisons with standard treatments, and published toxicity analyses of Lactobacillus spp. (Yakabe et al., 2009; Jia et al., 2011), B. xylanisolvens DSM23964 (Ulsemer et al., 2012a,b), B. uniformis CECT7771 (Fernandez-Murga and Sanz, 2016), and B. fragilis ZY312 (Wang et al., 2017), along with previous results which revealed that ELH-B2 had little effect on the production of secretory immunoglobulin A (sIgA) and chemokine (C-X-C motif) ligand 2 (CXCL2) or the balance of Treg and Th-17 cells, and even upregulated the diversity of intestinal microbiota (unpublished data).
Morphological analysis showed that B. ovatus ELH-B2 cells were non-motile, which excluded the pathogenic factor of flagella for inducing inflammation via activating the NF-κB pathway through Toll-like receptor 5 and secreting IL-8 (Neville et al., 2012), and facilitating nutrient acquisition, niche colonization (Lane et al., 2007; Neville et al., 2012), and biofilm formation (Houry et al., 2010). The translucent circles around ELH-B2 colonies on blood agar plates indicated the possible existence of hemolysin, which is a pore-forming toxin with cytolytic functions on various types of cells, such as keratinocytes, epithelial cells, and lymphocytes (Kennedy et al., 2010; Wilke and Wardenburg, 2010).
The virulence factors discovered via the Virulence Factor Database include genes facilitating protein secretion, carbohydrates degradation and maintaining cellular structures. These elements could be probiosis-related and contribute to bacterial adhesion and colonization, rather than pathogenicity (Wassenaar et al., 2015). The majority of the predict islands were discovered to be metabolism-related. Although TrbF of T4SS were identified, the rest preserved genes were absent. Thus B. ovatus ELH-B2 are not capable of producing the entire secretion systems for any possibilities of transfering virulence protein and antibiotic resistance genes (Aguilar et al., 2010). Moreover, toxin-antitoxin system are widely existed in natural bacteria strains for better adaptive ability to the environment resulted from evolution (Buts et al., 2005), the virulence characteristics of which requires further analysis.
Bacteroides species are, to some extent, considered to be opportunistic pathogens as some of them are carriers of virulence factors, such as the enterotoxigenic B. fragilis with bft (Sears, 2009) and B. caccae with ompW (Wei et al., 2001). The metalloprotease bft was not identified in ELH-B2, but ompW was found to be 95% identical. The protein encoded by ompW in B. caccae was discovered by pANCA monoclonal antibody in IBD patients and is closely associated with a pathogenic factor of Porphyromonas gingivalis that contributes to tissue damage. However, this TonB-linked structure of ompW is also conserved in the starch-utilization system of Bacteroides species that helps to break down resistant carbohydrates in the host. Hence, the underlying role of the ompW-like structure in ELH-B2 requires further investigation. Meanwhile, the blast results of the four genes essential for constructing PSA indicated the absence of the capsular polysaccharide, which suggests that ELH-B2 does not possess the PSA-associated anti-inflammatory character (Mazmanian et al., 2008), but also avoids the consequently potential abscess formation (Cohen-Poradosu et al., 2011).
Overall, the putative antibiotic resistance gene list based on the Comprehensive Antibiotic Resistance Database are almost in correspondence to the antibiotic resistance profiles of B. ovatus ELH-B2 acquired from MIC experiments. B. ovatus ELH-B2 was found to be less susceptible to antibiotics compared with the type strain. The three genes encoding β-lactamase, corresponding to cfxA for class A cephalosporinase, cfiA for class B metallo-β-lactamase and cepA for endogenous cephalosporinase (Garcia et al., 2008), were not or were only partially aligned in the genome of ELH-B2, which was consistent with the result that the novel strain was clinically resistant to penicillin G and cefoxitin. It is notable that neither plasmid nor antibiotic resistance-related genomic islands were found in the genome, indicating low chances of transferring the characteristics of antibiotic resistance to other intestinal commensals.
In general, the results demonstrated that B. ovatus ELH-B2 showed no oral pathogenicity in healthy animals with a daily dose of 109 cfu live cells, which is the appropriate concentration for commercial application that guarantees both the viability and integrity of bacterial cells after freeze-drying and restoration procedures (Miyamoto-Shinohara et al., 2000). Based on a mean body weight of 20 g for the mice, 3.5 × 1012 cfu of ELH-B2 cells are predicted to be safe for a 70 kg healthy human adult.
In the meantime, confidence in authorizing treatments for industrial and clinical applications would be enhanced once no side effects of the bacteria have been confirmed in immunodeficient animals (FAO/WHO, 2002). In this study, immunosuppressed condition was established via CTX injection, which is one of the most commonly used as a chemotherapeutic treatment for cancers and as an immunosuppressive agent before myeloablative therapies (Ehrke, 2003). Accordingly, the cytotoxicity was reflected in liver which is the first target organ engaging all toxic drugs (Singh et al., 2018), and spleen which is responsible for the immune status by controlling the proliferation of T cells, B cells and lymphocytes (Gong et al., 2015). CTX treatment led to distinctly upregulated liver parameter of alanine aminotransferase, injuries in spleen and disturbed hematological values.
Nevertheless, B. ovatus ELH-B2 did not accelerate the toxicities of CTX. Besides, the restoration of alkaline phosphatase revealed a recovery effect on the liver and pancreatic functions. Alkaline phosphatase contributes to the dephosphorylation of LPS from the gut by circulation (Moreira et al., 2012), and thereby the downregulation of which emphasized the capability of the Bacteroides to help reduce the threats of endotoxemia in the mice. This result corresponds to the previous characterization study of this novel strain. A reduction in the secretion of TNF-α was observed with B. ovatus ELH-B2 treatment, demonstrating its potential anti-inflammatory function, although in a non-significant way.
In summary, B. ovatus ELH-B2, a novel strain which was confirmed to be capable of attenuating LPS-induced inflammation in vivo, did not raise severe safety issues in either immunocompetent or immunosuppressed mice, and even partially relieved the side effects associated with the chemotherapeutic drug. Further assessments of viable doses and extreme conditions, such as intraperitoneal injection of bacteria, should be considered (Ulsemer et al., 2012b).
Author Contributions
HT and ZY carried out the experiment and drafted the manuscript. CW and QinZ participated in the analysis of the data. JZ and HZ provided the essential reagents and materials. QixZ conceived of the study and managed the project design. WC helped to revise the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (No. 31871773 and 31470161, and Key Program No. 31530056), the Natural Science Foundation of Jiangsu Province (BK20160175 and BK20141098), the National First-Class Discipline Program of Food Science and Technology (JUFSTR20180102), and the Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
References
- Aguilar J., Zupan J., Cameron T. A., Zambryski P. C. (2010). Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. U.S.A. 2 3758–3763. 10.1073/pnas.0914940107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashmi S., Hassan Z., Sadeghi B., Rozell B., Hassan M. (2011). Dynamics of early histopathological changes in GVHD after busulphan/cyclophosphamide conditioning regimen. Int. J. Clin. Exp. Pathol. 4 596–605. [PMC free article] [PubMed] [Google Scholar]
- Bottacini F., Motherway M. O., Kuczynski J., O’Connell K. J., Serafini F., Duranti S., et al. (2014). Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. 10.1186/1471-2164-15-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann T., Endo A., Gueimonde M., Vinderola G., Kneifel W., de Vos W. M., et al. (2017). Safety of novel microbes for human consumption: practical examples of assessment in the European Union. Front. Microbiol. 8:15. 10.3389/fmicb.2017.01725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005). Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 12 672–679. 10.1016/j.tibs.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Cano P. G., Santacruz A., Moya A., Sanz Y. (2012). Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One 7:e41079. 10.1371/journal.pone.0041079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. H., Xiong Z. H., Sun L. L., Yang J., Jin Q. (2012). VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40 D641–D645. 10.1093/nar/gkr989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Poradosu R., McLoughlin R. M., Lee J. C., Kasper D. L. (2011). Bacteroides fragilis-stimulated interleukin-10 contains expanding disease. J. Infect. Dis. 204 363–371. 10.1093/infdis/jir277 [DOI] [PubMed] [Google Scholar]
- Cousin F. J., Lynch S. M., Harris H. M. B., McCann A., Lynch D. B., Neville B. A., et al. (2015). Detection and genomic characterization of motility in Lactobacillus curvatus: confirmation of motility in a species outside the Lactobacillus salivarius clade. Appl. Environ. Microbiol. 81 1297–1308. 10.1128/aem.03594-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Tzianabos A. O., Mallory B. C., Carey V. J., Kasper D. L., Comstock L. E. (2001). Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 69 4342–4350. 10.1128/iai.69.7.4342-4350.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aimmo M. R., Modesto M., Biavati B. (2007). Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 115 35–42. 10.1016/j.ijfoodmicro.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Ehrke M. J. (2003). Immunomodulation in cancer therapeutics. Int. Immunopharmacol. 3 1105–1119. 10.1016/s1567-5769(03)00021-3 [DOI] [PubMed] [Google Scholar]
- FAO/WHO (2002). Guidelines for the Evaluation of Probiotics in Food: Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Manila: FAO/WHO. [Google Scholar]
- Fernandez-Murga M. L., Sanz Y. (2016). Safety assessment of Bacteroides uniformis CECT 7771 isolated from stools of healthy breast-fed infants. PLoS One 11:e0145503. 10.1371/journal.pone.0145503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia N., Gutierrez G., Lorenzo M., Garcia J. E., Piriz S., Quesada A. (2008). Genetic determinants for cfxA expression in Bacteroides strains isolated from human infections. J. Antimicrob. Chemother. 62 942–947. 10.1093/jac/dkn347 [DOI] [PubMed] [Google Scholar]
- Gong Y., Wu J., Li S. T. (2015). Immuno-enhancement effects of Lycium ruthenicum Murr. polysaccharide on cyclophosphamide-induced immunosuppression in mice. Int. J. Clin. Exp. Med. 8 20631–20637. [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Hordijk W., Lefort V., Gascuel O. (2009). PhyML: fast and accurate phylogeny reconstruction by maximum likelihood. Infect. Genet. Evol. 5 384–385. 14530136 [Google Scholar]
- Gutacker M., Valsangiacomo C., Bernasconi M. V., Piffaretti J. C. (2002). recA and glnA sequences separate the Bacteroides fragilis population into two genetic divisions associated with the antibiotic resistance genotypes cepA and cfiA. J. Med. Microbiol. 51 123–130. 10.1099/0022-1317-51-2-123 [DOI] [PubMed] [Google Scholar]
- Gutacker M., Valsangiacomo C., Piffaretti J. C. (2000). Identification of two genetic groups in Bacteroides fragilis by multilocus enzyme electrophoresis: distribution of antibiotic resistance (cfiA, cepA) and enterotoxin (bft) encoding genes. Microbiology 146 1241–1254. 10.1099/00221287-146-5-1241 [DOI] [PubMed] [Google Scholar]
- Hamady Z. Z. R., Scott N., Farrar M. D., Lodge J. P. A., Holland K. T., Whitehead T., et al. (2010). Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut 59 461–469. 10.1136/gut.2008.176131 [DOI] [PubMed] [Google Scholar]
- Hamady Z. Z. R., Scott N., Farrar M. D., Wadhwa M., Dilger P., Whitehead T. R., et al. (2011). Treatment of colitis with a commensal gut bacterium engineered to secrete human TGF-beta 1 under the control of dietary xylan. Inflamm. Bowel Dis. 17 1925–1935. 10.1002/ibd.21565 [DOI] [PubMed] [Google Scholar]
- Hirsh M., Carmel J., Kaplan V., Livne E., Krausz M. M. (2004). Activity of lung neutrophils and matrix metalloproteinases in cyclophosphamide-treated mice with experimental sepsis. Int. J. Exp. Pathol. 85 147–157. 10.1111/j.0959-9673.2004.00385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry A., Briandet R., Aymerich S., Gohar M. (2010). Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156 1009–1018. 10.1099/mic.0.034827-0 [DOI] [PubMed] [Google Scholar]
- Hsiao E. Y., McBride S. W., Hsien S., Sharon G., Hyde E. R., McCue T., et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155 1451–1463. 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X., Wang W., Song Y., Li N. (2011). A 90-day oral toxicity study on a new strain of Lactobacillus paracasei in rats. Food Chem. Toxicol. 49 1148–1151. 10.1016/j.fct.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 4 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. D., Wardenburg J. B., Gardner D. J., Long D., Whitney A. R., Braughton K. R., et al. (2010). Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J. Infect. Dis. 202 1050–1058. 10.1086/656043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg N. R., Ludwig W., Euzeby J., Whitman W. B. (2001). Bergey’s Manual of Systematic Bacteriology 2nd Edn Vol. 1 New York, NY: Springer [Google Scholar]
- Lane M. C., Alteri C. J., Smith S. N., Mobley H. L. T. (2007). Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U.S.A. 104 16669–16674. 10.1073/pnas.0607898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Q., Li Y. R., Kristiansen K., Wang J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24 713–714. 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- Li R. Q., Zhu H. M., Ruan J., Qian W. B., Fang X. D., Shi Z. B., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20 265–272. 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. X., Leong H. W. (2016). Computational methods for predicting genomic islands in microbial genomes. Comp. Struct. Biotechnol. J. 14 200–206. 10.1016/j.csbj.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453 620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- McArthur A. G., Waglechner N., Nizam F., Yan A., Azad M. A., Baylay A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57 3348–3357. 10.1128/aac.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto-Shinohara Y., Imaizumi T., Sukenobe J., Murakami Y., Kawamura S., Komatsu Y. (2000). Survival rate of microbes after freeze-drying and long-term storage. Cryobiology 41 251–255. 10.1006/cryo.2000.2282 [DOI] [PubMed] [Google Scholar]
- Moreira A. P., Texeira T. F., Ferreira A. B., Peluzio Mdo C., Alfenas Rde C. (2012). Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br. J. Nutr. 108 801–809. 10.1017/s0007114512001213 [DOI] [PubMed] [Google Scholar]
- Neville B. A., Forde B. M., Claesson M. J., Darby T., Coghlan A., Nally K., et al. (2012). Characterization of pro-inflammatory flagellin proteins produced by Lactobacillus ruminis and related motile Lactobacilli. PLoS One 7:40592. 10.1371/journal.pone.0040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole P. W., Marchesi J. R., Hill C. (2017). Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2:17057. 10.1038/nmicrobiol.2017.57 [DOI] [PubMed] [Google Scholar]
- Robertson K. P., Smith C. J., Gough A. M., Rocha E. R. (2006). Characterization of Bacteroides fragilis hemolysins and regulation and synergistic interactions of HlyA and HlyB. Infect. Immun. 74 2304–2316. 10.1128/iai.74.4.2304-2316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. B., Bennett T. K., Payne C. M., Smith C. J. (1994). Insertional activation of cepA leads to high-level beta-lactamase expression in Bacteroides fragilis clinical isolates. J. Bacteriol. 176 4376–4384. 10.1128/jb.176.14.4376-4384.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salva S., Marranzino G., Villena J., Aguero G., Alvarez S. (2014). Probiotic Lactobacillus strains protect against myelosuppression and immunosuppression in cyclophosphamide-treated mice. Int. Immunopharmacol. 22 209–221. 10.1016/j.intimp.2014.06.017 [DOI] [PubMed] [Google Scholar]
- Salvetti E., Orru L., Capozzi V., Martina A., Lamontanara A., Keller D., et al. (2016). Integrate genome-based assessment of safety for probiotic strains: Bacillus coagulans GBI-30, 6086 as a case study. Appl. Microbiol. Biotechnol. 100 4595–4605. 10.1007/s00253-016-7416-9 [DOI] [PubMed] [Google Scholar]
- Sears C. L. (2009). Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin. Microbiol. Rev. 22 349–366. 10.1128/cmr.00053-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C., Prakash C., Tiwari K. N., Mishra S. K., Kumar V. (2018). Premna integrifolia ameliorates cyclophosphamide-induced hepatotoxicity by modulation of oxidative stress and apoptosis. Biomed. Pharmacother. 107 634–643. 10.1016/j.biopha.2018.08.039 [DOI] [PubMed] [Google Scholar]
- Tan H. Z., Zhao J. X., Zhang H., Zhai Q. X., Chen W. (2018). Isolation of low-abundant Bacteroidales in the human intestine and the analysis of their differential utilization based on plant-derived polysaccharides. Front. Microbiol. 9:1319. 10.3389/fmicb.2018.01319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulsemer P., Henderson G., Toutounian K., Loffler A., Schmidt J., Karsten U., et al. (2013). Specific humoral immune response to the Thomsen-Friedenreich tumor antigen (CD176) in mice after vaccination with the commensal bacterium Bacteroides ovatus D-6. Cancer Immunol. Immunother. 62 875–887. 10.1007/s00262-013-1394-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulsemer P., Toutounian K., Schmidt J., Karsten U., Goletz S. (2012a). Preliminary safety evaluation of a new Bacteroides xylanisolvens isolate. Appl. Environ. Microbiol. 78 528–535. 10.1128/aem.06641-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulsemer P., Toutounian K., Schmidt J., Leuschner J., Karsten U., Goletz S. (2012b). Safety assessment of the commensal strain Bacteroides xylanisolvens DSM 23964. Regul. Toxicol. Pharmacol. 62 336–346. 10.1016/j.yrtph.2011.10.014 [DOI] [PubMed] [Google Scholar]
- Wang Y., Deng H. M., Li Z. C., Tan Y. F., Han Y. P., Wang X. Y., et al. (2017). Safety evaluation of a novel strain of Bacteroides fragilis. Front. Microbiol. 8:435. 10.3389/fmicb.2017.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. M., Zschüttig A., Beimfohr C., Geske T., Auerbach C., Cook H., et al. (2015). Virulence genes in a probiotic E. coli product with a recorded long history of safe use. Eur. J. Microbiol. Immunol. 5 81–93. 10.1556/EUJMI-D-14-00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Dalwadi H., Gordon L. K., Landers C., Bruckner D., Targan S. R., et al. (2001). Molecular cloning of a Bacteroides caccae TonB-linked outer membrane protein identified by an inflammatory bowel disease marker antibody. Infect. Immun. 69 6044–6054. 10.1128/iai.69.10.6044-6054.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke G. A., Wardenburg J. B. (2010). Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. U.S.A. 107 13473–13478. 10.1073/pnas.1001815107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakabe T., Moore E. L., Yokota S., Sui H., Nobuta Y., Fukao M., et al. (2009). Safety assessment of Lactobacillus brevis KB290 as a probiotic strain. Food Chem. Toxicol. 47 2450–2453. 10.1016/j.fct.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Yim S., Gwon S. Y., Hwang S., Kim N. H., Jung B. D., Rhee K. J. (2013). Enterotoxigenic Bacteroides fragilis causes lethal colitis in Mongolian gerbil. Anaerobe 21 64–66. 10.1016/j.anaerobe.2013.03.008 [DOI] [PubMed] [Google Scholar]


