Abstract
Aim
To develop an algorithm, based on convolutional neural network (CNN), for the classification of lung cancer lesions as T1-T2 or T3-T4 on staging fluorodeoxyglucose positron emission tomography (FDG-PET)/CT images.
Methods
We retrospectively selected a cohort of 472 patients (divided in the training, validation, and test sets) submitted to staging FDG-PET/CT within 60 days before biopsy or surgery. TNM system seventh edition was used as reference. Postprocessing was performed to generate an adequate dataset. The input of CNNs was a bounding box on both PET and CT images, cropped around the lesion centre. The results were classified as Correct (concordance between reference and prediction) and Incorrect (discordance between reference and prediction). Accuracy (Correct/[Correct + Incorrect]), recall (Correctly predicted T3-T4/[all T3-T4]), and specificity (Correctly predicted T1-T2/[all T1-T2]), as commonly defined in deep learning models, were used to evaluate CNN performance. The area under the curve (AUC) was calculated for the final model.
Results
The algorithm, composed of two networks (a “feature extractor” and a “classifier”), developed and tested achieved an accuracy, recall, specificity, and AUC of 87%, 69%, 69%, and 0.83; 86%, 77%, 70%, and 0.73; and 90%, 47%, 67%, and 0.68 in the training, validation, and test sets, respectively.
Conclusion
We obtained proof of concept that CNNs can be used as a tool to assist in the staging of patients affected by lung cancer.
1. Introduction
In recent years, advanced analysis of medical imaging using radiomics, machine, and deep-learning, including convolutional neural networks (CNNs), has been explored. These approaches offer great promise for future applications for both diagnostic and predictive purposes. CNNs are nonexplicitly programmed algorithms that identify relevant features on the images that allow them to classify an input object. They have been applied in various tasks such as detection (e.g., breast lesions on mammographic scans), segmentation (e.g., liver and liver lesions on computed tomography (CT)), and diagnosis (e.g., lung lesions on screening low-dose CT).
CNNs are a machine-learning technique based on an artificial neural network with deep architecture relying on convolution operations (the linear application of a filter or kernel to local neighbourhoods of pixel/voxels in an input image) and downsampling or pooling operations (grouping of feature map signals into a lower-resolution feature map). The final classification or regression task relies on higher-level features representative of a large receptive field that is flattened into a single vector. The development of an algorithm entails (a) selection of the hyperparameters, (b) training and validation, and (c) testing. The hyperparameters include the network topology, the number of filters per layer, and the optimisation parameters. During the training process, the dataset of input images (divided into training and validation sets) is repeatedly submitted to the network to capture the structure of the images that is salient for the task. Initially, the weights for each artificial neuron are randomly chosen. Then, they are adjusted at each iteration, targeting minimisation of the loss function, which quantifies how close the prediction is to the target class. The performance of the trained model is then evaluated using an independent test dataset. This is also aimed at assessing whether an “overfitting” has occurred. The overfitting problem can arise in the case of limited datasets with too many parameters compared with the dataset size, in which case a model “memorises” the training data rather than generalising from them [1].
In the field of lung imaging, CNNs have been tested in nodule segmentation from CT images. Average dice scores of 82% and 80% for the training and test datasets, respectively, have been reported [2]. CNNs have been demonstrated to achieve better results than conventional methods for the purpose of nodule detection [3, 4]. Moreover, a model for assessment of cancer probability in patients with pulmonary nodules has been proposed. The area under the curve (AUC) was found to be 0.90 and 0.87 for the training and test sets, respectively [5]. Stage assessment has not yet been described. The present study, as a first step towards complete TNM parameter assessment, aimed to develop an algorithm for the classification of lung cancer as T1-T2 or T3-T4 on staging fluorodeoxyglucose positron emission tomography (FDG-PET)/CT images.
2. Materials and Methods
2.1. Study Design and Patient Selection
In this retrospective single-centre investigation, we screened all patients who underwent FDG-PET/CT between 01/01/2011 and 27/06/2017 for the purpose of staging a suspected lung lesion, within 60 days before biopsy or surgical procedure. The inclusion criteria were (1) age > 18 years and (2) histological diagnosis of primary lung cancer. The exclusion criteria were (1) inconclusive histology due to inadequate biopsy sample and (2) diagnosis of nonmalignancy. The study was approved by the Institutional Ethics Committee.
2.2. Image Acquisition and Postprocessing
FDG-PET/CT was performed according to the standard institutional procedures, previously detailed [6]. Postacquisition processing was performed to generate an adequate dataset for the CNN. The original CT and PET image size was 512 × 512 × Nslices and 128 × 128 × Nslices, respectively, where Nslices is the number of slices in which the lesion appears. The CT images were clipped between −1000 and 400 Hounsfield units. PET images were resampled in the CT space. Then, both images were rescaled to lie between 0 and 1. Consequently, the dataset consisted of 3D bounding boxes on both PET and CT images, cropped around the lesion centre, identified by two nuclear medicine physicians (M.S. and M.K.) with dimension 128 × 128 × Nslices. Data augmentation, a strategy commonly used by deep-learning methods, was performed. Image patches were rotated in 2D space around the lesion centre about the z-axis by an angle randomly selected in a range of [−10°, 10°]. This processing step artificially expands the size of the training set and reduces the overfitting phenomena.
2.3. CNN Development and Analysis
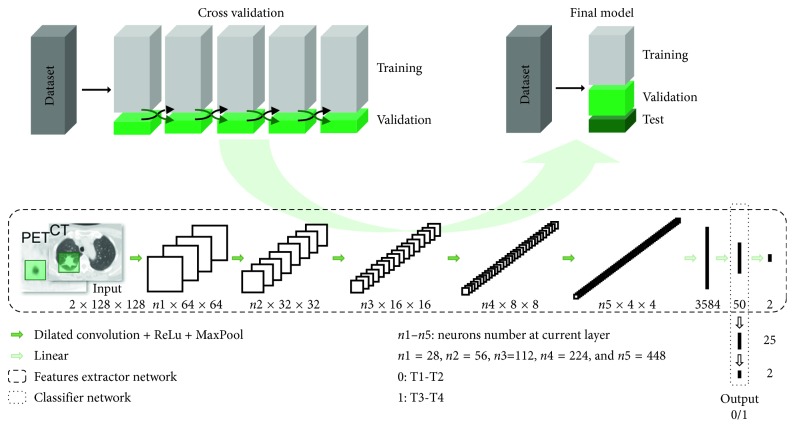
The study workflow and the networks' architecture are summarised in Figure 1. During the training phase, a fivefold cross-validation strategy was adopted by dividing the cohort into a training dataset and a validation dataset. To assess the performance of the final model, the cohort was divided into training, validation, and test datasets. The algorithm was composed of two networks: a feature extractor and a classifier. The feature extractor was a CNN that took a CT-PET image patch of 128 × 128 pixels as input and performed classification (T1-T2 with label = 0 and T3-T4 with label = 1) according to the appearance of the image patch. The feature extractor aimed to extract the most relevant features from a single patch. The classifier took as input the mean of the second to last layer of features extracted from all slices of a single patient and aimed to perform a classification (T1-T2 vs. T3-T4) for that patient. The softmax function was applied to the last layer of both networks, in order to obtain the probability of being T1-T2 and T3-T4. The class having the highest probability was assigned to each patient. Both models were trained with the Adam algorithm [7]. Table 1 summarises the parameters for the feature extractor and the classifier networks. The code was written in Python, using the PyTorch deep-learning library (http://pytorch.org/).
Figure 1.
Study workflow and networks' architecture. The network (consisting of two neural networks, the feature extractor, and the classifier) was trained using a cross-validation strategy. The entire dataset was divided into 5 randomly chosen parts. At each training run, 4/5 of the dataset were used as a training set and the remaining 1/5 was used as the validation set. Subsequently, the final model network was adjusted and tested for performance using the dataset divided into three sets: training, validation, and test. In the feature extractor CNN, the input images, both PET and CT, were submitted to a series of convolutions producing a stack of feature maps containing low-level features, rectified linear units (ReLU), and max pooling layers that downsample the feature maps (MaxPool), to produce higher-level features. In the classifier network, these higher-level features are used to perform the final classification T1-T2 (output label 0) vs T3-T4 (output label 1).
Table 1.
Summary of the parameters of the feature extractor and the classifier networks.
| Feature extractor | Classifier | |
|---|---|---|
| Loss function | Cross entropy | Weight cross entropy |
| Learning method | Adam | Adam |
| Learning rate | 1e − 6 | 1e − 3 |
| Weight decay | 1e − 6 | 1e − 6 |
| Epochs | 600 | 600 |
| Patients/batch | 2 | 2 |
| Slides/patient | 20 | n.a. |
n.a.: not applicable.
TNM classification system 7th edition [8] was used as reference.
The results were classified as Correct (concordance between reference and prediction) and Incorrect (discordance between reference and prediction). Accuracy (Correct/[Correct + Incorrect]), recall (Correctly predicted T3-T4/[All T3-T4]), and specificity (Correctly predicted T1-T2/[all T1-T2]) as commonly defined in deep-learning models, were used to evaluate CNN performance. The area under the curve (AUC) was calculated for the final model.
3. Results
From the institutional database, a cohort of 586 patients was selected by applying the abovementioned criteria. Patients with distant metastases or histology different from adenocarcinoma and squamous cell carcinoma were excluded from the present analysis. Therefore, 472 patients were included in the study (T1-T2 = 353 patients, T3-T4 = 119 patients). Staging was clinical and pathological in 97 and 375 of cases, respectively. Subsequently, the patients were randomly divided in training (n=303), validation (n=75), and test (n=94) sets. The patients' characteristics are summarised in Table 2.
Table 2.
Patients' characteristics.
| Characteristics | Patients, n (%) |
|---|---|
| Age (year): mean: 69 ± 9, median: 69 (range: 36–112) | |
|
| |
| Sex | |
| Male | 316 (67) |
| Female | 156 (33) |
|
| |
| Histology | |
| Adenocarcinoma | 317 (67) |
| Squamous cell carcinoma | 155 (33) |
|
| |
| TNM | |
| T1 | 159 (34) |
| T2 | 194 (41) |
| T3 | 79 (17) |
| T4 | 40 (8) |
| N0 | 259 (55) |
| N1 | 94 (20) |
| N2 | 103 (22) |
| N3 | 16 (3) |
| M0 | 472 (100) |
|
| |
| Stage ∗ | |
| I | 193 (41) |
| Ia | 115 (24) |
| Ib | 78 (17) |
|
| |
| II | 116 (25) |
| IIa | 66 (14) |
| IIb | 50 (11) |
|
| |
| III | 163 (34) |
| IIIa | 129 (27) |
| IIIb | 34 (7) |
∗The stage was clinically assessed in 97 patients (20%), while in the remaining 375 cases (80%), it was pathologically assessed.
Table 3 summarises the results of the cross-validation analysis.
Table 3.
Results of the cross-validation analysis.
| Training (n =378) | Validation (n =94) | |||||
|---|---|---|---|---|---|---|
| Accuracy (%) | Recall (%) | Specificity (%) | Accuracy (%) | Recall (%) | Specificity (%) | |
| CV 1 | 82.6 | 91.2 | 56.8 | 76.6 | 84.3 | 54.2 |
| CV 2 | 87.3 | 95.8 | 62.1 | 76.6 | 91.4 | 33.3 |
| CV 3 | 78.4 | 91.2 | 40.6 | 81.9 | 90.0 | 58.3 |
| CV 4 | 85.0 | 88.3 | 75.0 | 73.4 | 81.7 | 47.8 |
| CV 5 | 79.7 | 94.0 | 36.8 | 78.7 | 94.3 | 33.3 |
| Mean | 82.6 | 92.1 | 53.4 | 77.4 | 88.3 | 45.4 |
CV: cross validation.
The algorithm developed and tested in the present work achieved an accuracy of 69%, a recall of 70%, and a specificity of 67% in the test set, for the identification of T1-T2 and T3-T4 lung cancer, in the final model analysis. The AUC was 0.83, 0.73, and 0.68 in the training, validation, and test sets, respectively. Results of all metrics for the final model in the training, validation, and test sets are reported in Table 4.
Table 4.
Results of the final model.
| Accuracy (%) | Recall (%) | Specificity (%) | Area under the curve | |
|---|---|---|---|---|
| Training (n =303) | 86.8 | 85.9 | 89.5 | 0.83 |
| Validation (n =75) | 69.3 | 76.8 | 47.4 | 0.73 |
| Test (n =74) | 69.1 | 70.0 | 66.7 | 0.68 |
Figure 2 shows examples of patients classified by CNN as T1-T2 and T3-T4.
Figure 2.
Clinical examples of the network classification. Axial CT images (a, b) of a patient affected by lung adenocarcinoma pT1bN0 ((a) zoom on the lesion); the developed neural network classified the new input image ((b) green square on one of the slices in which the lesion is visible) as belonging to the class T1-T2 with 99% probability. Axial CT images (c, d) of a patient affected by lung adenocarcinoma pT4N1 ((c) zoom on the lesion); the developed neural network classified the new input image ((d) green square on one of the slices in which the lesion is visible) as belonging to the class T3-T4 with 92% probability.
4. Discussion
The algorithm developed and tested in the present work achieved an accuracy of 87%, 69%, and 69% in the training, validation, and test sets, respectively, for the classification of T1-T2 and T3-T4 lung cancer. The lower performances of the CNN in the validation and test sets compared with the training dataset are probably related to the sample size (n=75, n=94, and n=303, respectively).
The TNM staging system is the reference method for prognostication and treatment decision-making in cancer patients, including in those with lung cancer. Pathological assessment is considered the gold standard. However, patients with tumours of the same stage can experience variations in the incidence of recurrence and survival. These variations may be related to tumour biology and other factors, including potential differences in the pathological stage at diagnosis and at neoadjuvant treatment [9] and the possible effect of the number of surgically removed lymph nodes on the N-parameter [10]. Finally, pathological TNM staging is not feasible in advanced stages. Medical imaging is the reference when pathological assessment is not feasible. Hybrid FDG-PET/CT is noninvasive and provides whole-body assessment, resulting essential in baseline lung cancer staging. CT scan is the cornerstone of lung cancer imaging providing all the necessary information needed for clinical T staging. FDG-PET/CT outperforms other modalities in terms of diagnostic accuracy in mediastinal nodal involvement and extrathoracic disease detection [11, 12].
In lung CT imaging, deep-learning approaches have been used to detect lung nodules [3, 13], to segment [2], and to classify them as benign or malignant [5, 14]. Some preliminary data are available on radiation treatment planning [15] and outcome prediction [16]. Recently, the value of CNNs for classification of mediastinal lymph node metastasis on FDG-PET/CT images has been investigated in non-small cell lung cancer. CNNs proved promising in identifying lymph node involvement, with higher sensitivities but lower specificities compared with doctors [17].
The present work reports an innovative application of deep-learning providing an automated staging classification. Some limitations of this study have to be acknowledged. Firstly, the limited number of patients precluded the design of a network for a classification task with four outputs (T1, T2, T3, and T4). Moreover, better performance is expected in larger datasets. In future work, we plan to include a larger number of cases in a multicentre framework. Secondly, the reference to define the T-parameter was based on the pathological assessment in surgically treated patients (80% of cases), while in patients not suitable for surgery, clinical assessment was used as the standard. The choice to use the pathological staging when available instead of the clinical one was aimed to achieve higher algorithm performance. Finally, we did not test separately CT and PET image modality. It could be speculated that, in this first step investigation, the CNN nodes were mostly activated by the weighted features coming from the CT component, while for the N- and M-parameters, PET can be supposed to give a major contribution. The comparison between the performances of a CNN trained using either CT or PET images will be considered in future studies. However, our final objective is to develop a whole-body assessment tool processing both CT and PET images together.
In conclusion, the key result in the present preliminary investigation is the feasibility and promising performance of CNNs in assessing the T-parameter in lung cancer. The developed tool is able to provide in few minutes from baseline PET/CT the probability of a patient being T1-T2 or T3-T4. Further investigations are needed to develop robust algorithms for a complete TNM assessment. Compared with radiomics, CNNs have the advantage of eliminating the need for tumour segmentation, feature calculation and selection, which are even more critical issues in small lesions. Still, the possibility of a complementary role of radiomics and artificial intelligence techniques should be addressed. Moreover, improvement in risk stratification is foreseen with the incorporation of patients' clinical features in the neural network algorithm.
Acknowledgments
M. Kirienko was supported by the AIRC (Italian Association for Cancer Research) scholarship funded by the grant won by AC (IG-2016-18585).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Additional Points
Clinical Relevance. The development of an automated system based on neural networks, that is, the ability to perform a complete stage and risk assessment will ensure more accurate and reproducible use of imaging data and allow deeper integration of this information in the therapeutic plan for each individual patient.
Ethical Approval
The study was approved by the Institutional Ethics Committee. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflicts of Interest
A. Chiti received speaker honoraria from General Electric, Blue Earth Diagnostics, and Sirtex Medical System, acted as the scientific advisor for Blue Earth Diagnostics and Advanced Accelerator Applications, and benefited from an unconditional grant from Sanofi to Humanitas University. All honoraria and grants are outside the scope of the submitted work. All other authors have no conflicts of interest.
Authors' Contributions
MS, MK, and AC conceived the idea of the study; MS, MK, GS, and SM reviewed and contributed to the image processing; EV collected the data and selected the patients; GS, SM, and LA developed the network; LA and AR reviewed the images and selected the patients; MS and MK wrote the manuscript; all the authors discussed the results and commented on the manuscript. Margarita Kirienko and Martina Sollini contributed equally to this work.
References
- 1.Chartrand G., Cheng P. M., Vorontsov E., et al. Deep learning: a primer for radiologists. RadioGraphics. 2017;37(7):2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 2.Wang S., Zhou M., Liu Z., et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Medical Image Analysis. 2017;40:172–183. doi: 10.1016/j.media.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setio A. A. A., Ciompi F., Litjens G., et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Transactions on Medical Imaging. 2016;35(5):1160–1169. doi: 10.1109/tmi.2016.2536809. [DOI] [PubMed] [Google Scholar]
- 4.Dou Q., Chen H., Yu L., Qin J., Heng P. A. Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Transactions on Biomedical Engineering. 2017;64:1558–1567. doi: 10.1109/tbme.2016.2613502. [DOI] [PubMed] [Google Scholar]
- 5.Liao F., Liang M., Li Z., Hu X., Song S. 2017. Evaluate the malignancy of pulmonary nodules using the 3D deep leaky noisy-or network. http://arxiv.org/abs/1711.08324. [DOI] [PubMed] [Google Scholar]
- 6.Kirienko M., Cozzi L., Rossi A., et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. European Journal of Nuclear Medicine and Molecular Imaging. 2018;45(10):1649–1660. doi: 10.1007/s00259-018-3987-2. [DOI] [PubMed] [Google Scholar]
- 7.Kingma D. P., Ba J. Adam: a method for stochastic optimization. Proceedings of 3rd International Conference for Learning Representations; December 2014; San Diego, CA, USA. [Google Scholar]
- 8.Edge S., Byrd D., Compton C., Fritz A., Greene F., Trotti A. AJCC Cancer Staging Manual. 7th. Heidelberg, Germany: Springer; 2010. [Google Scholar]
- 9.Hellmann M. D., Chaft J. E., William W. N., et al. Pathological response after neoadjuvant chemotherapy in resectable non-small-cell lung cancers: proposal for the use of major pathological response as a surrogate endpoint. Lancet Oncology. 2014;15(1):e42–e50. doi: 10.1016/s1470-2045(13)70334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velez-Cubian F. O., Rodriguez K. L., Thau M. R., et al. Efficacy of lymph node dissection during robotic-assisted lobectomy for non-small cell lung cancer: retrospective review of 159 consecutive cases. Journal of Thoracic Disease. 2016;8(9):2454–2463. doi: 10.21037/jtd.2016.08.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purandare N. C., Rangarajan V. Imaging of lung cancer: implications on staging and management. Indian Journal of Radiology and Imaging. 2015;25(2):109–120. doi: 10.4103/0971-3026.155831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y., Li P., Zhang H., et al. Diagnostic value of fluorine 18 fluorodeoxyglucose positron emission tomography/computed tomography for the detection of metastases in non-small-cell lung cancer patients. International Journal of Cancer. 2013;132(2):E37–E47. doi: 10.1002/ijc.27779. [DOI] [PubMed] [Google Scholar]
- 13.Jin H., Li Z., Tong R., Lin L. A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Medical Physics. 2018;45(5):2097–2107. doi: 10.1002/mp.12846. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X., Liu L., Qi S., Teng Y., Li J., Qian W. Agile convolutional neural network for pulmonary nodule classification using CT images. International Journal of Computer Assisted Radiology and Surgery. 2018;13(4):585–595. doi: 10.1007/s11548-017-1696-0. [DOI] [PubMed] [Google Scholar]
- 15.Tseng H.-H., Luo Y., Cui S., Chien J.-T., Ten Haken R. K., Naqa I. E. l. Deep reinforcement learning for automated radiation adaptation in lung cancer. Medical Physics. 2017;44(12):6690–6705. doi: 10.1002/mp.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul R., Hawkins S. H., Balagurunathan Y., et al. Deep feature transfer learning in combination with traditional features predicts survival among patients with lung adenocarcinoma. Tomography. 2016;2(4):388–395. doi: 10.18383/j.tom.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Zhou Z., Li Y., et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Research. 2017;7(1):p. 11. doi: 10.1186/s13550-017-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.