Abstract
Pin1 belongs to the family of the peptidyl-prolyl cis-trans isomerase (PPIase), which is a class of enzymes that catalyze the cis/trans isomerization of the Proline residue. Pin1 is unique and only catalyzes the phosphorylated Serine/Threonine-Proline (S/T-P) motifs of a subset of proteins. Since the discovery of Pin1 as a key protein in cell cycle regulation, it has been implicated in numerous diseases, ranging from cancer to neurodegenerative diseases. The main features of Pin1 lies in its two main domains: the WW (two conserved tryptophan) domain and the PPIase domain. Despite extensive studies trying to understand the mechanisms of Pin1 functions, how these two domains contribute to the biological roles of Pin1 in cellular signaling requires more investigations. The WW domain of Pin1 is known to have a higher affinity to its substrate than that of the PPIase domain. Yet, the WW domain seems to prefer the trans configuration of phosphorylated S/T-P motif, while the PPIase catalyzes the cis to trans isomerasion. Such contradicting information has generated much confusion as to the actual mechanism of Pin1 function. In addition, dynamic allostery has been suggested to be important for Pin1 function. Henceforth, in this review, we will be looking at the progress made in understanding the function of Pin1, and how these understandings can aid us in overcoming the diseases implicated by Pin1 such as cancer during drug development.
Keywords: Pin1, WW domain, peptidyl-prolyl cis/trans isomerase (PPIase), phosphorylation, interdomain communication, cancer target, drug development
Introduction
Posttranslational modifications (PTMs) introduce diversity to the functions of many proteins in the cellular proteome. This allows the cells to exert more biological functions with lesser number of proteins. There exist many types of PTMs, of which, the reversible phosphorylation is widely studied for its role in regulating many signaling cascade (1). Initially, phosphorylation of a protein was thought to be the final step in activating or inhibiting signaling cascades until the discovery of the group IV WW domain proteins, notably the Pin1 protein (2, 3). Pin1 possesses two major domains, namely the WW domain and the peptidyl-prolyl cis/trans isomerase (PPIase) domain (3). The WW domain consists of two highly conserved tryptophan amino acids separated by ~20–22 residues, allowing Pin1 to bind to the phosphorylated consensus site ser/thr-pro (pS/T-P) motif (3–5). This allows Pin1 to exert its molecular function as an isomerase via the PPIase domain, leading to the cis/trans conversion of its substrate to elicit the intended biological outcomes (2, 6). This additional modification on the phosphorylation sites of multiple Pin1 substrates has provided an alternate view on how signaling cascades could be regulated under different cellular conditions (7, 8).
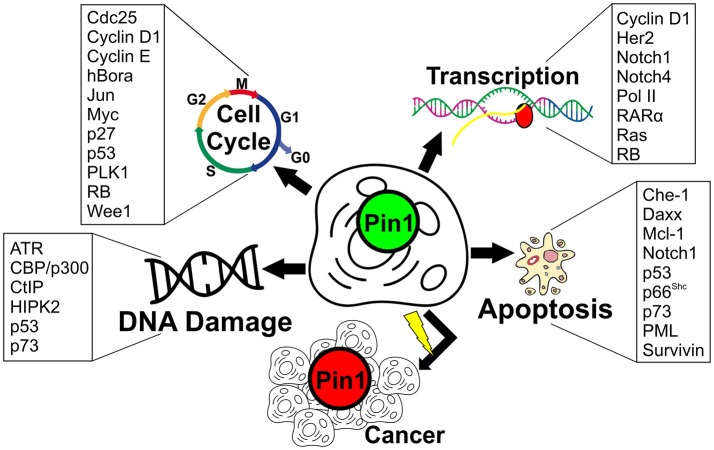
So far, Pin1 has been extensively studied for its role in various cellular functions, particularly in cell cycle regulation (2, 6, 9–11). Besides its importance in cell cycle progression, Pin1 has been further implicated in many biological processes such as embryonic development, cell motility, immune responses, gene transcription, and apoptosis (12–17). Due to its diverse role, perturbation to Pin1 expression levels has been implicated in many diseases such as cancer, and neurodegenerative diseases like Parkinson's and Alzheimer's disease (10, 15). Especially in the case of cancer, many functional substrates of Pin1 have been identified to potentially contribute to the manifestation of cancer via various biological processes as summarized in Figure 1. As such, Pin1 has been identified as an important target for therapeutic intervention for these diseases (48–50). Indeed, many Pin1 inhibitors have been identified, with the most recent ones being API-1, KPT-6566, compound 1 and 8 (51–54). Unfortunately, there remains much to do before any breakthrough in targeting Pin1 for the treatment of various diseases is achieved. This stems from our limited understanding of Pin1 mechanism in these biological processes, and how the unique WW and PPIase domains of Pin1 work to elicit its function.
Figure 1.
Pin1 substrates belonging to various cellular processes linked to cancer. Cell cycle – Cdc25 (18): Cell division cycle 25; Cyclin D1 (19–21); Cyclin E (22); hBora (23): Human protein aurora borealis; Jun (19, 24): Transcription factor AP-1; Myc (25): Myc proto-oncogene protein; p27 (26); p53 (27, 28); PLK1 (6, 29): Polo-like kinase 1; RB (30): Retinoblastoma-associated protein; Wee1 (31). Transcription – Cyclin D1 (19); Her2 (32, 33): Human epidermal growth factor receptor 2; Notch1 (34, 35): Neurogenic locus notch homolog protein 1; Notch4 (35): Neurogenic locus notch homolog protein 4; Pol II (36): RNA polymerase II; RARα (37): Retinoic acid receptor alpha; Ras (38); RB (30). DNA damage-ATR (39): Ataxia telangiectasia and Rad3-related protein; CBP/p300 (24): CREB-binding protein/p300; CtIP (40): C-terminal-binding protein (CtBP)-interacting protein; HIPK2 (41): Homeodomain-interacting protein kinase 2; p53 (27, 28); p73 (42). Apoptosis-Che-1 (43): Apoptosis-antagonizing transcription factor (AATF); Daxx (44): Death domain-associated protein 6; Mcl-1 (45): Induced myeloid leukemia cell differentiation protein; Notch1 (34, 35); p53 (27, 28); p66Shc; (46):66 kDa proto-oncogene Src homologous-collagen homolog (Shc) adaptor protein; p73 (42); PML (47): Promyelocytic leukemia protein; Survivin (47).
In addition, there have been many structural and protein dynamic studies to understand how Pin1 could target its biological substrates to exert its intrinsic cis/trans isomeric activity. This would then trigger its intended downstream cellular signals and effects. Many models have been introduced to explain the molecular mechanism of how Pin1 exert its catalytic activity. However, there has not been a model in agreement to truly explain how Pin1 acts on its biological substrates. Therefore, in this review, we will highlight the progress of Pin1 research in elucidating the actual mechanism of Pin1, with an emphasis on the structural importance of Pin1 on its function, and how the perturbation to this fundamental structure could explain for its roles in diseases such as cancer. We will also discuss how these structural features could be used in the drug development of Pin1 inhibitors.
Detailed structural features of human pin1
The human Pin1 consists of a total of 163 amino acid residues that forms two distinct domains, the WW domain and the PPIase domain as mentioned previously (Figure 2). The WW domain spans the first 39 amino acid residues of the Pin1 protein, while the PPIase domain spans residues 50–163. Both domains are known to be able to bind to the pS/T-P motifs, with the WW domain binding being noncatalytic in nature, while the PPIase domain possesses the sole catalytic site of the cis/trans isomerase activity in Pin1 (55–57).
Figure 2.
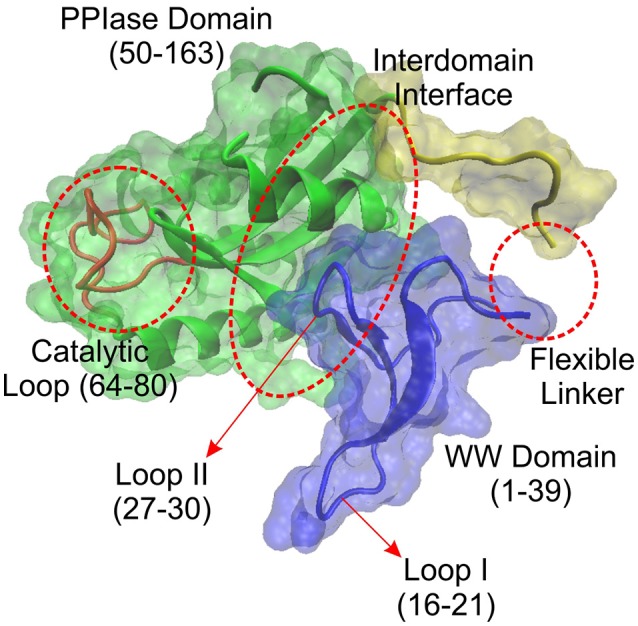
Molecular structure of human Pin1 (PDB: 1PIN) without ligand binding. Pin1 is predominantly made up of two major domains: WW domain and PPIase domain. Within the WW domain, there exist two loops, namely Loop I and II. As for the PPIase domain, there is a catalytic loop at the catalytic site. There also exists PPIase binding domain where it could bind to the pS/T-P motif apart from the WW domain. In addition, existing between the two domains, there lies an interdomain interface that has been found to be important in Pin1 allosteric regulation.
Apart from the two major domains, the Pin1 protein also consists of a flexible linker region consisting of approximately 17 amino acid residues spanning from residue E35 to A53 (58). In addition, within the WW domain, there exist two loops. Loop I is situated at residue S16 to R21, and Loop II at residue H27 to N30 (58). As for the PPIase domain, it also displays two main components. They are the substrate recognition segment (residue K63 to R80) where residues K63, R68, and R69 create a positive charged phosphate-binding loop to facilitate binding to the pS/T-P motif (58, 59). The other important segment of the PPIase domain is the catalytic active site that consists of amino acid residues such as H59, S115, C113, L122, M130, F134, T152, and H157 (58, 60, 61).
Besides these characteristics of Pin1, there exist the presence of a domain interface within Pin1, consisting of amino acid residues I28, the WW domain Loop II (H27 to N30), and part of the PPIase domain (S138 to R142). This domain interface has been suggested to play an important role in interdomain communication that regulates the function of Pin1 upon substrate binding (60–62). These minor features of Pin1 suggest the complexity of Pin1 function upon substrate binding and are a potential reason why Pin1 can interact with a large and diverse number of biological substrates to regulate cellular functions. However, how these characteristics can work hand-in-hand to confer the functions of Pin1 on its substrates remain a largely unfinished work.
The proposed models of the WW domain and PPIase domain in pin1 substrate interactions
As the two main domains of Pin1 protein, much research has been carried out on both WW and PPIase domains to understand their role in the interaction of Pin1 with its biological substrates. It has been reported in the early stages of Pin1 research that the WW domain has a ten-fold higher binding affinity with phosphorylated peptides in vitro as compared to the PPIase domain (3, 63). This property of the WW domain in Pin1 has led to a proposition that its major role is to aid Pin1 in specific targeting of its substrate, as well as to increase local concentration of Pin1 substrate to exert its catalytic function (7, 64). This scenario gave rise to a few proposed models of Pin1 substrate interactions. The first proposed model is the sequential binding model of Pin1 (Figure 3A) and remains widely accepted. In this model, the WW domain would first bind to the pS/T-P motif on its target substrate. This allows the PPIase domain to bind with another pS/T-P motif that is present on the same target substrate. Alternatively, the binding of the WW domain would lead to structural conformation change that allows the PPIase domain to displace the WW domain to bind to the same pS/T-P motif to exert its isomeric activity.
Figure 3.
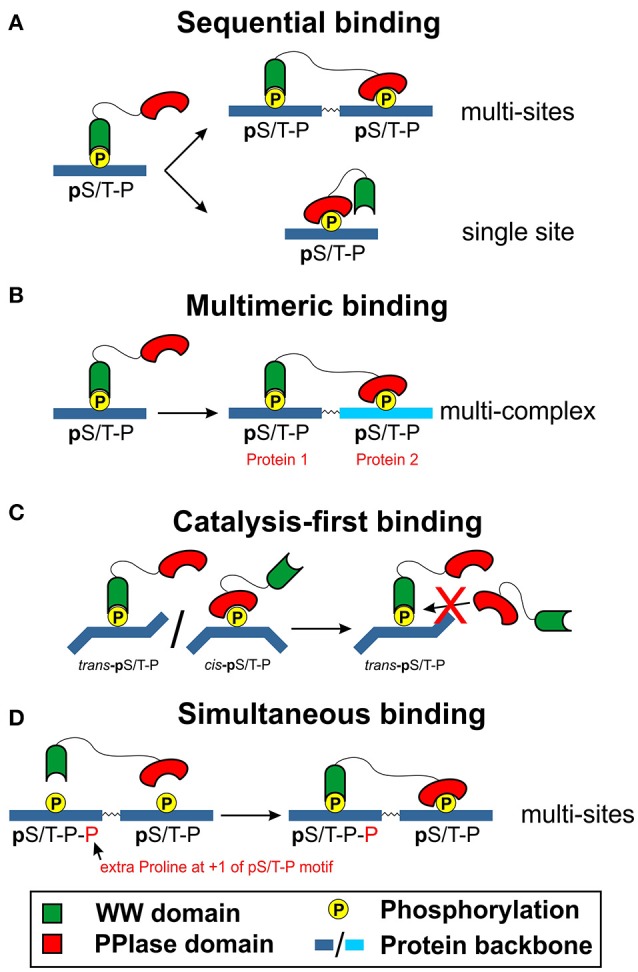
Proposed models of WW domain and PPIase domain interaction with the substrate of Pin1 [first highlighted by Innes et al. (65)]. Two decades of research has shed light on the potential mechanism as to how the WW and PPIase domains could be involved in substrate binding of Pin1. (A) Sequential binding model. The sequential binding model involves the initial binding of the WW domain of Pin1 before the PPIase domain could bind to the same pS/T-P motif or another pS/T-P motif on the same protein. (B) Multimeric binding model. The WW domain first binds to an active kinase with a pS/T-P motif before being brought close to a substrate of Pin1. The active kinase then phosphorylates S/T-P motif to allow the PPIase domain to bind to it to initiate the isomeric reaction. (C) Catalysis-first binding model. Existing trans pS/T-P motif will be bound to the WW domain of Pin1, while the PPIase domain could bind to the cis pS/T-P motif to catalyze the isomeric reaction to the trans configuration. Subsequently, the WW domain would then bind to the new trans pS/T-P motif to prevent the reverse conversion. (D) Simultaneous binding model. Both WW domain and PPIase domain can bind simultaneously, with the WW domain binding to pS/T-P-P motif and PPIase domain binding to the pS/T-P motif.
In the multimeric binding model (Figure 3B), it is applied in multi-protein complexes containing a Pin1 biological substrate and an active kinase that is present to phosphorylate the S/T-P motif of the Pin1 substrate (66). In this configuration, Pin1 first binds to the kinase at the pS/T-P motif via its WW domain. Subsequently, the active kinase would phosphorylate the S/T-P motif on the target substrate of Pin1. As Pin1 is already near its substrate, the PPIase domain would recognize and bind to the pS/T-P motif on the target substrate to exert its isomeric activity. This model is further supported by the identification of Pin1 interactors which are known kinases themselves as reflected in the IntAct Molecular Interaction database (67).
In another model termed the catalysis-first binding model (Figure 3C), it is believed that the phosphorylation of S/T-P motif on the target substrate for WW domain binding requires the PPIase function of Pin1 (57). This proposition was put forth by the observation that in all the known structures of the WW domain bound substrate peptides, the identified binding site is present in a trans configuration state (56, 57, 68). Moreover, a study conducted by Verdecia et al. (56) seems to suggest the strict preference of the wild type WW domain of Pin1 to bind to the trans configuration of its substrate peptide. Of interest, Pin1 also enhances the dephosphorylation activity of protein phosphatase such as PP2A, which too requires the pS/T-P motif to be in the trans configuration state. Therefore, the binding of the WW domain of Pin1 might be a stabilizing action for the Pin1 substrate to remain in its trans configuration state for dephosphorylation to occur (69, 70). In addition, a study by Namanja et al. (60) found the preference of WW domain binding to the trans but not the cis configuration of pS/T-P motif. Thus, if the WW domain prefers binding to a trans configured substrate peptide, the PPIase domain of Pin1 would first bind to the cis configured pS/T-P motif to catalyze the cis/trans isomerization to the trans configuration. This would lead to WW domain binding to the now trans configured pS/T-P motif to prevent the reverse isomerization to occur. This process would then allow the local concentration of the Pin1 PPIase to initiate further cis/trans isomerization to fast forward the propagation of downstream cellular signaling of the stabilized Pin1 substrate.
Indeed, previous studies have shown that the PPIase domain of Pin1 is able to bind to pS/T-P motif instead of just its WW domain (56, 58, 65). As mentioned earlier, this binding occurs at the substrate recognition segment, in particular, the phosphate-binding loop created by the three amino acid residues of K63, R68, and R69. Furthermore, a study done by Innes et al. (65) demonstrated the importance of this phosphate-binding loop in the target binding of Pin1 to initiate its catalytic isomerase function. This is despite the reported higher binding affinity of the WW domain of Pin1 and reinforced by the presence of PPIase domain-specific Pin1 inhibitors that do not interact with the WW domain (71). As such, the authors suggest that Pin1 could interact with its biological substrate via the simultaneous binding model (Figure 3D). In their study, they noticed that pS/T-P motifs that have an addition P residue in the +1 position, pS/T-P-P, seem to be targeted by the WW domain but not the PPIase domain of Pin1. This led them to believe that a substrate with multiple phosphate binding sites could allow for the simultaneous binding of Pin1 to its substrate. They observed this in the binding of Pin1 to Cdc25 and Serine/Threonine-protein kinase (PLK1). On the other hand, they identified that Pin1 binding via the PPIase domain is sufficient in proteins with only a single pS/T-P motif such as non-POU domain-containing octamer-binding (NONO) protein and splicing factor, proline-, and glutamine-rich (SFPQ) protein for its activity. As such, there seems to be two types of Pin1 interactors that requires either the binding of both WW and PPIase domains, or just the WW/PPIase domain alone.
These proposed four models of Pin1 interaction with its biological substrates for functional regulation might suggest two aspects. Firstly, there is still lack of concrete evidences to highlight which model(s) Pin1 may deploy for its interaction with its biological substrates. Secondly, these models demonstrate the potentially diverse interaction that Pin1 has on its already diverse biological interactors to play a role in the regulation of various biological functions. All these models do have their merits as well as potential doubts as to its suitability. For instant, the sequential binding model does fulfill the characteristics of the WW domain having a higher affinity of peptide binding to the pS/T-P motif than the PPIase domain. In proteins with multiple sites of pS/T-P motifs, this model could explain the use of the WW domain to localize the concentration of the PPIase domain for isomerization. However, for substrate with a single pS/T-P motif, the potential release of the WW domain from this motif, followed by the binding of the PPIase domain for catalytic activity does not seem to be energy favored. This is also highlighted by studies that we have mention previously (56, 57, 60, 68–70), where the WW domain favors the trans over cis configuration. As such, the PPIase isomerization would not have occurred with this sequential binding.
For multimeric binding to an active kinase via the WW domain of Pin1, the increased local concentration of Pin1 would allow the binding to its biological substrate in the vicinity to elicit its downstream biological processes. Furthermore, the pS/T-P motif of the kinase could be in a trans configuration state to allow for WW domain binding. In addition, the catalysis-first binding model could also be associated with the multimeric binding model where Pin1, bound to the active kinase via the WW domain, is brought to its target substrate by the active kinase. Subsequently, the active kinase would phosphorylate the Pin1 substrate, allowing for the PPIase domain of the Pin1 to bind and catalyze the cis-trans conformational change. Furthermore, the suggested simultaneous binding model could also fit the multimeric binding model, although this would occur on a single protein, instead of the proposed protein complex in the multimeric binding. Of interest, the binding of the WW domain in the trans configured pS/T-P motif could lead to conformational changes to the PPIase domain, increasing its binding capacity to the cis configured p-S/T-P motif and subsequently its catalytic efficiency. The potential role of interdomain communications between the WW domain and the PPIase domain of Pin1 has indeed been studied in recent years as discussed in the next section.
Interdomain communications between WW domain and PPIase domain of pin1
As highlighted earlier, Pin1 is made up of two distinct major domains in WW and PPIase domains. Besides these two domains, there exist an internal conduit of hydrophobic residues cluster that connects the interdomain interface and the catalytic site of the PPIase domain (60, 62). This has led to the suggestion of an allosteric mechanism present in Pin1. Classic allostery stems from the binding of a ligand to a part of a protein where it is distal to the active site. This binding would then lead to global conformation change of the protein, or localized changes to its active site, resulting in increased binding affinity of the active site or its enzymatic activities (72, 73). Recently, the concept of allostery, termed dynamic allostery, has been looked into to explain for the absence of structural conformation changes under allosteric effect of ligand binding on the main protein (74–76) since its first introduction more than three decades ago (77). This absence of structural change is replaced by the use of the protein's interdomain communications as demonstrated by computational studies (78, 79) and reviewed elsewhere (80). Indeed, a study by Behrsin et al. (59) showed that the WW domain is essential for the function of Pin1 isomeric activity in vivo, suggesting the presence of interdomain communication. In addition, studies conducted on Pin1 and tau interactions highlighted the need of WW domain interaction with the pS/T-P motif for the effective function of the PPIase activity (63, 81). Therefore, understanding the interdomain communication in Pin1, and how this could lead to the dynamic allostery of Pin1 has been of much focus in recent times.
Much work in understanding the presence of interdomain communication and dynamic allostery of Pin1 has been well studied using nuclear magnetic resonance (NMR) studies (82). As highlighted in a review by Sudol and Hunter (83), and studied by Verdecia et al. (56), both Cdc25c and FFpSPR [artificial substrate derived from peptide libraries screening for optimal PPIase efficiency (84)] have higher affinity to the WW domain of Pin1. This is also reflected in other Pin1 substrates that have been identified (66). This higher affinity of the WW domain for Pin1 substrate is contributed by the Loop I structure of this domain, where modification of the Loop I sequence led to the reduction of substrate binding affinity to Pin1 as demonstrated by Peng et al. (85) using the pCDC25C and FFpSPR peptides. Yet, by using a trans-locked inhibitor and a cis-locked inhibitor synthesized by Wang et al. (86, 87), Namanja et al. (60) found that the isolated PPIase domain has 2–4 times higher affinity to the cis-locked inhibitor than the full-length Pin1, while the trans-locked inhibitor can be bound by both domains as mentioned previously. This higher binding affinity of the PPIase domain without the WW domain suggests that the removal of the WW domain seems to remove the restriction imposed by the unbound WW domain on the PPIase domain for the cis configured pS/T-P motif. This observation clearly suggests a potential form of allostery present in Pin1.
Indeed, data obtained from studies by Namanja et al. (60, 62) showed that the changes in side chain dynamics instead of large structural changes are a means of allosteric communication within the PPIase domain (82). This was further supported using molecular dynamics (MD) simulations (79, 88–90). These changes in side chain dynamics could dictate the flexibility of the PPIase domain to allow for greater substrate binding or enzymatic activity. Base on this side chain dynamics study of Pin1, Peng and co-workers identified the I28 residue as an important amino acid within a conserved hydrophobic conduit of amino acid residues that is affected by substrate binding of Pin1 (62). The subsequent study based on the mutation of I28 to I28A by Wilson et al. (61) led to the discovery of interdomain communications governed by I28, the WW domain Loop II, and the PPIase domain residues S138 to R142. The authors highlighted the influence of this interdomain communications on the side chain dynamics of the conserved hydrophobic conduit region that would affect PPIase domain binding and catalytic activity. This study is further supported by a recent MD study from Barman and Hamelberg (91). They highlighted that the binding of the substrate compacts the WW domain closer to the catalytic domain via a hinge-like movement, an observation echoed by a more recent study done by Campitelli et al. (92) using their dynamic flexibility index analysis.
Collectively, these studies highlight the fact that the binding of WW domain could be crucial in transmitting an intra-protein signal to the PPIase domain and the catalytic site via the interdomain contact as highlighted previously. This transmission of signal could be the result of side chain dynamics of the conserved hydrophobic conduit region. This could subsequently lead to subtle hinge-shift mechanism induced by the bound WW domain that leads to improved PPIase domain binding and catalytic activity. Based on these evidences as well as the four binding models of Pin1 to its substrate, the multimeric and simultaneous binding models are the potential model of how Pin1 works in biological signaling cascades. This is due to the potential need of a bound WW domain for Pin1 binding and catalytic activity. Hence, having the WW domain bound to an active kinase or a simultaneous binding of a WW domain followed by the binding of the PPIase domain would support the presence of Pin1 interdomain allostery (Figure 4).
Figure 4.
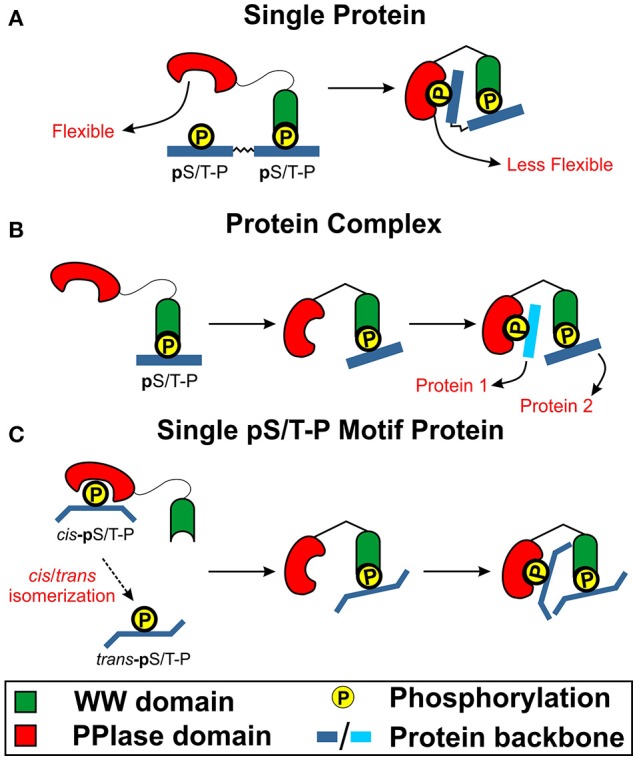
The role of interdomain communication in Pin1 function. Based on recent findings, WW domain could be required for initial binding to initiate conformation changes to the PPIase domain for its binding and catalytic function. (A) WW domain binds first to pS/T-P motif that leads to interdomain communication to trigger conformation change (decreased flexibility) that lead to PPIase binding to a distal pS/T-P motif on the same protein to initiate catalytic function. (B) WW domain first binds to the pS/T-P motif to initiate interdomain communication to trigger PPIase domain conformation change to bind a pS/T-P motif on another protein to induce catalytic function. (C) PPIase domain could first catalyze a few pS/T-P motifs from the cis to trans configuration. This would lead to WW domain binding and initiate interdomain communication to trigger PPIase conformation change. This would then increase the activity of PPIase domain activity to lead to more cis to trans isomerization of the pS/T-P motif of the same protein in the vicinity.
In contrast, there are known examples of Pin1 that only contains one half of the two domains structure. For instant, Pin1At, identified in the plant Arabidopsis thaliana, only contains the PPIase domain but not the WW domain as opposed to Pin1 (93). In addition, another single-domain Pin1, known as TbPin1, was identified from the parasite Trypanosoma brucei (94). TbPin1 is also known to only contain the PPIase domain. Despite the absence of the WW domain, Pin1At was enough to rescue the temperature-sensitive mutation of Pin1 homolog gene (ESS1/PTF1) in Saccharomyces cerevisiae from death (95). Furthermore, our lab previously showed that TbPin1 alone was also able to rescue the same temperature sensitive S. cerevisiae mutant from death (96). These studies highlight that the PPIase domain alone is enough to recover the loss of Pin1 function, brining into question the importance of WW domain in Pin1 function. In hindsight, the WW domain of Pin1 could possess an alternative role in maintaining Pin1 function and could be seen in a complementary role to accelerate the effect of Pin1 function. As discussed earlier in the review, the preference of WW domain for the trans-configured substrate of Pin1 potentially suggest that the WW domain of Pin1 seeks to stabilize the trans population of the target substrates, leading to increased concentration of the PPIase domain in the vicinity and accelerating the effect of Pin1. Nonetheless, the highlighted studies of TbPin1 and PinAt needs to be replicated in mammals before conclusions could be made to obsolete the role of WW domain in Pin1 function.
Implications of pin1 as a therapeutic target in cancer
As mentioned previously, Pin1 has been extensively studied due to its diverse involvement in many signaling pathways that has many implications in various diseases, with great emphasis on cancer and neurodegenerative diseases. Despite many studies done on understanding the mechanism of Pin1 interactions with its biological substrates, there remains a gap as to how the understanding of Pin1 mechanism could lead to clinical translation. The most obvious application would be to identify or synthesize inhibitors that could block the functions of Pin1. However, the fact that Pin1 has so many interactors brings the problem of off-target effects. Indeed, the regulation of Pin1 in cancer against that in neurodegenerative diseases is inversely related (97). Therefore, targeting just Pin1 might not be specific enough to reduce the pathogenesis of these diseases.
The development of Pin1 inhibitors has remained a challenge in the field of cancer studies as many known inhibitors of Pin1 remains unspecific enough to only block the effects of Pin1 despite a number of Pin1 inhibitors already found (49, 98, 99). Yet, we cannot rule out the fact that evidences point to the potential key role of Pin1 in tumourigenesis (49, 99, 100). As such, there are continued efforts to identify new and novel Pin1 inhibitors. This is observed from recent efforts to identify various types of Pin1 inhibitors that targets the PPIase catalytic domain (51–53, 101, 102). For instant, a study by Pu et al. (51) identified a chemically synthesized small molecule API-1 that has an Pin1 inhibition concentration (IC50) of 72.3 nM, which is 100× less than the IC50 of the first Pin1 inhibitor juglone. API-1 was found to bind to the PPIase domain of Pin1. The authors went on to show that API-1 was able to suppress the proliferation of hepatocellular carcinoma cells as well as inhibiting the tumor growth of xenografted mice. These evidences demonstrated the therapeutic value of API-1 as a cancer drug candidate. Another study by Cui et al. (102) demonstrated the use of a novel strategy to develop anticancer agents by inhibiting Pin1 activity via the synthesis of various pyrimidine derivatives. The authors were able to identify four potential Pin1 inhibitors with IC50 of <3 μM. Furthermore, this type of drug is only the second covalent binding Pin1 inhibitors to the phosphate-binding loop in the PPIase domain besides juglone. These studies clearly highlight the potential therapeutic value of Pin1 as a cancer drug target. Unfortunately, no drugs thus far has reached clinical trials (49, 98).
Of interest, most drugs identified thus far are mainly targeting the PPIase domain, especially the catalytic domain, with few drug candidates found to target the WW domain (98) as summarized in the list of known Pin1 inhibitors discovered thus far in Table 1. For instant, a series of studies conducted by Murray's group (107, 108, 115) set out to identify novel Pin1 inhibitors that are non-phosphorylated small molecule inhibitors that target the Pin1 active site at the PPIase domain using a structure-based design approach. They focused on finding a Pin1 inhibitor that binds to the phosphate-binding loop and were successful in first identifying benzothiophene with sub-micromolar inhibitory activity on Pin1 (107, 108). Unfortunately, this class of inhibitor showed low binding affinity to Pin1 when tested with whole cell assay. This led to the authors to suggest that its low cellular permeability could be the cause of such poor in vivo binding affinity to Pin1. The authors went on to improve the permeability of the benzothiophene class of drug to eventually yield dihydrothiazoles as a potent Pin1 inhibitor with low micromolar inhibitory activity toward cancer cell proliferation (115). However, the improvement of permeability led to reduced Pin1 inhibitory potency. This brings a question whether cellular permeability could be major limitation in their development of these structure-based Pin1 inhibitors targeting the PPIase domain.
Table 1.
Selected known Pin1 inhibitors shown to block Pin1 function by targeting various part of Pin1 protein [adapted from Zhou and Lu (49)].
| Inhibition Site | Pin1 inhibitor | Mode of discovery | IC50 | Mechanism of action | System tested | Limitation(s) |
|---|---|---|---|---|---|---|
| Pin1 active site | Juglone (103) | Low-throughput enzymatic (PPIase) assay | - | Covalent modification of Cys in the active site | in vitro | Low specificity |
| PiB (104) | Low-throughput enzymatic (PPIase) assay | 1.5 μM | - | in vitro; cell lines | Little evidence of Pin1 binding; insoluble in DMSO | |
| Pepticinnamin analogs (104) | Combinatorial synthesis | 600 nM | - | in vitro | Inconclusive evidence of Pin1 binding | |
| Cis-locked alkene peptidomimetics (87) | Structure-based design | 1.5 μM | Bind to Pin1 active site via substrate mimicking | in vitro; cell lines | - | |
| D-peptide inhibitor such as Ac-Phe-D-Thr(PO3H2)-Pip-Nal-Gln-NH2 (68, 105) | Solid phase peptide library synthesis | As low as 1 nM | Competitive inhibitor of Pin1 active site | in vitro; cell lines | Inactive in cell lines | |
| Aryl indanyl ketones (106) | Structure-based design | As low as 200 nM | Reversible inhibitor of Pin1 active site undergoing “twisted-amide” transition state | in vitro; cell lines | Binding to Pin1 not as well as hypothesized | |
| Benzothiophene (107, 108) | Structure-based design | 6 nM | Binds to Pin1 active site with high specificity | in vitro; cell lines | Potential low permeability; inactive in cell lines | |
| Phenyl imidazoles (109, 110) | Structure-based design | 830 nM | Binds to Pin1 active site | in vitro; cell lines | Some variant inactive in cell lines | |
| ATRA (all trans retinoic acid) (111) | High-throughput mechanism-based screening | 800 nM | Binds to Pin1 active site via substrate mimicking | in vitro; cell lines; mouse models; APL human patients | Moderate efficacy in humans; short half-life in humans | |
| KPT-6566 (52) | High-throughput structural- and mechanism-based screening | 1.2 μM | Covalent binding to Pin1 active site at C113 specifically | in vitro; cell lines | - | |
| Pyrimidine derivatives (102) | In-house library screening | As low as 1.68 μM | Covalent binding to the binding pocket of Pin1 active site | in vitro; cell lines | - | |
| PPIase domain | Dipentamethylene thiauram monosulfide (112) | Protease coupled enzymatic (PPIase) assay | 50 nM | Competitive inhibitor of Pin1 PPIase domain | in vitro; cell lines | Possible low specificity |
| Halogenated phenyl-isothiazolone TME-001 (113) | Real-time fluorescence detection method | 6.1 μM | Competitive inhibitor of Pin1 PPIase domain | in vitro; cell lines | Possible low specificity | |
| Cyclic peptide inhibitor Cys-Arg-Tyr-Pro- Glu-Val-Glu-Ile-Cys (113) | Phage display screening | 500 nM | Competitive inhibitor of Pin1 PPIase domain | in vitro | Cannot be used to inhibit intracellular Pin1 activity | |
| API-1 (51) | Computer-aided high-throughput virtual screening | 72.3 nM | Binds to Pin1 PPIase domain specifically | in vitro; cell lines; mouse models | - | |
| WW domain | EGCG (epigallo-catechin-3-gallate) (114) | Phenotypic association | 20 μM | Bind to both WW and PPIase domains | in vitro; cell lines; mouse models | No reports of inactivation on isolated PPIase domain |
Of importance, these studies only made use of the PPIase domain as the main consideration in designing of their structure-based Pin1 inhibitor. The authors did highlight some important points supporting intra-structure dynamics of the whole Pin1. Firstly, they noticed that despite the rich hydrogen bonding potential within the phosphate-binding loop in the PPIase domain of Pin1, hydrophobic interactions were central to benzothiophene binding to Pin1. Secondly, they also highlighted the presence of H-bond formation beyond the phosphate-binding loop that could be important for the Pin1 inhibitor binding infinity. Lastly, they highlighted the high degree of flexibility in Pin1 active site interaction with the inhibitors. All these observations do align with our previous discussion where interdomain communications between WW and PPIase domains could affect the side chain dynamics on the conserved hydrophobic conduit region and alters PPIase domain binding affinity (61, 91, 92). Moreover, we previously mentioned that the removal of the WW domain could alleviate the restriction imposed by the unbound WW domain on the PPIase domain for the cis configured pS/T-P motif (60). Henceforth, in line with these observations, much remains to be done to utilize the information from the studies of the actual roles by the WW and PPIase domains of Pin1, including the potential impact of interdomain communication could play in future drug screening for Pin1 inhibitors.
As suggested from all the studies on the two domains of Pin1 covered in this review, the WW domain binding displays a potentially important role in driving the signaling cascade triggered by Pin1 catalytic function. To advance our development of promising Pin1 inhibitors for treatment of diseases such as cancer, there are important considerations to be taken note of. Much interest has been put forth on identifying Pin1 inhibitors that target the PPIase domain as well as its active site. As suggested in this review, the binding affinity of Pin1 inhibitors could be affected by the substrate that is bound to the WW domain. Therefore, binding affinity studies for Pin1 inhibitors screening should consider the substrate effect upon its binding to the WW domain, and how this effect could alter the Pin1 inhibitors binding to the PPIase domain or its active site. Of interest, a very recent study by Momin et al. (116) using MD analysis suggests that substrate sequence can influence the eventual outcome of Pin1 function by differential triggering of Pin1 allosteric changes after substrate binding. This is explained by the substrate sequence-dependent allostery that affects the type of residue to residue contact within the interdomain interface. This led to the authors to suggest that drugs targeting the specific geometry of the interdomain interface at a specific Pin1 substrate binding could lead to better treatments of specific diseases. Whether these MD simulations could be translated to in vivo circumstances are yet to be determined and could be explored in future drug development for Pin1 inhibitor.
This potential substrate-dependent Pin1 activity could be important in future studies. This is highlighted in the lack of depth in the field of Pin1 research thus far. Most studies done on Pin1 mechanism focuses on just a few biological substrates of Pin1. As suggested in this review, the vast repertoire of Pin1 functional substrates and the different biological processes it could affect do point to the fact that Pin1 might behave differently in the presence of different substrate. This difference in behavior could affect its structural dynamics that makes identifying a functional Pin1 inhibitor difficult for different types of diseases. Therefore, to advance the research of the role of Pin1 in the pathogenesis of various diseases, more emphasis on the diversity of Pin1 substrates being tested must be done. As mentioned previously, Pin1 inhibitor screening could be done in the presence of its effector substrate to evaluate the efficacy of the Pin1 inhibitor in affecting the phenotype of the disease being studied. In addition, studies looking into how Pin1 cooperation with kinase in exerting its isomeric activity could be conducted to investigate if the proposed multimeric model holds true, and how the subsequent findings could affect the considerations when developing Pin1 inhibitors. Indeed, studies thus far do suggest that Pin1 is not a lone ranger, and it always needs a helping hand to exert its function. To advance the development of drugs to highlight the impact of Pin1 as a cancer therapy target, both domains should be considered during the drug development process for the field to realize the potential of Pin1 as a cancer therapeutic target.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to put forth their thanks to all the members of Y-CL's laboratory for their valuable suggestion and discussion. This work is financially supported by the Ministry of Education (MOE), Singapore Tier 2 (R-154-000-A61-112) and Tier1 (A51-114) grants awarded to Y-CL.
References
- 1.Seo J, Lee K-J. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol. (2004) 37:35–44. 10.5483/BMBRep.2004.37.1.035 [DOI] [PubMed] [Google Scholar]
- 2.Ping Lu K, Hanes SD, Hunter T. A human peptidyl–prolyl isomerase essential for regulation of mitosis. Nature (1996) 380:544–7. 10.1038/380544a0 [DOI] [PubMed] [Google Scholar]
- 3.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science (1999) 283:1325–8. [DOI] [PubMed] [Google Scholar]
- 4.Rotin D. WW (WWP) domains: from structure to function. Curr Top Microbiol Immunol. (1998) 228:115–33. [DOI] [PubMed] [Google Scholar]
- 5.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CSH, Yu J, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. (2005) 25:7092–106. 10.1128/MCB.25.16.7092-7106.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. (1998) 12:706–20. 10.1101/gad.12.5.706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol (2002) 12:164–72. 10.1016/S0962-8924(02)02253-5 [DOI] [PubMed] [Google Scholar]
- 8.Liou Y-C, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. (2011) 36:501–14. 10.1016/j.tibs.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu KP. Phosphorylation-dependent prolyl isomerization: a novel cell cycle regulatory mechanism. Prog Cell Cycle Res. (2000) 4:83–96. 10.1007/978-1-4615-4253-7_8 [DOI] [PubMed] [Google Scholar]
- 10.Yeh ES. Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer (2007) 7:381–8. 10.1038/nrc2107 [DOI] [PubMed] [Google Scholar]
- 11.Lin C-H, Li H-Y, Lee Y-C, Calkins MJ, Lee K-H, Yang C-N, et al. Landscape of Pin1 in the cell cycle. Exp Biol Med. (2015) 240:403–8. 10.1177/1535370215570829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. (2007) 3:619–29. 10.1038/nchembio.2007.35 [DOI] [PubMed] [Google Scholar]
- 13.Becker EBE. Bonni A. Pin1 in neuronal apoptosis. Cell Cycle (2007) 6:1332–5. 10.4161/cc.6.11.4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esnault S, Shen Z-J, Malter JS. Pinning down signaling in the immune system: the role of the peptidyl-prolyl isomerase Pin1 in immune cell function. Crit Rev Immunol. (2008) 28:45–60. 10.1615/CritRevImmunol.v28.i1.30 [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Uchida C, Shin R-W, Shimazaki K, Uchida T. Prolyl isomerase, Pin1: new findings of post-translational modifications and physiological substrates in cancer, asthma and Alzheimer's disease. Cell Mol Life Sci. (2008) 65:359–75. 10.1007/s00018-007-7270-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuura I, Chiang K-N, Lai C-Y, He D, Wang G, Ramkumar R, Uchida T, Ryo A, Lu K, Liu F. Pin1 promotes transforming growth factor-beta-induced migration and invasion. J Biol Chem. (2010) 285:1754–64. 10.1074/jbc.M109.063826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanes SD. Prolyl isomerases in gene transcription. Biochim Biophys Acta - Gen Subj (2015) 1850:2017–034. 10.1016/j.bbagen.2014.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crenshaw DG, Yang J, Means AR, Kornbluth S. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J (1998) 17:1315–27. 10.1093/emboj/17.5.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. (2001) 20:3459–72. 10.1093/emboj/20.13.3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liou Y-C, Ryo A, Huang H-K, Lu P-J, Bronson R, Fujimori F, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci USA. (2002) 99:1335–40. 10.1073/pnas.032404099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Wang S, Zhu T, Zhou J, Xu Q, Lu Y, et al. Pin1 contributes to cervical tumorigenesis by regulating cyclin D1 expression. Oncol Rep. (2006) 16:491–6. 10.3892/or.16.3.491 [DOI] [PubMed] [Google Scholar]
- 22.Yeh ES, Lew BO, Means AR. The loss of PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts to genomic instability. J Biol Chem. (2006) 281:241–51. 10.1074/jbc.M505770200 [DOI] [PubMed] [Google Scholar]
- 23.Lee Y-C, Que J, Chen Y-C, Lin J-T, Liou Y-C, Liao P-C, et al. Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A-Bora complex. J Cell Sci. (2013) 126:4862–72. 10.1242/jcs.121368 [DOI] [PubMed] [Google Scholar]
- 24.Pulikkan JA, Dengler V, Peer Zada AA, Kawasaki A, Geletu M, Pasalic Z, et al. Elevated PIN1 expression by C/EBPα-p30 blocks C/EBPα-induced granulocytic differentiation through c-Jun in AML. Leukemia (2010) 24:914–23. 10.1038/leu.2010.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y, et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol. (2013) 33:2930–49. 10.1128/MCB.01455-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W, Yang Q, Low CB, Karthik BC, Wang Y, Ryo A, et al. Pin1 catalyzes conformational changes of Thr-187 in p27Kip1 and mediates its stability through a polyubiquitination process. J Biol Chem. (2009) 284:23980–8. 10.1074/jbc.M109.022814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaydes JP, Luciani MG, Pospisilova S, Ball HM, Vojtesek B, Hupp TR. Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. J Biol Chem. (2001) 276:4699–708. 10.1074/jbc.M003485200 [DOI] [PubMed] [Google Scholar]
- 28.Wulf GM, Liou Y-C, Ryo A, Lee SW, Lu KP. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J Biol Chem. (2002) 277:47976–9. 10.1074/jbc.C200538200 [DOI] [PubMed] [Google Scholar]
- 29.Eckerdt F, Yuan J, Saxena K, Martin B, Kappel S, Lindenau C, et al. Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. J Biol Chem. (2005) 280:36575–83. 10.1074/jbc.M504548200 [DOI] [PubMed] [Google Scholar]
- 30.Rizzolio F, Lucchetti C, Caligiuri I, Marchesi I, Caputo M, Klein-Szanto AJ, et al. Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ. (2012) 19:1152–61. 10.1038/cdd.2011.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto K, Sagata N. Mechanism for inactivation of the mitotic inhibitory kinase Wee1 at M phase. Proc Natl Acad Sci USA. (2007) 104:3753–8. 10.1073/pnas.0607357104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam PB, Burga LN, Wu BP, Hofstatter EW, Lu K, Wulf GM. Prolyl isomerase Pin1 is highly expressed in Her2-positive breast cancer and regulates erbB2 protein stability. Mol Cancer (2008) 7:91. 10.1186/1476-4598-7-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanal P, Namgoong GM, Kang BS, Woo E-R, Choi HS. The prolyl isomerase Pin1 enhances HER-2 expression and cellular transformation via its interaction with mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1. Mol Cancer Ther. (2010) 9:606–16. 10.1158/1535-7163.MCT-09-0560 [DOI] [PubMed] [Google Scholar]
- 34.Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. (2009) 11:133–42. 10.1038/ncb1822 [DOI] [PubMed] [Google Scholar]
- 35.Rustighi A, Zannini A, Tiberi L, Sommaggio R, Piazza S, Sorrentino G, et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. (2014) 6:99–119. 10.1002/emmm.201302909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y-X, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. (2007) 21:2950–62. 10.1101/gad.1592807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brondani V, Schefer Q, Hamy F, Klimkait T. The peptidyl–prolyl isomerase Pin1 regulates phospho-Ser77 retinoic acid receptor α stability. Biochem Biophys Res Commun. (2005) 328:6–13. 10.1016/j.bbrc.2004.12.130 [DOI] [PubMed] [Google Scholar]
- 38.Ryo A, Liou Y-C, Wulf G, Nakamura M, Lee SW, Lu KP. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. (2002) 22:5281–95. 10.1128/MCB.22.15.5281-5295.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilton BA, Li Z, Musich PR, Wang H, Cartwright BM, Serrano M, et al. ATR plays a direct antiapoptotic role at mitochondria, which is regulated by prolyl isomerase Pin1. Mol Cell (2015) 60:35–46. 10.1016/j.molcel.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steger M, Murina O, Hühn D, Ferretti LP, Walser R, Hänggi K, et al. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell (2013) 50:333–43. 10.1016/j.molcel.2013.03.023 [DOI] [PubMed] [Google Scholar]
- 41.Bitomsky N, Conrad E, Moritz C, Polonio-Vallon T, Sombroek D, Schultheiss K, et al. Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc Natl Acad Sci USA. (2013) 110:E4203–12. 10.1073/pnas.1310001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell (2004) 14:625–36. 10.1016/j.molcel.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 43.De Nicola F, Bruno T, Iezzi S, Di Padova M, Floridi A, Passananti C, et al. The prolyl isomerase Pin1 affects Che-1 stability in response to apoptotic DNA damage. J Biol Chem. (2007) 282:19685–91. 10.1074/jbc.M610282200 [DOI] [PubMed] [Google Scholar]
- 44.Ryo A, Hirai A, Nishi M, Liou Y-C, Perrem K, Lin S-C, et al. A suppressive role of the prolyl isomerase Pin1 in cellular apoptosis mediated by the death-associated protein daxx. J Biol Chem. (2007) 282:36671–81. 10.1074/jbc.M704145200 [DOI] [PubMed] [Google Scholar]
- 45.Ding Q, Huo L, Yang J-Y, Xia W, Wei Y, Liao Y, et al. Down-regulation of myeloid cell leukemia-1 through Inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. (2008) 68:6109–17. 10.1158/0008-5472.CAN-08-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M, et al. Protein kinase C and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science (2007) 315:659–63. 10.1126/science.1135380 [DOI] [PubMed] [Google Scholar]
- 47.Reineke EL, Lam M, Liu Q, Liu Y, Stanya KJ, Chang K-S, et al. Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 Cells. Mol Cell Biol. (2008) 28:997–1006. 10.1128/MCB.01848-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Driver JA, Zhou XZ, Lu KP. Regulation of protein conformation by Pin1 offers novel disease mechanisms and therapeutic approaches in Alzheimer's disease. Discov Med. (2014) 17:93–9. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou XZ, Lu KP. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer (2016) 16:463–78. 10.1038/nrc.2016.49 [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Wu Y, Yang H, Li X, Jie M, Hu C, et al. Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis (2018) 9:883. 10.1038/s41419-018-0844-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pu W, Li J, Zheng Y, Shen X, Fan X, Zhou J-K, et al. Targeting Pin1 by inhibitor API-1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology (2018) 68:547–60. 10.1002/hep.29819 [DOI] [PubMed] [Google Scholar]
- 52.Campaner E, Rustighi A, Zannini A, Cristiani A, Piazza S, Ciani Y, et al. A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat Commun. (2017) 8:15772. 10.1038/ncomms15772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu K-J, Zhong H-J, Yang G, Wu C, Huang J-M, Li G, et al. Small molecule Pin1 inhibitor blocking NF-κB signaling in prostate cancer cells. Chem - An Asian J. (2018) 13:275–9. 10.1002/asia.201701216 [DOI] [PubMed] [Google Scholar]
- 54.Russo Spena C, De Stefano L, Palazzolo S, Salis B, Granchi C, Minutolo F, et al. Liposomal delivery of a Pin1 inhibitor complexed with cyclodextrins as new therapy for high-grade serous ovarian cancer. J Control Release (2018) 281:1–10. 10.1016/J.JCONREL.2018.04.055 [DOI] [PubMed] [Google Scholar]
- 55.Lu P-J, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature (1999) 399:784–8. 10.1038/21650 [DOI] [PubMed] [Google Scholar]
- 56.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. (2000) 7:639–43. 10.1038/77929 [DOI] [PubMed] [Google Scholar]
- 57.Wintjens R, Wieruszeski J-M, Drobecq H, Rousselot-Pailley P, Buée L, Lippens G, et al. 1 H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides. J Biol Chem. (2001) 276:25150–6. 10.1074/jbc.M010327200 [DOI] [PubMed] [Google Scholar]
- 58.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell (1997) 89:875–86. 10.1016/S0092-8674(00)80273-1 [DOI] [PubMed] [Google Scholar]
- 59.Behrsin CD, Bailey ML, Bateman KS, Hamilton KS, Wahl LM, Brandl CJ, et al. Functionally important residues in the peptidyl-prolyl Isomerase Pin1 revealed by unigenic evolution. J Mol Biol. (2007) 365:1143–62. 10.1016/j.jmb.2006.10.078 [DOI] [PubMed] [Google Scholar]
- 60.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson KA, Etzkorn FA, et al. Stereospecific gating of functional motions in Pin1. Proc Natl Acad Sci USA. (2011) 108:12289–94. 10.1073/pnas.1019382108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson KA, Bouchard JJ, Peng JW. Interdomain interactions support interdomain communication in human Pin1. Biochemistry (2013) 52:6968–81. 10.1021/bi401057x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Namanja AT, Peng T, Zintsmaster JS, Elson AC, Shakour MG, Peng JW. Substrate recognition reduces side-chain flexibility for conserved hydrophobic residues in human Pin1. Structure (2007) 15:313–27. 10.1016/j.str.2007.01.014 [DOI] [PubMed] [Google Scholar]
- 63.Smet C, Wieruszeski J-M, Buée L, Landrieu I, Lippens G. Regulation of Pin1 peptidyl-prolyl cis/trans isomerase activity by its WW binding module on a multi-phosphorylated peptide of Tau protein. FEBS Lett. (2005) 579:4159–64. 10.1016/j.febslet.2005.06.048 [DOI] [PubMed] [Google Scholar]
- 64.Zhou XZ, Lu PJ, Wulf G, Lu KP. Phosphorylation-dependent prolyl isomerization: a novel signaling regulatory mechanism. Cell Mol Life Sci. (1999) 56:788–806. 10.1007/s000180050026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Innes BT, Bailey ML, Brandl CJ, Shilton BH, Litchfield DW. Non-catalytic participation of the Pin1 peptidyl-prolyl isomerase domain in target binding. Front Physiol. (2013) 4:18. 10.3389/fphys.2013.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacobs DM, Saxena K, Vogtherr M, Bernadó P, Pons M, Fiebig KM. Peptide binding induces large scale changes in inter-domain mobility in human Pin1. J Biol Chem. (2003) 278:26174–82. 10.1074/jbc.M300796200 [DOI] [PubMed] [Google Scholar]
- 67.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. (2014) 42:D358–63. 10.1093/nar/gkt1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, Lücke C, Hunter T, Lu K-P, Fischer G, et al. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem Biol. (2007) 2:320–8. 10.1021/cb7000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Küllertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell (2000) 6:873–83. 10.1016/S1097-2765(05)00083-3 [DOI] [PubMed] [Google Scholar]
- 70.Kops O, Zhou XZ, Lu KP. Pin1 modulates the dephosphorylation of the RNA polymerase II C-terminal domain by yeast Fcp1. FEBS Lett. (2002) 513:305–11. 10.1016/S0014-5793(02)02288-3 [DOI] [PubMed] [Google Scholar]
- 71.Duncan KE, Dempsey BR, Killip LE, Adams J, Bailey ML, Lajoie GA, et al. Discovery and characterization of a nonphosphorylated cyclic peptide inhibitor of the peptidylprolyl isomerase, Pin1. J Med Chem. (2011) 54:3854–65. 10.1021/jm200156c [DOI] [PubMed] [Google Scholar]
- 72.Monod J, Wyman J, Changuex JP. On the nature of allosteric transitions: a plausible mdoel. J Mol Biol. (1965) 12:88–118. 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- 73.Fischer S, Olsen KW, Nam K, Karplus M. Unsuspected pathway of the allosteric transition in hemoglobin. Proc Natl Acad Sci USA. (2011) 108:5608–13. 10.1073/pnas.1011995108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci USA. (2009) 106:18249–54. 10.1073/pnas.0904492106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLeish TCB, Cann MJ, Rodgers TL. Dynamic transmission of protein allostery without structural change: spatial pathways or global modes? Biophys J (2015) 109:1240–50. 10.1016/j.bpj.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nussinov R, Tsai C-J. Allostery without a conformational change? Revisiting the paradigm. Curr Opin Struct Biol. (2015) 30:17–24. 10.1016/j.sbi.2014.11.005 [DOI] [PubMed] [Google Scholar]
- 77.Cooper A, Dryden DTF. Allostery without conformational change. Eur Biophys J. (1984) 11:103–9. 10.1007/BF00276625 [DOI] [PubMed] [Google Scholar]
- 78.Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning. PLoS Comput Biol. (2011) 7:e1002154. 10.1371/journal.pcbi.1002154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bowman GR, Geissler PL. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc Natl Acad Sci (2012) 109:11681–11686. 10.1073/pnas.1209309109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature (2014) 508:331–9. 10.1038/nature13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Landrieu I, Smet C, Wieruszeski J-M, Sambo A-V, Wintjens R, Buée L, et al. Exploring the molecular function of PIN1 by nuclear magnetic resonance. Curr Protein Pept Sci. (2006) 7:179–94. 10.2174/138920306777452303 [DOI] [PubMed] [Google Scholar]
- 82.Peng JW. Investigating dynamic interdomain allostery in Pin1. Biophys Rev. (2015) 7:239–49. 10.1007/s12551-015-0171-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell (2000) 103:1001–4. 10.1016/S0092-8674(00)00203-8 [DOI] [PubMed] [Google Scholar]
- 84.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science (1997) 278:1957–60. 10.1126/science.278.5345.1957 [DOI] [PubMed] [Google Scholar]
- 85.Peng T, Zintsmaster JS, Namanja AT, Peng JW. Sequence-specific dynamics modulate recognition specificity in WW domains. Nat Struct Mol Biol. (2007) 14:325–31. 10.1038/nsmb1207 [DOI] [PubMed] [Google Scholar]
- 86.Wang XJ, Hart SA, Xu B, Mason MD, Goodell JR, Etzkorn FA. Serine-cis-proline and serine-trans-proline isosteres: stereoselective synthesis of (Z)- and (E)-alkene mimics by Still-Wittig and Ireland-Claisen rearrangements. J Org Chem. (2003) 68:2343–9. 10.1021/jo026663b [DOI] [PubMed] [Google Scholar]
- 87.Wang XJ, Xu B, Mullins AB, Neiler FK, Etzkorn FA. Conformationally locked isostere of phosphoSer-cis-Pro inhibits Pin1 23-fold better than phosphoSer-trans-Pro isostere. J Am Chem Soc. (2004) 126:15533–42. 10.1021/ja046396m [DOI] [PubMed] [Google Scholar]
- 88.Davis IW, Arendall WB, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure (2006) 14:265–74. 10.1016/j.str.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 89.Dubay KH, Bothma JP, Geissler PL. Long-range intra-protein communication can be transmitted by correlated side-chain fluctuations alone. PLoS Comput Biol. (2011) 7:e1002168. 10.1371/journal.pcbi.1002168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo J, Pang X, Zhou H-X. Two Pathways mediate interdomain allosteric regulation in Pin1. Structure (2015) 23:237–47. 10.1016/j.str.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barman A, Hamelberg D. Coupled dynamics and entropic contribution to the allosteric mechanism of Pin1. J Phys Chem B (2016) 120:8405–15. 10.1021/acs.jpcb.6b02123 [DOI] [PubMed] [Google Scholar]
- 92.Campitelli P, Guo J, Zhou H-X, Ozkan SB. Hinge-shift mechanism modulates allosteric regulations in human Pin1. J Phys Chem B (2018) 122:5623–9. 10.1021/acs.jpcb.7b11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Landrieu I, De Veylder L, Fruchart JS, Odaert B, Casteels P, Portetelle D, et al. The Arabidopsis thaliana PIN1At gene encodes a single-domain phosphorylation-dependent peptidyl prolyl cis/trans isomerase. J Biol Chem. (2000) 275:10577–81. 10.1074/jbc.275.14.10577 [DOI] [PubMed] [Google Scholar]
- 94.Sun L, Wu X, Peng Y, Goh JY, Liou Y-C, Lin D, Zhao Y. Solution structural analysis of the single-domain parvulin TbPin1. PLoS One (2012) 7:e43017. 10.1371/journal.pone.0043017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yao J-L, Kops O, Lu P-J, Lu KP. Functional conservation of phosphorylation-specific Prolyl isomerases in plants. J Biol Chem. (2001) 276:13517–23. 10.1074/jbc.M007006200 [DOI] [PubMed] [Google Scholar]
- 96.Goh JY, Lai C-Y, Tan LC, Yang D, He CY, Liou Y-C. Functional characterization of two novel parvulins in Trypanosoma brucei. FEBS Lett. (2010) 584:2901–8. 10.1016/j.febslet.2010.04.077 [DOI] [PubMed] [Google Scholar]
- 97.Driver JA. Inverse association between cancer and neurodegenerative disease: review of the epidemiologic and biological evidence. Biogerontology (2014) 15:547–57. 10.1007/s10522-014-9523-2 [DOI] [PubMed] [Google Scholar]
- 98.Moore JD, Potter A. Pin1 inhibitors: Pitfalls, progress and cellular pharmacology. Bioorg Med Chem Lett. (2013) 23:4283–91. 10.1016/j.bmcl.2013.05.088 [DOI] [PubMed] [Google Scholar]
- 99.Min S-H, Zhou XZ, Lu KP. The role of Pin1 in the development and treatment of cancer. Arch Pharm Res. (2016) 39:1609–20. 10.1007/s12272-016-0821-x [DOI] [PubMed] [Google Scholar]
- 100.Lu Z, Hunter T. Prolyl isomerase Pin1 in cancer. Cell Res. (2014) 24:1033–49. 10.1038/cr.2014.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao H, Cui G, Jin J, Chen X, Xu B. Synthesis and Pin1 inhibitory activity of thiazole derivatives. Bioorg Med Chem. (2016) 24:5911–20. 10.1016/j.bmc.2016.09.049 [DOI] [PubMed] [Google Scholar]
- 102.Cui G, Jin J, Chen H, Cao R, Chen X, Xu B. Synthesis and biological evaluation of pyrimidine derivatives as novel human Pin1 inhibitors. Bioorg Med Chem. (2018) 26:2186–97. 10.1016/j.bmc.2018.03.024 [DOI] [PubMed] [Google Scholar]
- 103.Hennig L, Christner C, Kipping M, Schelbert B, Rücknagel KP, Grabley S, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry (1998) 37:5953–60. 10.1021/bi973162p [DOI] [PubMed] [Google Scholar]
- 104.Uchida T, Takamiya M, Takahashi M, Miyashita H, Ikeda H, Terada T, et al. Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem Biol. (2003) 10:15–24. 10.1016/S1074-5521(02)00310-1 [DOI] [PubMed] [Google Scholar]
- 105.Wildemann D, Erdmann F, Alvarez BH, Stoller G, Zhou XZ, Fanghänel J, et al. Nanomolar inhibitors of the peptidyl prolyl cis/trans isomerase Pin1 from combinatorial peptide libraries. J Med Chem. (2006) 49:2147–50. 10.1021/jm060036n [DOI] [PubMed] [Google Scholar]
- 106.Daum S, Erdmann F, Fischer G, Féaux de Lacroix B, Hessamian-Alinejad A, et al. Aryl indanyl ketones: efficient inhibitors of the human peptidyl prolylcis/trans isomerase Pin1. Angew Chemie Int Ed. (2006) 45:7454–8. 10.1002/anie.200601569 [DOI] [PubMed] [Google Scholar]
- 107.Guo C, Hou X, Dong L, Dagostino E, Greasley S, Ferre R, et al. Structure-based design of novel human Pin1 inhibitors (I). Bioorg Med Chem Lett. (2009) 19:5613–6. 10.1016/j.bmcl.2009.08.034 [DOI] [PubMed] [Google Scholar]
- 108.Dong L, Marakovits J, Hou X, Guo C, Greasley S, Dagostino E, et al. Structure-based design of novel human Pin1 inhibitors (II). Bioorg Med Chem Lett. (2010) 20:2210–4. 10.1016/j.bmcl.2010.02.033 [DOI] [PubMed] [Google Scholar]
- 109.Potter AJ, Ray S, Gueritz L, Nunns CL, Bryant CJ, Scrace SF, et al. Structure-guided design of alpha-amino acid-derived Pin1 inhibitors. Bioorg Med Chem Lett. (2010) 20:586–90. 10.1016/j.bmcl.2009.11.090 [DOI] [PubMed] [Google Scholar]
- 110.Potter A, Oldfield V, Nunns C, Fromont C, Ray S, Northfield CJ, et al. Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution. Bioorg Med Chem Lett. (2010) 20:6483–8. 10.1016/j.bmcl.2010.09.063 [DOI] [PubMed] [Google Scholar]
- 111.Wei S, Kozono S, Kats L, Nechama M, Li W, Guarnerio J, et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med. (2015) 21:457–66. 10.1038/nm.3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tatara Y, Lin Y-C, Bamba Y, Mori T, Uchida T. Dipentamethylene thiuram monosulfide is a novel inhibitor of Pin1. Biochem Biophys Res Commun. (2009) 384:394–8. 10.1016/j.bbrc.2009.04.144 [DOI] [PubMed] [Google Scholar]
- 113.Mori T, Hidaka M, Lin Y-C, Yoshizawa I, Okabe T, Egashira S, et al. A dual inhibitor against prolyl isomerase Pin1 and cyclophilin discovered by a novel real-time fluorescence detection method. Biochem Biophys Res Commun. (2011) 406:439–43. 10.1016/j.bbrc.2011.02.066 [DOI] [PubMed] [Google Scholar]
- 114.Urusova D V, Shim J-H, Kim DJ, Jung SK, Zykova TA, Carper A, et al. Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1. Cancer Prev Res. (2011) 4:1366–77. 10.1158/1940-6207.CAPR-11-0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo C, Hou X, Dong L, Marakovits J, Greasley S, Dagostino E, et al. Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket. Bioorg Med Chem Lett. (2014) 24:4187–91. 10.1016/j.bmcl.2014.07.044 [DOI] [PubMed] [Google Scholar]
- 116.Momin M, Yao X-Q, Thor W, Hamelberg D. Substrate sequence determines catalytic activities, domain-binding preferences, and allosteric mechanisms in Pin1. J Phys Chem B (2018) 122:6521–7. 10.1021/acs.jpcb.8b03819 [DOI] [PubMed] [Google Scholar]