Abstract
Immune cell recognition of antigens is a pivotal process in initiating immune responses against injury, pathogens, and cancers. Breakthroughs over the past decade support a major role for mechanical forces in immune responses, laying the foundation for the emerging field of mechanoimmunology. In this Perspective, we discuss the mechanical forces acting at the level of ligand–receptor interactions and how they underpin receptor triggering, signal initiation, and immune cell activation. We also highlight the novel biophysical tools and advanced imaging techniques that have afforded us the recent progress in our understanding of the role of forces in immune cell functions.
AN IMMUNE CELL’S JOURNEY THROUGH A MECHANICAL LANDSCAPE
To efficiently defend an organism against injury, infection, and cancer, leukocytes must orchestrate a complex multiscale chain of events. For decades, immunological research focused on identifying cellular and molecular players that mediate the intricate interplay between cells of the innate and adaptive arms during a concerted immune response. Innate immune cells, including neutrophils, macrophages, and antigen-presenting cells (APCs) such as dendritic cells (DCs), are early responders recruited to sites of inflammation, where they can phagocytose foreign and pathogenic substances themselves or coordinate a wider immune response by recruiting lymphocytes that then clear the threat.
However, it is only within the past decade that evidence has emerged highlighting the critical role of mechanical forces in immune cell functions. To mount effective immune responses, immune cells must rapidly migrate to and contact APCs, or pathogen-laden or transformed target cells. As for all animal cells, actin-mediated force generation is the main driver of migration in leukocytes, which navigate an array of barriers and tissues of differing architectures either by responding to complex guidance cues or by employing highly evolved search mechanisms (Munoz et al., 2014; Weninger et al., 2014). Leukocytes are therefore endowed with considerable plasticity in shape and migratory regulation (Renkawitz and Sixt, 2010), as they continually probe and respond to the geometry and mechanical cues provided by their environment (Hallmann et al., 2015). When an immune cell eventually encounters a target cell, it will physically “grasp” it and form a specialized synaptic interface, exerting forces on its conjugate in order to deliver its functions (Lim et al., 2011; Basu et al., 2016; Spillane and Tolar, 2017). In addition to these forces acting at the cellular level, recent progress in the field has demonstrated that immune receptors themselves respond to mechanical stimuli during antigen recognition, which is crucial for efficient discrimination of antigens. Indeed, mechanical forces acting directly on individual receptors influence receptor triggering and downstream intracellular signaling.
Given the multitude of functions carried out by immune cells within varying environments, it is not surprising that they experience mechanical forces ranging from piconewtons at the nanoscale to several orders of magnitude greater at the tissue level. Recent developments in imaging modalities and biophysical tools have finally allowed for the forces at the molecular scale to be probed. This Perspective explores recent progress in the emerging field of mechanoimmunology, focusing on immune receptor–ligand interactions and highlighting the novel biophysical tools that have afforded us hitherto inaccessible insights into the role of molecular-scale mechanical forces in immune cell functions (see Box 1).
Box 1: Force measurement techniques.
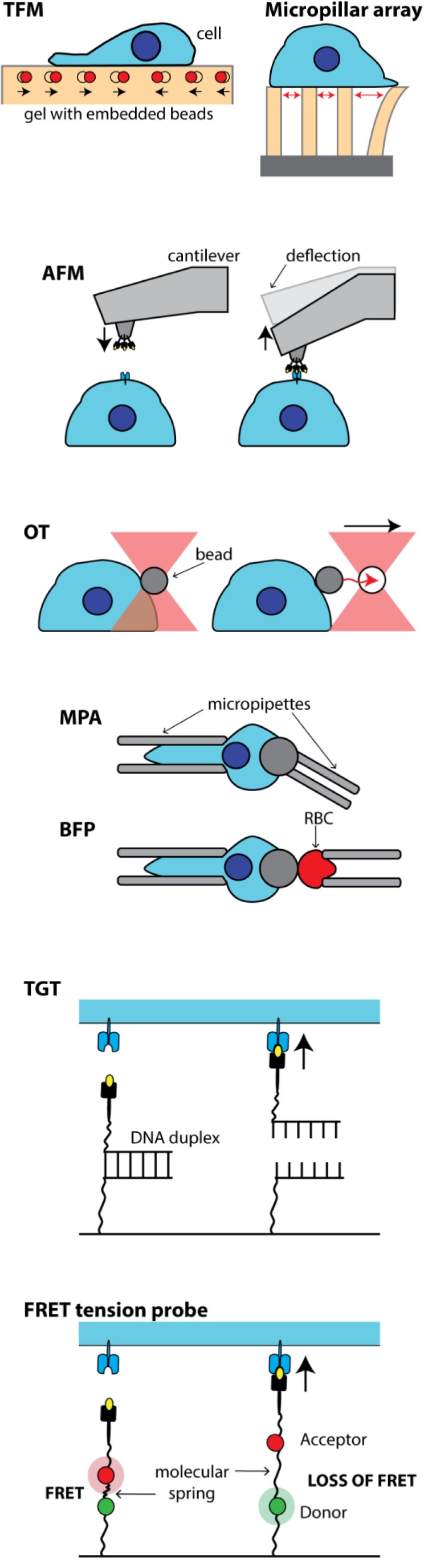
Traction force microscopy (TFM) relies on the principle that the elastic properties of a material relate the force per unit area (stress) it is subjected to and the ensuing fractional change in material length (strain) it experiences. Therefore, if the elastic properties of a substrate are known, and the strain it experiences can be measured, the forces exerted on it can be calculated. In TFM, induced deformations are usually determined by tracking the movement of tracer particles within a gel. Classically, cells are positioned on a layer of compliant material with an adhesion-functionalized surface (Style et al., 2014). Alternatively, TFM can also be realized using arrays of deformable micropillars, such as those made of polydimethylsiloxane (PDMS). If the elasticity of the micropillars is known, their deflection can be used to calculate the applied force (Tan et al., 2003).
Atomic force microscopy (AFM) employs a small scanning probe, consisting of a soft cantilever and a micrometer-scale tip, to determine minute tip–sample interaction forces (pN to nN). In close contact with the surface, attractive/repulsive forces induce deflection of the cantilever, which is tracked by a laser beam reflected from the back of the cantilever onto a photodiode detector. By functionalizing the cantilever tip with chemicals or biomolecules, or even replacing it with a live cell, interaction forces between biological systems can be measured (Müller and Dufrêne, 2011). Alternatively, AFM can be used to determine the mechanical properties (e.g., elasticity) of a sample by measuring the deflection of the cantilever, which reflects the loading force exerted on the sample, with increasing indentation depth (Kuznetsova et al., 2007).
Optical tweezers (OT) are created by focusing a laser beam to a diffraction-limited spot by means of a high–numerical aperture objective. In the vicinity of the focal point, particles of a refractive index greater than that of the bathing medium (nm to µm size, e.g., polystyrene beads or even cells) experience a pN force directed toward the focus. For small displacements from the focal point, the restoring force depends linearly on the displacement. If a molecule attached to a dielectric bead or a cell interacts with a binding partner, then its displacement from the focal spot is directly proportional to the magnitude of the interaction force (Neuman and Nagy, 2008).
Micropipette aspiration (MPA) techniques exploit negative pressure to aspirate lipid vesicles or cells partly into a micropipette to hold them in place or change their mechanical properties. Given Laplace’s law, if the pressure in the pipette and an aspirated spherical object are at equilibrium, the surface tension of the object can be calculated. In biomembrane force probe (BFP) measurements, a biotinylated red blood cell (RBC) is aspirated into a micropipette and a streptavidin ligand–coated glass bead is attached to the RBC. A cell is allowed to adhere to the ligand-coated bead and as cell and bead are separated, the deformation of the RBC is proportional to the interaction force (Gourier et al., 2008).
DNA-based tension gauge tethers (TGTs) utilize DNA duplexes to attach cell–receptor ligands to a surface. Depending on their length and sequence, DNA duplexes are characterized by a specific rupture force. Using TGTs with increasing rupture force, the force required for cell attachment or cell activation can be determined (Wang and Ha, 2013). When the DNA duplex is flanked with a fluorophore and a quencher, fluorescence is low prior to rupture due to Förster resonance energy transfer (FRET), and the time point of detachment can be visualized in real time via an increase in fluorescence. Alternatively, in multiplex TGT constructs, TGTs that are sensitive to different force thresholds are labeled with different fluorescent dyes and are used to simultaneously map different tension levels in cells (Wang and Wang, 2016). Employing the same principle, FRET-based tension probes are short, mainly alpha-helical peptidic structures that extend under applied force. A fluorescent protein FRET pair is attached to the ends of the sensor, and loss of FRET reflects extension of the linker (Stabley et al., 2011). In contrast to other force measurement methods, these fluorescent tension probes allow measurement of intracellular forces with molecular specificity at subcellular resolution. These techniques are sometimes grouped under the name molecular tension fluorescence microscopy (MTFM; Liu et al., 2017). The mechanical spring element in MTFM probes can consist of DNA hairpins, protein domains, or polymer chains such as polyethylene glycol or elastic polypeptide. This probe design builds on earlier FRET-based tension sensors from Martin Schwartz’s group, in which an elastic protein domain (e.g., TSMod, derived from the spider silk protein flagelliform) was incorporated within the protein of interest itself (Grashoff et al., 2010).
MECHANICAL CUES IN THE ENVIRONMENT REGULATE IMMUNE CELL BEHAVIOR
Within the past few years, it has become clear that immune cell activation is regulated not only by biochemical factors, but also by the stiffness of the environment the cells are interacting with. Cellular mechanosensing is the ability of cells to sense the physical characteristics of their environment (be it the extracellular matrix or adjoining cells). For this purpose, cells apply force on their environment and convert information about the resistance to this force into biochemical signaling through various mechanosensory proteins. Mechanosensing of substrate rigidity by macrophages has been shown to influence their phagocytic ability (Patel et al., 2012), cell morphology and elasticity (Blakney et al., 2012; Patel et al., 2012), and production of both proinflammatory and anti-inflammatory cytokines (Blakney et al., 2012; Previtera and Sengupta, 2015). Macrophages grown on stiff polyacrylamide gels (240 kPa) produced more proinflammatory mediators than those grown on soft substrates (∼0.3 kPa), and Toll-like receptor 4 activity enhanced this effect in response to lipopolysaccharide, a bacterial proinflammatory agent (Previtera and Sengupta, 2015). This suggests that biological stimulants and tissue elasticity can work synergistically to regulate the pro- or anti-inflammatory characteristics of macrophages during an infection.
It is not only cells of the innate immune system that respond to mechanical cues. T and B lymphocytes recognize a specific, so-called cognate antigen on the surface of an APC or an infected or cancerous cell. Both T- and B-cells have been shown to adapt their response to antigens based on the rigidity of the substrate they are presented on (Judokusumo et al., 2012; O’Connor et al., 2012; Wan et al., 2013; Zeng et al., 2015; Saitakis et al., 2017; Shaheen et al., 2017). Substrate stiffness modulates not only the level of activation, but also the nature of the cellular responses. In T-cells, this includes differentiation (O’Connor et al., 2012), gene expression, cell migration, morphology, and cytokine secretion (Saitakis et al., 2017), whereas in B-cells, proliferation, class switching, and antibody production are all influenced by substrate rigidity (Zeng et al., 2015). Experiments using a biomembrane force probe (BFP; see Box 1) revealed mechanical feedback between the substrate stiffness sensed by a T-cell and the force it generated, with greater force applied on stiffer targets (Husson et al., 2011). This was further confirmed with traction force microscopy (TFM; see Box 1), where Jurkat T-cells exerted stronger forces on stiffer hydrogels (Hui et al., 2015). In a key study by Morgan Huse’s group, stiffer target cells were shown to enhance the killing response of cytotoxic T lymphocytes (CTLs) (Basu et al., 2016). Interestingly, there is a strong negative correlation between cancer cell stiffness and metastatic potential (Swaminathan et al., 2011; Lekka, 2016). These studies revealed that metastatic cancer cells are softer than primary tumor cells, which raises the intriguing possibility of invasive cells evading CTL-mediated lysis by modulating their mechanical properties. Furthermore, Basu et al. demonstrated that mechanical force exerted on tumor cells by CTLs themselves facilitates perforin-mediated lysis. Using a micropillar array (see Box 1), they demonstrated that CTLs exert localized forces in areas into which they direct their lytic granules to deliver their cytolytic proteins, a process called degranulation (Basu et al., 2016). The relationships between a cell and its environment or interaction partners are thus bidirectional and, in the context of mechanical forces, can be characterized as displaying “mechanoreciprocity.”
IMMUNE RECEPTORS AS MECHANOSENSORS
The molecular mechanisms underlying the mechanosensitivity of immune cells are still being deciphered. Do immune receptors themselves have a mechanosensing capacity or do T- and B-cells perform mechanosensing through more conventional receptors such as integrins? Lymphocyte function–associated antigen 1 (LFA-1) is an integrin expressed on lymphocytes that binds to intercellular adhesion molecule 1 (ICAM-1) to promote adhesion during the formation of an immunological synapse at the interface between a lymphocyte and its conjugate. Previous studies suggested that lymphocytes are able to discern substrate stiffness independent of integrins (Judokusumo et al., 2012; O’Connor et al., 2012; Wan et al., 2013; Zeng et al., 2015), although the presence of adhesion molecules greatly enhances the ability of B-cells to discriminate between antigens (Shaheen et al., 2017). In T-cells, engagement of LFA-1 alone did not generate any measurable forces or intracellular signaling (Husson et al., 2011), suggesting that mechanosensitive receptors other than integrins are at play in lymphocytes.
The question of whether immune receptors are themselves inherently mechanosensitive has driven many new technological developments that experimentally uncouple force from antigen recognition. A T-cell will only recognize and respond to an APC or target cell if its T-cell receptor (TCR) binds to its cognate peptide bound to major histocompatibility complex (pMHC) on the surface of its interaction partner. The TCR itself is composed of an αβ heterodimer that has no intrinsic signaling domain, but is noncovalently associated with CD3 chains (εγ, εδ, and ζζ dimers) that together contain ten immunoreceptor tyrosine-based activation motifs (ITAMs) that can be phosphorylated to initiate signaling (Figure 1). Signaling can also be amplified through association with coreceptors such as CD4 or CD8, the expression of which specifies the function of the T-cell. Novel biophysical echniques have exploited surrogate conjugates (such as pMHC- or antibody-coated beads, bilayers, or surfaces) and/or artificial ligands to activate T-cells. Early evidence that the TCR functions as a mechanosensor came from Ellis Reinherz’s group (Kim et al., 2009). They used beads coated with engineered anti-CD3ε antibodies that bind only one site per TCR and are thus unable to cross-link it to trigger signals. By trapping the cells and beads in optical tweezers (OT; see Box 1), they were able to apply tangential forces on the TCR, which induced cytoplasmic Ca2+ mobilization, a widely adopted marker of lymphocyte activation. Furthermore, Li et al. stimulated T-cells with artificial APCs presenting modified elongated anti-CD3ε antibodies that were unable to trigger calcium influx. Only when shear stress was applied to the T-cells through buffer flow from a micropipette tip, or when T-cells were physically pulled away from the APC via micropipette aspiration (MPA; see Box 1), was Ca2+ signaling initiated (Li et al., 2010). In addition, optomechanical actuator nanoparticles that collapse upon near-infrared illumination, thus applying a mechanical load to the receptor–ligand complexes bound to the particles, were able to mechanically trigger calcium signaling in T-cells (Liu et al., 2016b). Overall, these studies suggest that physical forces acting on the TCR complex can directly initiate signaling in T-cells.
FIGURE 1:
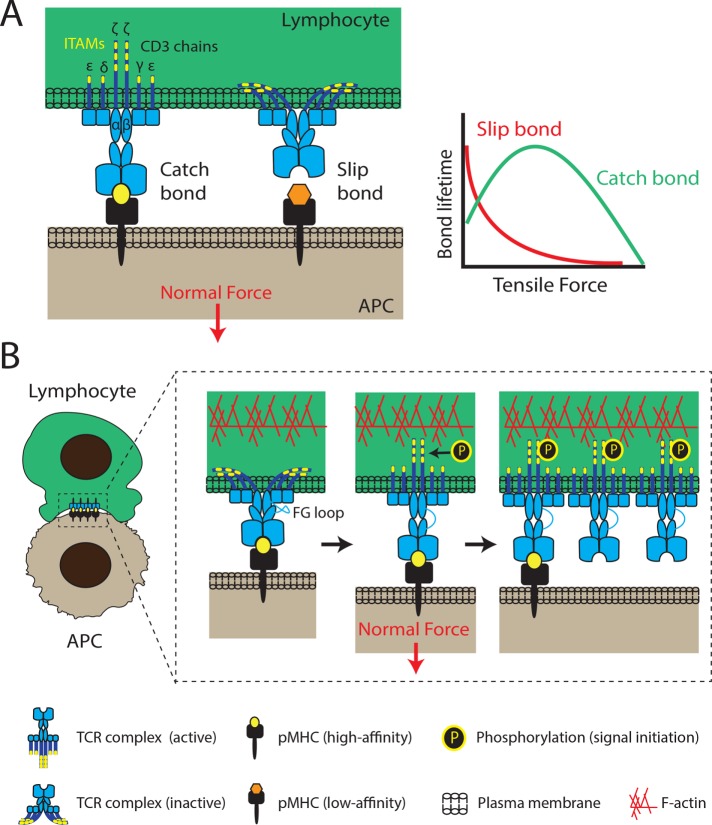
Mechanotransduction through the T-cell receptor. (A) The T-cell receptor complex consists of an αβ heterodimer that is noncovalently associated with ITAM-containing CD3 chains (εγ, εδ, and ζζ dimers). TCRs interact tightly with high-affinity pMHCs, forming catch bonds that are characterized by lifetimes that increase under load. Above a specific force threshold, lifetimes decrease. Longer TCR–pMHC interactions are more likely to lead to successful signal initiation and T-cell activation. Conversely, TCR interactions with low-affinity peptides exhibit slip-bond behavior, rupturing easily under low tensile forces. (B) Receptor deformation model of TCR activation. Force applied to the TCR upon pMHC binding triggers the unfolding of the FG loop region of the TCR and the exposure of its ITAMs (yellow bands) for phosphorylation and initiation of downstream signaling. FG loop unfolding facilitates extension of the TCR and its catch-bond behavior. Additionally, conformational changes of one TCR complex can propagate to its neighbors, producing clusters of active TCR complexes to amplify signaling. Both normal and tangential forces have been shown to initiate TCR signaling (normal forces shown here), with the contribution of each force component still under investigation. The F-actin cytoskeleton is thought to play a major role in both force generation and TCR clustering.
Intriguingly, a very recent study demonstrated that the mechanosensitive ion channel Piezo1 is critical for TCR triggering (Liu et al., 2018a). In their paper, Liu and colleagues propose a model in which membrane stretch induced by immunological synapse formation triggers Piezo1 activation, thereby causing Ca2+ influx, and in turn driving cytoskeletal rearrangements that regulate TCR signaling (Liu et al., 2018a). Although the mechanistic details underlying the role of Piezo1 in TCR activation require further study, these results bring into question the suitability of using calcium mobilization as an indicator of T-cell activation through TCR mechanosensing. Ca2+ fluxes are transient and heterogeneous between cells and involved in many different cellular processes, making it difficult to differentiate accurately between lymphocyte activation–associated calcium mobilizations and those resulting from other cellular phenomena.
There is long-standing evidence that reagents that disrupt the actin cytoskeleton abrogate TCR triggering (Campi et al., 2005; Choudhuri et al., 2005; Varma et al., 2006). In an elegant study using atomic force microscopy (AFM; see Box 1), Hu and Butte demonstrated that an intact filamentous actin (F-actin) network is needed for T-cells to generate force at the TCR and thus to trigger T-cell activation (Hu and Butte, 2016). Both Ca2+ flux and force generation were abrogated when T-cells were treated with latrunculin A, a drug that sequesters monomeric actin and thus prevents its polymerization into F-actin. However, the most important finding of this study was that the application of an external oscillating force to the AFM tip, mimicking cytoskeletal forces, rescued calcium signaling in latrunculin A–treated cells (Hu and Butte, 2016), providing direct evidence that the necessary force-sensing machinery is intrinsic to the TCR itself. Intriguingly, it is not only the actin cytoskeleton of the T-cell that is vital during T-cell priming. Highlighting the importance of APC-T-cell cross-talk at the immunological synapse, the cortical actin networks of professional APCs such as DCs have been shown to regulate the lateral mobility of ICAM-1 molecules at their surface, with constrained ICAM-1 mobility promoting the formation of mature immunological synapses and T-cell proliferation (Comrie et al., 2015). These data support a model in which ICAM-1 molecules with reduced mobility resist tensile forces exerted by the T-cell through ICAM-1/LFA-1 interactions more strongly, which in turn promotes firmer adhesion and maturation of the immunological synapse. Similarly, in natural killer (NK) cells, innate cytotoxic lymphocytes that recognize and destroy cancerous or virally infected cells, activation is regulated by the distribution and mobility of ICAM-1 molecules on the surfaces of target cells. Enhanced granule polarization and cytotoxicity were observed when ICAM-1 clusters were “tethered” to the actin cytoskeleton via ezrin and thus immobilized (Gross et al., 2010).
NOVEL MOLECULAR TENSION PROBES ILLUMINATE MECHANOIMMUNOLOGY
To enable quantification of the minute forces acting at the single-receptor level, innovative biophysical methods with improved sensitivity have been developed. In 2011, Khalid Salaita’s group pioneered the development of molecular tension fluorescence microscopy (MTFM; Stabley et al., 2011; see Box 1). Here each probe, consisting of a donor–acceptor fluorophore pair separated by a DNA-based molecular “ruler” and immobilized onto a surface, reports the force transmitted through receptors at single-molecule resolution via Förster resonance energy transfer (FRET). The Salaita group then further enhanced the sensitivity of these force sensors by immobilizing the FRET pair–decorated DNA hairpin onto a gold nanoparticle, with the probes now exhibiting dual quenching through both FRET and nanometal surface energy transfer (NSET) to the gold nanoparticle, providing a 100-fold increase in signal upon hairpin unfolding (Liu et al., 2016a). Using these tension probes, the authors were able to detect forces in the range of 12–19 pN experienced by individual TCR complexes during initial antigen recognition and preceding intracellular Ca2+ mobilization (Liu et al., 2016a). These results are in alignment with previous work in which single-molecule measurements showed that external forces in the range 10–20 pN are able to drive structural transitions in the TCR (Kim et al., 2009; Das et al., 2015) and increase the lifetime of TCR–pMHC interactions for strong agonists (Liu et al., 2014). When these tension sensors were incorporated into fluid lipid bilayers, the pN forces were shown to be sustained within TCR microclusters (Ma et al., 2016).
To measure forces applied on single receptor–ligand bonds accurately, Wang and Ha developed an alternative approach, known as the tension gauge tether (TGT; see Box 1), where a ligand is immobilized onto a solid surface through a DNA tether that ruptures at a critical force (Wang and Ha, 2013). By engineering a range of tethers with varying tension tolerances, this approach determines the forces required to activate cell signaling through single ligand–receptor bonds. Experiments combining TGTs with total internal reflection fluorescence (TIRF) microscopy were used to quantify B-cell receptor (BCR) accumulation and phosphorylation at the immune synapse as an indicator of B-cell signaling. Different classes of BCRs required different levels of mechanical force to induce activation, suggesting the existence of differing activation thresholds for different effector functions (Wan et al., 2015). The activation of the immunoglobulin (Ig) M (IgM)-BCR that is expressed on naïve B-cells before antigen encounter was dependent on the amount of mechanical force applied, with greater forces (>50 pN) resulting in more robust activation. In contrast, only limited mechanical force (<12 pN) was required for the activation of two other BCRs, IgG-BCR and IgE-BCR, which are expressed on differentiated memory B-cells that respond to a secondary challenge with the same antigen (Wan et al., 2015). This lower threshold could explain why memory B-cells exhibit faster and enhanced activation upon antigen reencounter.
Similarly, a recent study has shown that different B-cell subsets utilize different force thresholds to probe for antigen affinity (Nowosad et al., 2016). Using AFM, Pavel Tolar and colleagues demonstrated that B-cells use myosin-mediated contraction to pinch off part of the APC membrane to internalize antigen (Natkanski et al., 2013; Spillane and Tolar, 2017). In this process, the pushing and pulling of the membrane mediated by B-cell contractility contributed to antigen discrimination, with high-affinity antigens leading to stronger pulling forces and increased amounts of peptide being internalized (Natkanski et al., 2013; Spillane and Tolar, 2017). Using a degradation-sensitive DNA nanosensor, Spillane and Tolar then observed that in the situation where a B-cell could not mechanically remove the antigen, it secreted lysosomes that contained proteases capable of cleaving antigens from the APC surface (Spillane and Tolar, 2017). The mechanical threshold directing antigen internalization may be acting not solely at the level of individual molecules but rather at the level of antigen clusters. Naïve B-cells were found to gather antigen into large clusters, whereas germinal center B-cells undergoing affinity maturation formed smaller clusters of antigen that were trafficked to the cell periphery prior to endocytosis (Nowosad et al., 2016). A small cluster containing fewer antigens would require a higher affinity per molecule to surmount the mechanical threshold required for antigen extraction; thus only B-cells expressing BCRs with high affinity for an antigen will be selected for survival and antigen presentation to T-cells.
IMMUNE RECEPTORS EXHIBIT “CATCH BOND” BEHAVIOR
One of the most recent breakthroughs in the field of immune receptor triggering has been the discovery that the TCR forms ligand-induced “catch bonds” (Liu et al., 2014; Das et al., 2015). Catch bonds are characterized by lifetimes that lengthen with increasing force applied on the bond until a threshold force has been reached that results in increased frequency of bond rupture (Figure 1A). This is markedly different from “slip bonds,” which are immediately destabilized when they experience force. Catch bond behavior was first described for selectins (Marshall et al., 2003) and integrins (Kong et al., 2009) and shown to promote cellular adhesion.
The group of Cheng Zhu used an adaptation of the BFP method to show that force affects the dissociation kinetics of TCR-pMHC interactions in a peptide-specific manner (Liu et al., 2014; Pryshchep et al., 2014; Hong et al., 2015). The application of forces in the range of tens of pN prolonged the lifetimes of single TCR-pMHC bonds for agonists (catch bonds) but shortened those for antagonists (slip bonds; Liu et al., 2014). This could potentially be achieved through extrinsic forces arising due to relative cell–cell motion, or through intrinsic forces generated by the actomyosin network transporting TCR clusters, both of which lead to a pulling force on the TCR that mechanically tests the strength of the TCR–pMHC interaction. Selectively prolonging bond lifetimes for rare agonists compared with abundant self peptides enhances antigen discrimination, a mechanism that allows the cell to distinguish between an appropriate immune response and injurious autoimmunity, and is therefore a critical aspect of immune recognition.
To engulf particles, macrophages and other innate leukocytes form a phagocytic cup—a highly organized synapse that forms in response to activation through phagocytic receptors (Goodridge et al., 2011; Niedergang et al., 2016). As with the TCR, the BFP method was used to uncover the catch bond behavior of the phagocytic integrin MAC-1 (Rosetti et al., 2015). Macrophage uptake of Escherichia coli was shown to be dependent on catch bonds formed between the bacterial adhesin FimH and the glycoprotein CD48 on macrophage filopodia (Möller et al., 2013). Force-activated catch bonds enable the long-lived interaction between a filopodium and a bacterium required to initiate phagocytosis, whilst a “shovel”-like lamellipodium protruding from the macrophage directly breaks interactions between the bacterium and the substrate (Möller et al., 2013). As the field develops, it will be interesting to discover what other surface receptors exhibit catch-bond behavior during ligand recognition.
TRANSLATING MECHANICAL STIMULI INTO INTRACELLULAR SIGNALING
One of the main outstanding questions in immune-cell signaling is how an extracellular stimulus can be translated into an intracellular signaling cascade. In particular, a variety of models have been proposed to explain how TCR triggering can account for the selectivity, specificity, and speed of the T-cell response (van der Merwe and Dushek, 2011). When the effect of mechanical forces on the TCR complex is considered, a model involving conformational changes seems the most favorable. Indeed, it has been suggested that the TCR catch-bond behavior described above might be based on a force-induced allosteric change to generate additional intermolecular interactions that delay bond rupture. This idea is in agreement with the concept of the kinetic proofreading model, which postulates that completion of a series of reaction steps must occur during the TCR–pMHC bond lifetime in order to achieve T-cell activation (McKeithan, 1995), and an optimal dwell time of TCR–pMHC interactions has been identified for efficient T-cell activation (Kalergis et al., 2001). The implicit model is that a T-cell actively regulates forces transmitted to its TCR–pMHC complex to fine-tune bond lifetimes, thereby enhancing selective and differential levels of TCR activation. Emerging evidence supports a receptor deformation model (Figure 1B), in which mechanical force induces TCR-CD3 conformational changes to trigger signaling (Ma and Finkel, 2010). Forces generated through the actomyosin network and applied to the TCR would be the main driver, with any resistance to this force being converted into a conformational change in the TCR.
In a major tour de force, the combination of optical tweezers and DNA-based tether probes was used to show that the increased lifetime of TCR–pMHC bonds in response to tensile forces was dependent on a modification in the FG loop region of the TCR (Das et al., 2015). The FG loop is a well-structured element at the interface between the variable (Vβ) and constant (Cβ) domains of the TCR and has been shown to be stabilized through an interaction with the CD3γε dimer. Eliminating the FG loop abolished the catch bond behavior of the TCR, and conversely, stabilization of this domain resulted in enhanced bond lifetimes (Das et al., 2015, 2016). The FG loop is assumed to unfold upon experiencing force, leading to an extended conformation of the extracellular domain of the TCR complex (Figure 1B). This was also observed for pre-TCR–pMHC interactions, highlighting the novel role of mechanical forces during T-cell development (Das et al., 2016). The elongated conformation of the TCR favors catch-bond behavior and transmission of force across the TCR-CD3 domains, presumably leading to the release of CD3 chains from the membrane for phosphorylation. Indeed, the safety catch model postulates that the cytoplasmic portions of CD3ε and CD3ζ are embedded in the inner leaflet of the plasma membrane through a basic residue-rich sequence and that TCR engagement leads to dissociation of these chains from the membrane, exposing their ITAMs for phosphorylation by the kinase Lck (Xu et al., 2008; Zhang et al., 2011; Figure 1B). In support of this model, a recent study has shown that the cytoplasmic tails of CD3ε can exist in three conformational states with varying levels of association with the cell membrane (Guo et al., 2017). However, the mechanism by which force could be transduced from the extracellular region to the CD3 tails remains unclear due to a lack of structural information regarding the transmembrane organisation of the TCR complex. Despite the lack of direct experimental evidence, two models have been proposed: a first in which a pistonlike downward movement of the FG loop pushes on the cytoplasmic CD3 chains through the membrane (Kim et al., 2009; Wang and Reinherz, 2012), and a second in which the transmembrane domain of CD3ζ acts as a pivot point, resulting in ITAM exposure (Lee et al., 2015). The precise mechanical changes occurring in the TCR may depend on the direction of the force experienced by the TCR. Although both normal and tangential forces applied to T-cells were able to induce calcium signaling, tangential forces led to enhanced activation efficiency (Kim et al., 2009; Feng et al., 2017). The recent finding that T-cells laterally scan the surface of APCs or target cells for pMHC while extending dynamic microvilli (Cai et al., 2017) makes it even more difficult to determine the direction of physiological forces exerted on the TCR.
Ligand-induced conformational changes in CD3ε are thought to propagate to adjoining TCR complexes within the same cluster, and this cooperation between TCR complexes could explain the high sensitivity of T-cells to low numbers of pMHC antigens (Martinez-Martin et al., 2009). In addition, earlier work demonstrated that both receptor clustering and conformational changes in the CD3 chains were required for full TCR triggering (Minguet et al., 2007). Furthermore, Kuhns et al. have proposed that conformational changes that occur in the AB loop of the TCR Cα domain may regulate TCR oligomerization (Kuhns et al., 2010). Thus, it is not improbable that force-induced conformational changes lead to enhanced signaling by inducing TCR clustering (Blanco and Alarcón, 2012; Figure 1B). The positive relationship between TCR clustering and signaling efficiency at the nanometer scale was recently revealed using single-molecule localization microscopy. Only TCR complexes in dense clusters were phosphorylated and associated with downstream signaling molecules and the density of TCR clusters was dependent on the quantity and affinity of pMHC (Pageon et al., 2016). To bring these two concepts together, the Salaita group developed novel ratiometric tension probes that can simultaneously map receptor forces and clustering at the immunological synapse (Ma et al., 2016). The authors report colocalization between the ratiometric signal representing tension density and TCR clustering within the first minute of stimulation, showing that TCRs undergoing clustering are experiencing tension in the pN range (Ma et al., 2016). It is highly likely that TCR clustering is stabilized by the underlying F-actin network, potentially through membrane compartmentalization or direct tethering of the TCR complex to cortical actin. Additionally, receptor clustering may also be driven by the retrograde flow of actin (Yi et al., 2012) that is observed during cytotoxic synapse formation and target-cell engulfment (Ritter et al., 2015).
When these findings are considered as a whole, a unifying model emerges in which a force-induced mechanical switch occurs in the TCR upon receptor engagement, driving conformational changes and receptor clustering, and thus leading to robust intracellular signaling and effective cell activation. Receptor clustering has also been linked with activation in B-cells (Mattila et al., 2013) and NK cells (Pageon et al., 2013; Oszmiana et al., 2016). In macrophages and neutrophils, it is well established that phagocytosis is initiated by the lateral clustering of Fcγ receptors upon ligand binding (Sobota et al., 2005). It is likely that mechanical forces influence receptor clustering in these cell types too, although this has not yet been investigated and may involve different mechanisms. The cellular force-sensing machinery may also be involved in downstream signal-transduction events. For example, the force-sensing protein lymphocyte-specific Crk-associated substrate (Cas-L) has recently been implicated in physically linking TCR microclusters to the underlying actin network (Santos et al., 2016). Following initiation of TCR signaling, evidence suggests that Cas-L undergoes a conformational change in response to actin-induced stretch, leading to amplification of signaling, regulation of TCR microcluster transport, and inside-out integrin signaling, as well as actomyosin contraction (Santos et al., 2016). Further investigations will reveal the full extent of the involvement of mechanical forces in immune-cell signaling and effector functions acting at varying length scales and in different cellular compartments.
OUTLOOK
Within the past decade, pioneering biophysical approaches have contributed to our understanding of the mechanobiology at play during immune responses. The overall emerging picture is one where immune receptor signaling is governed by a complex regulatory network involving cross-talk and feedback loops between chemical and physical signals. We have discussed the key role of molecular-scale mechanical forces in effecting immune responses, but this probably also holds true for most receptor–ligand interactions (Chen et al., 2017). With the field of mechanoimmunology still in its infancy, further studies are required to elucidate the exact mechanisms that allow immune receptors to sense and regulate mechanical stimuli.
In recent years, immunotherapy has emerged as the biggest breakthrough in modern cancer treatment. With the development of chimeric antigen receptors (CARs), we are getting closer to achieving high specificity with reduced risks of off-tumor cytotoxicity, with clinical trials employing CAR T-cells achieving unprecedented remission rates (Frey and Porter, 2016). The role of mechanosensing in antigen discrimination is key to engineering improved CARs that will amplify minute differences in antigen structure to exclusively target tumor antigens. To this end, a deeper understanding of TCR-mediated mechanosensing is required. Key functional insights will no doubt continue to emerge with the design of ever-improving tension probes (Liu et al., 2017) and the development and refinement of novel biophysical tools. Improved in vivo imaging capabilities will likely be crucial, since immune cells move through and operate in such a variety of mechanically distinct 3D microenvironments within organisms. The ability to visualize cells in intact tissues directly will deepen our understanding of the unique mechanobiological mechanisms regulating immune cells and the influence of the mechanical landscape on their migration and functions. This is already becoming a reality, with Eric Betzig’s new adaptive optical-lattice light sheet microscope (AO-LLSM) allowing high-speed, high-resolution in vivo imaging of dynamic subcellular processes in 3D (Liu et al., 2018b). By combining this technology with genetically expressed force sensors, we may soon be able to map molecular-scale mechanical forces in and on cells deep within the complex tissues of living organisms.
Abbreviations used:
- AFM
atomic force microscopy
- APC
antigen-presenting cell
- BCR
B-cell receptor
- BFP
biomembrane force probe
- CAR
chimeric antigen receptor
- CTL
cytotoxic T lymphocyte
- DC
dendritic cell
- F-actin
filamentous actin
- FRET
Förster resonance energy transfer
- ICAM-1
intercellular adhesion molecule 1
- ITAM
immunoreceptor tyrosine-based activation motif
- LFA-1
lymphocyte function–associated antigen 1
- MPA
micropipette aspiration
- MTFM
molecular tension fluorescence microscopy
- NK
natural killer
- OT
optical tweezers
- pMHC
peptide–major histocompatibility complex
- TCR
T-cell receptor
- TFM
traction force microscopy
- TGT
tension gauge tether.
Footnotes
REFERENCES
- Basu R, Whitlock BM, Husson J, Le Floc’h A, Jin W, Oyler-Yaniv A, Dotiwala F, Giannone G, Hivroz C, Biais N, et al. (2016) Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell , 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakney AK, Swartzlander MD, Bryant SJ. (2012) The effects of substrate stiffness on the in vitro activation of macrophages and in vivo host response to poly(ethylene glycol)-based hydrogels. J Biomed Mater Res Part A , 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco R, Alarcón B. (2012) TCR nanoclusters as the framework for transmission of conformational changes and cooperativity. Front Immunol , 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, Gerard A, Liu TL, Chen BC, Betzig E, et al. (2017) Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science , 6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campi G, Varma R, Dustin ML. (2005) Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med , 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ju L, Rushdi M, Ge C, Zhu C. (2017) Receptor-mediated cell mechanosensing. Mol Biol Cell , 3134–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. (2005) T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature , 578–582. [DOI] [PubMed] [Google Scholar]
- Comrie WA, Li S, Boyle S, Burkhardt JK. (2015) The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J Cell Biol , 457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL, et al. (2015) Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA , 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DK, Mallis RJ, Duke-Cohan JS, Hussey RE, Tetteh PW, Hilton M, Wagner G, Lang MJ, Reinherz EL. (2016) Pre-T cell receptors (pre-TCRs) leverage Vβ complementarity determining regions (CDRs) and hydrophobic patch in mechanosensing thymic self-ligands. J Biol Chem , 25292–25305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Brazin KN, Kobayashi E, Mallis RJ, Reinherz EL, Lang MJ. (2017) Mechanosensing drives acuity of αβ T-cell recognition. Proc Natl Acad Sci USA , E8204–E8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey NV, Porter DL. (2016) CAR T-cells merge into the fast lane of cancer care. Am J Hematol , 146–150. [DOI] [PubMed] [Google Scholar]
- Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan ASH, Magee AS, Danielson ME, et al. (2011) Activation of the innate immune receptor Dectin-1 upon formation of a “phagocytic synapse.” Nature , 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourier C, Jegou A, Husson J, Pincet F. (2008) A nanospring named erythrocyte. the biomembrane force probe. Cell Mol Bioeng , 263–275. [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature , 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CC, Brzostowski JA, Liu D, Long EO. (2010) Tethering of intercellular adhesion molecule on target cells is required for LFA-1-dependent NK cell adhesion and granule polarization. J Immunol , 2918–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Yan C, Li H, Huang W, Shi X, Huang M, Wang Y, Pan W, Cai M, Li L, et al. (2017) Lipid-dependent conformational dynamics underlie the functional versatility of T-cell receptor. Cell Res , 505–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann R, Zhang X, Di Russo J, Li L, Song J, Hannocks MJ, Sorokin L. (2015) The regulation of immune cell trafficking by the extracellular matrix. Curr Opin Cell Biol , 54–61. [DOI] [PubMed] [Google Scholar]
- Hong J, Persaud SP, Horvath S, Allen PM, Evavold BD, Zhu C. (2015) Force-regulated in situ TCR–peptide-bound MHC class II kinetics determine functions of CD4+ T cells. J Immunol , 3557–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KH, Butte MJ. (2016) T cell activation requires force generation. J Cell Biol , 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. (2015) Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol Biol Cell , 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. (2011) Force generation upon T cell receptor engagement. PLoS One , e19680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. (2012) Mechanosensing in T lymphocyte activation. Biophys J , L5–L7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalergis AM, Boucheron N, Doucey M-A, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG. (2001) Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol , 229–234. [DOI] [PubMed] [Google Scholar]
- Kim ST, Takeuchi K, Sun Z-YJY, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL. (2009) The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem , 31028–31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. (2009) Demonstration of catch bonds between an integrin and its ligand. J Cell Biol , 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns MS, Girvin AT, Klein LO, Chen R, Jensen KDC, Newell EW, Huppa JB, Lillemeier BF, Huse M, Chien Y-H, et al. (2010) Evidence for a functional sidedness to the alphabetaTCR. Proc Natl Acad Sci USA , 5094–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA, Zhdanov RI. (2007) Atomic force microscopy probing of cell elasticity. Micron , 824–833. [DOI] [PubMed] [Google Scholar]
- Lee MS, Glassman CR, Deshpande NR, Badgandi HB, Parrish HL, Uttamapinant C, Stawski PS, Ting AY, Kuhns MS. (2015) A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3zetazeta. Immunity , 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekka M. (2016) Discrimination between normal and cancerous cells using AFM. Bionanoscience , 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-C, Chen B-M, Wu P-C, Cheng T-L, Kao L-S, Tao M-H, Lieber A, Roffler SR. (2010) Cutting edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol , 5959–5963. [DOI] [PubMed] [Google Scholar]
- Lim TS, Mortellaro A, Lim CT, Hämmerling GJ, Ricciardi-Castagnoli P, Hammerling GJ, Ricciardi-Castagnoli P. (2011) Mechanical interactions between dendritic cells and T cells correlate with T cell responsiveness. J Immunol , 258–265. [DOI] [PubMed] [Google Scholar]
- Liu B, Chen W, Evavold BD, Zhu C. (2014) Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell , 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CSC, Raychaudhuri D, Paul B, Chakrabarty Y, Ghosh AR, Rahaman O, Talukdar A, Ganguly D. (2018a) Cutting edge: Piezo1 mechanosensors optimize human T cell activation. J Immunol , ji1701118. [DOI] [PubMed] [Google Scholar]
- Liu T-L, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, et al. (2018b) Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science , eaaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K. (2016a) DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA , 5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Galior K, Ma VP-Y, Salaita K. (2017) Molecular tension probes for imaging forces at the cell surface. Acc Chem Res 7b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu Y, Chang Y, Seyf HR, Henry A, Mattheyses AL, Yehl K, Zhang Y, Huang Z, Salaita K. (2016b) Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nat Methods , 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma VP-Y, Liu Y, Blanchfield L, Su H, Evavold BD, Salaita K. (2016) Ratiometric tension probes for mapping receptor forces and clustering at intermembrane junctions. Nano Lett , 4552–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Finkel TH. (2010) T cell receptor triggering by force. Trends Immunol , 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. (2003) Direct observation of catch bonds involving cell-adhesion molecules. Nature , 190–193. [DOI] [PubMed] [Google Scholar]
- Martinez-Martin N, Risueno RM, Morreale A, Zaldivar I, Fernandez-Arenas E, Herranz F, Ortiz AR, Alarcon B. (2009) Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3. Sci Signal , ra43–ra43. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. (2013) The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity , 461–474. [DOI] [PubMed] [Google Scholar]
- McKeithan TW. (1995). Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA , 5042–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguet S, Swamy M, Alarcón B, Luescher IF, Schamel WWA. (2007) Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity , 43–54. [DOI] [PubMed] [Google Scholar]
- Möller J, Lühmann T, Chabria M, Hall H, Vogel V. (2013) Macrophages lift off surface-bound bacteria using a filopodium–lamellipodium hook-and-shovel mechanism. Sci Rep , 2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DJ, Dufrêne YF. (2011) Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol , 461–469. [DOI] [PubMed] [Google Scholar]
- Munoz MA, Biro M, Weninger W. (2014) T cell migration in intact lymph nodes in vivo. Curr Opin Cell Biol , 17–24. [DOI] [PubMed] [Google Scholar]
- Natkanski E, Lee W-Y, Mistry B, Casal A, Molloy JE, Tolar P. (2013) B cells use mechanical energy to discriminate antigen affinities. Science , 1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. (2008) Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods , 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedergang F, Di Bartolo V, Alcover A. (2016) Comparative anatomy of phagocytic and immunological synapses. Front Immunol , 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowosad CR, Spillane KM, Tolar P. (2016) Germinal center B cells recognize antigen through a specialized immune synapse architecture. Nat Immunol , 870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, Milone MC. (2012) Substrate rigidity regulates human T cell activation and proliferation. J Immunol , 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oszmiana A, Williamson DJ, Cordoba S-P, Morgan DJ, Kennedy PR, Stacey K, Davis DM. (2016) The size of activating and inhibitory killer Ig-like receptor nanoclusters is controlled by the transmembrane sequence and affects signaling. Cell Rep , 1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageon SV, Cordoba S-P, Owen DM, Rothery SM, Oszmiana A, Davis DM. (2013) Superresolution microscopy reveals nanometer-scale reorganization of inhibitory natural killer cell receptors upon activation of NKG2D. Sci Signal , ra62. [DOI] [PubMed] [Google Scholar]
- Pageon SV, Tabarin T, Yamamoto Y, Ma Y, Bridgeman JS, Cohnen A, Benzing C, Gao Y, Crowther MD, Tungatt K, et al. (2016) Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc Natl Acad Sci USA , E5454–E5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NR, Bole M, Chen C, Hardin CC, Kho AT, Mih J, Deng L, Butler J, Tschumperlin D, Fredberg JJ, et al. (2012) Cell elasticity determines macrophage function. PLoS One , e41024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previtera ML, Sengupta A. (2015) Substrate stiffness regulates proinflammatory mediator production through TLR4 activity in macrophages. PLoS One , e0145813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD, Zhu C. (2014) Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J Immunol , 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz J, Sixt M. (2010) Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep , 744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter AT, Asano Y, Stinchcombe JC, Dieckmann NM, Chen BC, Gawden-Bone C, van Engelenburg S, Legant W, Gao L, Davidson MW, et al. (2015) Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity , 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosetti F, Chen Y, Sen M, Thayer E, Azcutia V, Herter JM, Luscinskas FW, Cullere X, Zhu C, Mayadas TN. (2015) A lupus-associated Mac-1 variant has defects in integrin allostery and interaction with ligands under force. Cell Rep , 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitakis M, Dogniaux S, Goudot C, Bufi N, Asnacios S, Maurin M, Randriamampita C, Asnacios A, Hivroz C. (2017) Different TCR-induced T lymphocyte responses are potentiated by stiffness with variable sensitivity. Elife , e23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos LC, Blair DA, Kumari S, Cammer M, Iskratsch T, Herbin O, Alexandropoulos K, Dustin ML, Sheetz MP. (2016) Actin polymerization-dependent activation of Cas-L promotes immunological synapse stability. Immunol Cell Biol , 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen S, Wan Z, Li Z, Chau A, Li X, Zhang S, Liu Y, Yi J, Zeng Y, Wang J, et al. (2017) Substrate stiffness governs the initiation of B cell activation by the concerted signaling of PKCβ and focal adhesion kinase. Elife , e23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota A, Strzelecka-Kiliszek A, Gładkowska E, Yoshida K, Mrozin´ska K, Kwiatkowska K. (2005) Binding of IgG-opsonized particles to Fc gamma R is an active stage of phagocytosis that involves receptor clustering and phosphorylation. J Immunol , 4450–4457. [DOI] [PubMed] [Google Scholar]
- Spillane KM, Tolar P. (2017) B cell antigen extraction is regulated by physical properties of antigen-presenting cells. J Cell Biol , 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabley DR, Jurchenko C, Marshall SS, Salaita KS. (2011) Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat Methods , 64–67. [DOI] [PubMed] [Google Scholar]
- Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, Wilen LA, Xu Y, Dufresne ER. (2014) Traction force microscopy in physics and biology. Soft Matter , 4047. [DOI] [PubMed] [Google Scholar]
- Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R. (2011) Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res , 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. (2003) Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA , 1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe PA, Dushek O. (2011) Mechanisms for T cell receptor triggering. Nat Rev Immunol , 47–55. [DOI] [PubMed] [Google Scholar]
- Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. (2006) T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity , 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Chen X, Chen H, Ji Q, Chen Y, Wang J, Cao Y, Wang F, Lou J, Tang Z, et al. (2015) The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. Elife , e06925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Zhang S, Fan Y, Liu K, Du F, Davey AM, Zhang H, Han W, Xiong C, Liu W. (2013) B cell activation is regulated by the stiffness properties of the substrate presenting the antigens. J Immunol , 4661–4675. [DOI] [PubMed] [Google Scholar]
- Wang J, Reinherz EL. (2012) The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev , 102–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ha T. (2013) Defining single molecular forces required to activate integrin and notch signaling. Science , 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X. (2016) Integrins outside focal adhesions transmit tensions during stable cell adhesion. Sci Rep , 36959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weninger W, Biro M, Jain R. (2014) Leukocyte migration in the interstitial space of non-lymphoid organs. Nat Rev Immunol , 232–246. [DOI] [PubMed] [Google Scholar]
- Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, Wucherpfennig KW. (2008) Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell , 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Wu XS, Crites T, Hammer JA., 3rd (2012) Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol Biol Cell , 834–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi J, Wan Z, Liu K, Song P, Chau A, Wang F, Chang Z, Han W, Zheng W, et al. (2015) Substrate stiffness regulates B-cell activation, proliferation, class switch, and T-cell-independent antibody responses in vivo. Eur J Immunol , 1621–1634. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cordoba S-P, Dushek O, Anton van der Merwe P. (2011) Basic residues in the T-cell receptor cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci USA , 19323–19328. [DOI] [PMC free article] [PubMed] [Google Scholar]