We identify the Caenorhabditis elegans myosin light-chain kinase, MLCK-1, required for contraction of spermathecae. During contraction, MLCK-1 moves from the apical cell boundaries to the basal actomyosin bundles, where it stabilizes myosin downstream of calcium signaling. MLCK and ROCK act in distinct subsets of cells to coordinate the timing of contraction.
Abstract
Productive and coordinated tissue contraction requires spatiotemporal regulation of myosin activity. We use the contractile myoepithelial cells of the Caenorhabditis elegans spermatheca to elucidate the molecular mechanisms involved in contraction. Here, we identify and describe a novel myosin light-chain kinase, MLCK-1, that phosphorylates the myosin regulatory light chain and is required for contraction of the spermatheca and animal fertility. During contraction, MLCK-1 is recruited to basal actomyosin bundles and stabilizes myosin in these bundles downstream of phospholipase PLC-ε/PLC-1 and calcium signaling. MLCK and the Rho kinase ROCK are expressed in distinct subsets of spermathecal cells and act in concert to coordinate the timing of contraction. Our results suggest that MLCK-1 phosphorylates myosin primarily in the central bag cells of the spermatheca, while ROCK controls contractility in the distal neck and the valve connecting the spermatheca to the uterus.
INTRODUCTION
Contraction of the actomyosin cytoskeleton drives cell and tissue movements during morphogenesis (Siedlik and Nelson, 2015; Heer et al., 2017) and wound closure (Russo et al., 2005), and allows cells to perceive and respond to mechanical stress (Clark et al., 2007). In nonmuscle cells, cell contraction is driven by the interaction of the motor protein nonmuscle myosin II (NMII) with filamentous actin. NMII is activated by phosphorylation on its myosin regulatory light-chain subunits (MRLC; Sellers, 1981). Upon MRLC phosphorylation, myosin forms bipolar filaments and becomes catalytically active (Conti and Adelstein, 2008; Vicente-Manzanares et al., 2009). In mammalian cells, MRLC can be phosphorylated at Thr-18 and Ser-19 by several kinases, including myosin light-chain kinase (MLCK) (Sakurada et al., 1998; Hirata et al., 2009; Nakajima and Tanoue, 2010) and Rho kinase (ROCK) (Amano et al., 1996; Gally et al., 2009; Beach et al., 2017) and is dephosphorylated by myosin phosphatase (MYPT) (Ito et al., 2004) so that the level of p-MRLC in different tissues is determined by the balance of kinase and phosphatase activities (Kamm and Stull, 2011).
In addition to its role in nonmuscle cells, MLCK regulates contractility in skeletal muscle, cardiac muscle, and smooth muscle cells (Conti and Adelstein, 2008; Vicente-Manzanares et al., 2009; Hong et al., 2011; Zaidel-Bar et al., 2015). In humans, MLCK is encoded by four genes, MYLK1–4, that produce tissue-specific kinases with well-conserved kinase domains (Kamm and Stull, 2011; Chang et al., 2016). MYLK1 is the best characterized of the four MLCKs and encodes multiple isoforms, including smooth muscle myosin light-chain kinase (smMLCK) (Lazar and Garcia, 1999). Mutations in MYLK are associated with aortic aneurysms (Wang et al., 2010), inflammatory bowel disease (Du et al., 2016), asthma (Ammit et al., 2000; Flores et al., 2007; Ma et al., 2008; Wang et al., 2015), and sepsis (Gao et al., 2006). Proper MLCK function is therefore critical to the function of many contractile cell types.
Vertebrate MLCK contains a bilobate kinase domain that is inhibited by a short autoinhibitory or regulatory domain under low-Ca2+ conditions. Upon Ca2+/calmodulin (CaM) binding, autoinhibition is relieved, and the two lobes of the kinase rotate and close to facilitate ATP hydrolysis and MRLC phosphorylation (Knighton et al., 1992; Olah et al., 1993; Kobel et al., 1996; Stull et al., 1998). The structural domains and kinetics of MLCK have been studied well in vitro (reviewed in Stull et al., 1998; Hong et al., 2011) and in cell culture and ex vivo systems (Wadgaonkar et al., 2003; Isotani et al., 2004; Russo et al., 2005). Additionally, mammalian in vivo systems do not allow easy visualization of contractile tissue function in intact animals, limiting the ability to study the role of MLCKs in actomyosin contractility in real time.
Caenorhabditis elegans is small and transparent, allowing visualization of tissues in intact animals. The C. elegans gonad is an excellent in vivo model for the regulation of contraction in real time. Each hermaphrodite has two U-shaped gonad arms, surrounded by smooth-muscle-like sheath cells, which contract to ovulate mature oocytes into the spermatheca, where the oocyte is fertilized (Strome, 1986; McCarter et al., 1997; Hubbard and Greenstein, 2000). The spermatheca is made up of 24 myoepithelial cells that contract in coordination to propel the fertilized embryo into the uterus during ovulation (Strome, 1986; McCarter et al., 1999). Ovulation occurs ∼150 times per gonad arm during the reproductive lifespan of the animal (Hirsh et al., 1976).
Two pathways, a calcium-dependent pathway (Kariya et al., 2004; Kovacevic et al., 2013) and a Rho-regulated pathway (Wissmann et al., 1997, 1999; Gissendanner et al., 2008; Tan and Zaidel-Bar, 2015), are needed for contraction of the spermatheca. Rho regulates the degree of contraction and the timing of embryo transit through the spermatheca (Tan and Zaidel-Bar, 2015). The Rho kinase (ROCK/LET-502) regulates sheath cell contractility in C. elegans (Wissmann et al., 1999; Ono and Ono, 2016). In addition to phosphorylating MRLC, the Rho kinase ROCK elevates myosin activity by phosphorylating and inactivating myosin phosphatase (Kimura et al., 1996). Because ROCK/LET-502 is expressed primarily in the distal neck of the spermatheca and the spermatheca–uterine (sp-ut) valve (Wissmann et al., 1999), we hypothesized that an as yet unidentified MLCK might, in coordination with ROCK, regulate spermathecal contraction through phosphorylation of the MRLC.
In this study, we performed a candidate RNA interference (RNAi) screen to identify the kinase or kinases that might phosphorylate and activate myosin in the C. elegans spermatheca. We identified a previously uncharacterized gene, ZC373.4/mlck-1, that is required for contractility of the spermatheca. We show that loss of mlck-1 results in a failure of oocytes to exit the spermatheca and demonstrate that MRLC phosphorylation in the spermatheca depends on MLCK-1. MLCK-1 is also recruited to, and required for, maintenance of proper actomyosin bundles and dynamics. In addition to the role of MLCK-1 in phosphorylating the MRLC, we found that ROCK/LET-502 regulates MRLC phosphorylation in a subset of cells. Together, these two kinases coordinate spermathecal transit in the C. elegans spermatheca.
RESULTS
MLCK-1 is a putative myosin light-chain kinase required for spermathecal contractility
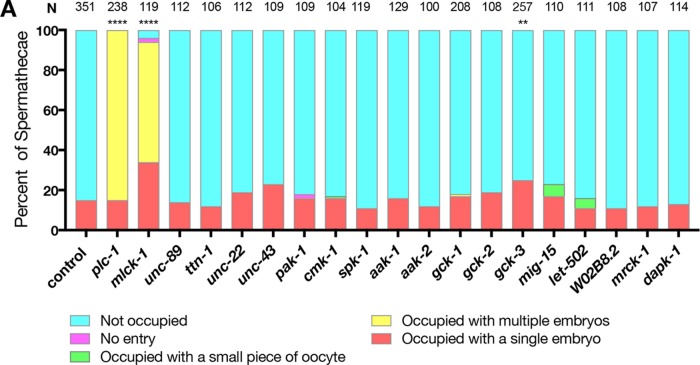
To identify potential kinases regulating spermathecal contractility, we performed a candidate RNAi screen. We screened C. elegans homologues of kinases that have previously been shown to phosphorylate MRLCs, including putative and known myosin light-chain kinases: ROCK (Amano et al., 1996), citron kinase (Yamashiro et al., 2003), myotonic dystrophy–related CDC42-binding kinase (MRCK) (Leung et al., 1998), zipper- interacting protein kinase (ZIPK) (Murata-Hori et al., 1999), and death-associated protein kinase (DAPK) (Shohat et al., 2001). We also screened several kinases that have been shown to have activity toward MRLC in C. elegans, including UNC-89 (Small et al., 2004), titin/TTN-1 (Flaherty et al., 2002), and twitchin/UNC-22 (Moerman et al., 1988; Benian et al., 1989). Additionally, we screened 11 other genes that had kinase domains similar to those of human MLCK, identified using the basic local alignment search tool (BLAST). To avoid disrupting embryonic development, animals were fed RNAi after hatching and scored for spermathecal contractility defects as adults. We have shown previously that disruption of spermathecal contractility can result in accumulation of one or more embryos in the spermatheca (Kovacevic and Cram, 2010). Animals that are mutants for the phospholipase plc-1, which is required for Ca2+ release in the spermatheca, display this phenotype (Kovacevic et al., 2013). We hypothesized that loss of the kinase that activates myosin in the spermatheca would result in comparable spermathecal contractility defects. Similarly to loss of plc-1, we found that knockdown of ZC373.4 significantly increases the percentage of spermathecae occupied by one or more embryos (Figure 1). Because ZC373.4, renamed MLCK-1, is the only kinase that displayed a strong defect in spermathecal contractility similar to that for plc-1, here we focus on characterizing the role of mlck-1 in spermathecal function.
FIGURE 1:
MLCK-1 is required for oocyte transit through the spermatheca. Wild-type animals were grown on empty vector (control), plc-1 (positive control), or candidate myosin kinase RNAi and scored as young adults for spermathecal occupancy in the following categories: unoccupied, occupied with a single embryo, more than one embryo, a small piece of embryo, or no entry, where oocytes fail to enter the spermatheca. MLCK-1 is required for WT ratios of occupied vs. unoccupied spermathecae. Statistics were performed for the total number of unoccupied spermathecae compared with the sum all other phenotypes. N is the total number of spermathecae counted. Fisher’s exact test: ****p < 0.0001, **p ≤ 0.01.
MLCK-1 is structurally similar to human MLCK
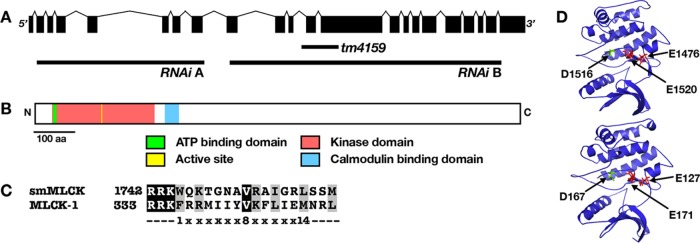
The mlck-1 gene is ∼7 kb and is predicted to encode a single 1211–amino acid protein with a kinase domain and an adjacent C-terminal 1-8-14 calmodulin binding domain (Figure 2, A–C; Yap et al., 2000). Additionally, the C-terminal end of the putative kinase domain contains a conserved HPW helix insertion, a complete linker sequence, and an autoregulatory helix with the three conserved hydrophobic residues necessary to anchor the autoregulatory helix to the kinase core in the absence of Ca2+/CaM binding (Supplemental Figure 1A; Chang et al., 2016). The location of the C-terminal CaM binding domain suggests that CaM binding might relieve autoinhibition and permit MLCK binding and phosphorylation of MRLC (Blumenthal and Stull, 1980). Additionally, MLCK-1 shares 50.2% homology in its kinase domain with Homo sapiens smMLCK (Uniprot ID Q15746; (Huang and Miller, 1991; Supplemental Figure 1B; Figure 2D). All four human MLCK protein kinase domains are structurally similar and show levels of homology similar to those for MLCK-1 (Supplemental Figure 1B). The predicted three-dimensional (3D) structures of the kinase domains are highly conserved (Zhang, 2008; Roy et al., 2010; Yang et al., 2015). Both have the glutamate residues, which bind the MRLC in the uninhibited conformation, and the active sites in similar locations (Figure 2D; Herring et al., 1992; Gallagher et al., 1993). MLCKs are known as dedicated kinases of the MRLC. This specificity is thought to be due to MRLC’s hydrophobic residues in the P+1, P+2, P+3, and P-3 positions (Stull et al., 1998) relative to the phosphorylatable serine, which are conserved in human and C. elegans MRLCs (Supplemental Figure 1C). Therefore, bioinformatic analysis suggests that MLCK-1 may act as a Ca2+/CaM–responsive myosin light-chain kinase.
FIGURE 2:
MLCK-1 has a serine/threonine kinase domain that is structurally similar to that in human MLCKs. (A) The mlck-1 gene spans ∼7 kb and is made up of 22 exons. The positions of two RNAi targeting constructs (A, B) and the tm4159 deletion are labeled. (B) MLCK-1 has an N-terminal kinase domain (pink), with its active site and ATP binding domain labeled in yellow and green, respectively. Putative calmodulin binding domains are labeled in blue. (C) MLCK-1 contains a putative 1-8-14 Ca2+-dependent calmodulin binding domain homologous to smMLCK. (D)The kinase domain–predicted structures (iTasser) are similar. Key glutamate residues (red) bind the RLC substrate. The active site aspartate residue is in green.
MLCK-1 is expressed in the spermatheca and is localized to contractile actomyosin bundles during contraction
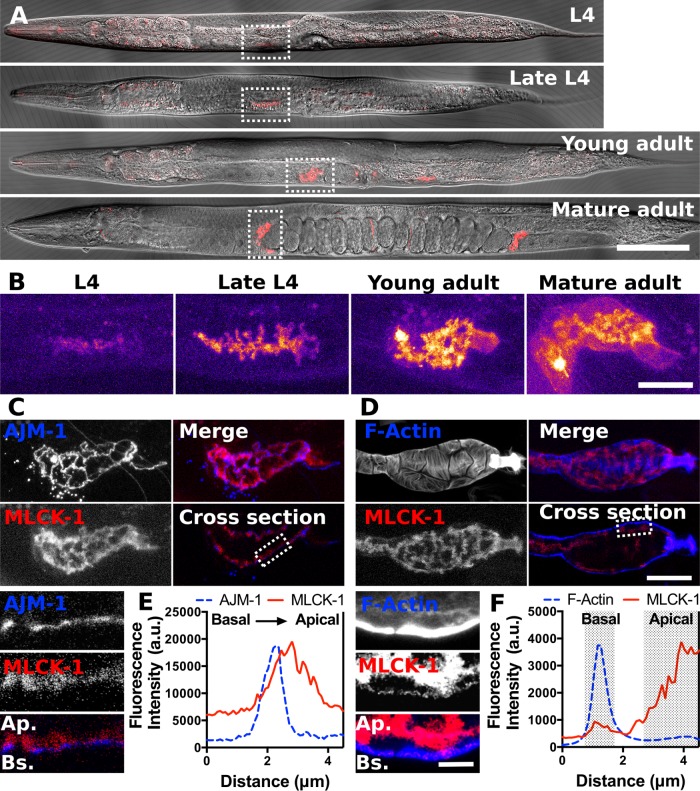
To characterize the MLCK-1 expression pattern, we generated a GFP-promoter transcriptional reporter and a CRISPR line in which the endogenous mlck-1 locus is labeled at the C-terminus with mKate2. The two lines displayed overlapping but not identical expression patterns (Figure 3 and Supplemental Figure 2). The transcriptional reporter line exhibited strong MLCK-1 expression in the pharynx, anal sphincter, vulval cells, spermatheca, and sp-ut valve (Supplemental Figure 2A), while the CRISPR line showed MLCK-1 expression in the pharynx, uterus, spermatheca, and sp-ut valve (Figure 3A). Importantly, both lines show expression in the spermatheca. There are no apparent phenotypes in the CRIPSR-tagged line, indicating that the MLCK-1::mKate2 fusion is likely functional (Supplemental Movie 1; Supplemental Figure 2B).
FIGURE 3:
MLCK-1 is expressed in the C. elegans spermatheca and localizes to apical cell boundaries and basal actin bundles. (A) Confocal images of animals from L4 to young adult expressing MLCK-1 tagged with mKate2 at the endogenous locus. MLCK-1 expression is seen in the spermatheca throughout development. (B) Magnified maximum-intensity projections of the spermathecae from the animals in A indicated with dashed boxes. The Fire color scale is used to highlight changes in fluorescence intensity. MLCK-1 expression increases from L4 to young adulthood and is primarily localized apically. In mature adults, a subpopulation of MLCK-1 is observed at the basal surface. (C) Confocal images of a spermatheca after the first ovulation from an animal coexpressing MLCK-1::mKate2 (red) and AJM-1::GFP (blue), which localizes near the apical surface of the spermathecal cells. Maximum-intensity projections show the proximity of MLCK-1 and AJM-1, with MLCK-1 immediately apical to AJM-1. The insert is a magnified cross-section from the area indicated with a dashed line, with basal at the bottom. (D) Confocal images of a WT excised spermatheca after the first ovulation expressing MLCK-1::mKate2 and stained with phalloidin to visualize F-actin. Maximum-intensity projections show actin bundles and MLCK-1 localization. The cross-section shows that prominent actin bundles are basal and a small population of MLCK-1 colocalizes with these bundles. Inserts are a magnified cross-section of the area indicated with a dashed line. Basal is at the bottom. Note that the brightness in the insert in D is enhanced to facilitate simultaneous visualization of thick basal actin bundles and the thin apical actin network. (E) Quantification of a line scan drawn across the apical cell surface in the area indicated by the insert in C. The fluorescence peak for AJM-1::GFP is just basal to MLCK-1::mKate2 peak. (F) Quantification of a line scan drawn across the area indicated in the insert in (D) from the basal to the apical cell surface. The majority of the MLCK-1::mKate2 signal is apical and a small peak aligns with the basal fluorescence peak for F-actin. Scale bar, 100 μm in A, 20 μm in B–D, and 5 μm in the inserts in C and D. Ap., apical; Bs., basal.
Movie S1.
MLCK-1 is recruited to basal actomyosin bundles during contraction in a PLC-1 dependent manner. Maximum intensity projections of a confocal 4-D ovulation movie of animals expressing MLCK-1::mKate2 treated with control (top) or plc-1 RNAi (bottom). In spermathecae of both wild type and plc-1(RNAi) animals, MLCK-1 is focused at apical cell boundaries prior to oocyte entry when the distal spermathecal neck first opens (0 s) and after the oocyte is completely within the spermathecae (100 s). In the wild type spermatheca, a subset of MLCK-1 diffuses away from cell boundaries during contraction (480 s) and remains there after oocyte exit (690 s). In the plc-1(RNAi) spermatheca, the embryo is retained in the spermatheca and MLCK-1::mKate2 remains exclusively at cell boundaries throughout the duration of imaging (1390 s). Ovulation was recorded by acquiring 20 confocal z-slices every 10 seconds and the frame rate of the movie is 10 frames per second. Scale bar is 20 μm.
By mid L4, faint MLCK-1 expression is evident in the cells of the developing spermatheca. Expression levels peak in adulthood (Figure 3B). During L4, MLCK-1 is localized to apical boundaries of the spermathecal cells (Figure 3, B and C), which face the lumen of the spermathecal bag. In adults, MLCK-1 is localized both apically and basally in the spermatheca (Figure 3B). We have previously shown that active myosin is recruited to basal actomyosin bundles concomitant with spermathecal contraction (Wirshing and Cram, 2017). Therefore, we anticipated that MLCK-1 would colocalize with its putative substrate, MLC-4, in these actomyosin bundles. To visualize the localization of the basal population of MLCK-1, gonads were excised from the labeled mlck-1::mKate2–expressing animals, and phalloidin was used to visualize F-actin. As expected, we observed thick bundles of actin at the outer, basal surface with a thinner actin network visible at the apical surface (Figure 3, D and F). MLCK-1 fluorescence intensity is brightest at the apical boundaries (Figure 3, C and E). However, some MLCK-1 is also localized to the basal cell surface, where it partially colocalizes with the contractile actomyosin bundles (Figure 3, D and F; Supplemental Figure 4).
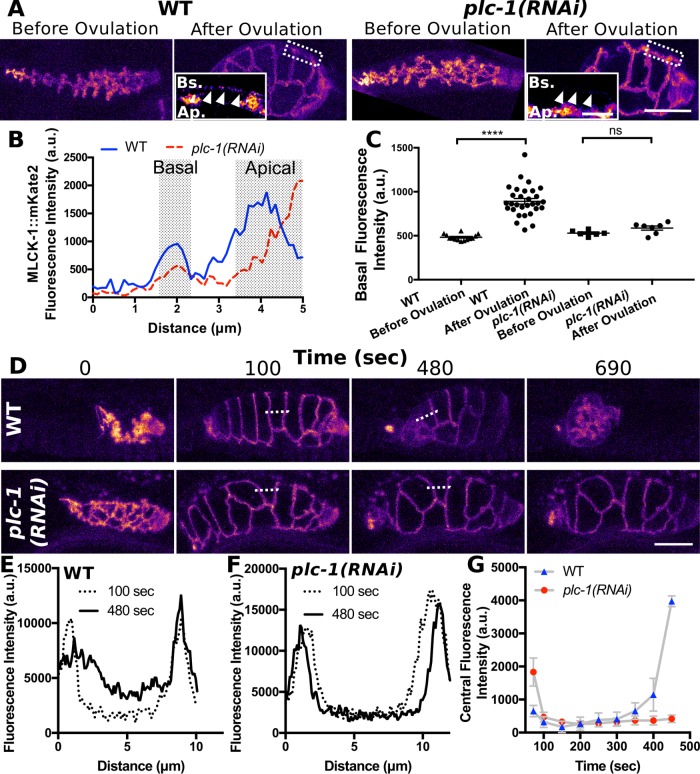
To determine whether the basal recruitment of MLCK-1 seen in adults requires ovulation and plc-1, we repeated the experiment on control and plc-1(RNAi) animals before and after the first ovulation. In spermathecae of control animals, MLCK-1::mKate2 fluorescence becomes visible at the basal cell surface after the first ovulation. In contrast, in spermathecae of plc-1(RNAi) animals, there is no significant increase MLCK-1 at the basal cell surface after ovulation (Figure 4, A–C). To understand when MLCK-1 localization changes during ovulation and to determine what drives MLCK-1 dynamics, we used four-dimensional (4D) confocal microscopy of live animals to capture time-lapse videos of ovulation. Before the first ovulation, MLCK-1 is restricted to apical boundaries (Figure 4, D–G). During oocyte entry, the spermatheca is stretched. However, this initial cell stretch is not sufficient to displace MLCK-1 from the apical boundaries because MLCK-1 localization remains unchanged until ∼350 s after oocyte entry (Figure 4A). MLCK-1 begins to move basally as the spermathecal cells contract and the embryo is expelled from the spermatheca, ∼480 s after opening of the distal neck (Figure 4). This partial relocalization of MLCK-1 to the basal surface is maintained throughout adulthood. In animals depleted of plc-1, the oocyte enters the spermatheca but becomes trapped as spermathecal cells fail to contract (Kovacevic et al., 2013). In these animals, MLCK-1 is retained at apical boundaries (Figure 4, D–G). These results suggest Ca2+ signaling may play a role in the initiation of contraction and the movement of MLCK-1 away from the apical boundaries.
FIGURE 4:
MLCK-1 is recruited basally during contraction in a PLC-1–dependent manner. (A) Confocal maximum-intensity projections of excised spermathecae from WT and plc-1(RNAi) animals before and after the first ovulation. Inserts are a magnified cross-section of the area indicated in A with a dashed line. Fluorescence intensity is increased in inserts to highlight the subset of basally located MLCK-1 present in spermathecae after the first ovulation, which is absent in plc-1(RNAi) spermathecae. White arrowheads indicate basal cell surface. (B) Quantification of MLCK-1::mKate2 fluorescence intensity across the cross-section shown in the inserts in A. (C) Fluorescence intensity at the basal cell surface measured from 14 WT spermathecae before the first ovulation, 30 after the first ovulation, 6 plc-1(RNAi) spermathecae before the first ovulation, and 7 after the first ovulation. (D) Still frames from a confocal 4D ovulation movie of animals expressing MLCK-1::mKate2 treated with control (WT) or plc-1 RNAi. In spermathecae of both WT and plc-1(RNAi) animals, MLCK-1 is localized to the apical surface of the spermatheca prior to oocyte entry when the distal spermathecal neck first opens (0 s) and after the oocyte is completely within the spermathecae (100 s). In WT spermathecae, a subset of MLCK-1 diffuses away from apical boundaries during contraction (480 s) and is retained basally even after oocyte exit (690 s). (E, F) Quantification of MLCK-1::mKate2 fluorescence intensity across a single cell, indicated by a dashed line in D, at 100 and 480 s for the WT (E) and plc-1(RNAi) (F) spermathecae. The peaks indicate MLCK-1 localized at cell boundaries. By 480 s, fluorescence intensity between the peaks increases in the WT cell (E). This is not seen in the plc-1(RNAi) cell (F). (G) Quantification of the average MLCK-1::mKate2 fluorescence intensity measured across the cell center, excluding MLCK-1 localized to cell boundaries, during ovulation. MLCK-1::mKate2 fluorescence is first detected at 350 s and increases during contraction in WT cells but not plc-1(RNAi) cells. Measurements were taken of individual cells from three different first ovulation movies for WT (nine cells total) and plc-1(RNAi) (nine cells total) animals at 50 s intervals. No more than three cells were measured from the same animal. Error bars represent SEM. Error bars indicated SEM. Unpaired t test: ns p > 0.05; ****p ≤ 0.0001. Scale bar, 20 μm and 5 μm for inserts. Ap., apical; Bs., basal.
MLCK-1 is required maternally for fertility
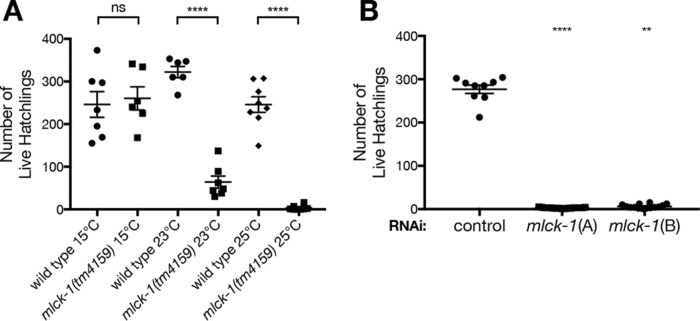
Disruption of genes required for regulating spermatheca contractility can result in the production of fewer live offspring (Kovacevic and Cram, 2010; Kovacevic et al., 2013). To test for fertility defects in mlck-1 loss of function, we used the hypomorphic allele mlck-1(tm4159) and two different mlck-1 RNAi clones that targeted distinct regions of the gene to disrupt MLCK-1 function (Figure 2A). The allele has a 514–base pair deletion that leaves the kinase domain intact (Figure 2A), while the RNAi depletes mlck-1 below detectable levels (Supplemental Figure 3). The tm4159 allele is temperature-sensitive, perhaps because of instability of the encoded protein (Varadarajan et al., 1996; Brown et al., 1997; Gidalevitz et al., 2006). The tm4159 animals grown at 25°C produce only a few live hatchlings. tm4159 animals grown at 23°C also produce significantly fewer live offspring than wild-type (WT) animals grown at the same temperature, while worms reared at 15°C produce WT numbers of live offspring (Figure 5A). Treatment with either of two nonoverlapping mlck-1 RNAi clones also resulted in significantly fewer live offspring (Figure 5B).
FIGURE 5:
MLCK-1 is required for fertility. (A) The mlck-1(tm4159) allele is temperature-sensitive and results in a reduction in number of live offspring. This reduction is more severe at higher temperatures. Each data point reflects the number of live F1 offspring from one individual. Error bars indicate SEM. Unpaired t test: ****p < 0.0001. (B) Depletion of mlck-1 using two RNAi clones that target distinct regions of the mlck-1 gene results in a significant reduction in numbers of live offspring compared with animals fed control RNAi. Error bars indicate SEM. One-way ANOVA with Dunnett’s multiple comparison test: ****p < 0.0001, **p ≤ 0.01.
To determine whether mlck-1 is required for embryonic development, we tracked the hatching of embryos from mlck-1 mothers at 23°C. We find that 18% of mlck-1(tm4159) embryos fail to hatch (n = 551) compared with only 2% of WT embryos (n = 821). This suggests a modest reduction in embryonic viability in mlck-1(tm4159) animals, which is insufficient to explain the observed reduction in live offspring (Supplemental Figure 5A). To explore the possibility tm4159 has a maternal effect, we mated mlck-1(tm4159) hermaphrodites with WT males and found that the paternally inherited WT mlck-1 allele is insufficient to rescue the reduction in numbers of live offspring (Supplemental Figure 5B). Conversely, when a WT hermaphrodite is crossed with a mlck-1(tm4159) male, the embryos are viable and WT numbers of offspring are observed (Supplemental Figure 5, A and B). This demonstrates that mlck-1/+ heterozygous offspring are viable and that mlck-1 is not required in sperm. Taken together, these results suggest that mlck-1 depletion reduces numbers of live offspring and embryonic viability through a maternal mechanism.
MLCK-1 is required for contraction of the spermatheca
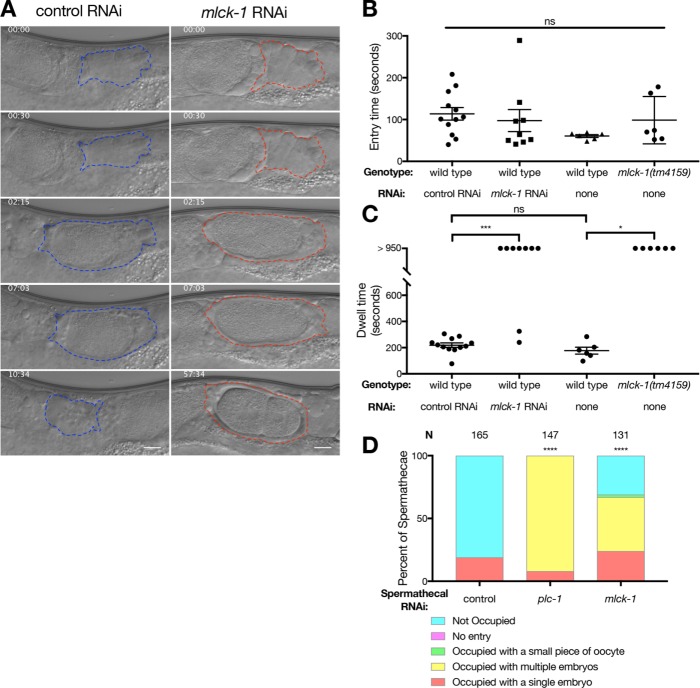
We next hypothesized that MLCK-1 may be required for contraction of the spermatheca and exit of the newly fertilized embryo into the uterus. To characterize the temperature sensitivity of transit defects in mlck-1(tm4159) animals, we performed an occupancy assay as described above on wild-type and mlck-1(tm4159) animals reared at 15, 23, or 25°C. Spermathecal contractility defects are more pronounced at the warmer temperatures, and animals grown at 23 and 25°C have a higher percentage of spermathecae occupied with multiple embryos (Supplemental Figure 5C). These results are in agreement with our mlck-1(RNAi) data (Figure 1), indicating that depletion of mlck-1 via RNAi or the mlck-1(tm4159) allele at 25°C produces similar results. To determine how spermathecal contraction is altered in real time, we imaged full ovulations of WT animals and animals depleted of mlck-1 via RNAi or the mlck-1(tm4159) allele (Supplemental Movies 2 and 3; Figure 6A). Three aspects of oocyte transit were quantified: entry time, dwell time, and exit time.
FIGURE 6:
MLCK-1 is required for contractility of the spermathecal bag cells. (A) Stills from time-lapse imaging of animals fed either control or mlck-1 RNAi, respectively. Embryos from hermaphrodites fed mlck-1 RNAi fail to exit the spermatheca. The spermatheca is outlined in either blue or red for animals fed control and mlck-1 (A) RNAi, respectively. Scale bar = 10 μm. (B) No significant difference in entry time is observed. One-way ANOVA with Tukey’s multiple comparison test. ns p > 0.05. (C) Dwell time in WT animals fed control RNAi is 218 ± 58.9 s (n = 12). In animals fed mlck-1 RNAi, 9 out of 11 ovulations imaged had indefinite dwell times. There is no significant difference between WT animals grown at 23°C and fed control RNAi and WT animals grown at 25°C and fed OP50. One hundred percent of mlck-1(tm4159) nematodes (n = 6) grown at the nonpermissive temperature, 25°C, fail to exit the spermatheca during the time imaged (>950 s). Error bars indicate SEM. One-way ANOVA with Tukey’s multiple comparison test: ns p > 0.05, *p ≤ 0.05, ***p ≤ 0.001. (D) Animals competent for RNAi in the spermatheca only fed mlck-1 RNAi have a significant number of spermathecae occupied compared with wild type. Fisher’s exact test (nonoccupied vs. all other outcomes): ****p < 0.0001.
Movie S2.
DIC movie of a wild type ovulation. The sheath contracts and the oocyte is squeezed into the spermatheca bag beginning at approximately 30sec. The oocyte remains in the spermatheca for several minutes until the sp-ut valve opens at approximately 4 minutes 30 seconds. The fertilized embryo is then pushed into the uterus through spermathecal contractions. Images were acquired at 1 image per second. Playback is 60 frames per second. Scale bar is 10μm.
Movie S3.
Depletion of mlck-1 results in embryos being trapped in the spermatheca. Contractions from the sheath cells coordinate with the distal neck to push the oocyte into the spermatheca bag. The oocyte is fertilized, but the spermatheca fails to contract and the embryo begins to develop and divide within the spermatheca for the length of imaging (>55 minutes). Images were acquired at 1 image per second. Playback is 60 frames per second. Scale bar is 10μm.
Entry time describes the time from the beginning of oocyte entry to when the distal neck cells close around the now fertilized embryo. Entry requires coordination between the proximal gonadal sheath cells and spermathecal distal neck cells. Loss of mlck-1 using either RNAi or the mlck-1(tm4159) allele at 25°C does not significantly slow entry time (Figure 6B). This result suggests that MLCK-1 may not play a critical role in sheath cell contraction.
Dwell time refers to the time between closure of the distal neck and the opening of the sp-ut valve. This is the amount of time that the embryo is completely enclosed by the spermatheca. Knockdown of mlck-1 with RNAi generally results in failure of embryos to exit the spermatheca (77%, n = 9; Supplemental Movie 3; Figure 6C; Supplemental Figure 6A). Occasionally, an mlck-1(RNAi) animal exhibits a successful transit, which may be due to variability in RNAi penetrance. In mlck-1(tm4159) animals grown at 25°C, oocytes enter the spermatheca but become trapped as the spermatheca fails to contract (100%, n = 6; Figure 6C). Trapping can result from defective sp-ut valve dilation. To address this, we constructed a spermathecal-specific RNAi strain. In this strain, rde-1, an Argonaute required for RNAi (Tabara et al., 1999), is rescued only in the spermathecal bag using the fkh-6 promoter (Hope et al., 2003). When we deplete mlck-1 in the spermathecal bag, leaving the sp-ut valve functional, embryos fail to exit properly (Figure 6D). This suggests that MLCK-1 in the spermathecal bag cells is needed for successful embryo exit.
The last metric we used is exit time, the time from the dilation of the spermatheca uterine valve to when the embryo has been pushed into the uterus and the sp-ut valve has fully reclosed. Although we cannot measure exit time when embryos become trapped in the spermatheca, as is seen in mlck-1(RNAi) and mlck-1(tm4159) animals at 25°C, mlck-1(tm4159) animals grown at 23°C exhibit significantly slower exits than WT animals (Supplemental Figure 6C). Taken together, these results suggest that MLCK-1 is required in the spermathecal bag cells, and that transit defects are mainly due to a decrease in spermathecal cell contractility and a failure to push the fertilized embryo out through the sp-ut valve and into the uterus.
MLCK-1 is required for production of p-MRLC in the spermatheca
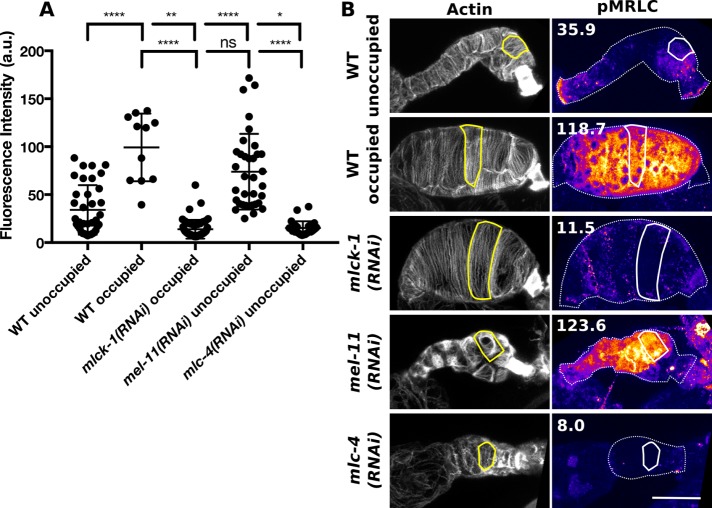
We hypothesized that transit defects in mlck-1 animals were due to failure to phosphorylate the MRLC during ovulation to activate myosin and initiate cell contraction. To determine the contribution of MLCK-1 to p-MRLC production in the spermatheca, we used immunohistochemistry to quantify p-MRLC levels (Figure 7). To confirm that our antibody accurately reports p-MRLC levels, we included mel-11(RNAi) and mlc-4(RNAi) spermathecae as controls. MEL-11 is the C. elegans ortholog of the myosin-associated phosphatase regulatory subunit (referred to here as myosin phosphatase) required to dephosphorylate MRLC during cell relaxation (Wissmann et al., 1999; Kovacevic et al., 2013; Ono and Ono, 2016; Wirshing and Cram, 2017). As expected, quantification of p-MRLC levels in mel-11(RNAi) spermathecal cells reveals that these cells have significantly elevated p-MRLC, compared with unoccupied WT spermatheca (Figure 7). MLC-4 is the only C. elegans nonmuscle MRLC (Shelton et al., 1999). As anticipated, we find that mlc-4(RNAi) spermathecal cells have very low levels of p-MRLC (Figure 7).
FIGURE 7:
MLCK-1 is required for pMRLC production in the spermatheca. (A) Quantification of pMRLC levels detected using a pMRLC-specific antibody. Measurements were taken from 37 WT unoccupied, 11 WT occupied, 53 mlck-1(RNAi), 35 mel-11(RNAi), and 28 mlc-4(RNAi) spermathecal cells with no more than three cells measured from the same animal. Dunnett’s multiple comparison test: ns p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001. Error bars represent SD. (B) Representative confocal maximum intensity projections of the data presented in (A) F-actin was stained using phalloidin to detect individual cells. An example cell is outlined in the image, showing actin and pMRLC levels. Numbers are the total fluorescence intensity values of the selected cells. Scale bar, 20 μm.
Comparison of p-MRLC levels in cells from unoccupied and occupied WT spermathecae reveals that occupied spermathecae tend to have higher p-MRLC levels (Figure 7). This suggests that p-MRLC levels may peak during ovulation. By comparison, cells of mlck-1(RNAi) spermathecae consistently have low p-MRLC levels (Figure 7). Even though all mlck-1(RNAi) spermathecal cells observed are from occupied spermathecae, these cells fail to achieve the peak p-MRLC levels seen in WT occupied spermathecae (Figure 7). In mlck-1(RNAi) spermatheca, only very low levels of p-MRLC can be seen at cell boundaries, while p-MRLC was detected at cell boundaries and throughout the cytosol in WT occupied spermatheca (Figure 7). p-MRLC levels in mlck-1(RNAi) and mlc-4(RNAi) spermathecal cells are not significantly different. In summary, we find that MLCK-1 is required for the production of peak p-MRLC levels during ovulation and that loss of mlck-1 results in reduced p-MRLC levels comparable to those from directly depleting MRLC.
MLCK-1 is required for development of parallel actomyosin bundles during ovulation
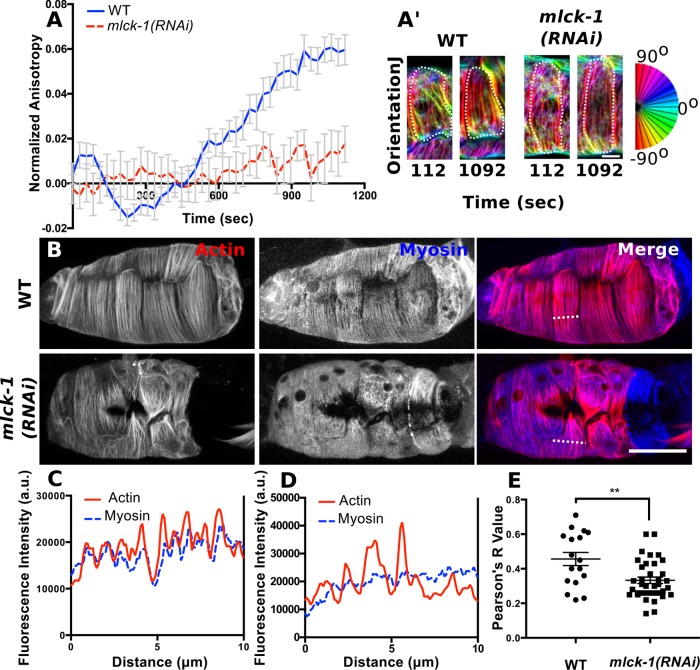
During spermathecal cell contraction, the actomyosin cytoskeleton reorganizes from an isotropic interconnected network to form highly organized, basal, parallel actomyosin bundles (Wirshing and Cram, 2017). Myosin activity plays a key role in spermathecal cell contractility and organization of these basal actomyosin bundles (Wirshing and Cram, 2017). Because MLCK-1 is required for spermathecal contractility and myosin activity, we hypothesized that MLCK-1 would also be required for WT actomyosin network organization. To determine the role of MLCK-1 in actomyosin bundle formation during ovulation, we used 4D confocal microscopy of live animals expressing GFP::ACT-1 driven by a spermathecal specific promoter to observe actin dynamics during ovulation. Actin network organization was quantified using the ImageJ plug-in FibrilTool (Boudaoud et al., 2014) to measure anisotropy. As previously reported, production of parallel actin bundles in WT spermathecal cells occurs after cell stretch due to oocyte entry and coincides with the onset of contraction (Figure 8, A and A’). In contrast, mlck-1(RNAi) spermathecal cells fail to produce WT actomyosin bundles. Instead, thin tortuous bundles that frequently fail to extend the whole length of the cell are present even after lengthy cell stretch (Figure 8). Simultaneous visualization of F-actin (moe-ABD::mCherry) and myosin (GFP::NMY-1) reveals that actin and myosin colocalize into parallel actomyosin bundles in WT spermathecae (Figure 8, B–E). Without mlck-1, myosin is poorly recruited to the basal actin bundles and a large fraction of myosin remains diffused throughout the cytosol (Figure 8, B–E). This indicates that MLCK-1 is required for production of the basally enriched parallel actomyosin bundles.
FIGURE 8:
MLCK-1 is required for formation of actomyosin bundles. (A) Quantification of anisotropy (degree of alignment) in individual spermathecal cells expressing GFP::ACT-1 to label actin during ovulation. FibrilTool was used to measure anisotropy in individual cells at 28-s intervals during ovulation. No more than two cells were measured from the same animal. For WT, n = 14 cells (7 animals); for mlck-1(RNAi), n = 6 cells (3 animals). (A’) Representative confocal maximum-intensity projections of spermathecal cells expressing GFP labeled actin before (112 s) and after (1092 s) development of actomyosin bundles analyzed in A. Each cell is outlined with a dotted line and OrientationJ was used to false color actin bundles according to their orientation. (B) Confocal maximum intensity projections of spermathecae expressing moeABD::mCherry to label F-actin and GFP::NMY-1 to label myosin. Note that myosin appears diffuse and is poorly recruited to actin bundles in the mlck-1(RNAi) spermatheca. (C, D) Fluorescence intensity across the line indicated in B for a WT (C) and an mlck-1(RNA1) (D) cell. Peaks indicate actomyosin bundles. Actin fluorescence peaks have corresponding myosin fluorescence peaks in the WT cell (C) but not in the mlck-1(RNAi) cell (D), where myosin fluoresces in uniform. (E) Colocalization analysis of actin and myosin in individual cells. Each point represents the Pearson’s R value for an individual cell with no more than three cells measured from the same spermatheca; WT, n = 17 cells (7 spermathecae); mlck-1(RNAi), n = 36 cells (12 spermathecae). Error bars represent SEM. Unpaired t test: **p ≤ 0.01. Scale bar, 5 μm (A) and 20 μm (B).
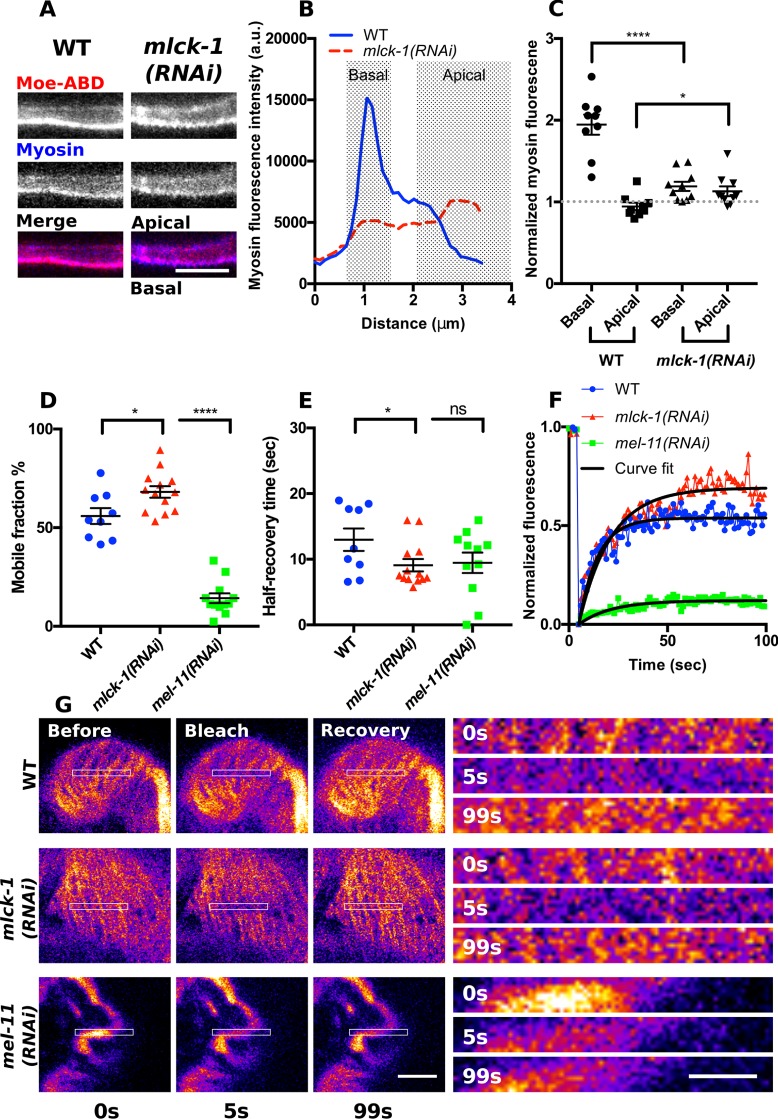
To further explore the effect of mlck-1 depletion on the distribution of myosin, we imaged GFP::NMY-1 in spermathecal cross-sections and observed a loss of basal myosin localization and an enrichment of myosin at the apical actin network when mlck-1 is depleted (Figure 9, A–C). Unphosphorylated nonmuscle myosin does not form bipolar filaments (Scholey et al., 1980) or stably associate with actin (Watanabe et al., 2007). To determine whether the GFP::NMY-1 distribution could be explained by a reduction in the stability of association of myosin with actin, we used fluorescence recovery after photobleaching (FRAP) to quantify myosin dynamics in actomyosin bundles. As has been seen in other systems (Watanabe et al., 2007; Kondo et al., 2011), depletion of mlck-1 resulted in enhanced myosin dynamics, increasing the mobile fraction and decreasing the recovery time compared with wild type (Figure 9, D–G). Conversely, increasing myosin activity by knocking down the myosin phosphatase mel-11 has the opposite effect on myosin dynamics. Depletion of mel-11 results in the formation of large myosin foci that form transverse bands extending across multiple actin bundles (Wirshing and Cram, 2017). These myosin foci are highly stable and show little recovery after bleaching (Figure 9, D–G). Overall, these results suggest that MLCK-1 is required for formation of basally enriched actomyosin bundles.
FIGURE 9:
MLCK-1 is required for stable recruitment of myosin into basal actomyosin bundles. (A) Confocal cross-section of a WT and an mlck-1(RNAi) spermathecal cell expressing moeABD::mCherry to label F-actin and GFP::NMY-1 to label myosin. In the WT cell, actin and myosin are primarily at the basal cell surface. In the mlck-1(RNAi) cell, basal actomyosin is less prominent and myosin is also enriched at the apical cell surface. (B) Measured myosin fluorescence intensity across the region shown in A. (C) Quantification of apical and basal myosin fluorescence intensity in WT and mlck-1(RNAi) cells normalized to the cytosolic myosin fluorescence intensity. Each point represents a measurement taken from a single cell, with only one cell measured per spermatheca; WT, n = 9; mlck-1(RNAi), n = 11. (D–G) FRAP analysis of myosin dynamics in WT, mlck-1(RNAi), and mel-11(RNAi) basal actomyosin bundles in spermathecal cells. Basal actomyosin bundles were bleached and recovery was monitored at 1-s intervals for 100 s to calculate the mobile fraction (D) and half recovery time (E). Each point is a measurement from a single cell with no more than three cells measured from the same animal; WT, n = 9 cells (7 animals); mlck-1(RNAi), n = 13 cells (6 animals); mel-11(RNAi), n = 12 cells (6 animals). (F, G) Representative myosin fluorescence recovery curves (F) for the cells shown in G. Unpaired t test: ns p > 0.05, *p ≤ 0.05, ****p ≤ 0.0001. Error bars represent SEM. Scale bar 5 μm and 2 μm for the inserts.
MLCK-1 and ROCK/LET-502 have distinct expression patterns and roles in spermathecal contractility
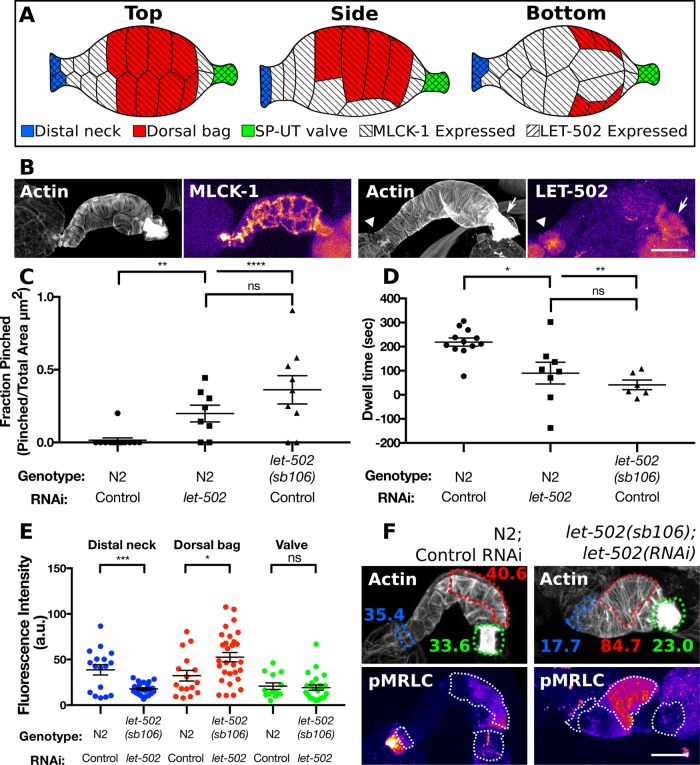
Given the importance of the Rho kinase ROCK/LET-502 in activating myosin and regulating tissue contractility, we sought to distinguish the roles of ROCK/LET-502 and MLCK-1 in the spermatheca. We used a line expressing LET-502 labeled at the endogenous locus with GFP using CRISPR to determine the expression pattern of let-502. As has been observed previously (Wissmann et al., 1999), LET-502 is only faintly expressed in the spermathecal bag cells and high levels of LET-502 are restricted to the distal neck and sp-ut valve cells (Figure 10, A and B). This is distinct from the expression pattern of MLCK-1, which is seen throughout the spermatheca including the distal neck, main bag cells, and sp-ut valve (Figures 3B and 10, A and B).
FIGURE 10:
MLCK-1 and ROCK/LET-502 have distinct spatial expression patterns and roles in the spermatheca. (A) Anatomy of the spermatheca highlighting differentially regulated cells and mlck-1 and let-502 expression patterns. (B) Confocal maximum projections of excised spermathecae stained with phalloidin to visualize F-actin expressing mlck-1 or let-502 labeled with mKate2 and GFP, respectively, at the endogenous locus. MLCK-1 is seen throughout the spermatheca, including the distal neck, bag, and valve cells, while LET-502 is prominently expressed only in the distal neck and valve. (C) Animals with depleted let-502 either through RNAi or the sb106 allele damage oocytes during entry into the spermatheca, resulting in a larger fraction of the pinched oocytes in the gonad arm. (D) LET-502 is also required for WT dwell time. Loss of let-502 results in the spermatheca uterine valve opening prior to distal neck closure and thus a negative dwell time. (E) Quantification of p-MRLC in three regions of the spermatheca using a p-MRLC–specific antibody. Each point represents a measurement taken from a single spermatheca; WT distal neck, dorsal bag, and valve, n = 17, 16, and 13, respectively; let-502(sb106);let-502(RNAi) distal neck, dorsal bag, and valve, n = 22, 29, and 24, respectively. (F) Representative confocal maximum intensity projections of the data presented in E. F-actin was stained using phalloidin to detect individual cells. The distal neck (blue), dorsal bag (red), and valve (green) cells are indicated with a dotted outline. Numbers in F indicate average fluorescence intensity of the outlined regions. The distal neck frequently has elevated p-MRLC, which is absent in animals depleted of let-502. Unpaired t test: ns p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. In C, the unpaired t test with Welch’s correction was used to account for differences in the SD. Scale bar, 20 μm.
To further characterize the role of LET-502 in oocyte entry and transit through the spermatheca, we depleted let-502 using RNAi and a temperature-sensitive allele, let-502(sb106), that has decreased function above 20°C (Piekny et al., 2000). Embryo transits in wild type, let-502(RNAi), and let-502(sb106) animals were observed using DIC microscopy. In agreement with previous work (Wissmann et al., 1999; Ono and Ono, 2016), we find that let-502 is required for oocyte entry, which is driven by sheath cell contraction (Figure 10, C and D). During oocyte entry, sheath cells pull the distal neck of the spermatheca over the incoming oocyte, and the distal neck cells contract to enclose the now-fertilized embryo. In WT animals, this process occurs without damaging the oocyte. In contrast, in let-502 depleted animals, the distal neck cells close on the incoming oocyte, pinching it in half, allowing part of it into the spermatheca while the other portion remains in the gonad arm (Figure 10C). In WT animals, the embryo remains completely enclosed by the spermatheca for a dwell time of ∼200 s. When let-502 is disrupted, the sp-ut valve does not remain closed. This results in short dwell times, where, in some cases, the valve opens to allow the oocyte into the uterus before the distal neck cells close (negative values in Figure 10D). These results, in conjunction with the LET-502 expression pattern, suggest that LET-502 is both expressed and functions to regulate the contractility of the distal neck cells and the sp-ut valve.
Because ROCK plays an important role in activating myosin (Amano et al., 1996; Gally et al., 2009; Beach et al., 2017), we used the same immunohistochemistry approach described above to quantify p-MRLC levels in spermathecae from let-502 depleted animals. In agreement with previous work, we find that let-502 RNAi produces a mild phenotype (Ono and Ono, 2016) and that the allele produces a stronger loss of function phenotype (Figure 10, C and D). For this reason, we used a combination of let-502 RNAi in the temperature sensitive background to deplete let-502 activity. Using this approach, we find that loss of let-502 does not result in a reduction in p-MRLC levels in the spermathecal bag cells compared with those in WT spermathecal bag cells (Figure 10, E and F). Rather, there is a slight but significant increase in p-MRLC in these cells in let-502 depleted animals (Figure 10, E and F). This differs from results obtained with mlck-1(RNAi) spermathecae where p-MRLC levels were strongly reduced (Figure 7). However, we do find that let-502 is required for p-MRLC levels in the distal neck. In WT spermathecae, the distal neck cells frequently have elevated p-MRLC levels (Figure 10, E and F). This is abolished by depletion of let-502 (Figure 10, E and F). Although our results indicate that let-502 is required for contractility of the valve, we observed low p-MRLC levels in valves from both WT and let-502 depleted spermathecae (Figure 10, E and F). This may indicate that basal p-MRLC levels are low in the valve so that we are unable to detect further depression upon let-502 depletion. Alternatively, it is possible that the compact actin structures of the valve preclude efficient antibody penetration. Overall, we find that let-502 appears to be required for maintaining p-MRLC levels and contractility of the distal neck and valve cells but plays less of a role in the main bag cells, where MLCK-1 is required for production of p-MRLC during contraction.
DISCUSSION
Here we characterize MLCK-1, a C. elegans Ca2+/CaM–dependent MLCK, and demonstrate its role in spermathecal contractility. We have shown previously that production of Ca2+ transients drives contraction (Kovacevic et al., 2013) and development of stress fiber–like actomyosin bundles (Wirshing and Cram, 2017). However, the kinase responsible for increasing myosin activity downstream of Ca2+ in the spermatheca was not known. MLCK-1 contains a highly conserved serine/threonine kinase domain, an autoregulatory helix, and a 1-8-14 CaM binding motif characteristic of Ca2+/CaM–dependent MLCKs (Rhoads and Friedberg, 1997; Yap et al., 2000; Chang et al., 2016) Animals depleted of MLCK-1 have flaccid spermathecae that fail to contract during ovulation, resulting in trapping of the embryo in the spermatheca. Using immunohistochemistry, we show that this decrease in contractility is due to a reduction in p-MRLC in MLCK-1–depleted spermathecal cells. Depressed spermathecal contractility reduces animal fertility and embryonic viability. The effect on embryonic viability is dependent on the maternal genotype, indicating that it may be due to altered mechanical stress experienced by the embryo during ovulation rather than to defects in embryogenesis. This effect has been reported previously in animals with decreased spermathecal contractility (Kovacevic and Cram, 2010).
MLCK-1 is prominently expressed in the spermatheca. Spermathecal expression is first detected at L4, as the spermatheca is developing, and increases as animals enter adulthood. We find that the subcellular localization of MLCK-1 changes during adulthood when animals begin to ovulate and the spermatheca is exposed to cyclic rounds of stretching and contraction. Prior to the first ovulation, MLCK-1 is found at apical cell boundaries, and after ovulation begins, a small fraction of MLCK-1 colocalizes with basal actomyosin bundles. Basal recruitment of MLCK-1 coincides with the onset of contraction, suggesting that basally recruited MLCK-1 is active and phosphorylating the MRLC in basal actomyosin bundles to drive contraction. This distribution is maintained throughout adulthood. Similarly, work in cell culture has shown that active smMLCK is recruited to contracting actin networks during cell migration (Chew et al., 2002) and contraction (Verin et al., 1998) and in vivo only a small fraction of the total MLCK is active during contraction (Isotani et al., 2004; Injeti et al., 2008). Consistent with this interpretation, we find that depletion of PLC-1 abolishes spermathecal Ca2+ signaling during ovulation (Kovacevic et al., 2013) and prevents basal MLCK-1 recruitment and actomyosin contraction. However, Ca2+ signaling alone does not appear to determine MLCK-1 localization. During ovulation, Ca2+ transients peak during contraction and then return to baseline preovulation levels (Kovacevic et al., 2013). Interestingly, we see that MLCK-1 is maintained at the basal actomyosin network throughout adulthood, suggesting that there is another mechanism that drives MLCK-1 localization. In addition to regulation by Ca2+/CaM, vertebrate MLCK is regulated by phosphorylation (Conti and Adelstein, 1981; Nishikawa et al., 1985; Klemke et al., 1997; Sanders et al., 1999; Birukov et al., 2001; Horman et al., 2008), interaction with cytoskeletal proteins (Sellers and Pato, 1984; Smith et al., 1999; Kudryashov et al., 2004; Takizawa et al., 2007), and mechanical stretch (Baumann et al., 2017). It is unclear whether similar regulatory mechanisms exist for MLCK-1, as MLCK-1 lacks the DXRXXL (Lin et al., 1999; Smith et al., 1999, 2002; Poperechnaya et al., 2000; Chen et al., 2014), telokin (Silver et al., 1997; Numata et al., 2001; Hong et al., 2009), and immunoglobulin G (IgG)-C2 (Kudryashov et al., 2004) domains that allow smMLCK to interact with actin, myosin, and other cytoskeletal components. However, other vertebrate MLCK isoforms expressed in skeletal and cardiac muscle also lack these conserved domains and yet are found associated with actin (Chan et al., 2008) and organized into sarcomeres (Cavadore et al., 1982), indicating that alternative uncharacterized mechanisms for localizing MLCKs at contractile actomyosin structures also exist in vertebrates. While the N-terminus of MLCK-1 contains the highly conserved serine/threonine kinase domain, the C-terminus is poorly conserved, and its function remains unclear.
We have shown previously that parallel actomyosin bundles develop during the first ovulation and are maintained throughout adulthood (Wirshing and Cram, 2017). Here, we show that MLCK-1 is required for parallel bundle production. Time-lapse microscopy of ovulation shows that loss of MLCK-1 results in persistence of the preovulation actin network characterized by webby, poorly oriented bundles. This phenotype has been described previously for plc-1– and nmy-1–depleted animals (Wirshing and Cram, 2017), indicating that the population of MLCK-1 at apical boundaries, retained in plc-1(RNAi), is not responsible for parallel bundle production. In addition to actin, myosin organization is also disrupted, with knockdown of mlck-1. In mlck-1–depleted cells, myosin is poorly recruited to basal actomyosin bundles. In cultured cells, myosin activity is required for stable association of myosin within actomyosin networks (Watanabe et al., 2007; Kondo et al., 2011; Juanes-Garcia et al., 2015). Our FRAP experiments show that myosin in basal actomyosin bundles is more dynamic in mlck-1–depleted than in WT cells. This suggests that localized MLCK-1 activity at the basal actomyosin network is one potential mechanism by which myosin is incorporated into basal actomyosin bundles.
Because ROCK/LET-502 is known to play a role in C. elegans somatic gonad contractility (Wissmann et al., 1999; Ono and Ono, 2016) and the formation of stress fibers (Amano et al., 1996; Gally et al., 2009; Beach et al., 2017), we sought to determine the contribution of this kinase to spermathecal function. Here we show that ROCK/LET-502 and MLCK-1 have distinct expression patterns and different roles in regulating spermathecal contraction. In agreement with previous work, we find that ROCK/LET-502 is strongly expressed in the distal neck and valve cells and only weakly in bag cells (Wissmann et al., 1999), while MLCK-1 is expressed throughout the spermatheca. Consistent with the ROCK/LET-502 expression pattern, we find that ROCK/LET-502 is predominantly required for regulating contractility and producing p-MRLC in the distal neck and valve. Unlike depletion of mlck-1, knockdown of let-502 does not decrease p-MRLC levels in spermathecal bag cells. In other systems, ROCK and MLCK play distinct and complementary roles to allow precise spatiotemporal regulation of myosin activity. This is seen in individual cells where MLCK activates myosin in peripheral stress fibers and ROCK is required for formation of central stress fibers (Totsukawa et al., 2000, 2004; Simoes and Fierro, 2005; Beach et al., 2017; Kassianidou et al., 2017) and in cell response to stretch (Lee et al., 2010), as well as at the tissue level, where MLCK and ROCK cooperate during wound closure (Russo et al., 2005). In the spermatheca, ROCK/LET-502 and MLCK cooperate to regulate contractility by acting primarily in a distinct subset of cells. This may have evolved as a mechanism for maintaining different tissue tone in the spermathecal distal neck and valve than in the bag. Tight closure of the spermathecal distal neck during ovulation is needed to prevent the embryo from sliding backward into the proximal gonad arm (Gissendanner et al., 2008; McGovern et al., 2018). However, hyperconstriction of the spermathecal bag cells can damage embryos (Tan and Zaidel-Bar, 2015), supporting the idea that distal neck and bag cells constrict to different degrees during ovulation. Our finding that ROCK/LET-502 is primarily required in a specific subset of somatic gonad cells may help explain why defects in somatic gonad contractility have been reported previously with let-502 depletion (Wissmann et al., 1999; Ono and Ono, 2016) without having a significant impact on actomyosin organization or p-MRLC levels in sheath cells (Ono and Ono, 2016).
Here, we describe a new myosin light-chain kinase, MLCK-1, required for contraction of the spermatheca and show that MLCK-1 and LET-502/ROCK act in distinct subsets of spermathecal cells to coordinate contraction during ovulation. While it is known that ROCK and MLCK share roles in regulating contractility of the gastrointestinal (Du et al., 2016; Rattan, 2017) and urinary tracts (Kirschstein et al., 2015) and integrity of the epithelia in airways (Olivera et al., 2007), the exact molecular mechanism by which these two kinases coregulate MRLC phosphorylation is less well understood. In addition to providing mechanistic insight into how the spermatheca functions, this study helps to establish an in vivo model of how cells coordinate tissue-level responses through the regulation of MLCK and ROCK.
MATERIALS AND METHODS
C. elegans strains and culture
All stains were grown on standard NGM (0.107 M NaCl, 0.25% wt/vol Peptone [Fisher Science Education], 1.7% wt/vol BD Bacto-Agar [Fisher Scientific], 0.5% Nyastatin [Sigma], 0.1 mM CaCl2, 0.1 mM MgSO4, 0.5% wt/vol cholesterol, 2.5 mM KPO4) agar plates seeded with OP50 Escherichia coli at 23°C (Hope, 1999) unless otherwise noted. Strains were generated by microinjection as described previously (Mello et al., 1991). The mlck-1p::GFP::unc-54 3′UTR reporter construct was injected at 20 ng/μl with the coinjection marker, rol-6, injected at 40 ng/μl. Lines expressing GFP::act-1 and moeABD::mCherry;GFP::nmy-1 were described previously (Wirshing and Cram, 2017). The MLCK-1 allele, mlck-1(tm4159), was obtained from the Japanese National Bioresource Project and outcrossed three times to WT worms to create the strain UN1604. The let-502(sb106) allele was previously described in (Piekny et al., 2000). The endogenously tagged strains, COP1510 mlck-1::degron::mKate2 and COP1226 let-502::degron::eGFP endogenously tagged with GFP strain, were generated by Nemametrix. COP1510 was outcrossed three times to create the strain UN1743. The strain was generated by crossing NK1069 with EU573. EU573 has been described previously (Gally et al., 2009). See Supplemental Table 1 for a list of strains and genotypes used in these experiments.
RNAi interference
RNAi feeding experiments were performed essentially as described previously (Kovacevic and Cram, 2010). To prepare seeded NGM-IPTG plates, HT115(DE3) bacteria transformed with the double-stranded RNA construct of interest were grown overnight at 37°C in Luria broth (LB) supplemented with 40 µg/ml ampicillin. The following day, 150 µl of the culture was seeded on NGM-IPTG agar (NGM supplemented with 25 µg/ml carbenicillin and 1 mM isopropylthio-β-galactoside, IPTG). Synchronized populations were obtained by alkaline lysis procedure (egg prep). For egg prep, starved dauer nematodes were allowed to recover for 48 h on NGM plates newly seeded with OP50. This produces young gravid adults for egg collection. Eggs were released using an alkaline hypochlorite solution as described in Hope (1999) and washed three times with filtered sterilized M9 buffer (22 mM KH2PO4, 42 mM NaHPO4, 86 mM NaCl, and 1 mM MgSO4; Hope, 1999). Clean eggs were then transferred to seeded NGM-IPTG plates. Animals were allowed to grow for ∼54 h at 23°C before being scored or imaged unless otherwise noted. Several of the RNAi constructs used were from the ORF-eome-RNAi v1.1 or the Ahringer Library. Some RNAi constructs were made by amplifying the coding region of the gene of interest off of mixed age N2 cDNA and cloning them into the empty vector pPD129.36 (Fire Vector Kit). Primers used to clone RNAi are available upon request. The empty vector was used as a negative control in all RNAi experiments.
Spermathecal occupancy assay
Synchronized populations obtained by egg prep, described above, were grown on NGM-IPTG plates with the indicated RNAi treatment, and worms were scored ∼54 h after egg prep when they were just beginning to ovulate. Worms were mounted on a 2% agarose–in–water pad and killed using 100 mM sodium azide. Scoring for occupancy of spermathecae was done using a 60× oil-immersion objective with a Nikon Eclipse 80i microscope.
Protein alignments
Kinase domains used in the alignments were identified using SMART (Schultz et al., 1998; Letunic et al., 2004, 2015; Letunic and Bork, 2018) and aligned using Clustal Omega (Sievers et al., 2011; McWilliam et al., 2013; Li et al., 2015). The 3D protein structures were predicted using iTasser (Zhang, 2008; Roy et al., 2010; Yang et al., 2015). The 3D structures were rotated, and residues were colored using the PyMol Molecular Graphics System (Version 1.74, Schrödinger). Calmodulin binding domains were identified using the Calmodulin Target Database (Yap et al., 2000), aligned using Clustal Omega (Sievers et al., 2011; McWilliam et al., 2013; Li et al., 2015), and made ready for publication using BoxShade.
Determination of number of live hatchlings
Embryos were egg-prepped onto an NGM plate seeded with OP50. The animals were grown at 23°C, unless otherwise noted, and at L4, animals were transferred to individual plates. Total live hatchlings were counted and aspirated every day until the mother died or there were two consecutive days of no new hatchlings on the plate.
Viability assay
Synchronized populations obtained by egg prep, described above, were grown on NGM at 23°C for ∼60 h. Worms were then moved to individual plates for 2-4 h and allowed to lay embryos on the plates. Hermaphrodites were removed, and all embryos laid were counted before incubation at 23°C for 24 h. Plates were scored for the number of worms that hatched from embryos (viable) and embryos that failed to hatch (not viable).
DIC imaging
Animals were immobilized either with 0.01% tetramisole and 0.1% tricaine in M9 buffer or with 0.05 μm Polybead microspheres diluted 1:1:1 in M9 and water on a 2% agarose pad. Imaging was performed on a Nikon Eclipse 80i microscope with a 60× oil-immersion lens using SPOT R3 software and a charge-coupled device camera. Frames were captured at a rate of 1 Hz. Image stacks were reassembled and analyzed using ImageJ software. To be considered “trapped,” worms were imaged for at least 950 s after entry. Raw dwell times for worms treated with mlck-1 RNAi can be found in Supplemental Figure 6.
Histochemistry
Visualization of F-actin in animals expressing MLCK-1 labeled with mKate2 or LET-502 labeled with GFP was done as described previously (Wirshing and Cram, 2017) except that 100 nM Acti-stain 488 (Cytoskeleton #PHDG1) was used in place of Texas Red-X phalloidin (Invitrogen) to allow simultaneous visualization of F-actin and mKate2. For quantification of p-MRLC, synchronized populations were obtained by egg prep and grown with the indicated RNAi treatment at 23°C for ∼54 h. Fixation and staining protocols were adapted from (Werner et al., 2007; Ono and Ono, 2016). Unless noted, all steps were done at room temperature. Animals were dissected using a 25-gauge hypodermic needle in phosphate-buffered saline (PBS), and dissected gonads were fixed with 4% formaldehyde and 0.2% glutaraldehyde in cytoskeleton buffer (10 mM MES pH 6.1, 138 mM KCl, 3 mM MgCl2, 10 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N,N-tetraacetic acid [EGTA], and 0.32 M sucrose) for 10 min. Fixative was removed by three washes with PBS and unreacted aldehydes were quenched by incubation in 0.1% sodium borohydride in PBS for 20 min. Quenching was followed by two PBS rinses before permeabilization with 0.25% Triton X-100 in PBS for 10 min. After three PBS rinses, dissected gonads were blocked in 1% BSA, 0.1% Tween-20, and 30 mM glycine in PBS for 1 h before addition of the primary antibody, anti-myosin light-chain (phospho S20) antibody (Abcam #ab2480), at 2.5 µg/ml in blocking buffer. Incubation with the primary was done overnight at 4°C followed by three PBS washes and incubation with the secondary, donkey anti-rabbit IgG H&L (DyLight 488) preadsorbed (Abcam #ab96919) diluted 1:1000 in blocking buffer, for 6 h at room temperature. After 6 h, 0.4 U/mL Texas Red-X phalloidin in PBS (InvitrogenA) was added, followed by incubation overnight at 4°C. Dissected gonads were washed three times in PBS and mounted in 90% glycerol, 20 mM Tris, pH 8.3, on 2% agarose pads.
Confocal microscopy
For time-lapse imaging of ovulation and observations of live or fixed animals, partially synchronized populations were obtained by egg prep and animals were grown at 23°C for ∼54 h, around the time of the first ovulation. Live animals were immobilized with 0.01% tetramisole and 0.1% tricaine in M9 buffer (Kirby et al., 1990; McCarter et al., 1997) and mounted on 2% agarose pads or with 0.05 μm Polybead microspheres (Polysciences) diluted 1:2 in water and mounted on 5% agarose pads (Wang and Audhya, 2014). Confocal microscopy was performed on an LSM 710 confocal microscope (Zeiss) equipped with Zen software (Zeiss) using a Plan-Apochromat 63×/1.40 oil DIC M27 objective. A 488-nm laser was used for GFP and DyLight 488, and a 561 nm laser was used for mKate2 and TexasRed. For movies, 40 or 20 z-slices were acquired at 14- or 10-s intervals for imaging actin labeled with GFP and MLCK-1 labeled with mKate2, respectively. Illumination of the spermathecae from animals expressing actin labeled with GFP (GFP::ACT-1) with the 488-nm laser for ∼5 min prior to oocyte entry frequently caused the valve to remain partially closed during ovulation, increasing oocyte dwell time. Live animals were imaged for ∼ 30 min total. For still images of live and fixed animals and tissue, z-slices were acquired at 0.38-μm intervals, with each slice representing the average of two or four scans.
FRAP
Live animals grown at 23°C for ∼54 h were immobilized as described above. Confocal microscopy was performed on an LSM 710 confocal microscope (Zeiss) equipped with Zen software (Zeiss) using the 63× oil objective and the 488-nm laser. Five confocal sections of the basal cell surface were acquired at 1-s intervals prior to the bleaching step to measure the starting fluorescence intensity, 10 bleaching scans were preformed using the 488-nm laser at 100% with a pixel dwell time of 0.95 μs in a 9 by 1.5–μm region across the center of the cell, and 95 images were acquired at 1-s intervals to monitor fluorescence recovery after bleaching. Analysis of FRAP data was done using a Jython script in ImageJ. This script allows selection of a bleached area, the 9 by 1.5–μm region, and a region outside of the bleached area. A region within this same cell was used. This nonbleached region is used to normalize measurements to account for photobleaching. With this experimental setup, however, we did not detect photobleaching of GFP.
Image analysis
ImageJ software was used for all image analysis. Unless otherwise noted, all images were background-subtracted prior to analysis. For consistency, all analysis was performed on cells of the main spermathecal bag, unless otherwise noted. For analysis of MLCK-1 dynamics during ovulation, MLCK-1::mKate2 fluorescence was measured from confocal maximum intensity projections of ovulation movies at 50- s intervals. For each measurement, a 20 pixel–wide line was drawn across one cell. The BAR script Find Peaks was used to detect the bright MLCK-1::mKate2 signal at the cell edges. The average distance between the peaks was used to find the cell center and 10 pixels above and below this central point were measured for the MLCK-1::mKate2 fluorescence signal, excluding the cell edges. For analysis of MLCK-1 distribution in fixed samples, a 100 pixel–wide line was drawn across a single sagittal confocal cross-section and the BAR script Find Peaks was used to determine fluorescence intensity at the basal cell surface. To measure p-MRLC intensities, maximum-intensity projections were generated and F-actin staining was used to distinguish individual cells. Individual cells were outlined and fluorescence intensity in the 488-nm channel corresponding to p-MRLC was measured. This same strategy was used to quantify p-MRLC in Figure 10, except that specific regions of the spermatheca were selected rather than individual cells. For measurements of actin network organization, FibrilTool (Boudaoud et al., 2014) was used to quantify actin anisotropy in selected cells. For movie analysis, maximum-intensity projections were generated and FibrilTool was used on individual cells at 28 s intervals (every other frame). Anisotropy measurements were normalized by taking the average anisotropy of the first 10 frames measured and subtracting this number from each measurement. OrientationJ (Rezakhaniha et al., 2012), configured using a Gaussian fit with a pixel size of 2, was used to measure the orientation distribution of actin bundles in individual cells and to generate color-coded images where color indicates orientation, hue indicates coherency, and brightness is the brightness of the original image. To determine actin and myosin distribution, a line with a pixel width of 10 was drawn across indicated cells to measure actin and myosin fluorescence intensity. For colocalization analysis, the ImageJ plug-in Coloc2 was used to measure the Pearson’s R value of selected cells. To measure the apical and basal distribution of GFP-labeled myosin, a 20 pixel–wide line was drawn across a single sagittal cross-section. The BAR script Find Peaks was used to detect bright GFP signals at the basal and apical cell surfaces. In spermathecae of WT animals, if the apical GFP signal was not above background and no peak was detected, the apical surface was determined manually. The average distance between the apical and basal peaks was used to find the center of the cell. The average of five pixels above and below this central point was measured for the cytosolic GFP signal, to which the apical and basal measurements were normalized.
Statistical analysis
All statistical analysis was performed with GraphPad Prism software. To compare two unpaired groups, the unpaired t test was used to determine whether the difference between the means of two data sets was significant when data had a normal distribution, and Welch’s correction was included if the different treatments were expected to have different standard deviations. To compare three or more unmatched groups, ordinary one-way analysis of variance (ANOVA) was performed with either a Dunnett’s multiple comparison or a Tukey’s multiple comparison test as necessary. The Mann–Whitney test was used when data did not have a normal distribution. All trapping data were analyzed using Fisher’s exact test. In all cases, the statistical test used and the resulting p values, indicated by symbols as ns (p > 0.05), *(p ≤ 0.05), **(p ≤ 0.01), ***(p ≤ 0.001), and ****(p ≤ 0.0001), are noted in each figure caption.
Supplementary Material
Acknowledgments
Some C. elegans strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources, National Institutes of Health. This work was supported by a grant from the National Institutes of Health National Institute of General Medical Sciences (GM110268) to E.J.C. and a grant from the Israel Science Foundation (1293/17) to R.Z.B.
Abbreviations used:
- CaM
calmodulin
- F-actin
filamentous-actin
- GFP
green fluorescent protein
- MLCK
myosin light-chain kinase
- MRLC
myosin regulatory light chain
- MYPT
myosin phosphatase
- NMII
nonmuscle myosin II
- pMRLC
phosphorylated myosin regulatory light chain
- RNAi
RNA interference
- ROCK
Rho kinase
- sp-ut
spermatheca–uterine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-01-0056) on April 5, 2018.
REFERENCES
- Amano M, Ito M, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. (1996). Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J Biol Chem , 20246–20249. [DOI] [PubMed] [Google Scholar]
- Ammit AJ, Armour CL, Black JL. (2000). Smooth-muscle myosin light-chain kinase content is increased in human sensitized airways. Am J Respir Crit Care Med , 257–263. [DOI] [PubMed] [Google Scholar]
- Baumann F, Bauer MS, Rees M, Alexandrovich A, Gautel M, Pippig DA, Gaub HE. (2017). Increasing evidence of mechanical force as a functional regulator in smooth muscle myosin light chain kinase. Elife , 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Bruun KS, Shao L, Li D, Swider Z, Remmert K, Zhang Y, Conti MA, Adelstein RS, Rusan NM, et al. (2017). Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat Cell Biol , 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian GM, Kiff JE, Neckelmann N, Moerman DG, Waterston RH. (1989). Sequence of an unusually large protein implicated in regulation of myosin activity in C. elegans. Nature , 45–50. [DOI] [PubMed] [Google Scholar]
- Birukov KG, Csortos C, Marzilli L, Dudek S, Ma S-F, Bresnick AR, Verin AD, Cotter RJ, Garcia JGN. (2001). Differential regulation of alternatively spliced endothelial cell myosin light chain kinase isoforms by p60 Src. J Biol Chem , 8567–8573. [DOI] [PubMed] [Google Scholar]
- Blumenthal DK, Stull JT. (1980). Activation of skeletal muscle myosin light chain kinase by calcium(2+) and calmodulin. Biochemistry , 5608–5614. [DOI] [PubMed] [Google Scholar]
- Boudaoud A, Burian A, Borowska-Wykret UM, Wrzalik R, Kwiatkowska D, Hamant O. (2014). FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc , 457–463. [DOI] [PubMed] [Google Scholar]
- Brown CR, Hong-Brown LQ, Welch WJ. (1997). Correcting temperature-sensitive protein folding defects. J Clin Invest , 1432–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavadore J-C, Molla A, Harricane M-C, Gabrion J, Benyamin Y, Demaille JG. (1982). Subcellular localization of myosin light chain kinase in skeletal, cardiac, and smooth muscles. Proc Natl Acad Sci USA , 3475–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Takeda M, Briggs LE, Graham ML, Lu JT, Horikoshi N, Weinberg EO, Aoki H, Sato N, Chien KR, et al. (2008). Identification of cardiac-specific myosin light chain kinase. Circ Res , 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AN, Mahajan P, Knapp S, Barton H, Sweeney HL, Kamm KE, Stull JT. (2016). Cardiac myosin light chain is phosphorylated by Ca2+/calmodulin-dependent and -independent kinase activities. Proc Natl Acad Sci USA , E3824–E3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tao T, Wen C, He W, Qiao Y, Gao Y, Chen X, Wang P, Chen C, Zhao W, et al. (2014). Myosin light chain kinase (MLCK) regulates cell migration in a myosin regulatory light chain phosphorylation-independent mechanism. J Biol Chem , 28478–28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew T-L, Wolf WA, Gallagher PJ, Matsumura F, Chisholm RL. (2002). A fluorescent resonant energy transfer-based biosensor reveals transient and regional myosin light chain kinase activation in lamella and cleavage furrows. J Cell Biol , 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Langeslag M, Figdor CG, van Leeuwen FN. (2007). Myosin II and mechanotransduction: a balancing act. Trends Cell Biol , 178–186. [DOI] [PubMed] [Google Scholar]
- Conti MA, Adelstein RS. (1981). The relationship between calmodulin binding and phosphorylation of smooth muscle myosin kinase by the catalytic subunit of 3’:5’ cAMP-dependent protein kinase. J Biol Chem , 3178–3181. [PubMed] [Google Scholar]
- Conti MA, Adelstein RS. (2008). Nonmuscle myosin II moves in new directions. J Cell Sci , 404–404. [DOI] [PubMed] [Google Scholar]
- Du L, Kim JJ, Shen J, Dai N. (2016). Crosstalk between inflammation and ROCK/MLCK signaling pathways in gastrointestinal disorders with intestinal hyperpermeability. Gastroenterol Res Pract , 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty DB, Gernert KM, Shmeleva N, Tang X, Mercer KB, Borodovsky M, Benian GM. (2002). Titins in C. elegans with unusual features: coiled-coil domains, novel regulation of kinase activity and two new possible elastic regions. J Mol Biol , 533–549. [DOI] [PubMed] [Google Scholar]
- Flores C, Ma S-F, Maresso K, Ober C, Garcia JG. (2007). A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol , 296–305. [DOI] [PubMed] [Google Scholar]
- Gallagher PJ, Herring BP, Trafny A, Sowadski J, Stull JT. (1993). A molecular mechanism for autoinhibition of myosin light chain kinases. J Biol Chem , 26578–26582. [PMC free article] [PubMed] [Google Scholar]
- Gally C, Wissler F, Zahreddine H, Quintin S, Landmann F, Labouesse M. (2009). Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development , 3109–3119. [DOI] [PubMed] [Google Scholar]
- Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, et al. (2006). Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol , 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. (2006). Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science , 1471–1475. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Kelley K, Nguyen TQ, Hoener MC, Sluder AE, Maina CV. (2008). The Caenorhabditis elegans NR4A nuclear receptor is required for spermatheca morphogenesis. Dev Biol , 767–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heer NC, Miller PW, Chanet S, Stoop N, Dunkel J, Martin AC. (2017). Actomyosin-based tissue folding requires a multicellular myosin gradient. Development , 1876–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BP, Gallagher PJ, Stull JT. (1992). Substrate specificity of myosin light chain kinases. J Biol Chem , 25945–25950. [PubMed] [Google Scholar]
- Hirata N, Takahashi M, Yazawa M. (2009). Diphosphorylation of regulatory light chain of myosin IIA is responsible for proper cell spreading. Biochem Biophys Res Commun , 682–687. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. (1976). Development of the reproductive system of Caenorhabditis. Dev Biol , 200–219. [DOI] [PubMed] [Google Scholar]
- Hong F, Haldeman BD, Jackson D, Carter M, Baker JE, Cremo CR. (2011). Biochemistry of smooth muscle myosin light chain kinase. Arch Biochem Biophys , 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Haldeman BD, John OA, Brewer PD, Wu Y-Y, Ni S, Wilson DP, Walsh MP, Baker JE, Cremo CR. (2009). Characterization of tightly associated smooth muscle myosin–myosin light-chain kinase–calmodulin complexes. J Mol Biol , 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA. (1999). C. elegans—A Practical Approach, Oxford, UK: Oxford University Press. [Google Scholar]
- Hope IA, Mounsey A, Bauer P, Aslam S. (2003). The forkhead gene family of Caenorhabditis elegans. Gene , 43–55. [DOI] [PubMed] [Google Scholar]
- Horman S, Morel N, Vertommen D, Hussain N, Neumann D, Beauloye C, Najjar NE, Forcet C, Viollet B, Walsh MP, et al. (2008). AMP-activated protein kinase phosphorylates and desensitizes smooth muscle myosin light chain kinase. J Biol Chem , 18505–18512. [DOI] [PubMed] [Google Scholar]
- Huang X, Miller W. (1991). A time-efficient, linear-space similarity algorithm. Adv Appl Math , 337–357. [Google Scholar]
- Hubbard EJA, Greenstein D. (2000). The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn , 2–22. [DOI] [PubMed] [Google Scholar]
- Injeti ER, Sandoval RJ, Williams JM, Smolensky AV, Ford LE, Pearce WJ. (2008). Maximal stimulation-induced in situ myosin light chain kinase activity is upregulated in fetal compared with adult ovine carotid arteries. Am J Physiol Heart Circ Physiol , H2289–H2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isotani E, Zhi G, Lau KS, Huang J, Mizuno Y, Persechini A, Geguchadze R, Kamm KE, Stull JT. (2004). Real-time evaluation of myosin light chain kinase activation in smooth muscle tissues from a transgenic calmodulin-biosensor mouse. Proc Natl Acad Sci USA , 6279–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Nakano T, Erdodi F, Hartshorne DJ. (2004). Myosin phosphatase: structure, regulation and function. Mol Cell Biochem , 197–209. [DOI] [PubMed] [Google Scholar]
- Juanes-Garcia A, Chapman JR, Aguilar-Cuenca R, Delgado-Arevalo C, Hodges J, Whitmore LA, Shabanowitz J, Hunt DF, Horwitz AR, Vicente-Manzanares M. (2015). A regulatory motif in nonmuscle myosin II-B regulates its role in migratory front–back polarity. J Cell Biol , 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm KE, Stull JT. (2011). Signaling to myosin regulatory light chain in sarcomeres. J Biol Chem , 9941–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya KI, Kim Bui Y, Gao X, Sternberg PW, Kataoka T. (2004). Phospholipase C epsilon regulate ovulation in Caenorhabditis elegans. Dev Biol , 201–210. [DOI] [PubMed] [Google Scholar]
- Kassianidou E, Brand CA, Schwarz US, Kumar S. (2017). Geometry and network connectivity govern the mechanics of stress fibers. Proc Natl Acad Sci USA , 2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. (1996). Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science , 245–248. [DOI] [PubMed] [Google Scholar]
- Kirby C, Kusch M, Kemphues K. (1990). Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev Biol , 203–215. [DOI] [PubMed] [Google Scholar]
- Kirschstein T, Sahre T, Kernig K, Protzel C, Porath K, Köhling R, Hakenberg OW. (2015). Inverse relationship of Rho kinase and myosin-light chain kinase expression in the aging human detrusor smooth muscle. BMC Urol , 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. (1997). Regulation of cell motility by mitogen-activated protein kinase. J Cell Biol , 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton DR, Pearson RB, Sowadski JM, Means AR, Ten Eyck LF, Taylor SS, Kemp BE. (1992). Structural basis of the intrasteric regulation of myosin light chain kinases. Science , 130–135. [DOI] [PubMed] [Google Scholar]
- Kobel B, Heierhorst J, Feill SC, Parker MW, Benian GM, Weiss KR, Kempl BE. (1996). Giant protein kinases: domain interactions and structural basis of autoregulation. EMBO J , 6810–6821. [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Hamao K, Kamijo K, Kimura H, Morita M, Takahashi M, Hosoya H. (2011). Enhancement of myosin II/actin turnover at the contractile ring induces slower furrowing in dividing HeLa cells. Biochem J , 569–576. [DOI] [PubMed] [Google Scholar]
- Kovacevic I, Cram EJ. (2010). FLN-1/FIlamin is required for maintenance of actin and exit of fertilized oocytes from the spermatheca in C. elegans. Dev Biol , 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic I, Orozco JM, Cram EJ. (2013). Filamin and phospholipase C-ε are required for calcium signaling in the Caenorhabditis elegans spermatheca. PLoS Genet , e1003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashov DS, Stepanova O V, Vilitkevich EL, Nikonenko TA, Nadezhdina ES, Shanina NA, Lukas TJ, Van Eldik LJ, Watterson DM, Shirinsky VP. (2004). Myosin light chain kinase (210 kDa) is a potential cytoskeleton integrator through its unique N-terminal domain. Exp Cell Res , 407–417. [DOI] [PubMed] [Google Scholar]
- Lazar V, Garcia JG. (1999). A single human myosin light chain kinase gene (MLCK; MYLK) transcribes multiple nonmuscle isoforms. Genomics , 256–267. [DOI] [PubMed] [Google Scholar]
- Lee C-F, Haase C, Deguchi S, Kaunas R. (2010). Cyclic stretch-induced stress fiber dynamics—dependence on strain rate, Rho-kinase and MLCK. Biochem Biophys Res Commun , 344–349. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res , 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res , D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res , D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Chen X-Q, Tan I, Manser E, Lim L. (1998). Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol , 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, Mcwilliam H, Squizzato S, Park YM, Buso N, Lopez R. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res , W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Luby-Phelps K, Stull JT. (1999). Properties of filament-bound myosin light chain kinase. J Biol Chem , 5987–5994. [DOI] [PubMed] [Google Scholar]
- Ma S-F, Flores C, Wade MS, Dudek SM, Nicolae DL, Ober C, Garcia JG. (2008). A common cortactin gene variation confers differential susceptibility to severe asthma. Genet Epidemiol , 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. (1997). Soma–germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol , 121–143. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. (1999). On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol , 111–128. [DOI] [PubMed] [Google Scholar]
- McGovern M, Castaneda PG, Pekar O, Vallier LG, Cram EJ, Hubbard EJA. (2018). The DSL ligand APX-1 is required for normal ovulation in C. elegans. Dev Biol , 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. (2013). Analysis tool Web services from the EMBL-EBI. Nucleic Acids Res , W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J , 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D, Benian G, Barstead R, Schriefer L, Waterston R. (1988). Identification and intracellular localization of the unc-22 gene product of C. elegans. Genes Dev , 93–105. [DOI] [PubMed] [Google Scholar]
- Murata-Hori M, Suizu F, Iwasaki T, Kikuchi A, Hosoya H. (1999). ZIP kinase identified as a novel myosin regulatory light chain kinase in HeLa cells. FEBS Lett , 81–84. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Tanoue T. (2010). Epithelial cell shape is regulated by Lulu proteins via myosin-II. J Cell Sci , 555–566. [DOI] [PubMed] [Google Scholar]
- Nishikawa M, Shirakawa S, Adelstein RS. (1985). Phosphorylation of smooth muscle myosin light chain kinase by protein kinase C. J Biol Chem , 8978–8983. [PubMed] [Google Scholar]
- Numata T, Katoh T, Yazawa M. (2001). Functional role of the C-terminal domain of smooth muscle myosin light chain kinase on the phosphorylation of smooth muscle myosin. J Biochem , 437–444. [DOI] [PubMed] [Google Scholar]
- Olah G, Mitchell R, Sosnick T, Walsh D, Trewhella J. (1993). Solution of the cAMP-dependent protein kinase catalytic subunit and its contraction upon binding the protein kinase inhibitor peptide. Biochemistry , 3649–3657. [DOI] [PubMed] [Google Scholar]
- Olivera DS, Boggs SE, Beenhouwer C, Aden J, Knall C. (2007). Cellular mechanisms of mainstream cigarette smoke-induced lung epithelial tight junction permeability changes in vitro. Inhal Toxicol , 13–22. [DOI] [PubMed] [Google Scholar]
- Ono K, Ono S. (2016). Two distinct myosin II populations coordinate ovulatory contraction of the myoepithelial sheath in the Caenorhabditis elegans somatic gonad. Mol Biol Cell , 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny AJ, Wissmann A, Mains PE. (2000). Embryonic morphogenesis in Caenorhabditis elegans integrates the activity of LET-502 Rho-binding kinase, MEL-11 myosin phosphatase, DAF-2 insulin receptor and FEM-2 PP2c phosphatase. Genetics , 1671–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poperechnaya A, Varlamova O, Lin PJ, Stull JT, Bresnick AR. (2000). Localization and activity of myosin light chain kinase isoforms during the cell cycle. J Cell Biol , 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan S. (2017). Ca 2+/calmodulin/MLCK pathway initiates, and RhoA/ROCK maintains, the internal anal sphincter smooth muscle tone. Am J Physiol Liver Physiol , G63–G66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezakhaniha R, Agianniotis A, Schrauwen JTC, Griffa A, Sage D, Bouten CVC, van de Vosse FN, Unser M, Stergiopulos N. (2012). Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol , 461–473. [DOI] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F. (1997). Sequence motifs for calmodulin recognition. FASEB J , 331–340. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc , 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JM, Florian P, Shen L, Graham WV, Tretiakova MA, Gitter AH, Mrsny RJ, Turner JR, Turner JR. (2005). Distinct temporal-spatial roles for Rho kinase and myosin light chain kinase in epithelial purse-string wound closure. Gastroenterology , 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurada K, Seto M, Sasaki Y. (1998). Dynamics of myosin light chain phosphorylation at Ser19 and Thr18/Ser19 in smooth muscle cells in culture. Am J Physiol , C1563–C1572. [DOI] [PubMed] [Google Scholar]
- Sanders LC, Matsumura F, Bokoch GM, de Lanerolle P. (1999). Inhibition of myosin light chain kinase by p21-activated kinase. Science , 2083–2086. [DOI] [PubMed] [Google Scholar]
- Scholey J, Taylor K, Kendrick-Jones J. (1980). Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature , 233–235. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA , 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. (1981). Phosphorylation-dependent regulation of Limulus myosin. J Biol Chem , 9274–9278. [PubMed] [Google Scholar]
- Sellers JR, Pato MD. (1984). The binding of smooth muscle myosin light chain kinase and phosphatases to actin and myosin. J Biol Chem , 7740–7746. [PubMed] [Google Scholar]
- Shelton CA, Carter JC, Ellis GC, Bowerman B. (1999). The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J Cell Biol , 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohat G, Spivak-Kroizman T, Cohen O, Bialik S, Shani G, Berrisi H, Eisenstein M, Kimchi A. (2001). The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism. J Biol Chem , 47460–47467. [DOI] [PubMed] [Google Scholar]
- Siedlik MJ, Nelson CM. (2015). Regulation of tissue morphodynamics: an important role for actomyosin contractility. Curr Opin Genet Dev , 80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm ADineen DGibson TJKarplus KLi WLopez RMcWilliam HRemmert MSöding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol , 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Vorotnikov AV, Watterson DM, Shirinsky VP, Sellers JR. (1997). Sites of interaction between kinase-related protein and smooth muscle myosin. J Biol Chem , 25353–25359. [DOI] [PubMed] [Google Scholar]
- Simoes RL, Fierro IM. (2005). Involvement of the Rho-kinase/myosin light chain kinase pathway on human monocyte chemotaxis induced by ATL-1, an aspirin-triggered lipoxin A4 synthetic analog. J Immunol , 1843–1850. [DOI] [PubMed] [Google Scholar]
- Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. (2004). Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J Mol Biol , 91–108. [DOI] [PubMed] [Google Scholar]
- Smith L, Parizi-Robinson M, Zhu M-S, Zhi G, Fukui R, Kamm KE, Stull JT. (2002). Properties of long myosin light chain kinase binding to F-actin in vitro and in vivo. J Biol Chem , 35597–35604. [DOI] [PubMed] [Google Scholar]
- Smith L, Su X, Lin P, Zhi G, Stull JT. (1999). Identification of a novel actin binding motif in smooth muscle myosin light chain kinase. J Biol Chem , 29433–29438. [DOI] [PubMed] [Google Scholar]
- Strome S. (1986). Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol , 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull JT, Lin PJ, Krueger JK, Trewhella J, Zhi G. (1998). Myosin light chain kinase: functional domains and structural motifs. Acta Physiol Scand , 471–482. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell , 123–132. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Ikebe R, Ikebe M, Luna EJ. (2007). Supervillin slows cell spreading by facilitating myosin II activation at the cell periphery. J Cell Sci , 3792–3803. [DOI] [PubMed] [Google Scholar]
- Tan PY, Zaidel-Bar R. (2015). Transient membrane localization of SPV-1 drives cyclical actomyosin contractions in the C. elegans spermatheca. Curr Biol , 141–151. [DOI] [PubMed] [Google Scholar]
- Totsukawa G, Wu Y, Sasaki Y, Hartshorne DJ, Yamakita Y, Yamashiro S, Matsumura F. (2004). Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J Cell Biol , 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. (2000). Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol , 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan R, Nagarajaram H, Ramakrishnan C. (1996). A procedure for the prediction of temperature-sensitive mutants of a globular protein based solely on the amino acid sequence. Proc Natl Acad Sci USA , 13908–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verin AD, Gilbert-McClain LI, Patterson CE, Garcia JGN. (1998). Biochemical regulation of the nonmuscle myosin light chain kinase isoform in bovine endothelium. Am J Respir Cell Mol Biol , 767–776. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol , 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadgaonkar R, Nurmukhambetova S, Zaiman AL, Garcia JGN. (2003). Mutation analysis of the non-muscle myosin light chain kinase (MLCK) deletion constructs on CV1 fibroblast contractile activity and proliferation. J Cell Biochem , 623–634. [DOI] [PubMed] [Google Scholar]
- Wang L, Audhya A. (2014). In vivo imaging of C. elegans endocytosis. Methods , 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo D, Cao J, Gong L, Kamm KE, Regalado E, Li L, Shete S, He W, Zhu M, et al. (2010). Mutations in myosin light chain kinase cause familial aortic dissections. Am J Hum Genet , 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhou T, Saadat L, Garcia JG. (2015). A MYLK variant regulates asthmatic inflammation via alterations in mRNA secondary structure. Eur J Hum Genet , 874–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hosoya H, Yonemura S. (2007). Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell , 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Munro E, Glotzer M. (2007). Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol , 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirshing ACE, Cram EJ. (2017). Myosin activity drives actomyosin bundle formation and organization in contractile cells of the Caenorhabditis elegans spermatheca. Mol Biol Cell , 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, Mains PE. (1999). The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol , 111–127. [DOI] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, McGhee JD, Mains PE. (1997). Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev , 409–422. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Totsukawa G, Yamakita Y, Sasaki Y, Madaule P, Ishizaki T, Narumiya S, Matsumura F. (2003). Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol Biol Cell , 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. (2015). The I-TASSER suite: protein structure and function prediction. Nat. Methods , 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. (2000). Calmodulin target database. J Struct Funct Genomics , 8–14. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Zhenhuan G, Luxenburg C. (2015). The contractome—a systems view of actomyosin contractility in non-muscle cells. J Cell Sci 1–9. [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics , 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.