Abstract
Introduction:
Mycobacterium leprae has a small genome and a tendency of persisting as a very low-grade infection. The authors have shown earlier, that the changes in TTC repeats, in M. leprae genome may contribute to the restriction of the pathogenicity of the bacterium and its survival strategy in case of pure neural Hansen's disease. We suspect, that a similar genomic reduction if happens in treated cases of Hansen's disease, can be a determining factor for developing persisters and relapse.
Aim:
The present study aimed to find out if there was any evidence of genomic reduction in treated cases of Hansen's disease that showed microbiological nonresponse.
Methods:
Skin biopsies were taken from treated cases of Hansen's disease at tertiary centers in Kolkata and at Raipur who had bacterial index (BI) unchanged or increased compared to their pretreatment BI. Analysis for the mutation in rpoB gene and folP1 gene were done to rule out rifampicin and dapsone resistance, respectively. The entire TTC repeat region of the bacteria was amplified by polymerase chain reaction and was subjected to sequencing. The obtained sequences were then analyzed by CLUSTALW.
Results:
A total of 127 patients were included in the study of which in 52 the BI remained same and 75 had an increase in BI, even after 6 months of completion of multidrug therapy. Among the samples, 2 had positive rpoB gene mutation. No mutation was found in the folP1 gene. The TTC repeat of both the rpoB-resistant samples was found to have 17 copies, which matched their pretreatment copy number. In other 125 cases, 60 cases showed no change from their pretreatment TTC number. Of those 65 samples that showed evidence of genomic reduction, 11 samples showed one copy, 41 showed 2 copies, and 13 showed 3 copies deletion. We also observed a significant regional variation.
Conclusion:
We concluded that there was evidence of genomic reduction, which might lead to microbiological nonresponse in treated cases of Hansen's disease. This indicated a possibility of future persistence and relapse.
KEY WORDS: Genomic reduction, hansen's disease, mycobacterium leprae, relapse, resistance, ttc repeats
Introduction
Mycobacterium leprae is unique in the sense that it is an obligate intracellular parasite and serves as excellent model to study how these bacteria exploits the functions of their host cells. Obligatory intracellular parasites possess a small genome and have a tendency of genome reduction.[1] This genome reduction is a common phenomenon of these obligatory intracellular parasites targeting overlapping subsets of potentially dispensable genes while adapting to the selective pressure of the different niches. Therefore, genes found in multiple copies may contribute to their specific adaptation, function and pathogenicity.[2]
TTC repeat is the variable number tandem repeat (VNTR) in M. leprae, reported first by Shin et al.[3] and has 21 repeats in some strain of M. leprae.[4] The stability of this TTC repeat was studied and was found to remain stable even after several serial passages in the footpads of nude mice.[5]
The change in the TTC repeat may contribute to strain differentiation and may also contribute to the restriction for the pathogenicity of the bacterium and their lifestyle.[5] In contrast, comparative genomic studies have revealed that in some cases, the genomes of bacteria, like in cases of Rickettsia or Mycobacteria spp., are reduced. It is assumed that for M. leprae, this tandem repeat is very specific and any deletion of genes in the TTC repeat sequence may have a detrimental effect on their fitness and on their ability to cause disease.[6,7,8] Furthermore, the occurrence of disease relapse or inability of multidrug therapy (MDT) to tame the bacterium proliferation is observed with an increasing trend.[9] Mycobacterial persisters have been reported in the treated cases of Hansen's disease and it was postulated that such persisters may contribute to relapse of Hansen's disease after completion of the World Health Organization (WHO) MDT.[10]
Previous reports suggested a few mutations in the rpoB and folP1 genes of the bacterium might contribute to its resistance against the drug therapy and might also have accounted for the relapse of the disease.[11] As the prevalence of drug-resistant bacterium is 1 in 1014 bacteria, it is imperative to consider another possible reason for the relapse. The authors have earlier shown that genomic reduction at TTC repeats can be a survival strategy for M. leprae in cases of pure neural Hansen's disease.[12,13]
In an attempt to find the most possible explanation for the microbiological nonresponsiveness, we tried to study the variable number of tandem repeats TTC, rpoB, and folP1 gene of M. leprae genome from clinical isolates from two different regions of the country. Our study aimed at finding evidence of genomic reduction and drug-resistant mutations in patients where the bacterial index (BI) remained refractory to treatment, even 6 months after completion of the MDT.
Materials and Methods
Patient selection criteria
In the tertiary leprosy centers of Kolkata and Raipur, patients who were diagnosed as multibacillary (MB) Hansen's disease and started with MB-MDT were followed up for microbiological response with slit skin smears. Of the 246 patients, who were initially enrolled for follow-up, 127 (51.6%) individuals who were found to have signs of “microbiological nonresponse” 6 months after completion of MB-MDT, were included in the study. “Microbiological non-response” was defined as the BI remaining at the same value as of corresponding pretreatment or an increase by at least 1+, 6 months after the completion of MB-MDT. Institutional ethical clearance was taken from all three study centers – Institute of Post Graduate Medical Education and Research, Kolkata; Medical College, Kolkata; and Pandit Jawaharlal Nehru Medical College, Raipur. Patients were explained properly about the study matter, and then, the duly signed informed consent form was collected from each study individual.
Collection of punch biopsy
A 4 mm punch biopsy was taken from the leprosy patch of the patient after proper sterilization of the site. The collected samples were stored at 4°C till being processed for the genomic DNA isolation.[14]
Genomic DNA isolation
Genomic DNA was isolated by standard phenol-chloroform method. The skin biopsy samples were homogenized using an automated mortar pestle (Sigma, USA). The homogenized samples were then incubated at 55°C for 2–4 h with lysis buffer (Tris and 0.25% Tween 20) and 20 mg/ml Proteinase K. Equal volume of phenol:chloroform (1:1) was added and centrifuged at 10000 rpm for 10 min. Equal volume of chloroform was added to the aqueous phase and again centrifuged at 10000 rpm for 10 min. One-tenth volume of 3M sodium acetate and 2 times the volume of ice-cold ethanol were added and the mixture was incubated at − 20°C. After incubation, the sample was centrifuged at 12,000 rpm for 10 min. The pellet was washed with ice-cold ethanol and air dried. Finally, the pellet was dissolved in 100 μl miliQ distilled water.[14]
Polymerase chain reaction amplification of TTC repeats
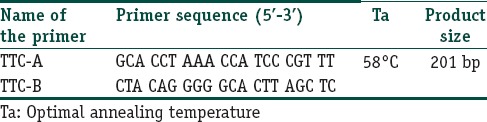
The primers used for region flanking entire 21 bp TTC repeat sequences were originally designed by Shin et al.[3] Both the primers were designed based on the M. leprae genome sequences flanking the TTC repeats to make sure that only the M. leprae genomic DNA would anneal with the primers [Table 1], by the polymerase chain reaction (PCR) conditions. Final DNA concentration of 100 ng was measured by spectrophotometer. Genomic DNA was amplified with AmpliTaq Gold, (Applied Biosystems, Inc., Foster City, CA) in a PCR reaction mixture, containing 1×PCR buffer (Applied Biosystems), 200 microM MgCl2, 2.5 mM each dNTP, and 20 picomoles of each primer TTC-A/TTC-B. The primer sequences, primer annealing temperature (Ta°), and PCR product sizes are given in Table 1. The PCRs were performed in the following conditions: 95°C 4 min, followed by 35 cycles of 95°C for 1 min, Ta° of the primer annealing [Table 1] for 1 min, 72°C for 1 min, and then 72°C for 10 min for the final extension. The amplified products were separated by electrophoresis on 2% agarose gel stained with 0.5 mg/mL ethidium bromide and visualized and photographed under an ultraviolet transilluminator.
Table 1.
Primer melting temperature value and primer sequences used in the amplification of TTC repeats
The amplified PCR products were then subjected to sequencing. Mutations in rpoB and folP1 gene were determined as described previously by Lavania et al.[15]
Statistical analysis
Data were summarized as percentages. The obtained sequences were analyzed for changes by CLUSTALW multiple sequence alignment using MAFFT (Version 7.214). Fisher's exact test was done to find out if there was any significant regional variation in TTC drop pattern. The cutoff for statistical significance was considered as P < 0.05.
Results
Of the 246 patients, who were initially enrolled for follow-up, 127 (51.6%) individuals were found to have signs of “microbiological nonresponse” 6 months after completion of MB-MDT and were included in the study. These 127 cases, who either had an increase in BI by at least 1+ or remained same in respect to their pretreatment BI value, even after 6 months of completing MDT, were selected for the study from three centers of Kolkata and Raipur. Of these, samples from 54 patients were received from Raipur, Chhattisgarh and the remaining 73 samples were received from patients of two centers in Kolkata. Of those, who were included in the study, 52 had BI remaining the same and 75 had an increase in BI at least by 1+, even after 6 months of completion of MDT.
Out of the 127 cases, 125 cases did not show any mutation in rpoB and folP1 gene of the M. leprae genome. The remaining two cases showed mutation at the base pair position 2,275,405 where G was replaced by C in the M. leprae genome, which corresponded to the coding region of rpoB gene (279 bp– 2275228 to 2275506).[15] Moreover, these patients were shown to have 17 copies of TTC repeats in the M. leprae genome after 6 months of follow-up, which were equal to their pretreatment value [Figure 1]. These two patients were reported from Kolkata. All other cases (n=125) had 13 copies of TTC repeat as their pretreatment value [Table 2].
Figure 1.
Electropherogram for the TTC repeat region with 17 copies
Table 2.
Regional variations in the genomic pattern of the Mycobacterium leprae isolated from the treated cases of Hansen's disease
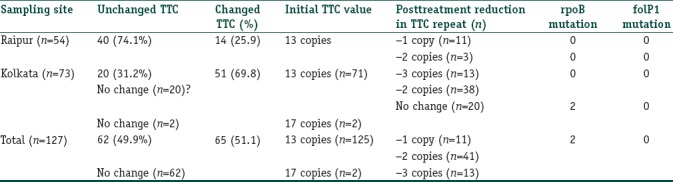
All the other remaining 125 cases were also subjected for TTC repeats analysis in follow-up after MDT. Out of the 125 cases, 60 cases showed no change in TTC repeats when subjected to PCR analysis (40 from Raipur and 20 cases from Kolkata) in comparison to their respective pretreatment TTC repeats. Out of the remaining 65 cases (14 cases from Raipur and 51 cases from Kolkata) PCR amplification of TTC repeats was analyzed. It showed changes in the amplified TTC repeats when compared to their respective pretreatment TTC repeats [Figure 2 and Table 3]. The amplified sequences of DNA are shown in Figure 3.
Figure 2.
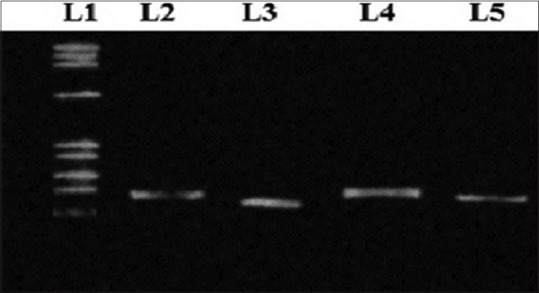
Polymerase chain reaction amplification of TTC repeat region of Mycobacterium leprae genome shows variations when separated in 2% agarose gel. L1– PhiX174 DNA ladder, L3 shows amplified TTC repeat region of Mycobacterium leprae in patient samples with loss in 2 copy numbers, L4 shows amplified TTC repeat region of Mycobacterium leprae in patient samples with loss in 1 copy number and L5 shows no change in TTC repeat region of the Mycobacterium leprae genome obtained from patient sample
Table 3.
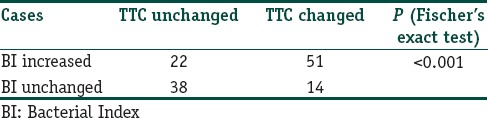
Correlation of changes in Bacterial Index with PCR findings
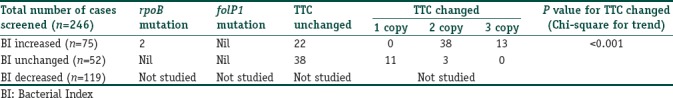
Figure 3.
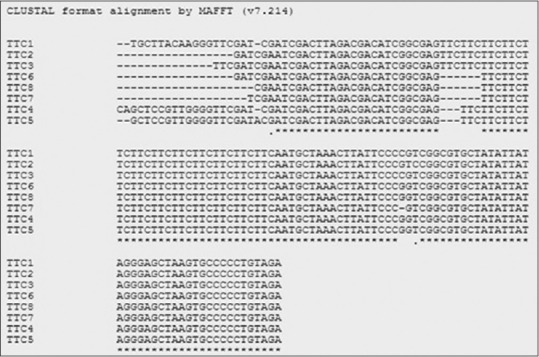
CLUSTALW sequence alignment of the TTC repeats obtained from patients samples of both Kolkata and Raipur. Multiple sequence alignment shows loss in 1 copy number of TTC repeat in the sample obtained from Kolkata (TTC4 and TTC5) and loss of 2 copy numbers of TTC repeats in samples obtained from Raipur (TTC6, TTC7, and TTC8). TTC2 and TTC3 sequences showed no changes in TTC repeats obtained from patients samples when aligned against TTC sequence obtained from nontreated case (TTC1)
CLUSTALW multiple sequence alignment analysis of 53 samples from Kolkata, showed in 38 samples, evidence of genomic reduction; they had 11 copies of TTC in follow-up visit with a deletion of 2 copy numbers from their pretreatment value [Figure 3]. The remaining 13 (excluding 2 with rpoB mutation) cases showed to have 10 copies of TTC repeats in their follow-up visit with a deletion of 3 copy numbers as compared with their initial copy numbers [Figure 3].
In 11 out of 14 cases collected from Raipur, we found to have 12 copies of TTC repeats in the genome of M. leprae isolate and showed a deletion in 1 copy number of TTC repeat region when compared with their initial copies. The remaining three cases found to have 11 copies of TTC repeats with a reduction of 2 copy numbers of TTC repeats when compared with their initial pretreatment TTC repeat counts as shown in Figure 3.
Fisher's exact test showed that there was significant regional variation in TTC drop pattern (P < 0.0001). There was a highly significant variation in the genomic drop pattern in the TTC repeat region [Tables 2 and 4].
Table 4.
Correlation of changes in Bacterial index with TTC changes
The trend for the changed TTC repeats showed that deletion of 2 copies of TTC repeat was significantly more common (P < 0.001) than deletion of single copy or 3 copies.
Discussion
Although the leprosy elimination program has seen major success in India and elsewhere, annual new case detection rate of leprosy has remained a cause of concern.[16] Studies have reported the problem of persistence of bacterium in treated cases of Hansen's disease even after the completion of WHO-MDT.[10] The authors have earlier shown that genomic reduction may be a survival strategy for M. leprae in cases of pure neural Hansen's disease.[12] In previous studies, a polymorphic site in the M. leprae genome was investigated particularly for distinguishing between relapse and reinfection.[17] It was[18,19,20] suggested that point mutations in rpoB and folP1 genes of the bacterium were responsible for the resistance against the two major drugs in MDT, namely rifampicin and dapsone, respectively. Hence, in the last phase of elimination program of Hansen's disease, a persisting infection of leprosy bacilli either by means of selective genomic reduction or by mutation in the drug resistance sites can be a real threat to the success of the WHO-MDT program. Our study aimed at finding evidence of genomic reduction and drug-resistant mutations in patients where the BI remained refractory to treatment, even 6 months after completion of the MDT.
Environmental effect on the bacterial genome can also make them act against the activity of the drugs, hence there is a possibility of relapse, reinfection, and resistance. Earlier, studies on VNTRs had demonstrated the effect of environment on the genome of both prokaryotic and eukaryotic cells. VNTRs based on 3, 5, or 6 nucleotide repeats were used earlier for the epidemiological investigation of amoxicillin resistance to Hemophillus influenza type B.[18]
The study by Shin et al. focused on TTC repeats in M. leprae from clinical isolates in Cebu City, Philippines. They reported evidence of variable numbers in TTC repeats of M. leprae obtained from MB patients and suggested a possible role in the adaptation of the bacterium against the environment of the hyperendemic regions of leprosy infection and also could be a useful mean to determine the source and transmission pattern of the bacterium.[3] In the present study, the posttreatment changes in TTC repeats were observed in more than half (51.1%) of clinical isolates of the patients from both Raipur and Kolkata, which clearly indicated a strong influence of the environmental factors including weather, presence of herd immunity, family history, prevalence, transmission rate, socioeconomic factors, and interventions such as MDT on the genomic structure of the organism. Furthermore, the 2 copies deletion of TTC repeat was significantly more than 1 copy deletion and 3 copies deletion. The exact mechanism of the evolution of M. leprae strain with different copy numbers of TTC repeats remained unexplained, but it could be postulated that it might be an adaptive response of the bacterium to survive difficult host conditions under antileprosy drugs, resulting in possible microbiological nonresponse. Our study also found that there were not many mutations in resistance determining genes (rpoB and folP1), so the microbiological nonresponse was possibly not linked with standard drug resistance.
Our study thus emphasized that though there was not much resistance of the individual bacillus against either dapsone or rifampicin; a substantial portion of the treated cases of MB Hansen, were still showing microbiological nonresponse with evidence of genomic reduction. We suggest this group may need a special attention, as there may be an increasing tendency of evasion of host defense, which may lead to future persisters and even relapse. We also suspect that truncated duration of MDT may have an impact on the final relapse rate if the otherwise drug sensitive bacilli are able to escape host defense due to genomic reduction. Although larger long-term prospective data with both clinical and microbiological parameters are needed for complete understanding of the impact of genomic reduction on leprosy; it can safely be concluded that to enhance the impact of leprosy elimination program a stronger vigilance is needed at least in a subset of patients where there are chances of bacterial persistence and genomic reduction.
Limitation of the study
This study is limited by the fact that principally we have conducted a cross-sectional study with only microbiological data. The time frame chosen for, doing BI and taking samples for bacterial DNA after 6 months of completion of MDT, is arbitrary. Moreover, due to lack of resources, our DNA analysis was restricted to the 127 samples where there were possibility of microbiological “nonresponse” and we were unable to compare them with the other group of patients where there was evidence of BI reduction. Our study can at best generate a hypothesis that there are signs of genomic reduction at the TTC repeat and this may be linked with possible bacterial survival or persistence even after complete course of MDT. However, a larger prospective cohort with longer follow-up comprising and comparing both clinical and microbiological data can throw more light on the actual impact of bacterial genomic reduction on the leprosy elimination process.
Conclusion
We conclude that there is evidence of genomic reduction, which may lead to microbiological nonresponse in treated cases of Hansen's disease. This signifies a possibility of future persistence and relapse. Further work is needed to be done with longitudinal studies to find conclusive evidence of genomic reduction and resultant treatment failure or relapse.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Andersson JO, Andersson SG. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev. 1999;9:664–71. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 2.Andersson JO, Andersson SG. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol. 2001;18:829–39. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- 3.Shin YC, Lee H, Lee H, Walsh GP, Kim JD, Cho SN, et al. Variable numbers of TTC repeats in Mycobacterium leprae DNA from leprosy patients and use in strain differentiation. J Clin Microbiol. 2000;38:4535–8. doi: 10.1128/jcm.38.12.4535-4538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, et al. Massive gene decay in the leprosy Bacillus. Nature. 2001;409:1007–11. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 5.Matsuoka M, Zhang L, Budiawan T, Saeki K, Izumi S. Genotyping of Mycobacterium leprae on the basis of the polymorphism of TTC repeats for analysis of leprosy transmission. J Clin Microbiol. 2004;42:741–5. doi: 10.1128/JCM.42.2.741-745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ten Bokum AM, Movahedzadeh F, Frita R, Bancroft GJ, Stoker NG. The case for hypervirulence through gene deletion in Mycobacterium tuberculosis. Trends Microbiol. 2008;16:436–41. doi: 10.1016/j.tim.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Wixon J. Featured organism: Reductive evolution in bacteria: Buchnera sp. Rickettsia prowazekii and Mycobacterium leprae. Comp Funct Genomics. 2001;2:44–8. doi: 10.1002/cfg.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakharkar KR, Dhar PK, Chow VT. Genome reduction in prokaryotic obligatory intracellular parasites of humans: A comparative analysis. Int J Syst Evol Microbiol. 2004;54:1937–41. doi: 10.1099/ijs.0.63090-0. [DOI] [PubMed] [Google Scholar]
- 9.Lavania M, Jadhav RS, Chaitanya VS, Turankar R, Selvasekhar A, Das L, et al. Drug resistance patterns in Mycobacterium leprae isolates from relapsed leprosy patients attending The Leprosy Mission (TLM) Hospitals in India. Lepr Rev. 2014;85:177–85. [PubMed] [Google Scholar]
- 10.Gupta UD, Katoch K, Singh HB, Natrajan M, Katoch VM. Persister studies in leprosy patients after multi-drug treatment. Int J Lepr Other Mycobact Dis. 2005;73:100–4. [PubMed] [Google Scholar]
- 11.Sapkota BR, Ranjit C, Neupane KD, Macdonald M. Development and evaluation of a novel multiple-primer PCR amplification refractory mutation system for the rapid detection of mutations conferring rifampicin resistance in codon 425 of the rpoB gene of Mycobacterium leprae. J Med Microbiol. 2008;57:179–84. doi: 10.1099/jmm.0.47534-0. [DOI] [PubMed] [Google Scholar]
- 12.De A, Reja AH, Biswas S, Bhattacharya B, Chatterjee G, Basu K, et al. Unique TTC repeat base pair loss mutation in cases of pure neural leprosy: A Survival strategy of Mycobacterium leprae? Indian J Dermatol. 2015;60:351–5. doi: 10.4103/0019-5154.160478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reja AH, De A, Biswas S, Chattopadhyay A, Chatterjee G, Bhattacharya B, et al. Use of fine needle aspirate from peripheral nerves of pure-neural leprosy for cytology and PCR to confirm the diagnosis: A pilot study. Indian J Dermatol Venereol Leprol. 2013;79:789–94. doi: 10.4103/0378-6323.120731. [DOI] [PubMed] [Google Scholar]
- 14.Reja AH, Biswas N, Biswas S, Dasgupta S, Chowdhury IH, Banerjee S, et al. Fite-Faraco staining in combination with multiplex polymerase chain reaction: A new approach to leprosy diagnosis. Indian J Dermatol Venereol Leprol. 2013;79:693–700. doi: 10.4103/0378-6323.116740. [DOI] [PubMed] [Google Scholar]
- 15.Lavania M, Hena A, Reja H, Nigam A, Biswas NK, Singh I, et al. Mutation at codon 442 in the rpoB gene of Mycobacterium leprae does not confer resistance to rifampicin. Lepr Rev. 2016;87:93–100. [PubMed] [Google Scholar]
- 16.Dogra S, Narang T, Kumar B. Leprosy – Evolution of the path to eradication. Indian J Med Res. 2013;137:15–35. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Fsihi H, Cole ST. The Mycobacterium leprae genome: Systematic sequence analysis identifies key catabolic enzymes, ATP-dependent transport systems and a novel polA locus associated with genomic variability. Mol Microbiol. 1995;16:909–19. doi: 10.1111/j.1365-2958.1995.tb02317.x. [DOI] [PubMed] [Google Scholar]
- 18.Williams DL, Gillis TP. Molecular detection of drug resistance in Mycobacterium leprae. Lepr Rev. 2004;75:118–30. [PubMed] [Google Scholar]
- 19.Bretagne S, Costa JM, Besmond C, Carsique R, Calderone R. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J Clin Microbiol. 1997;35:1777–80. doi: 10.1128/jcm.35.7.1777-1780.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Valero L, Rocha EP, Latorre A, Silva FJ. Reconstructing the ancestor of Mycobacterium leprae: The dynamics of gene loss and genome reduction. Genome Res. 2007;17:1178–85. doi: 10.1101/gr.6360207. [DOI] [PMC free article] [PubMed] [Google Scholar]



