Abstract
The in situ three-dimensional organization of chromatin at the nucleosome and oligonucleosome levels is unknown. Here we use cryo-electron tomography to determine the in situ structures of HeLa nucleosomes, which have canonical core structures and asymmetric, flexible linker DNA. Subtomogram remapping suggests that sequential nucleosomes in heterochromatin follow irregular paths at the oligonucleosome level. This basic principle of higher-order repressive chromatin folding is compatible with the conformational variability of the two linker DNAs at the single-nucleosome level.
INTRODUCTION
The fundamental unit of chromatin is the nucleosome, a 10-nm-diameter, 6-nm-thick cylindrical complex assembled from eight histone proteins and wrapped ∼1.65 times by 146 base pairs of DNA (Luger et al., 1997). In cells, many nucleosomes bind a linker histone, which stabilizes the two linker DNAs in a crossed conformation at the entry/exit position. When isolated or reconstituted, this larger nucleosome complex is called the chromatosome (Zhou et al., 2015; Bednar et al., 2017). Chemically fixed nucleosome chains can form highly ordered 30-nm fibers in vitro (Routh et al., 2008; Song et al., 2014), but these structures have not been detected inside cycling metazoan, plant, or yeast cells or isolated mitotic chromosomes (McDowall et al., 1986; Bouchet-Marquis et al., 2006; Eltsov et al., 2008, 2014, 2018; Fussner et al., 2011, 2012; Nishino et al., 2012; Gan et al., 2013; Chen et al., 2016; Ou et al., 2017; Cai et al., 2018a). While the consensus is that in situ chromatin structure is irregular (Hansen et al., 2018), the three-dimensional details of chromatin packing at the nucleosome level remain unknown.
Individual nucleosomes are challenging to identify in situ. The DNA-proximal negative-stain approach (ChromEMT) suffers distortions from chemical fixation, dehydration, and staining, resulting in large groups of nucleosomes appearing as amorphous elongated bodies instead of sets of discrete particles (Ou et al., 2017). For eukaryotic cellular cryo-electron microscopy (cryo-EM) samples, which must be thinned in cryogenic conditions, small high-contrast nonperturbative stains do not yet exist. Immuno-EM is also not suitable for protein identification in cryotomograms of thinned cells because 1) antibodies would freeze immediately on contact with any part of a cryo-EM sample, 2) the antibodies can access only the two surfaces of typical EM samples, and 3) antibody-gold complexes are too large (>10 nm) to unambiguously identify small complexes in crowded environments. Correlative cryo-light/cryo-superresolution microscopy can facilitate the localization of fluorescently tagged proteins, which can then be followed up by cryo-electron tomography (cryo-ET) to identify the densities that match the structure of the complex (Chang et al., 2014; Bauerlein et al., 2017; Hampton et al., 2017; Bharat et al., 2018). However, modern light cryomicroscopes cannot resolve individual nucleosomes in their densely packed nuclear environment.
Large multimegadalton complexes have been successfully identified by solely their structural signature (their size and shape) in crowded environments (Asano et al., 2015). This noninvasive structure-based approach could in principle be done for smaller complexes like nucleosomes. We have therefore taken advantage of sample-preparation, imaging-hardware, and image-processing advances to determine the structures, positions, and orientations of nucleosomes inside a HeLa cell. Our resultant mononucleosome subtomogram averages and their remapping reveal a first glimpse of higher-order heterochromatin structure and in situ architecture at the dinucleosome and trinucleosome levels.
RESULTS AND DISCUSSION
To determine how interphase mammalian chromatin is organized in situ, we performed Volta phase-contrast cryo-ET (Fukuda et al., 2015) on a HeLa cell that was thinned by a cryo-focused-ion-beam (cryo-FIB) milling workflow (Medeiros et al., 2018). The resultant cryotomogram shows exceptional detail, such as the clear delineation of membrane leaflets and a nucleoplasm densely populated with nucleosomes (Figure 1, A and B). Unlike interphase yeast cells, which have uniformly distributed nucleosomes (Chen et al., 2016; Cai et al., 2018a), mammalian cells have densely packed perinuclear heterochromatin (Figure 1C) flanking the nuclear pore and loosely packed euchromatin (Figure 1D) in the interior positions (Visser et al., 2000; van Steensel and Belmont, 2017).
FIGURE 1:
Volta cryotomogram of a cryo-FIB-thinned HeLa cell. (A) Tomographic slice (20 nm) of the nuclear periphery of a HeLa cell. The nuclear pore is indicated by the arrow. Three heterochromatic positions are delineated by purple dashed lines. (B) Segmentation of the mitochondrial outer membrane (brown), microtubules (green), and the nuclear envelope (dark blue). The in silico purified nucleosomes are also shown (blue and magenta puncta; see the text). (C, D) Tomographic slices (10 nm) of the (C) heterochromatin and (D) euchromatin positions boxed in A, enlarged 4.5-fold. Several nucleosomes are indicated by arrowheads.
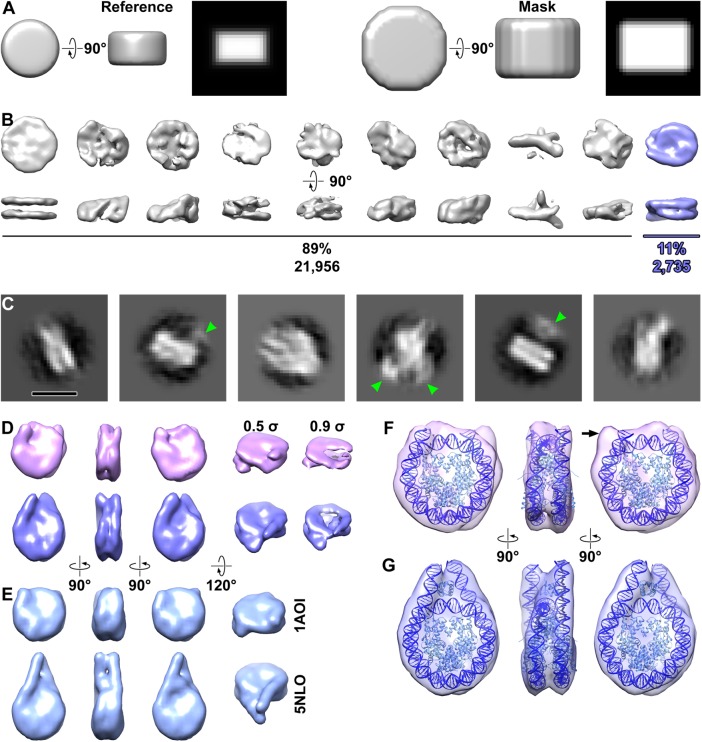
Multimegadalton complexes are straightforward to identify in cryotomograms (Briegel et al., 2009; Gan et al., 2011; Asano et al., 2015; Mahamid et al., 2016; Böck et al., 2017; Chaikeeratisak et al., 2017), but nucleosomes are not because they are only ∼200 kDa. In this study, we purify the nucleosomes “in silico” by combining template matching with three-dimensional classification (Bharat and Scheres, 2016), which we previously showed to be sensitive enough to identify nucleosomes of different linker-DNA conformations in nuclear lysates (Cai et al., 2018b). To minimize model bias, we used a featureless cylindrical reference (Figure 2A). This approach reveals a single three-dimensional class average that has the nucleosome’s unmistakable structural signatures: the left-handed wrapping of DNA, with the groove between the two gyres clearly visible at the position opposite of the DNA entry/exit site (Figure 2B). Even though no symmetry of any kind was enforced, the nucleosome class average has twofold symmetry around the dyad axis, which was seen in all previous crystal structures. To eliminate more model bias, we attempted to do three-dimensional classification using combinations of spheres and cylinders as masks and references. Our classification only succeeded when the mask was cylindrical (Supplemental Figure S1, A–C). This experiment suggests that alignment and classification convergence is adversely influenced by nucleosome-proximal densities. Negative control three-dimensional classification—using a cylindrical mask and reference—of cytoplasmic densities that were template matched the same way as for the nucleus did not produce any nucleosome-like class averages (Supplemental Figure S1D).
FIGURE 2:
Structural analysis of nucleosomes in situ. (A) The reference model (left half) and mask (right half) used for three-dimensional classification and averaging. The reference is 10 nm wide and 6 nm thick. Both the reference and mask have soft edges that slowly decay to zero. The rightmost subpanel (black background) for both the reference and mask are central slices through the side view. (B) Three-dimensional class averages of all template-matching hits. The nucleosome class average (blue) is oriented with its twofold dyad axis running horizontal. (C) Example two-dimensional class averages from the nucleosomes identified by three-dimensional classification. Some of the class averages have densities from nucleosome-associated complexes (green arrowheads). Bar, 10 nm. (D) Final three-dimensional class averages of nucleosomes, showing from left to right the front, side, and back and oblique views. One class (39%, magenta) has shorter linker-DNA densities that the other (61%, blue). Maps in all columns are contoured at 0.5 σ except in the rightmost column, which is set to 0.9 σ to better show the left-handed superhelical DNA path and the degree of linker-DNA heterogeneity. (E) Crystal structures of the nucleosome core (PDB 1AOI, top) and chromatosome (PDB 5NLO, bottom), rendered as 15-Å resolution density maps. (F, G) Refined maps of the two nucleosome classes, with the edited chromatosome crystal structure docked (F) without or (G) with the linker histone, and linker DNA appropriately truncated. The histones and DNA are light and dark blue, respectively.
Unlike our previous analysis of picoplankton nuclear lysates in which the nucleosomes were highly dispersed (Cai et al., 2018b), three-dimensional classification of HeLa nucleosomes in the crowded nucleus requires a cylindrical mask (Figure 2A). When we performed reference-free two-dimensional classification on the nucleosomes with a larger circular mask, we found that some class averages had extra densities in contact with the nucleosome (Figure 2C, green arrowheads). These densities are truncated by the mask, meaning that they belong to larger structures. Furthermore, these extra densities are weaker and featureless, consistent with their being averages of many different types of nucleosome-binding partners. Additional rounds of three-dimensional classification produced a final set of 1141 nucleosomes (see Materials and Methods).
Three-dimensional classification of the final nucleosome set into two classes yielded averages with either short or long linker-DNA densities (Figure 2D). These two classes refined to 24- and 21-Å resolution, respectively (Supplemental Figure S2), and resemble low-pass-filtered density maps calculated from crystal structures (Figure 2E). Indeed, the averages can accommodate the chromatosome crystal structure after rigid-body alignment and adjustment of the linker-DNA lengths (Figure 2, F and G) (Bednar et al., 2017). Note that the nucleosomes that contributed to a given class average have small variations in linker-DNA conformation that cannot yet be resolved. The class with the shorter linker DNA can be best fitted with a nucleosome core (151 base pairs). One of the linker-DNA densities cannot be adequately accounted for by the nucleosome crystal structure (Figure 2F and Supplemental Movie S1) and is instead consistent with partial unwrapping (Bilokapic et al., 2018a,b). The nucleosome class with the longer linker DNA is best fitted with ∼13 base pairs of DNA in each linker (172 base pairs total; Figure 2G and Supplemental Movie S2). The linker DNAs have a crossed conformation and remain visible when the density map’s contour level is raised (Figure 2D). This structural phenotype is consistent with the linker DNA’s conformational stabilization by a linker histone. Note that HeLa cells have a 183-base-pair nucleosome-repeat length (Lohr et al., 1977), which predicts that sequential nucleosomes are linked by an average ∼12-nm DNA (37 base pairs × 0.34 nm). Therefore, the short linker-DNA densities in one of the subtomogram averages arises from linker-DNA conformational heterogeneity in the individual nucleosomes. Finally, classification of the 1141 nucleosomes into four classes produces averages that show additional linker-DNA conformations, supporting the notion that the linker DNA is the most conformationally heterogeneous part of the nucleosome (Supplemental Figure S3).
Movie S1.
Density maps and docked atomic models of HeLa nucleosomes Subtomogram averages and models of the nucleosome classes with short (S1) and long (S2) linker DNAs are contoured at 0.5 σ. The color scheme is identical to Figure 2, F and G.
Movie S2.
Density maps and docked atomic models of HeLa nucleosomes Subtomogram averages and models of the nucleosome classes with short (S1) and long (S2) linker DNAs are contoured at 0.5 σ. The color scheme is identical to Figure 2, F and G.
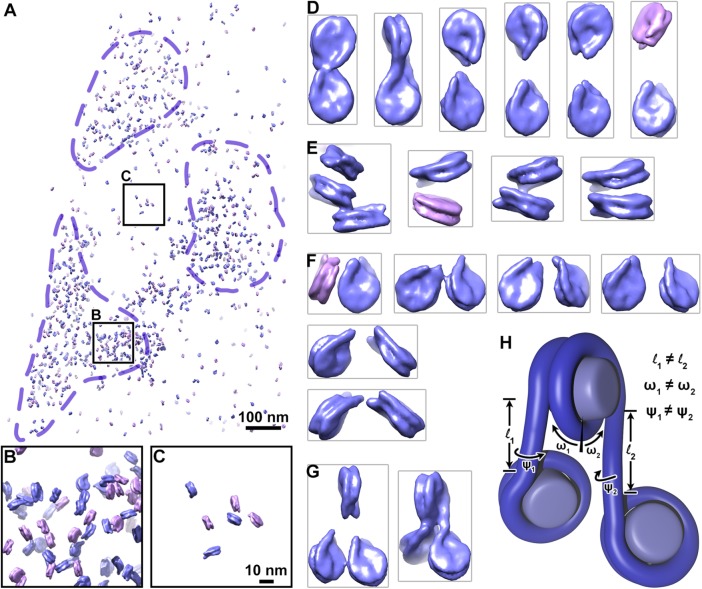
Our subtomogram averages presented an opportunity to visualize nucleosomes in the context of higher-order chromatin structure in situ. Using the three-dimensional refined orientations and positions, we remapped the nucleosomes back into an empty volume the size of the original cryotomogram (Figure 3). As expected from their appearance in the tomographic slices (Figure 1A), the nucleosomes are predominantly localized in the three heterochromatin clusters (Figure 3A). The heterochromatin and euchromatin contain both classes of nucleosomes (Figure 3, B and C). The nucleosomes in between the heterochromatin domains appear isolated instead of being parts of contiguous chains. Some nucleosomes must have been missed by our analysis. For example, nucleosomes oriented with their face parallel to the lamella surface were missed (Supplemental Figure S2, B and C); nucleosomes oriented this way are known to be challenging to locate in plunge-frozen samples (Chua et al., 2016). Our analysis would also have missed nucleosomes that make multiple contacts with large protein complexes (McGinty and Tan, 2015; Morgan et al., 2016; Wilson et al., 2016; Xu et al., 2016; Farnung et al., 2017; Liu et al., 2017; Ayala et al., 2018; Eustermann et al., 2018) and nucleosomes with unconventional structures such as partially unwrapped nucleosomes (Bilokapic et al., 2018a,b) and hexasomes (Kato et al., 2017).
FIGURE 3:
Chromatin is irregular at the oligonucleosome level in situ. (A) Model of short-linker (magenta) and long-linker (blue) nucleosomes remapped according to their positions and orientations in the nucleus. Dashed purple lines indicate approximate boundaries of heterochromatin. (B, C) Fourfold enlargements of the heterochromatin and euchromatin positions boxed in A. (D–G) Examples of (D) dinucleosomes connected by linker DNA. (E) face-to-face packed nucleosomes, (F) dinucleosomes not connected by linker DNA but likely to be in sequence with a third nucleosome that was missed by our analysis, and (G) trinucleosomes connected by linker DNA. For clarity, adjacent remapped nucleosomes were cropped out. (H) Schematic of a trinucleosome, showing histone octamers (light blue) and DNA (dark blue). The lengths (l1, l2), angles relative to the dyad axis (ω1, ω2), and rotation around the linker-DNA axes (ψ1, ψ2) are uncorrelated.
Many remapped nucleosomes are likely to be interacting with each other, because their linker-DNA densities are coaxial or because their cores are nearly stacked. We recognized four types of nucleosome–nucleosome arrangements (Figure 3, D–G, and Supplemental Figure S4, A–C): nucleosome pairs likely to be connected by linker DNA (Figure 3D); nucleosome pairs oriented with face-to-face interactions (Figure 3E); nucleosome pairs likely to share linker DNA with a third, unmapped nucleosome (Figure 3F); and trinucleosomes likely connected by linker DNA (Figure 3G). The visualization of linker-DNA densities in the subtomogram averages and remapped models provides the first clues about the path of DNA at the trinucleosome level (Figure 3, F and G). Nucleosomes in these examples are likely to follow an irregular zig-zag path. Periodic motifs such as those within tetranucleosomes were not found and therefore must be exceptionally rare (Schalch et al., 2005; Song et al., 2014). In support of this, our efforts to automatically identify dinucleosomes, which are found within tetranucleosomes, by three-dimensional classification failed to produce any meaningful averages (Supplemental Figure S4D).
Chromatin higher-order structure is extremely sensitive to linker-DNA parameters. For example, tetranucleosome face-to-face stacking can be abolished in vitro with a small change in linker-DNA length (Ekundayo et al., 2017). Recent cryo-EM studies showed that dinucleosomes have variable conformations even when they are reconstituted with a strong positioning sequence and are bound to either heterochromatin protein 1 or Polycomb repressive complex 2 (Machida et al., 2018; Poepsel et al., 2018). Our subtomogram averages and remapped nucleosomes are consistent with a model in which variations of linker-DNA length and orientation at the single-nucleosome level in situ give rise to irregular higher-order chromatin structure at the dinucleosome and trinucleosome levels (Figure 3H). Chromatin can therefore pack densely in heterochromatin without folding into periodic motifs. Future advances in cryo thinning, automation, subtomogram classification, and remapping will be important tools to dissect in situ chromatin structure in greater detail.
MATERIALS AND METHODS
Cell culture
HeLa CCL2 cells (American Type Culture Collection) were grown in DMEM (Life Technologies) supplemented with 10% inactivated fetal calf serum (Invitrogen) and 50 μg/ml streptomycin (AppliChem) at 37°C and 5% CO2. For EM imaging experiments, EM finder grids (gold NH2 R2/2, Quantifoil) were sterilized under UV light and then glow discharged. Grids were placed on the bottom of the wells of a 12-well plate (Nunc; Thermo Fisher) and equilibrated with DMEM for 30 min. Subsequently, 30,000 HeLa cells were seeded into each well and incubated overnight until vitrification.
Preparation of frozen-hydrated specimens
Plunge freezing was performed as previously reported (Weiss et al., 2017). Grids were removed from the wells using forceps. The forceps were then mounted in the Vitrobot, and the grid was blotted from the backside by installing a Teflon sheet on one of the blotting pads. Grids were plunge-frozen in liquid ethane/propane (37%/63%) (Tivol et al., 2008) using a Vitrobot Mk 4 (Thermo Fisher) and stored in liquid nitrogen.
Cryo-FIB milling
Cryo-FIB was used to cryo-thin samples of plunge-frozen HeLa cells so that they could be imaged by cryo-ET (Marko et al., 2007; Schaffer et al., 2017; Medeiros et al., 2018). Frozen grids with HeLa cells were first clipped into modified Autogrids (Thermo Fisher) and then transferred into the liquid-nitrogen bath of a loading station (Leica Microsystems). Grids were clamped onto a “40° pre-tilted TEM grid holder” (Leica Microsystems), and the holder was subsequently shuttled from the loading station to the dual-beam instrument using the VCT100 transfer system (Leica Microsystems). The holder was mounted on a custom-built cryo stage (Leica Microsystems) in a Helios NanoLab600i dual-beam FIB/SEM instrument (Thermo Fisher). The stage temperature was maintained below –154°C during the loading, milling, and unloading procedures. Grid quality was checked by scanning EM imaging (5 kV, 21 pA). Samples were coated with a platinum precursor gas using the Gas Injector System and a “cold deposition” technique (Hayles et al., 2007). Lamellae were milled in several steps. We first targeted two rectangular regions with the ion beam set to 30 kV and ∼400 pA to generate a ∼2-µm-thick lamella. The ion-beam current was then gradually decreased until the lamella reached a nominal thickness of ∼200 nm (ion beam set to ∼25 pA). After documentation of the lamellae by scanning EM imaging, the holder was brought back to the loading station using the VCT100 transfer system. The grids were unloaded and stored in liquid nitrogen.
Electron cryomicroscopy and electron cryotomography
The cryo-EM imaging details are listed in Supplemental Table S1. Cryo-FIB–processed HeLa cells were examined by both cryo-EM and cryo-ET (Weiss et al., 2017). Images were recorded on a Titan Krios transmission electron cryomicroscope (Thermo Fisher) equipped with a K2 Summit direct-detection camera (Gatan), a Quantum LS imaging filter (Gatan), and a Volta phase plate (Thermo Fisher). The microscope was operated at 300 kV with the imaging filter slit width set to 20 eV. Data were collected in focus using the Volta phase plate. The pixel size at the specimen level was 3.45 Å. Tilt series covered an angular range from -60° to +60° with 2° increments. The total dose of a tilt series was 120 e-/Å2. Tilt series and two-dimensional projection images were acquired automatically using SerialEM (Mastronarde, 2005). Three-dimensional reconstructions and segmentations were generated using the IMOD program suite (Mastronarde, 2008). To increase the contrast, the tilt series was binned twofold in the IMOD program Etomo, resulting in a final specimen-level pixel size of 6.9 Å.
Template matching
The subtomogram analysis strategy was to find as many candidate nucleosomes as possible and then remove the majority of false positives by three-dimensional classification (Cai et al., 2018b). Template matching was done with PEET (Nicastro et al., 2006; Heumann, 2016). To speed up the search, the tomogram was binned threefold, corresponding to a 10.35 Å voxels. A featureless 10-nm-diameter × 6-nm-thick cylinder was created with the Bsoft (Heymann and Belnap, 2007) program beditimg for use as the initial reference model. To emulate the effects of Volta phase contrast, this reference was corrupted with the Bsoft program bctf using a three-dimensional contrast transfer function with the fraction of amplitude contrast set to 0.5. To suppress the effects of nucleoplasmic background densities, the template was masked with a soft-edged cylinder. To minimize the number of false negatives, we used a very low cross-correlation cutoff of CC = 0.2. We also set the minimum interparticle spacing to 6 nm, which ensured that any face-to-face stacked nucleosomes would not be missed. To minimize model bias, only data up to ∼50 Å resolution were used. Using these criteria, ∼24,700 of ∼83,300 possible hits were retained. Visual inspection of the hits list confirmed that many nonnucleosome densities were also included.
Classification analysis and three-dimensional subtomogram remapping
All two- and three-dimensional classifications and three-dimensional autorefinements were done with RELION 2.1 (Kimanius et al., 2016) using default parameters except where noted below. No point symmetry was applied in either the classification or the refinement steps. A large box (∼2 × the nucleosome diameter) was used so that the particle center could be refined during classification. This box choice resulted in the introduction of new false positives, which were dealt with in a second round of template matching (see below). The template-matched particles were extracted using the subtomogram analysis routines (Bharat et al., 2015). Orientation information was discarded in this process. For two-dimensional classification, the mask diameter was 140 Å, and the regularization parameter T was set to 4. Three-dimensional classification was done with a featureless 10-nm-diameter × 6-nm-thick cylindrical reference and a larger cylindrical mask with a soft edge (Figure 2A). Sequential rounds of three-dimensional classification pruned the nucleosome class to 1883 particles. RELION performs classification and alignment simultaneously, meaning that it functions as another form of multiclass template matching in which the templates can change during the run. One consequence is that some nucleosome centers can translate to positions that either overlap neighboring nucleosomes or correspond to the nucleoplasm. To deal with the existence of new false positives, an additional round of template matching was performed in PEET, using only the refined positions of the 1883 classified particles that contributed to the nucleosome class averages. The refined nucleosome density map (including the 1883 particles) was used as the new template-matching reference. PEET removed the duplicated particles automatically. Next, the cross-correlation threshold relative to the template was incrementally increased until most of the spurious positions in the nucleoplasm were removed, yielding the final set of 1141 nucleosomes. A final three-dimensional classification was performed with two classes, resulting in one class average with long linker DNA and one with short linker DNA. Following three-dimensional autorefinement, the angular distribution was checked by loading the final .bild file and density maps together in UCSF Chimera (Pettersen et al., 2004).
To locate dinucleosomes, we performed three-dimensional classification using the coordinates of the nucleosome template-matching hits as a starting point. As a reference and mask, we used double-cylinder volumes, with separations approximating the dinucleosomes seen in our remapped models.
The nucleosome averages were remapped using the script ot_remap.py (https://github.com/anaphaze/ot-tools), which orients and positions each RELION class average into an empty volume the same size as the original tomogram using routines from EMAN2, IMOD, and Bsoft (Heymann and Belnap, 2007; Tang et al., 2007; Mastronarde, 2008; Cai et al., 2018b). One remapped model was created for each class (short and long linker DNA). The two models were then combined with the Bsoft program badd. Because the pairwise internucleosome distances and positions, that is, higher-order structure, was so heterogeneous, dinucleosomes and trinucleosomes had to be located manually in UCSF Chimera. To facilitate this manual search, the clipping planes were positioned so that the thickness along the view axis was <40 nm. Pairs of nucleosomes were considered to be interacting if their linker DNAs were aligned (sequential nucleosomes) or if any part of the two nucleosomes were within ∼2 nm.
Crystal structure docking
Because cryo-ET in situ subtomogram averages have much lower resolutions than crystal structures, the goal was to conservatively dock a chromatosome crystal structure into the subtomogram averages. Of the two chromatosome structures (Zhou et al., 2015; Bednar et al., 2017), 5NL0 fitted as a rigid body into the class with long linker DNA with minimal modification. This crystal structure was used as a starting point for further editing. For the nucleosome with longer linker DNA, 13 and 12 base pairs were removed from the linker-DNA termini, leaving 172 base pairs of DNA. For the nucleosome with shorter linkers, 24 and 22 base pairs of DNA were removed from the linker-DNA termini, leaving 151 base pairs of DNA. Next the chromatosome model was docked automatically with the UCSF Chimera fit-in-map routine, using a map simulated to 20-Å resolution. These produced map-to-model correlations of 0.95 (nucleosome with long linker) and 0.94 (nucleosome with short linker); see correlations for other linker lengths in Supplemental Table S2. Owing to the limited resolution, no further attempts were made to refine the atomic model.
Graphics
Figure panels were created in Adobe Illustrator CC, Google Sheets, or Blender 2.79 (www.blender.org) and then arranged in Adobe Photoshop CC.
Data availability
The unbinned frame-aligned tilt series was deposited in the Electron Microscopy Public Image Archive (Iudin et al., 2016) as EMPIAR-10179. The twofold binned tomogram and the nucleosome subtomogram averages with short and long linker DNA were deposited in the Electron Microscopy Data Bank as EMD-6948, EMD-6949, and EMD-6950, respectively.
Supplementary Material
Acknowledgments
We thank Duane Loh and Reza Khayat for discussions on heterogeneity and classification, John Heumann for advice on how to accelerate PEET template matching, and members of the Gan and Pilhofer teams and Alex Noble for feedback. ScopeM is acknowledged for instrument access at ETH Zürich. S.C. and L.G. were supported by the Singapore Ministry of Education (MOE) T2 R-154-000-624-112, MOE T1 R-154-000-A49-114, and National University of Singapore Young Investigator Award R-154-000-558-133. D.B. and M.P. were supported by the European Research Council, the Swiss National Science Foundation, and the Helmut Horten Foundation.
Abbreviations used:
- cryo-EM
cryo-electron microscopy
- cryo-ET
cryo-electron tomography
- cryo-FIB
cryo-focused-ion-beam
- EM
electron microscope/microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-05-0331) on August 9, 2018.
REFERENCES
- Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, Baumeister W. (2015). Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science , 439–442. [DOI] [PubMed] [Google Scholar]
- Ayala R, Willhoft O, Aramayo RJ, Wilkinson M, McCormack EA, Ocloo L, Wigley DB, Zhang X. (2018). Structure and regulation of the human INO80-nucleosome complex. Nature , 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W, Fernandez-Busnadiego R. (2017). In situ architecture and cellular interactions of PolyQ inclusions. Cell , 179–187 e110. [DOI] [PubMed] [Google Scholar]
- Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, et al (2017). Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol Cell , 384–397 e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Russo CJ, Lowe J, Passmore LA, Scheres SH. (2015). Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure , 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Scheres SH. (2016). Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc , 2054–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TAM, Hoffmann PC, Kukulski W. (2018). Correlative microscopy of vitreous sections provides insights into BAR-domain organization in situ. Structure , 879–886 e873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Strauss M, Halic M. (2018a). Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol , 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Strauss M, Halic M. (2018b). Structural rearrangements of the histone octamer translocate DNA. Nat Commun , 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck D, Medeiros JM, Tsao HF, Penz T, Weiss GL, Aistleitner K, Horn M, Pilhofer M. (2017). In situ architecture, function, and evolution of a contractile injection system. Science , 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Dubochet J, Fakan S. (2006). Cryoelectron microscopy of vitrified sections: a new challenge for the analysis of functional nuclear architecture. Histochem Cell Biol , 43–51. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, et al (2009). Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA , 17181–17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Chen C, Tan ZY, Huang Y, Shi J, Gan L. (2018a). Cryo-ET reveals the macromolecular reorganisation of S. pombe mitotic chromosomes in vivo. Proc Natl Acad Sci USA (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Song Y, Chen C, Shi J, Gan L. (2018b). Natural chromatin is heterogeneous and self-associates in vitro. Mol Biol Cell, 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikeeratisak V, Nguyen K, Khanna K, Brilot AF, Erb ML, Coker JK, Vavilina A, Newton GL, Buschauer R, Pogliano K, et al (2017). Assembly of a nucleus-like structure during viral replication in bacteria. Science , 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Chen S, Tocheva EI, Treuner-Lange A, Lobach S, Sogaard-Andersen L, Jensen GJ. (2014). Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat Methods , 737–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Lim HH, Shi J, Tamura S, Maeshima K, Surana U, Gan L. (2016). Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo. Mol Biol Cell , 3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EY, Vogirala VK, Inian O, Wong AS, Nordenskiold L, Plitzko JM, Danev R, Sandin S. (2016). 3.9 A structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res , 8013–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekundayo B, Richmond TJ, Schalch T. (2017). Capturing structural heterogeneity in chromatin fibers. J Mol Biol , 3031–3042. [DOI] [PubMed] [Google Scholar]
- Eltsov M, Grewe D, Lemercier N, Frangakis A, Livolant F, Leforestier A. (2018). Nucleosome conformational variability in solution and in interphase nuclei evidenced by cryo-electron microscopy of vitreous sections. Nucleic Acids Res, doi.org/10.1093/nar/gky670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. (2008). Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA , 19732–19737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M, Sosnovski S, Olins AL, Olins DE. (2014). ELCS in ice: cryo-electron microscopy of nuclear envelope-limited chromatin sheets. Chromosoma , 303–312. [DOI] [PubMed] [Google Scholar]
- Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP. (2018). Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature , 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung L, Vos SM, Wigge C, Cramer P. (2017). Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature , 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Laugks U, Lucic V, Baumeister W, Danev R. (2015). Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol , 143–154. [DOI] [PubMed] [Google Scholar]
- Fussner E, Djuric U, Strauss M, Hotta A, Perez-Iratxeta C, Lanner F, Dilworth FJ, Ellis J, Bazett-Jones DP. (2011). Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J , 1778–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussner E, Strauss M, Djuric U, Li R, Ahmed K, Hart M, Ellis J, Bazett-Jones DP. (2012). Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep , 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Ladinsky MS, Jensen GJ. (2011). Organization of the smallest eukaryotic spindle. Curr Biol , 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Ladinsky MS, Jensen GJ. (2013). Chromatin in a marine picoeukaryote is a disordered assemblage of nucleosomes. Chromosoma , 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton CM, Strauss JD, Ke Z, Dillard RS, Hammonds JE, Alonas E, Desai TM, Marin M, Storms RE, Leon F, et al (2017). Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat Protoc , 150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Connolly M, McDonald CJ, Pan A, Pryamkova A, Ray K, Seidel E, Tamura S, Rogge R, Maeshima K. (2018). The 10-nm chromatin fiber and its relationship to interphase chromosome organization. Biochem Soc Trans , 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles MF, Stokes DJ, Phifer D, Findlay KC. (2007). A technique for improved focused ion beam milling of cryo-prepared life science specimens. J Microsc , 263–269. [DOI] [PubMed] [Google Scholar]
- Heumann JM. (2016). PEET, University of Colorado Boulder. Retrieved from bio3d.colorado.edu/PEET/ (accessed 1 January, 2018).
- Heymann JB, Belnap DM. (2007). Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol , 3–18. [DOI] [PubMed] [Google Scholar]
- Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ, Patwardhan A. (2016). EMPIAR: a public archive for raw electron microscopy image data. Nat Methods , 387–388. [DOI] [PubMed] [Google Scholar]
- Kato D, Osakabe A, Arimura Y, Mizukami Y, Horikoshi N, Saikusa K, Akashi S, Nishimura Y, Park SY, Nogami J, et al (2017). Crystal structure of the overlapping dinucleosome composed of hexasome and octasome. Science , 205–208. [DOI] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, Lindahl E. (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife , e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li M, Xia X, Li X, Chen Z. (2017). Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature , 440–445. [DOI] [PubMed] [Google Scholar]
- Lohr D, Corden J, Tatchell K, Kovacic RT, Van Holde KE. (1977). Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci USA , 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature , 251–260. [DOI] [PubMed] [Google Scholar]
- Machida S, Takizawa Y, Ishimaru M, Sugita Y, Sekine S, Nakayama JI, Wolf M, Kurumizaka H. (2018). Structural basis of heterochromatin formation by human HP1. Mol Cell , 385–397.e8. [DOI] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Forster F, Hyman AA, Plitzko JM, Baumeister W. (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science , 969–972. [DOI] [PubMed] [Google Scholar]
- Marko M, Hsieh C, Schalek R, Frank J, Mannella C. (2007). Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods , 215–217. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol , 36–51. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. (2008). Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc , 212–217. [DOI] [PubMed] [Google Scholar]
- McDowall AW, Smith JM, Dubochet J. (1986). Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J , 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Tan S. (2015). Nucleosome structure and function. Chem Rev , 2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros JM, Bock D, Weiss GL, Kooger R, Wepf RA, Pilhofer M. (2018). Robust workflow and instrumentation for cryo-focused ion beam milling of samples for electron cryotomography. Ultramicroscopy , 1–11. [DOI] [PubMed] [Google Scholar]
- Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. (2016). Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science , 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science , 944–948. [DOI] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K. (2012). Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J , 1644–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. (2017). ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science , eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Poepsel S, Kasinath V, Nogales E. (2018). Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol , 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A, Sandin S, Rhodes D. (2008). Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA , 8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M, Mahamid J, Engel BD, Laugks T, Baumeister W, Plitzko JM. (2017). Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J Struct Biol , 73–82. [DOI] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ. (2005). X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature , 138–141. [DOI] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. (2014). Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science , 376–380. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. (2007). EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol , 38–46. [DOI] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ. (2008). An improved cryogen for plunge freezing. Microsc Microanal , 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell , 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser AE, Jaunin F, Fakan S, Aten JA. (2000). High resolution analysis of interphase chromosome domains. J Cell Sci (Pt 14), 2585–2593. [DOI] [PubMed] [Google Scholar]
- Weiss GL, Medeiros JM, Pilhofer M. (2017). In situ imaging of bacterial secretion systems by electron cryotomography. Methods Mol Biol , 353–375. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. (2016). The structural basis of modified nucleosome recognition by 53BP1. Nature , 100–103. [DOI] [PubMed] [Google Scholar]
- Xu P, Li C, Chen Z, Jiang S, Fan S, Wang J, Dai J, Zhu P, Chen Z. (2016). The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol Cell , 965–975. [DOI] [PubMed] [Google Scholar]
- Zhou BR, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. (2015). Structural mechanisms of nucleosome recognition by linker histones. Mol Cell , 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The unbinned frame-aligned tilt series was deposited in the Electron Microscopy Public Image Archive (Iudin et al., 2016) as EMPIAR-10179. The twofold binned tomogram and the nucleosome subtomogram averages with short and long linker DNA were deposited in the Electron Microscopy Data Bank as EMD-6948, EMD-6949, and EMD-6950, respectively.