Abstract
The endoplasmic reticulum (ER) and plasma membrane (PM) form junctions crucial to ion and lipid signaling and homeostasis. The Kv2.1 ion channel is localized at ER–PM junctions in brain neurons and is unique among PM proteins in its ability to remodel these specialized membrane contact sites. Here, we show that this function is conserved between Kv2.1 and Kv2.2, which differ in their biophysical properties, modulation, and cellular expression. Kv2.2 ER–PM junctions are present at sites deficient in the actin cytoskeleton, and disruption of the actin cytoskeleton affects their spatial organization. Kv2.2-containing ER–PM junctions overlap with those formed by canonical ER–PM tethers. The ability of Kv2 channels to remodel ER–PM junctions is unchanged by point mutations that eliminate their ion conduction but eliminated by point mutations within the Kv2-specific proximal restriction and clustering (PRC) domain that do not impact their ion channel function. The highly conserved PRC domain is sufficient to transfer the ER–PM junction–remodeling function to another PM protein. Last, brain neurons in Kv2 double-knockout mice have altered ER–PM junctions. Together, these findings demonstrate a conserved in vivo function for Kv2 family members in remodeling neuronal ER–PM junctions that is distinct from their canonical role as ion-conducting channels shaping neuronal excitability.
INTRODUCTION
Specialized membrane contact sites between the endoplasmic reticulum (ER) and plasma membrane (PM), or ER–PM junctions, are a ubiquitous feature of eukaryotic cells (Henne et al., 2015; Gallo et al., 2016; Chang et al., 2017; Saheki and De Camilli, 2017a). These specialized sites at which ER is held in close apposition (10–30 nm) to PM represent critical platforms for mediating Ca2+ homeostasis and signaling events and as ER and PM lipid metabolism and transport hubs (Dickson, 2017; Balla, 2018). ER–PM junctions are typically classified based on the resident ER protein serving as the PM tether as are members of the extended synaptotagmin or E-Syt (Saheki and De Camilli, 2017b), junctophilin or JP (Takeshima et al., 2015), or stromal interacting molecule or STIM (Prakriya and Lewis, 2015) families. These otherwise unrelated ER membrane proteins have a common membrane topology with a large cytoplasmic domain that mediates binding to specific classes of phospholipids in the inner leaflet of the PM (Carrasco and Meyer, 2011; Henne et al., 2015). The STIM proteins can also reversibly bind to PM Orai proteins in a process triggered by ER Ca2+ depletion (Prakriya and Lewis, 2015). While mRNA measurements have shown that many of these ER-localized tethering proteins have high levels of expression in the brain (e.g., Nishi et al., 2003; Min et al., 2007; Moccia et al., 2015; Takeshima et al., 2015), little is known of the cellular or subcellular localization of the corresponding proteins relative to the different classes of abundant ER–PM junctions observed in ultrastructural studies of brain neurons (Rosenbluth, 1962; Henkart et al., 1976; Wu et al., 2017).
Plasma membrane voltage-gated K+ or Kv channels play crucial and diverse roles in shaping neuronal function (Trimmer, 2015). Among these, the Kv2 family contains two members: Kv2.1 and Kv2.2. Like other Kv channels, Kv2.1 and Kv2.2 are key determinants of action potential characteristics and intrinsic electrical excitability in many types of mammalian brain neurons (Du et al., 2000; Malin and Nerbonne, 2002; Guan et al., 2007; Johnston et al., 2008; Liu and Bean, 2014; Pathak et al., 2016; Honigsperger et al., 2017; Palacio et al., 2017), and de novo mutations in Kv2.1 are associated with devastating neonatal encephalopathic epilepsies and neurodevelopmental delays (Torkamani et al., 2014; Saitsu et al., 2015; Thiffault et al., 2015; de Kovel et al., 2016, 2017). Kv2 channels are also prominently expressed in pancreatic islets (Jacobson et al., 2007; Li et al., 2013), smooth muscle cells (Patel et al., 1997; Schmalz et al., 1998), and other excitable and nonexcitable cell types. In brain neurons, Kv2 channels are distinct from other Kv channels (Trimmer, 2015) in being specifically localized to high-density micrometer-sized clusters prominent on the soma, proximal dendrites, and axon initial segment (Trimmer, 1991; Scannevin et al., 1996; Du et al., 1998; Murakoshi and Trimmer, 1999; Lim et al., 2000; Kihira et al., 2010; King et al., 2014; Mandikian et al., 2014; Bishop et al., 2015, 2018). Kv2 channels also form such clusters when exogenously expressed in cultured neurons and in heterologous cells (Scannevin et al., 1996; Lim et al., 2000; O’Connell and Tamkun, 2005; Mohapatra and Trimmer, 2006; O’Connell et al., 2006; Tamkun et al., 2007; Kihira et al., 2010; Bishop et al., 2015, 2018; Cobb et al., 2015). A short proximal restriction and clustering (PRC) domain within the relatively large cytoplasmic C-terminus of Kv2.1 is necessary for its clustered localization in neurons and heterologous cells (Scannevin et al., 1996; Lim et al., 2000) and is sufficient to transfer Kv2.1-like clustering to other Kv channels (Lim et al., 2000; Mohapatra and Trimmer, 2006). A point mutation within the highly conserved PRC motif also results in loss of Kv2.2 clustering (Bishop et al., 2015, 2018).
Immunoelectron microscopy-based studies have shown that immunoreactivity for PM Kv2.1 (Du et al., 1998; Mandikian et al., 2014; Bishop et al., 2018) and Kv2.2 (Bishop et al., 2015) is associated with subsurface cisternae, a class of ER–PM junctions that are prominent in somata of brain neurons (Rosenbluth, 1962; Henkart et al., 1976; Wu et al., 2017). In certain brain neurons, clusters of PM Kv2.1 channels overlie clusters of ER-localized ryanodine receptor (RyR) Ca2+ release channels (Antonucci et al., 2001; Misonou et al., 2005b; King et al., 2014; Mandikian et al., 2014), which are concentrated at ER–PM junctions to mediate local Ca2+ signaling events in diverse cell types (Franzini-Armstrong and Jorgensen, 1994; Sun et al., 1995). Recent studies reveal that in addition to being localized to ER–PM junctions, exogenous expression of Kv2.1 leads to recruitment and/or stabilization of ER–PM junctions in heterologous cells and cultured hippocampal neurons or CHNs (Fox et al., 2015). The ability of Kv2.1 to remodel ER–PM junctions exhibits the same phosphorylation-dependent regulation as Kv2.1 clustering (Cobb et al., 2015), which itself is regulated by numerous stimuli that impact Kv2.1 phosphorylation state (Misonou et al., 2004, 2005a; Cerda and Trimmer, 2011; Bishop et al., 2015, 2018). It is not currently known whether the remodeling of ER–PM junctions seen upon heterologous expression of Kv2.1 is a result of the impact of the expressed channel’s K+ conductance on the membrane potential and/or cellular ion homeostasis in the expressing cell (e.g., Felipe et al., 1993; Hegle et al., 2006; Jimenez-Perez et al., 2016), established mechanisms for remodeling ER–PM junctions (Carrasco and Meyer, 2011; Henne et al., 2015; Saheki and De Camilli, 2017b; Balla, 2018). Alternatively, Kv2.1 could remodel ER–PM junctions through a more direct structural role. Despite having a conserved clustered localization and a highly conserved PRC domain, Kv2.1 and Kv2.2 share only 61% overall amino acid (aa) identity, which drops to only 39% in their respective cytoplasmic C-termini that compose about half of their primary structure. Kv2.1 and Kv2.2 have distinct biophysical properties (e.g., Dong et al., 2013; Baver et al., 2014) and expression patterns (e.g., Hwang et al., 1992, 1993a,b; Johnston et al., 2008; Li et al., 2013; Bishop et al., 2015, 2018). Moreover, stimuli that trigger reversible modulation of voltage activation (e.g., Baver et al., 2014; Mandikian et al., 2014; Bishop et al., 2015) and dispersal of clustering (Bishop et al., 2015) of Kv2.1 do not have a detectable impact on Kv2.2 (Bishop et al., 2015), leading to questions as to whether Kv2.2 is also distinct from Kv2.1 in its ability to remodel ER–PM junctions. Last, it is not known how ablating expression of endogenous Kv2 channels impacts ER–-PM junctions in brain neurons in situ.
Here, we define the localization of Kv2.2 relative to ER–PM junctions in brain neurons in situ and in culture and determine whether, like Kv2.1, it also functions to remodel ER–PM junctions. We define the relationship of Kv2-containing ER–PM junctions to the actin cytoskeleton and to other classes of molecularly defined ER–PM junctions. We employ a strategic set of point mutations in Kv2.2 and Kv2.1 to dissect the respective contributions of K+ conduction and clustering to the Kv2-mediated remolding of ER–PM junctions, and also determine the domain necessary and sufficient for this function that among PM proteins is unique to Kv2.1. Finally, we use recently generated double-knockout mice lacking expression of both mammalian Kv2 channel family members to determine their in vivo role in regulating ER–PM junctions in brain neurons in situ. Our results provide compelling evidence for a conserved and noncanonical role for nonconducting Kv2 channels in impacting ER–PM junctions in brain neurons and other cell types in which these ion channels are abundantly expressed.
RESULTS
Plasma membrane clusters of Kv2.2 associate with ER–PM junctions in mammalian brain neurons in situ and in culture and in heterologous HEK293T cells
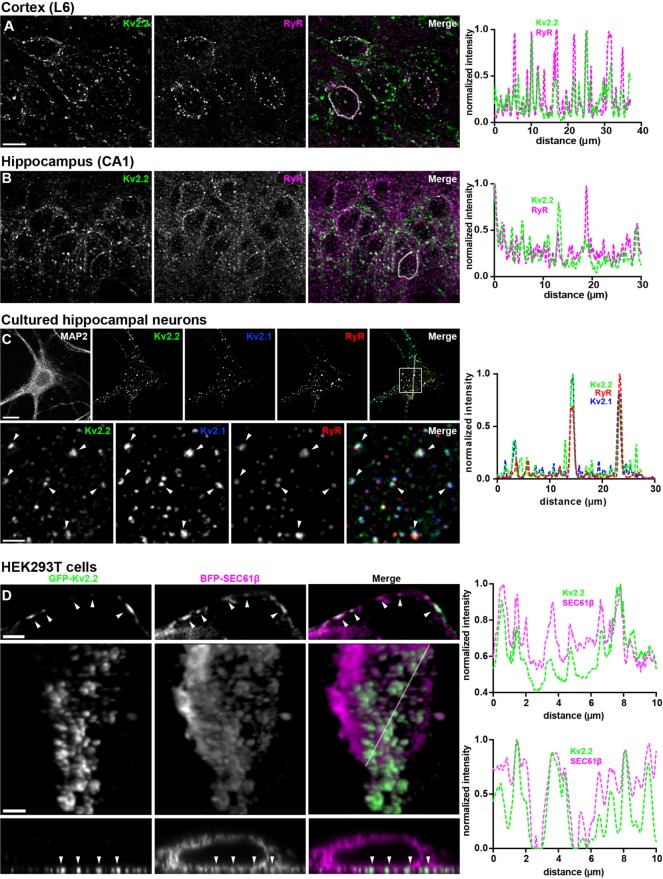
Kv2.2 is present in clusters on the somata, proximal dendrites, and axon initial segments of mammalian brain neurons (Johnston et al., 2008; Kihira et al., 2010; Bishop et al., 2015, 2018). To investigate the subcellular localization of these Kv2.2 clusters relative to native ER–PM junctions in brain neurons, we performed multiplex immunofluorescence labeling for PM Kv2.2 and ER-localized RyR Ca2+ release channels, which are concentrated at ER–PM junctions in many cell types, including certain types of brain neurons. In mouse brain sections, somatic Kv2.2 clusters were found at/near RyR clusters in specific neuron types, including hippocampal CA1 pyramidal neurons and layer 6 neocortical neurons (Figure 1, A and B). A similar juxtaposition of Kv2.2 and RyR clusters was seen in cultured hippocampal neurons (CHNs; Figure 1C). In these neurons, Kv2.2 was often found coclustered with Kv2.1 at ER–PM junctions (Figure 1C). Neurons in each preparation also contained RyR clusters that did not appear to colocalize with Kv2.2 or Kv2.1, suggesting the presence of other classes of ER–PM junctions in these cells (Figure 1, A–C). These findings demonstrate that Kv2.2 clusters localize to RyR-containing ER–PM junctions in intact mammalian brain neurons in situ and in culture.
FIGURE 1:
Kv2.2 associates with ER–PM junctions in mammalian brain neurons in situ and in culture and in heterologously expressing HEK293T cells. (A–C) Single z-stack images of multiplex immunofluorescence labeling of adult mouse brain sections showing neocortex (A) and hippocampal CA1 region (B) immunolabeled for Kv2.2 (green) and RyR (magenta), or cultured hippocampal neurons (C) immunolabeled for Kv2.2 (green), Kv2.1 (blue), and RyR (red), as indicated. Scale bar in Kv2.2 neocortex panel is 10 µm and holds for all brain panels. Scale bar in MAP2 CHN panel is 10 µm and holds for all CHN panels in that row. Scale bar in Kv2.2 magnified inset is 2.5 µm and holds for all panels in that row. Panels to the right of each set of images are the corresponding normalized fluorescence intensity values across the individual line scans depicted by the white line in the merged images. Image exposure time for brain sections was optimized for the labeling of each brain region independently. (D) Images of fixed HEK293T cells coexpressing GFP-Kv2.2 (green) and BFP-SEC61β (magenta). The top row shows a single N-SIM optical section taken through the center of a cell. The scale bar is 1.25 µm. The bottom rows show a 2D projection of a 3D reconstruction (middle row), and a single orthogonal slice through the 3D reconstruction (bottom row) of a cell imaged with a Zeiss Airyscan microscope. Scale bar in the GFP-Kv2.2 panel of the 3D reconstruction is 2.5 µm and holds for all panels in the bottom two rows. Panels to the right of each set of rows are the corresponding normalized fluorescence intensity values across the individual line scans depicted by the arrows (top) or white line (bottom) in the merged images.
We next determined whether heterologously expressed and clustered Kv2.2 localizes to ER–PM junctions. In HEK293T cells coexpressing GFP-tagged Kv2.2 and BFP-tagged SEC61β (a general ER marker; Zurek et al., 2011), optical sections taken through the centers of cells show fingerlike projections of SEC61β-positive ER, a subset of which were associated with PM Kv2.2 clusters that appear as discrete PM segments (Figure 1D). Three-dimensional reconstructions show that the ER projections terminating at Kv2.2-associated PM clusters were contiguous with bulk ER (Figure 1D; Supplemental Movie 1). Together, these results suggest that Kv2.2 localizes to ER–PM junctions in mammalian brain neurons and when heterologously expressed in HEK293T cells.
Movie S1.
Rotating 3D reconstruction of a fixed HEK293T cell expressing GFP-Kv2.2 (left panel, green) and BFP-SEC61β (middle panel, magenta). Merged image is shown in right panel.
Exogenous Kv2.2 expression remodels ER–PM junctions in cultured rat hippocampal neurons and heterologous cells
We next determined the impact of exogenous expression of recombinant Kv2.2 on ER–PM junctions in mammalian neurons and heterologous cells. We used total internal reflection fluorescence (TIRF) microscopy of living cells to selectively visualize fluorescence signals within ≈100 nm of the coverslip, including near-PM ER (i.e., ER at ER–PM junctions). In HEK293T cells expressing the fluorescent luminal ER marker DsRed2-ER5 (a general ER marker; Day and Davidson, 2009), the near-PM ER appeared as a highly ramified system of small reticular tubules and puncta (Figure 2A), the latter representing focal structures of cortical ER coincident with the PM or ER–PM junctions (Fox et al., 2015; Chang et al., 2017; Besprozvannaya et al., 2018). Exogenous expression of GFP-Kv2.2 led to a remodeling of the DsRed2-ER5–positive cortical ER to form larger foci that colocalized with the PM clusters of Kv2.2 (Figure 2B). Cells coexpressing GFP-Kv2.2 exhibited a significant increase in both the size of ER–PM junctions (Figure 2C) and the percentage of basal cell surface area with associated cortical ER (Figure 2D). No such changes were seen in cells expressing a related but distinct Kv channel, Kv1.4 (Figure 2, B–D), which is not localized to ER–PM junctions in neurons (Trimmer, 2015). Analysis of colocalization using Pearson’s correlation coefficient (PCC) revealed that DsRed2-ER5 was significantly more colocalized with Kv2.2 than it was with Kv1.4 (Figure 2E). We also found a nearly linear relationship between the sizes of Kv2.2 clusters and ER–PM junctions (Figure 2H, red points). As previously reported (Fox et al., 2015), significant increases in ER–PM junction size and ER-associated PM surface area were also observed in cells expressing Kv2.1 (Figure 2, B–E). Taken together, these data demonstrate that Kv2.2 can remodel ER–PM junctions and that this is a conserved function of Kv2 channels not shared with Kv1.4.
FIGURE 2:
Exogenous Kv2 expression remodels ER–PM junctions in HEK293T cells and cultured rat hippocampal neurons. (A, B) TIRF images of live HEK293T cells expressing DsRed2-ER5 (magenta) either alone (A) or coexpressed with (in green) GFP-Kv2.2, GFP-Kv2.1, or GFP-Kv1.4, as indicated (B). Scale bar in A is 5 µm and holds for all panels. (C) Graph of mean ER–PM junction (EPJ) size per cell measured from HEK293T cells coexpressing DsRed2-ER5 and GFP-Kv2.2, GFP-Kv2.1, or GFP-Kv1.4 or expressing DsRed2-ER5 alone (control). (D) Graph of percentage of the PM area per cell occupied by cortical ER measured from HEK293T cells coexpressing DsRed2-ER5 and GFP-Kv2.2, GFP-Kv2.1, or GFP-Kv1.4 or expressing DsRed2-ER5 alone (control). (E) Graph of PCC between DsRed2-ER5 and GFP-Kv2.2, GFP-Kv2.1, or GFP-Kv1.4 measured from HEK293T cells coexpressing DsRed2-ER5 and GFP-Kv constructs. The dashed line denotes a PCC of 0.5. Bars on all graphs are mean ± SD. See Supplemental Tables 1–3 for values and statistical analyses for C–E. (F) TIRF image of a live CHN expressing DsRed2-ER5 (magenta) alone. (G) TIRF image of a live CHN coexpressing DsRed2-ER5 (magenta) and GFP-Kv2.2 (green). Scale bar in DsRed2-ER5 panel is 10 µm and holds for all panels in that row. Magnified images are shown in bottom row. Scale bar in DsRed2-ER5 magnified inset panel is 2.5 µm and holds for all panels in that row. (H) Scatterplot shows sizes of Kv2.2 clusters and associated ER–PM junctions (EPJs, as indicated by DsRed2-ER5 in TIRF) in HEK293T cells (red points) and CHNs (black points). n = 3 cells each.
We next expressed DsRed2-ER5 alone or coexpressed DsRed2-ER5 with GFP-Kv2.2 in CHNs. TIRF imaging experiments revealed that GFP-Kv2.2 expression also remodeled neuronal ER–PM junctions (Figure 2, F and G). Similarly to HEK293T cells, we found a nearly linear relationship between the sizes of GFP-Kv2.2 clusters and ER–PM junctions in CHNs (Figure 2H, black points). These results demonstrate that exogenous expression of Kv2.2 in both HEK293T cells and CHNs is sufficient to remodel ER–PM junctions.
Kv2.2 channels associated with ER–PM junctions are on the cell surface
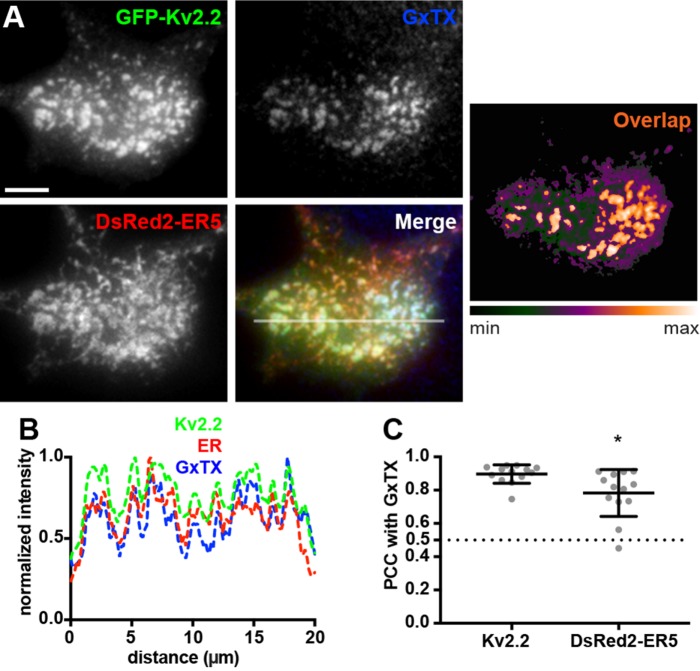
Given the extensive colocalization of Kv2.2 and these ER markers at ER–PM junctions, we next verified that the Kv2.2 present at these sites was in the PM by performing live-cell labeling with the membrane-impermeant and Kv2-specific tarantula toxin Guangxitoxin-1E or GxTX (Herrington et al., 2006). We used fluorescent GxTX conjugated to DyLight633 or GxTX-633 (Tilley et al., 2014) to label cell surface Kv2.2. We first validated this approach by coexpressing BFP-SEC61β with SEP-Kv2.1, a construct of Kv2.1 tagged with cytoplasmic mCherry and an extracellular pHluorin as a reporter of cell surface Kv2.1 (Jensen et al., 2017). We observed extensive colocalization of GxTX-633 and pHluorin signals (Supplemental Figure 1), showing that GxTX-633 is a reliable reporter for cell surface Kv2 channels. No detectable GxTX-633 labeling was observed in control HEK293T cells, or those expressing DsRed2-ER5 alone (unpublished data). GxTX-633 labeling of cells coexpressing GFP-Kv2.2 and DsRed2-ER5 showed a high degree of colocalization of all three signals (Figure 3, A and B). As expected, PCC measurements were slightly but significantly higher for the signals for GxTX-633 directly bound to GFP-Kv2.2 than for the GxTX-633 labeling indirectly associated with the DsRed2-ER5 present at Kv2.2-associated ER–PM junctions (Figure 3C). Taken together, these data demonstrate that the clusters of Kv2.2 associated with ER–PM junctions are on the cell surface.
FIGURE 3:
ER–PM junction-localized Kv2.2 channels are expressed on the cell surface. (A) TIRF images of a live HEK293T cell expressing GFP-Kv2.2 (green) and DsRed2-ER5 (red) and surface-labeled for Kv2 channels with GxTX-633 (blue). Heat map shows overlap of GFP-Kv2.2 and GxTX-633 pixels. Scale bar is 5 µm. (B) Fluorescence intensity values across the individual line scan depicted by the white line in the merged image. (C) Graph of PCC between GxTX and Kv2.2 or DsRed2-ER5 measured from live HEK293T cells surface labeled with GxTX-633 and coexpressing GFP-tagged Kv2.2 and DsRed2-ER5. Bars are mean ± SD. See Supplemental Table 5 for values and statistical analyses for C.
Kv2.2-containing ER–PM junctions are present at sites depleted in components of the cortical actin cytoskeleton
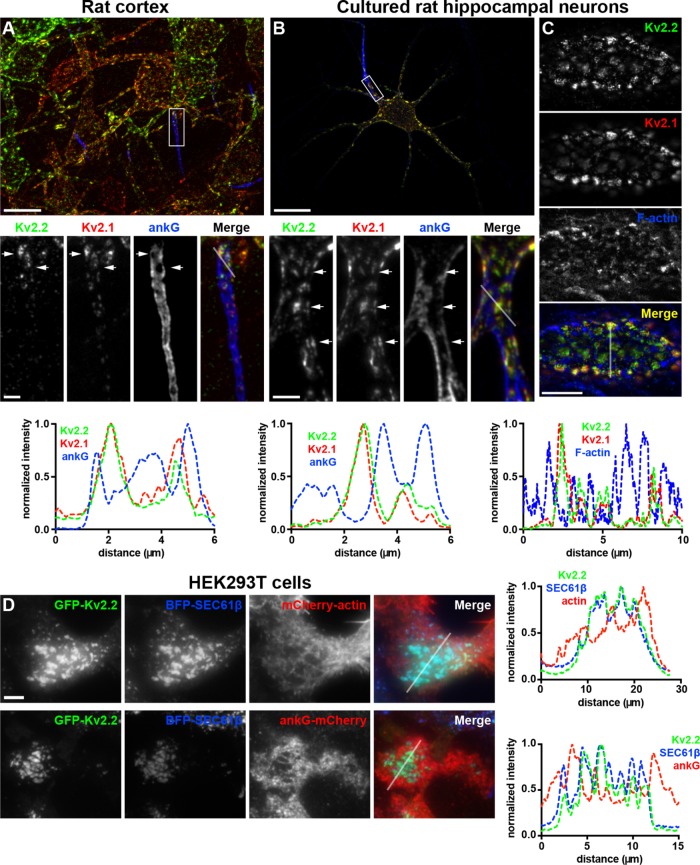
We next determined the relationship of Kv2.2-containing ER–PM junctions to the actin cytoskeleton. In addition to its prominent clustering on the soma and proximal dendrites, as shown in Figure 1, Kv2.2 is also localized to the axon initial segment or AIS (Johnston et al., 2008; Sanchez-Ponce et al., 2012), a subcellular compartment highly enriched in components of the actin cortical cytoskeleton, including a specialized complex of spectrins and ankyrins (Leterrier, 2016). We immunolabeled brain sections for Kv2.2 and ankyrin G (ankG), which is highly expressed at the AIS. We found that in neocortical layer 5 pyramidal neurons, Kv2.2 was present in robust clusters located on the AIS, as marked by ankG (Figure 4A). However, within the AIS, Kv2.2 clusters were present at sites deficient in ankG (Figure 4A). These ankG-deficient sites or “holes” represent locations at which the ER present in the AIS, termed the cisternal organelle, comes into close apposition to the PM (Sanchez-Ponce et al., 2011; King et al., 2014; Schluter et al., 2017).
FIGURE 4:
Kv2-mediated ER–PM junctions are located at sites depleted in components of the cortical actin cytoskeleton. (A) Mouse brain section immunolabeled for Kv2.2 (green), Kv2.1 (red), and ankG (blue). Scale bar for large image is 20 µm and for Kv2.2 inset is 3 µm and holds for all inset panels. (B) Projected z-stack of optical sections taken from a CHN immunolabeled for Kv2.2 (green), Kv2.1 (red), and ankG (blue). Scale bar for large image is 20 µm and for Kv2.2 inset is 3 µm and holds for all inset panels. (C) Single optical section taken from the cell body of a CHN immunolabeled for Kv2.2 (green) and Kv2.1 (red) and labeled for F-actin with phalloidin (blue). Scale bar for merged panel is 10 µm and holds for all panels in set. Panels below each set of images show the corresponding normalized fluorescence intensity values across the line scans indicated in the merged images in that column. (D) TIRF images of live HEK293T cells coexpressing GFP-Kv2.2 (green) and BFP-SEC61β (blue) in conjunction with (in red) mCherry-actin (top row) or ankG-mCherry (bottom row). Scale bar for GFP-Kv2.2 panel in top row is 5 µm and holds for all panels in set. Panels to the right of each row show the corresponding normalized fluorescence intensity values across the line scan depicted by the white line in the merged images.
We next immunolabeled for endogenous Kv2.2 and ankG in CHNs and found a similar relationship between the sites of Kv2.2 clustering on the AIS and regions deficient in both ankG and filamentous actin, the latter labeled with fluorescent phalloidin (Figure 4, B and C). This is apparent in line scan analyses, which revealed that the intensity profiles of the Kv2 immunolabeling and actin labeling were often negatively correlated (Figure 4). To determine whether this spatial relationship is also present in nonneuronal cells, we performed TIRF imaging on live HEK293T cells coexpressing GFP-Kv2.2, BFP-SEC61β, and mCherry-tagged actin. We found that GFP-Kv2.2 clusters and associated ER–PM junctions displayed a negatively correlated distribution with respect to cortical mCherry-actin (Figure 4D). We next coexpressed ankG-mCherry with BFP-SEC61β and Kv2.2 and found, similarly to what was observed on the AIS, a negatively correlated distribution of Kv2.2 clusters and their associated ER–PM junctions with ankG (Figure 4D), showing that the distinct localization of the endogenous proteins seen on the AIS of brain neurons could be recapitulated upon exogenous expression of these proteins in heterologous cells.
The actin cytoskeleton regulates the spatial organization of Kv2.2 clusters and associated ER–PM junctions
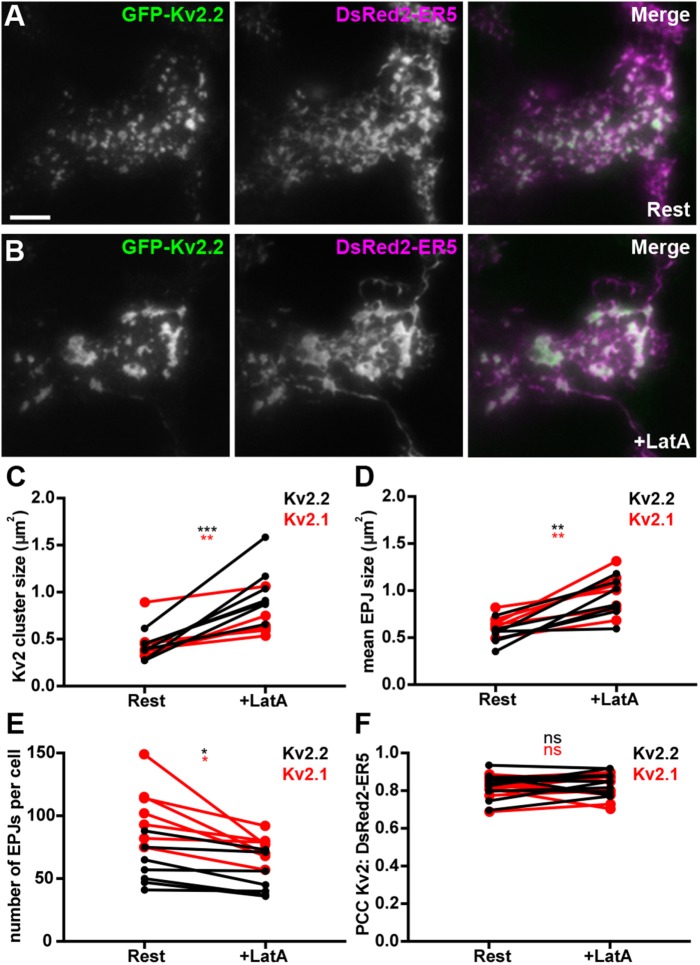
Given the distinct spatial relationship between Kv2.2-associated ER–PM junctions and the cortical actin cytoskeleton, we next determined the impact of disrupting the organization of the actin cytoskeleton on Kv2.2-mediated ER–PM junctions. We treated cells expressing Kv2.2 with latrunculin A (LatA), which disrupts the organization of filamentous actin (Spector et al., 1983). We found that LatA treatment led to a parallel reorganization of both Kv2.2 clusters and their associated ER–PM junctions (Figure 5, A and B; Supplemental Movie 2). This resulted in parallel significant increases in the sizes of both Kv2.2 clusters and ER–PM junctions (Figure 5, C and D), the latter reported by the DsRed2-ER5 signal coincident with the PM. The total number of ER–PM junctions in Kv2.2-expressing cells was also significantly reduced in response to LatA treatment (Figure 5E). Similar results were obtained upon LatA treatment of cells coexpressing GFP-Kv2.1 and DsRed2-ER5 (Figure 5, C–E), as suggested by a previous study (Fox et al., 2015). These changes were not observed in untreated cells over the course of 15 min (unpublished data). While LatA treatment significantly altered the spatial characteristics of Kv2.2 clusters and their associated ER–PM junctions, the extent of colocalization between GFP-Kv2.2 and DsRed2-ER5 was not significantly altered upon LatA treatment (Figure 5F). Similar results were obtained for Kv2.1 (Figure 5F). These results show that while LatA induced an apparent fusion of Kv2 clusters and associated ER–PM junctions, resulting in fewer, larger structures, these changes occurred in parallel and did not affect their association per se. These results also suggest that the distinct and mutually exclusive localization of Kv2.2 clusters and components of the cortical actin cytoskeleton seen in brain neurons likely contributes in the organization and maintenance of these clusters and their associated ER–PM junctions.
FIGURE 5:
Disrupting the actin cytoskeleton impacts spatial organization of Kv2.2-mediated ER–PM junctions. (A, B) TIRF images of a live HEK293T cell coexpressing GFP-Kv2.2 (green) and DsRed2-ER5 (magenta), prior to (Rest) and 15 min after latrunculin A (After LatA) treatment. Scale bar in GFP-Kv2.2 Rest panel is 5 µm and holds for all panels. Graphs show values measured from cells before (Rest) and after a 15-min treatment (After LatA) with 10 µM LatA. (C) Mean Kv2.2 cluster size per cell. (D) Mean ER–PM junction (EPJ) size per cell. (E) Number of ER–PM junctions per cell. (F) PCCs between Kv2.2 and DsRed2-ER5. See Supplemental Tables 6–9 for values and statistical analyses for C–F.
Movie S2.
Time lapse movie (15 minutes compressed into 10 seconds) of TIRF imaging of the effects of 10 μM LatA treatment of a live HEK293T cell expressing GFP-Kv2.2 (left panel, green) and DsRed2-ER5 (middle panel, red). Merged image is shown in the right panel.
Kv2.2-containing ER–PM junctions associate with ER–PM junctions formed by the known classes of ER–PM tethers
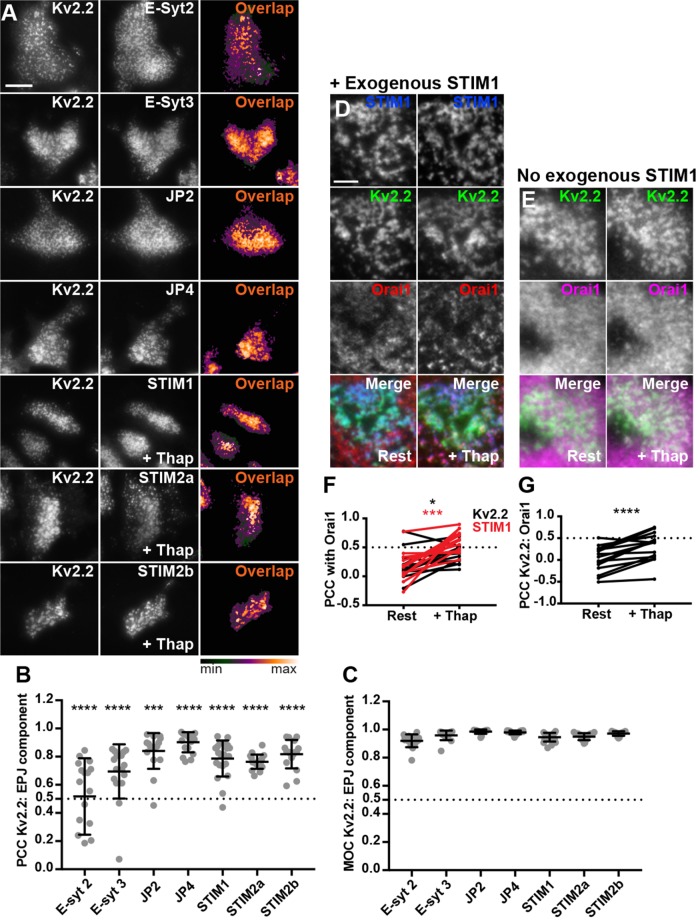
We next determined the relationship of Kv2.2 clusters and associated ER–PM junctions with those formed upon exogenous expression of the three other families of mammalian ER-localized ER–PM tethers. We coexpressed FP-tagged Kv2.2 and individual members of the E-Syt, JP, and STIM families in HEK293T cells. In cells coexpressing the STIMs, we also induced Ca2+ store depletion via treatment with 2 μM thapsigargin for 5 min. In all cases, we observed a high degree of colocalization between clusters of Kv2.2 and these ER–PM junction tethers (Figure 6A), as demonstrated by high values of PCC and Mander’s overlap coefficient (MOC) (Figure 6, B and C). In cells coexpressing STIM1, Kv2.2, and Orai1, store depletion resulted in a significant increase in colocalization not only of STIM1 and Orai1, but also of Orai1 and Kv2.2 (Figure 6, D and F). The store depletion–induced increase in colocalization of Kv2.2 and Orai1 also occurred in the absence of STIM1 coexpression (Figure 6, E and G), presumably due to endogenous STIM expression in HEK293T cells (Williams et al., 2001; Soboloff et al., 2006; Brandman et al., 2007; Shalygin et al., 2015). Together, these results show that Kv2.2 clusters can associate with ER–PM junctions formed by the three established families of ER–PM junction tethers. Interestingly, the PCCs between Kv2.2 and these ER tethers were significantly lower than the corresponding MOC values obtained from the same cells (Figure 6, B and C), suggesting that despite the extensive overlap in signal between Kv2.2 clusters and these established classes of ER–PM junctions, there are distinctions between them in their fine spatial organization.
FIGURE 6:
Kv2.2-containing ER–PM junctions colocalize with multiple components of mammalian ER–PM junctions. (A) TIRF images of live HEK293T cells coexpressing DsRed- or GFP-tagged Kv2.2 and various members of the E-Syt, JP, and STIM families of ER-localized PM tethers. Scale bar in top left GFP-Kv2.2 panel is 10 µm and holds for all panels. Heat maps show pixel overlap of Kv2.2 and ER–PM tether signals. STIM-containing cells were treated with 2 µM thapsigargin for 5 min prior to imaging. (B) Graph of PCC between Kv2.2 and ER–PM tethers. Bars are mean ± SD. (C) Graph of MOC values between Kv2.2 and ER–PM tether signals. Bars are mean ± SD. See Supplemental Table 10 for values and statistical analyses for B and C. (D) TIRF images of a live HEK293T cell coexpressing BFP-STIM1 (blue), DsRed-Kv2.2 (green), and GFP-Orai1 (red) prior to (Rest) and immediately after 5 min of treatment with 2 µM thapsigargin (+Thap). (E) TIRF images of a live HEK293T cell coexpressing DsRed-Kv2.2 (green) and GFP-Orai1 (magenta) prior to (Rest) and immediately after 5 min of treatment with 2 µM thapsigargin (+Thap). Scale bar in top left DsRed-Kv2.2 panel is 5 µm and holds for panels in D, E. (F) Graph of PCC between Orai1 and Kv2.2 (black) or STIM1 (red) measured from cells with BFP-STIM1 coexpression before (Rest) and after (+Thap) thapsigargin treatment. (G) Graph of PCC between Orai1 and Kv2.2 measured from cells without BFP-STIM1 coexpression before (Rest) and after (+Thap) thapsigargin treatment. Bars on all graphs are mean ± SD. See Supplemental Table 11 for values and statistical analyses for F and G.
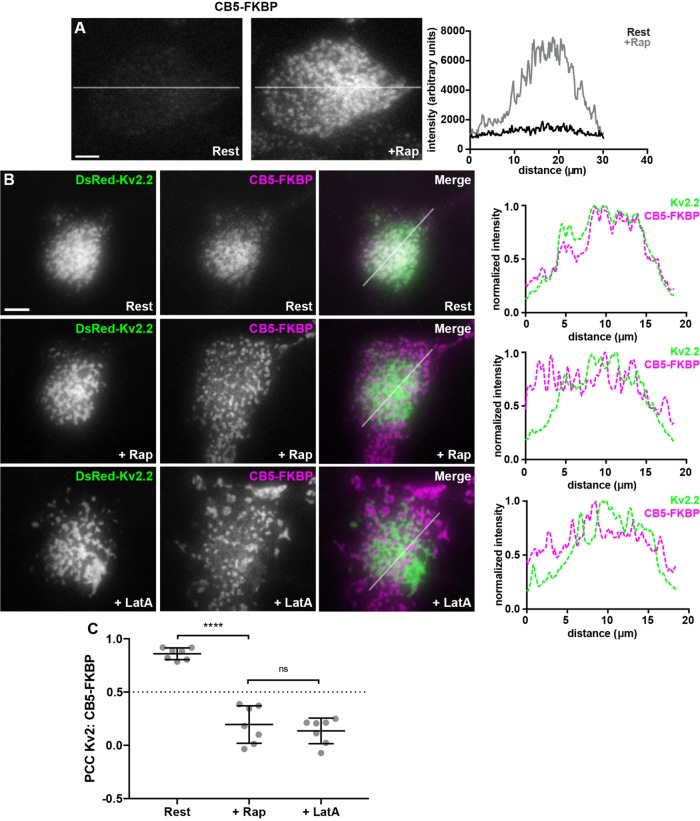
We next examined the relationship of Kv2-mediated ER–PM junctions to those formed acutely by a rapamycin-inducible system (Inoue et al., 2005; Dickson et al., 2016) employing ER-localized CB5-FKBP-CFP and PM-localized Lyn11-FRB (CB5/Lyn11). TIRF imaging reveals that acute treatment of HEK293T cells coexpressing CB5/Lyn11 with 5 μM rapamycin yields robust recruitment of ER to the cell cortex (Figure 7A). HEK293T cells coexpressing Kv2.2 and CB5/Lyn11 prior to rapamycin addition exhibited CB5-FKBP-CFP fluorescence similar to that of other ER reporters (e.g., BFP-SEC61β, DsRed2-ER5) in occurring throughout the ER, and also colocalized with clustered Kv2.2 at ER–PM junctions, the latter yielding a high degree of colocalization in TIRF imaging (Figure 7B). Surprisingly, unlike the other known classes of ER–PM junctions, the rapamycin-induced CB5/Lyn11 ER–PM junctions were largely distinct from and nonoverlapping with those associated with the Kv2.2 clusters (Figure 7B), as shown by the significant decrease in PCC upon rapamycin treatment (Figure 7C). Subsequent LatA treatment impacted the spatial organization of both the Kv2.2- and CB5/Lyn11-mediated ER–PM junctions (Figure 7B). However, they remained spatially segregated, so that there were no significant LatA-induced changes in PCC between Kv2.2 and CB5 (Figure 7C). These results, taken together, demonstrate that in heterologous cells Kv2-associated ER–PM junctions broadly overlap with those formed by the known classes of native ER–PM junction tethers, the exception being the unique artificial ER–PM junctions formed by the rapamycin-induced association of CB5 and Lyn1. Moreover, while the actin cytoskeleton plays a role in defining the spatial boundaries of both Kv2.2- and CB5/Lyn11-mediated ER–PM junctions, disrupting the actin cytoskeleton is not sufficient to homogenize these distinct membrane contact sites.
FIGURE 7:
Acutely formed ER–PM junctions are distinct from Kv2.2-containing ER–PM junctions. (A) TIRF images of CFP fluorescence in a live HEK293T cell coexpressing CFP-CB5-FKBP and Lyn11-FRB before (Rest) and immediately after treatment with 5 µM rapamycin (+Rap). Scale bar is 5 µm and holds for both panels. Graph to right of panels shows fluorescence intensity of CFP-CB5-FKBP across the individual line scan depicted by the white lines before (Rest) and immediately following treatment with 5 µM rapamycin (+Rap). (B) TIRF images of a live HEK293T cell coexpressing DsRed-Kv2.2 (green), CFP-CB5-FKBP (magenta), and Lyn11-FRB. Top row: Prior to rapamycin treatment (Rest). Middle row: Same cell immediately following 5 µM rapamycin treatment (+Rap). Bottom row: Same cell after subsequent 15-min treatment with 10 µM LatA (+LatA). Panels to the right of each row are the corresponding normalized fluorescence intensity values across the individual line scans depicted by the white line in the merged images. Scale bar is 10 µm and holds for all panels. (C) Graph of PCC between DsRed-Kv2.2 and CFP-CB5-FKBP. Bars are mean ± SD. See Supplemental Table 12 for values and statistical analysis.
Remodeling ER–PM junctions is a nonconducting function of Kv2 channels
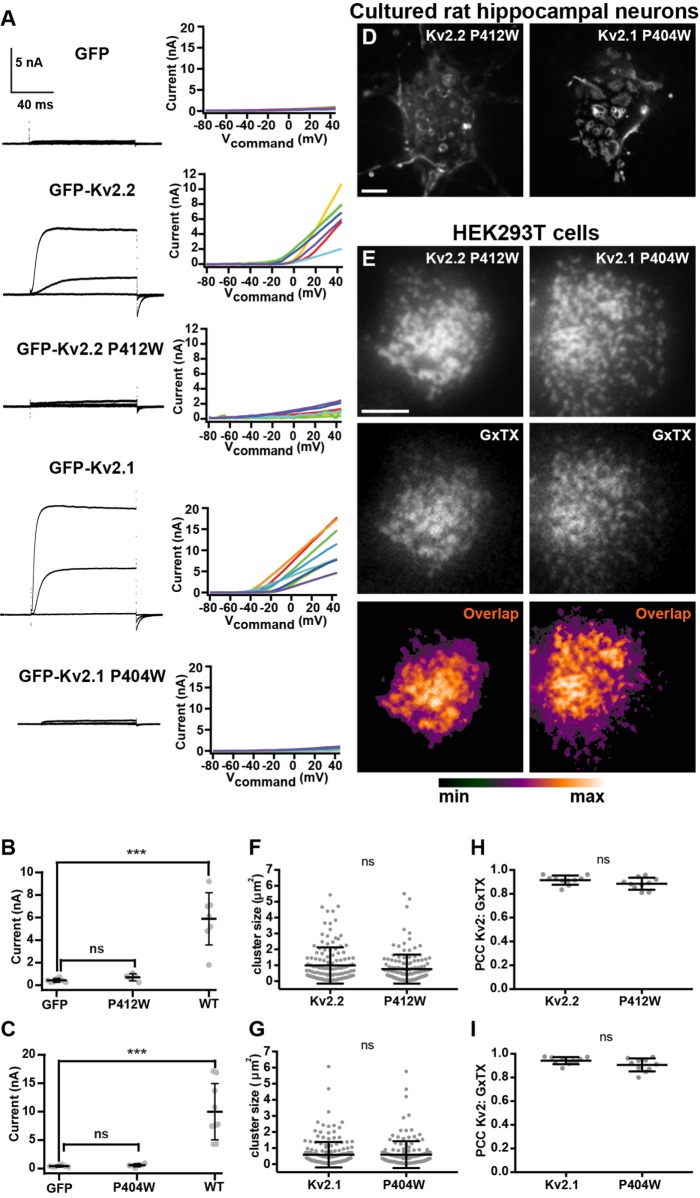
We next addressed whether the Kv2.2-mediated remodeling of ER–PM junctions is dependent on K+ flux through the channels. We generated a point mutation (P412W) in the S6 transmembrane helix of Kv2.2, a position analogous to a point mutation (P404W) previously shown to eliminate conductance through Kv2.1 channels heterologously expressed in Xenopus oocytes (Lee et al., 2003). We first expressed GFP-Kv2.2 P412W in HEK293T cells and evaluated conductance relative to wild-type GFP-Kv2.2 using voltage-clamp electrophysiology. HEK293T cells expressing GFP-Kv2 channels or GFP alone as a control were whole-cell patch clamped and held at a resting membrane potential of –80 mV. In response to positive voltage steps, delayed rectifier outward currents emerged from cells expressing GFP-Kv2.2, but not from cells expressing either GFP-Kv2.2 P412W or GFP (Figure 8, A and B). As expected from previous analyses in oocytes, GFP-Kv2.1 P404W was also nonconducting when expressed in HEK293T cells (Figure 8, A and C).
FIGURE 8:
Mutations that eliminate K+ conductance do not impact Kv2.2 channel clustering. (A) Exemplar whole-cell voltage clamp recordings (left) and corresponding graphs of current levels vs. command voltage (right) of HEK293T cells expressing GFP (control), GFP-Kv2.2, GFP-Kv2.2 P412W, GFP-Kv2.1, or GFP-Kv2.1 P404W. Recordings shown are representative responses to 100-ms steps from –100 mV to –40, 0, and +40 mV. Note the lack of outward currents in control, GFP-Kv2.2 P412W, and GFP-Kv2.1 P404W recordings. (B, C) Summary graphs showing whole cell current at +40 mV for cells expressing Kv2.2 (B) or Kv2.1 (C) isoforms. See Supplemental Table 13 for values and statistical analyses for B and C. (D) Deconvolved widefield image of a live CHN expressing GFP-Kv2.2 P412W or GFP-Kv2.1 P404W. Scale bar is 5 µm and holds for both panels. (E) TIRF images of live HEK293T cells expressing GFP-Kv2.2 P412W or GFP-Kv2.1 P404W and surface labeled with GxTX-633. Scale bar in the Kv2.2 P412W panel is 5 µm and hold for all panels. (F, G) Graphs of mean cluster size per cell measured from CHNs expressing GFP-Kv2.2 or GFP-Kv2.2 P412W (F) or GFP-Kv2.1 or GFP-Kv2.1 P404W (G). Bars are mean ± SD. See Supplemental Table 14 for values and statistical analyses for F and G. (H, I) Graph of PCC between Kv2 and GxTX from HEK293T cells expressing GFP-Kv2.2 or GFP-Kv2.2 P412W (H) or GFP-Kv2.1 or GFP-Kv2.1 P404W (I). Bars are mean ± SD. See Supplemental Table 15 for values and statistical analyses for H and I.
We next expressed GFP-Kv2.2 P412W in CHNs and found that it was localized in clusters indistinguishable from GFP-Kv2.2 (Figure 8D). The size of GFP-Kv2.2 P412W clusters was not significantly different from those of GFP-Kv2.2 (Figure 8F). We also found a lack of any significant differences in cluster sizes of wild-type GFP-Kv2.1 and nonconducting GFP-Kv2.1 P404W (Figure 8, D and G). We next surface labeled live HEK293T cells with GxTX-633 and found no significant differences in colocalization in GxTX-633 and GFP-Kv2.2 versus GFP-Kv2.2 P412W (Figure 8, E and H). A similar lack of significant differences was seen for GxTX labeling of GFP-Kv2.1 versus nonconducting GFP-Kv2.1 P404W (Figure 8, E and I). These data, taken together, demonstrate that these Kv2 mutants lack ionic conductance but exhibit cell surface expression and clustering indistinguishable from those in their wild-type counterparts.
We next addressed whether the clustered but nonconducting GFP-Kv2.2 P412W mutant retained its ability to recruit/stabilize cortical ER at ER–PM junctions. Live-cell TIRF imaging showed that GFP-Kv2.2 P412W remodeled the DsRed2-ER5–labeled cortical ER into ER–PM junctions (Figure 9A). We found no significant difference in cells expressing GFP-Kv2.2 P412W versus GFP-Kv2.2 in either the size of ER–PM junctions (Figure 9C) or the surface area of the PM occupied by the cortical ER (Figure 9D). The extent of colocalization of DsRed2-ER5 with GFP-Kv2.2 P412W was also not significantly different from that for GFP-Kv2.2 (Figure 9E). We next evaluated the lateral mobility of DsRed2-ER5–labeled cortical ER as an additional measure of its recruitment into ER–PM junctions (Wu et al., 2014; Fox et al., 2015). The mobility of PM-associated ER was significantly reduced in Kv2.2-expressing cells from that in control cells expressing DsRed2-ER5 alone (Supplemental Figure 2, A–C). Cortical ER mobility was not significantly different in cells expressing the nonconducting Kv2.2 P412W mutant versus those expressing WT Kv2.2 (Supplemental Figure 2, A–C). These parameters of cortical ER recruitment/stabilization were also not significantly different between WT Kv2.1 and the nonconducting Kv2.1 P404W mutant (Figure 9, B–E; Supplemental Figure 2, A–C). These data, taken together, demonstrate that the conserved function of Kv2 channels in localizing to and remodeling ER–PM junctions is independent of their canonical ion-conducting function and is a distinct nonconducting function.
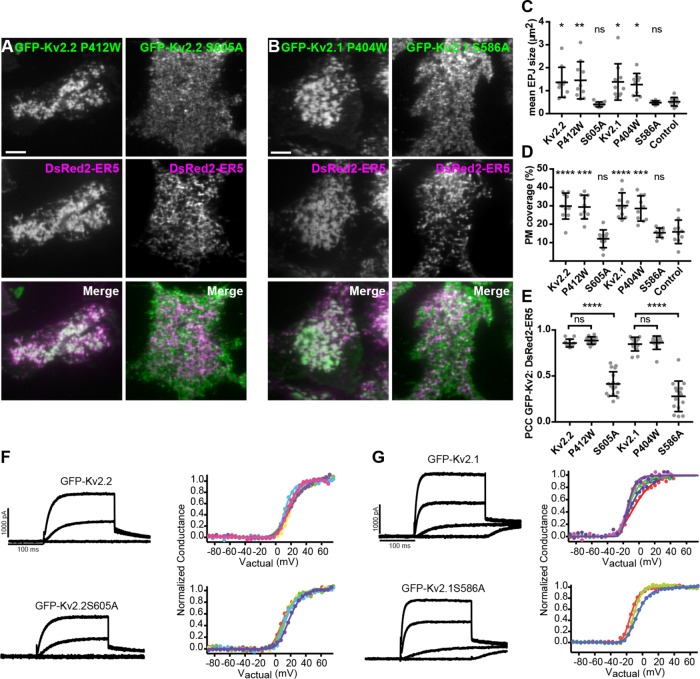
FIGURE 9:
Separation of function point mutations shows that clustering, but not conduction, is necessary for Kv2-mediated remodeling of ER–PM junctions. (A) TIRF images of live HEK293T cells expressing GFP-tagged Kv2.2 mutants (nonconducting P412W and nonclustering S605A in green) and DsRed2-ER5 (magenta). (B) TIRF images of live HEK293T cells expressing GFP-tagged Kv2.1 mutants (nonconducting P404W and nonclustering S586A in green) and DsRed2-ER5 (magenta). Scale bar in top left panels in A and B is 5 μm and holds for all panels in set. (C–E) Comparisons of cells expressing wild-type and mutant Kv2 isoforms (Kv2.2 P412W, Kv2.2 S605A, Kv2.1 P404W, or Kv2.1 S586A); control refers to cells expressing DsRed2-ER5 alone. (C) Mean ER–PM junction (EPJ) size per cell. (D) Percent PM per cell occupied by cortical ER. (E) PCC between DsRed2-ER5 and wild-type and mutant Kv2 isoforms. Bars on all graphs are mean ± SD. See Supplemental Tables 16–18 for values and statistical analyses. (F, G) Exemplar whole-cell voltage clamp recordings (left) and graphs of the corresponding normalized conductance–voltage relationship from HEK293T cells expressing GFP-Kv2.2 or GFP-Kv2.2 S605A (F) and GFP-Kv2.1 or GFP-Kv2.1 S586A (G). Different colors represent data from distinct cells. Recordings shown are representative responses to 200-ms steps from –100 mV to –40, 0, and +40 mV. Note the lack of a significant impact of the declustering point mutation on the properties of the whole cell currents. See Table 1 for values and statistical analyses for whole-cell current density and midpoint of voltage activation, respectively.
We next determined whether Kv2.2 clustering is necessary for remodeling of ER–PM junctions. We used a point mutant (S605A) in the Kv2.2 PRC domain that abolishes its clustering (Bishop et al., 2015). On the basis of previous analyses of large C-terminal truncation mutants in Kv2.1 that eliminate the entire PRC domain (e.g., VanDongen et al., 1990; Scannevin et al., 1996), we expected that this point mutant would not impact the ability of Kv2.2 to function as a conducting Kv channel. To verify this, we used whole-cell patch clamp recordings to compare currents from wild-type and nonclustered Kv2.2 channels in voltage-clamped cells. We found that expression of GFP-Kv2.2 S605A in HEK293T cells resulted in expression of voltage-activated outward currents (Figure 9F). Neither the overall K+ current density nor the conductance–voltage relationships of cells expressing GFP-Kv2.2 versus GFP-Kv2.2 S605A (Figure 9F; Table 1), or GFP-Kv2.1 versus GFP-Kv2.1 S586A (Figure 9G; Table 1) exhibited significant differences. Thus, these measurements of current density and the conductance–voltage relationship support the conclusion that Kv2 channels with these cytoplasmic point mutations that disrupt clustering do not affect the density of conducting channels on the cell surface or their gating.
TABLE 1:
Biophysical characteristics of Kv2 currents from nonclustering mutants.
| Sample | IK at +50 mV (pA/pF) | Vmid | n (cells) |
|---|---|---|---|
| GFP-Kv2.2 | 70.39 ± 41.67 | 17.34 ± 3.08 mV | 6 |
| GFP-Kv2.2 S605A | 51.11 ± 36.34a | 13.43 ± 3.10 mVb | 5 |
| GFP-Kv2.1 | 68.89 ± 17.95 | −10.09 ± 2.70 mV | 5 |
| GFP-Kv2.1 S586A | 80.89 ± 23.85c | −8.76 ± 4.90 mVd | 4 |
p values (two tailed, unpaired t-test):
a0.434 versus Kv2.2.
b0.067 versus Kv2.2.
c0.438 versus Kv2.1.
d0.649 versus Kv2.1.
We next determined the function of the nonclustering but conducting Kv2.2 S605A point mutant in remodeling ER–PM junctions. TIRF imaging revealed a diffuse localization of GFP-Kv2.2 S605A (Figure 9A). The ER–PM junction size (Figure 9C) and the percentage of PM surface area occupied by cortical ER (Figure 9D) were not significantly different between cells coexpressing GFP-Kv2.2 S605A and cells expressing DsRed2-ER5 alone. This nonclustered GFP-Kv2.2 S605A mutant also had significantly reduced colocalization with coexpressed DsRed2-ER5 relative to GFP-Kv2.2 (Figure 9E). We obtained similar results for Kv2.1, in that the ability to remodel ER–PM junctions was significantly reduced in the nonclustering but conducting GFP-Kv2.1 S586A point mutant (Figure 9, B–E). Taken together, these results using this set of separation-of-function point mutants demonstrate that Kv2 channel clustering, but not conduction, is necessary for the unique ability of PM Kv2 channels to localize to and remodel ER–PM junctions, and that this conserved function of Kv2 channels is distinct and separable from its role in conducting ions.
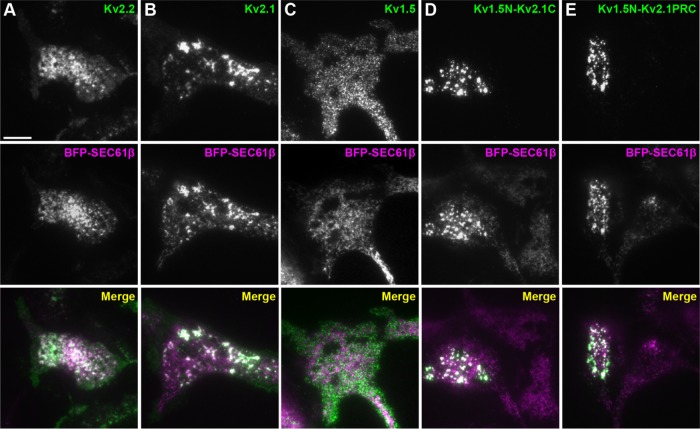
Finally, we determined whether the highly conserved PRC domain transfers the ability to remodel ER–PM junctions to another protein that lacks this function. We used a set of chimeras with C-terminal fragments of Kv2.1 containing the PRC domain appended to the C-terminus of the Kv1.5 channel (Lim et al., 2000), which, unlike Kv2 channels, does not form PM clusters or remodel ER–PM junctions (Figure 10C; also see Lim et al., 2000; Mohapatra and Trimmer, 2006). We found that transfer of the entire Kv2.1 C-terminus (aa 411–853) or a small fragment (aa 536–600) containing primarily the PRC domain to Kv1.5 was sufficient for the resultant Kv1.5-Kv2.1 chimeras to remodel ER–PM junctions (Figure 10, D and E). Taken together, these results show that the highly conserved PRC domain is both necessary and sufficient for the Kv2-mediated remodeling of ER–PM junctions.
FIGURE 10:
The PRC domain can act autonomously to transfer the ER–PM junction remodeling function of Kv2 channels to another PM protein. TIRF images of fixed and immunolabeled HEK293T cells coexpressing BFP-SEC61β (magenta) and in green either Kv2.2 (A), Kv2.1 (B), Kv1.5 (C), Kv1.5N-Kv2.1C (D), or Kv1.5N-Kv2.1PRC (E). Scale bar in A is 10 µm and holds for all panels.
Eliminating Kv2 channel expression in vivo impacts RyR-containing ER–PM junctions in brain neurons
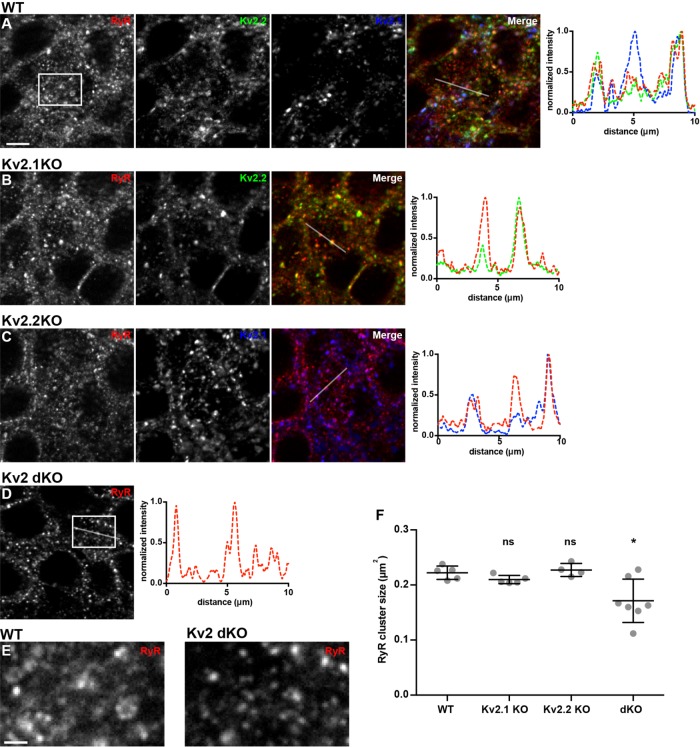
As detailed above, endogenously expressed Kv2.2 and Kv2.1 in brain neurons in situ and in culture colocalize with RyR-containing ER–PM junctions, and exogenously expressing either Kv2 channel remodels ER–PM junctions in CHNs and heterologous cells. We next tested whether eliminating Kv2 channel expression in knockout mice would impact the spatial organization of RyR-containing ER–PM junctions in brain neurons, taking advantage of the availability of Kv2.1 (Jacobson et al., 2007; Speca et al., 2014) and Kv2.2 (Hermanstyne et al., 2013) knockout mice, and Kv2 double-knockout mice (Bishop et al., 2018). We immunolabeled Kv2.2, Kv2.1, and RyR in brain sections from these mice and from wild-type controls and analyzed RyR clusters in hippocampal CA1 pyramidal neurons, which express both Kv2.2 and Kv2.1 (Speca et al., 2014; Bishop et al., 2015, 2018; Palacio et al., 2017). As shown in Figure 11, while there were no significant changes in the spatial characteristics of RyR clusters in the samples from the single Kv2.1 or Kv2.2 knockout mice from those from wild-type mice, the size of RyR clusters in CA1 pyramidal neurons was significantly reduced in the samples from the double Kv2 knockout mice (Figure 11, A–F). This supports an in vivo role for Kv2 channels in contributing to the spatial characteristics of RyR-containing ER–PM junctions in brain neurons.
FIGURE 11:
Genetic ablation of Kv2.2 and Kv2.1 alters RyR localization in mouse brain neurons. (A–D) Projected z-stack images of CA1 hippocampus from mouse brain sections from wild-type (A, WT), Kv2.1 knockout (B, Kv2.1KO), Kv2.2 knockout (C, Kv2.2KO), or Kv2.1 and Kv2.2 double-knockout (D, Kv2 dKO) mice immunolabeled for RyR (red), Kv2.2 (green), and Kv2.1 (blue). (A) RyR, Kv2.2, and Kv2.1 immunolabeling from WT mouse. (B) RyR and Kv2.2 immunolabeling from Kv2.1 KO mouse. (C) RyR and Kv2.1 immunolabeling from Kv2.2 KO mouse. (D) RyR immunolabeling from Kv2.1/Kv2.2 dKO mouse. Scale bar in A is 10 µm and holds for all panels in set. Panels to the right of each row are the corresponding normalized fluorescence intensity values across the individual line scans depicted by the white line in the merged images. (E) Enlarged selections of RyR-labeling of WT and Kv2.1/Kv2.2 dKO images as indicated by boxes in panels A and D, respectively. Scale bar in WT RyR inset panel is 1.25 µm and holds for both panels. (F) Graph of mean RyR cluster size measured from each genotype. Bars are mean ± SEM. See Supplemental Table 20 for values and statistical analyses.
DISCUSSION
Our results presented here demonstrate that both members of the Kv2 channel family have a conserved ability to remodel ER–PM junctions, which is unique among all PM proteins studied to date. We show that endogenous Kv2.2 ion channels localize to ER–PM junctions on somata, proximal dendrites, and the AIS in brain neurons. Experiments in CHNs and in heterologous HEK293T show that Kv2.2 channels can function to remodel ER–PM junctions, and that this is a conserved and nonconducting function of mammalian Kv2 ion channels that requires an intact PRC domain. We also show that when transferred to another protein, the PRC domain can act autonomously to remodel ER–PM junctions. Moreover, elimination of Kv2 expression in knockout mice leads to altered ER–PM junctions in brain neurons. The conserved function of Kv2.2 and Kv2.1 in remodeling ER–PM junctions makes the Kv2 channels the first family of PM proteins whose expression is sufficient to perform this function. Kv2-containing ER–PM junctions are found at sites deficient in components of the cortical actin cytoskeleton, which contributes to but is not the sole determinant of the overall spatial organization of Kv2 channel–containing ER–PM junctions. Kv2.2-containing ER–PM junctions are found associated with junctions containing diverse ER tethers that mediate ER-to-PM contacts, suggesting that ER–PM junctions formed by Kv2 channels and these ER tethers may structurally and functionally overlap in cells in which they are coexpressed. Separation-of-function mutants in Kv2.2 and Kv2.1 reveal that their conserved function in remodeling ER–PM junctions is independent of their well-established canonical function as ion-conducting channels regulating electrical signaling in neurons and nonneuronal cells, but entirely dependent on an intact PRC domain and their clustering in the PM. That Kv2.2 and Kv2.1 have distinct patterns of cellular expression suggests that the highly similar yet distinct functions of these mammalian Kv2 channel paralogs, including dynamic phosphorylation-dependent regulation of their clustering (Bishop et al., 2015), might distinctly impact the structure, function, and regulation of ER–PM junctions in the classes of neurons and nonneuronal cells in which they are differentially expressed.
In certain brain neurons in situ and in hippocampal neurons in culture, we found clusters of Kv2.2 at sites containing high densities of associated ER-localized RyRs, supporting the conclusions that these clusters represent native Kv2.2-containing ER–PM junctions and that these sites are associated with distinct neuronal Ca2+ signaling domains. Moreover, that elimination of expression of both Kv2 channels leads to changes in the spatial organization of RyR-containing ER–PM junctions in brain neurons suggests that Kv2 channels play a role in the structural organization of these Ca2+ signaling microdomains. Although Kv2.2 and Kv2.1 are unique among mammalian PM proteins in being capable of remodeling ER–PM junctions, their distinct cellular expression patterns in brain and in other mammalian tissues, together with their distinct phosphodependent regulation, may contribute to the unique phenotypes seen in mice upon knockout of either Kv2.2 (altered sleep wake cycles; Hermanstyne et al., 2010) or Kv2.1 (neuronal and behavioral hyperexcitability; Speca et al., 2014). The relative contributions of the separate functions of Kv2 channels, as ion-conducting channels shaping membrane excitability and in impacting the structure of ER–PM junctions, to the behavioral phenotypes of these mice are as yet unknown.
Our data from a strategically selected set of separation-of-function point mutants support the conclusion that recruitment/stabilization of ER–PM junctions is a nonconducting and physical function of Kv2 channels that relies on their clustering. Both Kv2.2 and Kv2.1 are bona fide PM voltage-gated K+ channels whose ion-conducting function underlies the bulk of the delayed rectifier K+ current in various classes of neurons (Murakoshi and Trimmer, 1999; Du et al., 2000; Malin and Nerbonne, 2002; Liu and Bean, 2014). Moreover, acute pharmacological inhibition of Kv2 channels impacts neuronal excitability and shapes the characteristics of action potentials (Guan et al., 2013; Liu and Bean, 2014; Kimm et al., 2015; Pathak et al., 2016; Honigsperger et al., 2017; Palacio et al., 2017). Our finding that the ability to remodel ER–PM junctions is a nonconducting function of Kv2 channels is intriguing, given previous findings that the bulk of exogenous Kv2.1 expressed in either heterologous cells or neurons may be present in a nonconducting state (Benndorf et al., 1994; O’Connell et al., 2010; Fox et al., 2013). That ion channels can have diverse nonconducting functions distinct from their canonical ion-conducting roles is an emerging theme in biology, with such roles as cell adhesion molecules, enzymes or scaffolds for enzymes and voltage sensors for intracellular events through conformational coupling (reviewed in Kaczmarek, 2006). Studies in pancreatic beta cells support a nonconducting function for Kv2.1 in regulating insulin secretion (Dai et al., 2012), which is dependent on Kv2.1 clustering (Fu et al., 2017). It is intriguing to speculate that the nonconducting role of Kv2.1 in beta cells is to impact the structure of ER–PM junctions, which have been proposed to play an important role in glucose-stimulated insulin secretion (Li et al., 2016; Lees et al., 2017).
The extent and nature of the physiological contribution of Kv2-mediated regulation of ER–PM junction organization in brain neurons is not known. However, recent studies employing whole exome sequencing have led to identification of encephalopathic epilepsy patients with de novo mutations in the KCNB1 gene that encodes Kv2.1. While the bulk of these disease-associated mutations are in the voltage-sensing and pore domains that are crucial to the canonical function of Kv2.1 as a bona fide Kv channel (e.g., Torkamani et al., 2014; Saitsu et al., 2015; Thiffault et al., 2015), a subset are nonsense mutations that result in a truncated cytoplasmic C-terminus (de Kovel et al., 2016, 2017; Marini et al., 2017). While the cytoplasmic C-terminus plays a modulatory role in regulating activation gating of Kv2.1 channels (Murakoshi et al., 1997; Park et al., 2006; Ikematsu et al., 2011), the most obvious effect of these nonsense mutations that eliminate the PRC domain is to disrupt the clustering of Kv2.1 (Scannevin et al., 1996; Lim et al., 2000; Bishop et al., 2015; Fox et al., 2015, 2018; Jensen et al., 2017) and presumably the organization of Kv2-containing ER–PM junctions. Generating mouse models that express the separation-of-function mutations used here to selectively disrupt Kv2.1 conduction and clustering may lead to insights into the relative contributions of the separable electrical and structural roles of Kv2 channels in normal physiology, and how these distinct classes of disease-associated mutations that should impact one or the other function contribute to pathophysiology.
Our results show that both members of the Kv2 family of ion channels can in themselves remodel ER–PM junctions. As this is the first family of mammalian PM proteins found to have this function, it suggests that Kv2 channels use a molecular mechanism distinct from all other known classes of endogenous ER–PM junction components (i.e., members of the E-Syt, JP, and STIM families), which are ER tethers that bind specific lipids present in the inner leaflet of the PM, although STIM family members also exhibit conditional interaction with PM Orai proteins (Carrasco and Meyer, 2011; Henne et al., 2015). That expression of either Kv2.2 and Kv2.1 is sufficient to remodel ER–PM junctions in the absence of their ion-conducting functions, and via a mechanism that requires an intact PRC domain, suggests that both Kv2 family members act through the same mechanism. We showed here that the PRC domain is sufficient to transfer the ability to remodel ER–PM junctions, which among PM proteins is unique to Kv2 channels, to another PM protein. This suggests a model where the PRC domain interacts directly with an ER-localized protein or lipid binding partner to tether the PM to the ER. That we have shown here that nonconducting Kv2 channels retain their ability to remodel ER–PM junctions is consistent with a mechanism involving direct protein–protein interactions, as opposed to less direct effects of Kv channel function on cell signaling pathways, for example, by influencing Ca2+ signaling events that lead to recruitment of ER–PM junctions, as occurs for junctions containing certain STIM and E-Syt family members (Carrasco and Meyer, 2011; Henne et al., 2015; Saheki and De Camilli, 2017b; Balla, 2018). In the case of Kv2 channels, these protein–protein interactions are mediated by the PRC domain, which is both necessary and sufficient for Kv2-mediated remodeling of ER–PM junctions. The abundant ER proteins VAPA and VAPB have recently been found by the Tamkun laboratory (Johnson et al., 2018) and ourselves (Kirmiz et al., 2018) to interact with Kv2.1 and Kv2.2. Our results here are consistent with a model where a direct interaction between these ER VAP proteins and PM Kv2 channels mediates this class of ER–PM junctions. That Kv2 channels form clusters at ER–PM junctions in diverse cell types including brain neurons of diverse mammalian species in situ and in culture (e.g., Trimmer, 1991; Scannevin et al., 1996; Murakoshi and Trimmer, 1999; Antonucci et al., 2001; Misonou et al., 2004, 2006, 2008; Kihira et al., 2010; Fox et al., 2013; Bishop et al., 2015, 2018; Frazzini et al., 2016), in spinal motor neurons (Muennich and Fyffe, 2004), and in nonneuronal heterologous cells such as human HEK293 (Bishop et al., 2015, 2018), monkey COS-1 (Cobb et al., 2015) and Madin-Darby canine kidney (Scannevin et al., 1996) cells, rat PC12 pheochromocytoma cells (Sharma et al., 1993), and CHO cells (Cobb et al., 2015), is consistent with such an underlying mechanism, as VAP proteins are highly conserved across diverse mammalian species and widely expressed in numerous cell types (Murphy and Levine, 2016). That VAPs are abundant ER proteins may be consistent with the observation that the formation of Kv2 clusters and recruitment of ER–PM junctions is not obviously saturable, so that the higher the level of Kv2.2 or Kv2.1 expression, the larger the clusters and associated ER–PM junctions (Antonucci et al., 2001; Cobb et al., 2015). However, it remains the case that other ER-localized proteins may also interact with PM Kv2 channels to mediate ER–PM junction formation in a cell type–specific manner. The conditional clustering of Kv2.1 is phosphorylation-dependent, supporting a model where the induction of ER–PM junction formation can be dynamically regulated and requires sufficient Kv2.1 phosphorylation, including in critical serine residues within the PRC domain itself (Lim et al., 2000; Cobb et al., 2015). That the PRC domain of Kv2.2, which does not exhibit phosphorylation-dependent clustering as does Kv2.1 (Bishop et al., 2015), contains these same serine residues suggests that should phosphorylation at these sites be required for Kv2.2 clustering and ER–PM junction remodeling, then in Kv2.2, this phosphorylation is more constitutive than the dynamically regulated phosphorylation of Kv2.1. It is intriguing that both STIM:Orai and Kv2-based ER–PM junctions are sensitive to Ca2+ signaling, but at a simplistic level in opposite directions and with distinct mechanisms, with the former regulated by Ca2+-dependent conformational changes in STIM proteins upon ER Ca2+ depletion leading to association with PM Orai proteins, and the latter by Ca2+-calcineurin-dependent changes in Kv2.1 phosphorylation state in response to elevated [Ca2+]I leading to dissociation of Kv2.1 but not Kv2.2 (Bishop et al., 2015) from ER–PM junctions.
That Kv2.2-containing ER–PM junctions can colocalize with members of the E-Syt, STIM, and JP families suggests potential overlap with these distinct classes of ER–PM junctions in coexpressing mammalian cells. One explanation of these findings is that these ER-localized PM tethers, by virtue of their ER localization, are passively recruited along with other ER proteins such as Sec61β to Kv2.2-containing ER–PM junctions. However, the lack of association of Kv2.2-containing ER–PM junctions and those generated via the rapamycin-triggered coupling of CB5-FKBP and Lyn11-FRB would argue against a promiscuous presence of Kv2.2 channels at any ER–PM junction, although these rapamycin-induced artificial junctions have certain characteristics, including having a narrower gap between ER and PM (Varnai et al., 2007; Dickson et al., 2016) than those formed by endogenous ER–PM junction components (e.g., Wu et al., 2017). As the tethering of E-Syts, JPs, and STIMs to the PM occurs at least in part on their binding to lipids on the PM inner leaflet (Henne et al., 2015), another possible explanation for the robust colocalization between Kv2.2-containing ER–PM junctions and these ER tethers is that Kv2.2 clustering results in a distinct lipid microenvironment in the PM inner leaflet at or near these clusters. Changes in the local lipid environment at/near Kv2.2 clusters could also underlie generation of ER–PM junctions at these sites, via recruitment of one or more lipid-binding ER–PM tethers. As noted above, these tethers in aggregate would need to have sufficiently robust expression across the numerous species and cell types in which endogenous and exogenous Kv2 channels are clustered. We note that our quantitative analyses of colocalization between Kv2.2-containing ER–PM junctions and these ER tethers suggest that despite the extensive overlap, as reported by high (≈1.0) MOC values, the intensity profiles of these proteins do not uniformly coincide, as shown by significantly lower paired PCC measurements (Dunn et al., 2011). That there is heterogeneity in ER–PM junctions within the same cell is consistent with the variable cooccurrence of Kv2.2 and Kv2.1 clusters with RyR clusters between and within different classes of mammalian brain neurons (Antonucci et al., 2001; Misonou et al., 2005b; Mandikian et al., 2014). This concept is further supported by the lack of colocalization between Kv2.2-containing ER–PM junctions and those formed via triggered coupling of CB5/Lyn11. Recent findings (Johnson et al., 2018; Kirmiz et al., 2018) suggest that formation of Kv2 channel–containing ER–PM junctions involves the direct interaction of PM Kv2 channels with ER VAP proteins. Future studies defining the subcellular localization of the different members of the E-Syt, JP, and STIM families endogenously expressed in mammalian brain neurons relative to those containing Kv2 channels and VAPs will lead to increased understanding of the relationship between the native ER–PM junctions formed by these ER tethers and those containing Kv2 channels.
That LatA treatment impacted the characteristics of both Kv2- and CB5/Lyn11-containing ER–PM junctions but did not lead to their fusion suggests that the actin cytoskeleton is not the only determinant of their distinct spatial organization. The effects of actin disruption on Kv2-containing ER–PM junctions, and that these junctions are localized to zones at the cell cortex depleted in actin and actin-interacting proteins, suggest a role for the actin cytoskeleton in shaping their spatial characteristics. This is consistent with previous studies demonstrating that Kv2.1 clusters on the axon initial segments of brain neurons are specifically localized to ankG-deficient “holes” (King et al., 2014) and that disruption of the actin cytoskeleton impacts clustering of Kv2.1 (O’Connell et al., 2006; Tamkun et al., 2007). Recent studies reveal that the STIM1:Orai1 complex at the immune synapse (Hartzell et al., 2016) and HeLa cell ER–PM junctions labeled with the reporter MAPPER (Hsieh et al., 2017) are also present in actin-poor zones, and that disruption of the actin cytoskeleton altered the distribution and dynamics of these HeLa cell ER–PM junctions (Hsieh et al., 2017). Depletion of ER Ca2+ stores can also impact ER–PM junctions via the conditional association of STIM1 and Orai1, which can then associate with those formed by Kv2.1 (Fox et al., 2015) or, as shown here, Kv2.2. That both ER (RyR) and PM (Orai1) Ca2+ channels colocalize with Kv2-containing ER–PM junctions suggests a potential structural role for Kv2 channels in regulating neuronal Ca2+ signaling and homeostasis above and beyond their established role in impacting intracellular Ca2+ through their ion-conducting effects on membrane excitability. Future studies will define the respective contributions of the separate yet highly conserved conducting and nonconducting roles of Kv2 channels in impacting cellular physiology and how this is disrupted under pathological conditions that may exert their effects through distinct impacts on these broadly and highly expressed ion channels.
MATERIALS AND METHODS
Preparation of mouse brain sections for immunohistochemistry
All procedures involving mice were approved by the University of California Davis Institutional Animal Care and Use Committee and were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). All mice were maintained under standard light–dark cycles and allowed to feed and drink ad libitum. Kv2.1-KO mice (RRID:IMSR_MGI:3806050) have been described previously (Jacobson et al., 2007; Speca et al., 2014), and were generated by breeding Kv2.1+/– mice that had been backcrossed on the C57/BL6J background (RRID:IMSR_JAX:000664). Kv2.2-KO mice (Hermanstyne et al., 2010, 2013) were obtained from Tracey Hermanstyne and Jeanne Nerbonne (Washington University School of Medicine). All Kv2.2-KO mice used here were obtained from heterozygotic crosses in the C57/BL6J background (RRID:IMSR_JAX:000664). Double-knockout mice for Kv2.1/Kv2.2 (Kv2 dKO) were generated by crossing Kv2.1+/– and Kv2.2–/– mice. Both male and female mice were used, and all were more than 12 wk old. Littermates were used when available. Mice were deeply anesthetized with 90 mg/kg Na-pentobarbital salt (Sigma Cat# P3761) in 0.9% NaCl solution through intraperitoneal injections, followed by boosts as needed. Once mice were completely anesthetized, they were transcardially perfused with 4.5 ml of ice-cold phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM sodium phosphate buffer [PB], pH 7.4) containing 10 U/ml heparin, followed by an ice-cold fixative solution of 4% formaldehyde (freshly prepared from paraformaldehyde; Sigma Cat# 158127) in 0.1 M sodium PB, pH 7.4 (0.1M PB), using 1 ml fixative solution per gram of mouse weight. Following perfusion, brains were removed from the skull and cryoprotected in 10% sucrose, 0.1 M PB overnight at 4°C, and then transferred to a solution of 30% sucrose, 0.1 M PB until they sank to the bottom of the tube (24–48 h). Following cryoprotection, all brains were frozen and cut on a freezing-stage sliding microtome (Richard Allen Scientific) to obtain 30 µm–thick sagittal sections. Sections were collected in 0.1 M PB and processed for immunohistochemistry (IHC) as free-floating sections.
Multiplexed fluorescence immunohistochemistry
Multiplex immunofluorescence labeling of mouse brain sections was performed essentially as previously described (Manning et al., 2012). Briefly, free-floating sections were washed three times in 0.1 M PB plus 10 mM sodium azide at room temperature with slow agitation. All subsequent incubations and washes were at room temperature with slow agitation on an orbital platform shaker, unless otherwise stated. Sections were incubated in blocking buffer (10% goat serum in 0.1 M PB, 0.3% Triton X-100, and 10 mM sodium azide) for 1 h. Immediately after blocking, sections were incubated with primary antibody combinations (diluted in blocking buffer) overnight at 4°C. All primary antibodies used in this study have been previously described (see Table 2 for a description of primary antibodies). Following incubation, sections were washed 3 × 10 min each in 0.1 M PB and incubated for 1 h with affinity-purified goat anti-rabbit and/or goat anti-mouse immunoglobulin G (IgG)-subclass-specific Alexa Fluor–conjugated secondary antibodies diluted in blocking buffer, using a 1:1500 dilution for Alexa Fluor 647 conjugates and a 1:2000 dilution for Alexa Fluor 488 and 555 conjugates, all secondary antibodies from ThermoFisher. Sections were also labeled with the DNA-specific dye Hoechst 33258 (200 ng/ml; ThermoFisher Cat# H1399) during the secondary antibody step. After three 10-min washes in 0.1 M PB, sections were mounted on gelatin-coated slides and dried, treated with 0.05% Sudan Black Sudan Black (EM Sciences Cat# 21610) in 70% ethanol for 1.5 min, extensively washed in water, and mounted with Prolong Gold (ThermoFisher Cat# P36930). All immunolabeling reported for quantification purposes is representative of three animals (biological replicates) per genotype, except for Kv2.2 KO, which included brain sections from two animals. Brain sections from all biological replicates within each experiment were labeled, treated, and mounted in parallel.
TABLE 2:
Antibody information.
| Antigen and antibody name | Immunogen | Manufacturer information | Concentration used | Figures |
|---|---|---|---|---|
| Kv2.2 (N372B/60) | Fusion protein aa 717–907 of rat Kv2.2 long isoform | Mouse IgG2a monoclonal antibody (mAb), NeuroMab catalogue #73-360, RRID:AB_2315867 | Purified, 10 μg/ml (1-cortex, 11), 20 μg/ml (1-CA1), 15 μg/ml (11) | 1, 4 (CHN), 10, 11 |
| Kv2.2 (N372C/51) | Fusion protein aa 717–907 of rat Kv2.2 long isoform | Mouse IgG1 mAb, NeuroMab catalogue #75-358, RRID:AB_2315866 | Purified, 10 μg/ml | 4 (IHC) |
| Kv2.1 (K89/34) | Synthetic peptide aa 837–853 of rat Kv2.1 | Mouse IgG1 mAb, NeuroMab catalogue #73-014, RRID:AB_1067225 | Tissue culture supernatant, 1:5 | 1, 4 (CHN), 10 |
| Kv2.1 (KC) | Synthetic peptide aa 837–853 of rat Kv2.1 | Rabbit polyclonal antibody (pAb), In-house (Trimmer Laboratory), RRID:AB_2315767 | Affinity-purified, 1:150 | 1, 4 (IHC), 11 |
| Ryanodine receptor (34C) | Partially purified chicken pectoral muscle ryanodine receptor | Mouse IgG1 mAb, Developmental Studies Hybridoma Bank, RRID:AB_528457 | Concentrated tissue culture supernatant, 1 μg/ml | 1 (CHN) |
| Kv1.5e | Synthetic peptide aa 271–284 of rat Kv1.5 | Rabbit pAb, In-house (Trimmer Laboratory), RRID:AB_2722698 | Affinity-purified, 1:100 | 10 |
| Ryanodine receptor (34C) | Partially purified chicken pectoral muscle ryanodine receptor | Mouse IgG1 mAb, ThermoFisher catalogue #MA3-925 RRID:AB_2254138 | Purified, 2.5 μg/ml (1-cortex), 1 μg/ml (1-CA1, 11) | 1 (IHC), 11 |
| AnkyrinG (N106/36) | Full-length recombinant human | Mouse IgG2b mAb, NeuroMab catalogue #75-146, RRID:AB_10673030 | Purified, 10 μg/ml | 4 (CHN) |
| AnkyrinG (N106/65) | Full-length recombinant human | Mouse IgG2b mAb, NeuroMab catalogue #75-147, RRID:AB_10675130 | Purified, 5 μg/ml | 4 (IHC) |
| MAP2 | Purified microtubule associated protein from rat brain | Chicken pAb, EnCor catalogue #CPCA-MAP2, RRID:AB_2138173 | Purified IgY fraction, 1:5000 | 1 |
All images were acquired on a Zeiss AxioObserver Z1 microscope with an X-Cite 120 lamp as the fluorescent light source and equipped with an AxioCam MRm digital camera. High-magnification optical sections were acquired using an ApoTome structured illumination system (Carl Zeiss MicroImaging) with a 63×/1.40 NA plan-Apochromat oil immersion objective. ApoTome z-stacks were acquired and processed with Axiovision 4.8.2 acquisition software (Carl Zeiss MicroImaging; RRID: SciRes_000111). All brain sections within a given experiment and immunolabeled with the same antibody cocktail were imaged under the same conditions (objective, exposure time, lamp settings, etc.). Image processing was performed in Axiovision (Carl Zeiss MicroImaging) and Fiji v2.0.0-rc-43/1.51 (NIH). All panels in a given figure were imaged and processed identically, unless otherwise noted. High-magnification ApoTome z-stacks were opened for analysis as raw image files in Fiji (NIH) using the Bio-Formats library importing plug-in (Linkert et al., 2010). Quantification was performed using single optical z-sections. All statistical analyses of immunolabeling were performed in Prism (GraphPad).
Quantification of RyR immunolabeling was performed in FIJI. Images were first background-subtracted; background levels were determined from “no primary antibody” immunolabeling controls for each animal and mathematically subtracted from paired images of RyR labeling, and images were converted to 8-bit. A region of interest (ROI) selection was made to include cell bodies of neurons in the pyramidal cell layer of hippocampal CA1, and the image was automatically converted into a binary mask using auto local thresholding (Bernsen, 1986). RyR cluster size was quantified automatically using the “analyze particles” function in FIJI. Particles smaller than 0.06 μm2 were excluded from this analysis.
Culture and transfection of rat hippocampal neurons
All procedures involving rats were approved by the University of California Davis Institutional Animal Care and Use Committee and were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals of the NIH. All rats were maintained under standard light–dark cycles and allowed to feed and drink ad libitum. Hippocampi were dissected from embryonic day 18 rat embryos, dissociated enzymatically for 20 min at 37°C in 0.25% (wt/vol) trypsin (ThermoFisher Cat# 15050065) in Hank’s balanced saline solution, and dissociated mechanically by triturating with polished glass Pasteur pipettes. Dissociated cells were suspended in Neurobasal (Invitrogen Cat# 21103-049) supplemented with 10% fetal bovine serum (FBS; Invitrogen Cat# 16140071), 2% B27 (Invitrogen Cat# 17504044), 1% GlutaMAX (Invitrogen Cat# 35050061), and 0.001% gentamicin (Life Technologies Cat #1570-064) and plated at 60,000 cells per dish in glass-bottomed dishes (MatTek Cat# P35G-1.5-14-C) or number 1.5 glass coverslips coated with poly-l-lysine (Sigma Cat# P2636). At 4–7 d in vitro (DIV), cytosine-d-arabinofuranoside (Millipore Cat# 251010) was added to inhibit nonneuronal cell growth. Immunofluorescence labeling was performed on CHNs at 14–18 DIV. For transfection experiments, CHNs were transiently transfected at DIV 5-10 using Lipofectamine 2000 (Invitrogen Cat# 11668019) for 1.5 h as previously described (Lim et al., 2000). Transfected CHNs were imaged 40–48 h posttransfection.
Heterologous cell culture, reagents, and transfection
HEK293T cells were maintained in DMEM supplemented with 10% Fetal Clone III (HyClone Cat# SH30109.03), 1% penicillin/streptomycin, and 1X GlutaMAX (ThermoFisher Cat# 35050061) in a humidified incubator at 37°C under 5% CO2. HEK293T cells were transfected with Lipofectamine as previously described (Bishop et al., 2015). Briefly, 6 × 104 HEK293T cells were plated onto 35-mm glass-bottomed dishes (MatTek Cat# P35G-1.5-14-C) coated with poly-l-lysine and incubated for 20–24 h. Cells were then transiently transfected using Lipofectamine following the manufacturer’s protocol in DMEM without supplements and then returned to regular growth media 4 h after transfection. HEK293T cells were imaged live or subjected to fixation 40–48 h posttransfection.
Cell fixation, immunolabeling, and fixed-cell imaging
For experiments involving imaging of fixed and immunolabeled HEK293T cells, fixation was performed as previously described (Dickson et al., 2016). Briefly, cells were fixed in 3.2% formaldehyde (freshly prepared from paraformaldehyde, Sigma Cat# 158127) and 0.1% glutaraldehyde (Ted Pella; Cat# 18426) for 30 min at room temperature, washed three times for 5 min in PBS, and quenched with 1% sodium borohydride in PBS for 15 min at room temperature. Cells were blocked and permeabilized in 4% nonfat milk powder in PBS containing 0.1% Triton X-100. Neurons (CHNs) were fixed in ice-cold 4% formaldehyde, 4% sucrose in PBS for 15 min at 4°C, washed three times for 5 min in PBS, and blocked and permeabilized in 4% nonfat milk powder in PBS containing 0.1% Triton X-100. Primary antibody incubations were performed in blocking solution for 1 h at room temperature. All antibodies used in this study have been described previously (see Table 2 for a description of primary antibodies). Following primary antibody incubation and three 5-min washes in blocking solution at room temperature, coverslips were immunolabeled with Alexa Fluor–conjugated goat anti-mouse IgG subclass-specific (Manning et al., 2012) or goat anti-rabbit IgG secondary antibodies (all secondary antibodies from ThermoFisher) at 1:1500 and Hoechst 33258 (200 ng/ml; ThermoFisher Cat# H1399) for 1 h in blocking solution, washed three times for 5 min in PBS, and mounted on microscope slides using Fluoromount G (Southern Biotech Cat# 0100-01), or for samples prepared for TIRF microscopy, imaged in PBS containing ascorbate.
For conventional fluorescence imaging (used in Figures 1, A–C, 4, A–C, and 11), images were acquired with an AxioCam MRm digital camera installed on a Zeiss AxioImager M2 microscope or with an AxioCam HRm digital camera installed on a Zeiss AxioObserver Z1 microscope with a 63×/1.40 NA plan-Apochromat oil immersion objective or a 20×/0.8 NA plan-Apochromat objective and an ApoTome coupled to Axiovision software (Zeiss, Oberkochen, Germany). For TIRF imaging of fixed cells, imaging was identical to that used in live-cell TIRF experiments but in the absence of a heated stage/objective heater. Images were obtained with an Andor iXon EMCCD camera installed on a TIRF/widefield equipped Nikon Eclipse Ti microscope using a Nikon LUA4 laser launch with 405-, 488-, 561-, and 647-nm lasers and a 100× PlanApo TIRF/1.49 NA objective run with NIS Elements software (Nikon). Images were collected within NIS Elements as ND2 images. For N-SIM imaging of fixed cells, images were acquired using a Hamamatsu ORCA-ER CCD camera installed on a SIM/widefield equipped Nikon Eclipse Ti microscope using an EXFO X-Cite metal halide light source and a 100× PlanApo TIRF/1.49 objective, run with NIS Elements software (Nikon). Images were collected within NIS Elements as ND2 images. SIM analysis was performed in NIS Elements. Airyscan imaging was performed with a Zeiss LSM 880 confocal laser scanning microscope (Carl Zeiss), equipped with an Airyscan detection unit, with a Plan-Apochromat 63×/1.40 Oil DIC M27 objective.
Plasmid constructs
All novel constructs used in this study (DsRed-Kv2.2, GFP-Kv2.2, GFP-Kv2.2 P412W, GFP-Kv2.2 S605A, GFP-Kv2.1 P404W, GFP-Kv2.1 S586A) were generated using standard molecular biology approaches and confirmed by sequencing. DsRed-Kv2.2 and GFP-Kv2.2 were generated using Gibson assembly to insert full-length rat Kv2.2, also termed Kv2.2long (Kihira et al., 2010) into the GFP-C1 or DsRed-C1 vector (ClonTech) resulting in fusion of DsRed or GFP to the N-terminus of full-length rat Kv2.2. GFP-Kv2.2 S605A, GFP-Kv2.1 P404W and GFP-Kv2.1 S586A were generated via site directed point mutagenesis utilizing a QuikChange PCR of GFP-Kv2.2 or GFP-Kv2.1 (Antonucci et al., 2001), respectively, or via Gibson assembly. GFP-Kv2.2 P412W was generated at Mutagenex. The plasmids encoding Kv2.1 in the RBG4 vector (Shi et al., 1994), Kv2.2 in the RBG4 vector (Bishop et al., 2015), Kv1.5N-Kv2.1C (Mohapatra and Trimmer, 2006), and Kv1.5N-Kv2.1PRC (Lim et al., 2000) have been described previously. Plasmids encoding DsRed2-ER5 and mCherry-actin were a generous gift from Michael Davidson (Florida State University; Addgene plasmids #55836 and #54965). The plasmid encoding ankG-mCherry was a generous gift from Benedicte Dargent (Aix Marseille University; Addgene plasmid #42566). The plasmids encoding BFP-SEC61β and BFP-STIM1 were a generous gift from Jodi Nunnari (University of California, Davis). The plasmid encoding GFP-JP2 was a generous gift from Fernando Santana (University of California, Davis). The plasmid encoding mCherry-E-Syt1-3 was a generous gift from Pietro De Camilli (Yale University School of Medicine). The plasmid encoding mCherry-JP4 was a generous gift from Yousang Gwack (University of California, Los Angeles). The plasmids encoding mCherry-STIM1, -STIM2α, and -STIM2β and GFP-Orai1 were a generous gift from Richard Lewis (Stanford University). The plasmids encoding CFP-CB5-FKBP and Lynn11-FRB (Inoue et al., 2005) were a generous gift from Eamonn Dickson (University of California, Davis).
Live cell Guangxitoxin labeling
The GxTX peptide used in surface labeling was synthesized at the Molecular Foundry of the Lawrence Berkeley National Laboratory under U.S. Department of Energy Contract DE-AC02-05CH11231. GxTX-633 was synthesized by conjugating GxTX to DyLight 633 Maleimide (ThermoFisher Scientific Cat# 46613) using methods for GxTX–maleimide conjugates described previously (Tilley et al., 2014). HEK293T cells were surface-labeled with 1 μM GxTX as described previously (Tilley et al., 2014) and imaged in TIRF as described below but in physiological saline solution (4.7 mM KCl, 146 mM NaCl, 2.5 mM CaCl2, 0.6 mM MgSO4, 1.6 mM NaHCO3, 0.15 mM NaH2PO4, 20 mM HEPES, pH 7.4) containing 8 mM glucose, 0.1 mM ascorbic acid, and 0.1% BSA.
Total internal reflection fluorescence imaging and drug treatments
TIRF imaging was performed at the UC Davis MCB Imaging Facility. Live transfected HEK293T cells cultured on glass-bottomed dishes were imaged in a physiological saline solution (4.7 mM KCl, 146 mM NaCl, 2.5 mM CaCl2, 0.6 mM MgSO4, 1.6 mM NaHCO3, 0.15 mM NaH2PO4, 20 mM HEPES, pH 7.4) containing 8 mM glucose and 0.1 mM ascorbic acid. Cells were maintained at 37°C during the course of imaging with a heated stage and objective heater. Fixed-cell TIRF imaging was performed identically but in the absence of a stage and objective heaters. For experiments involving latrunculin A (ThermoFisher Scientific Cat# 428021100UG) treatment, latrunculin A was diluted to 20 μM in imaging saline and added by pipette to glass-bottomed dishes already containing imaging saline to a final concentration of 10 μM. For experiments involving thapsigargin (Millipore Cat# 586005-1MG) treatment, thapsigargin was diluted to 4 μM in imaging saline and added by pipette to glass-bottomed dishes already containing imaging saline to a final concentration of 2 μM. For experiments involving rapamycin (Sigma Cat# R8781-200UL) treatment, rapamycin was diluted to 10 μM in imaging saline and added by pipette to glass-bottomed dishes already containing imaging saline to a final concentration of 5 μM. Images were obtained with an Andor iXon EMCCD camera installed on a TIRF/widefield-equipped Nikon Eclipse Ti microscope using a Nikon LUA4 laser launch with 405-, 488-, 561-, and 647-nm lasers and a 100× PlanApo TIRF, 1.49 NA objective run with NIS Elements software (Nikon). Images were collected within NIS Elements as ND2 images.
Cell culture and transfection for electrophysiology
All cell lines were grown in a humidified incubator at 37°C under 5% CO2. HEK293T cells were maintained in DMEM supplemented with 10% FBS (HyClone Cat # SH30109.02) and 1% penicillin/streptomycin. Transfections were performed with Lipofectamine. Cells were plated overnight prior to transfection and allowed to grow to ≈40% confluency. Lipofectamine was diluted, mixed, and incubated in Opti-MEM (Life Technologies Cat# 31965-062) at a 1:100 ratio for 5 min. Concurrently, 1 μg of plasmid DNA and Opti-MEM were mixed in the same manner. After incubation, the DNA and lipofectamine mixtures were combined, triturated, and allowed to incubate for 20 min. The transfection cocktail was added to cells for 5 h before the media was replaced. For experiments in Figure 8, 1 μg of GFP-Kv2 or a peGFP-C1 plasmid was used. For experiments in Figure 9, 0.2 μg of GFP-Kv2 plasmids was diluted with 0.8 μg of pcDNA3 plasmids.
Electrophysiology
Whole-cell voltage clamp was used to measure currents from HEK293T cells expressing GFP-Kv2.2, GFP-Kv2.2 P412W, GFP-Kv2.1, GFP-Kv2.1 P404W, or GFP as a control. On the day of the experiment (2 d after transfection), transiently transfected cells were detached with trypsin and plated onto cell culture–treated polystyrene dishes for electrophysiological measurements. The external (bath) solution contained (in mM) 3.5 KCl, 155 NaCl, 10 HEPES, 1.5 CaCl2, and 1 MgCl2, adjusted to pH 7.41 with NaOH. The internal (pipette) solution contained (in mM) 35 KOH, 70 KCl, 50 KF, 50 HEPES, and 5 EGTA adjusted to pH 7.2 with KOH. Liquid junction potential (calculated to be 7.8 mV) was not corrected for. Borosilicate glass pipettes (Sutter Instruments Cat #BF150-110-10HP) with resistance less that 3 MΩ were used to patch the cells. Recordings were at room temperature (22–24°C). Voltage clamp was achieved with an Axon Axopatch 200B amplifier (MDS Analytical Technologies) run by PATCHMASTER software, v2 × 90.2 (HEKA, Bellmore, NY). Holding potential was –80 mV. Capacitance and ohmic leak were subtracted using a P/5 protocol. Recordings were low-pass filtered at 10 kHz and digitized at 100 kHz. Voltage clamp data were analyzed and plotted with IgorPro software, version 7 (Wavemetrics). Current amplitudes at each voltage were the averages from 0.19–0.20 s after the voltage step. In the experiments plotted in Figure 8, series resistance compensation was not used. The estimated series resistance in these experiments ranged from 3 to 8 MΩ, which is predicted to result in substantial cell voltage errors for conducting channels. For quantitative comparison of current levels and voltage activation (Figure 9), we improved control of intracellular voltage by reducing the amount of DNA transfected (described above), partially blocking the K+ currents with tetraethylammonium (TEA), and using series resistance compensation. For experiments shown in Figure 9 on HEK293T cells expressing GFP-Kv2.2, GFP-Kv2.2 S605A, GFP-Kv2.1, or GFP-Kv2.1 S586A, the following modifications were made. The internal (pipette) solution contained (in mM) 140 KCl, 13.5 NaCl, 1.8 MgCl2, 0.09 EGTA, 4 Na-ATP, 0.3 Na-GTP, and 9 HEPES, adjusted to pH 7.2 with KOH. The external (bath) solution contained (in mM) 3.5 KCl, 155 TEA-Cl, 1.5 CaCl2, 1 MgCl2, 10 HEPES, and 10 glucose adjusted to pH 7.42 with NaOH. Extracellular TEA (155 mM) is predicted to inhibit at least 97% of Kv2.1 current at 0 mV (see Ikeda and Korn, 1995; Immke et al., 1999; Immke and Korn, 2000). A calculated liquid junction potential of 7.6 mV was corrected. Pipette tips were coated with Sylgard 184 (Dow Corning Cat #2010518) and fire-polished. Series resistance compensation with lag set to 10 µs was used to constrain calculated voltage error to ≤10 mV. Conductance was measured from the amplitude of outward tail currents averaged from the end of any capacitance transient until 2 ms after stepping to 0 mV from the indicated voltage. Fits with the fourth power of a Boltzmann distribution have been described previously, where Vmid is the voltage where the function reaches half-maximal conductance, and z is valence in units of elementary charge (e+) of each of the four independent voltage sensors (Sack et al., 2004). Conductance data shown are normalized to the maximal conductance of the Boltzmann fit.
Image analysis and statistics
All colocalization analyses were performed within Nikon NIS Elements using ND2 files. An ROI was drawn within a cell of interest and PCC and MOC values were collected. Measurements of structure sizes were quantified automatically within FIJI essentially as previously described (Dickson et al., 2016). ND2 files of DsRed2-ER5 or BFP-SEC61β collected in TIRF were imported directly into FIJI, background-subtracted, converted into an 8-bit image, and automatically converted into a binary mask using auto local thresholding (Bernsen, 1986). An ROI with identical dimensions and containing an area of 60.6 μm2 was drawn within each cell analyzed. The number of individual ER–PM junctions, average ER–PM junction size, and percent PM occupancy were quantified automatically using the “analyze particles” function in FIJI. Signals smaller than 0.04 μm2 were excluded from this analysis. An identical approach was taken in whole cell analysis.
Quantification of Kv2 cluster sizes was performed similarly. ND2 files of GFP-Kv2.2, GFP-Kv2.2 P412W, GFP-Kv2.1, or GFP-Kv2.1 P404W were collected in widefield and deconvolved in NIS Elements, imported directly into FIJI, converted into an 8-bit image, and automatically converted into a binary mask using auto local thresholding (Bernsen, 1986). Kv2 cluster size was quantified automatically using the “analyze particles” function in FIJI. For scatterplot generation of ER–PM junction and Kv2 cluster sizes (Figure 3J), ND2 files were imported directly into FIJI, background-subtracted using a rolling ball radius of 10 pixels, and converted into 8-bit images. Images were converted into binary masks and manually subjected to erosion operations designed to separate objects as described previously (Dickson et al., 2016). Care was taken to ensure that the resulting binary image was comparable to the original image. The areas of these structures were quantified automatically using the “analyze particles” function in FIJI. Areas from 10 to 20 overlapping structures from each cell were paired as coordinates. In cases where more than one structure overlapped, the areas of the overlapping structures were summed as single coordinates.
The coefficient of variation is defined as the SD of intensity divided by mean intensity as previously described (Bishop et al., 2015, 2018; Jensen et al., 2017). Quantification of coefficient of variation and intensity measurements were collected in FIJI. An ROI was drawn around a cell and SD of intensity and mean intensity values were collected.
For line scan analysis of fluorescence intensity, raw intensity values were collected within FIJI and normalized to the maximum value collected.
Analysis of DsRed2-ER5 velocity was performed in MATLAB (MathWorks) using the PIVlab toolkit (Thielicke and Stamhuis, 2014) as previously described (Fox et al., 2015). Briefly, successive frames (captured at 31.25 Hz) of DsRed2-ER5 expression in HEK293T cells transfected with DsRed2-ER5 alone or cotransfected with GFP-Kv2.2, GFP-Kv2.2 P412W, GFP-Kv2.1, or GFP-Kv2.1 P404W were collected in TIRF. Images were converted into BMP file format and 1 out of every 10 frames were imported into PIVlab (creating a time lapse of 320 ms). Contrast-limited adaptive histogram equalization (contrast enhancement) was engaged, and frame pairs were analyzed with three successive passes, utilizing interrogation areas of 64, 32, and 16 pixels. From an ROI drawn within the center of each cell analyzed, average velocity magnitude values (reported as pixels per frame) were collected.
For all analyses, values were imported into GraphPad Prism for presentation and statistical analysis as noted. For IHC experiments, we define biological replicates as individual animals. The data sets in this article involving IHC contain biological replicates. For experiments performed with cells in culture, we define biological replicates as experiments performed on different days and technical replicates as experiments performed on the same day. The data sets in this article involving cells in culture contain biological and/or technical replicates.
Supplementary Material
Acknowledgments
We thank Marina Besprozvannaya, Eamonn Dickson, Karl Murray, Jodi Nunnari, and Nicholas Vierra for helpful advice and critical discussion. We also thank Nicholas Vierra for critical reading of the manuscript. We acknowledge Yongam Lee and Steve Wiler for expert molecular biology technical assistance. We also thank Michael Paddy at the UC Davis MCB Imaging Facility for expert advice on imaging. We thank Grace Or for help in preparing cultured hippocampal neurons. GxTX was synthesized at the Molecular Foundry of the Lawrence Berkeley National Laboratory under U.S. Department of Energy Contract DE-AC02-05CH11231. This research was funded by NIH R01 NS042225 and NIH U01 NS090581 to J.S.T. and NIH R01 NS096317 to J.T.S. M.K. was supported by NIH T32 GM0007377. P.T. was supported by American Heart Association postdoctoral fellowship 17POST33670698.
Abbreviations used:
- AIS
axon initial segment
- ankG
ankyrin G
- CHNs
cultured hippocampal neurons
- DIV
days in vitro
- EPJ
ER–PM junction
- ER
endoplasmic reticulum
- E-Syt
extended synaptotagmin
- FBS
fetal bovine serum
- GxTX
Guangxitoxin-1E
- GxTX-633
Guangxitoxin–DyLight633 conjugate
- HBSS
Hank’s balanced saline solution
- IgG
immunoglobulin G
- IHC
immunohistochemistry
- JP
junctophilin
- Kv
voltage-gated K +
- Kv2 dkO
Kv2.1/Kv2.2 double KO
- LatA
latrunculin A
- mAb
monoclonal antibody
- MOC
Mander’s overlap coefficient
- NIH
National Institutes of Health
- pAb
polyclonal antibody
- PB
phosphate buffer
- PBS
phosphate-buffered saline
- PCC
Pearson’s correlation coefficient
- PM
plasma membrane
- PRC
proximal restriction and clustering
- Rap
rapamycin
- ROI
region of interest
- RyR
ryanodine receptor
- STIM
stromal interacting molecule
- TEA
tetraethylammonium
- Thap
thapsigargin
- TIRF
total internal reflection fluorescence
- VAPs
vesicle-associated membrane protein-associated proteins
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-05-0337) on August 9, 2018.
REFERENCES
- Antonucci DE, Lim ST, Vassanelli S, Trimmer JS. (2001). Dynamic localization and clustering of dendritic Kv2.1 voltage-dependent potassium channels in developing hippocampal neurons. Neuroscience , 69–81. [DOI] [PubMed] [Google Scholar]
- Balla T. (2018). Ca(2+) and lipid signals hold hands at endoplasmic reticulum–plasma membrane contact sites. J Physiol , 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baver SB, Hope K, Guyot S, Bjorbaek C, Kaczorowski C, O’Connell KM. (2014). Leptin modulates the intrinsic excitability of AgRP/NPY neurons in the arcuate nucleus of the hypothalamus. J Neurosci , 5486–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benndorf K, Koopmann R, Lorra C, Pongs O. (1994). Gating and conductance properties of a human delayed rectifier K+ channel expressed in frog oocytes. J Physiol (Pt 1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsen J. (1986). Dynamic thresholding of gray-level images. Proc 8th Int Conf Pattern Recogn Paris, 1251–1255.
- Besprozvannaya M, Dickson E, Li H, Ginburg KS, Bers DM, Auwerx J, Nunnari J. (2018). GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife , e31019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop HI, Cobb MM, Kirmiz M, Parajuli LK, Mandikian D, Philp AM, Melnik M, Kuja-Panula J, Rauvala H, Shigemoto R, et al. (2018). Kv2 ion channels determine the expression and localization of the associated AMIGO-1 cell adhesion molecule in adult brain neurons. Front Mol Neurosci , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop HI, Guan D, Bocksteins E, Parajuli LK, Murray KD, Cobb MM, Misonou H, Zito K, Foehring RC, Trimmer JS. (2015). Distinct cell- and layer-specific expression patterns and independent regulation of Kv2 channel subtypes in cortical pyramidal neurons. J Neurosci , 14922–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Liou J, Park WS, Meyer T. (2007). STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell , 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco S, Meyer T. (2011). STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem , 973–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O, Trimmer JS. (2011). Activity-dependent phosphorylation of neuronal Kv2.1 potassium channels by CDK5. J Biol Chem , 28738–28748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Chen YJ, Liou J. (2017). ER-plasma membrane junctions: why and how do we study them? Biochim Biophys Acta , 1494–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb MM, Austin DC, Sack JT, Trimmer JS. (2015). Cell cycle-dependent changes in localization and phosphorylation of the plasma membrane Kv2.1 K+ channel impact endoplasmic reticulum membrane contact sites in COS-1 cells. J Biol Chem , 29189–29201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XQ, Manning Fox JE, Chikvashvili D, Casimir M, Plummer G, Hajmrle C, Spigelman AF, Kin T, Singer-Lahat D, Kang Y, et al. (2012). The voltage-dependent potassium channel subunit Kv2.1 regulates insulin secretion from rodent and human islets independently of its electrical function. Diabetologia , 1709–1720. [DOI] [PubMed] [Google Scholar]
- Day RN, Davidson MW. (2009). The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev , 2887–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Brilstra EH, van Kempen MJ, Van’t Slot R, Nijman IJ, Afawi Z, De Jonghe P, Djemie T, Guerrini R, Hardies K, et al. (2016). Targeted sequencing of 351 candidate genes for epileptic encephalopathy in a large cohort of patients. Mol Genet Genomic Med , 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CGF, Syrbe S, Brilstra EH, Verbeek N, Kerr B, Dubbs H, Bayat A, Desai S, Naidu S, Srivastava S, et al. (2017). Neurodevelopmental disorders caused by de novo variants in KCNB1 genotypes and phenotypes. JAMA Neurol , 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson EJ. (2017). Endoplasmic reticulum-plasma membrane contacts regulate cellular excitability. Adv Exp Med Biol , 95–109. [DOI] [PubMed] [Google Scholar]
- Dickson EJ, Jensen JB, Vivas O, Kruse M, Traynor-Kaplan AE, Hille B. (2016). Dynamic formation of ER–PM junctions presents a lipid phosphatase to regulate phosphoinositides. J Cell Biol , 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong WH, Chen JC, He YL, Xu JJ, Mei YA. (2013). Resveratrol inhibits K(v)2.2 currents through the estrogen receptor GPR30-mediated PKC pathway. Am J Physiol Cell Physiol , C547–C557. [DOI] [PubMed] [Google Scholar]
- Du J, Haak LL, Phillips-Tansey E, Russell JT, McBain CJ. (2000). Frequency-dependent regulation of rat hippocampal somato-dendritic excitability by the K+ channel subunit Kv2.1. J Physiol , 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tao-Cheng JH, Zerfas P, McBain CJ. (1998). The K+ channel, Kv2.1, is apposed to astrocytic processes and is associated with inhibitory postsynaptic membranes in hippocampal and cortical principal neurons and inhibitory interneurons. Neuroscience , 37–48. [DOI] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. (2011). A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol , C723–C742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe A, Snyders DJ, Deal KK, Tamkun MM. (1993). Influence of cloned voltage-gated K+ channel expression on alanine transport, Rb+ uptake, and cell volume. Am J Physiol , C1230–C1238. [DOI] [PubMed] [Google Scholar]
- Fox PD, Haberkorn CJ, Akin EJ, Seel PJ, Krapf D, Tamkun MM. (2015). Induction of stable ER–plasma-membrane junctions by Kv2.1 potassium channels. J Cell Sci , 2096–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PD, Loftus RJ, Tamkun MM. (2013). Regulation of Kv2.1 K(+) conductance by cell surface channel density. J Neurosci , 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Jorgensen AO. (1994). Structure and development of E-C coupling units in skeletal muscle. Annu Rev Physiol , 509–534. [DOI] [PubMed] [Google Scholar]
- Frazzini V, Guarnieri S, Bomba M, Navarra R, Morabito C, Mariggio MA, Sensi SL. (2016). Altered Kv2.1 functioning promotes increased excitability in hippocampal neurons of an Alzheimer’s disease mouse model. Cell Death Dis , e2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Dai X, Plummer G, Suzuki K, Bautista A, Githaka JM, Senior L, Jensen M, Greitzer-Antes D, Manning Fox JE, et al. (2017). Kv2.1 clustering contributes to insulin exocytosis and rescues human beta-cell dysfunction. Diabetes , 1890–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Vannier C, Galli T. (2016). Endoplasmic reticulum–plasma membrane associations: structures and functions. Annu Rev Cell Dev Biol , 279–301. [DOI] [PubMed] [Google Scholar]
- Guan D, Armstrong WE, Foehring RC. (2013). Kv2 channels regulate firing rate in pyramidal neurons from rat sensorimotor cortex. J Physiol , 4807–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Tkatch T, Surmeier DJ, Armstrong WE, Foehring RC. (2007). Kv2 subunits underlie slowly inactivating potassium current in rat neocortical pyramidal neurons. J Physiol , 941–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell CA, Jankowska KI, Burkhardt JK, Lewis RS. (2016). Calcium influx through CRAC channels controls actin organization and dynamics at the immune synapse. Elife , e14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegle AP, Marble DD, Wilson GF. (2006). A voltage-driven switch for ion-independent signaling by ether-a-go-go K+ channels. Proc Natl Acad Sci USA , 2886–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkart M, Landis DM, Reese TS. (1976). Similarity of junctions between plasma membranes and endoplasmic reticulum in muscle and neurons. J Cell Biol , 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Liou J, Emr SD. (2015). Molecular mechanisms of inter-organelle ER–PM contact sites. Curr Opin Cell Biol , 123–130. [DOI] [PubMed] [Google Scholar]
- Hermanstyne TO, Kihira Y, Misono K, Deitchler A, Yanagawa Y, Misonou H. (2010). Immunolocalization of the voltage-gated potassium channel Kv2.2 in GABAergic neurons in the basal forebrain of rats and mice. J Comp Neurol , 4298–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanstyne TO, Subedi K, Le WW, Hoffman GE, Meredith AL, Mong JA, Misonou H. (2013). Kv2.2: a novel molecular target to study the role of basal forebrain GABAergic neurons in the sleep-wake cycle. Sleep , 1839–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J, Zhou YP, Bugianesi RM, Dulski PM, Feng Y, Warren VA, Smith MM, Kohler MG, Garsky VM, Sanchez M, et al. (2006). Blockers of the delayed-rectifier potassium current in pancreatic beta-cells enhance glucose-dependent insulin secretion. Diabetes , 1034–1042. [DOI] [PubMed] [Google Scholar]
- Honigsperger C, Nigro MJ, Storm JF. (2017). Physiological roles of Kv2 channels in entorhinal cortex layer II stellate cells revealed by Guangxitoxin-1E. J Physiol , 739–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TS, Chen YJ, Chang CL, Lee WR, Liou J. (2017). Cortical actin contributes to spatial organization of ER–PM junctions. Mol Biol Cell , 3171–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PM, Cunningham AM, Peng YW, Snyder SH. (1993a). CDRK and DRK1 K+ channels have contrasting localizations in sensory systems. Neuroscience , 613–620. [DOI] [PubMed] [Google Scholar]
- Hwang PM, Fotuhi M, Bredt DS, Cunningham AM, Snyder SH. (1993b). Contrasting immunohistochemical localizations in rat brain of two novel K+ channels of the Shab subfamily. J Neurosci , 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PM, Glatt CE, Bredt DS, Yellen G, Snyder SH. (1992). A novel K+ channel with unique localizations in mammalian brain: molecular cloning and characterization. Neuron , 473–481. [DOI] [PubMed] [Google Scholar]
- Ikeda SR, Korn SJ. (1995). Influence of permeating ions on potassium channel block by external tetraethylammonium. J Physiol (Pt 2), 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Dallas ML, Ross FA, Lewis RW, Rafferty JN, David JA, Suman R, Peers C, Hardie DG, Evans AM. (2011). Phosphorylation of the voltage-gated potassium channel Kv2.1 by AMP-activated protein kinase regulates membrane excitability. Proc Natl Acad Sci USA , 18132–18137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immke D, Korn SJ. (2000). Ion-Ion interactions at the selectivity filter. Evidence from K(+)-dependent modulation of tetraethylammonium efficacy in Kv2.1 potassium channels. J Gen Physiol , 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immke D, Wood M, Kiss L, Korn SJ. (1999). Potassium-dependent changes in the conformation of the Kv2.1 potassium channel pore. J Gen Physiol , 819–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. (2005). An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods , 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DA, Kuznetsov A, Lopez JP, Kash S, Ammala CE, Philipson LH. (2007). Kv2.1 ablation alters glucose-induced islet electrical activity, enhancing insulin secretion. Cell Metab , 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CS, Watanabe S, Stas JI, Klaphaak J, Yamane A, Schmitt N, Olesen SP, Trimmer JS, Rasmussen HB, Misonou H. (2017). Trafficking of Kv2.1 channels to the axon initial segment by a novel nonconventional secretory pathway. J Neurosci , 11523–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Perez L, Cidad P, Alvarez-Miguel I, Santos-Hipolito A, Torres-Merino R, Alonso E, de la Fuente MA, Lopez-Lopez JR, Perez-Garcia MT. (2016). Molecular determinants of Kv1.3 potassium channels-induced proliferation. J Biol Chem , 3569–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B, Leek AN, Sole L, Maverick EE, Levine TP, Tamkun MM. (2018). Kv2 potassium channels form endoplasmic reticulum/plasma membrane junctions via interaction with VAPA and VAPB. Proc Natl Acad Sci USA , E7331–E7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J, Griffin SJ, Baker C, Skrzypiec A, Chernova T, Forsythe ID. (2008). Initial segment Kv2.2 channels mediate a slow delayed rectifier and maintain high frequency action potential firing in medial nucleus of the trapezoid body neurons. J Physiol , 3493–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek LK. (2006). Non-conducting functions of voltage-gated ion channels. Nat Rev Neurosci , 761–771. [DOI] [PubMed] [Google Scholar]
- Kihira Y, Hermanstyne TO, Misonou H. (2010). Formation of heteromeric Kv2 channels in mammalian brain neurons. J Biol Chem , 15048–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimm T, Khaliq ZM, Bean BP. (2015). Differential regulation of action potential shape and burst-frequency firing by BK and Kv2 channels in substantia nigra dopaminergic neurons. J Neurosci , 16404–16417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AN, Manning CF, Trimmer JS. (2014). A unique ion channel clustering domain on the axon initial segment of mammalian neurons. J Comp Neurol , 2594–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmiz M, Vierra NC, Palacio S, Trimmer JS. (2018). Identification of VAPA and VAPB as Kv2 channel-interacting proteins defining endoplasmic reticulum–plasma membrane junctions in mammalian brain neurons. J Neurosci , 7562–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Wang JM, Swartz KJ. (2003). Interaction between extracellular Hanatoxin and the resting conformation of the voltage-sensor paddle in Kv channels. Neuron , 527–536. [DOI] [PubMed] [Google Scholar]
- Lees JA, Messa M, Sun EW, Wheeler H, Torta F, Wenk MR, De Camilli P, Reinisch KM. (2017). Lipid transport by TMEM24 at ER–plasma membrane contacts regulates pulsatile insulin secretion. Science . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier C. (2016). The axon initial segment, 50 years later: a nexus for neuronal organization and function. Curr Top Membr , 185–233. [DOI] [PubMed] [Google Scholar]
- Li L, Pan ZF, Huang X, Wu BW, Li T, Kang MX, Ge RS, Hu XY, Zhang YH, Ge LJ, et al. (2016). Junctophilin 3 expresses in pancreatic beta cells and is required for glucose-stimulated insulin secretion. Cell Death Dis , e2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XN, Herrington J, Petrov A, Ge L, Eiermann G, Xiong Y, Jensen MV, Hohmeier HE, Newgard CB, Garcia ML, et al. (2013). The role of voltage-gated potassium channels Kv2.1 and Kv2.2 in the regulation of insulin and somatostatin release from pancreatic islets. J Pharmacol Exp Ther , 407–416. [DOI] [PubMed] [Google Scholar]
- Lim ST, Antonucci DE, Scannevin RH, Trimmer JS. (2000). A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron , 385–397. [DOI] [PubMed] [Google Scholar]
- Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, Loranger B, Moore J, Neves C, Macdonald D, et al. (2010). Metadata matters: access to image data in the real world. J Cell Biol , 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PW, Bean BP. (2014). Kv2 channel regulation of action potential repolarization and firing patterns in superior cervical ganglion neurons and hippocampal CA1 pyramidal neurons. J Neurosci , 4991–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SA, Nerbonne JM. (2002). Delayed rectifier K+ currents, IK, are encoded by Kv2 alpha-subunits and regulate tonic firing in mammalian sympathetic neurons. J Neurosci , 10094–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandikian D, Bocksteins E, Parajuli LK, Bishop HI, Cerda O, Shigemoto R, Trimmer JS. (2014). Cell type-specific spatial and functional coupling between mammalian brain Kv2.1 K(+) channels and ryanodine receptors. J Comp Neurol , 3555–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning CF, Bundros AM, Trimmer JS. (2012). Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PLoS One , e38313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini C, Romoli M, Parrini E, Costa C, Mei D, Mari F, Parmeggiani L, Procopio E, Metitieri T, Cellini E, et al. (2017). Clinical features and outcome of 6 new patients carrying de novo KCNB1 gene mutations. Neurol Genet , e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SW, Chang WP, Sudhof TC. (2007). E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci USA , 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Menegola M, Mohapatra DP, Guy LK, Park KS, Trimmer JS. (2006). Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J Neurosci , 13505–13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Menegola M, Trimmer JS. (2005a). Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J Neurosci , 11184–11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, Anderson AE, Trimmer JS. (2004). Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci , 711–718. [DOI] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Trimmer JS. (2005b). Kv2.1: a voltage-gated K+ channel critical to dynamic control of neuronal excitability. Neurotoxicology , 743–752. [DOI] [PubMed] [Google Scholar]
- Misonou H, Thompson SM, Cai X. (2008). Dynamic regulation of the Kv2.1 voltage-gated potassium channel during brain ischemia through neuroglial interaction. J Neurosci , 8529–8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia F, Zuccolo E, Soda T, Tanzi F, Guerra G, Mapelli L, Lodola F, D’Angelo E. (2015). Stim and Orai proteins in neuronal Ca(2+) signaling and excitability. Front Cell Neurosci , 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra DP, Trimmer JS. (2006). The Kv2.1 C terminus can autonomously transfer Kv2.1-like phosphorylation-dependent localization, voltage-dependent gating, and muscarinic modulation to diverse Kv channels. J Neurosci , 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muennich EA, Fyffe RE. (2004). Focal aggregation of voltage-gated, Kv2.1 subunit-containing, potassium channels at synaptic sites in rat spinal motoneurones. J Physiol , 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shi G, Scannevin RH, Trimmer JS. (1997). Phosphorylation of the Kv2.1 K+ channel alters voltage-dependent activation. Mol Pharmacol , 821–828. [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Trimmer JS. (1999). Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J Neurosci , 1728–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Levine TP. (2016). VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta , 952–961. [DOI] [PubMed] [Google Scholar]
- Nishi M, Sakagami H, Komazaki S, Kondo H, Takeshima H. (2003). Coexpression of junctophilin type 3 and type 4 in brain. Brain Res Mol Brain Res , 102–110. [DOI] [PubMed] [Google Scholar]
- O’Connell KM, Loftus R, Tamkun MM. (2010). Localization-dependent activity of the Kv2.1 delayed-rectifier K+ channel. Proc Natl Acad Sci USA , 12351–12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell KM, Rolig AS, Whitesell JD, Tamkun MM. (2006). Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J Neurosci , 9609–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell KM, Tamkun MM. (2005). Targeting of voltage-gated potassium channel isoforms to distinct cell surface microdomains. J Cell Sci , 2155–2166. [DOI] [PubMed] [Google Scholar]
- Palacio S, Chevaleyre V, Brann DH, Murray KD, Piskorowski RA, Trimmer JS. (2017). Heterogeneity in Kv2 channel expression shapes action potential characteristics and firing patterns in CA1 versus CA2 hippocampal pyramidal neurons. eNeuro , doi: 10.1523/ENEURO.0267-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Mohapatra DP, Misonou H, Trimmer JS. (2006). Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science , 976–979. [DOI] [PubMed] [Google Scholar]
- Patel AJ, Lazdunski M, Honore E. (1997). Kv2.1/Kv9.3, a novel ATP-dependent delayed-rectifier K+ channel in oxygen-sensitive pulmonary artery myocytes. EMBO J , 6615–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak D, Guan D, Foehring RC. (2016). Roles of specific Kv channel types in repolarization of the action potential in genetically identified subclasses of pyramidal neurons in mouse neocortex. J Neurophysiol , 2317–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. (2015). Store-operated calcium channels. Physiol Rev , 1383–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J. (1962). Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol , 405–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack JT, Aldrich RW, Gilly WF. (2004). A gastropod toxin selectively slows early transitions in the Shaker K channel’s activation pathway. J Gen Physiol , 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P. (2017a). Endoplasmic reticulum–plasma membrane contact sites. Annu Rev Biochem , 659–684. [DOI] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P. (2017b). The extended-synaptotagmins. Biochim Biophys Acta , 1490–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Akita T, Tohyama J, Goldberg-Stern H, Kobayashi Y, Cohen R, Kato M, Ohba C, Miyatake S, Tsurusaki Y, et al. (2015). De novo KCNB1 mutations in infantile epilepsy inhibit repetitive neuronal firing. Sci Rep , 15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ponce D, DeFelipe J, Garrido JJ, Munoz A. (2011). In vitro maturation of the cisternal organelle in the hippocampal neuron’s axon initial segment. Mol Cell Neurosci , 104–116. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ponce D, DeFelipe J, Garrido JJ, Munoz A. (2012). Developmental expression of Kv potassium channels at the axon initial segment of cultured hippocampal neurons. PLoS One , e48557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin RH, Murakoshi H, Rhodes KJ, Trimmer JS. (1996). Identification of a cytoplasmic domain important in the polarized expression and clustering of the Kv2.1 K+ channel. J Cell Biol , 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter A, Del Turco D, Deller T, Gutzmann A, Schultz C, Engelhardt M. (2017). Structural plasticity of synaptopodin in the axon initial segment during visual cortex development. Cereb Cortex , 4662–4675. [DOI] [PubMed] [Google Scholar]
- Schmalz F, Kinsella J, Koh SD, Vogalis F, Schneider A, Flynn ER, Kenyon JL, Horowitz B. (1998). Molecular identification of a component of delayed rectifier current in gastrointestinal smooth muscles. Am J Physiol , G901–G911. [DOI] [PubMed] [Google Scholar]
- Shalygin A, Skopin A, Kalinina V, Zimina O, Glushankova L, Mozhayeva GN, Kaznacheyeva E. (2015). STIM1 and STIM2 proteins differently regulate endogenous store-operated channels in HEK293 cells. J Biol Chem , 4717–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, D’Arcangelo G, Kleinlaus A, Halegoua S, Trimmer JS. (1993). Nerve growth factor regulates the abundance and distribution of K+ channels in PC12 cells. J Cell Biol , 1835–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Kleinklaus AK, Marrion NV, Trimmer JS. (1994). Properties of Kv2.1 K+ channels expressed in transfected mammalian cells. J Biol Chem , 23204–23211. [PubMed] [Google Scholar]
- Soboloff J, Spassova MA, Hewavitharana T, He LP, Xu W, Johnstone LS, Dziadek MA, Gill DL. (2006). STIM2 is an inhibitor of STIM1-mediated store-operated Ca2+ Entry. Curr Biol , 1465–1470. [DOI] [PubMed] [Google Scholar]
- Speca DJ, Ogata G, Mandikian D, Bishop HI, Wiler SW, Eum K, Wenzel HJ, Doisy ET, Matt L, Campi KL, et al. (2014). Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav , 394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science , 493–495. [DOI] [PubMed] [Google Scholar]
- Sun XH, Protasi F, Takahashi M, Takeshima H, Ferguson DG, Franzini-Armstrong C. (1995). Molecular architecture of membranes involved in excitation–contraction coupling of cardiac muscle. J Cell Biol , 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Hoshijima M, Song LS. (2015). Ca(2)(+) microdomains organized by junctophilins. Cell Calcium , 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun MM, O’Connell KM, Rolig AS. (2007). A cytoskeletal-based perimeter fence selectively corrals a sub-population of cell surface Kv2.1 channels. J Cell Sci , 2413–2423. [DOI] [PubMed] [Google Scholar]
- Thielicke W, Stamhuis EJ. (2014). Towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J Open Res Software , e30. [Google Scholar]
- Thiffault I, Speca DJ, Austin DC, Cobb MM, Eum KS, Safina NP, Grote L, Farrow EG, Miller N, Soden S, et al. (2015). A novel epileptic encephalopathy mutation in KCNB1 disrupts Kv2.1 ion selectivity, expression, and localization. J Gen Physiol , 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley DC, Eum KS, Fletcher-Taylor S, Austin DC, Dupre C, Patron LA, Garcia RL, Lam K, Yarov-Yarovoy V, Cohen BE, Sack JT. (2014). Chemoselective tarantula toxins report voltage activation of wild-type ion channels in live cells. Proc Natl Acad Sci USA , E4789–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Bersell K, Jorge BS, Bjork RL, Jr, Friedman JR, Bloss CS, Cohen J, Gupta S, Naidu S, Vanoye CG, et al. (2014). De novo KCNB1 mutations in epileptic encephalopathy. Ann Neurol , 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS. (1991). Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc Natl Acad Sci USA , 10764–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS. (2015). Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron , 238–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDongen AM, Frech GC, Drewe JA, Joho RH, Brown AM. (1990). Alteration and restoration of K+ channel function by deletions at the N- and C-termini. Neuron , 433–443. [DOI] [PubMed] [Google Scholar]
- Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. (2007). Visualization and manipulation of plasma membrane–endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem , 29678–29690. [DOI] [PubMed] [Google Scholar]
- Williams RT, Manji SS, Parker NJ, Hancock MS, Van Stekelenburg L, Eid JP, Senior PV, Kazenwadel JS, Shandala T, Saint R, et al. (2001). Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem J , 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Covington ED, Lewis RS. (2014). Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol Biol Cell , 3672–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Whiteus C, Xu CS, Hayworth KJ, Weinberg RJ, Hess HF, De Camilli P. (2017). Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci USA , E4859–E4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek N, Sparks L, Voeltz G. (2011). Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic , 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.