Significance
It remains unclear how the structural properties of polyglutamine (polyQ) proteins, which underlie several neurodegenerative disorders, including Huntington’s disease and spinocerebellar ataxias (SCAs), translate into the toxicity of these proteins. Here, we demonstrate that coiled-coil structures in expanded polyQ regions of SCA type 3 (SCA3) proteins cause dendrite defects in Drosophila neurons, as well as behavioral abnormalities. Moreover, interactions of SCA3 with Foxo mediated by coiled-coil domains of these two proteins resulted in functional impairment of this transcription factor, whereas its overexpression significantly rescued the SCA3-induced defects. Our study expanded the current understanding of neuronal pathology mediated by polyQ proteins via the coiled-coil–mediated interactions. These results may have important implications in therapeutic strategies for polyQ protein-related diseases.
Keywords: polyQ, coiled coil, Foxo, dendrites, neurodegenerative diseases
Abstract
Neurodegenerative disorders, such as Huntington’s diseases and spinocerebellar ataxias (SCAs), are driven by proteins with expanded polyglutamine (polyQ) tracts. Recently, coiled-coil structures in polyQ regions of such proteins were shown to facilitate aggregate formation and ultimately lead to cell death. However, the molecular mechanism linking these structural domains to neuronal toxicity of polyQ proteins remains elusive. Here, we demonstrate that coiled-coil structures in the Q repeat region of SCA type 3 (SCA3) polyQ proteins confer protein toxicity in Drosophila neurons. To functionally characterize coiled-coil structures in the Q repeat regions, we generated three structural variants of SCA3 polyQ proteins: (i) MJDtr-76Q, containing both α-helical coiled-coil and β-sheet hairpin structures in the Q repeat region; (ii) MJDtr-70Q_cc0, possessing only α-helical coiled-coil structures due to the incorporation of β-sheet–breaking residues (Q-to-N or Q-to-E mutations); and (iii) MJDtr-70Q_pQp, with no secondary structure due to the introduced proline residues (Q-to-P mutations). Through comparative analysis of these variants, we found that coiled-coil structures facilitated nuclear localization of SCA3 polyQ proteins and induced dendrite defects in Drosophila dendritic arborization neurons. Furthermore, genetic and functional screening identified the transcription factor Foxo as a target of polyQ proteins, and coiled-coil–mediated interactions of Foxo and polyQ proteins in the nucleus resulted in the observed dendrite and behavioral defects in Drosophila. These results demonstrate that coiled-coil structures of polyQ proteins are crucial for their neuronal toxicity, which is conferred through coiled-coil to coiled-coil interactions with the nuclear targets of these proteins.
Polyglutamine (polyQ) diseases are neurodegenerative disorders that are characterized by the expansion of Q repeats in the associated proteins, such as those underlying spinocerebellar ataxias (SCAs) and the Huntingtin protein (HTT) (1, 2). The accumulation of misfolded polyQ proteins caused by Q expansion confers protein toxicity that results in neuropathic features, such as dendrite defects, leading to impaired neuronal functions and neuronal cell death (3, 4). Numerous studies (5, 6) have reported that aberrant dendritic morphologies are linked to defects in brain function, such as memory storage and cognition. Because pathological dendritic features are often observed in patients with polyQ diseases (7, 8), it is important to understand how misfolded polyQ proteins contribute to dendrite pathology.
Cellular and molecular mechanisms linking polyQ protein toxicity to dendrite pathology have been uncovered in models of several polyQ diseases. PolyQ protein toxicity can be conferred through the impairment of several cellular components, such as the endosomal machinery or cytoskeleton. For example, Richards et al. (9) showed that dendrite defects were associated with perturbations of the Rab11-dependent endosomal recycling system in a Drosophila model of Huntington’s disease. In addition, we previously demonstrated that dendrite defects were associated with perturbed actin cytoskeletal structure and impaired subcellular distribution of Golgi outposts in Drosophila models of SCA type 3 (SCA3), also known as Machado–Joseph disease (MJD) (10, 11). Despite these efforts, the molecular mechanism of polyQ protein toxicity and the link to the resulting dendrite pathology remain largely unknown.
The structure of polyQ proteins is one of the determining factors in the associated toxicity, but it is difficult to uncover the structural features of misfolded polyQ proteins using X-ray crystallography due to the high propensity of these proteins to form aggregates of random size. A number of previous studies have proposed β-sheets as the primary structural motif within expanded polyQ regions based on electron microscopy observations and computational predictions (12, 13). In contrast, Fiumara et al. (14) showed that substitution of several Qs with β-sheet–breaking residues did not hinder aggregation of HTT proteins, and that coiled-coil structures were present within the Q repeat regions. These results suggest that coiled-coil domains could contribute to the aggregation of HTT proteins. However, these data were obtained using yeast and HEK293T cells, and thus it is not clear whether these findings also apply to neurons.
Here, we demonstrate that coiled-coil domains of polyQ-expanded SCA3 proteins contribute to their toxicity in Drosophila dendritic arborization (da) sensory neurons, which share many morphological and functional features with some mammalian neurons (15). We identify a protein target of SCA3 for coiled-coil–mediated interaction, and thus present a molecular mechanism of how protein toxicity induced by coiled-coil structures leads to dendrite defects and behavioral abnormalities in this model system.
Results
Coiled-Coil Structures in the Q Repeat Region of SCA3 polyQ Proteins Contribute to Dendrite Defects.
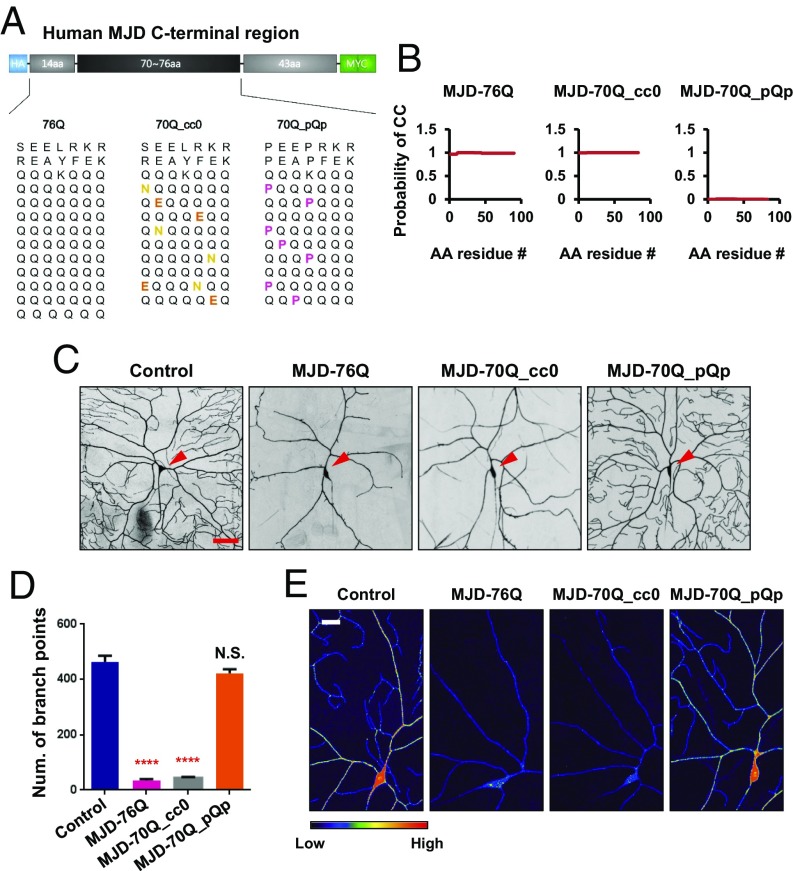
To find out whether coiled-coil structures of SCA3 polyQ proteins contribute to protein toxicity in neurons, we examined the effects of their coiled-coil structures on dendrite defects in Drosophila class IV da (C4da) neurons. We generated transgenic fly lines expressing the following three structural variants of the Q repeat region of truncated SCA3 polyQ proteins: (i) MJDtr-76Q, containing both α-helical coiled-coil and β-sheet hairpin structures in the Q repeat region; (ii) MJDtr-70Q_cc0, containing only α-helical coiled-coil structures and lacking β-sheet motifs due to the insertion of β-sheet–breaking mutations (Q-to-N or Q-to-E); and (iii) MJDtr-70Q_pQp, with no secondary structure due to the insertion of proline residues (Q-to-P) (Fig. 1A). The constructs for MJDtr-70Q_cc0 and MJDtr-70Q_pQp were designed based on the corresponding previously reported constructs of Q repeat regions of HTT protein, cc0 and pQp, respectively (14). We then estimated the supersecondary structures of these three structural variants in their Q repeat regions using the COILS program (16), as previously described (14). MJDtr-76Q and MJDtr-70Q_cc0 were predicted to have coiled-coil structures (P ≥ 0.967), while MJDtr-70Q_pQp was predicted to have no coiled-coil structures (P ≤ 0.007) (Fig. 1B). The prediction for MJDtr-70Q_pQp was consistent with the circular dichroism measurements for HTT proteins of similar composition (92% sequence similarity) within the Q repeat regions (14). Moreover, we evaluated the β-sheet formation ability of the three SCA3 variants expressed in fly brains using the thioflavin S staining method (17). Thioflavin S+ puncta were observed only in the fly brains expressing MJDtr-76Q proteins but not in those expressing the other two variants (SI Appendix, Fig. S1 A–C), suggesting that MJDtr-70Q_cc0 and MJDtr-70Q_pQp proteins may not contain β-sheet structures.
Fig. 1.
Coiled-coil structures of SCA3 polyQ proteins confer protein toxicity that induces dendrite defects. (A) Construction of three structural variants of truncated SCA3 proteins (MJDtr-76Q, MJDtr-70Q_cc0, and MJDtr-70Q_pQp). Sequences for the 14 N-terminal flanking amino acids and Q repeat regions are displayed in a heptad repeat pattern. Replaced amino acids are marked in yellow (Q-to-N, β-sheet–breaking mutation), orange (Q-to-E, β-sheet–breaking mutation), or pink (Q-to-P, secondary structure-breaking mutation). (B) Probabilities to form coiled-coil structure in three structural variants calculated by COILS. The y axes indicate the predicted probability to form coiled-coil structures, and x axes indicate the amino acid residue numbers. (C) Dendrite images of control C4da neurons (Leftmost panel) and C4da neurons overexpressing MJDtr-76Q (second from Left), MJDtr-70Q_cc0 (third from Left), or MJDtr-70Q_pQp (Rightmost panel). (UAS-CD4td-GFP/+;ppk-Gal4/+, UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-76Q, UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-70Q_cc0, and UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-70Q_pQp). Arrowheads (red) indicate cell bodies of C4da neurons. (Scale bar, 50 μm.) (D) Quantification of the number of dendrite branch points of C4da neurons expressing transgenes described in E. N.S., not significant; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey post hoc test; error bars, SEM; n = 6 neurons. (E) PM intensity of C4da neurons expressing transgenes described in E. Intensities of CD4-tdGFP are presented in pseudo color images converted by ImageJ. (Scale bar, 20 μm.)
Next, we compared terminal dendrite morphology and branch points of the control (w1118) C4da neurons and C4da neurons expressing the three structural SCA3 variants inserted in the same genomic locus. C4da neurons expressing MJDtr-76Q or MJDtr-70Q_cc0 showed a severe reduction in the number of terminal dendrites compared with the control, whereas C4da neurons expressing MJDtr-70Q_pQp exhibited no noticeable changes in dendrite morphology (Fig. 1C). Similar reductions were observed in the number of dendrite branch points (Fig. 1D). We then checked protein levels of these three transgenes after expressing them in adult fly brains using elav-Gal4. Western blot analysis showed that the total amounts of MJDtr-76Q and MJDtr-70Q_cc0 proteins were substantially higher than those of MJDtr-70Q_pQp proteins (SI Appendix, Fig. S1D). Thus, the lack of apparent dendrite defects in C4da neurons expressing MJDtr-70Q_pQp proteins could be due to the low total amount of the polyQ proteins. To test this possibility, we doubled the amount of the polyQ proteins by overexpressing two copies of UAS-MJDtr-70Q_pQp transgenes in C4da neurons. Dendrite morphology and the number of dendrite branch points were barely affected even by the increased amount of MJDtr-70Q_pQp proteins (SI Appendix, Fig. S1 E and F). These data suggest that coiled-coil structures in the Q repeat regions of MJDtr-76Q and MJDtr-70Q_cc0 proteins contribute to dendrite defects and also increase polyQ protein amounts, but without coiled-coil structures, the increased amount of polyQ proteins (MJDtr-70Q_pQp) seems to be not a critical factor contributing to dendrite defects.
Previously, we reported that F-actin structures were disrupted by pathogenic SCA3 polyQ proteins leading to dendrite defects (10). Thus, we examined whether expression of MJDtr-76Q or MJDtr-70Q_cc0 proteins induces the disruption of F-actin cytoskeletal structures in da neuronal clusters, and found that F-actin cytoskeletal structures of distal dendrites were substantially disrupted in da neuronal clusters expressing MJDtr-76Q and MJDtr-70Q_cc0 but not in those expressing MJDtr-70Q_pQp, compared with those in the control da neuronal clusters (SI Appendix, Fig. S1 G and H). In another previous study, we showed that the plasma membrane (PM) protein supply was severely reduced by pathogenic SCA3 polyQ proteins, leading to impaired elongation of dendrite terminals in C4da neurons (11). We thus examined alterations of the PM protein supply in C4da neurons expressing the three structural variants and found that this supply was diminished in the neurons expressing MJDtr-76Q or MJDtr-70Q_cc0 but not in those expressing MJDtr-70Q_pQp (Fig. 1E and SI Appendix, Fig. S1I). Taken together, these results suggest that coiled-coil structures in the Q repeat regions of MJDtr-76Q and MJDtr-70Q_cc0 proteins contribute to pathological dendrite phenotypes, including aberrant terminal dendrite morphology, impaired F-actin cytoskeleton, and diminished PM protein supply.
Coiled-Coil Structures of SCA3 polyQ Proteins Promote Their Nuclear Localization.
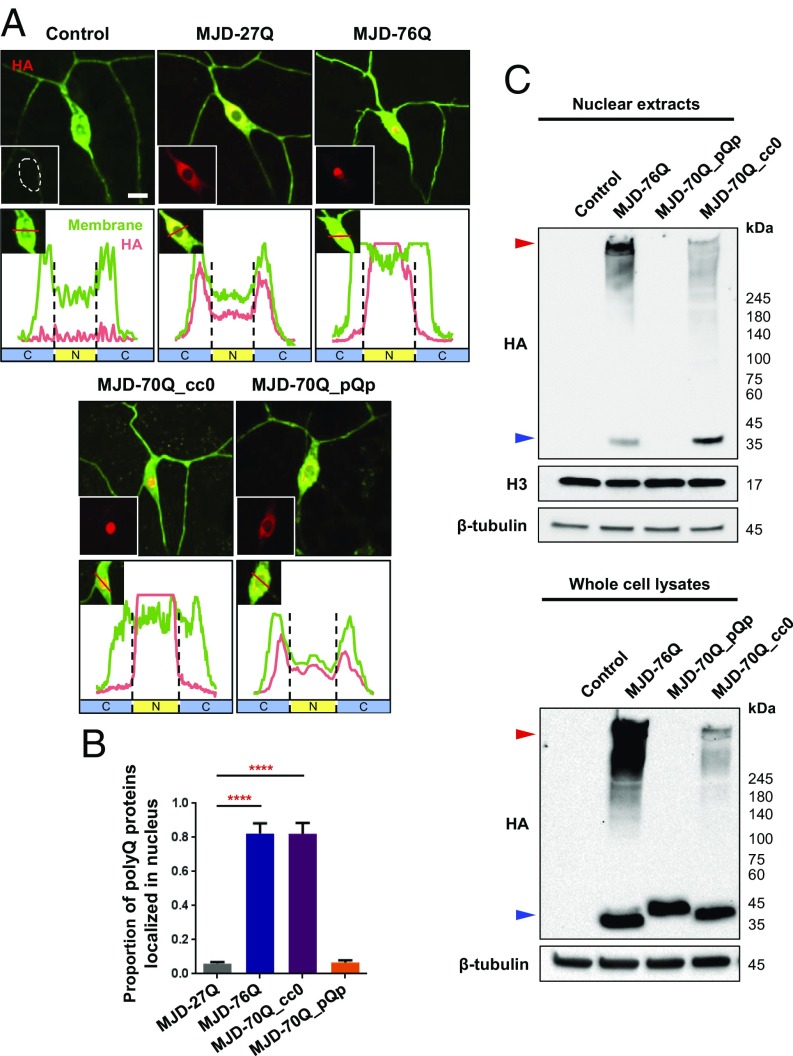
Nuclear localization of SCA3 polyQ proteins is critical for their toxicity and the associated dendrite defects in C4da neurons (10, 11). As the coiled-coil structures contributed to this toxicity, we next asked whether coiled-coil structures might facilitate the localization of SCA3 polyQ proteins to the nucleus. To this end, we examined the subcellular localization of the three SCA3 structural variants in C4da neurons, and found that MJDtr-76Q and MJDtr-70Q_cc0 proteins were primarily localized to the nucleus, whereas MJDtr-70Q_pQp proteins were diffusely distributed in the cytoplasm (Fig. 2A). The nuclear-localized proportions of MJDtr-76Q and MJDtr-70Q_cc0 proteins were significantly (P < 1.0 × 10−4) larger than those of MJDtr-70Q_pQp proteins (Fig. 2B). We also examined the subcellular localization of MJDtr-27Q proteins, as a nonpathogenic control, that have coiled-coil domains with a normal repeat of Qs (SI Appendix, Fig. S2A) (18). MJDtr-27Q proteins were diffusely distributed in the cytoplasm with a marginal nuclear-localized proportion, similar to MJDtr-70Q_pQp proteins (Fig. 2 A and B). These results suggest that coiled-coil domains with only expanded repeats of Qs (MJDtr-76Q and MJDtr-70Q_cc0) appear to facilitate the nuclear localization of SCA3 polyQ proteins, although other biophysical properties of polyQ proteins may also contribute to their subcellular localization. To corroborate this finding, we performed Western blot analysis of nuclear lysates from pan-neuronal cells expressing the three SCA3 structural variants using the elav-Gal4 driver. Consistently, significant amounts of MJDtr-70Q_cc0 and MJDtr-76Q proteins, but not MJDtr-70Q_pQp proteins, were detected in the nuclear lysates (Fig. 2C). MJDtr-70Q_pQp proteins were detected only in cytoplasmic lysates (SI Appendix, Fig. S2B). Certain amounts of monomers and insoluble aggregates of MJDtr-70Q_cc0 and MJDtr-76Q were detected in cytoplasmic lysates, suggesting that these two proteins are primarily enriched in the nucleus, but are not absent from the cytoplasm, as also indicated by the immunohistochemistry data (Fig. 2A).
Fig. 2.
Coiled-coil structures of SCA3 polyQ proteins promote their nuclear localization. (A) Immunohistochemical images representing subcellular localization of SCA3 proteins in C4da neurons expressing MJDtr-27Q, MJDtr-76Q, MJDtr-70Q_pQp, or MJDtr-70Q_cc0 proteins (UAS-CD4td-GFP/+;ppk-Gal4/+, UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-27Q, UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-76Q, UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-70Q_cc0, and UAS-CD4td-GFP/+;ppk-Gal4/UAS-MJDtr-70Q_pQp). (Upper five panels of each denoted genotype) Merged images of SCA3 proteins (red) with membrane marker, mCD8-GFP (green). Left Lower Insets represent cell body regions, and the white dashed line in the control C4da neuron indicates the outline of a cell body. (Scale bar, 5 μm.) (Lower five panels of each denoted genotype) Intensity profiles of fluorescent signals representing SCA3 proteins (red) and mCD8-GFP (green) across cell bodies along red lines (Left Upper Insets). Black dashed lines distinguish nucleus and cytoplasm. N (yellow) indicates nucleus. C (blue) indicates cytoplasm. (B) Comparison of nuclear proportions of SCA3 polyQ proteins among C4da neurons expressing transgenes described in A. ****P < 1.0 × 10−4 by one-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 6 neurons. (C) Western blot analyses using whole cell lysates and nuclear extracts of adult fly heads (elav-Gal4/+, elav-Gal4/MJDtr-76Q, elav-Gal4/MJDtr-70Q_cc0, and elav-Gal4/MJDtr-70Q_pQp). SCA3 variants were immunoblotted using anti-HA antibody. Histone 3 (H3) is used as the loading control for nuclear extracts, and β-tubulin is used as the loading controls for whole cell lysates and nuclear extracts. Red arrowheads indicate insoluble aggregates of SCA3 polyQ proteins. Blue arrowheads indicate monomers of SCA3 polyQ proteins. n = 3 independent experiments.
Interaction of Forkhead Box, Subgroup O with SCA3 polyQ Proteins in the Nucleus Leads to Its Functional Impairment and Ensuing Dendrite Defects.
Next, we searched for molecular targets of nuclear-localized MJDtr-76Q and MJDtr-70Q_cc0 proteins that mediated the observed dendrite defects. It has been suggested that polyQ proteins can target cellular proteins through coiled-coil to coiled-coil interactions (19). Furthermore, it was previously reported that the targets of mutant HTT proteins had a tendency to contain high amounts of coiled-coil structures (14). Based on these reports, we hypothesized that coiled-coil structures in the Q repeat region mediate interactions of nuclear MJDtr-76Q and MJDtr-70Q_cc0 proteins with other coiled-coil–rich proteins. Thus, we searched for transcription factors (TFs) that could regulate dendrite morphology and interact with MJDtr-76Q and MJDtr-70Q_cc0 proteins. To this end, we first identified 58 TFs that are involved in dendrite morphogenesis based on gene ontology biological processes and the previous literature (11, 20–26). We then selected 13 TFs with the largest numbers of amino acids in their coiled-coil domains (Table 1), and these TFs were tested for their effect on dendrite morphology. Among the 13 TFs, knockdown of 6 TFs [Forkhead box, subgroup O (Foxo), Cut, Kayak (Kay), Nubbin (Nub), Nejire (Nej or CBP), and Cap-n-collar (Cnc)] showed a significant (>25%) reduction in the number of dendrite branch points in C4da neurons (Fig. 3A). Moreover, overexpression of three TFs [Foxo, Cut, and E2F transcription factor 1 (E2f1)] led to the most significant (>fourfold) restoration in the number of dendrite branch points in C4da neurons expressing MJDtr-70Q_cc0 (Fig. 3B). Therefore, for further studies, we focused on Foxo and Cut as their knockdown and overexpression showed the opposite effects on dendrite morphology (Fig. 3C and SI Appendix, Fig. S3 A and B).
Table 1.
The 15 coiled-coil–rich transcription factor candidates were screened to identify the most potent interactor of SCA3 polyQ proteins
| Symbol | Name | Dendrite morphogenesis source | No. of amino acids in coiled-coil domain (maximum) | RNAi effect | Rescue effect | Interaction with polyQ proteins |
| Pros | Prospero | (21) | 67 | + | N.S. | E.L.N. |
| Acf | ATP-dependent chromatin assembly factor large subunit | (22) | 61 | N/A | N/A | N/A |
| Foxo | Forkhead box, subgroup O | (23) | 36 | +++ | +++ | YES |
| Cut | Cut | (20) | 35 | +++ | +++ | E.L.N. |
| Kay | Kayak | (23) | 28 | +++ | ++ | E.L.N. |
| Dsx | Doublesex | (24) | 28 | ++ | ++ | N/A |
| Nub | Nubbin | (23) | 28 | +++ | ++ | N/A |
| E2f1 | E2F transcription factor 1 | (22) | 25 | N.S. | +++ | E.L.N. |
| Nej | Nejire | (11) | 25 | +++ | N.S. | YES |
| Rel | Relish | (23) | 21 | ++ | ++ | N/A |
| Med | Medea | (25) | 21 | ++ | + | N/A |
| Exex | Extraextra | (26) | 21 | ++ | N.S. | N/A |
| Cnc | Cap-n-collar | (23) | 21 | +++ | ++ | N/A |
“RNAi effect” refers to the degree of change in the number of dendrite branch points in C4da neurons, when RNAi against TF was expressed by ppk1a-Gal4. RNAi effect was scaled as follows: “N.S.” (not significant), 0–5% decrease in the number of dendrite branch points of C4da neurons expressing RNAi compared with that in control C4da neurons; “+,” 5–15% decrease; “++,” 15–25% decrease; and “+++,” more than 25% decrease. N/A, Not available. “Rescue effect” refers to the degree of restoration in the number of dendrite branch points in C4da neurons, when each TF is overexpressed in larvae of UAS-CD4-tdGFP/+;ppk-Gal4,UAS-MJDtr-70Q_cc0/+. Rescue effect was scaled as follows: “N.S.,” one- to twofold restoration in the number of dendrite branch points in C4da neurons expressing a TF compared with that in C4da neurons expressing MJDtr-70Q_cc0; “+,” two- to threefold restoration; “++,” three- to fourfold restoration; and “+++,” more than fourfold restoration. “E.L.N” and “YES” in “Interaction with polyQ proteins” indicate that the TF is endogenously located in the nucleus and that the TF is colocalized with MJDtr-76Q and MJDtr-70Q_cc0 proteins in the nucleus, respectively.
Fig. 3.
Coiled-coil–rich Foxo interacts with SCA3 polyQ proteins, leading to its functional impairment. (A) Quantification of the number of dendrite branch points in C4da neurons expressing the denoted RNAi lines (see Materials and Methods). Each RNAi and UAS-CD4-tdGFP were expressed using ppk1a-Gal4. The yellow line indicates 25% reduction in the number of dendrite branch points compared with that in control C4da neurons. N.S., not significant; *P < 0.05; **P < 1.0 × 10−2; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 3 neurons. (B) Quantification of the number of dendrite branch points in C4da neurons overexpressing MJDtr-70Q_cc0 or MJD-70Q_cc0+ the denoted TFs (see Materials and Methods). Each TF and UAS-MJDtr-70Q_cc0 was coexpressed by ppk-Gal4. The yellow line indicates a fourfold increase in the number of dendrite branch points compared with that in C4da neurons expressing MJDtr-70Q_cc0 alone. N.S., not significant; *P < 0.05; **P < 1.0 × 10−2; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 4 neurons. (C) Dendrite images of C4da neurons expressing MJDtr-70Q_cc0 (Upper) or MJDtr-70Q_cc0 + Foxo (Lower) (UAS-CD4-tdGFP/+;ppk-Gal4,UAS-MJDtr-70Q_cc0/+ and UAS-CD4-tdGFP/UAS-Foxo;ppk-Gal4,UAS-MJDtr-70Q_cc0/+). Arrowheads (red) indicate cell bodies. (Scale bar, 50 μm.) (D) Subcellular localization of Foxo (green) and SCA3 structural variants (red) in C4da neurons overexpressing Foxo, MJDtr-27Q + Foxo, MJDtr-76Q + Foxo, MJDtr-70Q_cc0 + Foxo, or MJDtr-70Q_pQp + Foxo (ppk-Gal4,UASp-Foxo-GFP/+, ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-27Q, ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-76Q, ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-70Q_cc0, and ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-70Q_pQp). (Scale bar, 5 μm.) Outer and inner dashed lines (white) indicate the outlines of the cell body and nucleus. (E) Comparison of nuclear proportions of Foxo among C4da neurons expressing transgenes described in D. N.S., not significant; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 12 neurons. (F) Representative image of coimmunoprecipitation experiments using adult fly heads (UAS-Foxo-V5/+;elav-GeneSwitch-Gal4/+, UAS-Foxo-V5/+;elav-GeneSwitch-Gal4/UAS-MJDtr-76Q, UAS-Foxo-V5/+;elav-GeneSwitch-Gal4/UAS-MJDtr-70Q_cc0, and UAS-Foxo-V5/+;elav-GeneSwitch-Gal4/UAS-MJDtr-70Q_pQp). Top and Middle indicate immunoprecipitated insoluble aggregates and monomeric forms of HA-tagged SCA3 proteins, respectively. Bottom shows V5-tagged Foxo proteins. n = 3 independent experiments. (G) mRNA expression levels of Foxo target genes (Kay, Xnp, Dad, Khc, and Snama) measured by RT-PCR analysis in adult fly heads (elav-Gal4/+, elav-Gal4/UAS-MJDtr-76Q, elav-Gal4/UAS-MJDtr-70Q_cc0, and elav-Gal4/UAS-MJDtr-70Q_pQp). N.S., not significant; *P < 0.05; **P < 1.0 × 10−2; ***P < 1.0 × 10−3 by two-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 3 independent experiments.
We then examined whether Foxo and Cut are colocalized with MJDtr-76Q and MJDtr-70Q_cc0 proteins. Overexpressed Foxo proteins were located in both the cytoplasm and the nuclei of control C4da neurons (Discussion and Fig. 3D). However, when MJDtr-76Q or MJDtr-70Q_cc0 was co-overexpressed with Foxo, Foxo proteins were substantially localized in the nucleus, and the proportion of Foxo colocalized with MJDtr-76Q and MJDtr-70Q_cc0 proteins increased significantly (P < 1.0 × 10−4) (Fig. 3 D and E). In contrast, in C4da neurons expressing MJDtr-27Q or MJDtr-70Q_pQp, there was no apparent increase in nuclear localization of Foxo proteins (Fig. 3 D and E). These results suggest that MJDtr-76Q and MJDtr-70Q_cc0 proteins interact with Foxo and facilitate their nuclear localization in C4da neurons. Furthermore, in coimmunoprecipitation (co-IP) experiments on samples from heads of adult flies co-overexpressing Foxo and MJDtr-76Q, MJDtr-70Q_cc0, or MJDtr-70Q_pQp, respectively, we identified the interaction between Foxo and insoluble aggregates of MJDtr-76Q and MJDtr-70Q_cc0 proteins (Fig. 3F). Taken together, these data suggest that Foxo is one of the interactors of MJDtr-76Q and MJDtr-70Q_cc0, possibly through coiled-coil to coiled-coil interactions, with strong effects on dendrite morphology. For the other candidate Cut, we showed that Cut also genetically interacted with MJDtr-76Q and MJDtr-70Q_cc0 proteins, but were unable to measure its physical interactions with polyQ proteins due to the lack of anti-Cut antibodies for co-IP experiments (Discussion).
Our results suggest that the interaction of Foxo with MJDtr-76Q or MJDtr-70Q_cc0 proteins increased nuclear localization of this TF (Fig. 3 D–F). Thus, we determined whether interaction of Foxo with MJDtr-76Q or MJDtr-70Q_cc0 could affect its transcriptional activity in neurons. To this end, we selected five representative transcriptional target genes of Foxo [Kayak (Kay), Xnp, Daughters against dpp (Dad), Kinesin heavy chain (Khc), and Something that sticks like glue (Snama)] that are involved in regulation of dendrite morphogenesis or neurogenesis, and compared their transcription levels in the brains of control flies to those in the brains of flies expressing MJDtr-76Q, MJDtr-70Q_cc0, or MJDtr-70Q_pQp. The mRNA levels of these target genes were significantly (P < 0.05) reduced in flies expressing MJDtr-76Q or MJDtr-70Q_cc0 but not MJDtr-70Q_pQp, compared with those in control flies (Fig. 3G and SI Appendix, Fig. S3C). These results suggest that the interaction of nuclear Foxo with polyQ proteins containing coiled-coil domains impairs its transcriptional regulation of dendrite morphogenesis.
Coiled-Coil Domains of Foxo Are Required for Its Interaction with SCA3 polyQ Proteins.
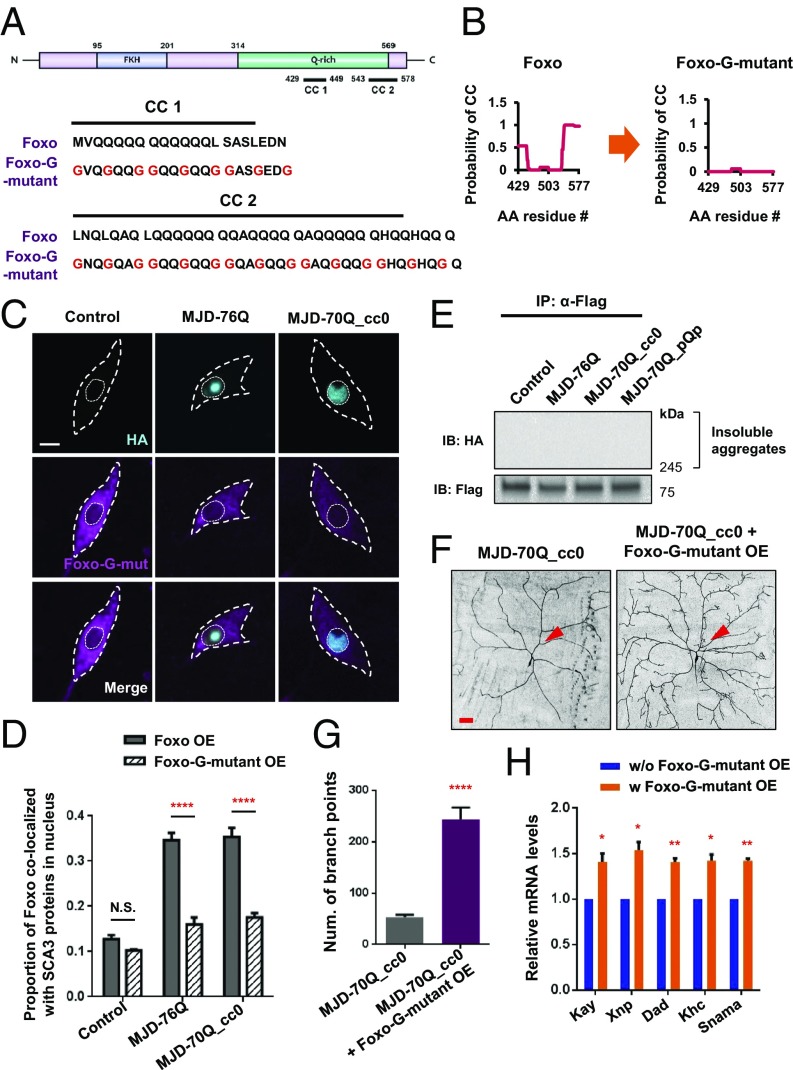
To investigate whether the interaction of Foxo with MJDtr-76Q and MJDtr-70Q_cc0 proteins involves coiled-coil to coiled-coil interactions, we generated the following two transgenic fly lines with disrupted coiled-coil domains in Foxo: (i) a fly line expressing Foxo-Glycine (Foxo-G) mutants with the substitution of three residues important for coiled-coil structure formation with glycine—a known breaker of this structure—in heptad repeats (Materials and Methods and Fig. 4A), and (ii) a fly line expressing Foxo-Deletion (Foxo-Del) mutants with the complete deletion of coiled-coil domains (SI Appendix, Fig. S4A). Using the COILS program, we found that Foxo-G and Foxo-Del mutants were unable to form coiled-coil structures (Fig. 4B and SI Appendix, Fig. S4A). We next examined the colocalization of Foxo mutants co-overexpressed with MJDtr-76Q or MJDtr-70Q_cc0 proteins in C4da neurons. The results showed that the nuclear localization of Foxo-G (Fig. 4 C and D) or Foxo-Del mutants (SI Appendix, Fig. S4 B and C) was significantly (P < 1.0 × 10−4) reduced compared with that of the native protein (Fig. 3D). Consistently, co-IP experiments demonstrated that the interaction of Foxo-G with MJDtr-76Q or MJDtr-70Q_cc0 was significantly reduced (Fig. 4E) compared with the native protein (Fig. 3F). Moreover, we examined whether Foxo-G mutants could restore dendrite defects caused by MJDtr-70Q_cc0 proteins. Overexpression of Foxo-G mutant proteins in C4da neurons expressing MJDtr-70Q_cc0 significantly (P < 1.0 × 10−4) restored the defects in dendrite morphology and the number of dendrite branch points (Fig. 4 F and G). We then examined whether the transcriptional activity of Foxo reduced by MJDtr-70Q_cc0 proteins could also be restored by overexpression of Foxo-G mutant proteins. mRNA levels of Foxo target genes (Kay, Xnp, Dad, Khc, and Snama) down-regulated by MJDtr-70Q_cc0 proteins were significantly (P < 0.05) restored by co-overexpression of Foxo-G mutant proteins (Fig. 4H). These data indicate that coiled-coil to coiled-coil interactions mediate the interaction of Foxo with MJDtr-76Q and MJDtr-70Q_cc0 proteins, contributing to dendrite pathology.
Fig. 4.
Disruption of coiled-coil structures in Foxo abolishes its interaction with SCA3 polyQ proteins. (A) Amino acid changes in Foxo-G mutants within the Q-rich domain of Foxo. Foxo contains a Forkhead (FKH) domain—a DNA-binding domain—in the N-terminal region, and a Q-rich domain including two coiled-coil domains in the C-terminal region. Amino acid sequences of two coiled-coil domains (CC1 and CC2) of wild-type Foxo and Foxo-G mutant proteins are shown in a heptad repeat pattern. a/d/g residues of coiled-coil heptad repeats are replaced with glycines (Gs), which are known as coiled-coil breakers. Replaced amino acids are marked in red. (B) Probabilities to form coiled-coil structure in Foxo-G mutants calculated by COILS. The y axis indicates probability to form coiled-coil structure, and x axis indicates the amino acid residue numbers. (C) Subcellular localization of Foxo-G mutant proteins (magenta) and SCA3 structural variants (cyan) in C4da neurons overexpressing Foxo-G mutant, MJDtr-76Q + Foxo-G mutant, or MJDtr-70Q_cc0 + Foxo-G mutant (UAS-Foxo-G mutant-Flag/UAS-CD4-tdGFP;ppk-Gal4/+, UAS-Foxo-G mutant-Flag/UAS-CD4-tdGFP;ppk-Gal4,UAS-MJDtr-76Q/+, and UAS-Foxo-G mutant-Flag/UAS-CD4-tdGFP;ppk-Gal4,UAS-MJDtr-70Q_cc0/+). (Scale bar, 5 μm.) Outer and inner dashed lines (white) indicate the outlines of the cell body and nucleus. (D) Comparison of nuclear proportions of wild-type Foxo and Foxo-G mutants colocalized with SCA3 proteins among C4da neurons expressing the transgenes described in C. N.S., not significant; ****P < 1.0 × 10−4 by two-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 12 neurons. (E) Representative image of co-IP experiments using adult fly heads (UAS-Foxo-G mutant-Flag/+;elav-GeneSwitch-Gal4/+, UAS-Foxo-G mutant-Flag/+;elav-GeneSwitch-Gal4/UAS-MJDtr-76Q, UAS-Foxo-G mutant-Flag/+;elav-GeneSwitch-Gal4/UAS-MJDtr-70Q_cc0, and UAS-Foxo-G mutant-Flag/+;elav-GeneSwitch-Gal4/UAS-MJDtr-70Q_pQp). Upper and Lower panels show insoluble aggregates of HA-tagged SCA3 proteins and Flag-tagged Foxo-G mutant proteins, respectively. n = 3 independent experiments. (F) Dendrite images of C4da neurons expressing MJDtr-70Q_cc0 (Left) or MJDtr-70Q_cc0 + Foxo-G mutant (Right) (UAS-CD4-tdGFP/+;ppk-Gal4,UAS-MJDtr-70Q_cc0/+ and UAS-CD4-tdGFP/UAS-Foxo-G mutant-Flag;ppk-Gal4,UAS-MJDtr-70Q_cc0/+). Arrowheads (red) indicate cell bodies. (Scale bar, 50 μm.) (G) Quantification of the number of dendrite branch points of C4da neurons expressing the transgenes described in F. ****P < 1.0 × 10−4 by Student’s t test; error bars, SEM; n ≥ 5 neurons. (H) mRNA levels of Foxo target genes (Kay, Xnp, Dad, Khc, and Snama) measured by RT-PCR analysis in adult fly heads (elav-Gal4/UAS-MJDtr-70Q_cc0 and UAS-Foxo-G mutant-Flag/+;elav-Gal4/UAS-MJDtr-70Q_cc0). *P < 0.05; **P < 1.0 × 10−2 by Student’s t test; error bars, SEM; n = 3 independent experiments.
Foxo Overexpression Restores the Behavioral Defects Induced by Coiled-Coil Structure-Mediated polyQ Protein Toxicity.
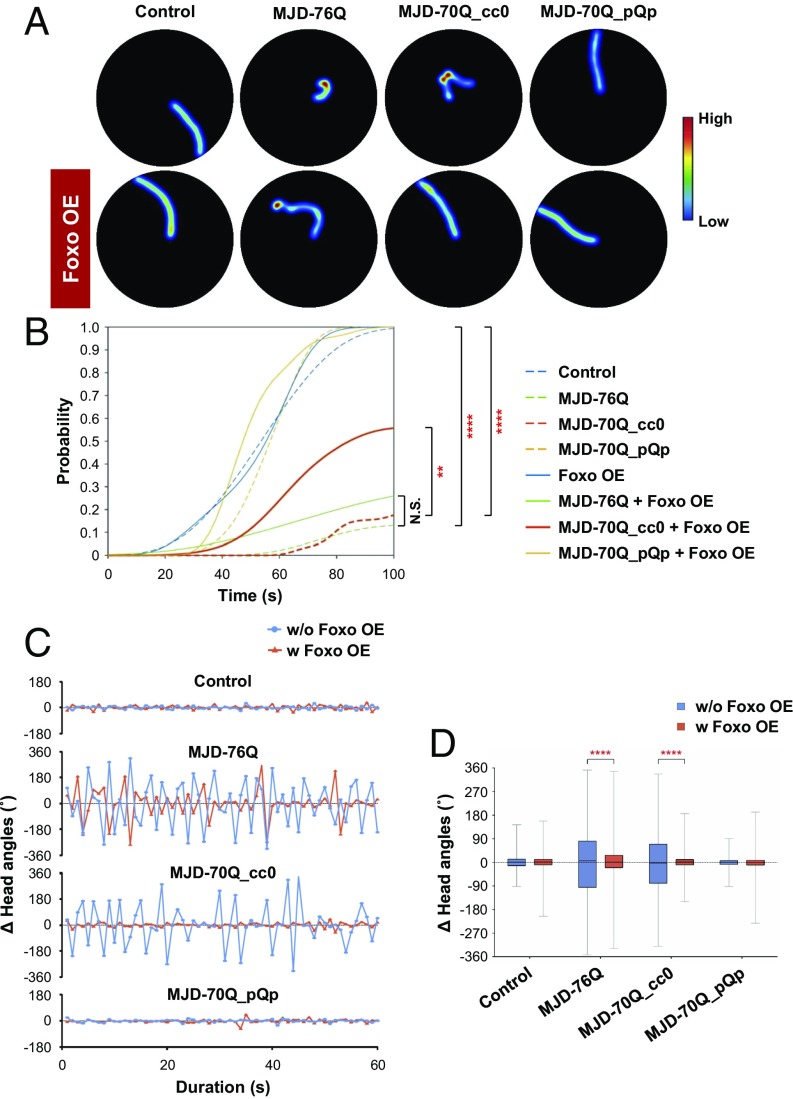
In addition to causing dendrite defects (Fig. 1C), polyQ proteins have been shown to affect neuronal survival, animal motility, and animal viability (3, 27). Thus, we investigated whether Foxo could rescue the motility defects induced by polyQ proteins in Drosophila larvae. Larval motility is assessed by measuring crawling and turning activities (28). We first tested the crawling capability using the following eight different lines of larvae: control larvae, and larvae expressing MJDtr-76Q, MJDtr-70Q_cc0, or MJDtr-70Q_pQp proteins in da neuronal clusters with or without co-overexpression of Foxo using the 109(2)80-gal4 driver. For each line, we placed a larva in the center of a Petri dish and monitored its movements until it reached the edge of the dish for up to 100 s (Fig. 5A) and repeated this experiment with more than 20 different larvae. We measured the time for the larvae to reach the dish edge and the cumulative fraction of larvae reaching the dish edge over time. All control larvae reached the dish edge within 100 s (Fig. 5B). However, only 13.3% and 19.2% of larvae expressing MJDtr-76Q and MJDtr-70Q_cc0 proteins, respectively, reached the dish edge within 100 s (Fig. 5B). The cumulative fraction of larvae co-overexpressing Foxo and MJDtr-70Q_cc0 proteins significantly (P < 1.0 × 10−4) increased to 56.3%, while larvae overexpressing Foxo alone showed no significant difference from the control, suggesting that Foxo is important in the regulation of larval motility affected by SCA3 polyQ proteins. In contrast, the cumulative fraction of larvae expressing MJDtr-70Q_pQp proteins was not significantly different from that of control larvae (Fig. 5B), suggesting that the crawling defects were induced by coiled-coil structures in polyQ proteins. Co-overexpression of Foxo and MJDtr-76Q resulted in only a partial (2.28-fold) increase in the cumulative fraction compared with larvae overexpressing MJDtr-76Q alone, although this increase was not statistically significant (Fig. 5B). This partial restoration suggests a potential additional toxicity mediated by coiled-coil structure-independent properties of polyQ proteins.
Fig. 5.
Foxo overexpression restores larval locomotion defects induced by coiled-coil structure-mediated polyQ toxicity. (A) Heat maps showing residence probability during traveling of larvae [109(2)80-Gal4/+;+/+, 109(2)80-Gal4/+;UAS-MJDtr-76Q/+, 109(2)80-Gal4/+;UAS-MJDtr-70Q_cc0/+, 109(2)80-Gal4/+;UAS-MJDtr-70Q_pQp/+, 109(2)80-Gal4/UAS-Foxo;+/+, 109(2)80-Gal4/UAS-Foxo;UAS-MJDtr-76Q/+, 109(2)80-Gal4/UAS-Foxo;UAS-MJDtr-70Q_cc0/+, and 109(2)80-Gal4/UAS-Foxo;UAS-MJDtr-70Q_pQp/+] monitored in the 90-mm Petri dish for up to 100 s. (B) Cumulative density functions showing fractions of larvae that reached the dish edge over time. The functions are estimated separately for larvae expressing each of the transgenes described in A. N.S., not significant; **P < 1.0 × 10−2; ****P < 1.0 × 10−4 by random permutation experiments; n ≥ 20 independent larvae. (C) Representative changing patterns of head angles of larvae, taken every second for 1 min. Turning the head to left and right is presented as positive (+) and negative (−) angles, respectively. The patterns are obtained separately for larvae expressing each of the transgenes described in A. (D) Box plots showing the variability of head angle changes in larvae expressing the transgenes described in A. ****P < 1.0 × 10−4 by random permutation experiments; n = 4 independent larvae.
We next used the same transgenic lines to examine another behavior: the larval head turning. To analyze the characteristics of head turning, we monitored the changes in the angle of larval heads for 1 min (SI Appendix, Fig. S5A). Control larvae rarely turned their heads, but larvae expressing MJD-70Q_cc0 showed significantly (P < 1.0 × 10−4) more frequent turning of their heads, while larvae expressing MJD-70Q_pQp showed patterns of head turning similar to those of control larvae (Fig. 5 C and D). The defects in head turning were significantly (P < 1.0 × 10−4) restored by Foxo overexpression in larvae expressing MJDtr-76Q or MJDtr-70Q_cc0, but the effect was stronger in larvae expressing MJDtr-70Q_cc0 (Fig. 5 C and D). The overexpression of Foxo alone had no effect on head turning (Discussion and Fig. 5). We next examined whether loss-of-function of Foxo could lead to behavioral defects similar to those induced by MJDtr-76Q and MJDtr-70Q_cc0 proteins. Indeed, larvae expressing Foxo RNAi showed similar defects in both crawling and turning (SI Appendix, Fig. S5 B–E). Taken together, these data suggest that Foxo can rescue the defects induced by coiled-coil structure-mediated polyQ protein toxicity in both dendrite morphology (Fig. 3 B and C) and larval locomotion.
Coiled-Coil Structure-Mediated Target Sequestration Affinity of polyQ Proteins Strongly Correlates with polyQ-Induced Dendrite Defects.
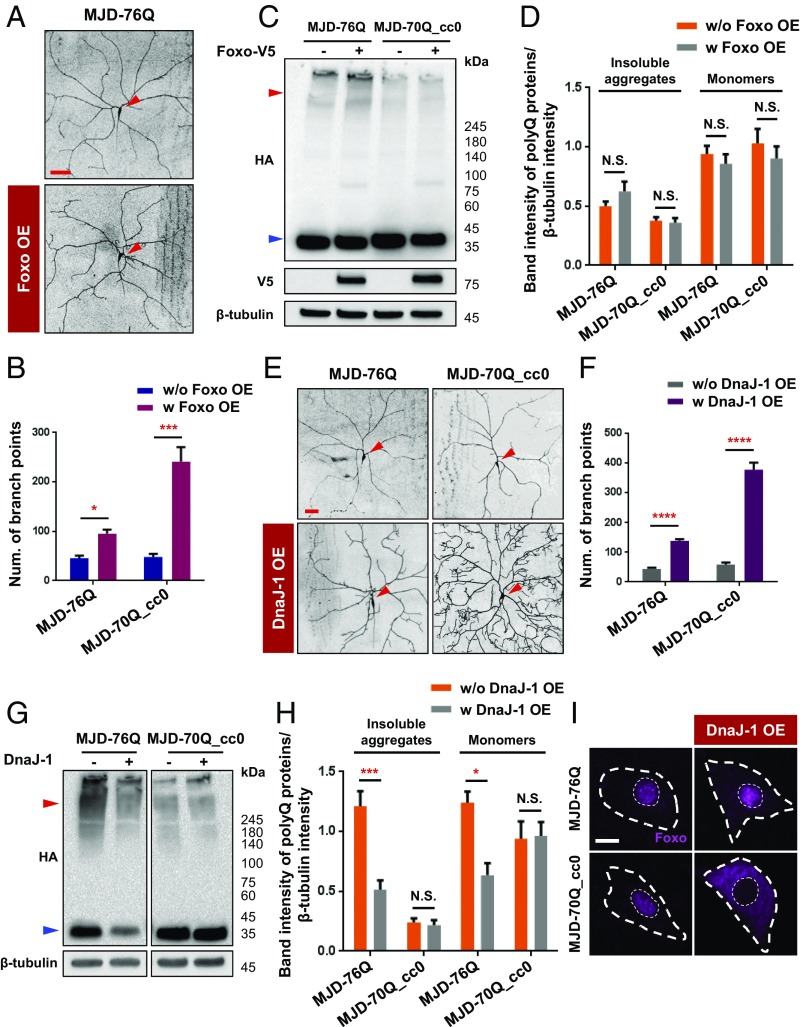
Next, we further investigated the ability of Foxo to restore dendrite defects caused by MJDtr-76Q and MJDtr-70Q_cc0 proteins containing coiled-coil domains (see also Fig. 3 B and C). Foxo overexpression significantly (P < 0.05) restored dendrite defects of C4da neurons expressing MJDtr-76Q, but the effect was weaker than that in C4da neurons expressing MJDtr-70Q_cc0 (Fig. 6 A and B), consistent with their efficacy in the restoration of larval locomotion defects. Foxo overexpression might affect the amounts of polyQ proteins. To test this possibility, we performed Western blot analysis of MJDtr-76Q and MJDtr-70Q_cc0 with or without Foxo overexpression, and found no significant effect of Foxo overexpression on the amounts of both monomers and insoluble aggregates of the polyQ proteins (Fig. 6 C and D).
Fig. 6.
Coiled-coil structure-mediated target sequestration affinity of polyQ proteins correlates with polyQ-induced dendrite defects. (A) Dendrite images of C4da neurons overexpressing MJDtr-76Q (Upper) or MJDtr-76Q + Foxo (Lower) (UAS-CD4td-GFP/+;ppk-Gal4,UAS-MJDtr-76Q/+ and UAS-CD4td-GFP/UAS-Foxo;ppk-Gal4,UAS-MJDtr-76Q/+). Arrowheads (red) indicate cell bodies of C4da neurons. (Scale bar, 50 μm.) (B) Quantification of the number of dendrite branch points in C4da neurons expressing the denoted transgenes (UAS-CD4td-GFP/+;ppk-Gal4,UAS-MJDtr-76Q/+, UAS-CD4td-GFP/UAS-Foxo;ppk-Gal4,UAS-MJDtr-76Q/+, UAS-CD4-tdGFP/+;ppk-Gal4,UAS-MJDtr-70Q_cc0/+, and UAS-CD4-tdGFP/UAS-Foxo;ppk-Gal4,UAS-MJDtr-70Q_cc0/+). The numbers of branch points are compared between C4da neurons expressing each SCA3 polyQ protein variant with and without Foxo overexpression. *P < 0.05; ***P < 1.0 × 10−3 by two-way ANOVA with Tukey post hoc test.; error bars, SEM; n ≥ 5 neurons. (C) Representative images obtained from Western blot analysis of adult fly heads (+/+;elav-GeneSwitch-Gal4/UAS-MJDtr-76Q, UAS-Foxo-V5/+;elav-GeneSwitch-Gal4/UAS-MJDtr-76Q, +/+;elav-GeneSwitch-Gal4/UAS-MJDtr-70Q_cc0, and UAS-Foxo-V5/+; elav-GeneSwitch-Gal4/UAS-MJDtr-70Q_cc0). SCA3 variants and Foxo-V5 were immunoblotted using anti-HA and anti-V5 antibodies, respectively. β-Tubulin is used as the loading control. Red and blue arrowheads indicate insoluble aggregates and monomers of SCA3 polyQ proteins, respectively. n = 3 independent experiments. (D) Quantification of band intensities obtained from Western blot analysis (C) for insoluble aggregates and monomers of MJDtr-76Q and MJDtr-70Q_cc0 proteins. Each band intensity was normalized with that of β-tubulin. N.S., not significant by two-way ANOVA with Tukey post hoc test; error bars, SEM; n = 3 independent experiments. (E) Dendrite images of C4da neurons overexpressing MJDtr-76Q (Upper Left), MJDtr-70Q_cc0 (Upper Right), MJDtr-76Q + DnaJ-1 (Lower Left), or MJDtr-70Q_cc0 + DnaJ-1 (Lower Right) (UAS-CD4td-GFP/+;ppk-Gal4,UAS-MJDtr-76Q/+, UAS-CD4td-GFP/+;ppk-Gal4,UAS-MJDtr-76Q/UAS-DnaJ-1.K, UAS-CD4td-GFP/+;ppk-Gal4,UAS-MJDtr-70Q_cc0/+, and UAS-CD4td-GFP/+;ppk-Gal4,UAS-MJDtr-70Q_cc0/UAS-DnaJ-1.K). Arrowheads (red) indicate cell bodies of C4da neurons. (Scale bar, 50 μm.) (F) Quantification of the number of dendrite branch points in C4da neurons expressing the transgenes described in D. ****P < 1.0 × 10−4 by two-way ANOVA with Tukey post hoc test; error bars, SEM; n ≥ 3 neurons. (G) Representative images obtained from Western blot analysis of adult fly heads (+/+;elav-Gal4/UAS-MJDtr-76Q, +/+;elav-Gal4,UAS-DnaJ-1/UAS-MJDtr-76Q, +/+;elav-Gal4/UAS-MJDtr-70Q_cc0, and +/+;elav-Gal4,UAS-DnaJ-1/UAS-MJDtr-70Q_cc0). SCA3 variants were immunoblotted using anti-HA antibody, and β-tubulin is used as the loading control. Red and blue arrowheads indicate insoluble aggregates and monomers of SCA3 polyQ proteins, respectively. n = 3 independent experiments. (H) Quantification of band intensities obtained from Western blot analysis (G) for insoluble aggregates and monomers of MJDtr-76Q and MJDtr-70Q_cc0 proteins. Each band intensity was normalized with that of β-tubulin. N.S., not significant; *P < 0.05; ***P < 1.0 × 10−3 by two-way ANOVA with Tukey post hoc test; error bars, SEM; n = 3 independent experiments. (I) Subcellular localization of Foxo (magenta) in C4da neurons overexpressing MJDtr-76Q (Upper Left), MJDtr-70Q_cc0 (Lower Left), MJDtr-76Q + DnaJ-1 (Upper Right), or MJDtr-70Q_cc0 + DnaJ-1 (Lower Right) (+/+;ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-76Q, UAS-DnaJ-1.K/+;ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-76Q, +/+;ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-70Q_cc0, and UAS-DnaJ-1.K/+;ppk-Gal4,UASp-Foxo-GFP/UAS-MJDtr-70Q_cc0). Outer and inner dashed lines (white) indicate the outlines of the cell body and nucleus, respectively. (Scale bar, 5 μm.)
The difference in the restoration capability of Foxo between MJDtr-76Q– and MJDtr-70Q_cc0–expressing C4da neurons might be due to the relative difference in polyQ toxicity. PolyQ toxicity is collectively determined by the amounts of toxic polyQ proteins and the affinity toward their sequestration targets. Previously, HSP-40, a mammalian homolog of DnaJ-1, was shown to affect both the amounts of toxic polyQ proteins and the interaction of polyQ proteins with their targets (29, 30). We first assessed the effect of DnaJ-1 overexpression on the extent of dendrite defects. Overexpression of DnaJ-1 in C4da neurons expressing MJDtr-76Q or MJDtr-70Q_cc0 significantly (P < 1.0 × 10−4) restored dendrite defects in both types of neurons (Fig. 6 E and F). However, this restoration was significantly stronger in C4da neurons expressing MJDtr-70Q_cc0 than in those expressing MJDtr-76Q, consistent with the restoration pattern observed after Foxo overexpression (Fig. 6B).
We then examined how DnaJ-1 overexpression altered the amounts of MJDtr-76Q and MJDtr-70Q_cc0 proteins. MJDtr-76Q proteins were significantly (P < 0.05) reduced in the amounts of insoluble aggregates and monomers by DnaJ-1 overexpression (Fig. 6 G and H), consistent with the previous observation (29, 31). In contrast, MJDtr-70Q_cc0 proteins showed no significant changes. We next attempted to assess how DnaJ-1 overexpression affected the target sequestration affinity of MJDtr-76Q and MJDtr-70Q_cc0 proteins using co-IP. However, our co-IP experiments (Fig. 3F) showed virtually no interactions of Foxo with monomers of MJDtr-76Q or MJDtr-70Q_cc0 proteins, making it difficult to accurately assess their target sequestration affinity. Thus, we instead measured Foxo subcellular localization to indirectly assess the target sequestration affinity of MJDtr-76Q or MJDtr-70Q_cc0 proteins after co-overexpressing DnaJ-1 and Foxo in C4da neurons expressing MJDtr-76Q or MJDtr-70Q_cc0 proteins. DnaJ-1 overexpression induced translocalization of Foxo proteins from the nucleus to the cytoplasm in C4da neurons expressing MJDtr-70Q_cc0, whereas only marginal changes in Foxo localization were observed in C4da neurons expressing MJDtr-76Q proteins (Fig. 6I).
In summary, DnaJ-1 overexpression resulted in a stronger decrease in the amount of MJDtr-76Q proteins, but a stronger decrease in target sequestration affinity of MJDtr-70Q_cc0 proteins. Considering a stronger restoration of dendrite defects by DnaJ-1 overexpression in C4da neurons expressing MJDtr-70Q_cc0 proteins than in those expressing MJDtr-76Q proteins, at least in the conditions tested in our study, these data suggest that the coiled-coil–mediated target sequestration affinity of toxic polyQ proteins appears to be more strongly associated with polyQ-induced dendrite defects, compared with the amount of toxic polyQ proteins.
Discussion
In this study, among the coiled-coil–rich TFs implicated in dendrite morphogenesis, Foxo and Cut were selected as the most likely candidates to interact with polyQ proteins (Table 1). Of the two TFs, we focused on Foxo because we previously found that Cut indirectly interacted with MJDtr-78Q proteins through CBP (11). Nevertheless, another mode of interaction between Cut and SCA3 polyQ proteins based on coiled-coil structures could not be excluded. According to a previous study by Grueber et al. (20), the overexpression of Cut significantly increased dendrite branching in class I da (C1da) neurons. Based on this observation, we further examined dendrite branching in C1da neurons co-overexpressing Cut and MJDtr-76Q, MJDtr-70Q_cc0, or MJDtr-70Q_pQp. The overexpression of Cut alone elongated terminal dendrites and significantly (P < 1.0 × 10−4) increased the number of dendrite branch points (SI Appendix, Fig. S6), consistent with the previous findings (20). These changes in dendrites, however, were significantly (P < 1.0 × 10−4) suppressed by co-overexpression of Cut with MJDtr-76Q or MJDtr-70Q_cc0, but not with MJDtr-70Q_pQp (SI Appendix, Fig. S6B). These results suggest that the function of Cut in dendrite branching is inhibited by SCA3 polyQ proteins containing coiled-coil structures, implying that such inhibition can be mediated by coiled-coil to coiled-coil interactions. Notably, CBP is also enriched with coiled-coil domains and thus likely to interact with polyQ proteins through the same interactions. Collectively, these data suggest that polyQ–CBP–Cut complex can be formed through coiled-coil to coiled-coil interactions between these proteins.
Our data showed that increased nuclear localization of Foxo is closely associated with polyQ protein toxicity (Fig. 3D). Foxo has been implicated in various neurodegenerative diseases, such as Alzheimer’s (32, 33), prion (34), and amyotrophic lateral sclerosis diseases (35), suggesting potential associations with toxic proteins accumulated in these disorders. Foxo proteins have been observed in both the cytoplasm and nucleus in C4da neurons (36). In CCL39 fibroblast cells, Foxo proteins are predominantly localized in the cytoplasm, but upon inhibition of the PI3K-Akt signaling, they are dephosphorylated and translocated into the nucleus to induce the expression of their target genes (37). In our study, we observed that the co-overexpression of Foxo with MJDtr-76Q or MJDtr-70Q_cc0 proteins increased the interaction of these proteins and the nuclear localization of Foxo (Fig. 3D), accompanied by reduced transcriptional activity of this TF (Fig. 3G and SI Appendix, Fig. S3C). The loss of Foxo function may indicate a reduced amount of the active TF—dephosphorylated Foxo—in the nucleus due to its interactions with MJDtr-76Q or MJDtr-70Q_cc0 proteins. Thus, our data suggest that MJDtr-76Q and MJDtr-70Q_cc0 proteins sequester Foxo in the nucleus, thereby reducing the amount of the active Foxo TF. Consistent with this model, Foxo RNAi led to larval locomotion defects (SI Appendix, Fig. S5 B–E). The overexpression of Foxo alone had no effect on larval locomotion (Fig. 5), suggesting that the amount of the active Foxo TF in the nucleus was not increased. Additional functional experiments should be carried out to test whether the overexpression of constitutively active Foxo TF contributes to larval locomotion.
We selected Foxo and Cut as the most likely TF targets of SCA3 polyQ proteins because they showed the strongest effects on dendrite branch points upon their knockdown or overexpression (Table 1). However, if the conventional statistical cut-off of P = 0.05 from the analysis of variance (Tukey post hoc test) was applied, four additional TFs [Doublesex (Dsx), Nub, Relish (Rel), and Cnc] with statistically significant effects on dendrite branch points could also be selected on the basis of the knockdown and overexpression experiments (Fig. 3 A and B). We did not pursue detailed functional experiments for these four TFs due to the lack of available antibodies or transgenic fly lines, and the difficulty in performing colocalization experiments because these TFs displayed the same nuclear localization as polyQ proteins. Nevertheless, the potential associations of these four TFs with neuronal processes impaired by protein toxicity have been previously reported. For example, Relish (NF-κB in mammals) was shown to regulate dendritic arborization (38), while the inhibition of NF-κB impaired synaptic signaling and learning in mice (39). Additionally, a number of studies have indicated the protective functions of Cnc (Nrf2 in mammals) against neuronal defects in various neurodegenerative diseases (40). Moreover, Dsx is involved in axonal projection in Drosophila (41), but its role in dendrite formation has been rarely studied. In addition, Nub is involved in the regulation of dendrite growth and branching (42). These data collectively suggest the potential roles of these coiled-coil–rich TFs in polyQ-induced neuronal toxicity. Additional functional experiments are needed to elucidate their potential roles.
Comparison of dendrite defect restoration in C4da neurons expressing MJDtr-76Q and MJDtr-70Q_cc0 proteins suggests that the coiled-coil–mediated target sequestration affinity of toxic polyQ proteins is more strongly associated with polyQ-induced dendrite defects, compared with the amount of toxic polyQ proteins (Fig. 6). The severity of Foxo-dependent dendrite defects should be determined based on the amount of Foxo proteins sequestered by toxic polyQ proteins through coiled-coil to coiled-coil interactions (Fig. 4). The sequestered Foxo amount can be then determined by both the total amount (monomers and insoluble aggregates) and the coiled-coil–mediated target sequestration affinity of toxic polyQ proteins. First, regarding the total amount of toxic polyQ proteins, DnaJ-1 overexpression showed no effects on the amounts of monomers and insoluble aggregates in fly brains expressing MJDtr-70Q_cc0 containing only coiled-coil structures, but significant decreases in fly brains expressing MJDtr-76Q containing both coiled-coil and β-sheet structures. These data suggest that DnaJ-1 appears to predominantly affect the amount of toxic polyQ proteins influenced by β-sheet structures, although other biophysical properties of polyQ proteins may also contribute to the amount. This issue can be verified using a polyQ structural variant containing only β-sheet structures; however, to our knowledge, such a variant has not been developed due to the difficulty in breaking coiled-coil structures exclusively. Second, regarding the coiled-coil–mediated target sequestration affinity, DnaJ-1 overexpression showed only marginal effects on the target sequestration affinity in C4da neurons expressing MJDtr-76Q, but significantly decreases in C4da neurons expressing MJDtr-70Q_cc0. These data suggest that DnaJ-1 appears to predominantly affect interactions of polyQ proteins with sequestrated targets mediated by the coiled-coil structures.
Taking these data together, our study suggests that coiled-coil structures of polyQ proteins may serve as another fundamental structural basis, an alternative to β-sheet structures, for their interactions with targets containing coiled-coil domains, which can lead to polyQ pathology. Specifically, in this study, we propose the polyQ–Foxo interaction as a molecular mechanism for coiled-coil structure-mediated polyQ protein toxicity, such as dendrite and larval locomotion defects. Our study adds to the current understanding of the structure-based polyQ protein toxicity that mediates neuronal pathology through coiled-coil to coiled-coil interactions of these proteins with their TF targets. Furthermore, our findings suggest the potential of therapeutic approaches based on the inhibition of coiled-coil to coiled-coil interactions between pathogenic polyQ proteins and coiled-coil–rich TFs.
Materials and Methods
Fly stocks used were as follows:
UAS-3xFLAG-Pros.S (BL32245), UASp-GFP-E2f1.WT (BL34058), UAS-Nej.WT-V5 (BL32573), UAS-Flag-Rel.68 (BL55778), UASp-Foxo-GFP (BL43633), UAS-Foxo.P (BL9575), UAS-Exex.B (BL9929), UAS-Pros RNAi (BL42538), UAS-Foxo RNAi (BL32427), UAS-Ct RNAi (BL33967), UAS-Kay RNAi (BL33379), UAS-Dsx RNAi (BL55646), UAS-Nub RNAi (BL55305), UAS-Nej RNAi (BL37489), UAS-Rel RNAi (BL33661), UAS-E2f1 RNAi (BL36126), UAS-Med RNAi (BL31928), UAS-Exex RNAi (BL57709), UAS-Cnc RNAi (BL40854), UAS-DnaJ-1.K (BL30553), UAS-MJDtr-27Q (BL8149) and elav-Gal4 (BL8760) were obtained from the Bloomington Drosophila Stock Center. UAS-Cut-3xHA (F001703), UAS-Dsx-3xHA (F001879), UAS-Cnc-3xHA (F000602), and UAS-Med-3xHA (F000591) were obtained from Fly ORF (43). The following lines were used as previously described: UAS-mCD8-GFP (III), ppk-gal4 (III), 109(2)80-gal4 (II), UAS-GMA, 221-gal4 (III) (10), UAS-Ct (II), ppk1a-gal4 (III), and UAS-CD4-tdGFP (II) (11). For pan-neuronal expression of transgenes, elav-Gal4 was used. 109(2)80-gal4 was used to drive the expression of transgenes in da neuronal clusters. ppk-gal4 and ppk1a-gal4 were used for the expression of transgenes in C4da neurons. 221-gal4 was used to expression of transgenes in C1da neurons. All flies were raised under standard conditions (25 °C and 60% humidity).
Supplementary Material
Acknowledgments
This work was supported by the Daegu Gyeongbuk Institute of Science and Technology Research and Development Program and Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science and Information and Communications Technology (ICT) (18-BD-0402, 18-BT-02, 18-01-HRSS-02, 2018R1A2B6001607); the Development of Platform Technology for Innovative Medical Measurements Program from the Korea Research Institute of Standards and Science Grant KRISS-2018-GP2018-0018 (to S.B.L.); and Institute for Basic Science Grant IBS-R013-A1 (to D.H.) funded by the Ministry of Science of ICT. Y.N.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807206115/-/DCSupplemental.
References
- 1.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 2.Paulson HL, Shakkottai VG, Clark HB, Orr HT. Polyglutamine spinocerebellar ataxias—From genes to potential treatments. Nat Rev Neurosci. 2017;18:613–626. doi: 10.1038/nrn.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates G. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni VA, Firestein BL. The dendritic tree and brain disorders. Mol Cell Neurosci. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Stuart GJ, Spruston N. Dendritic integration: 60 years of progress. Nat Neurosci. 2015;18:1713–1721. doi: 10.1038/nn.4157. [DOI] [PubMed] [Google Scholar]
- 7.Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 8.Sotrel A, Williams RS, Kaufmann WE, Myers RH. Evidence for neuronal degeneration and dendritic plasticity in cortical pyramidal neurons of Huntington’s disease: A quantitative Golgi study. Neurology. 1993;43:2088–2096. doi: 10.1212/wnl.43.10.2088. [DOI] [PubMed] [Google Scholar]
- 9.Richards P, et al. Dendritic spine loss and neurodegeneration is rescued by Rab11 in models of Huntington’s disease. Cell Death Differ. 2011;18:191–200. doi: 10.1038/cdd.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SB, Bagley JA, Lee HY, Jan LY, Jan YN. Pathogenic polyglutamine proteins cause dendrite defects associated with specific actin cytoskeletal alterations in Drosophila. Proc Natl Acad Sci USA. 2011;108:16795–16800. doi: 10.1073/pnas.1113573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung CG, et al. Golgi outpost synthesis impaired by toxic polyglutamine proteins contributes to dendritic pathology in neurons. Cell Rep. 2017;20:356–369. doi: 10.1016/j.celrep.2017.06.059. [DOI] [PubMed] [Google Scholar]
- 12.Nagai Y, et al. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 13.Buchanan LE, et al. Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proc Natl Acad Sci USA. 2014;111:5796–5801. doi: 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jan YN, Jan LY. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, Hirano A. A comparative study of modified Bielschowsky, Bodian and thioflavin S stains on Alzheimer’s neurofibrillary tangles. Neuropathol Appl Neurobiol. 1986;12:3–9. doi: 10.1111/j.1365-2990.1986.tb00677.x. [DOI] [PubMed] [Google Scholar]
- 18.Warrick JM, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer MH, Wanker EE, Andrade-Navarro MA. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 21.Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer EP, et al. Functional genomic analyses of two morphologically distinct classes of Drosophila sensory neurons: Post-mitotic roles of transcription factors in dendritic patterning. PLoS One. 2013;8:e72434. doi: 10.1371/journal.pone.0072434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirangi TR, Wong AM, Truman JW, Stern DL. Doublesex regulates the connectivity of a neural circuit controlling Drosophila male courtship song. Dev Cell. 2016;37:533–544. doi: 10.1016/j.devcel.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Majdazari A, et al. Dendrite complexity of sympathetic neurons is controlled during postnatal development by BMP signaling. J Neurosci. 2013;33:15132–15144. doi: 10.1523/JNEUROSCI.4748-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santiago C, Labrador JP, Bashaw GJ. The homeodomain transcription factor Hb9 controls axon guidance in Drosophila through the regulation of Robo receptors. Cell Rep. 2014;7:153–165. doi: 10.1016/j.celrep.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steele AD, Jackson WS, King OD, Lindquist S. The power of automated high-resolution behavior analysis revealed by its application to mouse models of Huntington’s and prion diseases. Proc Natl Acad Sci USA. 2007;104:1983–1988. doi: 10.1073/pnas.0610779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riedl J, Louis M. Behavioral neuroscience: Crawling is a no-brainer for fruit fly larvae. Curr Biol. 2012;22:R867–R869. doi: 10.1016/j.cub.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Gillis J, et al. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem. 2013;288:17225–17237. doi: 10.1074/jbc.M112.421685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YE, et al. Soluble oligomers of polyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol Cell. 2016;63:951–964. doi: 10.1016/j.molcel.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manolopoulos KN, Klotz LO, Korsten P, Bornstein SR, Barthel A. Linking Alzheimer’s disease to insulin resistance: The FoxO response to oxidative stress. Mol Psychiatry. 2010;15:1046–1052. doi: 10.1038/mp.2010.17. [DOI] [PubMed] [Google Scholar]
- 33.Maiese K. Forkhead transcription factors: New considerations for Alzheimer’s disease and dementia. J Transl Sci. 2016;2:241–247. doi: 10.15761/JTS.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Yi W, Feng B, Zhou Z, Xiao G. IGF-1-induced enhancement of PRNP expression depends on the negative regulation of transcription factor FOXO3a. PLoS One. 2013;8:e71896. doi: 10.1371/journal.pone.0071896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Baldie G, Periz G, Wang J. RNA-processing protein TDP-43 regulates FOXO-dependent protein quality control in stress response. PLoS Genet. 2014;10:e1004693. doi: 10.1371/journal.pgen.1004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears JC, Broihier HT. FoxO regulates microtubule dynamics and polarity to promote dendrite branching in Drosophila sensory neurons. Dev Biol. 2016;418:40–54. doi: 10.1016/j.ydbio.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez H, Hale VA, Dolcet X, Davies A. NF-kappaB signalling regulates the growth of neural processes in the developing PNS and CNS. Development. 2005;132:1713–1726. doi: 10.1242/dev.01702. [DOI] [PubMed] [Google Scholar]
- 39.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 40.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellert DJ, Knapp JM, Manoli DS, Meissner GW, Baker BS. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the roundabout receptors. Development. 2010;137:323–332. doi: 10.1242/dev.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corty MM, Tam J, Grueber WB. Dendritic diversification through transcription factor-mediated suppression of alternative morphologies. Development. 2016;143:1351–1362. doi: 10.1242/dev.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bischof J, et al. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.