Significance
The exogenous light signal and the endogenous circadian clock are two critical factors for directing growth and development in plants. In this study, we characterized a previously unidentified positive regulator of light signaling, CSU4. CSU4 interacts with CCA1, a key component of the circadian clock, to inhibit its transcriptional repression activity. Being a transcriptional regulator, CSU4 represses CCA1 in the early morning and PIF4 in the early evening in gating hypocotyl elongation. Thus, CSU4 likely acts as a key regulator of light- and circadian clock-mediated hypocotyl growth in plants.
Keywords: photomorphogenesis, CSU4, COP1, CCA1, PIF4
Abstract
CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) and DE-ETIOLATED 1 (DET1) are founding components of two central repressor complexes of photomorphogenesis that trigger the degradation of a larger number of photomorphogenic-promoting factors in darkness. Here, we identify COP1 SUPPRESSOR 4 (CSU4) as a genetic suppressor of the cop1-6 mutation. Mutations in CSU4 largely rescued the constitutively photomorphogenic phenotype of cop1-6 and det1-1 in darkness. Loss of CSU4 function resulted in significantly longer hypocotyl in the light. Further biochemical studies revealed that CSU4 physically interacts with CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and negatively regulates its transcriptional repression activity toward its targets. CSU4 represses the expression of CCA1 in the early morning and of PHYTOCHROME INTERACTING FACTOR 4 (PIF4) in the early evening. Our study suggests that CSU4 acts as a negative regulator of CCA1 via physically associating with CCA1, which in turn, likely serves to repress expression of CCA1 and PIF4 to promote photomorphogenesis.
Light is not only the major energy source for plants but also one of the most essential environmental factors affecting various physiological and developmental processes in plants. In nature, buried and germinated seeds undergo skotomorphogenesis to penetrate upward through the soil. Upon light exposure, etiolated seedlings initiate photomorphogenesis. These two distinct developmental processes are extremely critical for a germinated seed to become a healthy seedling and, subsequently, to complete its life cycle (1, 2).
An extensive number of components, including at least four classes of wavelength-specific photoreceptors—CONSTITUTIVELY PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) and transcription factors—work synergistically to ensure the proper skotomorphogenic and photomorphogenic development and their transition (3–5). In the dark, two classes of key repressors of photomorphogenesis—COP/DET/FUS and PHYTOCHROME INTERACTING FACTORs (PIF1, PIF3, PIF4, and PIF5)—are biologically functional and highly abundant in the nucleus (6–11). COP/DET/FUS function as E3 ubiquitin ligases and target numerous photomorphogenic-promoting factors such as ELONGATED HYPOCOTYL 5 (HY5) and HY5 HOMOLOG for ubiquitination and degradation (12, 13), while the basic helix-loop-helix transcription factors—PIFs—control a large group of target genes’ expression. Together, these COP/DET/FUS- and PIF-mediated molecular events inhibit photomorphogenesis and promote skotomorphogenesis. In the light, COP/DET/FUS activity is largely repressed through multiple regulatory mechanisms (8, 14–17), thus allowing the accumulation of their downstream substrates. Meanwhile, PIFs are rapidly phosphorylated, ubiquitinated, and degraded within minutes upon light illumination (9–11, 18–21). Consequently, the accumulation of COP/DET/FUS targets and the alternation of PIF-controlled gene expression synergistically contribute to the promotion of photomorphogenesis.
Circadian rhythm has been functionally linked to photomorphogenic development in plants. Consistently, a group of circadian clock factors function in the regulation of light signaling, either positively or negatively (22–26). The key components of the morning complex of the circadian clock—CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY)—positively regulate the expression of PIF4 to promote hypocotyl growth, thus acting as negative regulators of light-inhibited hypocotyl elongation. Consistently, loss of CCA1 or LHY function mutants display shortened hypocotyls, while CCA1 and LHY transgenic plants show elongated hypocotyls (22–24). In addition to those of the CCA1-LHY morning complex, evening-expressed circadian clock proteins are also involved in photomorphogenesis. The ELF3-ELF4-LUX evening complex directly binds to the promoter of PIF4 and represses its gene expression during early evening to gate hypocotyl elongation (26). In addition, light initiates the degradation of PIF4, and both ELF3 and TOC1 physically interact with PIF4 to repress its biochemical activity, consequently suppressing PIF4-mediated gene expression and hypocotyl elongation in the daytime (18, 27, 28). Both the PIF4 mRNA and protein levels are diurnally regulated by light and the circadian clock (26). These facts demonstrate that both light and the circadian clock converge on PIF4 to influence photomorphogenesis at both transcriptional and protein levels.
In this study, we report the identification and characterization of a previously uncharacterized positive regulator of light signaling, COP1 SUPPRESSOR 4 (CSU4). The csu4 gene largely suppresses the short hypocotyls, and partially rescues the opened cotyledon phenotypes of cop1-6 in darkness. Loss-of-function mutations in the CSU4 gene leads to significantly elongated hypocotyls in the light. CSU4 physically interacts with CCA1 and represses its transcriptional repression activity; it also negatively regulates CCA1 expression in the early morning and PIF4 transcript levels in the early evening. Collectively, CSU4 acts as a transcriptional coregulator in repressing CCA1 and PIF4 expression to inhibit hypocotyl elongation.
Results
Map-Based Cloning of CSU4.
To explore factors involved in light signaling, we performed a forward genetic screen for mutants that suppress cop1-6, which is a weak allele of cop1, and that exhibit constitutively photomorphogenic phenotype in darkness (29). Further genetic complementation tests recovered two extragenic and recessive alleles, namely csu4-1 and csu4-2, from this screen. To identify and characterize the CSU4 gene, we combined map-based cloning and whole-genome resequencing strategies as previously described (30, 31). CSU4 was mapped to an ∼300-kb region between 5-AB009053-9339 and 5-AB007646-9467 markers on chromosome 5 (SI Appendix, Fig. S1A). The analysis of genomic sequences in this region revealed a C-to-T change at the site of amino acid 667 from the start code (ATG) in At5g63440, causing a premature stop code at the 41st codon in csu4-1 (SI Appendix, Fig. S1). Next, we amplified the At5g63440 in csu4-2 cop1-6 using PCR. Next, the PCR products were subjected to sequencing, and a C-to-T substitution at amino acid 1147 from the ATG was identified, which results in a premature stop code at the 145th codon (SI Appendix, Fig. S1). Database searches revealed that CSU4 is a single-copy gene encoding a domain of unknown function (DUF)167 containing protein (SI Appendix, Fig. S1C).
Mutations in CSU4 Suppress cop1-6.
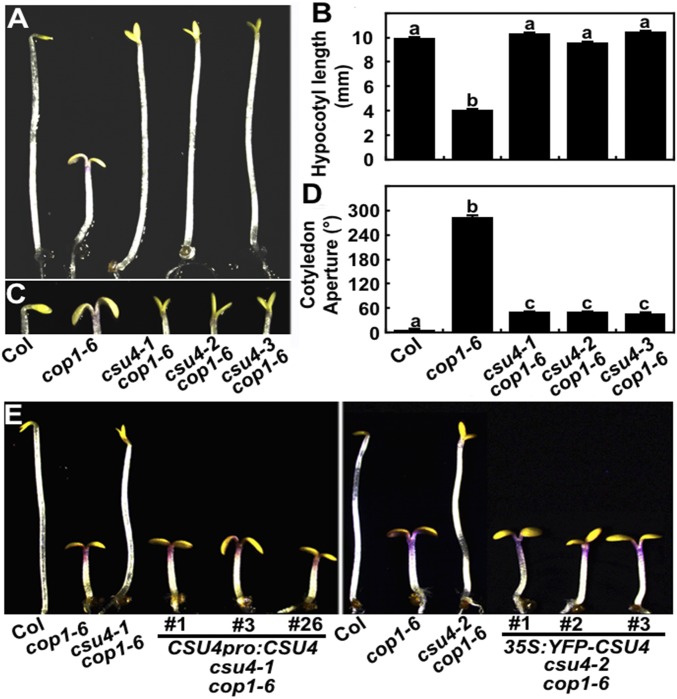
The cop1-6 mutant seedlings display constitutively phototmorphogenic phenotype in darkness (29). Each of the csu4 mutant alleles (csu4-1 and csu4-2) largely suppressed the cop1-6 in the dark (Fig. 1). Two independent csu4 cop1-6 double mutants displayed similar hypocotyl length compared with the wild type (WT), and drastically longer length compared with cop1-6 (Fig. 1 A and B). Although the cotyledons of csu4 cop1-6 mutant seedlings were obviously expanded, the opening angles were significantly smaller than that of cop1-6 in darkness (Fig. 1 C and D). The hypocotyl length of csu4 cop1-6 was longer than cop1-6 but shorter than WT when grown in various light conditions tested [white (W), blue (B), red (R), and far-red (FR)] (SI Appendix, Fig. S2 A–H). In addition, the csu4 cop1-6 adult plants grown in long-day (LD) conditions (16 h light/8 h dark) showed intermediated phenotype compared with WT and cop1-6 (SI Appendix, Fig. S2I). Together, these results indicate that csu4 largely suppressed the cop1-6 in the dark and partially suppressed cop1-6 in the light. Further, we isolated and characterized a T-DNA insertion csu4 mutant, namely csu4-3 (SALK_070923C) from the Arabidopsis Biological Resource Center, in which CSU4 gene expression was dramatically reduced (SI Appendix, Fig. S3). As expected, the constructed csu4-3 cop1-6 double-mutant plants displayed similar phenotype with csu4-1 cop1-6 and csu4-2 cop1-6 double mutants, both in the dark and in light conditions (Fig. 1 and SI Appendix, Fig. S2). Furthermore, csu4-1 cop1-6 transformed with genomic CSU4 driven by its own promoter (CSU4pro:CSU4 csu4-1 cop1-6) and csu4-2 cop1-6 transformed with CSU4 coding sequences fused with yellow fluorescent protein (YFP) at the N terminus driven by the cauliflower mosaic virus 35S promoter (35S:YFP-CSU4 csu4-2 cop1-6) both exhibited cop1-6 phenotype in darkness (Fig. 1E). These results demonstrate that disruption of CSU4 indeed rescues constitutively photomorphogenic phenotype of cop1-6 and that a functional CSU4 complements the suppressor phenotype conferred by csu4 in the cop1-6 background.
Fig. 1.
Mutations in CSU4 suppress cop1-6 in the dark. (A and B) Hypocotyl phenotypes and length of 4-d-old Col, cop1-6, and csu4 cop1-6 seedlings grown in the dark. Error bars represent SE (n ≥ 20). Letters above the bars indicate significant differences (P < 0.05) as determined by one-way ANOVA with Tukey’s post hoc analysis. (C and D) Cotyledon phenotypes and separation angle of 4-d-old Col, cop1-6, and csu4 cop1-6 seedlings grown in the dark. Error bars represent SE (n ≥ 30). Letters above the bars indicate significant differences (P < 0.05) as determined by one-way ANOVA with Tukey’s post hoc analysis. (E) Genomic complementation test. Phenotypes of Col, cop1-6, csu4-1 cop1-6, csu4-2 cop1-6, three independent CSU4pro:CSSU4 csu4-1 cop1-6, and three independent 35S:YFP-CSU4 csu4-2 cop1-6 transgenic lines grown in darkness for 4 d. The experiments were performed three times, with similar results. The graphs depict one of three experiments.
In addition to COP1, DET1 also acts as a central repressor of photomorphogenesis (32). Our genetic evidence revealed that csu4 det1-1 showed significantly longer hypocotyls compared with det1-1, suggesting that mutations in CSU4 not only suppress cop1-6 but also largely restore det1-1 phenotype in darkness (SI Appendix, Fig. S4).
At the 3′ end of the fourth intron, cop1-6 possesses a point mutation, “AG” to “GG”, leading to multiple cryptically spliced products at intron 4 (29, 31). Thus, we examined the splicing pattern of COP1-6 in csu4 cop1-6 double mutants, and found that mutations in CSU4 barely affect the splicing pattern and the transcript levels of COP1-6 mRNA (SI Appendix, Fig. S5).
The COP1-SPA Complexes and COP10-DDB1-DET1 Do Not Affect CSU4.
The COP1-SPA complexes and the COP10-DDB1-DET1 complex act as E3 ubiquitin ligases and promote degradation of a number of downstream targets (3, 4). We tested whether those types of E3 complexes play roles in regulating the CSU4 abundance in Arabidopsis seedlings. CSU4 protein levels were not significantly altered in cop1-4, cop1-6, multiple spa triple mutants, and det1-1 compared with WT in the dark and light (SI Appendix, Fig. S6A), indicating that the COP1-SPA complexes and the COP10-DDB1-DET1 complex do not affect CSU4 abundance. In addition, CSU4 protein accumulated at comparable levels in seedlings grown in various light conditions tested (continuous darkness, W, B, R, and FR) (SI Appendix, Fig. S6B). Seedlings grown in different intensities of W light also accumulated similar CSU4 protein levels (SI Appendix, Fig. S6B). Together, these data suggest that light signals might have little effect on CSU4 in plants.
In addition, the COP1 protein level in Columbia (Col) WT seedlings was indistinguishable from that in csu4 seedlings grown in the dark or in W light (SI Appendix, Fig. S6C), suggesting that CSU4 may not control COP1 abundance. COP1 exclusively localized in the nucleus, but not in the cytosol, in both etiolated Col and cus4 seedlings (SI Appendix, Fig. S6D), indicating that CSU4 may not affect the nuclear subcellular localization of COP1 in the dark.
csu4 Mutant Seedlings Are Hyposensitive to Light.
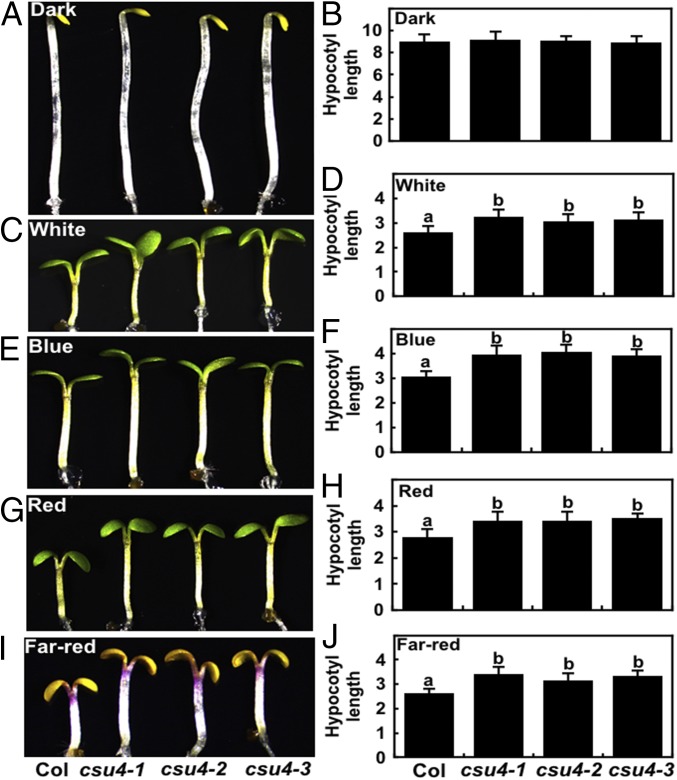
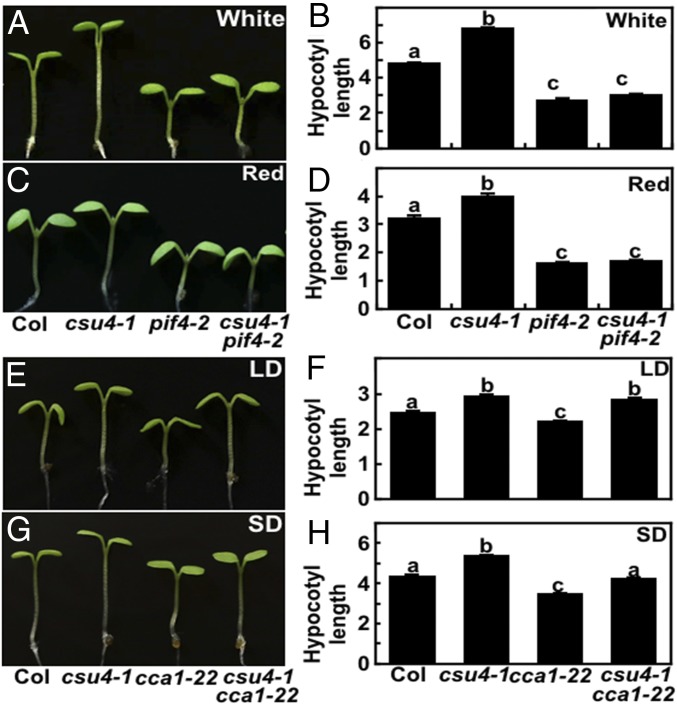
To substantiate CSU4’s role in light signaling, we analyzed the hypocotyl phenotypes of three independent csu4 (csu4-1 to csu4-3) single mutants (without the cop1-6 mutation) grown in various light conditions. All csu4 mutant seedlings showed indistinguishable phenotype with WT when grown in darkness for 4 d (Fig. 2 A and B). In all of the light conditions tested (W, B, R, and FR), all individual csu4 single-mutant seedlings displayed significantly longer hypocotyls than WT (Fig. 2 C–J), indicating that CSU4 promotes light-inhibited hypocotyl elongation and plays a positive role in light signaling.
Fig. 2.
The csu4 seedlings are hyposensitive to light. (A–J) Hypocotyl phenotypes and length of 5-d-old Col and three independent csu4 single-mutant seedlings grown in darkness (A and B) and in W (18.1 μmol⋅m−2⋅s−1) (C and D), B (3.88 μmol⋅m−2⋅s−1) (E and F), R (75.1 μmol⋅m−2⋅s−1) (G and H), and FR (2.05 μmol⋅m−2⋅s−1) (I and J) light conditions. The unit of hypocotyl length is millimeters. Error bars represent SE (n ≥ 20). Letters above the bars indicate significant differences (P < 0.05) as determined by one-way ANOVA with Tukey’s post hoc analysis. The experiments were performed three times, with similar results. The graphs depict one of three experiments.
CSU4 Physically Interacts with CCA1 and Represses Its Transcriptional Repression Activity.
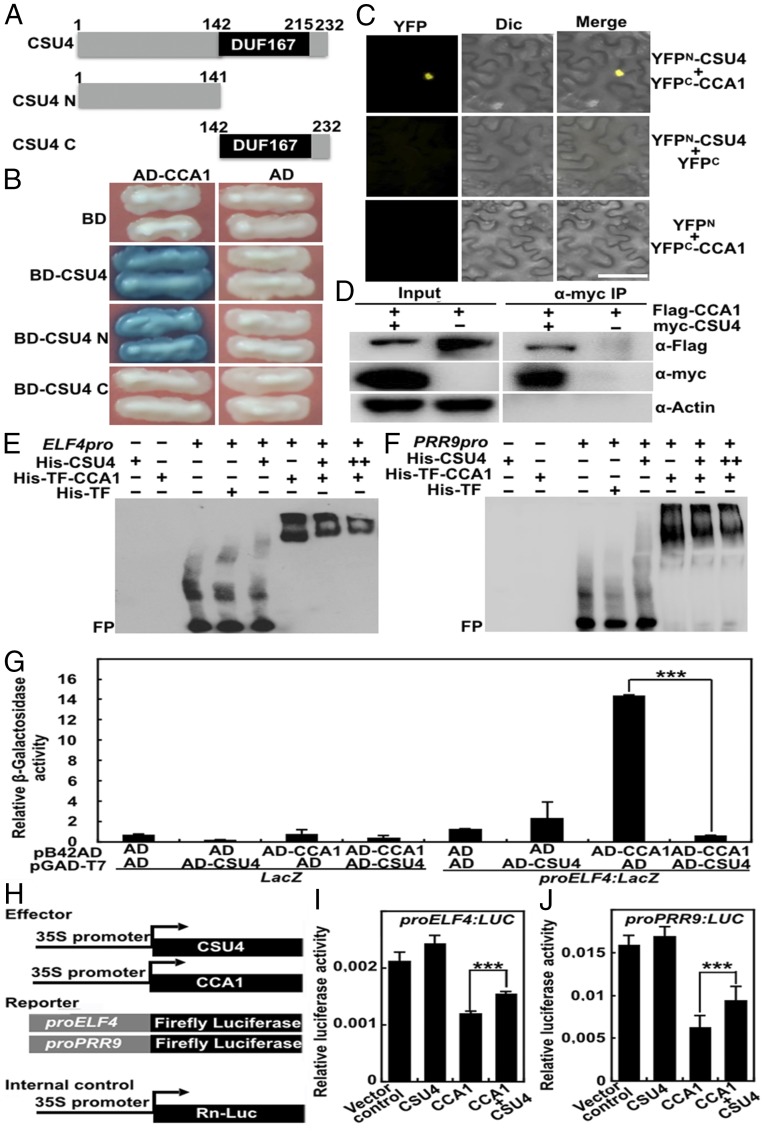
To understand the molecular base for CSU4 in promoting photomorphogenesis, we performed a yeast two-hybrid screen and revealed that CSU4 interacts with CCA1, a key component of the circadian clock and a promoter of hypocotyl growth (23) (Fig. 3 A and B). Further analysis showed that the N terminus of CSU4 (amino acids 1 to 141), but not the C terminus of CSU4 (amino acids 142 to 232) containing a DUF, interacted with CCA1 (Fig. 3 A and B). The data suggest that the N terminus of CSU4 is sufficient and necessary for its interaction with CCA1. To verify the interaction between CSU4 and CCA1, we performed a bimolecular fluorescence complementation (BiFC) assay. Strong YFP signals were clearly detected when CSU4, fused with the N terminus of YFP (YFPN-CSU4), and CCA1, fused with the C terminus of YFP (YFPC-CCA1), were transiently cotransfected into Nicotiana benthamiana leaves. However, no detectable YFP signals were produced when transiently coexpressing the controls—YFPN-CSU4 and YFPC or YFPN and YFPC-CCA1 (Fig. 3C). Next, we further employed a coimmunoprecipitation (co-IP) assay using N. benthamiana leaves transiently expressing 35S:myc-CSU4 and 35S:Flag-CCA1. As shown in Fig. 3D, myc-CSU4 coimmunoprecipitated with Flag-CCA1. Together, these data demonstrate that CSU4 physically interacts with CCA1 in planta.
Fig. 3.
CSU4 physically interacts with CCA1 and represses its transcriptional activity. (A) Schemes of the domain structure of CSU4 and the truncated CSU4 proteins. Numbers indicate the amino acid positions in CSU4. (B) Yeast two-hybrid interactions between the indicated CSU4 and CCA1. BD, binding domain. (C) BiFC assay showing the interaction of CSU4 with CCA1. Full-length CSU4 and CCA1 were fused to the split N- or C-terminal fragments of YFP (YFPN or YFPC, respectively). Unfused YFPN and YFPC fragments were used as negative controls. Merge, merged YFP channel and differential interference contrast (Dic) bright-field images. (Scale bar = 40 μm). (D) Co-IP analysis showing that myc-CSU4 interacts with Flag-CCA1. Total protein was extracted from tobacco leaves transiently expressing 35S:myc-CSU4 alone or together with 35S:Flag-CCA1; the immunoprecipitates were detected using anti-myc and anti-Flag antibodies, respectively. Actin served as a negative control. (E and F) EMSA showing that CSU4 interferes with the binding of CCA1 to ELF4 (E) or PRR9 (F) promoters. A (−) symbol indicates the absence of corresponding of probes or proteins. For His-CSU4, (+) and (++) indicate that 18 and 36 pmol is present, respectively. For His-TF, (+) indicates that 9.2 pmol is present. For His-TF-CSU4, (+) indicates that 8 pmol is present. TF (trigger factor), a prokaryotic ribosome 3-associated chaperone protein. FP indicates free probes. (G) Yeast one-hybrid analysis showing that CSU4 interferes with CCA1’s ability to bind to ELF4 promoter. Error bars represent the SD of four independent yeast cultures. ***P < 0.001 as determined by Student’s t test. The experiments were performed three times, with similar results. (H) Schematic representation of various constructs used in the transient transfection assay in Arabidopsis protoplasts. Arrows after the 35S promoters indicate the transcriptional start site. ELF4 or PRR9 promoter was fused to firefly luciferase to create the reporter constructs. (I and J) Bar graphs showing CSU4 represses the repression of CCA1 on the proELF4:LUC (I) and proPRR9:LUC (J) reporters. Error bars represent the SD of three independent transient transfections in Arabidopsis protoplasts. ***P < 0.001 as determined by Student’s t test. The experiments were performed three times, with similar results.
CCA1, a Myb-domain transcriptional factor, can bind to its numerous targets and is considered a bona fide transcriptional repressor (23). Thus, we examined whether CSU4 affects the binding of CCA1 to its targets using electrophoretic mobility shift assays (EMSAs). His-trigger factor (TF)-CCA1, but not His-TF (negative control), could directly bind to ELF4 or PRR9 promoters (two targets of CCA1) (33, 34) (Fig. 3 E and F). In the presence of His-CSU4, the binding of His-TF-CCA1 to ELF4 or PRR9 promoter DNA subfragments was obviously reduced. In addition, as the amount of His-CSU4 increased in the reactions, the His-TF-CCA1 binding affinity to ELF4 and PRR9 promoters was markedly decreased (Fig. 3 E and F), indicating that CSU4 may interfere with the binding of CCA1 to its target promoters. In yeast cells, activating domain (AD)-fused CCA1 could activate proELF4:LacZ reporter (Fig. 3G), indicating that CCA1 can directly bind to the ELF4 promoter, thus allowing AD to activate LacZ reporter in yeast cells. AD-CSU4 alone did not have a significant effect on proELF4:LacZ reporter; however, when cotransformed with AD-CCA1, activation of AD-CCA1 on proELF4:LacZ was dramatically reduced in yeast cells, as indicated by β-galactosidase activity (Fig. 3G). These results imply that CSU4 may interfere with CCA1’s ability to regulate promoter activity of ELF4. Similar to the previous finding (34), CCA1 alone could repress the transcription of proELF4:LUC or proPRR9:LUC when it was transiently expressed in Arabidopsis protoplasts (Fig. 3 H–J). CSU4 alone did not have any effect on these reporters; however, the repression of CCA1 on proELF4:LUC or proPRR9:LUC was significantly reduced when it was transiently coexpressed with CSU4 (Fig. 3 H–J). These results collectively suggest that CSU4 can reduce the transcriptional repression activity of CCA1 possibly through interfering with CCA1’s ability to bind its target promoters.
CSU4 Represses CCA1 in the Early Morning and PIF4 in the Early Evening.
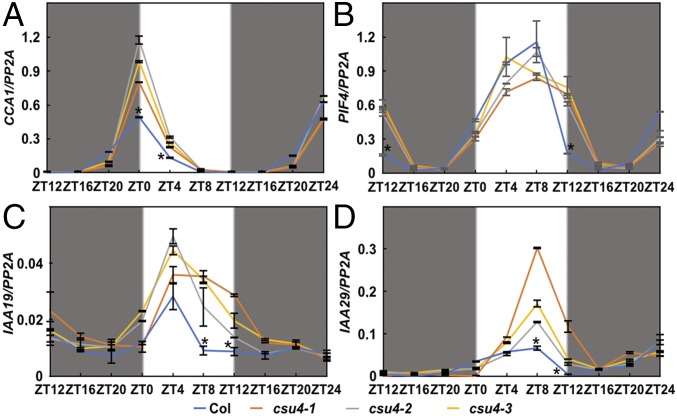
CCA1 is a central component of the circadian clock, which represses its own gene expression and phenotypically promotes hypocotyl elongation (23). Given that CSU4 inhibits hypocotyl elongation, interacts with CCA1, and negatively regulates CCA1 transcriptional repression activity (Figs. 2 and 3), we tested whether CSU4 affects the expression of CCA1. As shown in Fig. 4A, CCA1 transcript levels were clearly increased in csu4 mutants in the early morning [zeitgeber time (ZT)0 and ZT4], suggesting that CSU4 negatively regulates CCA1 at dawn.
Fig. 4.
CSU4 negatively regulates the expression of CCA1 in the early morning and PIF4 in the early evening. Real-time qPCR analysis of CCA1 (A), PIF4 (B), IAA19 (C), and IAA29 (D) transcript levels in Col and three independent csu4 mutant seedlings grown in 12 h light/12 h dark photoperiods. Error bars represent the SD of three technical replicates. *P < 0.05 as determined by Student’s t test. The experiments were performed three times, with similar results.
CCA1 promotes hypocotyl elongation predominantly through the activation of PIF4 expression (35). In agreement, PIF4 expression was markedly elevated in all three independent csu4 mutant seedlings at ZT12 (Fig. 4B), and IAA19 and IAA29 (PIF4-controlled genes) (36) were also increased in the csu4 mutants (Fig. 4 C and D), indicating that CSU4 represses the expression of PIF4 specifically in the early evening.
Considering that CSU4 affected the temporal regulation of CCA1 and PIF4 expression (Fig. 4 A and B), we examined whether CSU4 regulates the expression of other circadian clock genes. Expression levels of TOC1, GI, ELF3, ELF4, LUX, PRR5, PRR7, and PRR9 were changed at one or multiple time points in a 24-h period in the csu4 mutants compared with those in the WT (SI Appendix, Fig. S7), suggesting that CSU4 is a common regulator for the expression of these circadian clock genes. Although CSU4 affects the transcription of these clock genes, CSU4 itself was not regulated by the circadian clock at either the transcriptional or protein level (SI Appendix, Fig. S8).
Overexpression of Either CCA1 or PIF4 Partially Suppresses cop1 or det1.
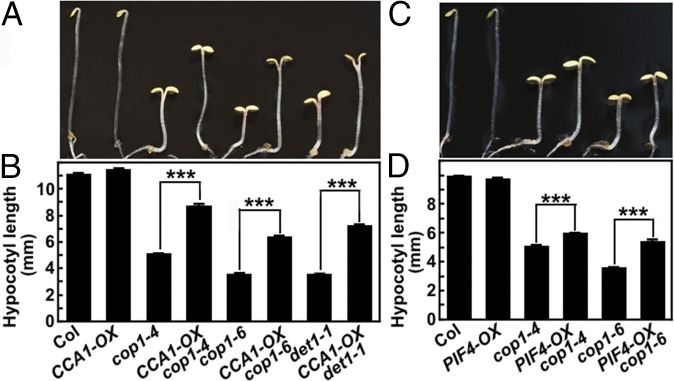
Mutations in CSU4 genetically suppressed cop1-6 or det1-1 (Fig. 1 and SI Appendix, Fig. S4), and CSU4 repressed the CCA1 and PIF4 transcript levels (Fig. 4 A and B). These results indicate that suppression of cop1-6 or det1-1 by csu4 is likely, at least in part, caused by elevation of CCA1 and PIF4. To test this, we generated CCA1-OX cop1-4, CCA1-OX cop1-6, CCA1-OX det1-1, PIF4-OX cop1-4, and PIF4-OX cop1-6 transgenic lines by genetic crossing. Overexpression of either CCA1 or PIF4 significantly suppressed the short hypocotyl phenotype of cop1-4, cop1-6, or det1-1 in the dark (Fig. 5). A previous study has shown that overexpression of PIF4 partially suppresses det1-1 (37). Together, these findings support the notion that elevation of CCA1 and PIF4 transcript levels due to csu4 mutations partially contributes to the suppression of cop1-6 or det1-1.
Fig. 5.
Overexpression of CCA1 or PIF4 partially suppress cop1 in darkness. (A and B) Hypocotyl phenotypes and length of 4-d-old Col, CCA1-OX, cop1-4, cop1-6, det1-1, CCA1-OX cop1-4, CCA1-OX cop1-6, and CCA1-OX det1-1 mutant seedlings grown in darkness. (C and D) Hypocotyl phenotype and length of 4-d-old Col, PIF4-OX, cop1-4, cop1-6, PIF4-OX cop1-4, and PIF4-OX cop1-6 mutant seedlings grown in darkness. Data are mean ± SE; n ≥ 20. ***P < 0.001 (Student’s t test) relative to Col. The experiments were performed three times, with similar results. The graphs depict one of three experiments.
Next, we examined the genetic interactions between CSU4 and CCA1 or PIF4. Consistent with previous studies, the pif4-2 mutant showed shorter hypocotyls as compared with WT in W and R light (38). The hypocotyl length of csu4-1 pif4-2 was similar to pif4-2 in W and R light (Fig. 6 A–D), suggesting that PIF4 acts genetically downstream of CSU4. Loss of CCA1 function resulted in shortened hypocotyls under light, which was consistent with results from previous studies (23, 39, 40). The csu4-1 cca1-22 double mutant showed indistinguishable hypocotyl length with csu4-1, which was significantly longer than WT and cca1-22 in LD conditions (Fig. 6 E and F). In short-day (SD) conditions (i.e., continuous W, B, R, and FR light conditions), the hypocotyl length of csu4-1 cca1-22 was shorter than that of csu4-1 but longer than those of WT and cca1-22 (Fig. 6 G and H and SI Appendix, Fig. S9). These data suggest that while the short hypocotyl phenotype of cca1-22 is dependent on a functional CSU4 in LD conditions, it is independent of CSU4 in SD W, B, R, and FR light conditions.
Fig. 6.
CSU4 acts upstream of PIF4. (A–D) Hypocotyl phenotypes and length of 5-d-old Col, csu4-1, pif4-2, and csu4-1 pif4-2 mutant seedlings grown in W (14.74 μmol⋅m−2⋅s−1) (A and B) or R (129 μmol⋅m−2⋅s−1) (C and D) conditions. (E–H) Hypocotyl phenotypes and length of 5-d-old Col, csu4-1, cca1-22, and csu4-1 cca1-22 mutant seedlings grown in LD (E and F) or SD (G and H) conditions. The unit of hypocotyl length is millimeters. Error bars represent SE (n ≥ 20). Letters above the bars indicate significant differences (P < 0.05) as determined by one-way ANOVA with Tukey’s post hoc analysis. The experiments were performed three times, with similar results. The graphs depict one of three experiments.
Discussion
Light and the circadian clock interact to control seedling photomorphogenic development in Arabidopsis seedlings. Although CSU4 is not circadian regulated at both the transcript and protein levels (SI Appendix, Fig. S8), it interacts with the circadian clock and controls the expression of several circadian clock genes in gating hypocotyl growth (Figs. 3 and 4), suggesting that CSU4 affects the circadian clock and its output pathway. CSU4, CCA1, and PIF4 coordinately modulate the hypocotyl under diurnal conditions. CSU4 interacted with CCA1 and reduced its DNA binding affinity, thereby repressing its transcriptional repression activity toward its targets (Fig. 3). It is possible, therefore, that CSU4 may form heterodimers with CCA1 to inhibit its ability to bind to target sites, thereby repressing CCA1 biochemical activity. Previous studies have demonstrated that CCA1 negatively regulates its own expression (23). Thus, suppression of CCA1 transcriptional repression activity by CSU4 likely leads to the elevation of its own expression, thereby inhibiting the PIF4 and PIF4-mediated gene expression and hypocotyl elongation. CSU4 acts as a transcriptional repressor, gating rhythmic expression of CCA1 and PIF4 diurnally (Fig. 4). Consequently, these molecular events contribute to the light-inhibited hypocotyl growth in plants.
Mutations in CSU4 largely rescued the cop1-6 or det1-1 phenotype, especially with respect to hypocotyl phenotypes in darkness (Fig. 1 and SI Appendix, Fig. S4). Both COP1 and DET1 function as E3 ubiquitin ligases in promoting degradation of numerous downstream targets in repressing photomorphogenesis in darkness (3, 4). CSU4 protein abundance is not controlled by COP1 and DET1 (SI Appendix, Fig. S6), suggesting that CSU4 is not likely a substrate of these two E3 ubiquitin ligases. COP1/DET/FUS contribute to the stabilization of PIF4 abundance (37), and overexpression of PIF4 in either a cop1 or det1-1 single-mutant background partially suppressed their shortened hypocotyl phenotypes (Fig. 5 C and D) (37). In csu4 mutant seedlings, both CCA1 and PIF4 transcript levels were significantly increased (Fig. 4 A and B). All this evidence supports a possibility that the suppression of cop1-6 and det1-1 phenotypes in the csu4 mutants is likely, at least in part, caused by the elevation of CCA1 and PIF4 transcript levels. DET1 physically interacts with the morning-phase components (CCA1 and LHY) and is recruited to their evening-phase gene targets, including TOC1 and GI, to repress their expression (41). Neither DET1 nor CSU4 is controlled by the circadian clock at both transcriptional and protein levels (SI Appendix, Fig. S8) (41). Like DET1, CSU4 associated with CCA1, and it negatively controlled CCA1 expression at dawn (Figs. 3 and 4A), suggesting a direct involvement of CSU4 as a transcriptional corepressor in mediating the circadian clock. Although the CSU4 constantly expressed under various light conditions or at different time points (SI Appendix, Fig. S8), it negatively controls CCA1 specifically in the early morning and PIF4 specifically in the early evening (Fig. 4). These facts indicate that additional or yet-unidentified factors are required for CSU4 in regulating CCA1 or PIF4 expression at specific time points.
PIF4 functions downstream of the circadian clock and gates circadian clock-mediated hypocotyl growth (26, 28). Both morning complex CCA1-LHY and evening complex ELF3-ELF4-LUX regulate PIF4 expression at different time points (26, 35). On the one hand, CCA1-LHY activates PIF4 expression to regulate hypocotyl growth during the day (35, 42) while ELF3 interacts with PIF4 independent of the evening complex to repress PIF4 biochemical activity in order to inhibit hypocotyl elongation in the daytime (28). On the other hand, the evening complex ELF3-ELF4-LUX directly associates with the promoter of PIF4 through transcription factor LUX to repress its gene expression, which in turn, serves to control PIF4-mediated gene expression and inhibit hypocotyl elongation in the evening (26). CSU4 repressed the CCA1 expression in the early morning and down-regulated PIF4 in the early night (Fig. 4 A and B), supporting that CSU4 plays an essential role in gating the timing of hypocotyl elongation probably through repression of CCA1 and PIF4 at dawn and dusk, respectively, to integrate photoperiod signals for growth and development.
Materials and Methods
Detailed descriptions of plant material and growth conditions, map-based cloning, sequencing by oligonucleotide ligation and detection, mutation identification, construction of plasmids, co-IP analysis, immunoblot analysis, EMSA analysis, yeast one- and two- hybrid assays, protoplast assays, BiFC assays, hypocotyl length measurements, production of specific CSU4 polyclonal antibody, and real-time qPCR assays are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Yong Ding for cca1-22 seeds. This work was supported by the National Key R&D Program of China (Grant 2017YFA0503800); the National Natural Science Foundation of China (Grants 31330048 and 31621001); the Peking-Tsinghua Center for Life Sciences; NIH Grants GM-47850, GM-109076, and GM-114297; the China Postdoctoral Science Foundation (X.Z.); open funds of the State Key Laboratory of Plant Physiology and Biochemistry (Grant SKLPPBKF1806); and Southern University of Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813171115/-/DCSupplemental.
References
- 1.Sullivan JA, Deng XW. From seed to seed: The role of photoreceptors in Arabidopsis development. Dev Biol. 2003;260:289–297. doi: 10.1016/s0012-1606(03)00212-4. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Ouyang X, Deng XW. Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol. 2014;21:96–103. doi: 10.1016/j.pbi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Shi H, et al. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc Natl Acad Sci USA. 2018;115:6482–6487. doi: 10.1073/pnas.1803861115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 7.Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- 8.von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 9.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen H, et al. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 13.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian HL, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng X, et al. Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell. 2013;25:115–133. doi: 10.1105/tpc.112.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernardo-García S, et al. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni W, et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong J, et al. Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr Biol. 2017;27:2420–2430.e6. doi: 10.1016/j.cub.2017.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni W, et al. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat Commun. 2017;8:15236. doi: 10.1038/ncomms15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaffer R, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 24.Seo PJ, et al. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–2442. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu JY, Oh E, Wang T, Wang ZY. TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun. 2016;7:13692. doi: 10.1038/ncomms13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieto C, López-Salmerón V, Davière JM, Prat S. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol. 2015;25:187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- 29.McNellis TW, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu D, et al. The RING-Finger E3 ubiquitin ligase COP1 SUPPRESSOR1 negatively regulates COP1 abundance in maintaining COP1 homeostasis in dark-grown Arabidopsis seedlings. Plant Cell. 2014;26:1981–1991. doi: 10.1105/tpc.114.124024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, et al. Arabidopsis COP1 SUPPRESSOR 2 Represses COP1 E3 ubiquitin ligase activity through their coiled-coil domains association. PLoS Genet. 2015;11:e1005747. doi: 10.1371/journal.pgen.1005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- 33.Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- 34.Kamioka M, et al. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell. 2016;28:696–711. doi: 10.1105/tpc.15.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa Y, Yamashino T, Mizuno T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:838–854. doi: 10.1093/pcp/pcp028. [DOI] [PubMed] [Google Scholar]
- 36.Sun J, Qi L, Li Y, Zhai Q, Li C. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell. 2013;25:2102–2114. doi: 10.1105/tpc.113.112417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong J, et al. Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell. 2014;26:3630–3645. doi: 10.1105/tpc.114.130666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Y, et al. Phosphorylation of histone H2A at serine 95: A plant-specific mark involved in flowering time regulation and H2A.Z deposition. Plant Cell. 2017;29:2197–2213. doi: 10.1105/tpc.17.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, et al. MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell. 2018;30:67–82. doi: 10.1105/tpc.17.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau OS, et al. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu SX, et al. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.