Significance
Icosahedral symmetry has generally been assumed during cryo-electron microscopy reconstructions of most spherical viruses, including flaviviruses. However, unlike other viruses, the flavivirus core appeared to lack an ordered structure. By removing symmetry constraints, it was found that in immature flaviviruses, the nucleocapsid core is asymmetrically positioned with respect to the lipid membrane and glycoprotein shell. Thus, the assembly and budding process leads to imperfect icosahedral symmetry, which may affect virus maturation. Similarly, in the study of many other viruses, icosahedral averaging may have hidden biologically significant events.
Keywords: flaviviruses, cryo-electron microscopy, assembly, budding, asymmetry
Abstract
Flaviviruses assemble initially in an immature, noninfectious state and undergo extensive conformational rearrangements to generate mature virus. Previous cryo-electron microscopy (cryo-EM) structural studies of flaviviruses assumed icosahedral symmetry and showed the concentric organization of the external glycoprotein shell, the lipid membrane, and the internal nucleocapsid core. We show here that when icosahedral symmetry constraints were excluded in calculating the cryo-EM reconstruction of an immature flavivirus, the nucleocapsid core was positioned asymmetrically with respect to the glycoprotein shell. The core was positioned closer to the lipid membrane at the proximal pole, and at the distal pole, the outer glycoprotein spikes and inner membrane leaflet were either perturbed or missing. In contrast, in the asymmetric reconstruction of a mature flavivirus, the core was positioned concentric with the glycoprotein shell. The deviations from icosahedral symmetry demonstrated that the core and glycoproteins have varied interactions, which likely promotes viral assembly and budding.
Flaviviruses, such as dengue virus (DENV), Zika virus (ZIKV), and West Nile virus (WNV), are enveloped positive-strand RNA viruses. These viruses can cause severe disease in humans. For example, DENV elicits muscle and joint pain referred to as breakbone fever, and ZIKV induces microcephaly in developing fetuses. WNV, first introduced to the United States in 1999, causes encephalitis in adults.
Flavivirus virions mature as they pass through the secretory system, acquiring their membrane when they bud into the endoplasmic reticulum (ER) (1). There are 60 copies of the trimeric glycoproteins present as spikes on the immature virus (2–4). After exposure to low pH in the trans-Golgi network, the glycoproteins undergo a conformational rearrangement from 60 trimers to 90 dimers, which lie flat on the virus surface (5, 6). In the structures of mature flaviviruses, the glycoproteins are arranged in a herringbone pattern that forms the outer shell of the virus (7–11). In contrast, flavivirus cores appear to lack a discernable organization, and icosahedrally averaged reconstructions of flaviviruses show no structure for the core other than a roughly spherical shell (2–4, 7–12). The crystal and NMR structures of the capsid protein suggest that the capsid associates with genomic RNA on one side and membrane on the other side (13–15).
Asymmetric cryo-EM reconstructions of bacteriophages have proven useful to elucidate the structure of portals and tails at unique vertices (16, 17). Asymmetric reconstructions have also been used to determine unique features of genomic and protein architecture inside virions (18–23). The antibody reactivity of flaviviruses suggests that virions exhibit conformational variability (24). To study unique structural features that may have been hidden by icosahedral averaging, an asymmetric reconstruction was performed of immature Kunjin virus (KUNV), a WNV strain with low virulence. It was found that the viral core was asymmetrically positioned with respect to the glycoprotein shell. In contrast, an asymmetric reconstruction of mature KUNV showed concentric positioning of the core. However, both structures showed perturbations in the glycoprotein organization.
Results
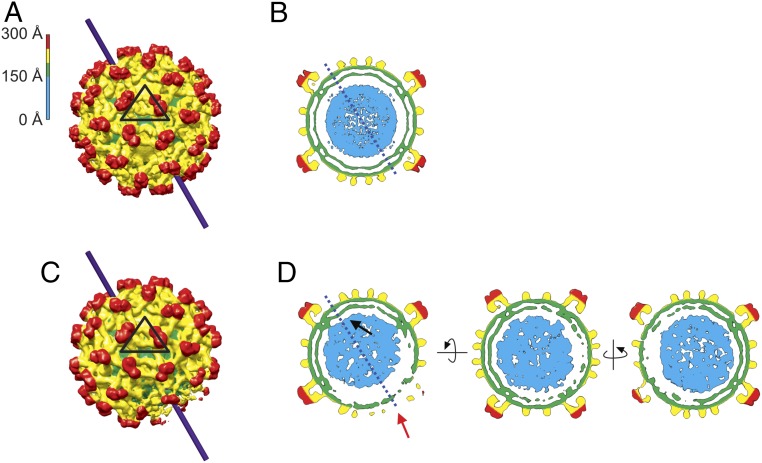
Cryo-EM movies of immature and mature KUNV were collected on an FEI Titan Krios equipped with a K2 direct electron detector. Immature and mature KUNV showed characteristic spiky and smooth particles, respectively (SI Appendix, Fig. S1 A and B). An asymmetric reconstruction (Materials and Methods) of immature KUNV was performed with the program RELION 2.1 (25), starting with an icosahedral reconstruction (Fig. 1 A and B) as an initial model. The asymmetric reconstruction was refined to ∼20 Å resolution (Fig. 1C and SI Appendix, Fig. S2). The position of the majority of the glycoprotein trimers in the asymmetric reconstruction of the immature virus correspond to their position in the icosahedral reconstruction, demonstrating that the glycoprotein shell has approximate icosahedral symmetry. In addition, the position of the transmembrane densities of these glycoproteins in the asymmetric reconstruction is essentially the same as in the icosahedral reconstruction.
Fig. 1.
Cryo-EM reconstructions of immature KUNV. (A) Surface view of the icosahedral reconstruction, colored radially as indicated in the color key. The icosahedral asymmetric unit is indicated by a black triangle. (B) Central cross-section of the icosahedral reconstruction. (C) Surface view of the asymmetric reconstruction. (D) Central cross-sections of the asymmetric reconstruction shown in three orthogonal views. The black arrow indicates the proximal pole, where the nucleocapsid core approaches closer to the glycoprotein shell, whereas the red arrow indicates the distal pole, where the core is farthest from the shell. The solid line in A and C and the dotted line in B and D are defined by joining the points where the proximal and distal poles would be in the asymmetric reconstruction, as shown in D. All reconstructions are contoured at ∼3 σ.
However, unlike the icosahedral reconstruction in which the nucleocapsid core density is situated concentric with the glycoprotein shell, in the asymmetric reconstruction, the nucleocapsid core is positioned eccentrically, coming closer to the membrane on one side (the proximal pole) than on the other side (the distal pole) of the glycoprotein shell. The eccentricity of the core was also observed in an independent asymmetric reconstruction of immature ZIKV (SI Appendix, Fig. S3). To verify that the asymmetric method did not induce artificial asymmetry into the reconstruction, simulations were performed in which projections of an icosahedral object were reconstructed without imposing symmetry. The reconstruction was icosahedral, as expected.
The core density in the asymmetric reconstruction is an oblate spheroid, with an approximate diameter of ∼260 Å stretching from pole to pole (SI Appendix, Fig. S4). The gap between the core and the inner leaflet of the membrane is ∼23 Å at the proximal pole, which is slightly smaller than the gap observed in the icosahedral reconstruction. The maximum external diameter of the glycoprotein shell is ∼560 Å. There are density protrusions from the core connecting it to density that represent presumably transmembrane helices of the prM and E glycoproteins.
At the distal pole, where the core is farthest from the glycoprotein shell, there are significant distortions in the glycoprotein organization (Fig. 1D). Density for the inner leaflet of the membrane and the transmembrane helices is missing in this region, resulting in a large gap between the core and membrane of ∼64 Å. A radial density plot shows there is a density peak at the distal pole, near the expected radius for the inner leaflet, which is not observed at the proximal pole (SI Appendix, Fig. S5). This suggests there is distinct density at the distal pole region.
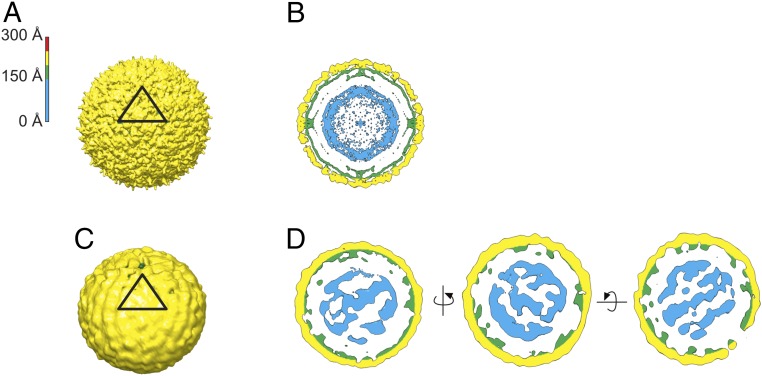
A ∼35-Å (Fig. 2C and SI Appendix, Fig. S6) asymmetric reconstruction of mature KUNV was calculated using an icosahedral reconstruction as an initial model (Fig. 2 A and B). This reconstruction has layers of membrane and glycoproteins at the same radial distance as in the icosahedral reconstruction. The virus is roughly spherical, and has an uneven surface, with a large patch of distorted density. The core in the asymmetric reconstruction of the mature virus (unlike in the immature virus) is positioned concentric with the glycoprotein shell (Fig. 2D). Protrusions extend from the core in the mature virus into the glycoprotein layer, but the density is noisy and less well resolved than for the immature virus.
Fig. 2.
Cryo-EM reconstructions of mature KUNV. (A) Icosahedral reconstruction of mature virus colored radially, as indicated by the color key, with the asymmetric unit indicated by a black triangle. (B) Cross-section of the icosahedral reconstruction. (C) Asymmetric reconstruction of the mature virus. (D) Cross-sections of the asymmetric reconstruction shown in three orthogonal views. All reconstructions are contoured at ∼3 σ.
Discussion
Cells infected with a flavivirus synthesize glycoproteins that probably form trimers in the membranes of the endoplasmic reticulum. As these trimers aggregate in the ER membrane, each with essentially the same structure and in identical environments, they will tend to form icosahedral structures, as predicted for the assembly of viruses by Crick and Watson (26). However, what determines the triangulation number and size of the final assembled virus is unclear. One determinant that may influence particle size is the nucleocapsid core. In the absence of a core, recombinant subviral particles containing prM and E have been reported to have a triangulation number of T = 1 (27). The core in immature KUNV contacts the glycoproteins at their transmembrane domains, as was also observed for immature ZIKV (4), and these interactions could influence particle assembly and size. There are likely multiple determinants of particle size, as subviral particles lacking cores have been observed that separate into two distinct size classes of approximately the same size as infectious virions, as well as the smaller T = 1 predicted particles (28).
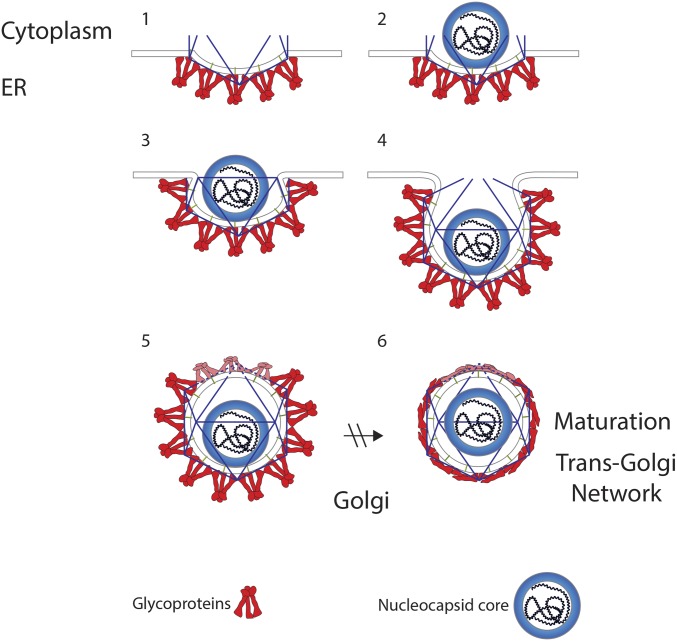
The tendency to form 3D icosahedral structures of any size in a membrane is inconsistent with the normal planar structure of a membrane. As more trimeric glycoproteins are recruited into the nascent icosahedral structure, there is pressure for the partially formed glycoprotein shell to grow and finally close at what will become the distal pole (Fig. 3). The closing of the shell is likely impeded by the high membrane curvature and steric clashes with adjacent spike glycoproteins at the bud neck, and probably results in missing trimers at the distal pole. In agreement with this hypothesis, a simulation of icosahedral virus budding determined that the final one to three subunits were not incorporated into nascent particles (29). Alternatively, one or more host proteins that might promote budding could be incorporated into the virus particle at the late stage of viral budding. It is unknown what viral and host machinery is involved in membrane scission during the final moments of budding, but the ESCRT proteins may play a role, as these were implicated in flavivirus budding (30).
Fig. 3.
Model for flavivirus budding. (1) The glycoproteins (red) assemble on the ER membrane and form the beginning of an icosahedron. (2) The nucleocapsid core (blue), which has an unknown structure, interacts with the glycoproteins on the ER membrane. (3) As the bud grows, more glycoprotein trimers are recruited into the virion. (4) At late-stage budding, virions are unable to incorporate the final copies of the trimeric glycoprotein spikes, which results in (5) fewer glycoproteins and distortions in the glycoprotein shell (top three trimers). The newly formed immature virus then leaves the ER and traffics through the Golgi and encounters low pH in the trans-Golgi network. (6) Maturation in the trans-Golgi network results in a large conformational rearrangement and distortions across the particle. As a result of maturation, the nucleocapsid is repositioned to be concentric with the glycoprotein shell.
The nucleocapsid core was found to be eccentrically positioned in an immature flavivirus, similar to its positioning in herpesvirus and HIV (31, 32). However, the flavivirus core is unique, in that it repositions as a result of maturation. During the maturation process, the glycoproteins undergo conformational rearrangements from trimers to dimers, possibly loosening capsid–glycoprotein interactions and leading to the release and centering of the nucleocapsid core. The connections between glycoprotein transmembrane domains and the core in immature ZIKV suggested that the core had icosahedral symmetry (4). On the basis of the asymmetric reconstructions of KUNV and ZIKV, there is, at a minimum, partial icosahedral symmetry in the core, at the capsid-glycoprotein contact sites. Partial icosahedral symmetry in nucleocapsid cores is not limited to flaviviruses. The cores of alphaviruses and hepatitis B virus were assumed to be icosahedral, but have been found to assemble with imperfect icosahedral symmetry (33, 34).
By avoiding the assumption of icosahedral symmetry, it has here been possible to determine the structure of an immature particle during early stages of assembly and budding. It would seem probable that other enveloped viruses might have a similar assembly process within a cell.
Materials and Methods
Cell Lines.
C6/36 were grown in Minimum Essential Media (MEM) supplemented with 25 mM Hepes at pH 7.3 and 10% FBS at 30 °C. Vero cells were grown in Dulbecco’s Modified Eagle Media (DMEM) supplemented with 1× nonessential amino acids and 10% FBS at 37 °C.
Immature KUNV Infection.
KUNV virus, strain MRM61C, was added to ∼7 × 107 C6/36 cells at a multiplicity of infection (MOI) of 8 in twenty, 150-mm dishes. The passage of the virus was P3, and of the cells was P11. The cells were infected with virus in 5 mL PBS with 1% FBS, 1% CaCl2, and 1% MgCl2, and rocked for 45 min at room temperature, followed by a 15-min incubation at 30 °C. Fifteen milliliters MEM, 25 mM Hepes at pH 7.3, and 2% FBS was added to the cells. The cells were then incubated at 30 °C overnight. At 17 h postinfection (hpi), the inoculum was removed, and the cells were washed twice with PBS and incubated with new media including 20 mM NH4Cl. The wash and media replacement was repeated at 19 hpi. Virus was harvested at 30, 48, and 70 hpi. Cytopathic effect was first observed at 48 hpi.
Mature KUNV Infection.
Approximately 1 × 107 Vero cells at P11 were infected by KUNV virus P3 at an MOI of 8. The cells were infected as described for immature virus, with the following changes: incubations were performed at 37 °C, and the media used during the infection was DMEM with 1× nonessential amino acids and 2% FBS. Virus was harvested at 24, 50, and 70 hpi, and cytopathic effect was first observed at 24 hpi.
Virus Purification.
The 400 mL of media containing virus was clarified at 9,500 rpm in the JA-10 rotor (Beckman Coulter, Inc.) for 20 min. Forty percent (wt/vol) polyethylene glycol (PEG) 8,000 was added to the media to a final concentration of 8%. The media was then stirred slowly at 4 °C overnight. The media was spun at 9,500 rpm in a JA-10 rotor (Beckman Coulter, Inc) for 50 min. The supernatant was removed, and precipitated virus was resuspended in 15 mL cold TNE at pH 8 (12 mM Tris at pH 8, 120 mM NaCl, 1 mM EDTA). The virus was then added to a polycarbonate bottle in which 2 mL of a 22% sucrose solution in TNE was underlaid with a Pasteur pipette. The virus was spun in a Type Ti50.2 rotor (Beckman Coulter, Inc.) at 32,000 rpm for 2 h. The supernatant was removed and the pellet was resuspended in 1 mL TNE. A potassium tartrate–glycerol gradient was made with 10–15–20–25–30–35% tartrate in TNE in an Ultra-Clear tube (Beckman Coulter, Inc). To make the gradient, 1.5 mL of each layer was overlaid using a 1-mL pipette tip that was cut with scissors to reduce mixing of the layers during layering. The virus was spun for 1 min at 3,000 × g to remove PEG aggregates, and then layered on top of the gradient. The layer interfaces were marked and the gradient was spun for 2 h at 32,000 rpm in a Type SW-41 rotor (Beckman Coulter, Inc.). The virus was visualized under a blue light. A blue band was observed near the 20%/25% interface, and was extracted with a 22-gauge needle and syringe. The virus was buffer exchanged and concentrated in a 100-kDa concentrator four times with TNE, to a final volume of ∼50 µL. Virus purity and the relative concentration of E protein was assessed by SDS/PAGE with BSA standards.
Cryo-EM.
Next, 2.5 µL of virus was added to the carbon side of nonglow discharged ultrathin lacey carbon grids No. 01824 (Ted Pella) and plunge-frozen into liquid ethane using a CP3 plunger (Gatan) at 85% humidity. Blotting times used were between 6 and 8 s.
Data were collected using a Titan Krios microscope (FEI) operated at 300 kV and equipped with a K2 Summit direct electron detector (Gatan). The magnification used was 18,000× in superresolution counting mode, which corresponded to a pixel size of 0.81 Å per pixel. The objective lens astigmatism was corrected with s2stigmator (35). Frames were recorded every 0.2 s for 8 s, with a dose rate of ∼8 electrons per physical pixel per second. The total dose for each image stack of 40 frames was ∼24 electrons per Å2. The defocus range used was −0.5 to −3 µm.
There were 387 and 369 movies collected for immature and mature virus, respectively. The Appion (36) and Leginon (37) systems were used for data collection and preprocessing. Individual frames of raw movies were aligned and distortion corrected with MotionCor2 (38). CTF values were estimated with CTFFIND4 (39). Particle picking was done with FindEM (40), resulting in 12,567 immature particles and 23,145 mature particles. A box size of 1,280 × 1,280 pixels was used for both particles, and the particles were binned 4×.
Icosahedral Reconstructions.
Reconstructions were performed with a beta release of the RELION 2.1 program (25). For the immature virus reconstruction, a particle mask of 650 Å in diameter was used. Two rounds of 2D alignment and classification were performed to remove “junk” particles from the dataset. The first round was run with 50 classes. Fifteen “good” classes were selected, which contained a total of 9,368 particles. A second round of 2D classification with 40 classes was run. Ten classes contained particles with defined features, which in total contained 7,396 particles. A random subset of 2,000 particles was selected from this dataset for initial model generation.
Five iterations of stochastic gradient descent were performed in RELION with the 2,000 particle subset to generate an initial model. Two rounds of refinement of the initial model were then performed using auto-refinement mode, which was run with all 7,396 particles while icosahedral symmetry was imposed. Sampling was initiated with 3.75° global sampling and local sampling starting at 0.9°. The reconstruction reached 9.3 Å, using the gold-standard Fourier shell correlation of 0.143.
The reconstruction for mature virus was performed similarly. A particle mask of 550 Å in diameter was used for the mature particle. Two rounds of 2D classification were performed to remove junk particles. In the first round, 50 classes were used, and 11 classes were selected that contained 16,938 particles. A second round of 2D classification resulted in two major classes that that were selected with a total of 9,206 particles. The particle set was used for stochastic gradient descent initial model generation in RELION. Refinement was carried out in auto-refinement mode, using the initial model low-pass filtered to 80 Å. The resolution was estimated to be 6.4 Å, using the gold-standard Fourier shell correlation.
Asymmetric Reconstructions.
Asymmetric reconstructions were performed with RELION 2.1. The icosahedral reconstruction of the immature virus was used as an initial model and low-pass filtered to 35 Å. Sampling was initiated with 3.75° global sampling and local sampling starting at 0.9°. The angles for the 9,638 particles used in the icosahedral reconstruction were matched to the icosahedral density without imposing symmetry. As the map is icosahedral, it would not be expected to change greatly, as each of the 60 icosahedral positions are equally valid for a given image. However, the nucleocapsid core repositioned itself from the center to one side of the glycoprotein shell within five iterations, which suggests its signal was strong enough to select a unique orientation. The reconstruction reached 19 Å resolution.
The refinement was repeated in 3D classification mode, with a T factor of 0.5. A low T factor was suggested to promote the emergence of asymmetric features in the refinement of other viruses (22). This structure also resulted in the same asymmetric positioning of the nucleocapsid core.
Mature virus was reconstructed using the icosahedral mature virus, low pass filtered to 40 Å resolution, as an initial model. The refinement, using 9,206 particles, reached 35 Å resolution. The asymmetric reconstruction of immature KUNV likely reached a higher resolution because the immature virus has recognizable spiky features compared with the smooth surface of the mature virus, which made alignment more accurate for the immature virus.
Asymmetric Reconstruction of Immature ZIKV.
An asymmetric reconstruction was calculated for immature ZIKV, using previously published data (4). The reconstruction was performed using the standard refinement procedure in cryoSPARC (41) and reached ∼20 Å resolution. Similar to the immature KUNV reconstruction, asymmetric features such as the eccentric positioning of the core were observed in the immature ZIKV reconstruction.
Simulations.
To validate the asymmetric reconstruction method, simulations were performed with RELION. The icosahedral reconstruction of immature KUNV was projected using relion_project for the 7,396 orientations determined for the icosahedral reconstruction. Noise was added to the simulated projections. The simulated projections were then used to refine the icosahedral reconstruction, without imposing symmetry. The reconstruction maintained its icosahedral symmetry and did not have an eccentrically positioned core. Thus, the reconstruction procedure did not introduce asymmetrical artifact.
A similar simulation was performed to confirm that known asymmetry was properly aligned and averaged during the asymmetric reconstruction. The asymmetric reconstruction of immature KUNV was projected at the orientations determined for the asymmetric reconstruction, and noise was added. The asymmetric refinement of an icosahedral initial model led to the emergence of features observed for the asymmetric reconstruction within five iterations.
Image Generation, Local Resolution Estimates, and Measurements.
Immature and mature reconstructions were contoured at 3 σ in UCSF Chimera (42). Local resolution maps of immature KUNV were generated using ResMap (43), with a maximum resolution of 12 Å for the icosahedral reconstruction and 45 Å for the asymmetric reconstruction. JSPR (44) was used to calculate radial density distributions. Figures were prepared with UCSF Chimera and Affinity Designer (Serif). UCSF Chimera and IMOD (45) were used to perform measurements of the particles.
Supplementary Material
Acknowledgments
We thank the members of the R.J.K. and M.G.R. laboratories for their help with experimental design. In particular, we acknowledge Yue Liu for many discussions on data processing. We thank Valorie Bowman, Steve Wilson, and the Purdue Cryo-EM Facility for cryo-EM training and computer support. We acknowledge Rodney McPhail for the artwork of Fig. 3 and Sheryl Kelly and Anita Robinson for administrative support. This work was supported by National Institutes of Health Grants R01AI076331 and R01AI073755 (to M.G.R. and R.J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The icosahedral and asymmetric immature KUNV reconstructions have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes 8982 and 8983, respectively. The icosahedral and asymmetric mature KUNV reconstructions have been deposited in the EMDB with accession codes 8984 and 8985.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809304115/-/DCSupplemental.
References
- 1.Hasan SS, Sevvana M, Kuhn RJ, Rossmann MG. Structural biology of Zika virus and other flaviviruses. Nat Struct Mol Biol. 2018;25:13–20. doi: 10.1038/s41594-017-0010-8. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostyuchenko VA, Zhang Q, Tan JL, Ng T-S, Lok S-M. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol. 2013;87:7700–7707. doi: 10.1128/JVI.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad VM, et al. Structure of the immature Zika virus at 9 Å resolution. Nat Struct Mol Biol. 2017;24:184–186. doi: 10.1038/nsmb.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu I-M, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 6.Li L, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn RJ, et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sirohi D, et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostyuchenko VA, et al. Structure of the thermally stable Zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Kostyuchenko VA, Rossmann MG. Structural analysis of viral nucleocapsids by subtraction of partial projections. J Struct Biol. 2007;157:356–364. doi: 10.1016/j.jsb.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dokland T, et al. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang Z, Song H, Shi Y, Qi J, Gao GF. Crystal structure of the capsid protein from Zika virus. J Mol Biol. 2018;430:948–962. doi: 10.1016/j.jmb.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo F, et al. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci USA. 2013;110:6811–6816. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorzelnik KV, et al. Asymmetric cryo-EM structure of the canonical Allolevivirus Qβ reveals a single maturation protein and the genomic ssRNA in situ. Proc Natl Acad Sci USA. 2016;113:11519–11524. doi: 10.1073/pnas.1609482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koning RI, et al. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun. 2016;7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai X, et al. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature. 2017;541:112–116. doi: 10.1038/nature20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Cheng L. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science. 2015;349:1347–1350. doi: 10.1126/science.aaa4938. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature. 2015;527:531–534. doi: 10.1038/nature15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, et al. The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci Adv. 2016;2:e1501929. doi: 10.1126/sciadv.1501929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn RJ, Dowd KA, Beth Post C, Pierson TC. Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology. 2015;479-480:508–517. doi: 10.1016/j.virol.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crick FH, Watson JD. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- 27.Ferlenghi I, et al. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell. 2001;7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- 28.Allison SL, et al. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J Virol. 2003;77:11357–11366. doi: 10.1128/JVI.77.21.11357-11366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lázaro GR, Mukhopadhyay S, Hagan MF. Why enveloped viruses need cores-the contribution of a nucleocapsid core to viral budding. Biophys J. 2018;114:619–630. doi: 10.1016/j.bpj.2017.11.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabata K, et al. Unique requirement for ESCRT factors in flavivirus particle formation on the endoplasmic reticulum. Cell Rep. 2016;16:2339–2347. doi: 10.1016/j.celrep.2016.07.068. [DOI] [PubMed] [Google Scholar]
- 31.Grünewald K, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 32.Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JC-Y, Chen C, Rayaprolu V, Mukhopadhyay S, Zlotnick A. Self-assembly of an alphavirus core-like particle is distinguished by strong intersubunit association energy and structural defects. ACS Nano. 2015;9:8898–8906. doi: 10.1021/acsnano.5b02632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierson EE, et al. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J Am Chem Soc. 2014;136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan R, Li K, Jiang W. Real-time detection and single-pass minimization of TEM objective lens astigmatism. J Struct Biol. 2017;197:210–219. doi: 10.1016/j.jsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roseman AM. FindEM–A fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mastronarde DN, Held SR. Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.