Significance
The plant hormone jasmonate promotes resistance to plant-eating organisms, ranging from pathogenic microbes to mammals. Jasmonate reprograms metabolism to fuel the production of diverse defense compounds and simultaneously inhibits plant growth. Understanding how growth is influenced across a range of defense levels remains unclear, but has important implications for optimizing crop productivity. Using a genetic approach to “tune” the jasmonate response, we assessed the physiological consequences of discrete levels of defense throughout the plant life cycle. Overactivation of jasmonate response led to carbon starvation, near loss of seed production and, under extreme conditions, lethality. Our findings explain the emergence of diverse strategies to keep jasmonate responses at bay and provide new insights into metabolic processes that underlie growth–defense trade-offs.
Keywords: jasmonate, growth–defense trade-off, plant immunity, plant-insect interaction, carbon starvation
Abstract
Plant immune responses mediated by the hormone jasmonoyl-l-isoleucine (JA-Ile) are metabolically costly and often linked to reduced growth. Although it is known that JA-Ile activates defense responses by triggering the degradation of JASMONATE ZIM DOMAIN (JAZ) transcriptional repressor proteins, expansion of the JAZ gene family in vascular plants has hampered efforts to understand how this hormone impacts growth and other physiological tasks over the course of ontogeny. Here, we combined mutations within the 13-member Arabidopsis JAZ gene family to investigate the effects of chronic JAZ deficiency on growth, defense, and reproductive output. A higher-order mutant (jaz decuple, jazD) defective in 10 JAZ genes (JAZ1–7, -9, -10, and -13) exhibited robust resistance to insect herbivores and fungal pathogens, which was accompanied by slow vegetative growth and poor reproductive performance. Metabolic phenotypes of jazD discerned from global transcript and protein profiling were indicative of elevated carbon partitioning to amino acid-, protein-, and endoplasmic reticulum body-based defenses controlled by the JA-Ile and ethylene branches of immunity. Resource allocation to a strong defense sink in jazD leaves was associated with increased respiration and hallmarks of carbon starvation but no overt changes in photosynthetic rate. Depletion of the remaining JAZ repressors in jazD further exaggerated growth stunting, nearly abolished seed production and, under extreme conditions, caused spreading necrotic lesions and tissue death. Our results demonstrate that JAZ proteins promote growth and reproductive success at least in part by preventing catastrophic metabolic effects of an unrestrained immune response.
As sessile organisms, plants continuously adjust their growth, development, and metabolism in response to environmental stress. Complex regulatory networks involving plant hormones play a central part in linking stress perception to transcriptional responses that permit acclimation to harsh environments (1, 2). The lipid-derived hormone jasmonoyl-l-isoleucine (JA-Ile) and its metabolic precursors and derivatives, collectively known as jasmonates (JAs), perform a critical role in plant resilience to many environmental challenges (3, 4). JAs are perhaps best known for orchestrating local and systemic immunity to organisms that exploit plants as a source of food and shelter (5). The hormone controls the expression of large sets of genes that specify a myriad of defense traits, including the biosynthesis of specialized metabolites that thwart attack by diverse organisms, ranging from microbes to mammals (6–8). Interestingly, transcriptional responses triggered by JA-Ile also result in growth inhibition (9–14). The dual role of JA-Ile in promoting defense and restricting growth provides an attractive opportunity to better understand the antagonistic relationship between growth and immunity, with implications for improving crop productivity (15, 16). Many gaps remain, however, in understanding how defense hormones reconfigure metabolism within the constraints of available resources to achieve an optimal balance between immunity and other physiological tasks (17).
In cells containing low JA-Ile levels, JASMONATE ZIM-DOMAIN (JAZ) proteins bind directly to and repress the activity of various transcription factors (TFs) (9, 18, 19). The most thoroughly studied JAZ-interacting TFs are MYC2 and its closely related paralogs (20–23). JAZ proteins repress MYC activity by providing a scaffold on which to recruit corepressors, such as NINJA and TOPLESS (24, 25), as well as by impeding the association of the coactivator protein MED25 with the transcription initiation complex (26–28). In addition to recruiting transcriptional repression complexes to the promoters of JA-responsive genes, JAZ proteins participate in the primary JA-Ile perception event leading to ubiquitin-dependent JAZ degradation. When intracellular levels of JA-Ile rise above a threshold concentration, the hormone promotes binding of JAZ to the F-box protein CORONATINE INSENSITIVE 1 (COI1), which is a component of the E3 ubiquitin ligase complex SCFCOI1 (18, 29, 30). JAZ proteins tagged with polyubiquitin chains by SCFCOI1 are destined for proteolytic destruction by the 26S proteasome, thereby relieving repression on MYC activity. Genetic epistasis analyses in Arabidopsis are consistent with biochemical and structural studies showing that COI1 and JA-Ile comprise a functional module dedicated to JAZ degradation, and that JAZ depletion is sufficient to derepress the expression of target genes controlled by MYC and other TFs (12).
Positive regulators of the core JA-Ile signaling pathway in Arabidopsis, including MYC TFs and components (e.g., COI1) of the JAZ degradation machinery, have been thoroughly studied through characterization of the corresponding loss-of-function mutants (31). In contrast, an understanding of how JAZ proteins negatively regulate JA responses has been hindered by the multimembered nature of JAZ gene families, which in Arabidopsis consists of 13 members (JAZ1–13) (3, 8, 32). Although there is evidence that individual JAZ genes control JA responses in specific tissues and cell types (33, 34), the absence of strong phenotypes in most jaz single mutants described to date suggests some degree of redundancy among JAZ family members (8, 18, 32, 35). Analysis of Arabidopsis mutants defective in multiple JAZ genes supports this interpretation. For example, constitutive JA responses in a jaz quintuple (jazQ) mutant defective in JAZ1, -3, -4, -9, and -10 are relatively mild in comparison with the effects of exogenous JA treatment, and are fully suppressed by mutations that block JA-Ile biosynthesis or perception (12, 35). Indeed, treatment of plants with methyl-JA (MeJA) and other precursors of JA-Ile has been used extensively to study short-term responses (hours to days) to the hormone. This approach, however, is limited in its ability to target specific JAZ proteins or the tissues in which they are expressed. Multiple negative feedback circuits involving JA-Ile catabolism and JAZ repressors that are recalcitrant to JA-Ile–mediated degradation further mask the full range of responses to the hormone (reviewed in ref. 3). These considerations provide a rationale for using extreme higher-order jaz mutants to investigate the physiological, metabolic, and developmental consequences of derepressing JA responses over the course of ontogeny.
Here, we developed a series of higher-order jaz mutants with which to interrogate the effects of chronic overactivation of JA responses. Progressive mutation of JAZ genes was positively correlated with the strength of defense traits and inversely associated with growth and fertility. Detailed characterization of a jaz decuple (jazD) mutant defective in JAZ1–7, -9, -10, and -13 revealed constitutive activation of both JA and ethylene responses, leading to resistance to caterpillar feeding and infection by a necrotrophic fungal pathogen. Metabolic phenotypes of jazD were indicative of elevated carbon and sulfur partitioning to chemical defense traits controlled by the MYC and ethylene response factor (ERF) branches of immunity. The strong defense sink in jazD leaves was also associated with increased cellular respiration and carbon starvation, but not reductions in photosynthetic rate. Introduction of a jaz8 mutation into the jazD background further exacerbated growth defects and nearly abolished seed production in the resulting jaz undecuple mutant. Similarly, elicitation of JA responses in the sensitized jazD background caused spreading necrotic lesions and tissue death. Collectively, our data indicate that JAZ proteins promote growth and reproductive success by attenuating the harmful metabolic effects of an unfettered JA response.
Results
Reduced Growth and Fertility of a jazD Mutant Is Associated with Extreme Sensitivity to JA.
We used insertion mutations to construct a series of higher-order jaz mutants with which to interrogate the biological consequences of chronic JAZ deficiency in Arabidopsis (SI Appendix, Fig. S1). The 13-member JAZ family in Arabidopsis is comprised of five phylogenetic groups (I–V) that are common to angiosperms (SI Appendix, Fig. S2A). The previously described jazQ mutant harbors mutations in the sole member (JAZ10) of group III, all three members of group V (JAZ3, -4, -9), and one member (JAZ1) of the largest group I clade. Building on the jazQ chassis, we used genetic crosses to introduce five additional mutations that target the remaining group I members (JAZ2, -5, -6) and two genes (JAZ7 and -13) within group IV (SI Appendix, Fig. S2). The resulting homozygous jaz1–7, -9, -10, -13 decuple mutant, referred to hereafter as jazD, thus targets all JAZs except for JAZ8 and the two group II genes (JAZ11 and JAZ12).
Cultivation of plants in the absence of exogenous JA showed that, whereas jazQ roots and leaves grow more slowly than WT Col-0, growth of jazD was even slower than jazQ (Fig. 1 A and B). Soil-grown jazD plants displayed less leaf area and shorter petioles than jazQ, and also accumulated more anthocyanins (Fig. 1B, “mock,” and SI Appendix, Fig. S3). Leaf biomass measurements taken over a 20-d time course confirmed that the relative growth rate (RGR) of jazD rosettes during this developmental stage was significantly less than WT (Fig. 1C). That the RGR of jazQ was comparable to WT, despite the reduced biomass of jazQ rosettes at later times in development, may reflect growth changes occurring before the first time point of sampling (11 d after sowing) or the lack of statistical power needed to resolve small differences in RGR that are compounded over time into larger differences in rosette size. Although bulk protein, lipid, and cell wall content of rosette leaves was similar between all three genotypes under our growth conditions, the ratio of leaf dry weight (DW) to fresh weight was increased in jazD relative to WT and jazQ (SI Appendix, Fig. S4). The restricted growth of jazD roots and leaves was associated with changes in flowering time under long-day growth conditions. jazD plants were delayed in their time-to-flowering compared with jazQ but contained a comparable number of leaves at the time of bolting (SI Appendix, Fig. S3).
Fig. 1.
A jaz decuple mutant (jazD) is highly sensitive to jasmonate and exhibits reduced growth and fertility. (A) Root length of 8-d-old WT Col-0 (WT), jazQ, and jazD seedlings grown in the presence of 0, 5, or 25 µM MeJA. Data show the mean ± SD of 30 plants per genotype at each concentration. Capital letters denote significant differences according to Tukey’s honest significant difference (HSD) test (P < 0.05). (B) jazD leaves are hypersensitive to COR. The eighth leaf of 40-d-old plants grown under 12-h light/12-h dark photoperiod was treated with 5 µL water (mock) or 50 µM COR. Leaves were excised and photographed after 2 or 4 d of treatment. Arrows denote location of visible anthocyanin accumulation at the site of COR application. Inset, Right is enlargement of photograph of the COR-treated jazD. (Scale bars: 1 cm.) (C) RGR of soil-grown WT, jazQ, and jazD plants. (D) Total fatty acid content in seeds from the indicated genotype. Data show the mean ± SD of seeds obtained from five plants per genotype. (E) Time course of seed germination. Colored bars indicate the percentage of germinated seeds at various times after sowing on water agar: white, day 1; gray, day 2; black, day 3 and all later times; red, nongerminated seeds.
We next compared the response of jazQ and jazD mutants to exogenous JA. Root growth assays showed that the extent of JAZ deficiency (jazD > jazQ > WT) inversely correlated with root length under a range of MeJA concentrations, with growth of jazD roots effectively arrested in the presence of 5 μM MeJA (Fig. 1A and SI Appendix, Fig. S5). Shoot responsiveness to the hormone was assessed by treating intact leaves with coronatine (COR), which is a potent agonist of the JA-Ile receptor (29). WT and jazQ leaves showed visible accumulation of anthocyanin pigments at the site of COR application (i.e., midvein) within 4 d of the treatment, with no apparent signs of chlorosis (Fig. 1B). In contrast, jazD leaves exhibited visible chlorosis at the site of COR application within 2 d of treatment and, strikingly, near complete loss of chlorophyll and spreading of necrosis-like symptoms throughout the leaf 4 d after treatment, leading to tissue death (Fig. 1B). These data indicate that progressive loss of JAZ genes in jazQ and jazD results in both quantitative (e.g., root growth inhibition) and qualitative (e.g., COR-induced tissue necrosis) differences in JA responsiveness, and are consistent with the notion that the hypersensitivity of jazD results, at least in part, from loss of JAZ-mediated negative-feedback control of JA responses.
Measurements of reproductive output showed that, whereas the total seed yield of jazQ was only marginally affected, seed production by jazD plumented to about one-third of WT levels (Table 1). The reduced fecundity of jazD resulted from a combination of decreased average mass per seed and lower total seed number per plant; mutant plants produced fewer seeds per silique, and the size and number of siliques per plant were reduced as well (Table 1). The reduced size of jazD seeds correlated with a reduction in total fatty acid per seed (Fig. 1D). Analysis of seed fatty acid profiles showed that jazQ and jazD seeds contain less oleic acid (18:1) and more linoleic acid (18:2) (SI Appendix, Fig. S6), suggesting alterations in fatty acid metabolism during seed development. The effect of jazD on seed size and lipid abundance was associated with reduced rates of seed germination (Fig. 1E). These findings indicate that constitutive JA responses resulting from JAZ depletion are associated with poor reproductive performance.
Table 1.
Seed and fruit production in higher-order jaz mutants
| Genotype | Seed yield per plant† (mg) | Average seed mass‡ (μg) | Silique length§ (cm) | No. seed per silique§ | No. silique per plant§ |
| WT | 608.3 ± 103.8 | 21.6 ± 1.3 | 1.59 ± 0.07 | 63 ± 11 | 451 ± 77 |
| jazQ | 524.3 ± 98.5 | 17.3 ± 0.9* | 1.70 ± 0.06 | 58 ± 6 | 533 ± 100 |
| jazD | 192.7 ± 70.0* | 16.6 ± 0.7* | 1.45 ± 0.08* | 37 ± 4* | 329 ± 119* |
Data show the mean ± SD of at least 10 plants per genotype. Asterisks denote significant difference compared with WT plants according to Tukey’s HSD test (*P < 0.05).
Seed yield was determined by collecting all seeds from individual WT Col-0 and jaz mutant plants.
Average seed mass was determined by weighing batches of 200 seeds.
Fully elongated 7th, 9th, and 11th siliques were collected for measurements of silique traits. These traits were used to calculate the estimated number of siliques per plant.
Constitutive Activation of JA- and Ethylene-Mediated Defense Pathways in jazD Plants.
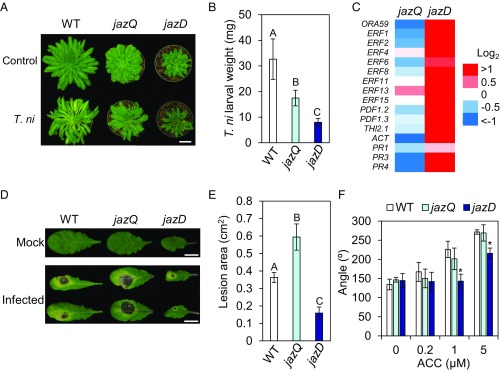
Having established the effects of jazQ and jazD on growth and reproduction, we next assessed how these mutations impact JA-mediated signaling pathways for defense. Similar to results obtained with long-day–grown plants, jaz-mediated leaf growth restriction was observed under short-day conditions (Fig. 2A), which we used to promote leaf biomass and delay flowering in plants used for insect bioassays. In tests performed with the generalist herbivore Trichoplusia ni, we found that the strength of host resistance to insect feeding positively correlated with the severity of jaz mutation (jazD > jazQ > WT), consistent with a role for JAZs in the negative regulation of defense (Fig. 2 A and B).
Fig. 2.
jazD plants are highly resistant to insect herbivores and necrotrophic pathogens. (A) Representative short-day grown WT Col-0 (WT), jazQ, and jazD plants before and after challenge with four T. ni larvae for 12 d. (Scale bar: 3 cm.) (B) Weight gain of T. ni larvae reared on plants shown in A. Data show the mean ± SD of at least 30 larvae per genotype. Capital letters denote significant differences according to Tukey’s HSD test (P < 0.05). (C) Heat map displaying the expression level of various jasmonate/ethylene-responsive genes in leaves of jazQ and jazD normalized to WT. ACT, agmatine coumaroyltransferase (At5g61160). (D) Representative leaf symptoms following 5 d treatment with B. cinerea spores or mock solution. (Scale bars: 2 cm.) (E) Disease lesion size on leaves of the indicated genotype. Data show the mean ± SD of at least 19 leaves per genotype. Capital letters denote significant differences (Tukey’s HSD test, P < 0.05). (F) Apical hook angle of seedlings grown in the presence of various concentrations of the ethylene precursor ACC. Data show the mean ± SD of at least 21 seedlings per genotype. Asterisks denote significant difference compared with WT (Tukey’s HSD test, *P < 0.05).
Messenger RNA sequencing (RNA-seq) was used to investigate the molecular basis of the enhanced antiinsect resistance. Global transcript profiles revealed that the total number of differentially expressed genes in jazD leaves (relative to WT) was more than 10-fold greater than that in jazQ (2,107 and 186 for jazD and jazQ, respectively) (SI Appendix, Fig. S7 and Dataset S1). Among the 186 genes whose expression was statistically different in the jazQ vs. WT comparison, the majority (59%) of these were also differentially expressed in jazD. Gene Ontology (GO) analysis of 1,290 genes expressed to higher levels in jazD than WT showed that “response to JA/wounding,” as well as “defense response,” were among the biological processes most statistically overrepresented in this comparison (SI Appendix, Fig. S7 and Dataset S1). These results, together with analysis of metabolic pathways that are differentially activated in jaz mutants (see below), indicate that the strength of antiinsect resistance correlates with the extent of JAZ deficiency and concomitant reprogramming of gene expression.
Analysis of the RNA-seq data also revealed that ethylene-response genes were highly expressed in jazD but not jazQ. In particular, antifungal defense genes controlled by the synergistic action of JA and ethylene were modestly repressed in jazQ but highly induced in jazD (Fig. 2C and Dataset S1). Among these were genes encoding the AP2/ERFs ERF1 and ORA59, which integrate JA and ethylene signals to promote the expression of antimicrobial compounds, including various defensins (PDFs), pathogenesis-related (PR) proteins, and hydroxycinnamic acid amides (HCAAs) (Fig. 2C) (36–38). Strikingly, several PDF transcripts (e.g., PDF1.2) were among the most abundant of all mRNAs in jazD leaves, with expression levels comparable to that of the most highly expressed photosynthesis transcripts (Dataset S1). In agreement with the RNA-seq data, jazQ plants were slightly more susceptible than WT to the necrotrophic pathogen Botrytis cinerea, whereas jazD leaves were more resistant to the spread of disease lesions (Fig. 2 D and E). To determine whether jazQ and jazD differentially affect other ethylene responses, we assessed apical hook formation in ethylene-elicited seedlings. Consistent with studies showing that apical hook formation is attenuated by JA signaling (39), we found that stimulation of hook curvature in response to treatment with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) was reduced in jazD but not jazQ seedlings (Fig. 2F). These data indicate that whereas jazQ moderately activates JA responses and increases resistance to insect feeding, jazD strongly induces both the JA and ethylene branches of immunity to confer robust resistance to insect feeding and infection by B. cinerea.
To validate the RNA-seq results and gain additional insight how jazD promotes leaf defense, we used quantitative tandem mass spectrometry to quantify global changes in protein abundance in jazD vs. WT in leaves. Among a total of 4,850 unique proteins identified in both genotypes, 149 and 120 proteins accumulated to higher and lower levels, respectively, in jazD (Dataset S2) (threshold fold-change >1.2, P < 0.05). GO analysis of the 120 down-regulated proteins revealed enrichment of functional categories related to cytokinin response, cold response, and various functional domains of photosynthesis (SI Appendix, Table S1). Analysis of proteins that were more abundant in jazD showed there was good agreement with the corresponding mRNA levels determined by RNA-seq; transcripts encoding 78% of these 149 proteins were also induced in jazD plants (Dataset S2). As expected, there was strong enrichment in this protein set of GO categories associated with response to JA, herbivore, and fungal attack, among other defense-related processes (SI Appendix, Table S1). For example, the proteomic analysis revealed that jazD coordinately up-regulated the abundance of most JA biosynthetic enzymes, as well as canonical JA marker proteins, such as VSP1 and VSP2 (SI Appendix, Fig. S8). jazD leaves exhibited high expression of an agmatine coumaroyltransferase (At5g61160) and associated transporter (At3g23550) involved in the production of antifungal HCAAs (40, 41) (Datasets S1 and S2). Transcripts encoding the acyl-CoA N-acyltransferase NATA1 (At2g39030), which catalyzes the formation of the defense compound N(δ)-acetylornithine (42), were 50-fold higher in jazD leaves compared with WT and jazQ and were accompanied by increased NATA1 protein abundance (Datasets S1 and S2). Perhaps most striking was the coordinate up-regulation in jazD leaves, at both the mRNA and protein levels, of most known structural and enzymatic components of the endoplasmic reticulum (ER)-derived ER body (SI Appendix, Fig. S9), which is implicated in induced immunity (43, 44). These findings establish a central role for JAZ proteins as negative regulators of diverse leaf defense traits.
Reprogramming of Primary and Specialized Metabolism in jazD Plants.
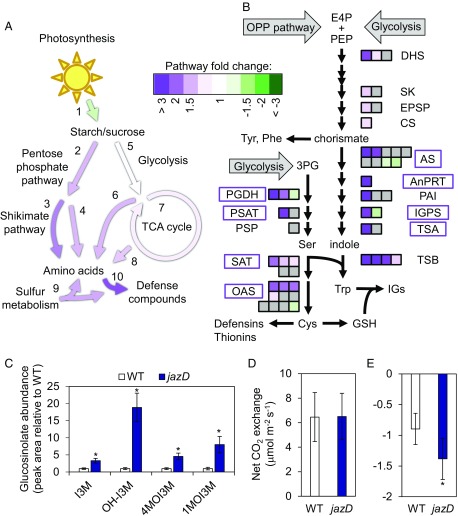
To investigate how the activation of multiple defense pathways influences primary metabolism, we used the RNA-seq and proteomics data to infer metabolic pathways that are altered in jazD leaves. Mapping of differentially expressed genes to Kyoto Encyclopedia of Genes and Genomes pathway databases showed that the tricarboxylic acid (TCA) cycle, oxidative pentose phosphate pathway, sulfur assimilation and metabolism, and various amino acid biosynthetic pathways were among the processes most highly induced in jazD, whereas photosynthesis components were generally down-regulated (Fig. 3A and Dataset S3).
Fig. 3.
Reconfiguration of primary and secondary metabolism in jazD. (A) Mapping of differentially regulated genes in jazD to various metabolic pathways implicates elevated production of defense metabolites derived from amino acids. Mapped pathways include photosynthesis (1), pentose phosphate pathway (2), shikimate pathway (3), amino acids from pentose phosphate intermediates (4), glycolysis (5), amino acids from glycolysis intermediates (6), TCA cycle (7), amino acids from TCA intermediates (8), sulfur metabolism (9), and defense metabolites from amino acids (10). Colored arrows denote the average fold-change of differentially expressed transcripts mapping to a particular pathway (P < 0.05) (Dataset S3). (B) Schematic of tryptophan biosynthesis from erythrose 4-phosphate (E4P), phosphoenolpyruvate (PEP), and 3-phosphoglycerate (3PG) illustrates up-regulation of genes and proteins in jazD. Each arrow represents an enzymatic reaction in the pathway. Boxes represent individual genes, colored by fold-change of jazD relative to WT according to RNA-seq data (Dataset S3), whereas gray boxes denote genes with no significant change in expression. Gene names within boxes denote significantly increased protein levels according to proteomics data. Gene abbreviations: AnPRT, anthranilate phosphoribosyltransferase; AS, anthranilate synthase; CS, chorismate synthase; DHQS, 3-dehydroquinate synthase; DHS, 3-deoxy-7-phosphoheptulonate synthase; DQD/SDH, 3-dehydroquinate dehydratase/shikimate dehydrogenase; EPSP, 5-enolpyruvylshikimate-3-phosphate synthase; IGPS, indole-3-glycerol-phosphate synthase; IGs, indole glucosinolates; OAS, O-acetylserine lyase; PAI, phosphoribosylanthranilate isomerase; PGDH, phosphoglycerate dehydrogenase; PSAT, phosphoserine aminotransferase; PSP, phosphoserine phosphatase; SAT, serine acetyltransferase; SK, shikimate kinase; TSA, tryptophan synthase alpha subunit; TSB, tryptophan synthase β-subunit. (C) Indole glucosinolate levels in jazD leaves relative to that in WT leaves. Asterisks denote significant differences in comparison with WT (Student’s t test, *P < 0.05). Abbreviations: 1MOI3M, 1-methoxyindol-3-ylmethyl (neoglucobrassicin); 4MOI3M, 4-methoxyindol-3-ylmethyl (methoxyglucobrassicin); I3M, indol-3-ylmethyl (glucobrassicin); OH-I3M, 4-hydroxyindol-3-ylmethyl (hydroxyglucobrassicin). (D and E) Net gas exchange rate in WT and jazD rosette leaves measured at 400 µmol CO2 and 20 °C after acclimation in 500 μmol m−2 s−1 in light (D) or dark (E).
One prominent example of a metabolic pathway that was up-regulated in jazD was the shikimate pathway for the biosynthesis of aromatic amino acids. Trp biosynthetic enzymes involved in the production of indole glucosinolates (IGs) showed particularly high expression at the mRNA and protein levels (Fig. 3B and Dataset S3). Consistent with this finding, genes encoding enzymes in the phosphoserine pathway that supplies Ser for the biosynthesis of Trp and Cys (45) were highly up-regulated in jazD, as was the abundance of the corresponding enzymes as determined from proteomics data (Fig. 3B, SI Appendix, Fig. S7, and Datasets S2 and S3). LC-MS analysis of leaf extracts showed that several IGs accumulate to high levels in jazD (Fig. 3C and SI Appendix, Fig. S10), thereby validating the omics data. In agreement with previous studies employing JA elicitation (46–48), we also found that pathways involved in sulfur assimilation and cysteine biosynthesis, as well as ascorbate and glutathione metabolic pathways that protect against oxidative stress, were strongly up-regulated in jazD (Fig. 3B and SI Appendix, Fig. S11). These data indicate that genetic depletion of JAZ proteins recapitulates the transcriptional effects of exogenous JA, and demonstrate that JAZ proteins exert control over pathways that operate at the interface of primary and specialized metabolism.
We next addressed the question of whether jazD modulates net carbon assimilation. Despite the down-regulation of photosynthetic mRNAs and proteins in jazD, modeling of photosynthetic parameters derived from gas-exchange data indicated that the leaf area-based photosynthetic rate of jazD plants was comparable to WT (Fig. 3D and SI Appendix, Fig. S12 A and B). This finding was confirmed by 13C isotope discrimination measurements, which showed that the degree of CO2 resistance through mesophyll cells was similar in WT, jazQ, and jazD leaves (SI Appendix, Fig. S12 C and D). In contrast to photosynthesis, the net loss of CO2 from jazD leaves in the dark exceeded that of WT by ∼50% (Fig. 3E). Increased cellular respiration in jazD was confirmed by experiments showing that the mutant had increased respiration in both the day and night portions of the photoperiod (SI Appendix, Fig. S12 E and F). These findings are consistent with the notion that increased cellular respiration is associated with high-level production of defense compounds (49). GO analysis of the 817 down-regulated genes in jazD leaves showed enrichment for growth-related processes, including “response to light stimulus,” “cell wall organization,” “response to abiotic stimulus,” “carbohydrate biosynthetic process,” and “lipid biosynthetic process” (SI Appendix, Fig. S7).
jazD Plants Exhibit Symptoms of Carbon Starvation.
Increased respiration and partitioning of carbon to metabolic defense pathways, in the absence of compensatory changes in photosynthesis, raised the possibility that jazD plants have a carbon deficit. Time-course studies showed that the rates of starch accumulation (WT: 0.103 µmol Glc g−1 DW h−1; jazD: 0.113 µmol Glc g−1 DW h−1) and degradation (WT: −0.220 g−1 DW h−1; jazD: −0.186 µmol Glc g−1 DW h−1) were comparable between WT and jazD (Fig. 4A). However, starch levels in jazD leaves were slightly lower than WT at all times of the diel cycle except at the end of the night, when starch was mostly depleted but modestly elevated in jazD relative to WT. jazD leaves also had consistently lower sucrose levels (Fig. 4B). We also found that genes involved in starch and sucrose metabolism were generally down-regulated in jazD, including the mRNA and protein abundance of the plastidic starch biosynthetic enzyme phosphoglucomutase (PGM1, At5g51820) (Datasets S1 and S2).
Fig. 4.
jazD plants exhibit symptoms of carbon starvation. (A and B) Time course of starch (A) and sucrose (B) levels in WT Col-0 (WT) and jazD plants during long-day photoperiod. Asterisks denote significant differences in comparison with WT (Student’s t test, *P < 0.05). (C) Heat map showing the expression level of SSM genes in jazQ and jazD leaves. Gene-expression levels determined by RNA-seq are represented as fold-change (log2) over WT. (D and E) Photograph (D) and DW (E) of 16-d-old WT, jazQ, and jazD seedlings grown horizontally on MS medium containing the indicated concentration of sucrose. (Scale bar: D, 0.5 cm.) (F) Root length of 11-d-old WT, jazQ, and jazD seedlings grown vertically on MS medium lacking sucrose (open bar) or containing 23 mM sucrose (filled bar). Two-way ANOVA was used to test the effect of sucrose on growth (E and F) and showed that, whereas genotype (P < 0.001 for both WT vs. jazQ and WT vs. jazD) and sucrose (P < 0.001 for both WT vs. jazQ and WT vs. jazD) significantly affect shoot and root growth, the genotype × sucrose interaction was significant only for jazD comparisons.
To test whether these changes in central metabolism are associated with carbon deficit, we used the RNA-seq data to query the expression of genes that are induced by conditions (e.g., prolonged darkness) leading to carbon starvation. We found that 42 of 278 (15%) sugar starvation marker (SSM) genes defined by Baena-González et al. (50), including several DARK INDUCIBLE (DIN) genes that respond to reduced energy status (51), were expressed to much higher levels in jazD than WT and jazQ (Fig. 4C). We also examined the expression of EIN3-regulated glutamate dehydrogenases (GDH) that replenish 2-oxoglutarate for the TCA cycle and are considered metabolic markers of carbon deficiency (52–54). Both the transcript and protein abundance of GDH1 (At5g18170) and GDH2 (At5g07440) were statistically increased in jazD in comparison with WT (Datasets S1 and S2), consistent with a carbon deficit in this mutant.
To test the hypothesis that carbon limitation contributes to the slow growth of jaz mutants, we compared the growth of WT, jazQ, and jazD seedlings on agar medium supplemented with sucrose. The data showed that although exogenous sucrose promotes increased biomass in all genotypes tested, the stimulatory effect on the growth of jazD shoots was statistically greater than that of WT and jazQ (Fig. 4 D and E). Exogenous sucrose also enhanced the root growth of jazD in comparison with WT and jazQ (Fig. 4F). Control experiments with sorbitol showed that the growth-promoting effect of sucrose was not attributed to changes in osmotic strength of the growth medium (SI Appendix, Fig. S13). These data provide evidence that the reduced growth of jazD but not jazQ results in part from a limitation in carbon supply.
A jaz1–jaz10 and jaz13 Undecuple Mutant Produces Few Viable Seeds.
The ability of jazD plants to perceive and respond to exogenous JA suggested that the remaining JAZ proteins in the mutant can actively repress JA-responsive genes. We hypothesized that mutation of these remaining JAZ loci (i.e., JAZ8, JAZ11, and JAZ12) in the jazD background may further enhance the level of growth-defense antagonism. To test this, we focused on JAZ8 because of its established role in repressing JA responses (25) and the availability of a naturally occurring jaz8-null allele (32). Moreover, the increased expression of JAZ8 in jazD leaves (>15-fold relative to WT) (SI Appendix, Fig. S8) was consistent with a role in negative-feedback control of JA responses. Screening of progeny derived from genetic crosses between jazD and jaz8 resulted in the identification of an undecuple mutant (jazU) homozygous for mutations in JAZ1–JAZ10 and JAZ13 (SI Appendix, Fig. S14A). Root growth assays showed that jazU roots were even shorter than jazD in the presence of very low concentrations (e.g., 1 µM) of MeJA (Fig. 5A). When grown on JA-free medium, jazU showed an even stronger constitutive short-root phenotype than jazD (Fig. 5A). Similarly, the rosette morphology of jazU confirmed the progressive effect of JAZ depletion on restriction of rosette growth, including reduced biomass, leaf area, and petiole length (Fig. 5B and SI Appendix, Fig. S14 B–D). Most strikingly, jazU plants exhibited near complete loss of viable seed production (Fig. 5C). We estimated that less than 3% of jazU flowers set fruit; although jazU pollen was viable in crosses, among flowers that produced fruit, most senesced and aborted during silique filling (SI Appendix, Fig. S14E). Among the few jazU flowers that did produce seeds, seed set per silique was severely reduced, with recovery of only a few viable seeds per plant (SI Appendix, Fig. S14 F–H). The collective seed-yield phenotypes of jazQ, jazD, and jazU strongly support a key role for JAZ proteins in promoting reproductive vigor.
Fig. 5.
Genetic combination of jaz8 and jazD further restricts growth and nearly abolishes seed production in the resulting undecuple mutant. (A) Root length of 10-d-old WT Col-0 (WT), jazD, and jaz undecuple (jazU) seedlings grown in the presence of 0, 0.2, or 1 µM MeJA. Data show the mean ± SD of 14–20 seedlings per genotype at each concentration. Capital letters denote significant differences according to Tukey’s HSD test (P < 0.05). (B) Photograph of WT, jazQ, jazD, and jazU rosettes of 28-d-old plants. (C) Photograph of WT, jazD, and jazU inflorescence of 8-wk-old plants.
Discussion
A major objective of this study was to employ higher-order jaz mutants as tools to achieve a deeper understanding of JA responses throughout the plant life cycle. Our data show that moderate and severe JAZ deficiency in jazQ and jazD, respectively, recapitulates many of the effects of exogenous JA on growth, defense, and metabolism. Comparison of leaf transcriptomes revealed that the degree of JAZ depletion correlated both with the extent of transcriptional reprogramming and the strength of growth–defense antagonism. The complexity of growth- and immune-related phenotypes in higher-order jaz mutants is thus consistent with the capacity of JAZ proteins to directly control the activity of diverse TFs (3). Based on current annotations of the Arabidopsis TF repertoire (55), we identified 218 (147 up- and 71 down-regulated) and 20 (11 up- and 9 down-regulated) TF-encoding genes that are differentially expressed in jazD and jazQ, respectively (Dataset S1). These findings support a general model in which JAZ proteins reside at the apex of a transcriptional hierarchy that promotes defense and simultaneously inhibits growth (3, 8). jaz loss-of-function mutations may also indirectly influence gene expression through changes in the expression of TF-encoding genes that promote subsequent waves of transcription or compensatory responses. A future challenge will be to link specific phenotypes of jaz mutants to defined JAZ-TF regulatory modules that exert temporal and spatial control over JA responses (12, 33, 34).
A significant finding of our work was that jazQ and jazD mutants show not only quantitative variation in JA-related phenotypes but also qualitative differences in select defense traits. This is exemplified by the observation that jazD constitutively activates the ethylene branch of immune signaling to increase resistance to B. cinerea, whereas jazQ moderately inhibits ethylene-dependent defense responses to this pathogen. The elevated JA/ethylene-dependent resistance of jazD is consistent with the ability of some JAZ proteins to repress the activity of two master regulators of ethylene signaling, EIN3 and EIL1, which in turn activate the expression of TFs (e.g., ERF1) that mediate resistance to necrotrophic pathogens (36, 37, 56–58). Thus, although jazQ moderately relieves repression on MYC TFs to enhance resistance to insect feeding, this combination of jaz mutations (jaz1, -3, -4, -9, -10) may be insufficient to derepress EIN3/EIL1. In this scenario, antagonistic interactions between the MYC2 and ERF1 branches of JA signaling (59) would explain the increased susceptibility of jazQ to B. cinerea. In contrast, we propose that the severe JAZ deficiency in jazD simultaneously derepresses both the MYC2 and EIN3/EIL1 branches to confer strong resistance to chewing insects and necrotrophic pathogens. Given that EIN3 physically interacts with MYC2 to inhibit JA-regulated defense against insect herbivores (39), robust activation of insect and fungal pathogen defenses in jazD suggests that MYC2-EIN3/EIL1 antagonism may require certain JAZ proteins to control the balance between transcriptional repression and activation (3). The distinct biotic stress phenotypes of jazQ and jazD may reflect a larger pool of JAZs in jazQ, or may be attributed specifically to one or more JAZ proteins (i.e., JAZ2, -5, -6, -7, -13) present in jazQ but absent in jazD. Complementation of jazD with WT JAZ genes may help to address this hypothesis. It is noteworthy that many phenotypes of jazD, including high expression of PDF genes and enhanced resistance to both caterpillar feeding and pathogen infection, are also observed in mutants defective in JA catabolism (60, 61). These findings suggest that changes in JA homeostasis may deplete JAZ abundance to phenocopy the effects of jaz mutants.
JAZ depletion in higher-order jaz mutants may propagate changes in protein–protein interaction networks that shape the diversity of JA responses and hormone cross-talk (3). The strong induction of IG biosynthesis in jazD is consistent with the role of MYC-MYB heterodimers in JA-mediated activation of IG biosynthetic genes (62, 63), such that genetic depletion of JAZ favors MYC–MYB interaction. A similar argument applies to the role of JAZ–DELLA interactions in the control of growth–defense balance; JAZ depletion could restrict growth through increased DELLA activity and attenuation of gibberellin-mediated responses (64–66). It remains to be determined whether changes in DELLA abundance or other components of the gibberellin pathway contribute to the reduced growth of higher-order jaz mutants. In fact, the critical role of MYC2, -3, and -4 in restricting leaf growth and biomass in jazQ (12) suggests that these TFs contribute to the growth phenotype of jazD. Other potential factors contributing to the reduced growth rate of jazD include down-regulation of genes involved in cell wall organization (14), and perturbation in auxin levels resulting from altered Trp metabolism.
Lifetime viable seed production is a reliable measure of the cost of defense. Consistent with studies showing that induced defense responses in the absence of herbivore pressure curtail seed production (67, 68), we found that jazQ, jazD, and jazU mutants have weak, moderate, and severe negative effects on seed production, respectively. It therefore appears that either too little (e.g., coi1 mutation) or too much (e.g., jazU mutation) JA response can lead to reproductive failure, albeit for different physiological reasons. Similarly, the reduction in seed size and quality in higher-order jaz mutants is consistent with the increased seed size in JA-deficient mutants (69). These findings suggest that unrestrained JA responses give rise to trade-offs in which increased partitioning of resources to defense in source tissues reduces nutrient availability in sink tissues. It has been shown, for example, that carbon starvation in leaves has direct negative effects on seed development and quality in Arabidopsis (70). In demonstrating a critical role for JAZ proteins in promoting reproductive performance, our results provide a fitness-based explanation for the emergence of multitiered mechanisms to restrain JA responses, with implications for understanding the evolution of JA signaling systems (3, 17).
Several features of higher-order jaz mutants, including constitutive production of defense compounds, slow growth, and reduced fecundity, are indicative of hyperactive immunity (71). These effects were exaggerated under conditions designed to further deplete JAZ in the sensitized jazD background, either by treatment with exogenous hormone or addition of jaz8 mutation. The necrosis of COR-treated jazD leaves provides strong evidence that JAZ proteins protect against JA-mediated cell death resulting from a runaway immune response. Although the underlying cause of tissue necrosis remains to be determined, we note that JAs have long been known to promote senescence-like symptoms and to elicit the production of reactive oxygen species (72–75). Moreover, silencing of the JAZh paralog in Nicotiana attenuata caused spontaneous necrosis at late stages of leaf development, which was associated with increased reactive oxygen species production (76). It is possible that the growth inhibitory and senescence-like symptoms of jaz mutants reflects autotoxicity of defense compounds, as recently proposed for IGs in Arabidopsis (77). The emerging link between IG production and ER body formation (43), coupled with our discovery that JAZ proteins suppress the expression of both IGs and ER body components, provide the impetus for future studies aimed at understanding how JA coordinates the proliferation of specialized organelles with defense metabolism. The striking diversity of chemical defense pathways expressed in jazD indicates that jaz mutants, potentially from any plant species, may be useful tools for discovery of novel antimicrobial and antiinsect defense compounds.
Our analysis of the effects of chronic JAZ deficiency provides insights into how vegetative growth rate is coordinated with changes in primary and specialized metabolism. In general, our data support the acclimatory response hypothesis to explain how plant growth rate is modulated in response to changes in the supply and demand for assimilated carbon (17, 70). Several independent lines of evidence indicate that partitioning of resources to a strong defense sink in jazD leaves is associated with carbon starvation and downward adjustment of growth rate. First, the expression of genes and protein markers of carbon starvation were up-regulated in jazD leaves. Consistent with the concept of resource allocation costs, exogenous sucrose partially restored the growth of jazD leaves and roots. Second, the area-based respiration rate of jazD leaves was increased without a corresponding change in photosynthetic rate, potentially contributing to an energy deficit. Third, the sucrose and starch content in jazD leaves was modestly depleted throughout most times of the diel cycle, consistent with studies showing that JA elicitation reduces photoassimilate levels (78). Given that the rates of starch synthesis and degradation were similar in jazD and WT leaves, it appears that the control mechanisms underlying the precise pacing of starch reserves (70, 79) are largely operational under conditions where JA signaling strongly diverts carbon to defense. These findings suggest that JAZ proteins flexibly adjust growth–defense balance to match anticipated changes in resource availability, thereby avoiding the detrimental effects of hyperimmunity.
In summary, our results demonstrate that JA-triggered immunity imposes major metabolic demands that, if not properly restrained, are detrimental to plant fitness. In environments where assimilated carbon, nitrogen, water, and other essential nutrients may be limiting, the bioenergetic and biosynthetic demands associated with increased defense must be tightly coordinated with other physiological tasks. We propose that JAZ proteins assist in balancing the growth–defense continuum not as a binary on–off switch but rather by matching the biotic stress level to available resources. In particular, our results indicate that growth and reproductive penalties of immunity become evident only at high levels of defense; a condition that is buffered by multiple negative-feedback mechanisms. The intermediate level of antiinsect defense exhibited by jazQ does not appear to impose major allocation costs under laboratory growth conditions (35). Rather, the level of growth restriction exhibited by jazQ may reflect other adaptive roles of growth–defense antagonism. Downward adjustment of growth may, for example, provide a mechanism to concentrate defense compounds in affected tissues, promote cross-resilience to potential future stress by reserving resources, or optimize the timing of vegetative-to-reproductive transition (11, 17, 80). In contrast to jazQ, severe JAZ depletion in jazD activates transcriptional programs that dictate much stronger allocation of central metabolites to defense, with significant negative effects on growth and reproduction. A high energetic expense associated with synthesis of defense proteins, provision of amino acid precursors and biosynthetic enzymes, transport, and storage of defense compounds in jazD is supported by hallmarks of carbon starvation. The diversion of resources to defense in jazD likely extends beyond carbon to other facets of intermediary metabolism. The induced production of defensins and other sulfur-rich defense compounds, for example, provides a plausible explanation for up-regulation of enzymes involved in sulfate assimilation and the biosynthesis of cysteine, consistent with the importance of sulfur metabolism in plant defense (46–48). A challenge for future studies will be to elucidate the mechanisms by which the status of carbon and other nutrients is sensed and adjusted during growth-to-defense transitions.
Materials and Methods
The Columbia accession (Col-0) of Arabidopsis thaliana was used as WT for all experiments. jazD was constructed by crossing jazQ (35) to other transfer-DNA or transposon insertion mutants obtained from the Arabidopsis Biological Research Center (Ohio State University). Detailed information on the construction of jazD and jazU are described in SI Appendix. Details of plant growth conditions, chemical treatments, insect and pathogen bioassays, physiological assays, including gas-exchange measurements, RNA-seq and proteomic analyses, and metabolite quantification are provided in the SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tom Sharkey, Sean Weise, Sarathi Weraduwage, and John Froehlich for helpful assistance with measurements of photosynthesis, respiration, starch and sucrose content, and total leaf protein; Douglas Whitten in the Michigan State University (MSU) Proteomics Core Facility for proteomic analysis; Tony Schilmiller in the MSU Mass Spectrometry Facility for assistance with glucosinolate measurements; and the MSU Genomics Facility for RNA sequencing. This work was primarily funded by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy through Grant DE-FG02-91ER20021. Construction of mutant lines was supported by National Institutes of Health Award GM57795 (to G.A.H.). K.S. was supported in part by the Japan Society for Promotion of Science Research Fellowship for Young Scientists Award (24-824). We also acknowledge support from the Michigan AgBioResearch Project MICL02278 and the Discretionary Funding Initiative from Michigan State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE116681).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811828115/-/DCSupplemental.
References
- 1.Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 2.Santner A, Calderon-Villalobos LIA, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- 3.Howe GA, Major IT, Koo AJ. Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol. 2018;69:387–415. doi: 10.1146/annurev-arplant-042817-040047. [DOI] [PubMed] [Google Scholar]
- 4.Wasternack C, Hause B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos ML, Kang J-H, Howe GA. Jasmonate-triggered plant immunity. J Chem Ecol. 2014;40:657–675. doi: 10.1007/s10886-014-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Annu Rev Genet. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- 8.Chini A, Gimenez-Ibanez S, Goossens A, Solano R. Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol. 2016;33:147–156. doi: 10.1016/j.pbi.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havko NE, et al. Control of carbon assimilation and partitioning by jasmonate: An accounting of growth-defense tradeoffs. Plants (Basel) 2016;5:E7. doi: 10.3390/plants5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Major IT, et al. Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017;215:1533–1547. doi: 10.1111/nph.14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attaran E, et al. Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 2014;165:1302–1314. doi: 10.1104/pp.114.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bömer M, et al. COI1-dependent jasmonate signalling affects growth, metabolite production and cell wall protein composition in Arabidopsis. Ann Bot. June 19, 2018 doi: 10.1093/aob/mcy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Züst T, Agrawal AA. Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annu Rev Plant Biol. 2017;68:513–534. doi: 10.1146/annurev-arplant-042916-040856. [DOI] [PubMed] [Google Scholar]
- 16.Karasov TL, Chae E, Herman JJ, Bergelson J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell. 2017;29:666–680. doi: 10.1105/tpc.16.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Q, Major IT, Howe GA. Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr Opin Plant Biol. 2018;44:72–81. doi: 10.1016/j.pbi.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 19.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 20.Kazan K, Manners JM. MYC2: The master in action. Mol Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 21.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi T, Huang H, Song S, Xie D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell. 2015;27:1620–1633. doi: 10.1105/tpc.15.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueroa P, Browse J. Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J. 2015;81:849–860. doi: 10.1111/tpj.12776. [DOI] [PubMed] [Google Scholar]
- 24.Pauwels L, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shyu C, et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. 2012;24:536–550. doi: 10.1105/tpc.111.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature. 2015;525:269–273. doi: 10.1038/nature14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Çevik V, et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012;160:541–555. doi: 10.1104/pp.112.202697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An C, et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc Natl Acad Sci USA. 2017;114:E8930–E8939. doi: 10.1073/pnas.1710885114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J, et al. Dynamic perception of jasmonates by the F-box protein COI1. Mol Plant. August 7, 2018 doi: 10.1016/j.molp.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Browse J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 32.Thireault C, et al. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015;82:669–679. doi: 10.1111/tpj.12841. [DOI] [PubMed] [Google Scholar]
- 33.Gimenez-Ibanez S, et al. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017;213:1378–1392. doi: 10.1111/nph.14354. [DOI] [PubMed] [Google Scholar]
- 34.Li R, et al. Flower-specific jasmonate signaling regulates constitutive floral defenses in wild tobacco. Proc Natl Acad Sci USA. 2017;114:E7205–E7214. doi: 10.1073/pnas.1703463114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campos ML, et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun. 2016;7:12570. doi: 10.1038/ncomms12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- 37.Pré M, et al. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, et al. Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA59 transcription factor. Plant J. 2018;95:444–457. doi: 10.1111/tpj.13960. [DOI] [PubMed] [Google Scholar]
- 39.Song S, et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell. 2014;26:263–279. doi: 10.1105/tpc.113.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muroi A, et al. Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta. 2009;230:517–527. doi: 10.1007/s00425-009-0960-0. [DOI] [PubMed] [Google Scholar]
- 41.Dobritzsch M, et al. MATE transporter-dependent export of hydroxycinnamic acid amides. Plant Cell. 2016;28:583–596. doi: 10.1105/tpc.15.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adio AM, et al. Biosynthesis and defensive function of Nδ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell. 2011;23:3303–3318. doi: 10.1105/tpc.111.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakano RT, et al. PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana. Plant J. 2017;89:204–220. doi: 10.1111/tpj.13377. [DOI] [PubMed] [Google Scholar]
- 44.Yamada K, Hara-Nishimura I, Nishimura M. Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol. 2011;52:2039–2049. doi: 10.1093/pcp/pcr156. [DOI] [PubMed] [Google Scholar]
- 45.Benstein RM, et al. Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell. 2013;25:5011–5029. doi: 10.1105/tpc.113.118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kruse C, et al. Sulfur-enhanced defence: Effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biol (Stuttg) 2007;9:608–619. doi: 10.1055/s-2007-965432. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki-Sekimoto Y, et al. Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 2005;44:653–668. doi: 10.1111/j.1365-313X.2005.02560.x. [DOI] [PubMed] [Google Scholar]
- 48.Yatusevich R, et al. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 2010;62:1–11. doi: 10.1111/j.1365-313X.2009.04118.x. [DOI] [PubMed] [Google Scholar]
- 49.Bolton MD. Primary metabolism and plant defense—Fuel for the fire. Mol Plant Microbe Interact. 2009;22:487–497. doi: 10.1094/MPMI-22-5-0487. [DOI] [PubMed] [Google Scholar]
- 50.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 51.Fujiki Y, et al. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol Plant. 2001;111:345–352. doi: 10.1034/j.1399-3054.2001.1110312.x. [DOI] [PubMed] [Google Scholar]
- 52.Gibon Y, et al. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 2009;32:859–874. doi: 10.1111/j.1365-3040.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- 53.Tsai K-J, Lin C-Y, Ting C-Y, Shih M-C. Ethylene-regulated glutamate dehydrogenase fine-tunes metabolism during anoxia reoxygenation. Plant Physiol. 2016;172:1548–1562. doi: 10.1104/pp.16.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyashita Y, Good AG. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot. 2008;59:667–680. doi: 10.1093/jxb/erm340. [DOI] [PubMed] [Google Scholar]
- 55.Jin J, et al. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Müller M, Munné-Bosch S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169:32–41. doi: 10.1104/pp.15.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caarls L, et al. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc Natl Acad Sci USA. 2017;114:6388–6393. doi: 10.1073/pnas.1701101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smirnova E, et al. Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol Plant. 2017;10:1159–1173. doi: 10.1016/j.molp.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 62.Schweizer F, et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013;25:3117–3132. doi: 10.1105/tpc.113.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gigolashvili T, et al. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 64.Yang D-L, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 66.Machado RAR, Baldwin IT, Erb M. Herbivory-induced jasmonates constrain plant sugar accumulation and growth by antagonizing gibberellin signaling and not by promoting secondary metabolite production. New Phytol. 2017;215:803–812. doi: 10.1111/nph.14597. [DOI] [PubMed] [Google Scholar]
- 67.Agrawal AA. Induced responses to herbivory and increased plant performance. Science. 1998;279:1201–1202. doi: 10.1126/science.279.5354.1201. [DOI] [PubMed] [Google Scholar]
- 68.Baldwin IT. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA. 1998;95:8113–8118. doi: 10.1073/pnas.95.14.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farmer EE, Dubugnon L. Detritivorous crustaceans become herbivores on jasmonate-deficient plants. Proc Natl Acad Sci USA. 2009;106:935–940. doi: 10.1073/pnas.0812182106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 71.Bomblies K, Weigel D. Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet. 2007;8:382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- 72.Qi T, et al. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell. 2015;27:1634–1649. doi: 10.1105/tpc.15.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shan X, et al. The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 2011;155:751–764. doi: 10.1104/pp.110.166595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiol. 1980;66:246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orozco-Cárdenas ML, Narváez-Vásquez J, Ryan CA. Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell. 2001;13:179–191. [PMC free article] [PubMed] [Google Scholar]
- 76.Oh Y, Baldwin IT, Gális I. NaJAZh regulates a subset of defense responses against herbivores and spontaneous leaf necrosis in Nicotiana attenuata plants. Plant Physiol. 2012;159:769–788. doi: 10.1104/pp.112.193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, et al. Salt and methyl jasmonate aggravate growth inhibition and senescence in Arabidopsis seedlings via the JA signaling pathway. Plant Sci. 2017;261:1–9. doi: 10.1016/j.plantsci.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 78.Machado RAR, et al. Leaf-herbivore attack reduces carbon reserves and regrowth from the roots via jasmonate and auxin signaling. New Phytol. 2013;200:1234–1246. doi: 10.1111/nph.12438. [DOI] [PubMed] [Google Scholar]
- 79.Sulpice R, et al. Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol Plant. 2014;7:137–155. doi: 10.1093/mp/sst127. [DOI] [PubMed] [Google Scholar]
- 80.Wang K, et al. Two abscisic acid-responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell. 2018;30:1006–1022. doi: 10.1105/tpc.18.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.