Significance
In this study, we provide insight into the mechanism of effector recognition by plant nucleotide-binding domain and leucine-rich repeat proteins (NLRs), a ubiquitous class of immune receptors that plays a central role in crop protection. By structural and functional analysis of a complex between a fungal effector and an integrated decoy domain (ID) from a rice NLR, we demonstrate the importance of IDs in effector recognition and generate crucial knowledge for future engineering of NLRs to expand their recognition specificities. In addition, we propose, as a hypothesis regarding the diversity of fungal effectors, that in structurally conserved effector families, the molecular mechanisms of host target protein-binding are conserved but not the host target proteins themselves.
Keywords: plant immunity, immune receptor, rice, Magnaporthe oryzae, effector
Abstract
The structurally conserved but sequence-unrelated MAX (Magnaporthe oryzae avirulence and ToxB-like) effectors AVR1-CO39 and AVR-PikD from the blast fungus M. oryzae are recognized by the rice nucleotide-binding domain and leucine-rich repeat proteins (NLRs) RGA5 and Pikp-1, respectively. This involves, in both cases, direct interaction of the effector with a heavy metal-associated (HMA) integrated domain (ID) in the NLR. Here, we solved the crystal structures of a C-terminal fragment of RGA5 carrying the HMA ID (RGA5_S), alone, and in complex with AVR1-CO39 and compared it to the structure of the Pikp1HMA/AVR-PikD complex. In both complexes, HMA ID/MAX effector interactions involve antiparallel alignment of β-sheets from each partner. However, effector-binding occurs at different surfaces in Pikp1HMA and RGA5HMA, indicating that these interactions evolved independently by convergence of these two MAX effectors to the same type of plant target proteins. Interestingly, the effector-binding surface in RGA5HMA overlaps with the surface that mediates RGA5HMA self-interaction. Mutations in the HMA-binding interface of AVR1-CO39 perturb RGA5HMA-binding, in vitro and in vivo, and affect the recognition of M. oryzae in a rice cultivar containing Pi-CO39. Our study provides detailed insight into the mechanisms of effector recognition by NLRs, which has substantial implications for future engineering of NLRs to expand their recognition specificities. In addition, we propose, as a hypothesis for the understanding of effector diversity, that in the structurally conserved MAX effectors the molecular mechanism of host target protein-binding is conserved rather than the host target proteins themselves.
Intracellular nucleotide-binding domain and leucine-rich repeat protein (NLR) receptors are key elements of plant immunity. They are characterized by a central nucleotide-binding (NB-ARC) and a C-terminal leucine-rich repeat (LRR) domain (1–3). Plant NLRs detect pathogens by recognizing virulence effectors inside host cells and activate defense responses and immunity. Since they confer immunity against many crop diseases, which represent a major problem in agriculture, NLR-coding genes are widely used in crop resistance breeding.
The specific recognition of pathogen effectors by NLRs relies on several mechanisms (4). Some NLRs directly interact with effectors, while others recognize the effector-mediated modification of a host protein that is either a virulence target of the effector (guardee) or a mimic of such a target (decoy) (5, 6). Decoys are thought to stem from the duplication of virulence target genes and to have lost their original cellular function to only serve as effector traps. Recently, an integrated decoy model for effector recognition has been proposed for NLRs carrying noncanonical integrated domains (IDs) and relying on a second, genetically clustered NLR (7). This mechanism was confirmed for Arabidopsis thaliana RRS1 that recognizes, through its WRKY ID, the bacterial effector PopP2, which targets plant WRKY transcription factors (8, 9).
In rice, the paired NLR genes RGA4 and RGA5 confer resistance to Magnaporthe oryzae isolates carrying the effector genes AVR1-CO39 or AVR-Pia, while Pikp-1 and Pikp-2 recognize isolates expressing AVR-PikD (10, 11). AVR1-CO39, AVR-Pia, and AVR-PikD are sequence-unrelated, but possess highly similar β-sandwich structures characteristic of the M. oryzae AVRs and ToxB-like (MAX) effector family in plant pathogenic Ascomycete fungi that has specifically expanded in M. oryzae (12, 13). Both RGA5 and Pik-1 contain a heavy metal-associated ID (HMA, also called the Related to yeast ATX1 or RATX1 domain) that is crucial for specific effector recognition through direct binding (10, 13). Structure–function analyses provided detailed insight into Pikp-1HMA/AVR-PikD and RGA5HMA/AVR-Pia–binding and established a causal link between these interactions and recognition specificities (13–15).
To further investigate the recognition of MAX effectors by HMA IDs in rice NLRs, we have resolved and functionally validated the crystal structures of RGA5_S (RGA5982–1116), a small C-terminal fragment of RGA5 carrying the HMA domain (SI Appendix, Fig. S1), alone and in complex with AVR1-CO39. This shows that specific recognition of the two MAX effectors, AVR1-CO39 and AVR-PikD, by the HMA-IDs of two different rice NLRs, RGA5 and Pikp-1, involves distinct binding surfaces in both partners of the complex. These findings have substantial implications for future engineering of immune receptors carrying HMA IDs to expand their effector recognition specificities.
Results
Previously, C-terminal fragments of RGA5 carrying the HMA ID have been shown to self-interact and to interact with AVR1-CO39 in yeast (10, 16). ITC (isothermal titration calorimetry) measurements with recombinant RGA5982–1116 (RGA5_S) and AVR1-CO39 with its signal peptide deleted (dSP-AVR1-CO39) (SI Appendix, Fig. S1) showed that their interaction relies on direct binding with an equilibrium dissociation constant (Kd) of 5.4 µM (17). This Kd is on the same order of magnitude as the one measured for the RGA5_S/dSP-AVR-Pia complex (Kd = 1.8 µM) (SI Appendix, Fig. S2A). To reveal the molecular and structural details of AVR1-CO39–binding to the HMA ID of RGA5, we generated crystals for RGA5_S and RGA5_S/AVR1-CO39 diffracting at 1.78 and 2.19 Å resolutions, respectively (17, 18).
RGA5HMA Domain Has a Canonical HMA Fold and Forms a Dimer.
The RGA5_S structure was determined by molecular replacement using Phaser (19) with the homologous structure of the copper-binding domain of HMA7 (3DXS) as a search model. This provided a model (PDB accession 5ZNE) containing 73 residues (997–1069). They adopt, as expected, an HMA domain fold characterized by two α-helices and a four-stranded antiparallel β-sheet (topology β1-αA-β2-β3-αB-β4) (Fig. 1A and SI Appendix, Table S1). This structure is highly similar to that of the Pikp-1 HMA domain [PDB: 5A6P; sequence identity: 55.1%, rmsd 0.90 Å for 66 (997–1007 and 1010–1064) of 73 residues (997–1069) aligned by Coot (20)] and other HMAs from various organisms (SI Appendix, Fig. S3 A and B). The metal-binding motif, characteristic of HMAs, is degenerated in RGA5HMA, as only the first Cys is conserved, while the second is replaced by a Ser.
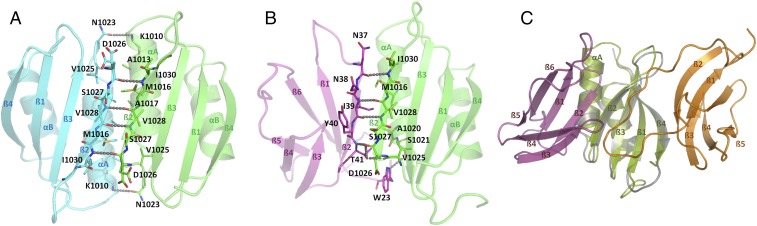
Fig. 1.
AVR1-CO39 and AVR-PikD bind to opposite surfaces of the HMA-IDs. (A) The structures of the RGA5HMA homodimer and (B) the RGA5HMA/dSP-AVR1-CO39 complex are shown as cartoons with the RGA5HMA molecules in cyan or green and the effector in magenta. Key residues in the binding interface are shown as sticks with carbon residues in the color of the cartoon. Hydrogen bonds are depicted as dashed lines. (C) Overlay of the structures of the RGA5HMA/dSP-AVR1-CO39 (green and magenta, respectively) and the Pikp-1HMA/dSP-AVR-PikD complexes (gray and orange, respectively) shown as cartoons.
RGA5_S monomers dimerize through an intimate binding interface that buries 11% of the exposed surface area of RGA5HMA (492 of 4,600 Å2) and that is formed by the secondary structure elements αA, β2, and the loop linking them. Binding occurs through hydrophobic interactions and 10 hydrogen bonds distributed on three interaction sites (Fig. 1A and SI Appendix, Fig. S4A). The first site is characterized by the antiparallel alignment of the β2s of the two RGA5HMA molecules and leads to the formation of one continuous β-sheet comprising the two four-stranded β-sheets of the monomers (Fig. 1A and SI Appendix, Fig. S4A). The other two sites are formed by the antiparallel alignment of the αAs and by the αA/β2 turns of the two RGA5HMA molecules.
A Single Interface Within RGA5HMA Mediates AVR1-CO39–Binding and Self-Interaction.
Diffraction data obtained with RGA5_S/dSP-AVR1-CO39 crystals (17) were used to solve the structure of the RGA5_S/dSP-AVR1-CO39 complex using molecular replacement of the RGA5_S structure and the NMR structure of dSP-AVR1-CO39 (PDB: 2MYV). The final refined model contained residues 993–1069 of RGA5 and residues 23–81 of AVR1-CO39 (PDB accession 5ZNG, Fig. 1B). The protein structures in the complex are highly similar to the structures of the isolated proteins (RMSD for backbone atoms 0.62 and 1.2 Å, residue range 997–1069 and 23–81, for the RGA5_S and AVR1-CO39, respectively). RGA5HMA and dSP-AVR1-CO39 form a complex with a 1:1 stoichiometry where β2 of AVR1-CO39 is aligned in an antiparallel manner to β2 of RGA5HMA, resulting in a continuous antiparallel β-sheet comprising the four-stranded β-sheet of RGA5HMA and β1, 2, and 6 of AVR1-CO39 (Fig. 1B). Complex stability is mediated by an intimate binding interface that buries 12% and 10%, respectively, of the solvent-exposed surface area of the effector (495 Å2) and RGA5HMA (455 Å2) and that is dominated by hydrophobic interactions. Hydrogen bonds, involving residues of AVR1-CO39, mainly from β2 and the N terminus, and residues in RGA5HMA, mainly from αA and β2, are distributed on three binding areas. (Fig. 1B and SI Appendix, Fig. S4B).
The first important binding area is formed by W23, K24, and T41 of AVR1-CO39 and V1025 and D1026 of RGA5 and dominated by main- and side-chain hydrogen-bonding between AVR1-CO39T41 and RGA5D1026 (SI Appendix, Figs. S4B and S5A). The second area involves N38 and I39 of AVR1-CO39 and V1028 and E1029 of RGA5 (SI Appendix, Fig. S5B). The third interaction site is centered on R1012 and I1030 of RGA5 and D35, V36, N37 of AVR1-CO39 (SI Appendix, Fig. S5C).
The binding of AVR1-CO39 to RGA5HMA involves the same interface and many of the same residues as the RGA5HMA homodimerization, suggesting that binding of AVR1-CO39 to RGA5_S competes with RGA5HMA self-interaction and explaining why such RGA5HMA homodimers do not occur in the RGA5_S/dSP-AVR1-CO39 complex (Fig. 1 and SI Appendix, Figs. S4 and S5).
Binding of AVR1-CO39 and AVR-Pia to RGA5_S Compete with RGA5_S Self-Interaction.
Analytical gel filtration (GF) was performed to test the competition between RGA5_S/dSP-AVR1-CO39 complex formation and RGA5_S self-interaction. This showed that dSP-AVR1-CO39 is, as previously described, monomeric in solution (12), while RGA5_S forms homodimers (Fig. 2A). When dSP-AVR1-CO39 and RGA5_S were combined at a 1:1 ratio, two elution peaks with retention times indistinguishable from the dSP-AVR1-CO39 monomer peak and the RGA5_S dimer peak were detected (Fig. 2A). SDS/PAGE analysis of the elution fractions showed coelution of dSP-AVR1-CO39 with RGA5_S in the first peak, indicating formation of an RGA5_S/dSP-AVR1-CO39 heterocomplex with a 1:1 stoichiometry (Fig. 2A).
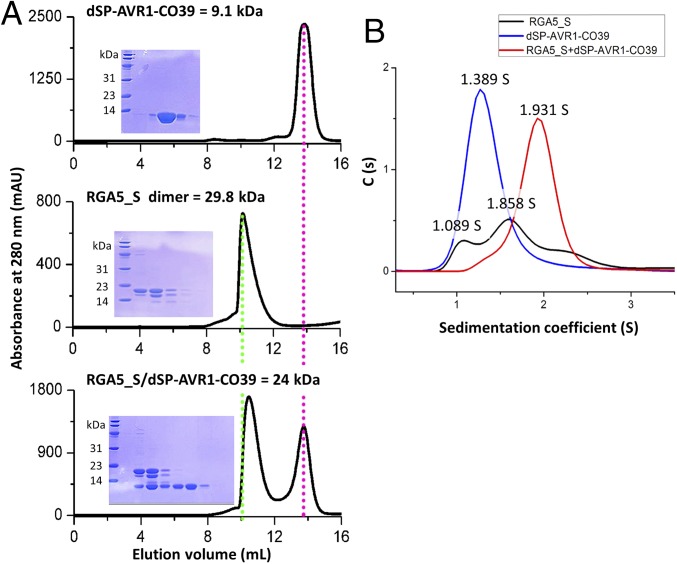
Fig. 2.
AVR1-CO39 competes with RGA5HMA self-interaction for RGA5_S-binding in vitro. (A) Gel filtration traces showing the retention volume and SDS/PAGE gels of relevant elution fractions of dSP-AVR1-CO39, RGA5_S, and a 1:1 mixture of both proteins. The molecular weight of the recombinant proteins is depicted on the Top of the panels. (B) Sedimentation coefficient distributions calculated from sedimentation velocity analytical ultracentrifugation with dSP-AVR1-CO39, RGA5_S, and a 1:1 mixture of both proteins. These GF (A) and SV-AUC (B) experiments were performed three times with similar results.
In sedimentation velocity analytical ultracentrifugation (SV-AUC), dSP-AVR1-CO39 occurred in one uniform state with a sedimentation coefficient of 1.389 S, while RGA5_S occurred in two main states: a small fraction (8.5%) with a low sedimentation coefficient (1.089 S) that contains likely monomeric RGA5_S and a major fraction (85%) with a sedimentation coefficient of 1.858 S that probably corresponds to the dimer (Fig. 2B). A small fraction of RGA5_S with even higher sedimentation coefficients corresponds potentially to higher-order oligomers. When dSP-AVR1-CO39 and RGA5_S were mixed, we detected only one predominant peak, whose sedimentation coefficient differs from those obtained with single proteins and that probably corresponds to the RGA5_S/dSP-AVR1-CO39 complex (Fig. 2B). This further confirms that RGA5_S forms homodimers in solution and that there is competition between RGA5_S/dSP-AVR1-CO39–binding and RGA5_S self-interaction.
GF and SV-AUC analysis were also performed with dSP-AVR-Pia and RGA5_S and showed that this MAX effector also competes with RGA5_S self-interaction, suggesting that the AVR-Pia–binding interface in RGA5HMA overlaps with the αA-β2 homodimerization surface (SI Appendix, Fig. S2 B and C).
ITC Measurements and NMR Titration Support the RGA5_S/dSP-AVR1-CO39 Structure and Validate the RGA5HMA-Binding Surface in AVR1-CO39.
The RGA5_S/dSP-AVR1-CO39 structure indicates that residues of RGA5_S outside the HMA domain are not involved in AVR1-CO39–binding since they are not resolved. To test this, the binding strength of dSP-AVR1-CO39 to a minimal RGA5HMA construct (RGA5991–1072, SI Appendix, Fig. S1) was determined by ITC. A Kd of 7.2 µM, almost identical to the Kd for RGA5_S/dSP-AVR1-CO39 (5.4 µM), was found (17), indicating that residues of RGA5_S outside of the HMA domain do not contribute to AVR1-CO39–binding (SI Appendix, Fig. S6).
To further investigate complex formation in solution, NMR titration experiments were performed where 1H- 15N-HSQC NMR spectra of 15N-labeled dSP-AVR1-CO39 were recorded in the absence and presence of unlabeled RGA5HMA. Mapping of the NH chemical shifts identified two major sequence stretches where most of the conformational rearrangements occur (Fig. 3 A–C): the first at the N terminus corresponds to the beginning of β1 and the second to the β1-β2 turn and β2. Together, these residues form the three main binding areas detected in the crystal structure of the RGA5_S/dSP-AVR1-CO39 complex (SI Appendix, Fig. S5). The low chemical shift differences in the rest of the molecule indicate that residues in the strands β3, β4, β5, and β6 are not perturbed and not involved in the interaction with RGA5HMA. NMR titration therefore defines the same binding interface as the crystal structure.
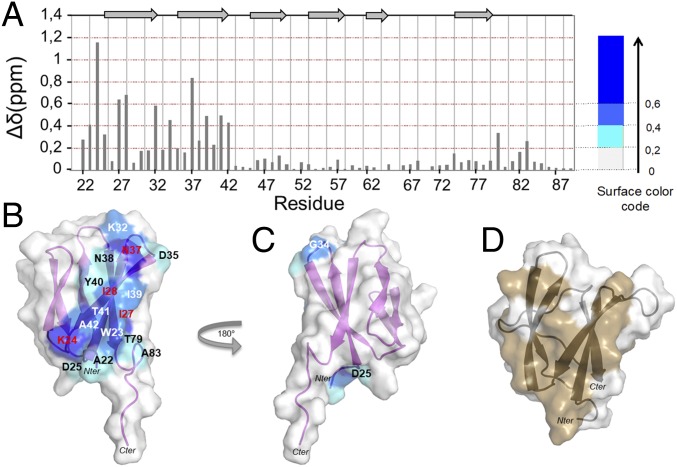
Fig. 3.
AVR1-CO39 binds RGA5HMA through the interface defined by the crystal structure of the complex. (A) Plot of the chemical shift differences (∆ppm) between unbound (R = 0) and RGA5HMA-bound dSP-AVR1-CO39 (R = 2). Chemical shift differences were calculated as the Hamming distance (31), ∆δ(ppm) = I∆δ(1H)ijI + 0.102 × I∆δ(15N)ijI, where ∆δ(1H)ij and ∆δ(15N)ij are the differences of the 1H and 15N chemical shifts at R = 0 and R = 2, respectively. (B) Structure of dSP-AVR1-CO39 showing the chemical shift differences from NMR titration with the following color code: surfaces of residues with ∆δ(ppm) ≥ 0.6 in dark blue (residues in red letter), 0,6 > ∆δ(ppm) ≥ 0.4 in light blue (residues in white letters), and for 0.2 > ∆δ(ppm) ≥ 0.4 in cyan (residues in black letters). (C) 180° rotation of B. (D) Structure of the WT dSP-AVR-Pia with the RGA5HMA interaction surface previously determined by NMR (14) in brown.
Structure-Guided Mutations of Key Residues in the RGA5HMA-Binding Surface of dSP-AVR1-CO39 Disrupt Complex Formation in Vitro and in Vivo.
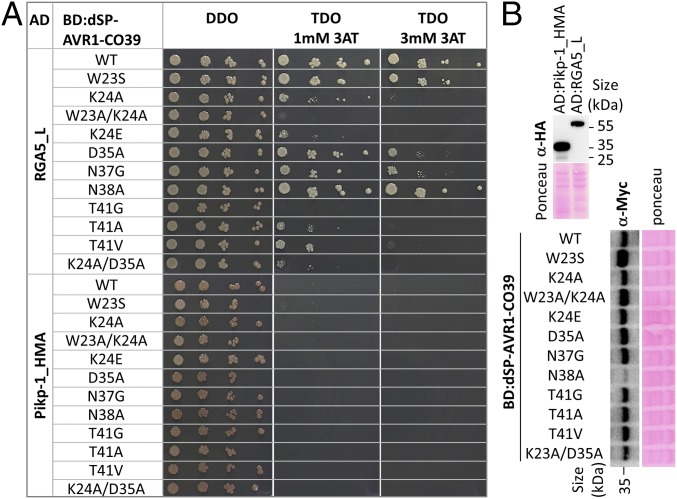
Based on the RGA5HMA/dSP-AVR1-CO39 structure, we designed mutations in the HMA-binding surface of AVR1-CO39 to perturb complex formation: seven dSP-AVR1-CO39 mutants with substitutions in the main RGA5HMA-binding area (W23S, K24A, K24E, W23A/K24A, T41G, T41A, and T41V), one mutation in the second area (N38A), two in the third area (D35A, N37G), and one double mutation targeting both the first and third area (K24A/D35A) (SI Appendix, Fig. S4). These mutants were tested in yeast two-hybrid assay for interaction with RGA5_L (Fig. 4A and SI Appendix, Fig. S1). A complete loss of interaction was observed for dSP-AVR1-CO39W23A/K24A and dSP-AVR1-CO39T41G, which both affect the main RGA5HMA-binding area (Fig. 4A). A severe reduction of binding to RGA5_L was caused by the mutations K24A, K24E, T41A, T41V, and K24A/D35A, while the D35A and N37G mutations in the third binding area only mildly reduced the interaction. The W23S and N38A mutations did not affect binding to RGA5_L. Pikp-1HMA was used as a control and showed no interactions. All Gal4 DNA-binding domain (BD)- or Gal4 activation domain (AD)-fused proteins were expressed according to immunoblotting (Fig. 4B).
Fig. 4.
Key residues within the HMA-binding surface of AVR1-CO39 are required for in vivo association with RGA5_L. (A) Interaction of RGA5_L (residues 883–1116, AD fusion) with dSP-AVR1-CO39 wild type and variants carrying point mutations in the HMA-binding surface (BD fusion) in yeast two-hybrid experiments. The HMA domain of Pikp-1 (AD:Pikp-1_HMA) was used as a control. Four dilutions of diploid yeast clones (1/1, 1/10, 1/100, 1/1000) were spotted on TDO medium (-Trp/-Leu/-His supplemented with 1 or 3 mM of 3-amino–1,2,4-triazole) to assay for interactions and on DDO (-Trp/-Leu) to monitor proper growth. Pictures were taken after 5 d of growth. The experiment was performed three times with consistent results. (B) Yeast protein extracts were analyzed by immunoblotting using anti-myc or anti-HA antibodies.
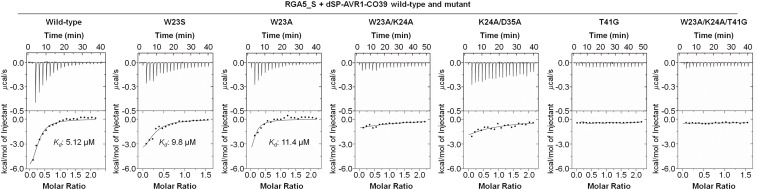
To test the interaction of the dSP-AVR1-CO39 mutants with RGA5_S in vitro, recombinant proteins were produced (except K24E, T41A, and T41V). The dSP-AVR1-CO39W23A and dSP-AVR1-CO39W23A/K24A/T41G mutants were also expressed to further assess the contribution of binding area 1 to RGA5HMA-binding. Soluble proteins were obtained for all mutants except K24A, D35A, N37G, and N38A and were analyzed by circular dichroism for proper folding (SI Appendix, Fig. S7A). ITC analyses showed that replacement of W23 by S or A only slightly reduced the affinity of dSP-AVR1-CO39 to RGA5HMA (Kds of 9.8 µM and 11.4 µM), while the double mutants W23A/K24A and K24A/D35A had drastically reduced affinities (Fig. 5). Replacement of T41 by G in dSP-AVR1-CO39T41G or dSP-AVR1-CO39K24A/D35A/T41G completely abolished binding.
Fig. 5.
In vitro binding assays demonstrate the importance of binding area 1 in the HMA-binding interface of AVR1-CO39 for RGA5_S/dSP-AVR1-CO39 complex formation. ITC curves for the titration of RGA5_S by dSP-AVR1-CO39 variants carrying point mutations in the HMA-binding surface. To ensure easy comparison, mutant proteins were used in the same concentration range as the AVR1-CO39 wild-type protein. Bottom shows the integrated heats of injection (squares) and the best fit (solid line) to a single-site–binding model. The assays were performed at least three times with similar results.
Further investigation by GF confirmed that the dSP-AVR1-CO39 mutants carrying the W23A or W23S substitution still bind RGA5_S, while the T41G and W23A/K24A/T41G mutations abolished dSP-AVR1-CO39–binding to RGA5_S (SI Appendix, Fig. S8).
Since the T41G mutation so strongly affects RGA5HMA-binding, dSP-AVR1-CO39T41G and dSP-AVR1-CO39W23A/K24A/T41G structures were reassigned using 15N-NOESY-HSQC and 15N-TOCSY-HSQC 3D NMR experiments. The chemical shift differences between the mutants and the wild-type dSP-AVR1-CO39 (SI Appendix, Fig. S7 B and C) show that replacement of T41 by a glycine propagates to the residues close in space in particular I27 and I28 in β1 that show strong chemical shift variation. Interresidue contacts observed in the 15N-NOESY-HSQC 3D NMR spectra indicate that while β-strands are essentially conserved, several interstrand long-range contacts involving residues in β1 are missing in the mutants. For example, the contact between HN-I28 (β1) and Hα-T41 (β2), found in the wild-type protein, is missing in both dSP-AVR1-CO39T41G and dSP-AVR1-CO39W23A/K24A/T41G. Since β2 of dSP-AVR1-CO39 is central for RGA5HMA-binding, the local distortion and probable flexibility at the C terminus of β2 caused by the T41G mutation is believed to be the main factor that destabilizes the complex.
To determine whether dSP-AVR1-CO39 associates with RGA5 through the same interface in planta as in vitro and in yeast, we performed coimmunoprecipitation (Co-IP) experiments in Nicotiana benthamiana using YFP-tagged dSP-AVR1-CO39 mutants, and HA-tagged RGA5_L. HA:RGA5_L was coprecipitated with all YFP:dSP-AVR1-CO39 constructs except the T41G, W23A/K24A, K24E and W23S mutants (SI Appendix, Fig. S9). For T41G and W23A/K24A, this is consistent with the Y2H, ITC, and GF data. For the W23S mutant, loss of association with RGA5_L was not observed in other assays. The K24A/D35A mutation drastically reduced RGA5HMA interaction in ITC and Y2H, but not in planta. As in the Y2H assay, the in planta association of RGA5_L with AVR1-CO39 was reduced in the T41A and D35A mutants and not affected by N37G or N38A.
These results show that key residues in the main RGA5HMA-binding area of AVR1-CO39 (in particular, W23, K24, and T41) are crucial for the interaction with RGA5HMA. Residues D35 and N37 within the second interaction area contribute weakly to RGA5HMA-binding, while N38 in the third area does not play a major role.
RGA5HMA/AVR1-CO39 Complex Formation Is Required for Resistance.
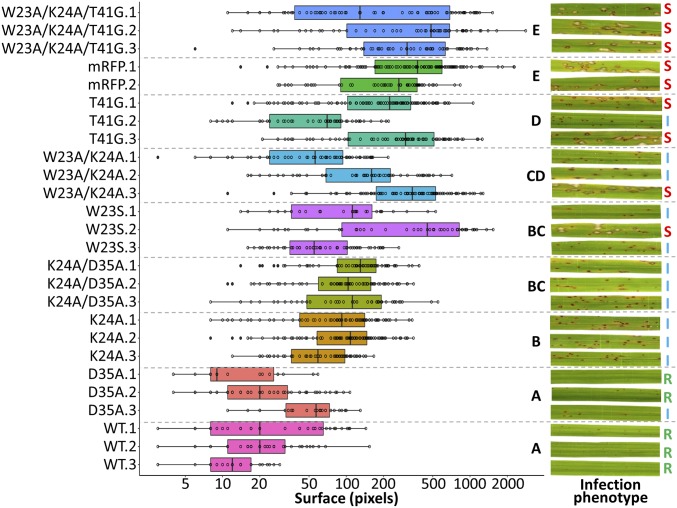
To evaluate whether binding of AVR1-CO39 to RGA5HMA is required for immunity, transgenic M. oryzae isolates carrying AVR1-CO39 variants affected at various degrees in their interaction with RGA5HMA were generated together with positive (wild-type AVR1-CO39) and negative (mRFP) controls. Infection experiments with the susceptible rice variety Nipponbare (pi-co39) demonstrated that all transformants possess full virulence (SI Appendix, Fig. S10). Quantification of transgene expression by qRT-PCR showed that AVR1-CO39 was expressed in the transgenic isolates (SI Appendix, Fig. S11). On the resistant Kitaake variety (Pi-CO39), only AVR1-CO39D35A triggered immunity at the same level as wild-type AVR1-CO39, which is consistent with the low impact of this residue on interaction with RGA5_L in Y2H and Co-IP (Fig. 6). All other mutants induced disease symptoms ranging from small lesions, characteristic of mild recognition, to fully expanded disease lesions, identical to the virulent mRFP control isolate and indicating loss of effector recognition.
Fig. 6.
Key residues within the HMA-binding surface of AVR1-CO39 are important for RGA5-mediated effector recognition in rice. Transgenic M. oryzae isolates carrying wild-type AVR1-CO39 (WT), AVR1-CO39 variants affected in the HMA-binding surface or mRFP (control), were spray-inoculated on plants of the rice cultivar Kitaake (Pi-CO39). Three independent transgenic isolates were generated for each construct (two isolates for mRFP). Leaves from 10 different plants for each isolate were scanned 7 d after inoculation, and areas of disease lesions were measured and plotted. The boxes represent the first quartile, median, and third quartile. Difference of lesion areas was assessed by an ANOVA followed by a Tukey HSD test. Groups with the same letter (A to E) are not significantly different at level 0.05. Representative disease phenotypes are shown (Right) for each strain (I, intermediate; R, resistant; S, susceptible). The experiment was performed two times with similar results.
Overall, these data highlight an important role of the HMA-binding surface of AVR1-CO39 in its recognition by the RGA5 immune receptor. However, AVR1-CO39 recognition seems to be quite resilient to modifications in this interaction surface, as only the triple mutant, affected in three important residues of the main area, showed full virulence and complete loss of recognition.
Discussion
The identification of unconventional IDs in NLRs, as effector detector modules, was a major breakthrough and led to the development of the integrated decoy model for effector recognition (7, 21, 22). This model states that IDs are paralogous to and act as mimics of true effector target proteins and that the presence of pathogens is sensed either by effector-mediated modification of the ID or by its interaction with the effector. In the present study, we demonstrate that binding of the M. oryzae effector AVR1-CO39 to the HMA ID of the rice NLR RGA5 contributes to effector recognition. Indeed, the binding strength of mutants of this MAX effector to the isolated RGA5HMA ID is correlated with the level of immunity against M. oryzae elicited on a resistant rice variety. Similar results have also been observed in the case of RGA5HMA/AVR-Pia and Pikp-1HMA/AVR-PikD (13–15). We therefore provide an additional demonstration of the important role of direct ID/effector interaction in the specific recognition of effectors.
By solving the RGA5HMA/AVR1-CO39 crystal structure and through NMR titration experiments, we found that the RGA5HMA-binding surface in AVR1-CO39 overlaps with the RGA5HMA-binding surface in AVR-Pia (14). Indeed, as in AVR1-CO39, N-terminal residues of AVR-Pia located before β1, as well as residues in β2, mediate the interaction (14). However, in AVR-Pia, the HMA-binding surface is more extended than in AVR1-CO39 and also involves the loop between β2 and β3 as well as the beginning of β3 (Fig. 3D). GF and AUC data indicate that, like AVR1-CO39, AVR-Pia competes with RGA5HMA self-interaction, suggesting that it also binds to the αA/β2 surface of the HMA. Further elucidation of this complex awaits a crystal structure of the RGA5HMA/AVR-Pia complex that we could not yet obtain.
Our analysis reveals that the HMA IDs of Pikp-1 and RGA5, which both interact with structurally conserved MAX effectors and are important for NLR receptor activation, are structurally highly similar and self-interact through almost identical interfaces formed by αA and β2. Besides, the formation of the RGA5HMA/AVR1-CO39 and Pikp-1HMA/AVR-PikD complexes shows, at the molecular level, important similarities. In both complexes, binding involves antiparallel alignment of the β-sheet of the HMA with one of the two β-sheets of the MAX effector, resulting in one antiparallel continuous β-sheet in the heterocomplex. However, there are also fundamental differences. Pikp-1HMA/AVR-PikD interaction relies very much on the N-terminal extension of AVR-PikD that is not part of the core MAX effector fold and makes strong and highly specific interactions with a defined area of the Pikp-1HMA surface (13). Conversely, RGA5HMA/AVR1-CO39–binding exclusively relies on interactions with the core MAX effector domain and does not involve the C-terminal unstructured extension of AVR1-CO39. In addition, in the two complexes, effector-binding is mediated by different surfaces of both the HMA-ID and the MAX effector. In the RGA5HMA/AVR1-CO39 complex, the β2s of both partners are aligned, while in the Pikp-1HMA/AVR-PikD complex, β4 of the HMA is aligned with β3 of the effector. Therefore, effector-binding involves different β-sheets of the effectors and occurs on opposite sides of the HMAs. AVR1-CO39 disrupts the RGA5HMA homodimer by competing for β2/αA-binding, while in the Pikp-1HMA/AVR-PikD complex, the β2/αA surface is free for self-interaction and, consequently, Pikp-1HMA occurs as a dimer in the Pikp-1HMA/AVR-PikD crystal structure. The fact that HMA IDs in different NLRs interact through completely different interfaces with sequence-unrelated but structurally conserved MAX effectors has major implications. It suggests that these interactions do not rely on conservation but evolved independently by convergence of the different MAX effectors to the same type of plant target proteins.
An exciting perspective of this finding, from an applied point of view, is that it may be possible to engineer these HMA IDs for extended recognition specificity, allowing them to recognize both AVR1-CO39 and AVR-PikD. Indeed, introduction of the additional binding site by changing the relevant surface-exposed residues should, in each case, not interfere with the original binding site located at the opposing site of the HMA.
The relevance of HMA self-interactions for RGA5 function is unclear, and it is unknown whether it occurs in the context of the full length RGA5 receptor. RGA5 forms heterocomplexes of unknown stoichiometry with RGA4, a constitutively active immune activator that is repressed by RGA5 in the resting state and triggers immunity upon RGA5-mediated AVR1-CO39 recognition (10, 16). RGA5 was also shown to form homocomplexes in planta, suggesting that the RGA4/RGA5 inactive complex may incorporate more than one RGA5 protein (16). In this context, RGA5HMA self-interaction could be relevant for RGA4 repression, and a switch to “activated state” might be triggered upon effector-mediated disruption of the RGA5HMA dimer. Alternatively, RGA5HMA self-interaction may not be relevant for RGA5 activity, either because the αA/β2 HMA surface is unoccupied in the inactive receptor complex or because it is engaged in other intra- or intermolecular interactions, potentially required for RGA4 repression. In this case, AVR1-CO39–binding to RGA5HMA could disrupt such interactions, resulting in conformational changes that would lead to RGA4 de-repression and immune activation. To test these hypotheses, it will be important to identify RGA5HMA mutations that uncouple self-interaction from AVR1-CO39–binding and to further search for alternative inter- or intramolecular interactions of the HMA ID.
Virulence targets of AVR1-CO39, AVR-Pia, or AVR-PikD have not been described yet. However, the integrated decoy model suggests that small HMA proteins (sHMAs), paralogous to Pikp-1HMA and RGA5HMA, are among them. The differential interaction of the two MAX effectors with their cognate HMA IDs suggests that AVR1-CO39 and AVR-PikD evolved independently toward these targets and target them in different manners. Whether AVR1-CO39 acts by competing with sHMA homo- or heterodimerization through the αA-β2 surface remains to be investigated since homocomplex formation of plant sHMAs has not been reported, and knowledge on sHMAs, which represent in plants a huge and rapidly evolving protein family, is extremely limited (23, 24). The importance of sHMAs for plant–pathogen interactions is strongly supported by the finding that the rice sHMA Pi21 is required for full susceptibility to the blast fungus (25). Therefore, there is a strong need to test whether sHMA proteins are virulence targets of MAX effectors and to elucidate their molecular and cellular functions.
Our finding that the interaction of AVR1-CO39 and AVR-PikD with HMA domains is presumably a consequence of convergent evolution toward similar host proteins suggests that MAX effectors potentially target other host proteins besides sHMAs. This hypothesis is supported by work on the M. oryzae MAX effector AvrPiz-t, for which five different host target proteins have been described (i.e., two E3 ubiquitin ligases, a K+ transporter, a b-ZIP transcription factor, and a nucleoporin protein Nup98) (26–30). These observations raise a fundamental question: How can these distinct MAX effectors target such a wide diversity of host proteins? Our study suggests that the molecular mechanism by which AVR1-CO39 and AVR-PikD interact with HMA domains is conserved. In both cases, binding relies on antiparallel alignment of one β-sheet of the MAX effector with the β-sheet of the HMA domain. This involves either β2 or β3 of the MAX effector, engaged in multiple backbone and side-chain interactions with the target β-structure. In addition, variable additional interactions by residues in β1 or from sequences outside the core MAX effector fold, such as the short and long N-terminal extensions of AVR1-CO39 and AVR-PikD, respectively, stabilize the interaction. Therefore, we hypothesize that, in the structurally conserved MAX effector family, the structural determinants and molecular mechanism of host target protein-binding are conserved rather than the host target proteins themselves. This establishes a framework for the understanding of the evolution and diversification of fungal effectors that can be tested by identifying additional host target proteins of MAX effectors and elucidating the structural details of MAX effector/target protein-binding.
Methods
Experimental procedures can be found in SI Appendix, including molecular biology, protein expression in Escherichia coli and N. benthamiana, yeast two-hybrid analysis, biochemical and physical characterization of proteins, and their complexes and infection assays.
Supplementary Material
Acknowledgments
The work was supported by the National Natural Science Foundation of China (NSFC) (Grant 31571990); the Agence Nationale de la Recherche (ANR) (Grant ANR-15-CE20-0007); the National Key Research and Development Program (Grant 2016YFD0300700) from the Ministry of Science and Technology, China; the 111 Project (B13006) and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) Project (IRT1042) from the Ministry of Education, China; the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (Grant 2017SKLAB7-9); and the French Infrastructure for Integrated Structural Biology (Grant ANR-10-INBS-0005). This work also benefited from interactions promoted by COST (European Cooperation in Science and Technology Program) Action FA 1208.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5ZNG and 5ZNE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810705115/-/DCSupplemental.
References
- 1.Takken FL, Goverse A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr Opin Plant Biol. 2012;15:375–384. doi: 10.1016/j.pbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013;4:297. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol. 2013;4:348. doi: 10.3389/fimmu.2013.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesari S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018;219:17–24. doi: 10.1111/nph.14877. [DOI] [PubMed] [Google Scholar]
- 5.Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 6.van der Hoorn RAL, Kamoun S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN. A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Front Plant Sci. 2014;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Roux C, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 9.Sarris PF, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 10.Cesari S, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida K, et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell. 2009;21:1573–1591. doi: 10.1105/tpc.109.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Guillen K, et al. Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 2015;11:e1005228. doi: 10.1371/journal.ppat.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maqbool A, et al. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife. 2015;4:e08709. doi: 10.7554/eLife.08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortiz D, et al. Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell. 2017;29:156–168. doi: 10.1105/tpc.16.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De la Concepcion JC, et al. Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat Plants. 2018;4:476–485. doi: 10.1038/s41477-018-0194-x. [DOI] [PubMed] [Google Scholar]
- 16.Césari S, et al. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33:1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L, et al. Crystallization of the rice immune receptor RGA5A_S with the rice blast fungus effector AVR1-CO39 prepared via mixture and tandem strategies. Acta Crystallogr F Struct Biol Commun. 2018;74:262–267. doi: 10.1107/S2053230X18003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D, Zhang Y, Zhao Y, Liu J, Peng YL. Expression, purification, crystallization and preliminary X-ray diffraction analysis of the effector-interaction domain of the resistance protein RGA5-A from Oryza sativa L. japonica. Acta Crystallogr F Struct Biol Commun. 2015;71:171–174. doi: 10.1107/S2053230X14028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroj T, Chanclud E, Michel-Romiti C, Grand X, Morel JB. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016;210:618–626. doi: 10.1111/nph.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarris PF, Cevik V, Dagdas G, Jones JD, Krasileva KV. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016;14:8. doi: 10.1186/s12915-016-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Abreu-Neto JB, Turchetto-Zolet AC, de Oliveira LF, Zanettini MH, Margis-Pinheiro M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013;280:1604–1616. doi: 10.1111/febs.12159. [DOI] [PubMed] [Google Scholar]
- 24.Tehseen M, Cairns N, Sherson S, Cobbett CS. Metallochaperone-like genes in Arabidopsis thaliana. Metallomics. 2010;2:556–564. doi: 10.1039/c003484c. [DOI] [PubMed] [Google Scholar]
- 25.Fukuoka S, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- 26.Shi X, et al. The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel. PLoS Pathog. 2018;14:e1006878. doi: 10.1371/journal.ppat.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang M, et al. The Nup98 homolog APIP12 targeted by the effector AvrPiz-t is involved in rice basal resistance against Magnaporthe oryzae. Rice. 2017;10:5. doi: 10.1186/s12284-017-0144-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C-H, et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park CH, et al. The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog. 2016;12:e1005529. doi: 10.1371/journal.ppat.1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, et al. Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr Biol. 2016;26:2399–2411. doi: 10.1016/j.cub.2016.06.072. [DOI] [PubMed] [Google Scholar]
- 31.Schumann FH, et al. Combined chemical shift changes and amino acid specific chemical shift mapping of protein–protein interactions. J Biomol NMR. 2007;39:275–289. doi: 10.1007/s10858-007-9197-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.