Significance
Class XIV myosins are promising therapeutic targets for the treatment of apicomplexan disease because of their importance in parasite fitness and divergence from conventional human myosins. In this study, we report the crystal structure of a class XIV myosin, MyoA from Toxoplasma gondii. Structural analysis complemented with mutagenesis, biochemical, and functional data support a model whereby unique sequence elements in class XIV motors result in unique mechanisms of force production and chemomechanical coupling. Notably, many of these elements are located in known binding sites for allosteric inhibitors of myosin function, highlighting the potential for the design of class-specific myosin inhibitors as treatments for apicomplexan disease. With the established structural and biochemical platforms, we are poised to advance detailed functional studies and support inhibitor development.
Keywords: myosin, Apicomplexa, Toxoplasma gondii, motility, X-ray crystallography
Abstract
Parasites of the phylum Apicomplexa are responsible for significant morbidity and mortality on a global scale. Central to the virulence of these pathogens are the phylum-specific, unconventional class XIV myosins that power the essential processes of parasite motility and host cell invasion. Notably, class XIV myosins differ from human myosins in key functional regions, yet they are capable of fast movement along actin filaments with kinetics rivaling previously studied myosins. Toward establishing a detailed molecular mechanism of class XIV motility, we determined the 2.6-Å resolution crystal structure of the Toxoplasma gondii MyoA (TgMyoA) motor domain. Structural analysis reveals intriguing strategies for force transduction and chemomechanical coupling that rely on a divergent SH1/SH2 region, the class-defining “HYAG”-site polymorphism, and the actin-binding surface. In vitro motility assays and hydrogen–deuterium exchange coupled with MS further reveal the mechanistic underpinnings of phosphorylation-dependent modulation of TgMyoA motility whereby localized regions of increased stability and order correlate with enhanced motility. Analysis of solvent-accessible pockets reveals striking differences between apicomplexan class XIV and human myosins. Extending these analyses to high-confidence homology models of Plasmodium and Cryptosporidium MyoA motor domains supports the intriguing potential of designing class-specific, yet broadly active, apicomplexan myosin inhibitors. The successful expression of the functional TgMyoA complex combined with our crystal structure of the motor domain provides a strong foundation in support of detailed structure–function studies and enables the development of small-molecule inhibitors targeting these devastating global pathogens.
The phylum Apicomplexa comprises several thousand parasitic protozoan species that cause significant morbidity and mortality worldwide. Of particular relevance to human health are Plasmodium spp. (which cause malaria), Cryptosporidium spp. (cryptosporidiosis), and Toxoplasma gondii (toxoplasmosis). As obligate intracellular parasites, apicomplexans’ life cycle critically depends on their ability to traverse the environment and actively invade host cells (1). These processes are accomplished via “gliding motility,” a unique form of locomotion whereby membrane adhesins anchored to intracellular actin filaments are thought to be translocated rearward by parasite myosin motors interacting with these actin filaments to generate fast, forward motion of the parasite (2, 3).
T. gondii has the largest repertoire of myosins among Apicomplexa, with 11 unique isoforms that are implicated in numerous essential processes including cell division, organellar inheritance, motility, and host cell invasion (4, 5). The major myosin driving parasite motility and host cell invasion, MyoA, belongs to class XIV of the myosin superfamily, which exists only in apicomplexans and select ciliates (4, 5). Of the class XIV isoforms, MyoA is the most well conserved across the phylum, likely because of its central role in motility and invasion (6); depletion of MyoA in T. gondii severely impairs both of these processes (7–10).
Myosins adopt a generally conserved architecture that includes a force-generating motor domain, a light chain/calmodulin-binding neck domain that acts as a lever arm, followed by a tail region that serves as a cargo-binding domain or mediates dimerization, which, for class II myosins, leads to filament formation. As a result of the lack of a tail region, apicomplexan MyoA is among the smallest known myosin isoforms, yet it maintains a functional neck domain that coordinates the ELC1 and MLC1 light chains, the latter of which anchors the myosin via a unique N-terminal extension (11). The myosin motor domain itself typically comprises several subdomains including the upper and lower 50-kDa subdomains (U50 and L50, respectively), the transducer, the converter, and, depending on the myosin class, an src homology 3 (SH3) subdomain (12). The U50 and L50 subdomains are separated by a large cleft that opens and closes in a nucleotide-dependent manner to mediate actin binding, and the transducer plays a key role in integrating structural changes throughout the motor domain. The converter then amplifies structural changes associated with the myosin ATPase cycle to generate the force-producing powerstroke and motion. Each subdomain critically relies on conserved motifs that couple conformational changes in the active site with large-scale force production (13). Intriguingly, sequence analysis of class XIV myosins reveals substitutions in key sites that are likely to have profound effects on force generation, transduction, and motor modulation (14, 15). In class II myosins, for example, substitution of a conserved Gly for Ala or Val in the hinge connecting helices SH1 and SH2 uncouples this mechanical communication pathway to the lever arm, impairing force transduction, ATPase activity, actin binding, and in vitro motility (16). Strikingly, the SH helices of class XIV myosins, which are structurally analogous to the reactive thiol region of class II muscle myosins, possess a Ser instead of the conserved Gly at this position, yet the kinetic and mechanical properties of the motor remain similar to conventional myosins (15, 17, 18). Furthermore, class XIV myosins rely on phosphorylation-dependent modulatory processes (19, 20) and possess divergent actin-binding motifs with implications for actin-binding dynamics (15, 17) and a key substitution in the “HYAG” site where a Phe or Tyr in almost all other myosin classes is substituted for a Ser or Thr, which is considered a defining trait of the class (4). Ultimately, the mechanistic details of how class XIV myosins function with kinetics rivaling previously studied myosins are largely unresolved.
Toward establishing a more comprehensive model of myosin-dependent apicomplexan motility, we report the 2.6-Å resolution crystal structure of the T. gondii MyoA (TgMyoA) motor domain in complex with the nucleotide analog ADP-AlF4. Notably, structural analysis of this class XIV myosin, complemented with mutagenesis. Structural analysis complemented with mutagenesis and functional data reveals mechanisms of force transduction and chemomechanical coupling, in addition to the functional modulation resulting from phosphorylation sites, and surface pocket analysis highlights the potential for the design of novel class XIV-specific myosin inhibitors.
Results
Structural Analysis of the Class XIV TgMyoA.
Sequence analysis of TgMyoA predicts a central motor domain framed by smaller ancillary domains. To probe the structural modularity of TgMyoA and identify a stable crystallization construct, we generated truncations at the N and C termini (Fig. 1A) that were coexpressed in insect cells with the recently identified MyoA chaperone, UNC (18). Of the expression constructs screened, only TgMyoA full length (residues 1–831) and lacking the neck region (residues 1–778) yielded soluble protein, and only the latter could be crystallized. TgMyoA 1–778 eluted as a monomer by size-exclusion chromatography (SEC; Fig. 1B) and was functional to bind and release actin in a nucleotide-dependent fashion (Fig. 1C). Diffraction-quality crystals were obtained with TgMyoA 1–778 in complex with MgADP-aluminum fluoride (i.e., MgADP-AlF4), a stable analog of posthydrolysis ADP and inorganic phosphate associated with the priming of the converter and lever arm for force generation, known as the prepowerstroke state (PPS).
Fig. 1.
Structural analysis of the class XIV TgMyoA. (A) Schematic representation of TgMyoA expression and crystallization (blue line) constructs. (B) Sx200 SEC profile and SDS/PAGE gel showing homogenous preparation of monomeric TgMyoA 1–778. (C) Schematic illustration of actin cosedimentation assay (Top) showing that TgMyoA 1–778 is able to bind actin and is responsive to ATP (Bottom). F-actin with bound TgMyoA was pelleted at 340,000 × g. Supernatant containing excess globular actin and non–actin-binding MyoA (lane 1). ATP was added to the resuspended pellet, dissociating functional TgMyoA from the actin, and the F-actin was pelleted again, leaving only functional myosin in the supernatant (lane 2). The pellet containing F-actin and ATP nonresponsive myosin was then resuspended (lane 3). (D) Overview of TgMyoA 1–778 structure, highlighting the upper (orange) and lower (purple) 50-kDa domains and the putative SH3 (green), transducer (blue), and converter (red) subdomains. The region encompassing 779–831 (hashed lines) is not present in the crystal construct. (E) Topology map of TgMyoA motor domain with subdomains colored as in D.
The structure of PPS TgMyoA 1–778 was solved by molecular replacement by using the pruned, core motor domain of PPS human myosin 1c [Protein Data Bank (PDB) ID code 4BYF] (21) and refined to a resolution of 2.6 Å. Through iterative refinements, electron density emerged for the N- and C-terminal ancillary domains resulting in a final model extending from Y33 through S777 with residues M632–K639 (loop 2) remaining unmodeled (Fig. 1 and SI Appendix, Fig. S1). Overall, TgMyoA adopts a modular architecture with the predominantly α-helical upper (U50; Fig. 1 D and E, orange) and lower (L50; Fig. 1 D and E, magenta) 50-kDa subdomains separated by the actin-binding cleft in the open position. Located at the base of the actin-binding cleft is the active site with bound MgADP-AlF4 and the central transducer subdomain that comprises seven sequence-distal β-strands (Fig. 1 D and E, blue). The U50, L50, and transducer domains constitute the core of the motor, which most closely resembles that of Dictyostelium discoideum class I myosin (PDB ID code 1LKX), aligning with an rmsd of 0.99 Å over 394 Cαs (22). The TgMyoA core is remarkably well conserved, with increasing divergence radiating outward (SI Appendix, Fig. S2). Divergent actin-binding loops (SI Appendix, Fig. S2) likely represent adaptations required to bind the unconventional apicomplexan actin (23–26). The sequence-divergent N terminus (M1–N111) comprises a disordered N-terminal extension (M1–V32) and a more domain-like region (Y33–L95). Iterative building cycles of the Y33–L95 region revealed a partial β-barrel topology consistent with an SH3 domain (green) found in class II, V, and VI myosins (12) (Fig. 1 D and E). The lack of sequence identity and partial disorder, however, made it difficult to unambiguously classify the domain and register the primary sequence. The final model of the putative SH3 domain was based on simulated annealing and omit maps with bulky side chains such as Trp-39, Met-53, and Trp-90 used to anchor the sequence register (SI Appendix, Fig. S1). Importantly, none of the key divergent sequences of class XIV myosins are found within Y33–L95, and therefore the localized structural ambiguity does not impact the mechanistic analysis herein. The C-terminal ancillary domain (TgMyoA residues G704–E774), also known as the converter subdomain (red), is oriented in the upward “primed” position, typical of PPS myosins (Fig. 1 D and E). In contrast to the motor core, the converter domain most closely resembles that of class II myosin from Drosophila melanogaster (PDB ID code 4QBD), aligning with an rmsd of 0.80 Å over 43 Cαs. Overall, the modular architecture of TgMyoA is functionalized through a network of loops, switches, and relays that govern myosin force generation (Fig. 1E).
Interactions with the SH Helices Maintain Chemomechanical Coupling.
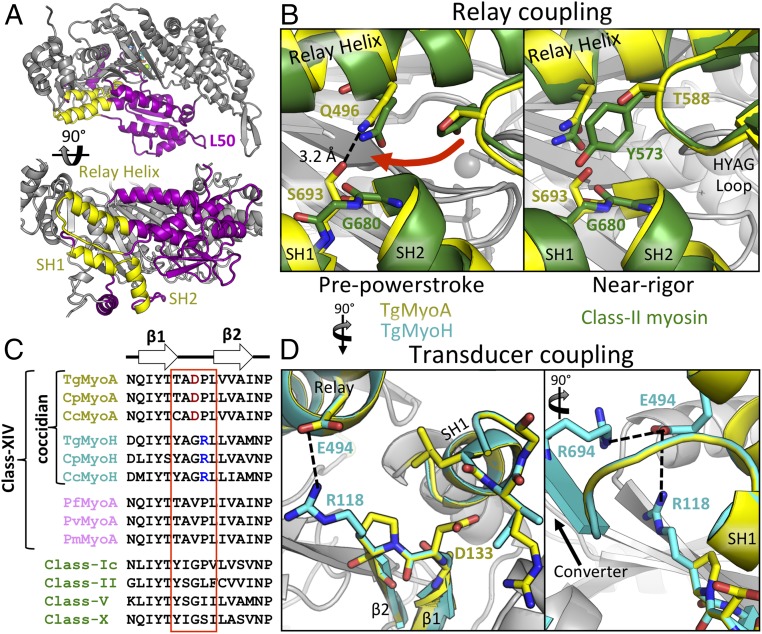
The SH helices constitute a structurally conserved motif composed of adjacent α-helices, SH1 (TgMyoA L695-Q702) and SH2 (TgMyoA P682-A691), connected by a short linker (Figs. 1E and 2A). Together with the adjacent relay helix and transducer elements, the SH helices participate in coupling the conformation of the active site to the converter and lever arm position, thereby playing a key role in force generation. In class II myosins, substitution of a conserved Gly in the SH1/SH2 linker (G680 in D. discoideum class II myosin) with Ala or Val displaces the SH helices from the relay helix and transducer, effectively uncoupling force transduction, which causes severe motor defects (27–29). In TgMyoA, a Ser (S693) is substituted for the conserved Gly. The result is a stabilizing hydrogen bond between S693 and the side chain of Q496 in the relay helix (Fig. 2B, Left), which likely serves to maintain, and possibly enhance, transduction of force from the active site to the lever arm via the SH helices. Interpreted in the context of class II myosins, this interaction may facilitate class XIV myosin motility by enhancing the kinetics of the myosin ATPase cycle, particularly by reducing the duration of the rate-limiting ADP release step following the force-generating powerstroke (29). Serine 693 also appears to play a role in supporting the class XIV-defining HYAG site polymorphism, where the conserved Tyr or Phe of most other myosin classes is substituted for Thr or Ser in class XIV myosins (T588 in TgMyoA). To further assess the scope of these observations, we simulated the near-rigor state of the class XIV SH helices by aligning the relevant substructures with their counterparts in the near-rigor state (i.e., ADP-bound) of D. discoideum myosin II (PDB ID code 1MMA; Fig. 2B, Right) (30). In this state, the HYAG-containing loop shifts toward the SH1/SH2 linker by ∼6.0 Å. The bulky side chains of Tyr or Phe in other myosin classes, which are normally accommodated by an opposing Gly, are unlikely to be tolerated in class XIV motors as a result of steric clashes with S693. Thus, it appears that the class XIV HYAG sequence divergence (Y/F→T/S) is required to accommodate the Gly→Ser change of the class XIV SH1/SH2 linker.
Fig. 2.
Interactions in the SH helices maintain chemomechanical coupling. (A) Overview showing the position of the SH helices and relay helix (yellow) within the TgMyoA L50 domain (purple). (B) SH helices of TgMyoA (yellow) and class II myosin (green) highlight unique interactions between the SH1/SH2 pivot point, the relay helix, and the HYAG site in the PPS (Left) and near-rigor (Right) states. (C) Sequence alignments of β1/2 linker regions from various coccidian myosins (yellow and cyan), Plasmodium MyoA (pink), and non-class XIV myosins (green). (D) Unique interactions between the β1/2 linker, SH helices, and relay helix in TgMyoA and TgMyoH colored as in C.
Next, we evaluated the interactions between the class XIV SH helices and the transducer subdomain. TgMyoA D133, located in the loop connecting β1 and β2 of the transducer, takes the place of a conserved Gly in previously studied myosins (G118 in chicken class II myosin; PDB ID code 1BR1; Fig. 2C) and is oriented toward the SH1 helix (Fig. 2D). The side chain of D133 is positioned within 3.3 Å and 4.0 Å, respectively, from the backbone amides of SH1 helix residues L698 and R701, suggesting the potential for coccidian-specific stabilizing interactions between the transducer and SH1 helix, further strengthening the coupling between these substructural elements. To interrogate the structural implications of divergence in the β1/β2 loop across different class XIV myosins, we generated a high-confidence homology model of T. gondii myosin H (MyoH; TgMyoH). MyoH is a coccidian-specific class XIV myosin that is localized to the parasite conoid region and is thought to initiate motility/invasion before “handing off” actin filaments to MyoA at the border between apical complex and the inner membrane complex (IMC) (31, 32). Intriguingly, TgMyoH does not share the divergent D133 with TgMyoA and instead retains the Gly (G117) of previously studied myosins, which is unable to bridge the transducer and SH1 helix. However, an Arg (R118) in the β1/β2 loop of TgMyoH that is conserved in all class XIV MyoH isoforms (Fig. 2C) forms a salt bridge with the side-chain carboxylate of E494 of the relay helix (Fig. 2D), which also interacts with R694 of the converter subdomain, potentially providing an alternative pathway for force transduction or tighter coupling of this region to the swing of the converter/lever arm throughout the TgMyoH ATPase cycle.
Phosphomimetic Mutations Alter the Stability and Motility of TgMyoA.
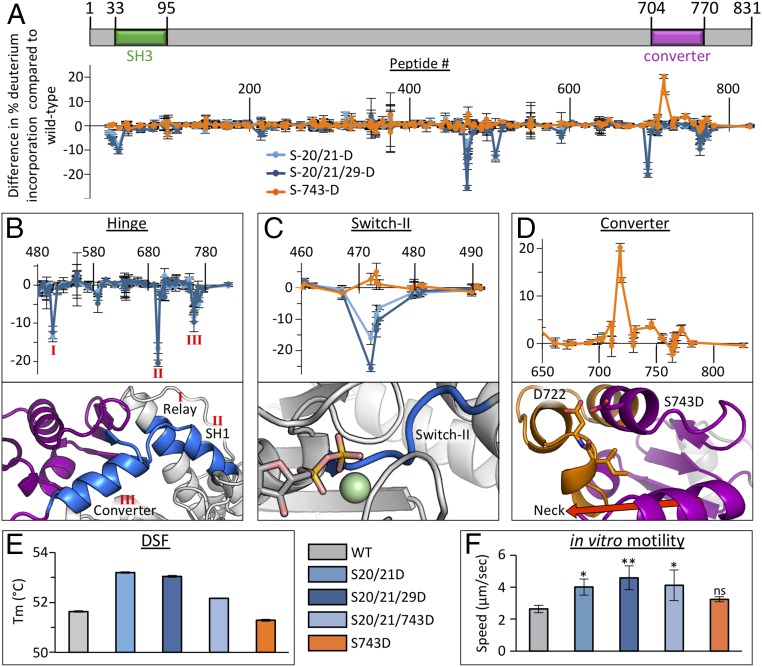
Phosphorylation of the TgMyoA N-terminal extension that precedes the putative SH3 domain (S20, S21, and S29) and converter subdomain (S743) are important for fast motility and host cell egress of T. gondii tachyzoites (19, 20). To interrogate the associated molecular mechanisms, we generated a series of phosphomimetic mutants and measured the associated dynamics of altered solvent accessibility [hydrogen–deuterium exchange coupled with MS (HDX-MS)], thermostability [differential scanning fluorimetry (DSF)], and functional response to actin (in vitro motility assays; Fig. 3). In previously studied myosins, the flexible N-terminal extension interacts with the converter/lever arm, L50 subdomain, and first light chain when the lever arm is in the downward position (33–35) associated with the near-rigor state, but not in the upward primed position associated with the PPS (36). Notably, we previously reported HDX-MS data showing that binding of TgELC1 to the neck region of MgADP-bound TgMyoA stabilized the TgMyoA N terminus, in addition to the neck and converter (37). Presuming that interactions between the TgMyoA N terminus and lever arm likely form the basis for modulation of TgMyoA motility by N-terminal phosphorylation, double (S20/21D) and triple (S20/21/29D) phosphomimetic mutants of full-length TgMyoA were generated and characterized in the near-rigor (i.e., MgADP-bound) state, with both light chains (MLC1 and ELC1) bound. HDX-MS data show that not only is the N terminus stabilized by these mutations, several sequence-distal regions within TgMyoA also showed significant increases in order and solvent protection (Fig. 3A). Mapping these changes onto our crystal structure of the PPS TgMyoA motor reveals protected regions around the hinge between the motor domain and converter subdomain, including the converter itself, the relay helix, and the SH1 helix (Fig. 3B). Although we are unable to draw conclusions regarding the specific conformational changes induced in these regions because of a lack of an MgADP-bound crystal structure, the HDX-MS data support a model whereby the phosphorylated N terminus of TgMyoA modulates motility via interactions formed with the motor and converter when the lever arm is in the downward position. Intriguingly, phosphomimetic mutations in the N terminus also resulted in solvent protection of the switch II loop in the TgMyoA active site, ∼30 Å away from the aforementioned cluster of interactions (Fig. 3C). The significant distance between the active site and other affected regions indicates an allosterically driven effect, likely mediated via the relay helix. Next, we measured the effects of the phosphomimetic mutations by using DSF and in vitro motility assays, which showed significant increases in thermal stability (Fig. 3E) and actin-based motility (Fig. 3F), consistent with previously reported cell-based assays (19). In contrast, mutation of S743 in the converter subdomain, another phosphorylation site shown to be crucial for parasite motility (19, 20), to Asp significantly reduced solvent protection (Fig. 3D) and overall stability (Fig. 3E) in the converter subdomain. When analyzed in the context of the structure, it appears that destabilization may result from electrostatic and steric repulsion formed between the longer, more negative side chain of D743, and the closely packed helices that constitute the remainder of the converter subdomain, particularly the nearby residue D722 (Fig. 3D). Notably, there was no statistically significant change in actin-based motility relative to WT (Fig. 3F), suggesting that, if S743 phosphorylation plays a role in modulating motility in cell-based assays (19), it may do so by stabilizing the glideosome complex rather than directly affecting motor activity. To assess the potential for synergistic effects arising from phosphorylation at the N and C termini, we generated the hybrid phosphomimetic construct S20/21/743D. No cooperativity was observed by DSF, in vitro motility assays, or HDX-MS (Fig. 3 E and F and Dataset S1).
Fig. 3.
Phosphomimetic mutations affect motility of TgMyoA by stabilizing or destabilizing key functional regions. (A) Schematic representation of TgMyoA aligned with HDX difference map showing change in percent deuterium incorporation of full-length TgMyoA phosphomimetic mutants in the near-rigor state relative to WT over 300 s. Error bars (SD) represent independent triplicates. Peptide number corresponds to the centroid amino acid of the peptide from which a data point is obtained. (B–D). Expanded views of HDX difference map showing change in percent deuterium incorporation in different TgMyoA subdomains by the phosphomimetic mutants (Top) with the stabilized (blue) and destabilized (orange) regions mapped onto the TgMyoA crystal structure (Bottom). The protected regions in the relay, SH1, and converter associated with the S20/21D and S20/21/29D mutations are indicated as I, II, and III, respectively, in B; change in percentage incorporation in the switch II loop associated with the S-20/21D, S20/21/29D, and S743D mutations are shown in C (AlF4 is omitted in figure to prevent obstruction of switch II); and change in percentage incorporation in the converter subdomain associated with the S743D mutation is shown in D. (E) DSF thermal shift assay of TgMyoA comparing WT vs. phosphomimetic mutants. Error bars (SD) represent independent triplicates. (F) Actin filament displacement velocities of WT and phosphomimetic mutants in in vitro motility assays. Error bars (SD) represent five independent experiments. Mutant speeds were compared with WT by one-way ANOVA and Šídák’s multiple comparison tests: *P < 0.05 and **P < 0.005; “ns” indicates P > 0.05.
Surface Pocket Analysis Supports the Potential for Designing Class XIV-Specific Inhibitors.
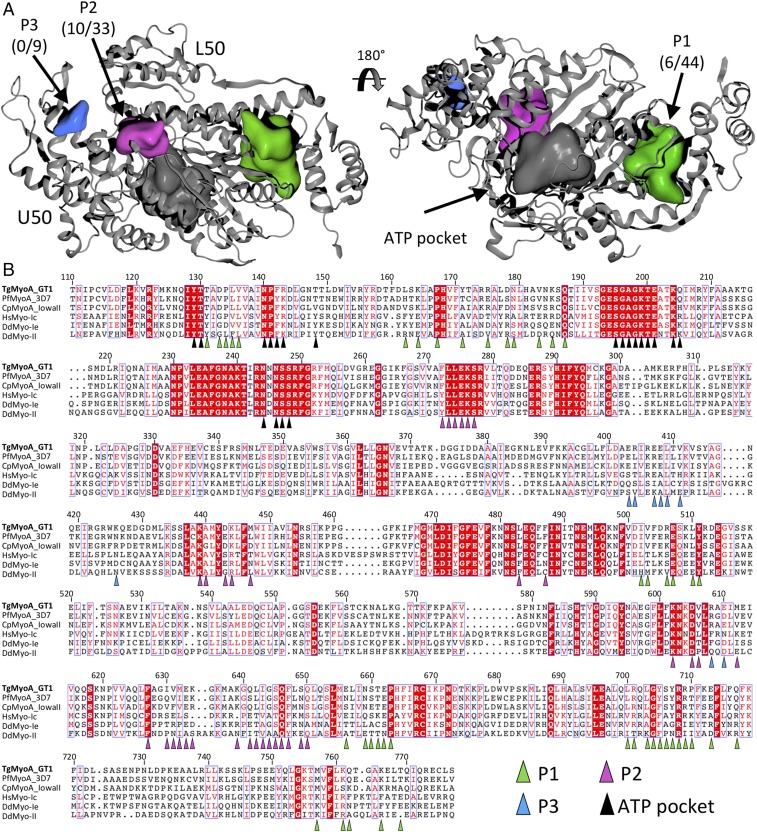
Class XIV myosins are promising targets for the development of small-molecule inhibitors of apicomplexan parasites because of their divergence from animal myosins and their essential role in the apicomplexan life cycle. A comprehensive pocket analysis combined with a structure-guided sequence alignment was performed to identify possible binding sites for allosteric inhibitors (Fig. 4). In addition to the conserved active site, a large pocket (“P1,” 2,700 Å3) was identified between the TgMyoA core and the converter subdomain (Fig. 4A). The residues lining the P1 pocket are largely conserved between the MyoA homologs [78% ID, 84% similar over 44 amino acids (AA)] but diverge between the myosin classes (14% ID, 33% similar over 44 AA; Fig. 4B). Notably, the P1 pocket is already known as the binding site for omecamtiv mecarbil (OM), a specific allosteric inhibitor of human cardiac myosin (38, 39). We next investigated the halogenated pseudilin class of small molecules that bind deep in the actin-binding cleft of other myosin classes, including class Ie and class II from D. discoideum (40, 41). In the absence of inhibitor, these motors present a clear solvent-accessible pseudilin pocket with a volume of ∼2,900 Å3 in PPS (SI Appendix, Fig. S4). In contrast, the corresponding pocket is significantly smaller in our PPS TgMyoA structure (“P2,” 1,070 Å3; Fig. 4 and SI Appendix, Fig. S4). In fact, P2 was not detected in TgMyoA until the missing residues of loop 2 (M632–K639) were modeled. We next aligned our structure with several previous PPS myosin structures via the L50 domains, which revealed a ∼4.0-Å shift in the position of the class XIV U50 domain toward the L50 domain (SI Appendix, Fig. S4). The result is a class XIV actin-binding cleft that, while in the open conformation associated with the PPS, remains significantly more closed than previous PPS myosin structures, accounting for the reduced volume of the pseudilin-binding pocket relative to other myosin classes for which there are structural data available. Similar to the P1 pocket, the residues lining P2 are largely conserved between MyoA homologs (76% ID, 91% similar over 33 AA) but diverge with respect to other myosins (30% ID, 60% similar over 33 AA; Fig. 4). Strikingly, analysis of our crystal structure also revealed a smaller pocket (“P3,” 550 Å3), which is not observed in other myosin crystal structures examined (Fig. 4) and therefore may be ideally suited for selective inhibitors. The success of OM and halogenated pseudilins as specific myosin inhibitors supports the viability of P1 and P2 as druggable sites, and the divergence of pocket-lining residues between class XIV and human myosins could form the basis of specificity for a targeted class XIV inhibitor.
Fig. 4.
Pocket analysis of TgMyoA supports potential for class XIV-specific inhibitors. (A) Surface pocket analysis of the TgMyoA crystal structure. Solvent-accessible pockets, shown in green (P1), purple (P2), and blue (P3), were identified by using default parameters in OEDocking software, allowing the shape of the pockets to extend toward the solvent by one solvent radius. Pocket labels include the number of identical residues between TgMyoA and sequence-aligned myosins (B) among the total pocket residues. (B) Structure-guided sequence alignment of the core motor domains from class XIV, class I, and class II myosins. Highly variable N- and C-terminal regions have been omitted for clarity. Residues belonging to each of the solvent-accessible pockets are indicated with colored triangles.
Discussion
To accomplish the essential processes of motility and host cell invasion, apicomplexan parasites rely on force generated by the unconventional class XIV myosins such as the glideosome-associated MyoA. This dependence is highlighted by severe impairments in host cell invasion and parasite motility upon depletion of these myosins (7–10), which, along with their divergence from human myosins, supports the potential for targeted therapeutic intervention. Here we present a structural characterization of a class XIV myosin in the form of MyoA from the apicomplexan parasite T. gondii. Structural analyses reveal unique spatial relationships within key functional regions, providing important mechanistic insight into the molecular basis of motility for these global pathogens.
A central question in apicomplexan motility is how class XIV myosins maintain fast motility despite numerous divergent sequence motifs in functionally important regions. Mutation of a key Gly to Ala or Val in the hinge between the SH1/SH2 helices of class II myosins impairs force production by uncoupling the allosteric communication pathway that connects the active site to the lever arm by way of the SH helices. These mutations result in a myosin that binds actin filaments, hydrolyzes ATP, but proceeds very slowly through the subsequent force-generating steps, effectively blocking motility (42). Strikingly, structural analysis of TgMyoA revealed a unique network of interactions that are able to maintain chemomechanical coupling between the transducer subdomain, the SH helices, and the relay helix despite sequence differences. Specifically, unlike Ala or Val substitutions for the conserved Gly in class II myosins, the polar side chain of TgMyoA S693, at the equivalent Gly site, forms a stabilizing hydrogen bond with Q496 of the relay helix. The functional significance of the class-defining HYAG polymorphism was also revealed, as the smaller Ser and Thr residues that define class XIV motors are required to accommodate S693 in the SH1/SH2 linker. Therefore, a potential outcome of S693 in the class XIV myosins is the strengthening of the coupling of the SH helices with neighboring structural elements, which would allow strain through the lever arm to be more effectively transmitted to the active site, in turn impacting the kinetics of the ATPase cycle (43) so that fast actin filament motility by TgMyoA is possible (44). A similar effect may result from potential H-bond formation between TgMyoA D133 in the transducer domain and backbone amides of the SH1 helix. Conversely, R118 in the TgMyoH transducer domain is ideally positioned to form a salt bridge with E494 of the relay helix, which is likely important for coupling the transducer, relay, and SH helices’ communication pathway to the converter, as is the conserved salt bridge already formed between E494 and R694 of the converter subdomain. This could enhance force transduction to counteract the greater compliance expected in the significantly longer lever arm of TgMyoH, or could result in different actin affinities between TgMyoA and TgMyoH, as may be necessary for actin filaments to be “handed off” from TgMyoH to TgMyoA at the apical complex/IMC border in coccidians (31, 32).
Class XIV myosin-dependent motility and host cell egress (19, 20) in apicomplexan parasites is enhanced by phosphorylation of the TgMyoA N terminus (S20, S21, and S29) and converter subdomain (S743). To interrogate the underlying molecular mechanisms, we measured the effects of corresponding phosphomimetic mutations on class XIV myosin function. Compared with the unphosphorylated motor, which is active and supports actin-filament motility, the phosphomimetic mutants have as much as twofold faster actin filament speeds (Fig. 3F). Thus, TgMyoA phosphorylation appears to modulate rather than activate the motor. This twofold increase in actin filament speeds in vitro corresponds well with the approximate threefold increase in parasite motility initiation and gliding speed seen in parasites expressing similar TgMyoA phosphomimetic mutations (19). From a structural standpoint, HDX-MS analyses support a model whereby a phosphorylated N-terminal extension (encompassing S20, S21, and S29) is stabilized through contacts with a groove formed between the motor domain and the converter subdomain as observed in previously characterized myosins (12, 34, 35). Not only do these interactions stabilize the interface between the TgMyoA motor and lever arm, they also alter the state of the switch II loop in the active site, which influences coordination of the γ-phosphate of the bound nucleotide (45). This allosteric change is likely mediated via the long relay helix, which extends from the C terminus of the switch II loop to the hinge between the converter and core motor domain, where the majority of stabilization by N-terminal phosphomimetics was observed. The relay helix is crucial for transforming small conformational changes in the switch II loop into large movements of the lever arm throughout the ATPase cycle. Intriguingly, the HDX-MS analysis presented here supports a mechanism whereby structural information is communicated to the active site, via the relay helix, that may modulate affinity for nucleotides or the kinetics of the ATPase cycle in a phosphorylation-dependent fashion. Based on a previous study of class I myosin, we speculate that phosphorylation of the TgMyoA N terminus may enhance motility via a faster rate of ADP release, in an inverse fashion to that observed upon deletion of the class Ib N-terminal region (35). Furthermore, TgMyoA residues S21 [the most frequently phosphorylated site on the protein (20)] and S29 are completely conserved in the MyoA, C, and D isoforms of all apicomplexans, suggesting that mechanisms of phosphorylation-dependent regulation are broadly conserved across the phylum. A notable difference, however, is observed in one or two polymorphisms in the T. gondii GT1 strain characterized here. Residue A725, oriented toward the core of the converter subdomain, is replaced with Val in type II and III strains (e.g., ME49 and VEG, respectively) and is proximal to the region destabilized by the S743D phosphomimetic mutation. The larger side chain of a Val residue may result in additional steric clashes, further enhancing the effects of phosphorylated S743. Despite the divergence of this residue within different strains of TgMyoA, the Ala at this position is conserved with MyoA from Plasmodium falciparum and Cryptosporidium parvum, as well as TgMyoH.
The divergence of class XIV from human myosins, combined with their crucial role in apicomplexan biology, makes them attractive potential targets for therapeutic agent development. Structural analysis reveals a highly conserved active site, indicating that development of class XIV-specific small-molecule inhibitors is likely to be restricted to allosteric pockets. Based on our surface pocket analysis of the TgMyoA crystal structure, P1 and P2 pockets emerged as the most intriguing candidate sites. Although the P3 pocket is notably smaller, its spatial proximity to P2 may support the design of a linked small molecule capable of sampling both pockets. Sequence variation between the P1 pockets of class XIV motors and human homologs suggests that a structure-based in silico approach, coupled with biochemical validation, is a promising avenue toward the development of novel class XIV-specific myosin inhibitors. Intriguingly, the relative conservation of P1 residues within class XIV myosins suggests a partial overlap of drug space, perhaps allowing the identification of a single, shared pharmacophore that can be used to target the class XIV myosins of multiple apicomplexan parasites.
The structural characterization of a class XIV MyoA reported here reveals a detailed network of divergent features that govern regulation and maintain chemomechanical coupling in these unconventional myosins. The insight gained from these studies will continue to support structural and functional dissection of these crucial molecular motors to better understand motility in apicomplexan parasites and support structure-guided inhibitor design.
Materials and Methods
TgMyoA 1–831/ELC1/MLC1 complex and TgMyoA 1–778 were produced recombinantly in insect cells as previously reported (37). Diffraction data for TgMyoA 1–778 were collected at the Stanford Synchrotron Radiation Lightsource. The structure of TgMyoA 1–778 was solved by molecular replacement by using the motor domain of human myosin 1c (PDB ID code 4BYF, chain A) as the search model. Data collection and refinement statistics are presented in SI Appendix, Table S1. Atomic coordinates and structure factors have been deposited in PDB under ID code 6DUE. HDX reactions were conducted in triplicate with Mg2+ and ADP to lock TgMyoA in near-rigor state and carried out for 3 s and 300 s at 23 °C. Full HDX-MS data are provided in SI Appendix, Dataset S1. Detailed methods are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff at the Stanford Synchrotron Radiation Lightsource. This work was supported by Canadian Institutes of Health Research Grant 148596 (to M.J.B.), Natural Sciences and Engineering Research Council of Canada Discovery Grant 2014-05218 (to J.E.B.), US Public Health Service Grants AI054961 (to G.E.W.) and GM094229 (to D.M.W.), National Institutes of Health/National Heart, Lung, and Blood Institute Grant 1P01HL128192-01A1 (to J.B.), and the Canada Research Chair Program (M.J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.M.O. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6DUE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811167115/-/DCSupplemental.
References
- 1.Harker KS, Ueno N, Lodoen MB. Toxoplasma gondii dissemination: A parasite’s journey through the infected host. Parasite Immunol. 2015;37:141–149. doi: 10.1111/pim.12163. [DOI] [PubMed] [Google Scholar]
- 2.Frenal K, Dubremetz JF, Lebrun M, Soldati-Favre D. Gliding motility powers invasion and egress in Apicomplexa. Nat Rev Microbiol. 2017;15:645–660. doi: 10.1038/nrmicro.2017.86. [DOI] [PubMed] [Google Scholar]
- 3.Boucher LE, Bosch J. The apicomplexan glideosome and adhesins–Structures and function. J Struct Biol. 2015;190:93–114. doi: 10.1016/j.jsb.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenal K, Foth BJ, Soldati-Favre D. Myosin class XIV and other myosins inprotists. In: Coluccio LM, editor. Myosins: A Superfamily of Molecular Motors, Proteins and Cell Regulation. Vol 7. Springer; Dordrecht, The Netherlands: 2008. pp. 421–440. [Google Scholar]
- 5.Mueller C, Graindorge A, Soldati-Favre D. Functions of myosin motors tailored for parasitism. Curr Opin Microbiol. 2017;40:113–122. doi: 10.1016/j.mib.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Heintzelman MB. Gliding motility in apicomplexan parasites. Semin Cell Dev Biol. 2015;46:135–142. doi: 10.1016/j.semcdb.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Meissner M, Schlüter D, Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- 8.Andenmatten N, et al. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat Methods. 2013;10:125–127. doi: 10.1038/nmeth.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egarter S, et al. The toxoplasma Acto-MyoA motor complex is important but not essential for gliding motility and host cell invasion. PLoS One. 2014;9:e91819. doi: 10.1371/journal.pone.0091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frénal K, Marq JB, Jacot D, Polonais V, Soldati-Favre D. Plasticity between MyoC- and MyoA-glideosomes: An example of functional compensation in Toxoplasma gondii invasion. PLoS Pathog. 2014;10:e1004504. doi: 10.1371/journal.ppat.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frénal K, et al. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Fujita-Becker S, et al. Functional characterization of the N-terminal region of myosin-2. J Biol Chem. 2006;281:36102–36109. doi: 10.1074/jbc.M605171200. [DOI] [PubMed] [Google Scholar]
- 13.Preller M, Holmes KC. The myosin start-of-power stroke state and how actin binding drives the power stroke. Cytoskeleton (Hoboken) 2013;70:651–660. doi: 10.1002/cm.21125. [DOI] [PubMed] [Google Scholar]
- 14.Heintzelman MB, Schwartzman JD. A novel class of unconventional myosins from Toxoplasma gondii. J Mol Biol. 1997;271:139–146. doi: 10.1006/jmbi.1997.1167. [DOI] [PubMed] [Google Scholar]
- 15.Herm-Götz A, et al. Toxoplasma gondii myosin A and its light chain: A fast, single-headed, plus-end-directed motor. EMBO J. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson B, Ruppel KM, Wu Y, Spudich JA. Cold-sensitive mutants G680V and G691C of Dictyostelium myosin II confer dramatically different biochemical defects. J Biol Chem. 1997;272:27612–27617. doi: 10.1074/jbc.272.44.27612. [DOI] [PubMed] [Google Scholar]
- 17.Bookwalter CS, et al. Reconstitution of the core of the malaria parasite glideosome with recombinant Plasmodium class XIV myosin A and Plasmodium actin. J Biol Chem. 2017;292:19290–19303. doi: 10.1074/jbc.M117.813972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bookwalter CS, Kelsen A, Leung JM, Ward GE, Trybus KM. A Toxoplasma gondii class XIV myosin, expressed in Sf9 cells with a parasite co-chaperone, requires two light chains for fast motility. J Biol Chem. 2014;289:30832–30841. doi: 10.1074/jbc.M114.572453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaji RY, et al. Phosphorylation of a myosin motor by TgCDPK3 facilitates rapid initiation of motility during Toxoplasma gondii egress. PLoS Pathog. 2015;11:e1005268. doi: 10.1371/journal.ppat.1005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Q, et al. Calcium-dependent phosphorylation alters class XIVa myosin function in the protozoan parasite Toxoplasma gondii. Mol Biol Cell. 2014;25:2579–2591. doi: 10.1091/mbc.E13-11-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Münnich S, Taft MH, Manstein DJ. Crystal structure of human myosin 1c–The motor in GLUT4 exocytosis: Implications for Ca2+ regulation and 14-3-3 binding. J Mol Biol. 2014;426:2070–2081. doi: 10.1016/j.jmb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz S, et al. Malaria parasite actin filaments are very short. J Mol Biol. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- 24.Schüler H, Mueller AK, Matuschewski K. Unusual properties of Plasmodium falciparum actin: New insights into microfilament dynamics of apicomplexan parasites. FEBS Lett. 2005;579:655–660. doi: 10.1016/j.febslet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 25.Sahoo N, Beatty W, Heuser J, Sept D, Sibley LD. Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Mol Biol Cell. 2006;17:895–906. doi: 10.1091/mbc.E05-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skillman KM, et al. Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites. PLoS Pathog. 2011;7:e1002280. doi: 10.1371/journal.ppat.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preller M, et al. Structural basis for the allosteric interference of myosin function by reactive thiol region mutations G680A and G680V. J Biol Chem. 2011;286:35051–35060. doi: 10.1074/jbc.M111.265298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kad NM, Patlak JB, Fagnant PM, Trybus KM, Warshaw DM. Mutation of a conserved glycine in the SH1-SH2 helix affects the load-dependent kinetics of myosin. Biophys J. 2007;92:1623–1631. doi: 10.1529/biophysj.106.097618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batra R, Geeves MA, Manstein DJ. Kinetic analysis of Dictyostelium discoideum myosin motor domains with glycine-to-alanine mutations in the reactive thiol region. Biochemistry. 1999;38:6126–6134. doi: 10.1021/bi982251e. [DOI] [PubMed] [Google Scholar]
- 30.Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- 31.Graindorge A, et al. The conoid associated motor MyoH is indispensable for Toxoplasma gondii entry and exit from host cells. PLoS Pathog. 2016;12:e1005388. doi: 10.1371/journal.ppat.1005388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacot D, et al. An apicomplexan actin-binding protein serves as a connector and lipid sensor to coordinate motility and invasion. Cell Host Microbe. 2016;20:731–743. doi: 10.1016/j.chom.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 34.Mentes A, et al. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci USA. 2018;115:1292–1297. doi: 10.1073/pnas.1718316115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuman H, et al. A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning. Proc Natl Acad Sci USA. 2014;111:2116–2121. doi: 10.1073/pnas.1321022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 37.Powell CJ, et al. Dissecting the molecular assembly of the Toxoplasma gondii MyoA motility complex. J Biol Chem. 2017;292:19469–19477. doi: 10.1074/jbc.M117.809632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson AM, et al. Omecamtiv mecarbil enhances the duty ratio of human β-cardiac myosin resulting in increased calcium sensitivity and slowed force development in cardiac muscle. J Biol Chem. 2017;292:3768–3778. doi: 10.1074/jbc.M116.748780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woody MS, et al. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat Commun. 2018;9:3838. doi: 10.1038/s41467-018-06193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preller M, Chinthalapudi K, Martin R, Knolker HJ, Manstein DJ. Inhibition of myosin ATPase activity by halogenated pseudilins: A structure-activity study. J Med Chem. 2011;54:3675–3685. doi: 10.1021/jm200259f. [DOI] [PubMed] [Google Scholar]
- 41.Chinthalapudi K, et al. Mechanism and specificity of pentachloropseudilin-mediated inhibition of myosin motor activity. J Biol Chem. 2011;286:29700–29708. doi: 10.1074/jbc.M111.239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uyeda TQ, Tokuraku K, Kaseda K, Webb MR, Patterson B. Evidence for a novel, strongly bound acto-S1 complex carrying ADP and phosphate stabilized in the G680V mutant of Dictyostelium myosin II. Biochemistry. 2002;41:9525–9534. doi: 10.1021/bi026177i. [DOI] [PubMed] [Google Scholar]
- 43.Chinthalapudi K, Heissler SM, Preller M, Sellers JR, Manstein DJ. Mechanistic insights into the active site and allosteric communication pathways in human nonmuscle myosin-2C. eLife. 2017;6:e32742. doi: 10.7554/eLife.32742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 45.Houdusse A, Kalabokis VN, Himmel D, Szent-Györgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: A novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.